SUMMARY
Proline-directed phosphorylation is regulated by the prolyl isomerase Pin1, which plays a fundamental role in driving breast cancer stem-like cells (BCSCs). Rab2A is a small GTPase critical for vesicle trafficking. Here, we show that Pin1 increases Rab2A transcription to promote BCSC expansion and tumorigenesis in vitro and in vivo. Mechanistically, Rab2A directly interacts with and prevents dephosphorylation/inactivation of Erk1/2 by the MKP3 phosphatase, resulting in Zeb1 upregulation and β-catenin nuclear translocation. In cancer cells, Rab2A is activated via gene amplification, mutation or Pin1 overexpression. Rab2A overexpression or mutation endows BCSC traits to primary normal human breast epithelial cells, whereas silencing Rab2A potently inhibits the expansion and tumorigenesis of freshly isolated BCSCs. Finally, Rab2A overexpression correlates with poor clinical outcome in breast cancer patients. Thus, Pin1/Rab2A/Erk drives BCSC expansion and tumorigenicity, suggesting potential drug targets.
INTRODUCTION
Cancer stem cells (CSCs) or tumor-initiating cells (TICs) have been hypothesized to retain the capacity of self-renewal to regenerate the bulk of a heterogeneous tumor. The CSC concept has important implications for understanding the molecular mechanisms of cancer progression and identifying targets for cancer therapeutics, since CSCs are thought to be responsible for tumor initiation, progression, metastasis, relapse, and drug resistance (Liu and Wicha, 2010). Thus, the elucidation of CSC regulatory mechanisms and the identification of targets to eradicate the CSC compartment in a tumor may be essential to achieve long-term remission of cancer (Liu and Wicha, 2010).
An increasing number of regulators of breast cancer stem-like cells (BCSCs), notably transcription factors including Zeb1 and β-catenin, have been identified (Reya and Clevers, 2005; Wellner et al., 2009). These transcription modulators are further regulated by upstream signaling pathways. For example, Erk signaling has been shown to regulate BCSCs by increasing transcription of Zeb1 and nuclear accumulation of unphosphorylated (active) β-catenin (Chang et al., 2011; Shin et al., 2010). However, regulatory pathways upstream of Erk signaling that regulate BCSCs are still not fully understood.
Among the small GTPase superfamily, Ras has been shown to induce the epithelial-mesenchymal transition (EMT) and confer CSC traits to breast cells in vitro and in vivo (Liu et al., 2009; Shin et al., 2010). Rac1 is involved in CSC maintenance in non-small cell lung adenocarcinoma and glioma, as well as in intestinal progenitor and stem cell expansion (Akunuru et al., 2011; Myant et al., 2013; Yoon et al., 2011). However, the roles of other GTPase family members in CSCs in solid tumors are yet to be elucidated.
Protein phosphorylation on certain serine or threonine residues preceding a proline (pSer/Thr-Pro) is a central signaling mechanism in cell proliferation and transformation (Blume-Jensen and Hunter, 2001). We have shown that certain pSer/Thr-Pro motifs exist in two distinct conformations, cis and trans, and identified Pin1, a unique prolyl isomerase that binds to and catalyzes cis/trans isomerization of specific pSer/Thr-Pro motifs (Lu et al., 1996; Lu and Zhou, 2007; Yaffe et al., 1997). Pin1 induces conformational changes of these Ser/Thr-Pro motifs after phosphorylation, which now can be visualized by proline isomer-specific antibodies (Nakamura et al., 2012). Importantly, Pin1 is overexpressed and/or activated in human cancers and plays a critical role in breast cancer development in vitro and in vivo (Chen et al., 2013; Lee et al., 2011; Lu and Zhou, 2007; Lu and Hunter, 2014). Recently, we and others have found that Pin1 is increased in human BCSCs and plays a fundamental role in driving BCSCs and tumorigenesis (Luo et al., 2014; Rustighi et al., 2014). Although Pin1 has been reported to activate and inactivate a large subset of key oncogenes and tumor suppressors, respectively (Lu and Zhou, 2007; Lu and Hunter, 2014), the downstream target of Pin1 in BCSCs is largely unknown.
In searching for Pin1 downstream targets in BCSCs using genome-wide expression profiling, we identified Rab2A, a small GTPase mainly localized to the ER-Golgi intermediate compartment (ERGIC) that is essential for membrane trafficking between the ER and Golgi apparatus, but with no known function in cancer or CSCs (Stenmark, 2009; Tisdale and Balch, 1996). We show that as a Pin1 transcriptional target, Rab2A is a BCSC-promoting gene that enhances tumorigenesis via activating Erk signaling. Thus, the Pin1/Rab2A/Erk axis drives BCSC expansion and tumorigenicity, providing attractive targets in BCSCs for cancer therapy.
RESULTS
Genomic Profiling Analyses Identifies Rab2A as a Pin1 Downstream Gene
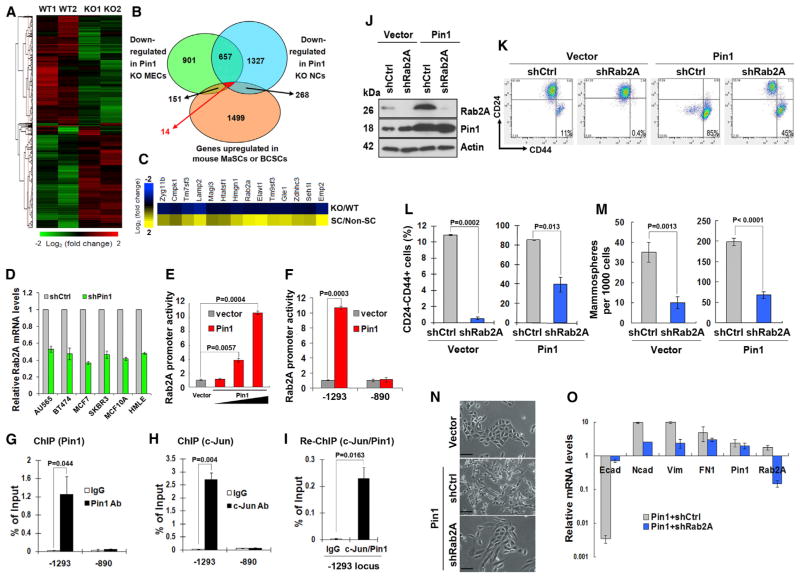
We have previously demonstrated a fundamental role of Pin1 in regulating human BCSCs and mouse mammary stem cells (MaSCs) (Luo et al., 2014). To elucidate the underlying molecular mechanisms, we analyzed the effects of Pin1 knockout (KO) on gene expression in mouse mammary epithelial cells (MECs). Global expression profiling of Lin− MECs from Pin1 KO and wild-type (WT) littermates identified 1,723 genes that were downregulated in both Pin1 KO mice (Figures 1A and 1B; Tables S1 and S2). To narrow down the list of Pin1-regulated genes, we compared MEC gene expression with that of neurospheres prepared from the same mice (Figure 1B; Table S3). Although comparing expression profiles of stem cells from WT and Pin1 KO mice may be a better approach, the MaSC-enriched Lin−CD24+CD29+ or Lin−CD24medCD49fhi populations are very small in Pin1 KO mice (Luo et al., 2014), which made it difficult to get enough RNA from each mouse. As an alternative approach, we re-analyzed two published expression profiling datasets of mouse MaSCs and BCSCs (Stingl et al., 2006; Zhang et al., 2008) and compared them with our expression profiling of Lin− MECs and neurons from Pin1 KO and WT mice. We identified 14 genes downregulated in both Pin1 KO MECs and neurons and upregulated in either MaSCs or BCSCs (Figure 1C; Table S4).
Figure 1. Genome-wide Expression Profiling Identifies Rab2A as a Pin1 Target that Promotes BCSC Expansion and EMT.
(A) Hierarchical cluster of the microarray data of the Lin− population of mammary epithelial cells in two pairs of WT and Pin1 KO littermates.
(B) Genomic profiling identified 14 potential target genes that were downregulated in Pin1 KO MECs and neuron cells (NCs) but upregulated in mouse MaSCs or BCSCs.
(C) Heatmap depicting the fold changes of 14 candidate genes, which were downregulated in Pin1 KO cells (presented by KO/WT ratio) but upregulated in either mouse MaSCs or BCSCs (presented by SC/non-SC ratio).
(D) Pin1 KD reduced Rab2A mRNA in human breast cancer lines, as assayed by real-time PCR.
(E and F) Pin1 activated the Rab2A promoter in a dose-dependent manner.
(G–I) Both Pin1 and c-Jun bound to the Rab2A promoter, as shown by ChIP and Re-ChIP analyses. Real-time PCR data were calibrated to immunoglobulin G (IgG) control and normalized with sample inputs of chromatin harvested prior to immunoprecipitation.
(J) Rab2A was knocked down in vector control and Pin1-overexpressing HMLE cells, as confirmed by immunoblot.
(K and L) Rab2A KD in HMLE cells reduced the CD24−CD44+ population and suppressed the ability of Pin1 overexpression to increase the CD24−CD44+ population.
(M) Rab2A KD in HMLE cells reduced mammosphere-forming activity and impaired the ability of Pin1 overexpression to increase mammosphere-forming activity.
(N and O) Rab2A KD impaired the ability of Pin1 overexpression to induce the EMT in HMLE cells, as shown by cell morphology (N) or upregulation of E-cadherin and downregulation of N-cadherin, fibronectin, and vimentin by qRT-PCR (O). Scale bar represents 100 μm.
In all panels, error bars represent SD of three independent experiments. See also Figures S1 and S2.
To validate these candidate genes, we used qRT-PCR to determine the effects of Pin1 knockdown (KD) on their expression in six human breast cell lines. Rab2A, Lamp2, and Magi3 were downregulated in more than five KD cell lines (Figures 1D and S1A). Pin1 KD also reduced Rab2A protein in all six different cell lines (Figure S1B). To test the effects of these genes in BCSCs, we silenced their expression using two different small hairpin RNAs (shRNAs) in MCF10A cells and examined the CD24−CD44+ population, which enriched human BCSCs (Al-Hajj et al., 2003). Rab2A KD consistently decreased the CD24−CD44+ population (Figure S1C), suggesting a requirement of Rab2A for BCSC maintenance. Thus, we focused on Rab2A as a Pin1 target.
In the promoter region of Rab2A, there are two putative AP-1 binding sites (−1,293 and −890) (Figure S1D). Notably, Pin1 activates transcription factors c-Jun and c-Fos to increase AP-1 activity (Monje et al., 2005; Wulf et al., 2001). We therefore tested whether Pin1 regulated Rab2A transcription. In the reporter assay, we found that Pin1 overexpression enhanced Rab2A transcription in a dose-dependent manner (Figure 1E). Deletion analysis suggested that Pin1 appeared to act on the distal AP-1 site, but not the proximal site (Figure 1F). To confirm that Pin1 regulates Rab2A transcription through AP-1, we first examined whether Pin1 bound to Rab2A promoter by chromatin immunoprecipitation (ChIP) using cells transfected with Pin1 expression plasmid. Compared to control immunoglobulin G, anti-Pin1 antibodies showed appreciable binding to the −1,293 locus, as assayed by qPCR (Figure 1G). Next, we used a c-Jun antibody to perform the ChIP assay in human mammary large-T and hTERT transformed epithelial cells (HMLE)-Ras cells, because Pin1 binds to c-Jun, which is phosphorylated by JNK and cooperates with Ras to increase the transcriptional activity of c-Jun toward its target genes (Wulf et al., 2001). Indeed, c-Jun specifically associated with the −1,293 locus in the Rab2A promoter (Figure 1H). Moreover, a sequential ChIP (re-ChIP) analysis using c-Jun antibody followed by Pin1 antibody demonstrated that both proteins were present in the same complex on the −1,293 locus (Figure 1I). Thus, Pin1 activates Rab2A transcription through AP-1 and increases its protein levels in breast cancer cells.
Rab2A Is a Major Mediator of Pin1 in Regulating BCSC Function
To investigate whether Rab2A is a functional downstream target of Pin1, we knocked down Rab2A in control or Pin1-overexpressing HMLE cells to examine whether Rab2A mediates the action of Pin1 in BCSCs (Figure 1J). As shown previously (Luo et al., 2014), Pin1 overexpression drastically increased the population of BCSC-enriched CD24−CD44+ cells by 8- to 9-fold above that of the vector control-infected HMLE cells (Figures 1K and 1L). Rab2A KD greatly reduced the size of the CD24−CD44+ population in vector control HMLE cells (Figures 1K and 1L), as did Pin1 KD (Luo et al., 2014). In Pin1-overexpressing cells, Rab2A KD partially decreased the abundance of CD24−CD44+ cells (Figures 1K and 1L). We then performed a mammosphere-forming assay, which measures the frequency of early progenitor/stem cells and BCSCs (Dontu and Wicha, 2005). Rab2A KD decreased mammosphere formation by both vector control and Pin1-overexpressing HMLE cells (Figure 1M). Thus, Rab2A was required to sustain the BCSC-enriched population in control cells and Pin1-overexpressing cells.
Recently, we showed that Pin1 overexpression induces EMT in HMLE cells (Luo et al., 2014). Strikingly, Rab2A KD in Pin1-overexpressing cells reverted the EMT phenotype. After Rab2A KD, Pin1-overexpressing HMLE cells changed to the epithelial morphology (Figure 1N), with increased E-cadherin and decreased N-cadherin, vimentin, and fibronectin levels (Figure 1O). Cell migration, a property associated with the EMT, was also greatly attenuated by Rab2A KD in Pin1-overexpressing cells in wound-healing (Figure S2A) and transwell migration assays (Figure S2B). Thus, Rab2A is a major mediator of Pin1 in regulating BCSC function.
Rab2A Is Amplified in Human Cancers and Its Overexpression Increases the BCSC-Enriched Population and Tumorigenicity
To determine more directly the role of Rab2A in breast cancer, we first checked Rab2A gene alterations in cancers in the cBio Cancer Genomics Portal (Cerami et al., 2012). Significantly, Rab2A gene amplification occurs in a wide range of human cancers, with the highest frequency of ~9.5% (72 of 760) in invasive breast carcinoma patients (Figure 2A). Importantly, Rab2A mRNA levels increase significantly with increasing gene copy number in invasive breast carcinomas (p = 1.56E−84) and others (Figure S3A). Moreover, Rab2A amplification is independent of MYC on 8q, according to Tumorscape software (Beroukhim et al., 2010).
Figure 2. Rab2A Is Amplified in Human Cancers, Its Overexpression Expands BCSCs and Tumorigenicity, and the Cancer-Derived Rab2A Q58H Mutant Has Reduced GTPase Activity.
(A) Rab2A gene amplification in a wide range of human cancers reported in cBioPortal.
(B) Stable overexpression of Rab2A in Pin1 KD or control HMLE cells using retrovirus-mediated gene transfer, as determined by immunoblot.
(C) Overexpression of Rab2A in HMLE cells potently induced the CD24−CD44+ population and rescued the phenotypes inhibited by Pin1 KD.
(D) Overexpression of Rab2A increased the mammosphere formation in control shRNA (shCtrl) HMLE cells and rescued the phenotypes inhibited by Pin1 KD.
(E and F) Overexpression of Rab2A potently induced the EMT in HMLE cells, as assayed by cell morphology (E) and real-time RT-PCR of the marker expressions (F).
(G and H) Rab2A overexpression increased tumorigenicity of BCSCs, while its KD impaired the ability of Pin1 overexpression to increase tumorigenicity of BCSCs, as measured by a limiting-dilution tumor-initiation assay in nude mice. Two months later, mice were sacrificed and evaluated for tumor weight (G) and tumor incidence (H).
(I) Q58 in Rab2A is evolutionally conserved across species.
(J and K) The Q58H mutant displayed decreased GTP hydrolysis activity, compared to the WT Rab2A protein in the in vitro GTPase assay, as monitored by α-32P-labeled GTP hydrolysis (J), and quantified by densitometry of three independent experiments (K).
(L) HMLE-Ras cells infected with Rab2A Q58H were more potent in forming tumors than those infected with WT Rab2A when overexpressed at the endogenous levels.
In all panels, error bars represent SD of three independent experiments. See also Figure S3.
We then overexpressed Rab2A in control shRNA or Pin1 KD HMLE cells (Figure 2B). Moderate Rab2A overexpression (two to three times the endogenous level) not only strongly increased the CD24−CD44+ population (Figures 2C and S3B) and mammosphere formation (Figure 2D) in control HMLE cells but also significantly rescued the defects in BCSC-associated properties in Pin1 KD cells (Figures 2C and 2D). Like Pin1 overexpression (Luo et al., 2014), ectopic Rab2A expression also induced EMT in HMLE cells, which developed an elongated fibroblast-like morphology with decreased cell-cell contact (Figure 2E). The EMT phenotype was further confirmed by decreased E-cadherin and increased N-cadherin, vimentin, and fibronectin expression in Rab2A-overexpressing cells (Figure 2F). Rab2A overexpression also enhanced cell migration in wound-healing (Figures S3C and S3D) and transwell (Figures S3E and S3F) assays. To further investigate whether Rab2A is sufficient to induce transformation, we performed soft-agar colony formation assay on Rab2A-overexpressing and control vector cells. Whereas control cells could hardly form colonies, Rab2A-overexpressing cells robustly formed colonies (Figures S3G and S3H), further supporting the oncogenic activity of Rab2A.
To evaluate the impact of Rab2A on tumor initiation, we assessed the effects of Rab2A overexpression on tumor formation by limiting dilution transplantation assays in nude mice. We used HMLER cells, HMLE cells transformed with V12H-Ras to form tumors in nude mice (Elenbaas et al., 2001). No mice inoculated with 1 × 104 control HMLER cells developed tumors, while tumors developed in only two of eight mice inoculated with 105 control cells and three of six mice injected with 106 control cells (Figures 2G and 2H). To examine whether endogenous Rab2A is necessary for Pin1 to promote tumorigenicity of BCSCs, we knocked down Rab2A in Pin1-overexpressing HMLER cells. No tumors arose when 1 × 104 Pin1-expressing cells infected with Rab2A shRNA (shRab2A) were injected into mice (Figures 2G and 2H). Although four of eight mice inoculated with 105 Pin1-shRab2A HMLER cells formed tumors, seven mice injected with an equal number of Pin1 cells developed tumors. Similarly, with Pin1 overexpression, 106 Rab2A KD cells formed fewer tumors than control Pin1-overexpressing HMLER cells. Thus, Rab2A inhibition potently impairs the ability of Pin1 to promote BCSC tumorigenicity.
Rab2A Is Mutated in Human Cancers and the Q58H Mutation Activates Rab2A
In addition to gene amplification, the Rab2A Q58H mutation has been repeatedly identified in lung cancer patients in the cBio Cancer Genomics Portal (Cerami et al., 2012). Given that Q58 is highly conserved in Rab2A genes across species (Figure 2I) and most of the oncogenic mutants in the Ras superfamily affect the enzyme’s ability to hydrolyze GTP (Schubbert et al., 2007), we examined whether this mutation might affect the intrinsic ability of Rab2A to hydrolyze GTP. Indeed, Rab2A Q58H hydrolyzed [α-32P]GTP to [α-32P]GDP more slowly than the WT protein (Figures 2J and 2K), resulting in more protein in the GTP-bound state.
We then asked whether the Q58H mutation might increase the potency of Rab2A to expand the BCSC-enriched population. We stably expressed Flag-Rab2A and its mutant using lentiviruses with a less optimal Kozak sequence, resulting in proteins expressed similar to the endogenous level (Figure S3I). Interestingly, Rab2A increased the CD24−CD44+ percentage to 59%, but Rab2A Q58H increased this population to 79% (Figure S3J). To examine whether the Q58H mutation increased tumorigenicity, we examined tumor formation by injecting 1 × 106 HMLER cells infected with vector control or Flag-Rab2A and Q58H mutant expressing at the endogenous level into nude mice. Although cells expressing WT Rab2A or its Q58H mutant formed tumors in all mice, the Q58H mutant tumors grew significantly faster than WT controls (Figures 2L and S3K), suggesting that the Rab2A Q58H mutant is more active in promoting tumor growth than WT protein.
Erk1/2 Activation Is Essential for Rab2A to Regulate BCSC Expansion
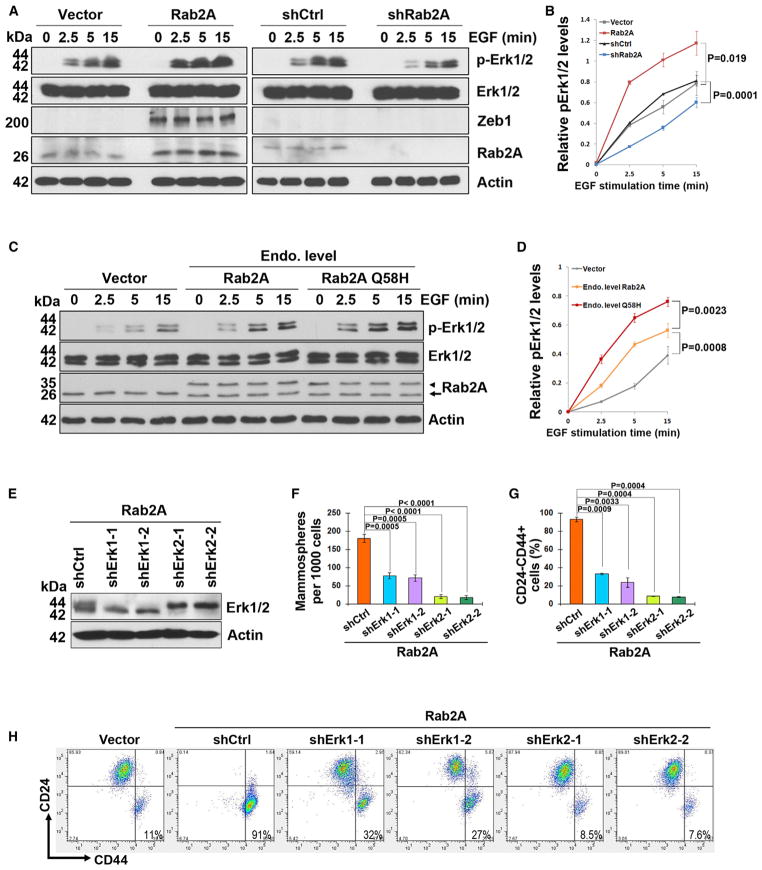
To understand how Rab2A expands the BCSC-enriched population, we examined whether Rab2A activates Erk1/2, which is crucial for Ras to induce EMT and increase the CD24−CD44+ population (Shin et al., 2010). After serum starvation and EGF stimulation, Rab2A overexpression significantly increased Erk1/2 activation monitored by p-Erk1/2 in a time-dependent manner and also increased expression of Zeb1 (Figures 3A and 3B), a transcription factor critical for inducing the EMT and CD24−CD44+ population (Shin et al., 2010; Wellner et al., 2009). In contrast, Rab2A KD substantially impaired Erk1/2 activation (Figures 3A and 3B). We then asked whether the Q58H mutation might increase Erk1/2 phosphorylation. When expressed at the endogenous level, the Q58H mutant induced Erk1/2 activation even faster than WT Rab2A after EGF stimulation (Figures 3C and 3D). Thus, Rab2A and its Q58H mutant promote Erk1/2 activation. Next, we silenced Erk1 or 2 in Rab2A-overexpressing HMLE cells (Figure 3E). Erk1 KD only partially inhibited, but Erk2 KD substantially decreased, the ability of Rab2A overexpression to induce mammosphere formation (Figure 3F) and the CD24−CD44+ population (Figures 3G and 3H), indicating that Rab2A acts through Erk1/2 to maintain BCSC-associated properties.
Figure 3. Rab2A and Its Q58H Mutant Drive BCSC Expansion via Activating Erk1/2.
(A and B) Rab2A regulated Erk1/2 phosphorylation and downstream Zeb1 expression. HMLE cells stably expressing Rab2A or shRNA or control vectors were treated with EGF after serum starvation for the indicated time points to activate Erk1/2 and analyzed by immunoblot.
(C and D) Rab2A Q58H mutant activated Erk1/2 faster than WT Rab2A when overexpressed at the endogenous levels after EGF treatment. Arrowhead, exogenous Flag-Rab2A; arrow, endogenous Rab2A. immunoprecipitated by Rab2A in a dose-dependent manner (Figure 4G), suggesting that Rab2A competed with MKP3 to bind Erk1/2 in vivo. However, unlike the MKP3 competition results, similar amounts of Erk1/2 were immunoprecipitated by Flag-Rab2A even though decreasing amounts of MEK1 were expressed (Figure S5B). These results may be expected, because although the docking motif of MEK is important for the ERK-MEK interaction, there are other mechanisms to ensure the activation of Erk by MEK1, such as scaffold protein facilitation and the MEK catalytic site interacting with the Erk activation loop (Roskoski, 2012).
(E) Erk1 or Erk2 was knocked down by two independent lentivirus-mediated shRNAs in Rab2A-overexpressing cells.
(F) KD of Erk1/2, especially Erk2, prevented Rab2A from increasing the mammosphere-forming capability.
(G and H) KD of Erk1/2, especially Erk2, prevented Rab2A from increasing the CD24−CD44+ population. In all panels, error bars represent SD of three independent experiments.
Rab2A Directly Interacts with and Prevents Erk1/2 Inactivation by MKP3
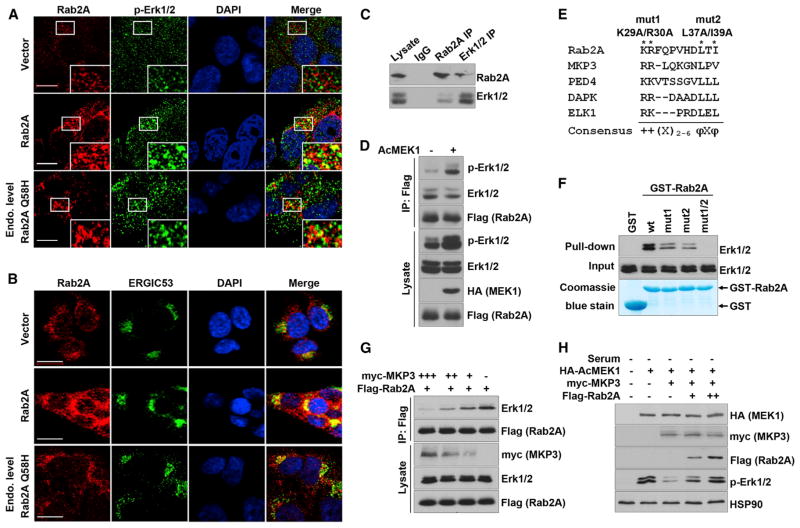
To elucidate how Rab2A overexpression or its Q58H mutation activates Erk1/2, we first examined whether Rab2A co-localized with Erk1/2. Overexpressing WT Rab2A not only activated Erk1/2 but also surprisingly colocalized with activated Erk1/2 at the perinuclear region at 5 min (Figure 4A) and 1 hr (Figure S4A) after EGF stimulation. Overexpressing Rab2A Q58H at levels similar to the endogenous level also activated and colocalized with Erk1/2 (Figures 4A and S4A). Importantly, Rab2A and its Q58H mutant colocalized with Erk1/2 at the ERGIC, as assayed by ERGIC53 staining (Figure 4B). To examine whether Rab2A’s vesicular trafficking function is associated with Erk activation, we used brefeldin A (BFA) to block the trafficking from the ERGIC to ER. As shown previously (Hauri et al., 2000), BFA treatment damaged the ERGIC structure (Figure S4B) but did not obviously affect Erk phosphorylation (Figure S4C), suggesting that Erk1/2 activation is likely to be independent of Rab2A’s trafficking function.
Figure 4. Rab2A Interacts with and Prevents Inactivation of Erk1/2 by MKP3.
(A) Overexpressed Rab2A and its Q58H mutant co-localized with p-Erk1/2. Stable HMLE cells were serum starved and then treated with 10 ng/ml EGF for 5 min. Scale bar represents 10 μm.
(B) Rab2A and its Q58H mutant co-localized with ERGIC53. Scale bar represents 20 μm.
(C) Reciprocal coIP of endogenous Rab2A with Erk1/2. Lysates of HMLE cells were immunoprecipitated with Rab2A or Erk1/2 antibodies, followed by western blot for Rab2A and Erk1/2, respectively.
(D) Rab2A immunoprecipitated with total Erk1/2 and p-Erk1/2 in HEK293 cells co-transfected with Flag-Rab2A and constitutive activated MEK1 (AcMEK1).
(E) The consensus Erk docking motifs were found in Rab2A and several other Erk binding partners. + and φ represent basic and hydrophobic amino acids, respectively. X represents any amino acids.
(F) Mutations in the Erk docking motif in Rab2A impaired its binding to Erk1/2.
(G) Rab2A and MKP3 competed to bind Erk1/2. Lysates of 293T cells transfected with decreasing doses of myc-MKP3 and a constant dose of Flag-Rab2A were immunoprecipitated with M2 (Flag) antibody, followed by western blot for Erk1/2 and Flag-Rab2A.
(H) Rab2A competed with MKP3 and kept Erk1/2 in the phosphorylated status. 293T cells were transfected to express Rab2A, MKP3, and AcMEK1, which induced p-Erk1/2 in serum-starved cells and was largely reversed by Myc-MKP3 expression, whereas Flag-Rab2A expression dose-dependently restored Erk1/2 phosphorylation.
See also Figures S4 and S5.
The unexpected finding that Rab2A colocalizes with activated Erk1/2 at the ERGIC suggested that Rab2A might directly interact with Erk1/2. Indeed, we detected co-immunoprecipitation of the endogenous Rab2A with total Erk1/2 in HMLE cells by reciprocal co-immunoprecipitation (coIP) (Figure 4C). To see whether Rab2A interacted with p-Erk1/2, we transfected constitutively active MEK1 (AcMEK1) to increase p-Erk1/2 levels and found that Rab2A interacted with p-Erk1/2 in the coIP assay (Figure 4D). These results were further supported by our findings that Rab2A contains a conserved common docking motif for binding Erk (Tanoue et al., 2000) (Figure 4E) and that GST-Rab2A pulled down Erk1/2 in cells (Figure 4F), as well as recombinant Erk1 or Erk2 in vitro (Figure S5A). To examine whether the integrity of this docking motif is required for Rab2A to bind to Erk, we substituted the known critical residues KR (mut1) or LXI (mut2) or both residues (mut1/2) with Ala residues. Compared to WT Rab2A, while either mut1 or mut2 markedly reduced binding to Erk, mutating both sequences completely abolished the ability of Rab2A to bind to Erk (Figure 4F). Thus, Rab2A directly interacts with Erk through the specific Erk docking sequence in Rab2A.
Interestingly, the above conserved docking motif is also found in MKP3 and MEK1. To examine whether Rab2A and MKP3 or MEK1 compete to interact with Erk, HEK293 cells were co-transfected with decreasing doses of myc-MKP3 or the constitutively active hemagglutinin-MEK1 and a constant dose of Flag-Rab2A. With decreasing amounts of MKP3 expressed, more Erk1/2 were immunoprecipitated by Rab2A in a dose-dependent manner (Figure 4G), suggesting that Rab2A competed with MKP3 to bind Erk1/2 in vivo. However, unlike the MKP3 competition results, similar amounts of Erk1/2 were immunoprecipitated by Flag-Rab2A even though decreasing amounts of MEK1 were expressed (Figure S5B). These results may be expected, because although the docking motif of MEK is important for the ERK-MEK interaction, there are other mechanisms to ensure the activation of Erk by MEK1, such as scaffold protein facilitation and the MEK catalytic site interacting with the Erk activation loop (Roskoski, 2012).
The above results suggest that Rab2A might prevent Erk1/2 dephosphorylation by competing with MKP3 for Erk1/2 binding. To test this possibility, we transfected HEK293 cells with MKP3 and the constitutively active MEK1 as well as different amounts of epitope-tagged Rab2A. Expression of the active MEK1 induced Erk1/2 phosphorylation even in serum-starved cells, and this was largely reversed by myc-MKP3 expression (Figure 4H). However, Flag-Rab2A expression restored Erk1/2 phosphorylation in a dose-dependent manner (Figure 4H). Thus, Rab2A directly binds to Erk1/2 and keeps it in an active form by competing with MKP3.
To further demonstrate whether this Rab2A-Erk interaction is functionally important for Rab2A to regulate BCSC, we infected HMLE cells with Flag-tagged Rab2A or its mutants defective in binding to Erk (Figure S4C). Rab2A markedly increased the CD24−CD44+ population, but none of its Rab2A mutants altered this population (Figure S5D). In addition, overexpression of Rab1A, the small GTPase that is highly related to Rab2A with over 70% similarity and also localized to the ERGIC but without a conserved docking motif for binding to Erk, had no effect on either Erk activation or the BCSC phenotype (Figures S5G–S5K). Thus, Rab2A directly interacts with and prevents Erk1/2 inactivation by MKP3 to regulate BCSCs.
Rab2A Promotes the Nuclear Translocation of Erk1/2 Downstream β-Catenin
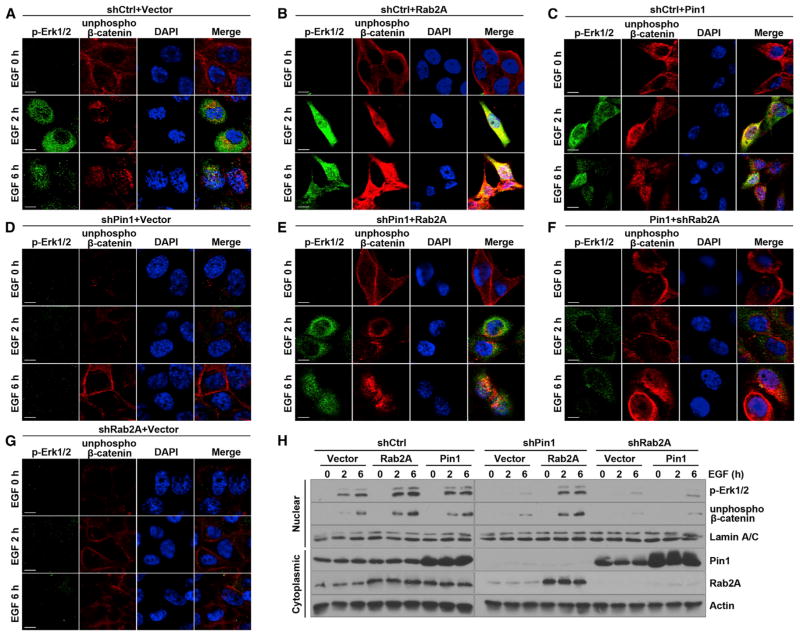
As Erk1/2 signaling increases the nuclear accumulation of unphosphorylated (active) β-catenin (Chang et al., 2011), and because Pin1 also has a similar effect on β-catenin in breast cancer cells (Ryo et al., 2001), we examined whether Pin1/Rab2A/p-Erk signaling regulates nuclear β-catenin levels. Confocal analysis showed that most unphosphorylated β-catenin localized at the plasma membrane in starved HMLE cells but translocated into the nucleus, along with increased p-Erk1/2 6 hr after EGF stimulation (Figure 5A). However, in Rab2A-overexpressing and Pin1-overexpressing cells, not only was p-Erk1/2 obviously increased but also unphosphorylated β-catenin was readily detected in the nucleus as early as 2 hr and accumulated further with time after EGF stimulation (Figures 5B and 5C). In contrast, in Rab2A or Pin1 KD cells, not only was p-Erk1/2 not increased but also nuclear unphosphorylated β-catenin was hardly detectable even 6 hr after stimulation (Figures 5D and 5G). Notably, overexpression of Rab2A in Pin1 KD cells caused Erk1/2 activation and nuclear translocation and, importantly, unphosphorylated β-catenin localization to the nucleus (Figure 5E). Conversely, Rab2A KD in Pin1-overexpressing cells prevented Erk1/2 activation and nuclear translocation of unphosphorylated β-catenin (Figure 5F). Western blot analysis with nuclear fraction further confirmed these results (Figure 5H). These data together support a model in which the Pin1/Rab2A/Erk1/2 pathway activates β-catenin.
Figure 5. Rab2A/p-Erk Signaling Promotes the Nuclear Translocation of Activeβ-Catenin.
(A and B) Rab2A promoted the nuclear translocation of unphosphorylated β-catenin (active form). HMLE cells were serum starved and then stimulated by EGF for the indicated time points.
(C–E) Pin1 also promoted the nuclear translocation of unphosphorylated β-catenin and Rab2A overexpression in Pin1 KD cells rescued Erk1/2 activation and β-catenin translocation from the cell membrane to the nucleus.
(F and G) Rab2A KD in Pin1-overexpressing or vector control cells inhibited p-Erk1/2 activation and β-catenin nuclear translocation. Scale bars represent 10 μm.
(H) Rab2A promoted the nuclear accumulation of p-Erk1/2 and unphosphorylated β-catenin. Nuclear and total proteins were extracted after EGF stimulation following serum starvation.
Rab2A Endows BCSC Traits to Normal Human MECs and Is Necessary for Tumorigenesis of Human Primary BCSCs
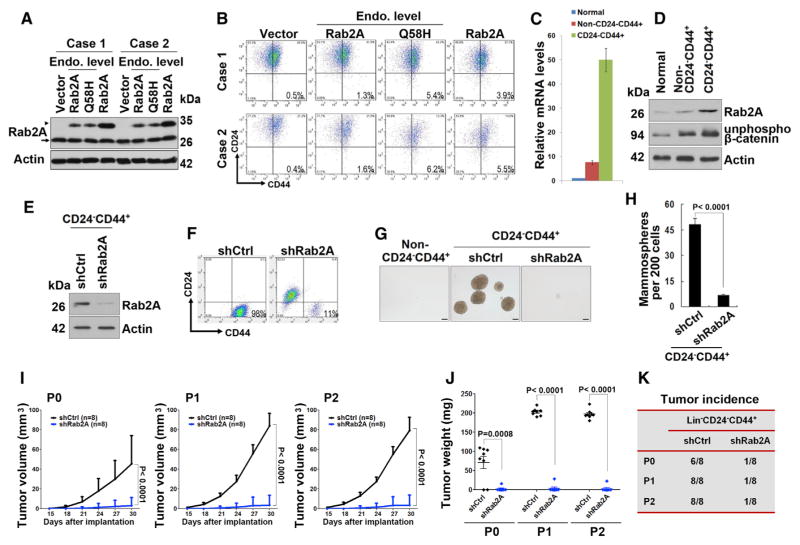
To extend our findings to primary human cells, we sorted Lin− MECs isolated from reduction mammoplasty tissue from two human donors and infected them with lentiviruses expressing Flag-Rab2A, Flag-Rab2A Q58H at levels similar to or three times the endogenous level (Figures 6A and S6A). Rab2A overexpression led to a dose-dependent increase in the CD24−CD44+ population (Figure 6B). Overexpressing Rab2A Q58H similar to the endogenous level increased the CD24−CD44+ population more than overexpressing Rab2A at three-times-higher levels (Figure 6B). Thus, increasing Rab2A activity by either overexpression or using naturally occurring cancer-derived mutation endows BCSC traits to normal human MECs.
Figure 6. Rab2A and Its Q58H Mutant Endow BCSC Traits to Normal Primary Human MECs, whereas Silencing Rab2A Inhibits Primary Human BCSC Expansion and Tumorigenesis.
(A) Western blot showed lentivirus-mediated overexpression of Rab2A and Q58H mutant in two cases of human normal Lin− MECs. Arrowhead, exogenous Flag tagged protein; arrow, endogenous protein.
(B) Rab2A or Rab2A Q58H mutant increased the CD24−CD44+ population in primary human MECs.
(C) Real-time PCR showed that expression of Rab2A mRNA was markedly increased in the Lin−CD24−CD44+ population, compared to Lin−non-CD24−CD44+ or normal epithelial cells.
(D) Expression of Rab2A and unphosphorylated β-catenin protein was markedly increased in Lin−CD24−CD44+ cells compared to Lin−non-CD24−CD44+ cells in human breast cancer tissue and those in normal breast tissue from the same patient.
(E) Rab2A was knocked down in Lin−CD24−CD44+ cells sorted from human breast cancer tissue.
(F) Rab2A KD in Lin−CD24−CD44+ breast cancer cells decreased the CD24−CD44+ population.
(G and H) Rab2A KD in Lin−CD24−CD44+ breast cancer cells decreased mammosphere formation. Scale bar represents 100 μm.
(I–K) Rab2A KD interfered with both tumor initiation and growth of primary BCSCs in vivo, as shown by tumor growth curve (I), tumor weights (J), and tumor incidence (K). 2,000 lentivirus-transduced Lin−CD24−CD44+ cells isolated from eight breast cancer patients were serially transplanted as xenografts into eight nude mice. P0, freshly isolated primary cells; P1, passage 1; P2, passage 2.
In (C) and (H), error bars represent SD of three independent experiments. In (I) and (J), error bars represent SD of eight mice. See also Figure S6.
To assess whether Rab2A is also important for tumorigenesis of BCSCs in human primary breast cancer, we sorted Lin−CD24−CD44+ cells from freshly isolated breast cancer cells of eight patients (Table S5) and analyzed Rab2A expression and its impact on BCSCs in vitro and in vivo (Figure S6B). As compared with those in Lin−non-CD24−CD44+ cancer cells, Rab2A mRNA levels were approximately seven times higher in Lin−CD24−CD44+cells, and five to seven times lower in normal breast epithelial cells (Figure 6C). Consistent with these results, Rab2A protein and unphosphorylated β-catenin were higher in the Lin−CD24−CD44+ cells than those in Lin−non-CD24−CD44+ cancer cells or normal MECs (Figure 6D). We then knocked down Rab2A in Lin−CD24−CD44+ primary breast cancer cells (Figure 6E). The CD24−CD44+ population was significantly reduced in Rab2A KD cells, being only one-ninth of that in control cells (Figure 6F). Rab2A KD also significantly decreased the mammosphere-forming activity of the CD24−CD44+ cells (Figures 6G and 6H). Thus, Rab2A is required for sustaining the BCSC-associated properties of human primary breast cancer cells.
We finally investigated whether Rab2A was required for the tumorigenicity of the Lin−CD24−CD44+ population. We injected 2,000 control or Rab2A shRNA-transduced Lin−CD24−CD44+ cells, or Lin−non-CD24−CD44+ cells isolated from eight breast cancer patients, into nude mice. While no tumors developed in mice injected with the cells that were not CD24−CD44+, 2,000 control Lin−CD24−CD44+ cells generated six tumors in eight injected mice (Figures 6I–6K). Lentivirus-mediated KD of Rab2A not only drastically reduced tumor incidence (Figure 6K) but also potently reduced tumor growth, as measured by tumor volumes and weights (Figures 6I and 6J). We then dissociated the tumors and sorted again for CD24−CD44+ cells for the serial transplantation. When control tumors were passaged in nude mice, they were serially transplanted at least for two more passages without reduced tumorigenicity (Figure 6K), but Rab2A KD cells had substantially decreased frequency of tumor formation and reduced tumor growth (Figures 6I–6K). Thus, Rab2A is highly expressed in BCSC-enriched population in human breast cancer, and silencing Rab2A potently inhibits BCSC expansion and tumorigenesis.
Rab2A Overexpression Correlates with Poor Clinical Outcomes in Breast Cancer Patients
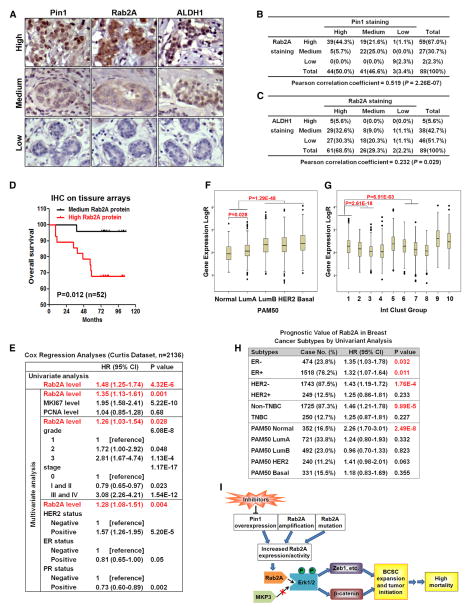
We analyzed expression of Rab2A, Pin1, and ALDH1, a marker for stem and progenitor cells as well as BCSCs (Ginestier et al., 2007), in normal and cancerous breast tissue arrays using immunohistochemistry. Pin1 and Rab2A were undetectable or low in all 24 human normal breast tissue, but their expression was dramatically increased in many of 65 human breast cancer tissue (Figures 7A and 7B). Remarkably, Rab2A expression was highly correlated with Pin1 and ALDH1 expression in normal and cancerous breast tissue (Figures 7A–7C). We next analyzed the correlation of Rab2A expression and clinical outcome in the subset of 52 breast cancer patients, for which clinical data were available. Higher Rab2A expression was significantly associated with higher mortality in breast cancer patients, as shown by Kaplan-Meier survival curves (p = 0.012) (Figure 7D).
Figure 7. Rab2A Expression Correlates with Pin1 and ALDH1 Expression and with Poor Clinical Outcome in Breast Cancer Patients.
(A–C) Rab2A expression correlated with Pin1 and ALDH1 expression in the tissue array of normal and cancerous breast tissue.
(D) High Rab2A expression correlated with poor overall survival in the tissue array dataset of breast cancer patients.
(E) Rab2A was a strong and independent biomarker to predict breast cancer specific survival in the Curtis breast cancer dataset by Cox regression analyses.
(F) Boxplots of Rab2A expression stratified by the PAM50 classifier in the Curtis breast cancer dataset.
(G) Boxplots of Rab2A expression stratified by the IntClust subtypes in the Curtis breast cancer dataset.
(H) Univariate Cox regression analysis showed that HER2-negative, non-triple-negative, or PAM50-normal subtypes of breast cancer patients with higher Rab2A mRNA levels had a higher risk of breast cancer mortality.
(I) A schematic model for how the Pin1/Rab2A/Erk signal pathway regulates tumor initiation via CSC regulators, contributing to high mortality in breast cancer. See also Figure S7.
To expand our immunohistochemistry findings on limited samples, we studied multiple independent breast cancer datasets from Oncomine (Rhodes et al., 2007), which collectively link clinical data with Rab2A mRNA expression in ~3,000 patients. Rab2A overexpression was closely associated with advanced stage in the Bittner dataset, with metastasis in the Schmidt data-set, and with death at 3 or 5 years in the Bild and Kao datasets (Bild et al., 2006; Kao et al., 2011; Schmidt et al., 2008) (Figures S7A–S7D).
Giving that the microarray experiments and the methods to normalize data vary among different datasets, making it difficult to pool the data from different datasets, we chose to further analyze the Curtis dataset, which has over 2,000 patients (Curtis et al., 2012). The Rab2A mRNA level was a strong prognostic factor for survival by univariate Cox regression analysis (Figure 7E). A high Rab2A level was still independently associated with high mortality, even using multivariate analysis adjusted for proliferation markers (MKI67 and PCNA), or tumor grade and stage, or the status of HER2, ER, and PR (Figure 7E). We next analyzed Rab2A expression in the PAM50 intrinsic subtypes (Parker et al., 2009) and integrative subgroups (Curtis et al., 2012). Strikingly, high Rab2A levels were found in the poor-prognosis subtypes, defined as PAM50 intrinsic subtypes, luminal B, HER2-enriched, and basal-like, and in the IntClust5, IntClust6, IntClust9, and IntClust10 integrative subgroups (Figures 7F and 7G), whereas low Rab2A levels were mostly observed in the better-prognosis subtypes (normal-like PAM50 intrinsic subtype and integrative subgroups IntClust3 and IntClust4) (Figures 7F and 7G). Notably, a high Rab2A level was tightly linked to high mortality in the most common subgroups of breast cancer patients, defined as HER2-negative or non-TNBC (triple-negative) patients (Figure 7H), which account for 87.5% and 87.3% of all cases, respectively. Thus, Rab2A plays a key oncogenic role in promoting BCSCs and aggravating breast cancer malignancy.
DISCUSSION
Our profiling of Pin1-regulated genes led to the discovery that Pin1 promotes BCSCs by increasing Rab2A transcription. We suggest that in breast cancer, Pin1 overexpression, Rab2A gene amplification, or genetic mutation activates Rab2A, which drives BCSCs and tumorigenesis in large part by preventing Erk1/2 inactivation via MKP3, leading to activation of known BCSC regulators and contributing to high mortality in breast cancer patients (Figure 7I). This pathway may offer attractive BCSC targets for cancer therapy. Notably, ATRA has been identified to bind and ultimately degrade active Pin1 selectively in cancer cells (Wei et al., 2015).
Emerging evidence has shown that Rab family members play important roles in epithelial neoplasia (Goldenring, 2013). Although Rab25 has opposing functions in cancer, promoting tumor aggressiveness (Cheng et al., 2004) and suppressing tumor development (Nam et al., 2010; Tong et al., 2012), most Rab members with increased expression in cancer are positively associated with cancer progression, especially with invasion and metastasis (Hou et al., 2008; Jacob et al., 2013). We report here that Rab2A plays a major oncogenic role in human breast cancer via promoting BCSC expansion and tumorigenicity. Furthermore, we have provided strong genetic evidence that Rab2A is abnormally activated in human cancer by either gene amplification or activating Rab2A point mutation, both of which enhance its ability to activate Erk1/2 signaling. Finally, gene expression profiling data in five large datasets containing ~3,000 breast cancer patients strongly link Rab2A to poor patient outcome. Notably, in the >2,000-patient Curtis dataset, high Rab2A levels were found in poor-prognosis subtypes, whereas low Rab2A levels were mostly observed in better-prognosis subtypes. Significantly, a high Rab2A level was strongly associated with high mortality, even in multivariate analysis. It is worth noting that the association between Rab2A and cancer mortality was even more striking in the most common groups (~87%) of breast cancer patients, defined as HER2-negative or non-TNBC patients. Since it is challenging to predict poor prognosis in these common subgroups of breast cancer patients without profiling many genes, Rab2A expression might represent a useful prognosis biomarker.
Rab2A has previously been implicated in intercellular vesicle trafficking regulation (Stenmark, 2009; Tisdale and Balch, 1996). Our findings highlight the role of Rab GTPases in altering cell signaling. Previously, Rab5 was shown to transduce cell-survival signals by facilitating vesicular translocation to the nucleus of multiple signaling kinases in neurons, including Akt and Erk (Delcroix et al., 2003). The endosomal Rab23 is an essential negative regulator that attenuates the mouse sonic hedgehog signaling pathway downstream of the sonic hedgehog receptor and its effector (Eggenschwiler et al., 2001). Our results that Erk signaling is critical for Rab2A to regulate BCSCs are reminiscent of previous reports showing that the Erk pathway is required for BCSC maintenance. For example, Ras signals through the Erk pathway (Chiu et al., 2002), and Erk2 activation is essential for Ras to induce the EMT and to promote the CD24−CD44+ population (Shin et al., 2010). SHP2 influences BCSCs and enhances breast tumor maintenance and progression through activation of the Erk pathway (Aceto et al., 2012).
It is interesting to note that the docking motif in Erk1/2 is common to its activators, substrates, and regulators, suggesting that they may compete for binding to Erk1/2 in vivo (Bardwell et al., 2003; Tanoue et al., 2000). Indeed, these docking interactions are crucial for the specificity of Erk1/2 signaling and the duration of the activation-inactivation cycles of Erk1/2. In the absence of activation, Erk1/2 is mainly localized to the cytoplasm. Activated Erk1/2 translocates to the nucleus and phosphorylates nuclear targets. MKP3 shuttles between the cytoplasm and nucleus but is largely localized in the cytoplasm (Karlsson et al., 2004). It is possible that Erk1/2 bound by Rab2A is temporarily protected from the action of the phosphatase MKP3. Thus, the competitive docking of Rab2A and phosphatase to Erk1/2 may play an important role in enhancing Erk signaling. Moreover, we found that Erk1/2 and Rab2A colocalized at the ERGIC. Although Erk3 is principally localized to the Golgi and ERGIC in several cell lines (Bind et al., 2004), the role of Erks at the ERGIC is largely unknown. It would be interesting to define the molecular details of the Erk1/2 and Rab2A interactions and their regulation at the ERGIC for Erk signaling.
Small GTPases act through cycling between an active, GTP-bound and an inactive, GDP-bound state. Activating Ras mutations occur in ~30% of human cancers. Most of the activating mutations impair intrinsic GTPase activity or confer resistance to GTPase activating proteins (GAPs), causing mutant Ras to accumulate in the active GTP-bound form (Schubbert et al., 2007). The Rab2A Q58H mutation, a naturally occurring genetic mutation found in human cancers, not only reduced GTP hydrolysis but also was more potent in activating Erk signaling and driving BCSCs than the WT protein. Therefore, the Q58H protein might be a hyperactive form of Rab2A in human cancers.
EXPERIMENTAL PROCEDURES
Microarray Analysis
RNA from Lin− MECs and neuron cells of Pin1 KO and WT mice was extracted with the total RNA isolation kit (Agilent). Microarray expression profiles were collected using Affymetrix GeneChip Mouse Expression Array 430A.
Serial Transplantation Assay
Lin−CD24−CD44+ cells were sorted from eight breast cancer specimens and cultured as a single-cell suspension in ultra-low-attachment dishes, then infected with lentivirus expressing control vector or Rab2A shRNA. After 1 week of puromycin selection, 2,000 transduced cells from each patient were injected into the mammary fat pads of 5-week-old nude mice. For serial passaging, cells from the primary tumors were sorted again for Lin−CD24−CD44+ cells. Among the six primary tumors formed in the control shRNA group, four tumors were randomly selected and passaged into eight mice (two mice per tumor). For the one tumor formed from 2,000 shRab2A cells, this tumor’s cells were injected into eight mice for serial passaging. The same procedure was applied to the second passage of xenograft cells, as described previously (Yu et al., 2007). All studies involving human subjects were approved by the institutional review board at Beth Israel Deaconess Medical Center or Sun Yat-Sen Memorial Hospital. All studies involving mice were approved by the institutional animal care and use committee at Beth Israel Deaconess Medical Center and performed in accordance with the relevant protocols.
Statistical Analysis
All data are presented as the means ± SD, followed by determining significant differences using the two-tailed t test or ANOVA test. Limiting-dilution data were analyzed by the single-hit Poisson model using a complementary log-log generalized linear model (Bonnefoix et al., 2001) with L-Calc Software (STEMCELL Technologies). Correlations of Rab2A expression with other gene expression were analyzed with the Pearson correlation test. For survival analysis, Kaplan-Meier analysis and univariate and multivariate Cox regression analysis were used. All tests of significance were set at p < 0.05.
Supplementary Material
Highlights.
Pin1 increases Rab2A transcription, promoting BCSC expansion and tumorigenesis
Rab2A blocks Erk1/2 inactivation by MKP3, leading to Zeb1 and β-catenin activation
Rab2A amplification or mutation in human cancer activates Erk1/2 and increases BCSCs
Rab2A overexpression is tightly linked to high mortality in human breast cancer
Acknowledgments
We are very grateful to Drs. Robert Weinberg and Wenjun Guo for providing expert advice and critical reagents, and we thank Drs. John Blenis and William C. Hahn for reagents and/or advice and members of K.P.L. and X.Z.Z. laboratories for constructive discussions. C.-H.C. is a DOD Breast Cancer Research Program Postdoctoral Fellow and a NIH T32 training grant awardee. This work was supported by a Komen for Cure grant (to X.Z.Z.), the Ministry of Science and Technology of China 973 project (to E.S.), and the National Natural Science Foundation of China (U1205024) and NIH (grants CA167677, AG039405, and DA031663 to K.P.L.). K.P.L. and X.Z.Z. are inventors of Pin1 technology, which was licensed by BIDMC to Pinteon Therapeutics. Both own equity in and consult for Pinteon. K.P.L. also serves on its board of directors. Their interests were reviewed and are managed by BIDMC in accordance with its conflict of interest policy.
Footnotes
ACCESSION NUMBERS
Microarray data have been deposited to the NCBI GEO and are available under accession number GSE49971.
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2015.03.002.
References
- Aceto N, Sausgruber N, Brinkhaus H, Gaidatzis D, Martiny-Baron G, Mazzarol G, Confalonieri S, Quarto M, Hu G, Balwierz PJ, et al. Tyrosine phosphatase SHP2 promotes breast cancer progression and maintains tumor-initiating cells via activation of key transcription factors and a positive feedback signaling loop. Nat Med. 2012;18:529–537. doi: 10.1038/nm.2645. [DOI] [PubMed] [Google Scholar]
- Akunuru S, Palumbo J, Zhai QJ, Zheng Y. Rac1 targeting suppresses human non-small cell lung adenocarcinoma cancer stem cell activity. PLoS ONE. 2011;6:e16951. doi: 10.1371/journal.pone.0016951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell AJ, Abdollahi M, Bardwell L. Docking sites on mitogen-activated protein kinase (MAPK) kinases, MAPK phosphatases and the Elk-1 transcription factor compete for MAPK binding and are crucial for enzymic activity. Biochem J. 2003;370:1077–1085. doi: 10.1042/BJ20021806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- Bind E, Kleyner Y, Skowronska-Krawczyk D, Bien E, Dynlacht BD, Sánchez I. A novel mechanism for mitogen-activated protein kinase localization. Mol Biol Cell. 2004;15:4457–4466. doi: 10.1091/mbc.E04-03-0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- Bonnefoix T, Bonnefoix P, Callanan M, Verdiel P, Sotto JJ. Graphical representation of a generalized linear model-based statistical test estimating the fit of the single-hit Poisson model to limiting dilution assays. J Immunol. 2001;167:5725–5730. doi: 10.4049/jimmunol.167.10.5725. [DOI] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CJ, Yang JY, Xia W, Chen CT, Xie X, Chao CH, Woodward WA, Hsu JM, Hortobagyi GN, Hung MC. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-β-catenin signaling. Cancer Cell. 2011;19:86–100. doi: 10.1016/j.ccr.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Chang CC, Lee TH, Luo M, Huang P, Liao PH, Wei S, Li FA, Chen RH, Zhou XZ, et al. SENP1 deSUMOylates and regulates Pin1 protein activity and cellular function. Cancer Res. 2013;73:3951–3962. doi: 10.1158/0008-5472.CAN-12-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng KW, Lahad JP, Kuo WL, Lapuk A, Yamada K, Auersperg N, Liu J, Smith-McCune K, Lu KH, Fishman D, et al. The RAB25 small GTPase determines aggressiveness of ovarian and breast cancers. Nat Med. 2004;10:1251–1256. doi: 10.1038/nm1125. [DOI] [PubMed] [Google Scholar]
- Chiu VK, Bivona T, Hach A, Sajous JB, Silletti J, Wiener H, Johnson RL, 2nd, Cox AD, Philips MR. Ras signalling on the endoplasmic reticulum and the Golgi. Nat Cell Biol. 2002;4:343–350. doi: 10.1038/ncb783. [DOI] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcroix JD, Valletta JS, Wu C, Hunt SJ, Kowal AS, Mobley WC. NGF signaling in sensory neurons: evidence that early endosomes carry NGF retrograde signals. Neuron. 2003;39:69–84. doi: 10.1016/s0896-6273(03)00397-0. [DOI] [PubMed] [Google Scholar]
- Dontu G, Wicha MS. Survival of mammary stem cells in suspension culture: implications for stem cell biology and neoplasia. J Mammary Gland Biol Neoplasia. 2005;10:75–86. doi: 10.1007/s10911-005-2542-5. [DOI] [PubMed] [Google Scholar]
- Eggenschwiler JT, Espinoza E, Anderson KV. Rab23 is an essential negative regulator of the mouse Sonic hedgehog signalling pathway. Nature. 2001;412:194–198. doi: 10.1038/35084089. [DOI] [PubMed] [Google Scholar]
- Elenbaas B, Spirio L, Koerner F, Fleming MD, Zimonjic DB, Donaher JL, Popescu NC, Hahn WC, Weinberg RA. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenring JR. A central role for vesicle trafficking in epithelial neoplasia: intracellular highways to carcinogenesis. Nat Rev Cancer. 2013;13:813–820. doi: 10.1038/nrc3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauri HP, Kappeler F, Andersson H, Appenzeller C. ERGIC-53 and traffic in the secretory pathway. J Cell Sci. 2000;113:587–596. doi: 10.1242/jcs.113.4.587. [DOI] [PubMed] [Google Scholar]
- Hou Q, Wu YH, Grabsch H, Zhu Y, Leong SH, Ganesan K, Cross D, Tan LK, Tao J, Gopalakrishnan V, et al. Integrative genomics identifies RAB23 as an invasion mediator gene in diffuse-type gastric cancer. Cancer Res. 2008;68:4623–4630. doi: 10.1158/0008-5472.CAN-07-5870. [DOI] [PubMed] [Google Scholar]
- Jacob A, Jing J, Lee J, Schedin P, Gilbert SM, Peden AA, Junutula JR, Prekeris R. Rab40b regulates trafficking of MMP2 and MMP9 during invadopodia formation and invasion of breast cancer cells. J Cell Sci. 2013;126:4647–4658. doi: 10.1242/jcs.126573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao KJ, Chang KM, Hsu HC, Huang AT. Correlation of microarray-based breast cancer molecular subtypes and clinical outcomes: implications for treatment optimization. BMC Cancer. 2011;11:143. doi: 10.1186/1471-2407-11-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson M, Mathers J, Dickinson RJ, Mandl M, Keyse SM. Both nuclear-cytoplasmic shuttling of the dual specificity phosphatase MKP-3 and its ability to anchor MAP kinase in the cytoplasm are mediated by a conserved nuclear export signal. J Biol Chem. 2004;279:41882–41891. doi: 10.1074/jbc.M406720200. [DOI] [PubMed] [Google Scholar]
- Lee TH, Chen CH, Suizu F, Huang P, Schiene-Fischer C, Daum S, Zhang YJ, Goate A, Chen RH, Zhou XZ, Lu KP. Death-associated protein kinase 1 phosphorylates Pin1 and inhibits its prolyl isomerase activity and cellular function. Mol Cell. 2011;42:147–159. doi: 10.1016/j.molcel.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Wicha MS. Targeting breast cancer stem cells. J Clin Oncol. 2010;28:4006–4012. doi: 10.1200/JCO.2009.27.5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Casimiro MC, Wang C, Shirley LA, Jiao X, Katiyar S, Ju X, Li Z, Yu Z, Zhou J, et al. p21CIP1 attenuates Ras- and c-Myc-dependent breast tumor epithelial mesenchymal transition and cancer stem cell-like gene expression in vivo. Proc Natl Acad Sci USA. 2009;106:19035–19039. doi: 10.1073/pnas.0910009106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Hunter T. Prolyl isomerase Pin1 in cancer. Cell Res. 2014;24:1033–1049. doi: 10.1038/cr.2014.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu KP, Zhou XZ. The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nat Rev Mol Cell Biol. 2007;8:904–916. doi: 10.1038/nrm2261. [DOI] [PubMed] [Google Scholar]
- Lu KP, Hanes SD, Hunter T. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature. 1996;380:544–547. doi: 10.1038/380544a0. [DOI] [PubMed] [Google Scholar]
- Luo ML, Gong C, Chen CH, Lee DY, Hu H, Huang P, Yao Y, Guo W, Reinhardt F, Wulf G, et al. Prolyl isomerase Pin1 acts downstream of miR200c to promote cancer stem-like cell traits in breast cancer. Cancer Res. 2014;74:3603–3616. doi: 10.1158/0008-5472.CAN-13-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monje P, Hernández-Losa J, Lyons RJ, Castellone MD, Gutkind JS. Regulation of the transcriptional activity of c-Fos by ERK. A novel role for the prolyl isomerase PIN1. J Biol Chem. 2005;280:35081–35084. doi: 10.1074/jbc.C500353200. [DOI] [PubMed] [Google Scholar]
- Myant KB, Cammareri P, McGhee EJ, Ridgway RA, Huels DJ, Cordero JB, Schwitalla S, Kalna G, Ogg EL, Athineos D, et al. ROS production and NF-κB activation triggered by RAC1 facilitate WNT-driven intestinal stem cell proliferation and colorectal cancer initiation. Cell Stem Cell. 2013;12:761–773. doi: 10.1016/j.stem.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Greenwood A, Binder L, Bigio EH, Denial S, Nicholson L, Zhou XZ, Lu KP. Proline isomer-specific antibodies reveal the early pathogenic tau conformation in Alzheimer’s disease. Cell. 2012;149:232–244. doi: 10.1016/j.cell.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam KT, Lee HJ, Smith JJ, Lapierre LA, Kamath VP, Chen X, Aronow BJ, Yeatman TJ, Bhartur SG, Calhoun BC, et al. Loss of Rab25 promotes the development of intestinal neoplasia in mice and is associated with human colorectal adenocarcinomas. J Clin Invest. 2010;120:840–849. doi: 10.1172/JCI40728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskoski R., Jr ERK1/2 MAP kinases: structure, function, and regulation. Pharmacol Res. 2012;66:105–143. doi: 10.1016/j.phrs.2012.04.005. [DOI] [PubMed] [Google Scholar]
- Rustighi A, Zannini A, Tiberi L, Sommaggio R, Piazza S, Sorrentino G, Nuzzo S, Tuscano A, Eterno V, Benvenuti F, et al. Prolyl-isomerase Pin1 controls normal and cancer stem cells of the breast. EMBO Mol Med. 2014;6:99–119. doi: 10.1002/emmm.201302909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryo A, Nakamura M, Wulf G, Liou YC, Lu KP. Pin1 regulates turnover and subcellular localization of beta-catenin by inhibiting its interaction with APC. Nat Cell Biol. 2001;3:793–801. doi: 10.1038/ncb0901-793. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Böhm D, von Törne C, Steiner E, Puhl A, Pilch H, Lehr HA, Hengstler JG, Kölbl H, Gehrmann M. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res. 2008;68:5405–5413. doi: 10.1158/0008-5472.CAN-07-5206. [DOI] [PubMed] [Google Scholar]
- Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- Shin S, Dimitri CA, Yoon SO, Dowdle W, Blenis J. ERK2 but not ERK1 induces epithelial-to-mesenchymal transformation via DEF motif-dependent signaling events. Mol Cell. 2010;38:114–127. doi: 10.1016/j.molcel.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, Li HI, Eaves CJ. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- Tanoue T, Adachi M, Moriguchi T, Nishida E. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat Cell Biol. 2000;2:110–116. doi: 10.1038/35000065. [DOI] [PubMed] [Google Scholar]
- Tisdale EJ, Balch WE. Rab2 is essential for the maturation of pre-Golgi intermediates. J Biol Chem. 1996;271:29372–29379. doi: 10.1074/jbc.271.46.29372. [DOI] [PubMed] [Google Scholar]
- Tong M, Chan KW, Bao JY, Wong KY, Chen JN, Kwan PS, Tang KH, Fu L, Qin YR, Lok S, et al. Rab25 is a tumor suppressor gene with antiangiogenic and anti-invasive activities in esophageal squamous cell carcinoma. Cancer Res. 2012;72:6024–6035. doi: 10.1158/0008-5472.CAN-12-1269. [DOI] [PubMed] [Google Scholar]
- Wei S, Kozono S, Kats L, Nechama M, Li W, Guarnerio J, Bozkurt G, Luo ML, You MH, Yao Y, et al. Active Pin1 as a key target of ATRA anticancer activity against acute promyelocytic leukemia and breast cancer through suppression of multiple cancer-driving pathways. Nat Med. 2015 doi: 10.1038/nm.3839. Published online ▪ ▪ ▪–▪ ▪ ▪. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- Wulf GM, Ryo A, Wulf GG, Lee SW, Niu T, Petkova V, Lu KP. Pin1 is overexpressed in breast cancer and cooperates with Ras signaling in increasing the transcriptional activity of c-Jun towards cyclin D1. EMBO J. 2001;20:3459–3472. doi: 10.1093/emboj/20.13.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB, Schutkowski M, Shen M, Zhou XZ, Stukenberg PT, Rahfeld JU, Xu J, Kuang J, Kirschner MW, Fischer G, et al. Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science. 1997;278:1957–1960. doi: 10.1126/science.278.5345.1957. [DOI] [PubMed] [Google Scholar]
- Yoon CH, Hyun KH, Kim RK, Lee H, Lim EJ, Chung HY, An S, Park MJ, Suh Y, Kim MJ, Lee SJ. The small GTPase Rac1 is involved in the maintenance of stemness and malignancies in glioma stem-like cells. FEBS Lett. 2011;585:2331–2338. doi: 10.1016/j.febslet.2011.05.070. [DOI] [PubMed] [Google Scholar]
- Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, Song E. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- Zhang M, Behbod F, Atkinson RL, Landis MD, Kittrell F, Edwards D, Medina D, Tsimelzon A, Hilsenbeck S, Green JE, et al. Identification of tumor-initiating cells in a p53-null mouse model of breast cancer. Cancer Res. 2008;68:4674–4682. doi: 10.1158/0008-5472.CAN-07-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.