SUMMARY
Although casein kinase 1δ (CK1δ) is at the center of multiple signaling pathways, its role in the expansion of central nervous system progenitor cells is unknown. Using mouse cerebellar granule cell progenitors (GCPs) as a model for brain neurogenesis, we demonstrate that the loss of CK1δ or treatment of GCPs with a highly selective small molecule inhibits GCP expansion. In contrast, CK1δ overexpression increases GCP proliferation. Thus, CK1δ appears to regulate GCP neurogenesis. CK1δ is targeted for proteolysis via the anaphase-promoting complex/cyclosome (APC/CCdh1) ubiquitin ligase, and conditional deletion of the APC/CCdh1 activator Cdh1 in cerebellar GCPs results in higher levels of CK1δ. APC/CCdh1 also downregulates CK1δ during cell cycle exit. Therefore, we conclude that APC/CCdh1 controls CK1δ levels to balance proliferation and cell cycle exit in the developing central nervous system. Similar studies in medulloblastoma cells showed that CK1δ holds promise as a new therapeutic target.
INTRODUCTION
The casein kinase 1 (CK1) family of monomeric serine/threonine protein kinases is evolutionarily conserved in eukaryotes. Seven members have been identified in mammals: α, β, δ, ε, γ1, γ2, and γ3 (Gross and Anderson, 1998; Knippschild et al., 2005; Rowles et al., 1991; Zhai et al., 1995). These kinases target a broad spectrum of substrates to control diverse biological processes, e.g., signal transduction, circadian rhythms, nuclear import, DNA repair, apoptosis, spindle assembly, vesicle trafficking, neurite outgrowth, and primary cilia formation (Behrend et al., 2000; Beyaert et al., 1995; Cheong and Virshup, 2011; Desagher et al., 2001; Gault et al., 2012; Gross and Anderson, 1998; Knippschild et al., 2005; Petronczki et al., 2006; Price, 2006; Vielhaber and Virshup, 2001). However, whether CK1 mediates the generation of specific classes of central nervous system (CNS) neurons is unknown. (Lohler et al., 2009)
During brain development, cerebellar granule cell progenitors (GCPs) expand to produce the most numerous neuronal population in the brain. This proliferation is followed by cell cycle exit and differentiation. Thus, we predict that drivers of GCP expansion and proliferation are downregulated during cell cycle exit. However, others have postulated that CK1 isoforms are unregulated (Knippschild et al., 2005). Whether CK1δ is downregulated during GCP cell cycle exit is unknown. CK1δ is targeted for ubiquitin-mediated proteolysis via the anaphase-promoting complex/cyclosome (APC/CCdh1). Conditional deletion of the APC/C activator Cdh1 in the developing cerebellum increases CK1δ levels in vivo. Furthermore, CK1δ stabilization increases GCP proliferation, suggesting a crucial role of APC-dependent CK1δ degradation during cell cycle exit. Moreover, downregulation of CK1δ in GCPs increases the level of Wee1 a cell cycle inhibitory kinase. Wee1 turnover increases Cdk1 activity and mitotic entry (Owens et al., 2010; Smith et al., 2007; Watanabe et al., 2005; Watanabe et al., 2004). We previously demonstrated that CK1δ controls Wee1 degradation (Penas et al., 2014), which is important for cell proliferation.
APC/CCdh1 is a tumor suppressor; thus, APC/C-dependent degradation of CK1δ is most likely deregulated in some cancers. GCPs are thought to give rise to medulloblastoma, the most common malignant pediatric brain tumor. Several GCP developmental pathways are deregulated in medulloblastoma, including WNT, SHH, MYC, and some undefined pathways (Hatten and Roussel, 2011). Mutations in the SHH receptors Patched (PTCH1), Suppressor of fused (SUFU), and Smoothened (SMO) are associated with medulloblastoma and other malignancies (Evans et al., 1991; Hallahan et al., 2004; Svard et al., 2006; Taylor et al., 2002; Yauch et al., 2009). Group 3 (G3) medulloblastoma, the most aggressive form of the disease, is associated with MYC overexpression (Cho et al., 2011; Ellison et al., 2011; Northcott et al., 2011; Pfister et al., 2009). Recent sequencing studies have demonstrated CK1δ overexpression in G3 medulloblastoma, suggesting a role for CK1 isoforms in some medulloblastoma subgroups (Gibson et al., 2010; Jones et al., 2012; Northcott et al., 2012; Pugh et al., 2012; Robinson et al., 2012).
CK1δ is expressed in mouse cerebellum (Lohler et al., 2009), an opportune model for CNS neurogenesis. Here we investigated the role of CK1δ in GCP expansion in the developing CNS. We also examined whether proteolytic degradation via APC/CCdh1 regulates CK1δ in vitro and in vivo. Finally, we measured the levels of CK1δ in medulloblastoma cells relative to that in control GCPs and determined whether the cells are responsive to CK1δ inhibition in vivo in allograft and intracranial xenograft mouse models. Our results indicate that CK1δ may be a novel therapeutic target in medulloblastoma.
RESULTS
CK1δ Is Required for Cerebellar GCP Proliferation
During normal brain development, GCPs expand to generate 45 billion granule neurons; the adult human brain contains 100 billion neurons (Roussel and Hatten, 2011). Because CK1δ is expressed postnatally in cerebellar GCPs (Figure 1A-B), we examined whether it is involved in GCP neurogenesis and cell cycle exit . Purified GCPs are used to study proliferation and differentiation because they proliferate effectively in cell aggregates in suspension. Conversely, they exit the cell cycle and differentiate when plated on poly-D-lysine/laminin–coated plates.
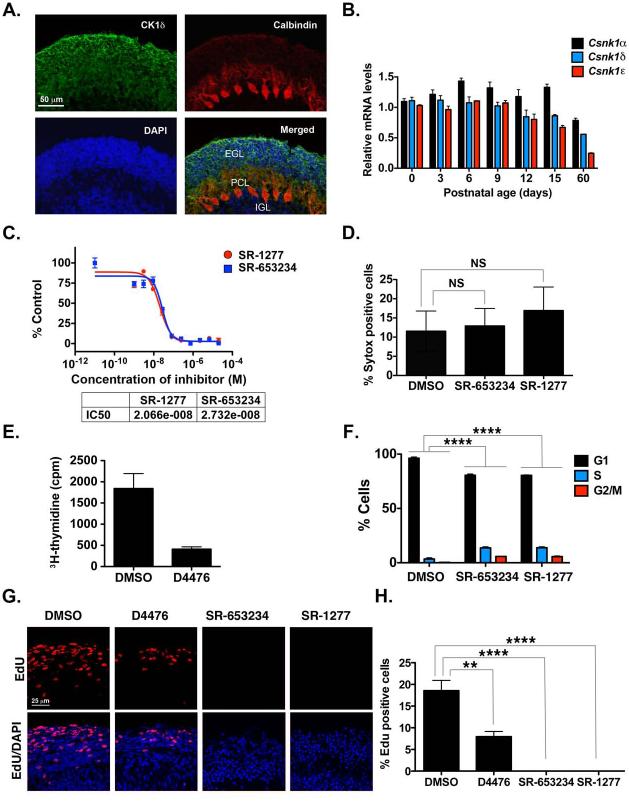
Figure 1. CK1δ Expression in Postnatal GCPs and Control of GCP Proliferation in vitro and ex vivo.
(A) Cerebellar sections from P8 pups were stained with antibodies against CK1δ (green) or calbindin (red) and DAPI (blue). (B) CK1α , CK1δ, and CK1ε mRNA was amplified by qRT-PCR, and fold-change in gene expression in postnatal mouse cerebellum was determined by normalizing to GAPDH values relative to control. (C) GCPs were incubated for 24 h with increasing concentrations of SR-653234 or SR-1277, and the amount of proliferation was determined by 3H-thymidine incorporation. Results were plotted relative to that seen in the DMSO control. (D) GCPs were treated with 100 nM SR-653234 or SR-1277 for 24 h, and then Sytox and Hoechst staining was performed (NS, not significant, as determined by one-way ANOVA and Dunnett multiple comparisions test). (E) D4476 (20 μM) reduces GCP proliferation, and (F) SR-653234 (100 nM) and SR-1277 (100 nM) increase the percentage of GCPs in the S or G2/M phase. GCPs were treated for 24 h with the indicated compounds or DMSO, and the proportion of cells in each cell cycle phase was determined by PI-FACS. (G) Organotypic cerebellar slices were treated with SR-1277 (100 nM), SR-653234 (100 nM), D4476 (20 μM), or DMSO for 1 h, after which EdU was added to the media for 20 h. Slices were stained with EdU (red) and the nuclear marker DAPI (blue). (H) Quantification of (G). Results are shown as the average values of three independent experiments and are represented as the mean and standard error of the mean (± SEM) (*p <0.05, **p <0.001, ***p <0.001, ****p <0.0001).
To determine whether CK1δ inhibition affects GCP proliferation, we treated cells in suspension with SR-653234 or SR-1277, two highly specific, potent small-molecule inhibitors of CK1δ (Bibian et al., 2013; Penas et al., 2014). We measured the rate of proliferation of purified GCPs in the presence and absence of SR-653234 or SR-1277 by 3H-thymidine uptake (Figure 1C). Both compounds inhibited GCP proliferation with a similar IC50 (Figure 1C), but neither caused cell death (Figure 1D). Treatment with the well-characterized CK1δ inhibitor D4476 also reduced GCP proliferation in vitro (Figure 1E). The proportion of GCPs in S phase was higher in cells treated with SR-653234 or SR-1277 (13.7% and 18.9%, respectively) than it was in DMSO-treated controls (3.4%), as were those in G2/M (SR-653234, 5.8%; SR-1277, 5.6%; DMSO, 0.1%), as determined by PI-FACS (Figure 1F). We did not observe cells containing DNA content lower than 2N (sub-G1 phase), which further confirmed that the inhibitor concentrations used did not kill the cells (Figure 1F). These results suggest that pharmacologic inhibition of CK1δ induces GCP cell cycle arrest in the S or G2/M phases.
To test whether CK1δ is required for GCP proliferation in an ex vivo model, we treated slices of postnatal cerebellar tissue with D4476, SR-653234, or SR-1277 (Figure 1G). The cerebellar GCP is a well-studied model of proliferation; dividing cells are restricted to the external germinal layer. Postmitotic GCPs localize beneath mitotic cells, initiate differentiation by extending parallel fiber axons, and migrate along the radial fibers of Bergmann glia (Edmondson and Hatten, 1987; Rakic, 1972). Thus, the position of labeled GCPs in organotypic slices of developing cerebellum indicates their proliferation status. EdU assays of organotypic slices of postnatal cerebellum in culture (Tomoda et al., 1999) showed less EdU uptake in D4476-treated slices and dramatically less in SR-653234– or SR-1277–treated cells relative to the DMSO-treated control (Figure 1G). EdU incorporation is a measure of proliferation; therefore, these results suggest that CK1δ inhibition disrupts GCP proliferation ex vivo.
CK1δ Knockdown Reduces Cerebellar GCP Proliferation
To validate the requirement of CK1δ in GCP proliferation, we depleted CK1δ levels by siRNA-mediated knockdown. Electroporation of purified GCPs with two different siRNAs effectively decreased CK1δ mRNA and protein expression without affecting CK1ε or CK1α levels (Figure 2A). SHH is a potent mitogen of GCP proliferation; therefore, we tested whether depleting the level of CK1δ affected the rate of SHH-mediated incorporation of EdU into GCPs. CK1δ knockdown decreased the levels of the proliferative markers phospho-Histone H3 and cyclin B1 in the absence or presence of SHH (Figure 2B-C). Furthermore, EdU incorporation was reduced in GCPs electroporated with CK1δ-specific siRNAs, relative to that in control siRNA (Figure 2E-F). In contrast, CK1ε depletion did not affect EdU incorporation (Figure 2D). These results indicate that CK1δ is required for GCP proliferation in vitro, and reducing its levels attenuates SHH-induced mitogenesis.
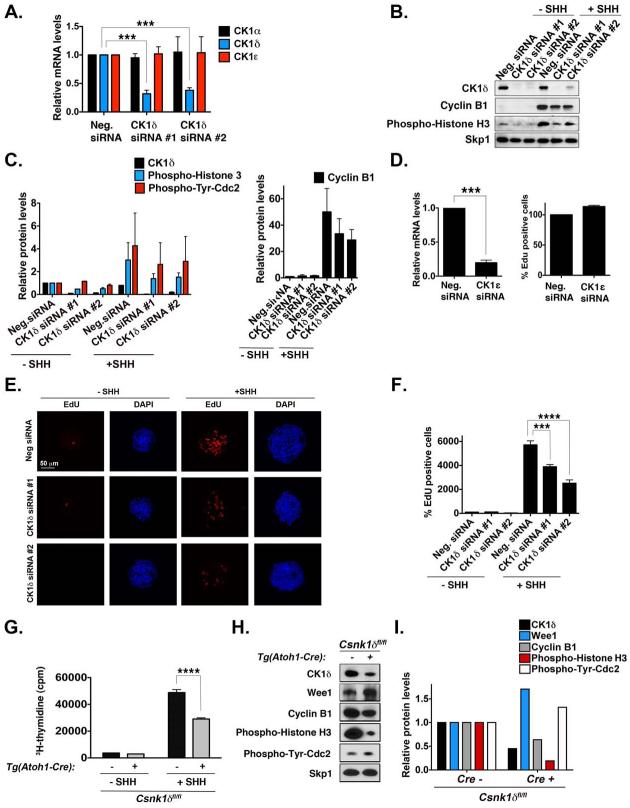
Figure 2. CK1δ Knockdown Reduces GCP Proliferation in vitro and ex vivo.
(A) GCP cells electroporated with two different CK1δ siRNAs were analyzed by qRT-PCR. CK1δ was significantly knocked down, but CK1α and CK1ε levels were not altered. The mRNA was amplified by qRT-PCR, and the fold-change in gene expression was determined by normalizing to GAPDH values relative to controls. (B) After electroporation of GPCs with or without SHH (75 ng/mL), the levels of CK1δ, cyclin B1, and phospho–histone H3 were analyzed by immunoblotting with antibodies against the proteins. Skp1 served as a loading control. CK1δ was knocked down by both siRNAs. (C) Quantification of the amount of CK1δ, phospho-Histone H3, phospho-Tyr-Cdk1 (a measure of Wee1 inhibition of Cdk1), and cyclin B1, relative to the loading control Skp1, after CK1δ siRNA electroporation in GCPs from (B). (D) GCPs electroporated with CK1ε siRNA were analyzed by qRT-PCR. CK1ε was significantly knocked down, but EdU uptake was not reduced in the presence of SHH. Levels are expressed as the percentage of EdU uptake in cells treated with the negative siRNA. (E) CK1δ siRNA electroporation reduces EdU uptake of GCPs in the presence of SHH. Proliferative GCP aggregates were stained with EdU (red) and the nuclear marker DAPI (blue). (F) Quantification of the EdU incorporation in (E). Levels are expressed as a percentage compared to that in cells treated with the negative siRNA without SHH. (G) GCPs isolated from Csnk1d-deleted mice proliferate less in the presence or absence of SHH. Purified GCPs were treated for 24 h with the compounds, and then 3H-thymidine was added to the media for an additional 24 h. Plots representing the amounts of 3H-thymidine incorporated by GCPs from CK1δ-deleted or control mice are shown. (H) GCPs purified from conditional Csnk1d-deleted mice have lower levels of CK1δ, cyclin B1, and phospho-Histone H3, indicating decreased proliferation. Representative immunoblots of CK1δ, Wee1, cyclin B1, phospho-Histone H3, and phospho-Tyr-cdc2 relative to Skp1 are shown. (I) Quantification of the amount of CK1δ , Wee1, phospho-Histone H3, phospho-Tyr-Cdc2, and cyclin B1 protein, relative to the loading control Skp1, in GCPs isolated from Tg(Atoh1-Cre)+;Csnk1dfl/fl or Tg (Atoh1-Cre)-;Csnk1dfl/fl mice from (H). Results are shown as the averages of three independent experiments and are represented as the mean ± SEM (***p <0.001, ****p <0.0001).
To determine whether CK1δ is important for GCP proliferation ex vivo, we conditionally deleted CK1δ in cerebellar GCPs by using Atoh1-Cre, a GCP-specific Cre-driver, and measured 3H-thymidine incorporation in purified GCPs. GCPs purified from Tg(Atoh1-Cre)+;Csnk1dfl/fl mice had a slightly lower rate of 3H-thymidine incorporation than did GCPs purified from Tg(Atoh1-Cre)–;Csnk1dfl/fl mice. However, we observed a more pronounced decrease in proliferation in GCPs from Tg(Atoh1-Cre)+;Csnk1dfl/fl mice treated with SHH (Figure 2G). Cyclin B1 and phospho-Histone 3 levels were also lower after CK1δ deletion (Figure 2H), suggesting that GCP expansion is reduced upon CK1δ deletion ex vivo. Wee1 levels were upregulated after CK1δ deletion (Figure 2H-I); thus, increased Wee1 levels may also limit GCP expansion. Together, these results demonstrate that CK1δ functions in cerebellar GCP proliferation in vitro and ex vivo.
CK1δ Inhibition Affects GCP Cell Cycle Progression
To better understand the role of CK1δ in GCP proliferation, we analyzed the levels of cell cycle regulators after pharmacologic inhibition of CK1δ in purified GCPs. GCPs were treated with SHH, SR-1277, or both for 24 or 48 h and processed for qRT-PCR analysis. We first analyzed the levels of various cyclins that are essential regulators of cyclin-dependent kinases and cell cycle transitions in multiple model systems, including GCPs. SR-1277 decreased the mRNA levels of cyclins A1 (Ccna1), B1 (Ccnb1), D2 (Ccnd2) and E1 (Ccne1) induced by SHH (Figure 3A), but did not alter that of the cyclin-dependent kinase inhibitors p21Cip1 (Cdkn1a) and p27Kip1 (Cdkn1b). Similar results were found after electroporation of purified GCPs with specific CK1δ siRNAs (Figure 3B). Incubation with SR-1277 or electroporation with CK1δ siRNAs also decreased the levels of the main effectors of the SHH pathway, Gli1 and Gli2, in GCPs (Figure S1A-B). These results confirmed that specific inhibition or decreased levels of CK1δ arrest the GCP cell cycle.
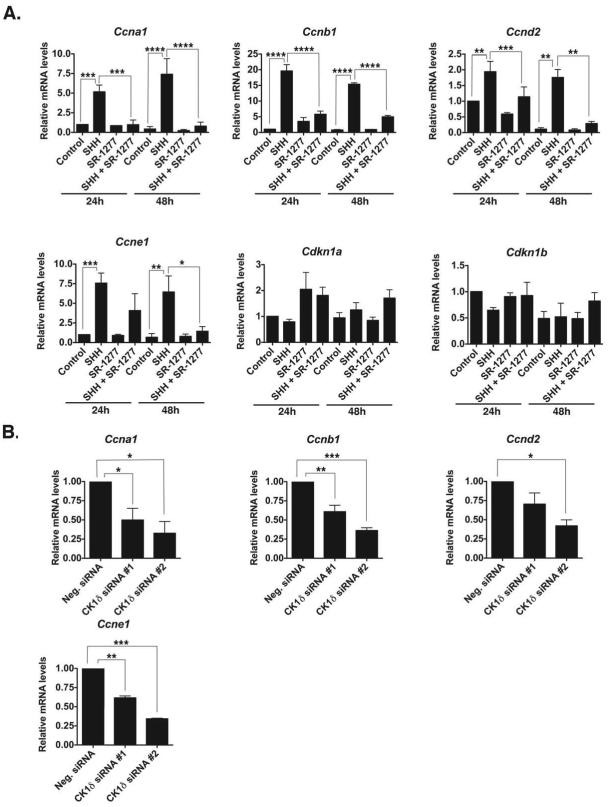
Figure 3. Inhibition or Knockdown of CK1δ Reduces the mRNA Levels of Cell Cycle Components.
(A) SR-1277 (100 nM) decreases the expression of SHH-induced levels of Ccna1, Ccnb1, Ccnd2, Ccne1, Cdkn1a, and Cdkn1b mRNA in GCPs. GCPs were treated with SHH (75 ng/mL) and/or SR-1277 for 24 or 48 h. The mRNA was amplified by qRT-PCR, and fold-change in gene expression was determined by normalizing to GAPDH values relative to control samples. (B) CK1δ knockdown reduces the expression of Ccna1, Ccnb1, Ccnd2, and Ccne1 mRNA levels in the presence of SHH. GCPs were electroporated with two different siRNAs against CK1δ, and the mRNA levels were analyzed after 72 h in vitro. Results shown are the averages of three independent experiments and are represented as the mean ± SEM (*p <0.05, **p <0.001, ***p <0.001, ****p <0.0001).
APC/CCdh1 Specifically Targets CK1δ for Proteolysis
Many cell cycle regulators are subject to ubiquitin-dependent proteolysis; therefore, we asked whether CK1δ is degraded via this process. APC/CCdh1 recognizes many substrates via a canonical destruction (D-box) motif that contains a minimal consensus sequence of RXXL, where X is any amino acid. Cdh1 binding to RXXL motifs initiates ubiquitin transfer and subsequent ubiquitin-dependent substrate degradation (Barford, 2011; Owens and Hoyt, 2005; Song and Rape, 2011). We examined the protein sequence of all human CK1 isoforms for putative RXXL motifs. CK1δ has two RXXL motifs, one at position 8 (RYRL; DB1) and one at position 193 (RDDL; DB2), that are evolutionarily conserved (Figure 4A).
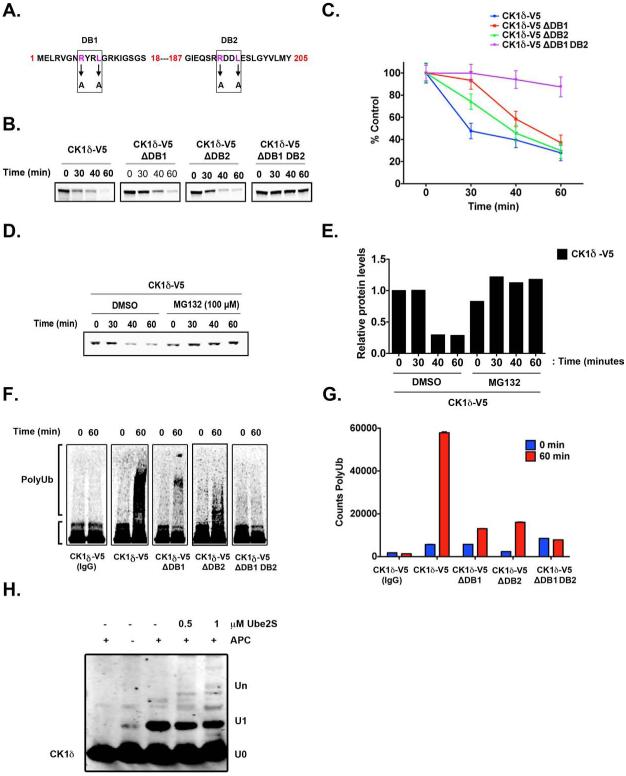
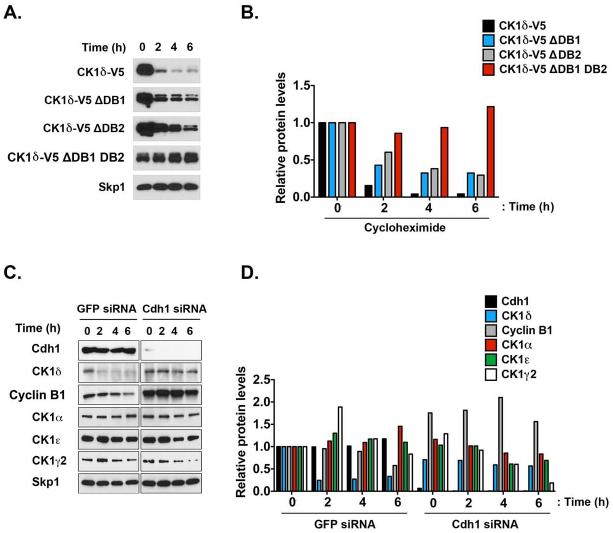
Figure 4. APC/CCdh1 induces CK1δ Ubiquitination and Degradation in vitro.
(A) Two destruction boxes (D-boxes) in human CK1δ, DB1 and DB2, were mutated by substituting alanine (A) for the corresponding arginine (R) and leucine (L) residues. (B-C) Both D-boxes in CK1δ are required for proteolysis. In vitro degradation assay showing 35S-labeled wild-type CK1δ-V5, DB1 mutant (CK1δ-V5 ΔDB1), DB2 mutant (CK1δ-V5 ΔDB2), and DB1 DB2 double-mutant (CK1δ-V5 ΔDB1 DB2) after incubation in extracts prepared from HeLa cells in G1. Samples collected at the indicated time points were analyzed by autoradiography. (C) The quantification of (B); protein levels were measured in three separate experiments using Quantity One image analysis software (Bio-Rad). An unpaired t test was performed, and a p-value of 0.01 was obtained. (D) Autoradiogram showing in vitro degradation of 35S-labeled wild-type CK1δ-V5 in HeLa cell extracts at G1, in the presence or absence of the proteasome 26S inhibitor MG132 (100 μM). (E) Quantification of CK1δ-V5 from (D). (F-G) Both D-boxes in CK1δ are required for efficient ubiquitination. Autoradiogram of 35S-labeled wild-type CK1δ-V5, ΔDB1, ΔDB2, and ΔDB1 DB2 mutants after in vitro ubiquitination by anti-Cdc27 immunoprecipitates from HeLa cell extracts at G1. (G) The extent of polyubiquitination was quantified for the entire lane above the inputs by using Quantity One image analysis software. From three separate experiments, an unpaired t test was performed, and a p-value of 0.005 was obtained. Results shown are the averages of three independent experiments and are represented as the mean ± SEM. (H) Purified CK1δ and immunoprecipitated APC/C were incubated together in vitro, and the extent of ubiquitination was determined after SDS-PAGE and anti-CK1δ autoradiography.
We hypothesized that the putative D-box motifs in CK1δ are functional and mediate recognition by APC/CCdh1. Deletion or mutation of bona fide D-boxes in previously reported APC/CCdh1 substrates decrease Cdh1-dependent ubiquitination and degradation (Penas et al., 2011); therefore, we performed site-directed mutagenesis to produce versions of CK1δ-V5 that had mutations in DB1 (ΔDB1), DB2 (ΔDB2), or both (ΔDB1 DB2). Both D-boxes were mutated with alanine substitutions of their respective arginine (R) and leucine (L) residues (Figure 4A). Several studies have shown that the D-box–dependent destruction of substrates can be ablated with RXXL-to-AXXA substitutions (Choi et al., 2008; King et al., 1996; Listovsky et al., 2004; Stewart and Fang, 2005; Zur and Brandeis, 2002).
We measured the in vitro degradation of wild-type or D-box–mutated CK1δ in somatic HeLa cell extracts that were isolated from cells in early G1 phase, when APC/CCdh1 is most active. Mutating DB1 or DB2 reduced CK1δ destruction, but inactivating both D-boxes profoundly stabilized the protein (Figure 4B-C). Furthermore, the degradation of wild-type CK1δ was similarly inhibited by the 26S proteasome inhibitor MG132 (Kisselev et al., 2012), suggesting that D-box–mediated degradation of CK1δ is ubiquitin pathway dependent (Figure 4D-E).
To test whether CK1δ is an in vitro substrate of APC/CCdh1, we incubated immunopurified APC/CCdh1 with CK1δ, Ube2s, and ubiquitin. CK1δ was robustly ubiquitinated via APC/CCdh1 (Figure 4F-H). To assess whether mutation of CK1 D-boxes reduced APC/CCdh1-mediated ubiquitination, we performed in vitro ubiquitination assays with purified G1 APC/CCdh1 and in vitro–translated, 35S-labeled CK1δ-V5 or CK1δ-V5 D-box mutants as substrates. Each single mutant significantly reduced CK1δ ubiquitination (Figure 4F-G), and the double-mutant nearly abolished polyubiquitination. Consistent with this finding, mutating both D-boxes stabilized CK1δ more than inactivating either one independently (Figure 5A-B).
Figure 5. CK1δ is Degraded by APC/CCdh1 in a D-box–dependent Manner.
(A) CK1δ D-box mutations reduce the turnover of the protein in HeLa cells. HeLa cells transfected with the wild-type CK1δ-V5 or D-box mutants were treated with cycloheximide (100 μg/mL). Samples were then collected at the indicated time points and analyzed by immunoblotting. (B) Quantification of (A). (C) Cdh1 is required for CK1δ degradation. HeLa cells were transfected with the indicated siRNA, treated with cycloheximide for the indicated times, retrieved at the time points shown, and analyzed by immunoblotting. (D) Quantification of (C).
To determine whether Cdh1 depletion reduces CK1δ turnover, we measured the degradation of CK1 isoforms in HeLa cells transfected with Cdh1 siRNA or control GFP siRNA. Although CK1δ and cyclin B1 were degraded in cells transfected with GFP siRNA, they were stabilized in Cdh1-depleted cells; other CK1 isoforms did not degrade in the same manner (Figure 5C-D), suggesting that Cdh1 specifically targets CK1δ. We previously described CK1δ-dependent Wee1 turnover (Penas et al., 2014); thus, we predicted that Cdh1 controls the level of Wee1. Wee1 levels were upregulated when CK1δ was downregulated (Figure S2A-B), and they were reduced even more when the CK1δ D-box mutant was overexpressed (Figure S2C-D). Cdh1 knockdown–mediated increase of CK1δ also decreased Wee1 levels (Figure S2A). Thus, the relationships between Cdh1 and CK1δ or Cdh1 and Wee1 were inverse.
APC/CCdh1 substrate levels oscillate during the cell cycle, reaching a minimum during G1, when APC/CCdh1 is most active (Penas et al., 2011). Thus, we predicted that if CK1δ were an APC/CCdh1 substrate, its levels would also decrease. To test this directly, we synchronized HeLa cells in mitosis via a well-established thymidine/nocodazole protocol, released them into G1 by washing away nocodazole, and monitored CK1δ levels via Western blot analysis (Figure S2E-F). Cell cycle progression was monitored by PI-FACS (Figure S2G). CK1δ levels were stable through mitosis and early G1 but decreased late in G1, 6 to 7 h after release from nocodazole-induced arrest (Figure S2E). CK1δ was undetectable before cells entered the S phase (Figure S2E-F). In contrast, CK1α, ε, and γ2 levels did not decrease during G1, suggesting that CK1δ is a unique APC/CCdh1 substrate among CK1 isoforms. Cyclin B1 levels decreased upon exit from mitosis (2 h after nocodazole release), as determined by phospho-Histone H3 staining and PI-FACS (Figure S2E-G).
APC/CCdh1 Controls CK1δ in Cerebellar GPCs in vivo
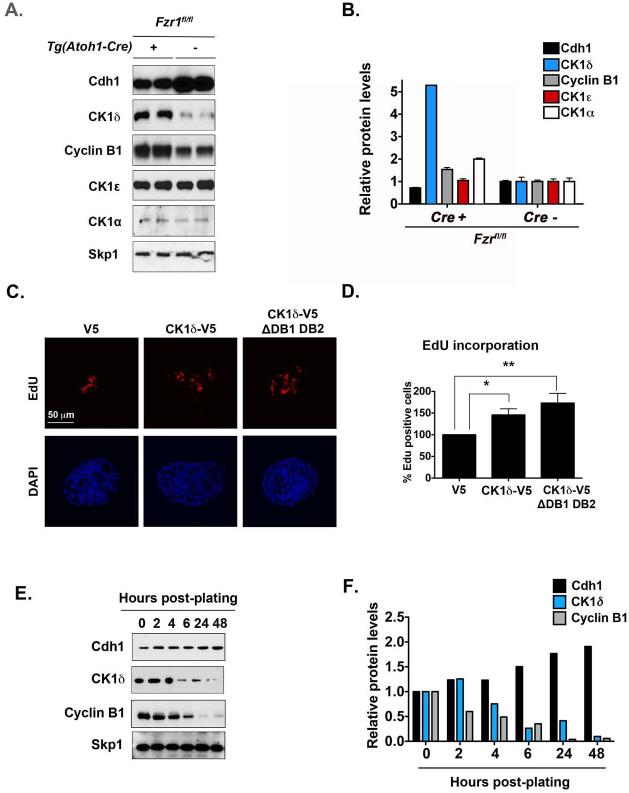
To test whether APC/CCdh1 regulates CK1δ ubiquitination and degradation in vivo, we conditionally deleted Cdh1 (or Fzr1) in cerebellar GCPs by crossing Tg(Atoh1-Cre)+ mice with Fzr1fl/fl mice (Garcia-Higuera et al., 2008b; Schuller et al., 2007). Atoh1 is a bHLH transcription factor required for GCP neurogenesis (Ben-Arie et al., 1997); thus, Tg(Atoh1-Cre)+;Fzr1fl/fl mice should have lower Cdh1 levels in GCPs relative to their wild-type or Cre– littermates. Cdh1 protein level was lower in GCPs purified from postnatal day (P) 7 Tg(Atoh1-Cre)+;Fzr1fl/fl mice than in Tg(Atoh1-Cre)–;Fzr1fl/fl mice (Figure 6A-B). Lower Cdh1 levels were also associated with increased CK1δ protein in Tg(Atoh1-Cre)+;Fzr1fl/fl mice. These results suggest that CK1δ is degraded via APC/CCdh1 in GCPs in the developing mouse cerebellum. Although GCPs from Tg(Atoh1-Cre)+;Fzr1fl/fl mice express higher levels of cell cycle regulators (e.g., cyclin B1), their cerebella develop normally. This could be attributed to the incomplete knockout by Atoh1-Cre or compensatory mechanisms of Cdc20, another APC/C activator in GCPs.
Figure 6. Conditional Deletion of Fzr1 in the Cerebellum Increases CK1δ Levels.
(A) Immunoblot analysis showing that the levels of cyclin B1 and CK1δ, but not CK1ε, are higher in GCPs purified from Fzr1-knockout mice than in those from control mice. Protein extracts were made directly after GCP purification. GCPs were not maintained in culture. (B) Quantification of (A). (C) Overexpression of CK1δ-V5 in purified GCPs increases cell proliferation, as indicated by the amount of EdU-positive cells (red) in the presence of SHH (75 ng/mL). EdU incorporation into cells electroporated with the CK1δ-V5 or CK1δ-V5 ΔDB1 DB2 construct was normalized to that of cells electroporated with the empty control vector (V5). (D) Quantification of (C). (E) CK1δ levels decrease during GCP cell cycle exit. Representative Western blotting of CK1δ, Cdh1, cyclin B1, and the loading control Skp1. (F) Quantification of (E). Results shown are averages of three independent experiments and are represented as the mean ± SEM (*p <0.05, **p <0.001).
We previously showed that APC/CCdh1 targets substrates for degradation during the GCP cell cycle (Harmey et al., 2009). Because reducing CK1δ levels or activity suppressed GCP expansion and APC/CCdh1 substrates often induce cell cycle transition, we asked whether CK1δ overexpression would stimulate GCP proliferation. Relative to V5 empty control vector, CK1δ-V5 or CK1δ-V5 ?DB1 DB2 overexpression increased GCP proliferation (Figure 6C-D). These results suggest that controlling CK1δ levels is key to GCP cell cycle transition. When GCPs were plated on poly-D-lysine/laminin–coated dishes, their CK1δ levels decreased. The cells then exited the cell cycle and differentiated. This reduction was similar to that observed for cyclin B1 (Figure 6E-F).
CK1δ Inhibition Decreases Medulloblastoma Growth ex vivo
CK1δ controls GCP proliferation in vitro and ex vivo, and GCPs are thought to give rise to some forms of medulloblastoma (Gibson et al., 2010; Kawauchi et al., 2012; Schuller et al., 2008; Yang et al., 1999). Therefore, we tested whether CK1δ is a possible therapeutic target for medulloblastoma. First we measured CK1δ protein and mRNA levels in tumors obtained from mouse models of medulloblastoma (Figures 7A-B and S3A-B). CK1δ protein levels were higher in tumors derived from
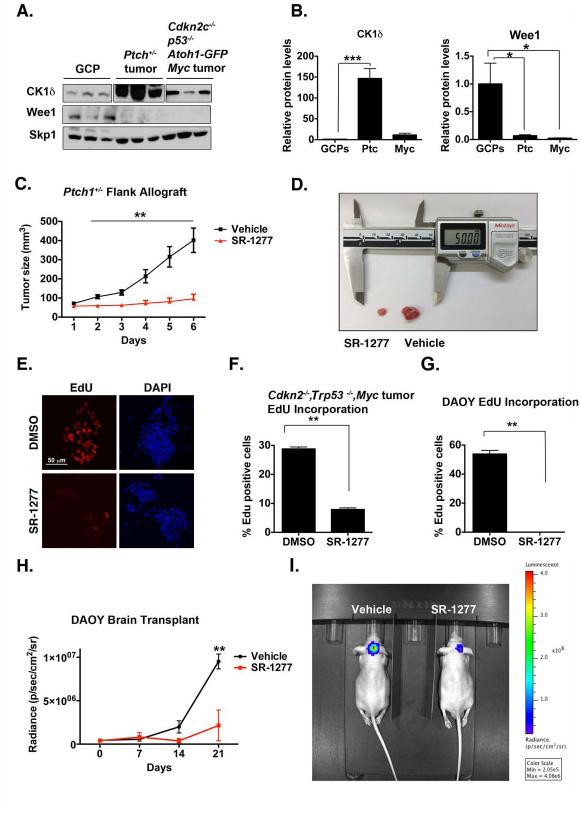
Figure 7. Murine Medulloblastoma Cells Express Elevated Levels of CK1δ, the Inhibition of which Reduces Tumor Growth in vivo.
(A) CK1δ protein is overexpressed in Ptch1+/−, Cdkn2−/−, Trp53−/−, and c-Myc tumors, whereas Wee1 is downregulated. Skp1 was used as a loading control. (B) Quantification of (A). (C-D) SR-1277 decreases proliferation of Ptch1+/− allograft tumors. Ptch1+/− tumor cells were injected subcutaneously into mice. Once the tumor reached a volume of 50 to 90 mm3, treatment with vehicle or SR-1277 (20 mg/kg, twice daily) was initiated. Tumor size was quantified in four samples for each time point, and the averages are shown. (D) An image showing representative SR-1277– treated (left) and vehicle-treated (right) tumors. (E) Proliferation of Cdkn2−/−, Trp53−/−, and c-Myc tumor cells is reduced in the presence of SR-1277. (F) Quantification of EdU incorporation into Cdkn2−/−, Trp53−/−, or c-Myc tumor cells after DMSO or SR-1277 treatment. (G) EdU-incorporation assay shows that proliferation of DAOY cells is reduced in the presence of SR-1277 (500 nM). (H-I) SR-1277 also reduces the intracranial growth of DAOY cells. (H) Twelve days after mice were transplanted with DAOY tumor cells, D-luciferin was administered intraperitoneally, and bioluminescence was measured. (I) Fluorescence imaging of representative mice in which DAOY cells were implanted intracraniallly and then treated with SR-1277 (20 mg/kg, twice daily) or vehicle for 21 days. Bioluminescence was quantified from the encircled regions that enclose the entire tumor. Results shown are the means ± SEM of three independent experiments (*p <0.05, **p <0.001, ***p <0.001).
Ptch1+/− or Myc mice (Goodrich et al., 1997; Kimura et al., 2005) than in untransformed GCPs (Figure 7A). Higher protein levels were not accompanied by increased Csnk1d (CK1δ) mRNA, indicating possible differential regulation of CK1δ in medulloblastoma relative to GCPs. Consistent with this notion, the level of the APC/C repressor Emi1 (FBXO31) in Myc-derived tumors was higher (Figure S3C-D), indicating altered APC/C activity that could contribute to the difference in protein and RNA levels. Furthermore, increased CK1δ protein levels corresponded with decreased Wee1 levels (Figure 7A-B), suggesting that CK1δ-dependent control of Wee1 turnover also mediates medulloblastoma cell proliferation.
Ptch1 functions as an antagonist of SHH, which is a potent mitogen for cerebellar medulloblastoma (Wechsler-Reya and Scott, 2001; Wechsler-Reya and Scott, 1999). Ptch1 mutation constitutively activates the SHH pathway and induces medulloblastoma tumors in 14% to 20% of mice. Ptch1+/− mice have been used extensively to model human SHH-subgroup medulloblastoma. CK1δ upregulation in mouse models of medulloblastoma suggests that it might be an attractive therapeutic target. To test this directly, we assessed the effectiveness of SR-1277 in reducing tumor growth in vivo. We implanted allografts from Ptch1+/− mice into immunocompromised recipients and started treatment when the tumors reached a volume of 50 to 90 mm3. SR-1277 treatment significantly inhibited tumor growth (Figure 7C-D).
Human G3 medulloblastoma has been recently modeled in mice by overexpressing Myc (c-Myc) in neural progenitors purified from the cerebellum of P7 Cdkn2c−/−;Trp53−/− mice and transplanting those cells into the cortices of naïve CD1 nude mice (Kawauchi et al., 2012). CK1δ protein was upregulated in G3 medulloblastoma cells (Figure 7A-B); therefore, we tested whether its inhibition reduces proliferation in this model. We treated mouse G3 medulloblastoma neurospheres with SR-1277 and measured the proliferation via an EdU-incorporation assay in vitro. SR-1277 inhibited proliferation, suggesting that CK1δ inhibition has potential as a therapeutic strategy for multiple human tumors. These results further indicate that human medulloblastoma cells may also respond to SR-1277. Treatment of two human medulloblastoma cell lines, DAOY and D283, with SR-1277 reduced proliferation (Figure 7G, Figure S3E-F). SR-1277 inhibited DAOY and D283 cell proliferation with the same efficacy as multiple compounds currently in clinical trials for cancer (Figure S3E-F). SR-1277 is 24% brain penetrant (Bibian et al., 2013); thus, it also reduced DAOY cell proliferation intracranially (Figure 7H-I). Collectively, these results validate CK1δ as a therapeutic target for human medulloblastoma.
DISCUSSION
In the present study, three lines of evidence demonstrated that CK1δ regulates granule cell neurogenesis during normal cerebellar development. First, conditional loss of CK1δ in GCPs or siRNA knockdown in wild-type GCPs reduced proliferation, as measured by EdU and 3H-thymidine incorporation. The loss of CK1δ also diminished SHH-induced GCP proliferation. Second, treatment of GCPs with specific CK1δ inhibitors dramatically reduced proliferation in vitro and ex vivo. Third, CK1δ overexpression had the opposite effect, namely it stimulated GCP proliferation. Our studies further showed that CK1δ is targeted for proteolysis via the APC/CCdh1 ubiquitin ligase, and conditional deletion of the APC/C activator Cdh1 in cerebellar GCPs increased CK1δ levels. These findings also increase our understanding of developmental brain tumor formation. We observed high levels of CK1δ in a mouse model of medulloblastoma, and treatment with specific inhibitors of CK1δ dramatically reduced tumor growth. Together, these results suggest that CK1δ regulates normal GCP neurogenesis in the developing brain and medulloblastoma growth and that APC/CCdh1-dependent degradation of CK1δ controls the proliferation rate of normal cells and tumor cells.
Although CK1δ is expressed in several tissues (Lohler et al., 2009), its role in development has not been elucidated. Here we demonstrate that CK1δ is required for the proliferation and expansion of GCPs, one of two principal classes of neurons in the developing cerebellum. Decreasing CK1δ levels lowered cyclin levels. Furthermore, consistent with decreased cell cycle transition in the absence of CK1δ, inhibition or knockdown of CK1δ decreased the levels of the main effectors of the SHH pathway, which is an important mitogenic pathway for GCP expansion during cerebellar development (Salero and Hatten, 2007; Wechsler-Reya and Scott, 1999).
Centrosomal CK1δ mediates the formation of primary cilia, an organelle that functions in WNT- and SHH-signal transduction (Greer et al., 2014). In fact, several proteins that localize to primary cilia or are involved in ciliogenesis restrict cell proliferation by arresting cells at G1/S, G2/M, or both phases. Therefore, CK1δ deletion or inhibition may affect the cell cycle by disrupting ciliogenesis. Another possibility is that after CK1δ deletion or inhibition, increased levels of Wee1 induce cell cycle arrest (Penas et al., 2014). We found that CK1δ overexpression and depletion had the opposite effects on Wee1 levels. Namely, CK1δ overexpression reduced the level of Wee1, and CK1δ depletion increased it. Thus, modulating CK1δ levels appears to directly control Wee1 turnover, which is important for transitioning through the S and G2/M phases.
The present study shows that APC/CCdh1 complex–mediated degradation controls CK1δ levels in GCPs. Conditional deletion of Fzr1, which encodes Cdh1, in GCPs of the developing cerebellum increased the levels of CK1δ but not CK1α or CK1ε, and overexpression of CK1δ increased GCP proliferation. Although we demonstrated that APC/CCdh1 regulates CK1δ levels, we did not detect a difference in EdU incorporation in the Fzr1-knockout mice relative to their wild-type littermates, which may be due to incomplete deletion of Fzr1 or compensation from Cdc20, another APC/C activator. CK1δ is the only CK1 isoform that is targeted by APC/CCdh1 in the developing cerebellum. How APC/CCdh1 acquires specificity for the CK1δ isoform in the context of GCP proliferation is unknown, since other CK1 isoforms also contain D-boxes that could potentially mediate turnover via APC/CCdh1. However, CK1δ is the only CK1 isoform that contains an N-terminal D-box motif; the other CK1 members may require activation of upstream signaling pathways to be recognized by APC/CCdh1. The identification of GCP-specific interactors or substrates may also shed light on the mechanism by which APC/CCdh1 regulates CK1δ levels during cell cycle progression in this system.
CK1δ may be deregulated in medulloblastoma. CK1δ protein level was higher in two different types of medulloblastoma, Ptch1−/−-driven and Myc-driven medulloblastomas, which model SHH and G3 subtypes of human medulloblastoma, respectively. Increased CK1δ expression in medulloblastoma is consistent with previous findings of elevated CK1δ levels in adenocarcinoma (Brockschmidt et al., 2008) and breast cancer (Knippschild et al., 2005). Thus, CK1δ is an attractive therapeutic target because highly specific, small-molecule inhibitors can be generated against it (Bischof et al., 2012; Rena et al., 2004). We developed and characterized the highly selective CK1δ small-molecule inhibitor SR-1277, which reduced medulloblastoma tumor growth in vivo. SR-1277 decreased the proliferation of medulloblastoma cells that either contained alterations in SHH signaling or overexpressed Myc. C-MYC expression has been linked to poor outcome in multiple studies of patients with medulloblastoma (Gilbertson and Ellison, 2008; Hatten and Roussel, 2011); thus, it will be important to study the effectiveness of SR-1277 treatment for human G3 medulloblastoma.
In summary, the present study showed that CK1δ regulates cerebellar GCP proliferation, suggesting an important role of CK1 in brain development. In addition, CK1δ may influence the malignant transformation of GCPs, as medulloblastoma growth was also related to CK1δ levels. Furthermore, our results demonstrate that CK1δ is targeted for proteolysis via the APC/CCdh1 ubiquitin ligase, suggesting that APC/CCdh1 regulates CK1δ in proliferating cells. They also indicate that measuring CK1δ protein levels in tumors will be essential for determining responsiveness to CK1δ-inhibitor treatment, as APC/CCdh1 is deregulated in various cancers (Garcia-Higuera et al., 2008a; Penas et al., 2011).
EXPERIMENTAL PROCEDURES
Animal Husbandry
This study was approved by the Institutional Animal Care and Use Committees of the University of Miami, The Rockefeller University, Scripps Florida, and St. Jude Children’s Research Hospital. (See Supplemental Information).
GCP Culture System
GCPs were purified from cerebellar cortices of P6 CD-1 (Jackson Laboratory), Tg(Atoh1-Cre);Csnk1dfl/fl, or Tg(Atoh1-Cre);Fzr1fl/fl mice by using Percoll gradient sedimentation (see Supplemental Information). For proliferation assays, GCPs were suspended in culture medium; for cell cycle exit and differentiation assays, GCPs were plated in poly-D-lysine/laminin–coated plates. GCPs were treated with compounds and then subsequently used for apoptosis, 3H-thymidine–incorporation, or EdU-proliferation assays; fixed for FACS analysis; or lysed to obtain protein for Western blot analysis or RNA for qRT-PCR analysis (see Supplemental Information).
Plasmids, siRNAs, and Site-directed Mutagenesis
The CK1δ-V5 construct was generated by cloning the full-length Csnk1d gene from the Gateway donor vector pDONR223-CSNK1D (Addgene) into the Gateway destination vector pcDNA-DEST40 (Invitrogen). The siRNAs and primers used for cloning are listed in the Supplemental Information.
HeLa Cell Culture System
HeLa cells were transfected with plasmids by using TransIT-LT1 transfection reagent (Mirus Bio) or with siRNAs using DharmaFECT 1 transfection reagent (Thermo Scientific) per the manufacturers’ instructions. HeLa cells were lysed for in vitro cyclohexamide degradation and ubiquitination assays, to obtain protein for Western blot analysis or RNA for qRT-PCR analysis, or synchronized and fixed for flow cytometric analysis (see Supplemental Information).
Organotypic Slice Cultures and Proliferation Assays
Cerebella were isolated from P8 mice, and 250-μm sagittal slices of cerebellar cortex were cut using a Leica VT1000S vibratome. Slices were then plated on Millipore culture inserts in 6-well culture dishes (Falcon) containing 1.5 mL serum-free medium. Slices were treated with compounds, and EdU-incorporation assay or immunohistochemical analyses were performed (see Supplemental Information).
In vivo Allograft
Ptch1+/− tumor cells in matrigel (BD Biosciences) solution were injected subcutaneously into the right flank of a NU-Foxn1nu mouse (Charles River Laboratories). Treatment with SR-1277 began when tumors reached 50 to 90 mm3 (see Supplemental Information).
Murine G3 Medulloblastoma Neurospheres in Culture
Tumor cells were maintained in culture as previously described (Kawauchi et al., 2012) (see Supplemental Information). They were treated with SR-1277, and EdU-proliferation assays were performed.
Transduction and Transplantation of DAOY Cells
DAOY cells were transduced with firefly luciferase lentivirus (Capital Biosciences). Stable clones were then selected with puromycin (see Supplemental Information), and 105 labeled DAOY cells were injected into the ventral pallidum of NCr nude mice (Taconic). After 10 days, tumor growth was monitored weekly by bioluminiscence imaging of the pallidum (see Supplemental Information).
Statistical Analyses
All experiments were conducted independently and at least in triplicate. Statistical analysis was performed with Prism software (Graphpad). Data in Figures 1D, 1E, 1G, 2F, 3A, 3B, S1A, S1B, and 6D were analyzed via one-way ANOVA followed by Bonferroni multiple comparisions test (p <0.5); that in Figures 1F and 2G, two-way ANOVA followed by Bonferroni multiple comparisions test (p <0.5); that in Figures 2D, 7F, and 7G, paired t test (p <0.5); and that in Figures 1B, 7B, 7C, and 7H, one-way ANOVA followed by Dunnett multiple comparisions test (p <0.5).
Supplementary Material
HIGHLIGHTS.
CK1δ is required for cerebellar granule cell progenitor neurogenesis.
CK1δ inhibition or CK1δ knockdown induces cell cycle arrest.
CK1δ is targeted for degradation via the anaphase-promoting complex/cyclosome.
CK1δ destruction is required for cell cycle exit.
ACKNOWLEDGMENTS
We thank Dr. David Rowitch (UCSF) for providing the Atoh1-Cre deleter line and Dr. Marc Kirschner for helpful discussions and critical reading of the manuscript. We also thank all members of the Center for Therapeutic Innovation (University of Miami) and Department of Cancer Biology (Scripps Florida) for helpful suggestions. We thank Dr. Angela MacArthur and the Department of Scientific Editing at St. Jude Children’s Research Hospital for helpful suggestions. This work was supported by R21 NS056991 (N.A.), R01NS067289 (N.A. and M.E.H.), NIH grants NCI CA-096832 (M.F.R.), Core Grant CA-021765 (M.F.R., D.K., and D.F.), the Anderson Fellowship (D.K.), and ALSAC (M.F.R.); The Spanish Ministerio de Economía y Competitividad (MINECO; SAF2012-38215), Fundación Ramón Areces, Comunidad de Madrid (S2010/BMD-2470), and the European Union Seventh Framework Programme (MitoSys project; HEALTH-F5-2010-241548) (M.M.). The authors have no financial interests to declare.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Barford D. Structure, function and mechanism of the anaphase promoting complex (APC/C) Q. Rev. Biophys. 2011;44:153–190. doi: 10.1017/S0033583510000259. [DOI] [PubMed] [Google Scholar]
- Behrend L, Stoter M, Kurth M, Rutter G, Heukeshoven J, Deppert W, Knippschild U. Interaction of casein kinase 1 delta (CK1delta) with post-Golgi structures, microtubules and the spindle apparatus. Eur. J. C.ell Biol. 2000;79:240–251. doi: 10.1078/s0171-9335(04)70027-8. [DOI] [PubMed] [Google Scholar]
- Ben-Arie N, Bellen HJ, Armstrong DL, McCall AE, Gordadze PR, Guo Q, Matzuk MM, Zoghbi HY. Math1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390:169–172. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- Beyaert R, Vanhaesebroeck B, Declercq W, Van Lint J, Vandenabele P, Agostinis P, Vandenheede JR, Fiers W. Casein kinase-1 phosphorylates the p75 tumor necrosis factor receptor and negatively regulates tumor necrosis factor signaling for apoptosis. J. Biol. Chem. 1995;270:23293–23299. doi: 10.1074/jbc.270.40.23293. [DOI] [PubMed] [Google Scholar]
- Bibian M, Rahaim RJ, Choi JY, Noguchi Y, Schurer S, Chen W, Nakanishi S, Licht K, Rosenberg LH, Li L, et al. Development of highly selective casein kinase 1delta/1epsilon (CK1delta/epsilon) inhibitors with potent antiproliferative properties. Bioorg. Med. Chem. Lett. 2013;23:4374–4380. doi: 10.1016/j.bmcl.2013.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J, Leban J, Zaja M, Grothey A, Radunsky B, Othersen O, Strobl S, Vitt D, Knippschild U. 2-Benzamido-N-(1H-benzo[d]imidazol-2-yl)thiazole-4-carboxamide derivatives as potent inhibitors of CK1δ/ε. Amino Acids. 2012;43:1577–1591. doi: 10.1007/s00726-012-1234-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockschmidt C, Hirner H, Huber N, Eismann T, Hillenbrand A, Giamas G, Radunsky B, Ammerpohl O, Bohm B, Henne-Bruns D, et al. Anti-apoptotic and growth-stimulatory functions of CK1 delta and epsilon in ductal adenocarcinoma of the pancreas are inhibited by IC261 in vitro and in vivo. Gut. 2008;57:799–806. doi: 10.1136/gut.2007.123695. [DOI] [PubMed] [Google Scholar]
- Cheong JK, Virshup DM. Casein kinase 1: Complexity in the family. Int. J. Biochem. Cell Biol. 2011;43:465–469. doi: 10.1016/j.biocel.2010.12.004. [DOI] [PubMed] [Google Scholar]
- Cho YJ, Tsherniak A, Tamayo P, Santagata S, Ligon A, Greulich H, Berhoukim R, Amani V, Goumnerova L, Eberhart CG, et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J. Clin. Oncol. 2011;29:1424–1430. doi: 10.1200/JCO.2010.28.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E, Dial JM, Jeong DE, Hall MC. Unique D box and KEN box sequences limit ubiquitination of Acm1 and promote pseudosubstrate inhibition of the anaphase-promoting complex. J. Biol. Chem. 2008;283:23701–23710. doi: 10.1074/jbc.M803695200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desagher S, Osen-Sand A, Montessuit S, Magnenat E, Vilbois F, Hochmann A, Journot L, Antonsson B, Martinou JC. Phosphorylation of bid by casein kinases I and II regulates its cleavage by caspase 8. Mol. Cell. 2001;8:601–611. doi: 10.1016/s1097-2765(01)00335-5. [DOI] [PubMed] [Google Scholar]
- Edmondson JC, Hatten ME. Glial-guided granule neuron migration in vitro: a high-resolution time-lapse video microscopic study. J. Neurosci. 1987;7:1928–1934. doi: 10.1523/JNEUROSCI.07-06-01928.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison DW, Dalton J, Kocak M, Nicholson SL, Fraga C, Neale G, Kenney AM, Brat DJ, Perry A, Yong WH, et al. Medulloblastoma: clinicopathological correlates of SHH, WNT, and non-SHH/WNT molecular subgroups. Acta. Neuropathol. 2011;121:381–396. doi: 10.1007/s00401-011-0800-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DG, Farndon PA, Burnell LD, Gattamaneni HR, Birch JM. The incidence of Gorlin syndrome in 173 consecutive cases of medulloblastoma. Br J Cancer. 1991;64:959–961. doi: 10.1038/bjc.1991.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Higuera I, Manchado E, Dubus P, Canamero M, Mendez J, Moreno S, Malumbres M. Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat. Cell Biol. 2008a;10:802–811. doi: 10.1038/ncb1742. [DOI] [PubMed] [Google Scholar]
- Garcia-Higuera I, Manchado E, Dubus P, Canamero M, Mendez J, Moreno S, Malumbres M. Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat. Cell Biol. 2008b;10:802–811. doi: 10.1038/ncb1742. [DOI] [PubMed] [Google Scholar]
- Gault WJ, Olguin P, Weber U, Mlodzik M. Drosophila CK1-gamma, gilgamesh, controls PCP-mediated morphogenesis through regulation of vesicle trafficking. J. Cell Biol. 2012;196:605–621. doi: 10.1083/jcb.201107137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson P, Tong Y, Robinson G, Thompson MC, Currle DS, Eden C, Kranenburg TA, Hogg T, Poppleton H, Martin J, et al. Subtypes of medulloblastoma have distinct developmental origins. Nature. 2010;468:1095–1099. doi: 10.1038/nature09587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbertson RJ, Ellison DW. The origins of medulloblastoma subtypes. Ann. Rev. Pathol. 2008;3:341–365. doi: 10.1146/annurev.pathmechdis.3.121806.151518. [DOI] [PubMed] [Google Scholar]
- Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- Greer YE, Westlake CJ, Gao B, Bharti K, Shiba Y, Xavier CP, Pazour GJ, Yang Y, Rubin JS. Casein kinase 1delta functions at the centrosome and Golgi to promote ciliogenesis. Mol. Biol. Cell. 2014;25:1629–1640. doi: 10.1091/mbc.E13-10-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross SD, Anderson RA. Casein kinase I: spatial organization and positioning of a multifunctional protein kinase family. Cell. Signal. 1998;10:699–711. doi: 10.1016/s0898-6568(98)00042-4. [DOI] [PubMed] [Google Scholar]
- Hallahan AR, Pritchard JI, Hansen S, Benson M, Stoeck J, Hatton BA, Russell TL, Ellenbogen RG, Bernstein ID, Beachy PA, et al. The SmoA1 mouse model reveals that notch signaling is critical for the growth and survival of sonic hedgehog-induced medulloblastomas. Cancer Res. 2004;64:7794–7800. doi: 10.1158/0008-5472.CAN-04-1813. [DOI] [PubMed] [Google Scholar]
- Harmey D, Smith A, Simanski S, Moussa CZ, Ayad NG. The anaphase promoting complex induces substrate degradation during neuronal differentiation. J. Biol. Chem. 2009;284:4317–4323. doi: 10.1074/jbc.M804944200. [DOI] [PubMed] [Google Scholar]
- Hatten ME, Roussel MF. Development and cancer of the cerebellum. Trends Neurosci. 2011;34:134–142. doi: 10.1016/j.tins.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Jager N, Kool M, Zichner T, Hutter B, Sultan M, Cho YJ, Pugh TJ, Hovestadt V, Stutz AM, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi D, Robinson G, Uziel T, Gibson P, Rehg J, Gao C, Finkelstein D, Qu C, Pounds S, Ellison DW, et al. A mouse model of the most aggressive subgroup of human medulloblastoma. Cancer Cell. 2012;21:168–180. doi: 10.1016/j.ccr.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H, Stephen D, Joyner A, Curran T. Gli1 is important for medulloblastoma formation in Ptc1+/− mice. Oncogene. 2005;24:4026–4036. doi: 10.1038/sj.onc.1208567. [DOI] [PubMed] [Google Scholar]
- King RW, Glotzer M, Kirschner MW. Mutagenic analysis of the destruction signal of mitotic cyclins and structural characterization of ubiquitinated intermediates. Mol. Biol. Cell. 1996;7:1343–1357. doi: 10.1091/mbc.7.9.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisselev AF, van der Linden WA, Overkleeft HS. Proteasome inhibitors: an expanding army attacking a unique target. Chem. Biol. 2012;19:99–115. doi: 10.1016/j.chembiol.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knippschild U, Gocht A, Wolff S, Huber N, Lohler J, Stoter M. The casein kinase 1 family: participation in multiple cellular processes in eukaryotes. Cell. Signal. 2005;17:675–689. doi: 10.1016/j.cellsig.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Listovsky T, Oren YS, Yudkovsky Y, Mahbubani HM, Weiss AM, Lebendiker M, Brandeis M. Mammalian Cdh1/Fzr mediates its own degradation. EMBO J. 2004;23:1619–1626. doi: 10.1038/sj.emboj.7600149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohler J, Hirner H, Schmidt B, Kramer K, Fischer D, Thal DR, Leithauser F, Knippschild U. Immunohistochemical characterisation of cell-type specific expression of CK1delta in various tissues of young adult BALB/c mice. PLoS One. 2009;4:e4174. doi: 10.1371/journal.pone.0004174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, Mack S, Bouffet E, Clifford SC, Hawkins CE, French P, et al. Medulloblastoma comprises four distinct molecular variants. J. Clin. Oncol. 2011;29:1408–1414. doi: 10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Shih DJ, Peacock J, Garzia L, Morrissy AS, Zichner T, Stutz AM, Korshunov A, Reimand J, Schumacher SE, et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature. 2012;488:49–56. doi: 10.1038/nature11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens L, Simanski S, Squire C, Smith A, Cartzendafner J, Cavett V, Caldwell Busby J, Sato T, Ayad NG. Activation domain-dependent degradation of somatic Wee1 kinase. J. Biol. Chem. 2010;285:6761–6769. doi: 10.1074/jbc.M109.093237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens TJ, Hoyt MA. The D box asserts itself. Mol. Cell. 2005;18:611–612. doi: 10.1016/j.molcel.2005.05.019. [DOI] [PubMed] [Google Scholar]
- Penas C, Ramachandran V, Ayad NG. The APC/C ubiquitin ligase: from cell biology to tumorigenesis. Front. Oncol. 2011;1:60. doi: 10.3389/fonc.2011.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penas C, Ramachandran V, Simanski S, Lee C, Madoux F, Rahaim RJ, Chauhan R, Barnaby O, Schurer S, Hodder P, et al. Casein kinase 1delta dependent Wee1 degradation. J. Biol. Chem. 2014;289:18893–18903. doi: 10.1074/jbc.M114.547661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M, Matos J, Mori S, Gregan J, Bogdanova A, Schwickart M, Mechtler K, Shirahige K, Zachariae W, Nasmyth K. Monopolar attachment of sister kinetochores at meiosis I requires casein kinase 1. Cell. 2006;126:1049–1064. doi: 10.1016/j.cell.2006.07.029. [DOI] [PubMed] [Google Scholar]
- Pfister S, Remke M, Benner A, Mendrzyk F, Toedt G, Felsberg J, Wittmann A, Devens F, Gerber NU, Joos S, et al. Outcome prediction in pediatric medulloblastoma based on DNA copy-number aberrations of chromosomes 6q and 17q and the MYC and MYCN loci. J. Clin. Oncol. 2009;27:1627–1636. doi: 10.1200/JCO.2008.17.9432. [DOI] [PubMed] [Google Scholar]
- Price MA. CKI, there's more than one: casein kinase I family members in Wnt and Hedgehog signaling. Genes Dev. 2006;20:399–410. doi: 10.1101/gad.1394306. [DOI] [PubMed] [Google Scholar]
- Pugh TJ, Weeraratne SD, Archer TC, Pomeranz Krummel DA, Auclair D, Bochicchio J, Carneiro MO, Carter SL, Cibulskis K, Erlich RL, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106–110. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. Mode of cell migration to the superficial layers of fetal monkey neocortex. J. Comp. Neurol. 1972;145:61–83. doi: 10.1002/cne.901450105. [DOI] [PubMed] [Google Scholar]
- Rena G, Bain J, Elliott M, Cohen P. D4476, a cell-permeant inhibitor of CK1, suppresses the site-specific phosphorylation and nuclear exclusion of FOXO1a. EMBO Rep. 2004;5:60–65. doi: 10.1038/sj.embor.7400048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L, Phoenix TN, Hedlund E, Wei L, Zhu X, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488:43–48. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel MF, Hatten ME. Cerebellum development and medulloblastoma. Curr. Top. Dev. Biol. 2011;94:235–282. doi: 10.1016/B978-0-12-380916-2.00008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowles J, Slaughter C, Moomaw C, Hsu J, Cobb MH. Purification of casein kinase I and isolation of cDNAs encoding multiple casein kinase I-like enzymes. Proc. Natl. Acad. Sci. U. S. A. 1991;88:9548–9552. doi: 10.1073/pnas.88.21.9548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salero E, Hatten ME. Differentiation of ES cells into cerebellar neurons. Proc. Natl. Acad. Sci. U. S. A. 2007;104:2997–3002. doi: 10.1073/pnas.0610879104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller U, Heine VM, Mao J, Kho AT, Dillon AK, Han YG, Huillard E, Sun T, Ligon AH, Qian Y, et al. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell. 2008;14:123–134. doi: 10.1016/j.ccr.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller U, Zhao Q, Godinho SA, Heine VM, Medema RH, Pellman D, Rowitch DH. Forkhead transcription factor FoxM1 regulates mitotic entry and prevents spindle defects in cerebellar granule neuron precursors. Mol. Cell Biol. 2007;27:8259–8270. doi: 10.1128/MCB.00707-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A, Simanski S, Fallahi M, Ayad NG. Redundant ubiquitin ligase activities regulate wee1 degradation and mitotic entry. Cell Cycle. 2007;6:2795–2799. doi: 10.4161/cc.6.22.4919. [DOI] [PubMed] [Google Scholar]
- Song L, Rape M. Substrate-specific regulation of ubiquitination by the anaphase-promoting complex. Cell Cycle. 2011;10:52–56. doi: 10.4161/cc.10.1.14387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart S, Fang G. Destruction box-dependent degradation of aurora B is mediated by the anaphase-promoting complex/cyclosome and Cdh1. Cancer Res. 2005;65:8730–8735. doi: 10.1158/0008-5472.CAN-05-1500. [DOI] [PubMed] [Google Scholar]
- Svard J, Heby-Henricson K, Persson-Lek M, Rozell B, Lauth M, Bergstrom A, Ericson J, Toftgard R, Teglund S. Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev. Cell. 2006;10:187–197. doi: 10.1016/j.devcel.2005.12.013. [DOI] [PubMed] [Google Scholar]
- Taylor MD, Liu L, Raffel C, Hui CC, Mainprize TG, Zhang X, Agatep R, Chiappa S, Gao L, Lowrance A, et al. Mutations in SUFU predispose to medulloblastoma. Nat Genet. 2002;31:306–310. doi: 10.1038/ng916. [DOI] [PubMed] [Google Scholar]
- Tomoda T, Bhatt RS, Kuroyanagi H, Shirasawa T, Hatten ME. A mouse serine/threonine kinase homologous to C. elegans UNC51 functions in parallel fiber formation of cerebellar granule neurons. Neuron. 1999;24:833–846. doi: 10.1016/s0896-6273(00)81031-4. [DOI] [PubMed] [Google Scholar]
- Vielhaber E, Virshup DM. Casein kinase I: from obscurity to center stage. IUBMB Life. 2001;51:73–78. doi: 10.1080/15216540117461. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Arai H, Iwasaki J, Shiina M, Ogata K, Hunter T, Osada H. Cyclin-dependent kinase (CDK) phosphorylation destabilizes somatic Wee1 via multiple pathways. Proc. Natl. Acad. Sci. U. S. A. 2005;102:11663–11668. doi: 10.1073/pnas.0500410102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N, Arai H, Nishihara Y, Taniguchi M, Watanabe N, Hunter T, Osada H. M-phase kinases induce phospho-dependent ubiquitination of somatic Wee1 by SCFbeta-TrCP. Proc. Natl. Acad. Sci. U. S. A. 2004;101:4419–4424. doi: 10.1073/pnas.0307700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler-Reya R, Scott MP. The developmental biology of brain tumors. Annu. Rev. Neurosci. 2001;24:385–428. doi: 10.1146/annurev.neuro.24.1.385. [DOI] [PubMed] [Google Scholar]
- Wechsler-Reya RJ, Scott MP. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron. 1999;22:103–114. doi: 10.1016/s0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- Yang XW, Wynder C, Doughty ML, Heintz N. BAC-mediated gene-dosage analysis reveals a role for Zipro1 (Ru49/Zfp38) in progenitor cell proliferation in cerebellum and skin. Nat. Genet. 1999;22:327–335. doi: 10.1038/11896. [DOI] [PubMed] [Google Scholar]
- Yauch RL, Dijkgraaf GJ, Alicke B, Januario T, Ahn CP, Holcomb T, Pujara K, Stinson J, Callahan CA, Tang T, et al. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science. 2009;326:572–574. doi: 10.1126/science.1179386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai L, Graves PR, Robinson LC, Italiano M, Culbertson MR, Rowles J, Cobb MH, DePaoli-Roach AA, Roach PJ. Casein kinase I gamma subfamily. Molecular cloning, expression, and characterization of three mammalian isoforms and complementation of defects in the Saccharomyces cerevisiae YCK genes. J. Biol. Chem. 1995;270:12717–12724. doi: 10.1074/jbc.270.21.12717. [DOI] [PubMed] [Google Scholar]
- Zur A, Brandeis M. Timing of APC/C substrate degradation is determined by fzy/fzr specificity of destruction boxes. EMBO J. 2002;21:4500–4510. doi: 10.1093/emboj/cdf452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.