Abstract
Objective
Temporal lobe epilepsy (TLE) patients exhibit signs of memory impairments even when seizures are pharmacologically controlled. Surprisingly, the underlying molecular mechanisms involved in TLE-associated memory impairments remain elusive. Memory consolidation requires epigenetic transcriptional regulation of genes in the hippocampus; therefore, we aimed to determine how epigenetic DNA methylation mechanisms affect learning-induced transcription of memory-permissive genes in the epileptic hippocampus.
Methods
Using the kainate rodent model of TLE and focusing on the brain-derived neurotrophic factor (Bdnf) gene as a candidate of DNA methylation-mediated transcription, we analyzed DNA methylation levels in epileptic rats following learning. After detection of aberrant DNA methylation at the Bdnf gene, we investigated functional effects of altered DNA methylation on hippocampus-dependent memory formation in our TLE rodent model.
Results
We found that behaviorally driven BdnfDNA methylation was associated with hippocampus-dependent memory deficits. Bisulfite sequencing revealed that decreased BdnfDNA methylation levels strongly correlated with abnormally high levels of BdnfmRNA in the epileptic hippocampus during memory consolidation. Methyl supplementation via methionine (Met) increased BdnfDNA methylation and reduced BdnfmRNA levels in the epileptic hippocampus during memory consolidation. Met administration reduced interictal spike activity, increased theta rhythm power, and reversed memory deficits in epileptic animals. The rescue effect of Met treatment on learning-induced BdnfDNA methylation, Bdnf gene expression, and hippocampus-dependent memory, were attenuated by DNA methyltransferase blockade.
Interpretation
Our findings suggest that manipulation of DNA methylation in the epileptic hippocampus should be considered as a viable treatment option to ameliorate memory impairments associated with TLE.
Introduction
Temporal lobe epilepsy (TLE) is a partial adult onset form of human epilepsy that is commonly associated with memory deficits.1 However, the underlying molecular mechanisms responsible for memory loss with TLE are unclear. DNA methylation, typically associated with gene silencing, is a potent epigenetic regulator of gene transcription involved in central nervous system development, synaptic plasticity, and long-term memory formation.2–5 DNA methylation is catalyzed by DNA methyltransferases (DNMT)6 and has been shown to be involved in TLE.7–11 Furthermore, interference with DNMT-mediated global and loci-specific DNA methylation changes increased field excitatory postsynaptic potentials in the epileptic hippocampus and lowered seizure threshold in a rodent TLE model,10 indicating that DNA methylation may play an important role in seizure susceptibility and possibly the maintenance of the disorder. It is tempting to speculate, therefore, that global and gene-specific elevations in DNA methylation with TLE may serve as a compensatory mechanism to control seizure activity by decreasing proepileptic neuronal gene expression.10
Alterations in memory-permissive genes, such as brain-derived neurotrophic factor (Bdnf), correlate with TLE and may contribute to the development, maintenance, and severity of the disorder.12–14 Chronic overexpression or downregulation in Bdnf expression has been linked to memory impairments.15,16 Additionally, activity-dependent Bdnf gene transcription in the hippocampus is controlled by DNA methylation mechanisms during memory formation3,17,18 and Bdnf DNA methylation is abnormally regulated in the epileptic hippocampus.10,17 Therefore, we hypothesize that a consequence of TLE-associated DNA methylation changes is that normal transcription of neuronal genes required for proper memory formation, such as Bdnf, are severely compromised in the epileptic hippocampus and contribute to epilepsy-associated memory deficits.
Using a rodent hippocampus-dependent memory paradigm, we found that Bdnf DNA methylation levels significantly decreased while Bdnf mRNA levels increased in the epileptic hippocampus during memory consolidation. Methyl supplementation with Met significantly increased Bdnf DNA methylation levels, restored Bdnf mRNA levels in the epileptic hippocampus, reversed hippocampus-dependent memory deficits, and in electroencephalography (EEG) studies, decreased interictal spike activity while increasing theta rhythm power. Inhibition of DNMT activity blocked the effect of methyl supplementation with Met on Bdnf DNA methylation and mRNA levels in the epileptic hippocampus, and prevented the effects on memory enhancements. Collectively, these results suggest that aberrant DNA methylation-mediated gene transcription contributes to TLE-associated memory deficits, and that methyl supplementation via Met may be an effective therapeutic option for reversing hippocampus-dependent memory impairments.
Materials and Methods
Animals
Adult male Sprague Dawley rats (250–300 g) were used for all experiments. Animals were double housed in a 12 h light/dark cycle and allowed access to food and water ad libitum. Procedures were performed with the approval of the University of Alabama at Birmingham Institutional Animal Care and Use Committee and according to the national policies and guidelines.
Kainate treatment
Animals were injected with kainic acid (KA) (10 mg/kg; Tocris Cookson Inc., Ellisville, MO) or saline (vehicle) intraperitoneally (i.p.). Behavioral seizures following KA injection were scored following the Racine scale.19 Animals were considered in status epilepticus (SE) when they reached a score of 4 or 5 on the Racine scale. Vehicle-treated animals were handled in the same manner as the kainate-treated animals, except for KA administration. All animals were sacrificed 3 weeks post-SE and all kainate-treated animals used in the study had observable seizures. The hippocampus was removed and placed in ice-cold oxygenated (95%/5% O2/CO2) cutting solution (110 mmol/L sucrose, 60 mmol/L NaCl, 3 mmol/L KCl, 1.25 mmol/L NaH2PO4, 28 mmol/L NaHCO3, 0.5 mmol/L CaCl2, 7 mmol/L MgCl2, 5 mmol/L glucose, 0.6 mmol/L ascorbate). Area CA1 was microdissected and frozen immediately on dry ice. The tissue was stored at −80°C.
Drug treatments
Animals were i.p. injected with saline (0.9% NaCl, pH 7.4), methionine (100 mg/kg; Sigma-Aldrich, St. Louis, Missouri, USA), or 5-aza-2′-deoxycytidine (0.4 mg/kg; Sigma-Aldrich) 1 h before behavioral testing, 1.5 h before sacrifice for amino acid array experiments, or 3 h before sacrificing for all other molecular experiments. These time points were selected because optimal bdnf expression levels are reached at 2 h post fear training.3 Importantly, prior to behavioral studies, we confirmed that methionine administration resulted in increased methionine levels in the hippocampus at 1.5 h postmethionine treatment as described previously,20 which precede its effect on DNA methylation and subsequent gene expression. Thus, methionine was administered 1 h prior to behavioral training and animals were sacrificed at 2 h posttraining. As controls for these experiments, homecaged/untrained animals (controls) were sacrificed at 3 h postmethionine administration.
Real time PCR
RNA was extracted from hippocampal area CA1 (AllPrep DNA/RNA Mini Kit, Qiagen, Hilden, Germany), then 0.3 μg of RNA was converted to cDNA (iSript RT-PCR iQ SYBR Green Supermix, Bio-Rad, Hercules, CA). Q-PCR amplifications were performed on the iQ5 real time PCR system (Bio-Rad) at 95.0°C for 3 min, 40 repeats of 95.0°C for 15 sec, followed by 58.0°C for 1 min, 95.0°C for 1 min, 55.0°C for 1 min, 81 repeats of 55.0°C for 10 sec each, and, finally, held at 4.0°C, using primer sets to amplify cDNA designed for Bdnf IX (sense 5′–GAGAAGAGTGATGACCATCCT–3′; antisense 5′–TCACGTGCTCAAAAGTGTCAG–3′) and Gapdh (sense 5′–ACCTTTGATGCTGGGGCTGGC–3′; antisense 5′–GGGCTGAGTTGGGATGGGGACT–3′). All samples were run in duplicate at a primer molarity of 10 μmol/L and Bdnf was compared to Gapdh. Expression of Gapdh was unchanged across all treatment groups. Cycle threshold (Ct) values were analyzed using the comparative Ct method to calculate differences in gene expression between samples.
Direct bisulfite DNA sequencing
DNA was extracted from hippocampal area CA1 (AllPrep DNA/RNA Mini Kit, Qiagen), with 1 μg of DNA used for bisulfite modification (EpiTect Bisulfite Kit, Qiagen). Bisulfite-treated DNA was then amplified for a primer targeting 12 CpG sites within promoter 4 and exon IV of the Bdnf gene for sequencing with the sense strand as (5′–GGTAGAGGAGGTATTATATGATAGTTTA–3′); and the antisense strand as (5′–TACTCCTATTCTTCAACAAAAAAATTAAAT–3′). The thermocycler protocol used to amplify bisulfite-modified DNA was as follows: 5 min at 95°C, 50 repeats at 95°C for 1 min, followed by 60°C for 1 min, followed by 72°C for 1 min, which was then followed by a final cycle of 5 min at 72°C and then terminated at 4°C. The PCR products were cleaned (ExoSAP-IT (Affymetrix, Santa Clara, CA) and sequenced using the Bdnf reverse primer (UAB Heflin Center for Human Genetics, http://www.uab.edu/hcgs). The percentage of CpG site methylation was then calculated from the electropherogram by determining the ratio between peak values of guanine (G) and adenine (A) (G/[G + A]) using Chromas software (Technelysium Pty Ltd., South Brisbane, Australia). Methylation percentage for each CpG site within the DNA amplicon was quantified by measuring the ratio between peak height values of cytosine (C) and thymine (T), yielding the basic equation for the methylation percentage to be (C/[C + T] × 100) when the forward primer is used for DNA sequencing. If the reverse primer was used, the guanine (G) and adenine (A) peak heights were used instead, yielding the equation (G/[G + A] × 100). Sequencing was performed using the reverse primer because it resulted in a cleaner chromatogram and more consistent analysis of DNA methylation as described previously.21–23
Western blotting
To quantify BDNF levels, protein extracts (10 μg) were separated on 4–12% Nupage BisTris gels (Invitrogen, Carlsbad, CA). The proteins were transferred onto a Polyvinylidene fluoride membrane using a semidry transblotter (Bio-Rad). Blots were then probed with a BDNF antibody (1:1000, Cat. No. sc-546, Santa Cruz Biotechnology Inc., Dallas, Texas, USA). Secondary goat anti-rabbit 800CW antibody was used for protein detection (Licor Odyssey System, LiCor, Lincoln, NE). Quantifications were normalized to actin (1:1000, Cat. No. ab1801, Abcam, Cambridge, UK) and valosin-containing protein (1:5000, #ab11433, Abcam).
Amino acid array
Hippocampal tissue was isolated, flash frozen in 2-methylbutane, and amino acid levels measured using a high-performance liquid chromatography (HPLC) system with autosamplers and a 474 scanning fluorescence detector (Vanderbilt Brain Institute Neurochemistry core at Vanderbilt University, http://braininstitute.vanderbilt.edu).
EEG recordings
Two weeks post-SE, animals were anesthetized and maintained with 2.5% isofluorane. A dental drill was used to drill six holes through the skull, bilaterally, ~2–3 mm posterior and lateral to Bregma, ~4 mm posterior to Bregma and 5 mm lateral to midline, and approximately −6 mm posterior to Bregma and 3–4 mm lateral to midline. Three 2.4-mm stainless steel screws (Plastics One, Inc., Roanoke, Virginia, USA) were screwed halfway into the two holes closest to Bregma and in one hole farthest from Bregma. Then, an EEG electrode (Plastics One, Inc.) with two lead wires and a ground wire, cut to a length that would touch but not penetrate the dural surface (~1.7 mm), were inserted into the remaining three drill holes, with the ground wire positioned in one of the holes farthest from Bregma. The lead wires were placed bilaterally on the cortical surface of the parietal hemispheres in the cortical region over the underlying hippocampi. This two-electrode system does not allow us to identify the anatomical origin of epileptic activity. Once the electrode was positioned, dental acrylic (Darby Dental Supply, Memphis, TN) was applied to form a stable cap on the skull. When the acrylic had dried, the scalp was closed with skin glue (3M Vetbond 3M, Saint Paul, Minnesota, USA). After the animals were sacrificed, electrodes were confirmed to be on the surface of the brain. Macroscopically, there appeared to be no damage to the underlying brain tissue. One week after electrode placement surgery, rats were individually housed in specially constructed EEG monitoring cages. EEG data were acquired using Biopac Systems amplifiers (Biopac EEG100C) and AcqKnowlege 4.2 EEG Acquisition and Reader Software (Biopac Systems, Inc., Goleta, CA). EEG data were stored and analyzed in digital format. Cages were also equipped with IR Digital Color CCD cameras (Digimerge Technologies, Markham, Ontario, Canada) that recorded each animal concurrently with EEG monitoring; recordings were acquired for review using security system hardware and software (L20WD800 Series, Lorex Technology, Inc., Linthicum, MD). All collected data were manually analyzed for abnormalities by an experienced observer blinded to genotype. All EEG recordings were scored for the presence of isolated spikes, repetitive spiking, and seizures while the animals were awake. Spikes were defined as having a duration of <200 msec with 5× baseline amplitude, whereas repetitive spiking activity was defined as ≥3 spikes lasting ≤5 sec. Seizures were defined as high-frequency and high-amplitude repetitive spiking, or low-frequency, high-amplitude spike-wave patterns ≥10 sec. The corresponding video of the animal was analyzed for associated behavior during these abnormal events. EEG rhythm power analysis was performed offline in the MATLAB environment. We obtained the absolute power, and the mean power of brain alpha (8–13 Hz) and theta (4–8 Hz) frequency bands every 30 sec from all implanted electrodes (AcqKnowlege Software BIOPAC, Goleta, California, USA). The absolute power of a band is the integral of all of the power values within its frequency range. To calculate the absolute power and mean power, we used 5 × 30 sec periods for each rat, 15 sec before and 15 sec after the interictal discharge.
Behavioral tests
Animals were handled for three consecutive days prior and transported 2 h before behavioral experiments in the open field, contextual fear conditioning, and object location.
Open field
The testing room was dimly lit, and the open-field chamber had a white floor and white walls. The animals were placed into the chamber for 20 min and allowed to move freely around the apparatus. Total movement time and time spent in the center of the open area was calculated for each animal. Animals were considered to be in the open area if they were 10 cm away from the walls.
Contextual fear conditioning
The animals were placed in a chamber, allowed to explore for 2 min, and subjected to a 1-sec, 0.5 mA subthreshold footshock. This 2 min, 1 sec pairing was repeated three times over the course of 6 min, ending with a 1-min exploratory period. Vehicle naive animals were left untouched in their home cages. For behavioral testing, animals were placed back in the same chamber 24 h later, and freezing behavior was monitored for a period of 5 min.
Object location
The animals were placed in an open-field chamber for five consecutive days. Days 1–3 consisted of habituation to the chamber for 5 min. On the training day (day 4), two identical objects (100 mL Pyrex bottles) were placed 10 cm from the corners of the chamber. The rats were allowed to explore the objects for 15 min. Testing day (day 5) was 24 h after the training trial. For testing, one of the objects was placed in the middle of the box, and the other object was placed in the same location as during the training trial. Animals were tested for 5 min. Time spent exploring the objects was recorded and expressed by a discrimination index (DI = [tnovel − tfamiliar]/[tnovel + tfamiliar] × 100%). Time spent with the objects was calculated and the discrimination indices between treatment groups were compared.
Statistical analysis
Relative mRNA fold changes for all genes and brain regions were analyzed using the comparative Ct method. Due to a lack of homogeneity between groups (Levene test), mRNA and protein expression data were first transformed using a log10 base transformation. Log10 transformation of mRNA and protein data resulted in a nonsignificant Levene test. Transformed data were then analyzed using either a Student's t-test or one-way analysis of variance (ANOVA), with a Dunnett or Tukey post hoc test, where appropriate. DNA methylation and behavioral data were analyzed using either a Student's t-test or one-way ANOVA with a Tukey post hoc test. IBM SPSS Statistics, Armonk, New York, USA was used for all statistical analysis. Significance was set at P ≤ 0.05 for all analyses. Figures were made in GraphPad Prism GraphPad Software, Inc., La Jolla, California, USA.
Results
Bdnf mRNA and DNA methylation levels are altered in the epileptic hippocampus during memory consolidation
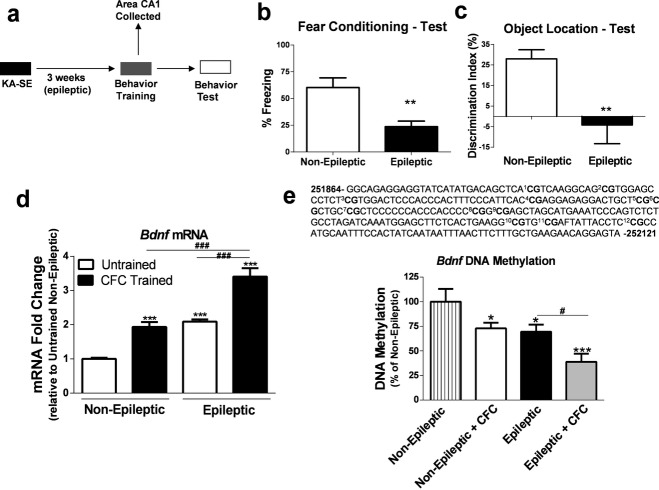
To confirm memory deficits in the kainate rodent model of TLE, epileptic rats (3 weeks post-SE) were trained in two hippocampus-dependent memory tasks: (1) CFC or (2) OL (Fig.1a).24–27 As expected, we found that epileptic rats displayed markedly reduced freezing behavior in the CFC memory task (Fig.1b), suggesting that hippocampus-dependent memory formation was compromised in our TLE animal model. This finding was further supported by a lack of memory retention in the OL memory task (Fig.1c).
Figure 1.
Altered brain-derived neurotrophic factor (Bdnf) gene expression and DNA methylation levels in the epileptic hippocampus are associated with memory deficits. (a) Diagram of experimental setup. (b, c) Epileptic animals displayed hippocampus-dependent memory deficits on test day at 24 h posttraining in the contextual fear conditioning (CFC) and the object location (OL) memory paradigms (CFC:t(17) = 3.41, P < 0.01, n = 10–11; OL:t(9) = 3.37, P < 0.01, n = 5–6, t-test, *significance relative to non-epileptic group). (d) BdnfmRNA levels were significantly increased in the hippocampus of the non-epileptic, epileptic untrained, and epileptic CFC animals following CFC training compared to non-epileptic untrained controls. BdnfmRNA levels were significantly increased in the hippocampus of the epileptic CFC animals compared to all other groups (F3,18 = 67.58, P < 0.001, n = 5–6, one-way analysis of variance [ANOVA] with post hoc test, *significance relative to non-epileptic group; #significance between experimental groups). (e) Bisulfite sequencing analysis of 12 CpG sites within promoter 4 and the noncoding exon IV of the Bdnf gene revealed a significant decrease in DNA methylation levels in the non-epileptic CFC-trained, epileptic untrained, and epileptic CFC-trained animals compared to the non-epileptic untrained controls. The epileptic CFC group had significantly decreased DNA methylation compared to all other groups (F3,15 = 11.64, P < 0.001, n = 4–5, one-way ANOVA with post hoc test, *significance relative to non-epileptic group; #significance between experimental groups). Error bars are SEM.
BDNF activity is crucial for proper memory formation and is significantly increased with epilepsy in both rodent models and in humans.15,16,18,28 Furthermore, Bdnf transcriptional activation in area CA1 of the hippocampus is necessary for proper memory formation and CA1 is also a region affected with chronic seizures in TLE.29–32 Therefore, we assessed Bdnf mRNA levels in area CA1 of epileptic animals during memory consolidation. At 2 h after CFC, Bdnf mRNA levels were significantly increased in area CA1 of the untrained epileptic group and the CFC-trained non-epileptic group compared to non-epileptic untrained group (Fig.1d). Interestingly, Bdnf mRNA levels in area CA1 of epileptic CFC-trained group was significantly greater than levels observed in both the epileptic untrained group and the CFC-trained non-epileptic group (Fig.1d), suggesting that Bdnf is further elevated with epilepsy during memory consolidation.
We next assessed Bdnf DNA methylation levels in area CA1 of the epileptic hippocampus during memory consolidation. DNA methylation levels at Bdnf promoter 4 were assessed due to the fact that exon IV is the primary Bdnf exon responsible for activity-dependent Bdnf mRNA expression.17,33 In Figure1e, a schematic of the 12 CpG dinucleotide sites we analyzed by bisulfite sequencing are shown that encompass part of promoter 4 and the noncoding exon IV region of the Bdnf gene. At 2 h, non-epileptic CFC-trained and epileptic untrained groups displayed reduced Bdnf DNA methylation levels, which were inversely correlated with Bdnf mRNA levels in area CA1 of the hippocampus compared to non-epileptic untrained controls (Fig.1e). Bdnf DNA methylation levels were significantly reduced in area CA1 of epileptic CFC-trained animals compared to all groups assessed, suggesting that Bdnf DNA methylation was abnormally regulated during memory consolidation with epilepsy (Fig.1e). Together, these data indicate that DNA methylation mechanisms may contribute to aberrant activity-dependent Bdnf gene transcription in the epileptic hippocampus during memory consolidation.
Effect of methyl supplementation with methionine on Bdnf DNA methylation and mRNA levels in the epileptic hippocampus during memory consolidation
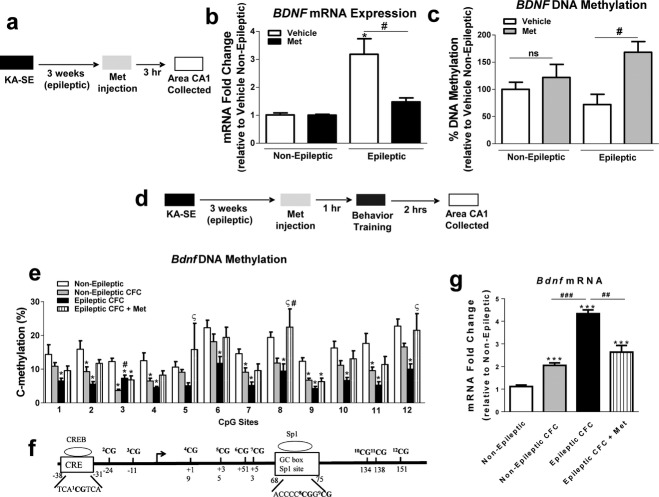
Methionine (Met) administration has been shown to increase DNA methylation, and significantly affect neuronal function and behavior.34–38 Therefore, we treated epileptic rats with Met and determined whether manipulating DNA methylation could affect Bdnf DNA methylation and mRNA levels in the epileptic brain (Fig.2a). First, we determined whether i.p. injection of Met (100 mg/kg) was sufficient to elevate Met levels in the rat hippocampus, as this dose has previously been reported to significantly elevate Met levels in the rat brain at 1.5 h after Met i.p. administration.20 An amino acid array analysis revealed that Met i.p. injection resulted in a sevenfold increase in hippocampal Met levels compared to nonepileptic and non–Met-treated epileptic animals (Table1). Met treatment significantly decreased Bdnf mRNA levels in the epileptic hippocampus and had no effect on Bdnf mRNA levels of the nonepileptic animals (Fig.2b). Additionally, Met administration significantly increased Bdnf DNA methylation in the epileptic hippocampus (Fig.2c). These results suggest that methyl supplementation with Met can decrease Bdnf gene transcription and increase Bdnf DNA methylation in the epileptic hippocampus.
Figure 2.
Methyl supplementation with methionine (Met) alters hippocampal brain-derived neurotrophic factor (Bdnf) gene DNA methylation and mRNA levels in epileptic animals. (a) Diagram of experimental setup. (b) Methyl supplementation with Met significantly reduced BdnfmRNA levels in the hippocampus of the epileptic animals (F3,24 = 9.99, P < 0.001, n = 6–8, one-way analysis of variance [ANOVA] with post hoc test, *significance relative to non-epileptic group; #significance between experimental groups). (c) Methyl supplementation with Met significantly increased BdnfDNA methylation in the epileptic hippocampus (F3,12 = 4.26, P < 0.05, n = 4, one-way ANOVA with post hoc test, *significance relative to nonepileptic group; #significance between experimental groups). (d) Experimental design setup. (e) Decreased BdnfDNA methylation in the epileptic contextual fear conditioning (CFC) group was rescued by Met supplementation (F3,22 = 15.06, P < 0.001, n = 5–9, one-way ANOVA with post hoc test, *significance relative to non-epileptic; #significance relative to non-epileptic CFC; §significance relative to epileptic CFC). (f) A schematic of transcription factor (TF)-binding regulatory elements within the rat Bdnf exon IV transcription start site (TSS). The cAMP response element-binding (CREB) TF is known to bind to the depicted sequence within the Bdnf gene while the depicted Sp1-binding site is considered to be a putative site. (g) Methyl supplementation with Met significantly reduced BdnfmRNA levels in the hippocampus of CFC-trained epileptic animals (F3,19 = 48.69, P < 0.001, n = 5–6, one-way ANOVA with post hoc test, *significance relative to non-epileptic group; #significance between experimental groups). Error bars are SEM.
Table 1.
Amino acid levels in the epileptic hippocampus following methionine treatment
| Amino acids (pmol/μg protein) | Non-epileptic | Epileptic | Epileptic + Met |
|---|---|---|---|
| Methionine | 1.4 | 1.3 | 8.3***### |
| Aspartic | 59.5 | 52.6 | 44.0 |
| Serine | 36.3 | 29.2 | 27.6* |
| Glutamic acid | 261.8 | 214.3 | 187.5* |
| Glycine | 22.0 | 12.9*** | 10.6*** |
| Histidine | 106.4 | 92.6 | 90.2 |
| Taurine | 219.8 | 194.6 | 194.5 |
| Arginine | 3.1 | 3.9 | 6.3* |
| Threonine | 17.2 | 15.6 | 15.3 |
| Alanine | 21.0 | 17.8 | 16.8 |
| Proline | 15.2 | 13.2 | 11.3 |
| GABA | 41.2 | 34.5 | 36.5 |
| Cystine | 2.3 | 2.3 | 2.5 |
| Tyrosine | 2.9 | 2.2* | 2.0* |
| Valine | 1.8 | 1.7 | 1.6 |
| Lysine | 4.8 | 5.1 | 5.7 |
| Isoleucine | 129.5 | 118.2 | 125.0 |
| Phenylalanine | 1.6 | 1.4 | 1.4 |
| Leucine | 2.5 | 2.4 | 2.4 |
Epileptic animals show significant deficits in glycine and tyrosine levels compared to vehicle (saline)-treated non-epileptic controls. Met-treated epileptic animals showed a sevenfold increase in hippocampal Met levels compared to non-epileptic and epileptic groups. Met-treated epileptic animals had significantly lower serine, glutamic acid, glycine, and tyrosine levels compared to the non-epileptic group. Arginine levels were significantly elevated in the hippocampus of epileptic Met-treated animals compared to the non-epileptic group. (*P < 0.05, ***P < 0.001, ###P < 0.001; n = 4, one-way ANOVA with post hoc test. *Significance relative to non-epileptic animals; #significance relative to epileptic animals). ANOVA, analysis of variance; GABA, γ-aminobutyric acid.
Next, we assessed the effects of methyl supplementation with Met on Bdnf DNA methylation and mRNA levels during memory consolidation (Fig.2d). We found that Met administration significantly increased DNA methylation levels in area CA1 of epileptic CFC-trained animals, and elevated Bdnf DNA methylation to levels similar to the nonepileptic CFC-trained group (Fig.2e). Interestingly, Met had its strongest effect at CpG sites 5, 8, and 12 of the Bdnf CpG sties assayed, suggesting that these sites may be of particular importance in Bdnf transcription. In fact, CpG site 8 encompasses a transcription factor-binding site for specificity protein 1 (Sp1) that might have been disrupted due to physical hindrance by the methyl functional group found within the Sp1 DNA consensus sequence (Fig.2f). In response to Met treatment, Bdnf DNA methylation changes correlated with decreased Bdnf mRNA levels in area CA1 of epileptic animals during memory consolidation (Fig.2g). These results suggest that in the epileptic hippocampus, methyl supplementation via Met affects activity-dependent Bdnf transcription during memory consolidation.
Methyl supplementation via methionine reduces interictal spike activity, improves theta rhythm power, and reverses hippocampus-dependent memory impairments with epilepsy
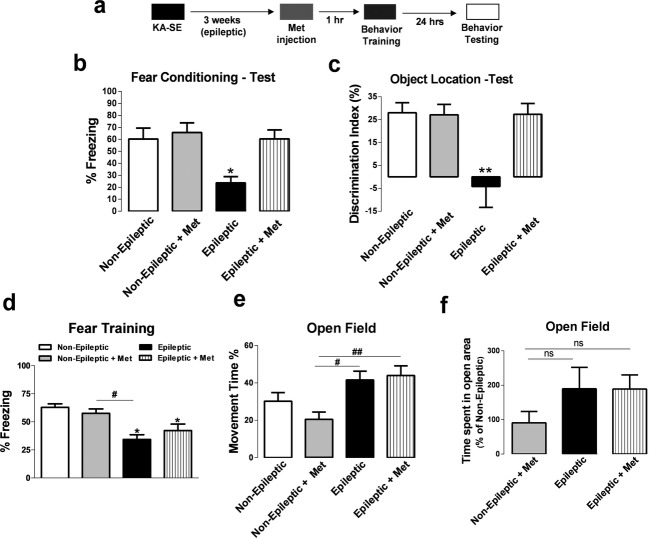
Due to the robust effects of methyl supplementation with Met on Bdnf mRNA levels in the epileptic hippocampus during memory consolidation, we next investigated whether Met administration could alter hippocampus-dependent memory formation in epileptic animals (Fig.3a). Met treatment did not significantly alter freezing behavior in nonepileptic CFC-trained animals; however, Met administration reversed memory impairments in epileptic CFC-trained animals (Fig.3b). We confirmed the effect of Met administration on hippocampus-dependent memory using the OL memory task and found that Met reversed memory deficits observed in epileptic animals during OL memory consolidation (Fig.3c). Importantly, methyl supplementation via Met had no adverse effects on freezing behavior during CFC training (Fig.3d) or on anxiety and mobility activity using the open-field test (Fig.3e), suggesting that the rescue of epilepsy-related memory deficits via Met occurred independent of changes in locomotor activity, exploration, or anxiety.
Figure 3.
Methyl supplementation reverses hippocampal memory deficits associated with epilepsy. (a) Diagram of experimental setup. (b) Methyl supplementation rescued memory impairments in the epileptic animals in the contextual fear conditioning (CFC) paradigm (F3,36 = 5.95, P < 0.01, n = 10–11, one-way analysis of variance [ANOVA] with post hoc test, *significance relative to non-epileptic group). (c) Methyl supplementation rescued memory deficits in the epileptic animals in the object location (OL) paradigm (F3,19 = 7.06, P < 0.01, n = 5–6, one-way ANOVA with post hoc test, *significance relative to non-epileptic group). (d) Methyl supplementation had no effect on fear conditioning training (F3,26 = 7.68, P < 0.01, n = 7–9, one-way ANOVA with post hoc test, *significance relative to non-epileptic group; #significance between experimental groups). (e) Methyl supplementation with Met did not significantly alter movement time in the open-field paradigm. Epileptic animals showed greater mobility compared to non-epileptic rats (F3,24 = 5.53, P < 0.01, n = 6–8, one-way ANOVA with post hoc test, *significance relative to non-epileptic group; #significance between experimental groups). (f) Methyl supplementation with Met did not alter the time spent in the open arena during the open-field paradigm (F3,24 = 1.46, P > 0.05, n = 6–8, one-way ANOVA with post hoc test). Error bars are SEM.
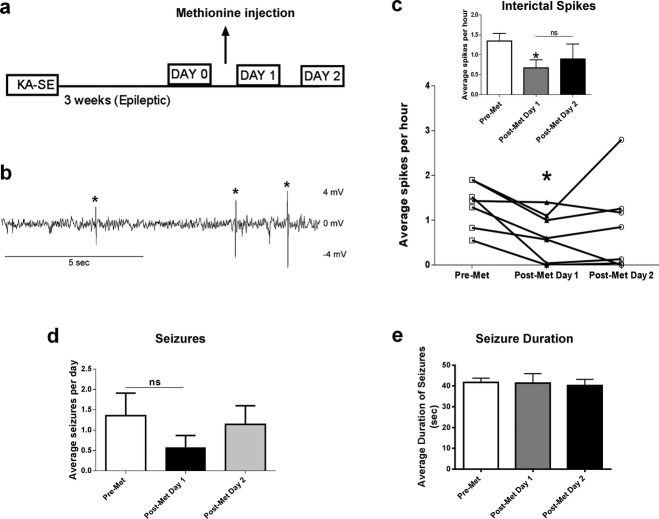
Recent studies have demonstrated that interictal spiking may contribute to epilepsy-related memory deficits.39,40 Therefore, we investigated whether methyl supplementation via Met could affect interictal spike activity using continuous EEG-video monitoring on epileptic animals (Fig.4a). In Figure4b, a representative trace of interictal spikes are shown. In the first 24 h following Met treatment, interictal spike rates were significantly reduced, suggesting that methyl supplementation with Met has a strong, yet short-acting influence on interictal spiking, as EEG activity showed a trend toward returning back to baseline 24 h later (Fig.4c). No difference was found in number of seizures per day or seizure duration due to Met treatment in the epileptic animals (Fig.4d and e), suggesting that Met treatment does not directly affect seizure activity.
Figure 4.
Methyl supplementation decreases interictal spiking. (a) Diagram of experimental setup. (b) Representative trace of interictal spikes observed in epileptic rats 3 weeks post-KA administration (* represents what was counted as a spike). (c) Methyl supplementation significantly reduced interictal spike number in the first 24 h postinjection. Interictal spiking rate showed a trend toward returning back to baseline at 24–48 h post-Met supplementation (F2,6 = 4.06, P < 0.05, n = 7, one-way analysis of variance [ANOVA] with post hoc test, *significance relative to pre-Met). (d) Methyl supplementation did not have a significant effect on number of seizures per day (F2,18 = 0.82, P > 0.05, n = 7, one-way ANOVA with post hoc test). (e) Methyl supplementation did no significantly affect duration of seizures (F2,23 = 0.91, P > 0.05, n = 3–16, one-way ANOVA with post hoc test, ns = not significant). Error bars are SEM.
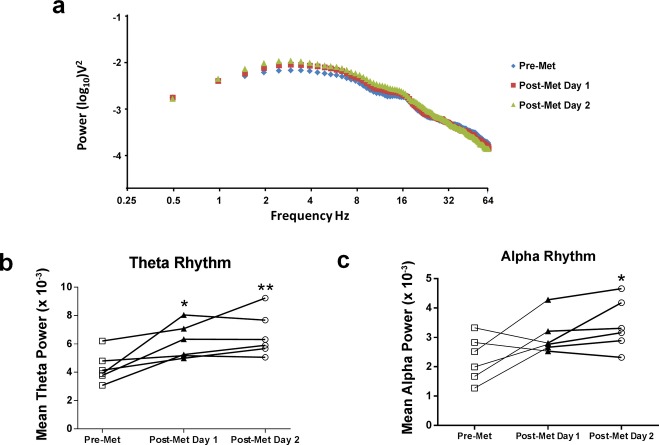
We next performed an analysis of the power spectrum surrounding spiking events, focusing in particular on the theta band, since this has been shown to be important in a number of cognitive processes and may contribute to epilepsy-related memory deficits.41–43 Additionally, methyl supplementation has been show to improve theta rhythm power in humans.44,45 The power spectrum revealed that there was a broad increase in rhythm power at the lower frequencies, between 2 and 18 Hz post-Met treatment (Fig.5a), which encompasses the theta and alpha rhythms. The mean power of theta rhythm was significantly increased in epileptic animals post-Met treatment (Fig.5d). In addition, on the second day post-Met treatment, the mean power of alpha was significantly increased in the epileptic animals (Fig.5c). It is therefore possible that the effect of methionine on interictal activity and theta rhythm power contributed to the restoration of memory deficits in our rodent model of TLE.
Figure 5.
Methyl supplementation increased theta and alpha rhythm power. (a) Power spectrum density surrounding interictal spikes. (b) Methyl supplementation significantly increased theta rhythm power (F2,5 = 12.30, P < 0.01, n = 6, one-way analysis of variance [ANOVA] with post hoc test, *significance relative to pre-Met). (c) Methyl supplementation significantly increased alpha rhythm power on day 2 post-Met treatment (F2,5 = 5.90, P < 0.05, n = 6, one-way ANOVA with post hoc test, *significance relative to pre-Met). Error bars are SEM.
DNMT inhibition prevented the effects of methionine
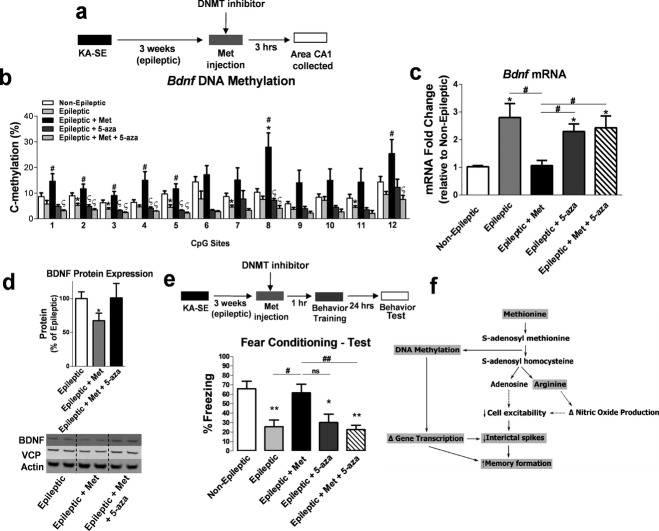
Next, we considered whether DNMT inhibition would block outcomes of methyl supplementation with Met including Bdnf DNA methylation and mRNA expression, and reversal of memory deficits (Fig.6a). 5-Aza-2′-deoxycytidine (5-AZA) was chosen for DNMT inhibition because it can be i.p. injected and at the administered dose, has been previously reported to alter behavior, DNA methylation, and BDNF expression in the hippocampus.46 DNMT inhibition with 5-AZA blocked the effect of methyl supplementation via Met on Bdnf DNA methylation, specifically at CpG sites 1, 2, 3, 4, 5, 8, and 12 within the Bdnf gene (Fig.6b). Interestingly, CpG site 1 encompasses a binding site for the transcription factor cAMP response element-binding (CREB) protein,47 while CpG site 8 encompasses a putative Sp1 transcription factor-binding site as mentioned earlier, suggesting that aberrant methylation at these sites could affect Bdnf expression by disrupting transcription factor binding (Fig.2f). Additionally, DNMT blockade partially attenuated the decrease in Bdnf mRNA (Fig.6c) and protein expression observed with methyl supplementation (Fig.6d). These findings strongly support that the effect of methyl supplementation on Bdnf gene and protein expression is partly due to DNA methylation mechanisms in the epileptic hippocampus during memory consolidation. Importantly, DNMT inhibition also prevented the memory rescue observed with Met treatment of epileptic CFC-trained animals (Fig.6e), suggesting that methyl supplementation reversed memory impairments in a rodent model of TLE via DNA methylation mechanisms.
Figure 6.
DNA methyltransferases (DNMT) inhibition prevents the effect of methyl supplementation on hippocampal brain-derived neurotrophic factor (Bdnf) DNA methylation, Bdnf expression and memory formation. (a) Diagram of experimental setup. (b) The increase in BdnfDNA methylation due to methyl supplementation is blocked by DNMT inhibition (F4,18 = 6.93, P < 0.01, n = 3–6, one-way analysis of variance [ANOVA] with post hoc test, *significance relative to non-epileptic group; # significance relative to epileptic group; §significance relative to epileptic + Met group). (c) DNMT inhibition attenuated the effect of methyl supplementation with Met on BdnfmRNA levels in the epileptic hippocampus (F4,24 = 11.96, P < 0.001, n = 5–8, one-way ANOVA with post hoc test, *significance relative to non-epileptic group; #significance between experimental groups). (d) BDNF protein expression was significantly reduced in Met-treated epileptic animals compared to nontreated epileptic animals. DNMT inhibition attenuated the decrease in BDNF protein expression due to methyl supplementation with Met. Duplicates of each sample were run and normalized to valosin-containing protein (VCP) and beta-actin 4 as within-lane loading controls. Representative blots of samples from each treatment group are shown (F3,14 = 3.84, P < 0.05, n = 5–6, one-way ANOVA with post hoc test, *significance relative to epileptic group). (e) DNMT inhibition blocked Met-induced memory enhancement (F4,31 = 7.81, P < 0.001, n = 7–8, one-way ANOVA with post hoc test, *significance relative to non-epileptic group; #significance between experimental groups). Error bars are SEM. (f) Potential mechanism of the effect of methionine treatment on memory restoration in epileptic animals. Shaded words represent the molecular mechanisms demonstrated in the present study. Unshaded words and dotted lines are potential pathways that could also be involved in the memory restoration process with methionine. Thus, methionine can be rapidly converted to S-adenosyl methionine (SAM), the universal methyl donor in the brain. SAM donates a CH3 group that leads to increased DNA methylation and gene transcription changes in the epileptic hippocampus. Once SAM donates a methyl group, it is converted to S-adenosyl-l-homocysteine (SAH), which leads to the production of adenosine. Adenosine production can lead to decreases in cell excitability that could subsequently decrease interictal spike activity in the epileptic hippocampus. Arginine is also a downstream by product of SAH hydrolysis that can lead to changes in nitric oxide production and can affect cell excitability.
Discussion
TLE is a debilitating disorder with many associated comorbidities including declarative memory loss. While pharmacological treatments may reduce the incidence of seizures, memory deficits often go untreated and the underlying mechanisms remain poorly understood. Persistent, aberrant changes in gene expression that occur in the epileptic brain are believed to have proepileptic implications.14 Paradoxically, memory consolidation requires de novo transcription of numerous genes in the hippocampus that have been implicated in epilepsy, suggesting that gene expression changes in the epileptic hippocampus can have enduring effects on long-term memory formation. For example, Bdnf, a gene widely implicated in the process of memory formation,18 is known to be abnormally increased in the epileptic hippocampus12,48,49 and partial knockdown or overexpression of brain Bdnf interferes with memory formation,15,16 further demonstrating that alterations of memory-permissive genes like Bdnf expression, results in memory deficits. Therefore, abnormal increases in hippocampal Bdnf gene expression with epilepsy may contribute to memory impairments associated with the disorder.
For the present studies, we chose to focus on one candidate gene, Bdnf, because of its well-established role in memory formation and because BDNF expression and function is significantly altered in TLE.12,18,48 However, it is noteworthy to mention that many genes are impacted by DNA methylation mechanisms in the hippocampus during memory formation.2,3,50 Thus, our present studies do not exclude the possibility that DNA methylation changes at other genes may have also been impacted by Met supplementation.35,37
Currently, the literature is controversial as to the exact role that BDNF alone would play in effectively restoring memory formation in disease states such as in epilepsy. Previous studies support that activity-dependent increases in BDNF levels are necessary for normal memory formation. However, epilepsy results in exacerbated levels of BDNF, which may contribute to memory deficits with epilepsy.16,51 Furthermore, other studies have made important links between increased BDNF and posttraumatic stress disorder,52 which is associated with memory deficits. Additionally, infusion of BDNF in the hippocampus increases seizure activity.12 Thus, an interesting idea is that overexpression of BDNF may contribute to memory deficits by increasing cortical activity and seizure susceptibility.51 It is reasonable to speculate that decreasing BDNF expression in an epileptic brain might decrease hyperexcitability to allow for proper memory formation with epilepsy. Our results suggest that aberrant regulation of the Bdnf gene via DNA methylation mechanisms is strongly correlated with epilepsy-related memory deficits, however, further work is needed to elucidate the direct role of Bdnf DNA methylation alone on cognitive deficits in epilepsy.
Recent work has established that DNA methylation is altered in epilepsy and that these molecular changes contribute to persistent gene regulation in the hippocampus.7,9–11 However, whether DNA methylation in the epileptic hippocampus is a pro- or antiepileptic mechanism remains undetermined. For example, prior studies suggest that in the epileptic hippocampus, global increases in DNA methylation may act to dampen hyperexcitability through downregulation of gene expression, as DNMT inhibition in area CA1 with zebularine resulted in a decreased latency to seizures when administered prior to SE and increased cell excitability in epileptic animals.10 Interestingly, another report suggests that DNMT inhibition using 5-AZA, administered in an electrical kindling model of epilepsy decreased seizure severity.11 It is important to note that differences in research approach, design, and experimental epilepsy animal model used may have contributed to the conclusions drawn from these DNMT inhibitor studies. Thus, additional studies are warranted to fully elucidate the role of DNA methylation in epilepsy.
One mechanism for elevating DNA methylation levels in the brain is through methyl supplementation with Met or its derivative S-adenosyl methionine (SAM), the universal methyl donor in the brain. Met or SAM administration has been shown to alter global and gene-specific DNA methylation levels in the brain and with experience-driven behavioral change.11,35–38 Additionally, methyl supplementation with Met has been shown to reverse lead-induced memory deficits and to reduce seizures in a transgenic mouse model.34,53,54 Therefore, global modification of the methylome with epilepsy compromises transcriptional regulation of memory-permissive genes in the hippocampus that is required for proper memory formation. In consideration that DNA methylation at the Bdnf gene is significantly decreased in the epileptic hippocampus,10 we studied the effect of promoting methylation events such as DNA methylation at this gene promoter and on TLE-associated hippocampus-dependent memory deficits. We hypothesized that increasing DNA methylation mechanisms via methyl supplementation with Met might serve as an ideal therapeutic approach for restoring Bdnf gene regulation and proper hippocampus-dependent memory formation in a rodent TLE model.
In the present study, we found that Met administration significantly elevated Bdnf DNA methylation levels in the hippocampus of untrained epileptic animals and returned hippocampal Bdnf mRNA levels back to levels comparable to untrained non-epileptic animals. In response to a learning experience methyl supplementation with Met significantly increased hippocampal Bdnf DNA methylation and reduced chronic overexpression of Bdnf mRNA levels observed in the hippocampus of epileptic animals during memory consolidation. These findings strongly support that methyl supplementation with Met restored hippocampus-dependent memory formation via a DNA methylation-regulatory mechanism. These findings are further supported by previous studies demonstrating that Met treatment can restore proper DNA methylation levels at other genes such as the reelin and glucocorticoid receptor (GR) promoters while altering reelin and GR transcript expression in the hippocampus of rodent models of schizophrenia and maternal care, respectively.37,55 These prior findings and our present studies suggest that promoting DNA methylation may serve to restore proper gene expression, not only with epilepsy disorders but also with other neurological disorders.
Interestingly, we found that Met treatment did not significantly alter global DNA methylation levels in non-epileptic animals. The following factors could have contributed to these results. First, in the present study we administered only a single, acute injection of Met, whereas prior studies administered Met, chronically.35–38 Therefore, future studies should focus on further evaluating the effect of chronic or intermittent treatment of Met in experimental TLE animals compared to controls. Second, because fluctuations in DNA methylation levels are intimately linked to the degree of neuronal function and seizure activity results from intense, synchronized, neuronal network hyperactivity,5 it is plausible to consider that there may be changes in DNA methylation levels at other genes in the epileptic hippocampus in response to methyl supplementation. Finally, increased DNMT expression is observed with epilepsy, suggesting that a greater rate of de novo DNA methylation might occur in an epileptic brain and thus a greater demand for methyl turnover than in a non-epileptic brain.56 Nonetheless, we found that methyl supplementation with Met could reverse memory deficits associated with epilepsy using two hippocampus-dependent rodent memory paradigms. These findings are in strong agreement with previous studies demonstrating that methyl supplementation can alter behavior and reverse memory impairments.34–37,53,57
Furthermore, it has been hypothesized that interictal spike activity is a major contributor to epilepsy-related memory deficits.39,40,58 Where previous findings demonstrated that daily methyl supplementation reduced seizures in a line of transgenic mice,54 we found that one acute administration of methyl supplementation was not sufficient to reduce number of seizures or seizure frequency. However, we did observe that Met treatment decreased interictal spike activity in epileptic rats. Moreover, theta rhythm power has been linked to memory formation and long-term potentiation59–61 and degradation of theta frequency oscillations are believed to contribute to memory deficits in epilepsy.41–43 Based on these previous studies and a report that demonstrated that methyl supplementation increases theta rhythm power in humans,45 we sought to determine whether oscillatory power was altered in epileptic animals following supplementation of Met. We found that methyl supplementation with Met resulted in a marked increase in theta rhythm power. There was also an increase in alpha power on the second day following Met treatment, but this effect was only present after the behavioral testing and is therefore unlikely to have impacted our behavior results. However, due to alpha's rhythms presumed role in cognitive inhibition,62 increased alpha power may have an effect on seizure activity but is beyond the scope of this study. We, therefore, hypothesize that another benefit of methyl supplementation is a reduction in interictal spike activity and an increase in theta rhythm power that may help promote proper memory consolidation in epileptic animals. The pathogenetic mechanism of this intriguing finding remains to be elucidated; however, we speculate that decreasing BDNF alone may result in decreased cortical network activity. Importantly, several studies have demonstrated that increasing BDNF results in seizure activity while partial knockdown of Bdnf is neuroprotective against seizure activity.12,49,51,63
The effect of BDNF on interictal spiking activity in epilepsy is well documented. For example, prior studies demonstrate that an antagonist (K252a) of BDNF-TrkB signaling effectively reduced the probability of multiple population spike peaks, delayed the latency of spontaneous spikes, and reduced the burst frequency induced by subcutaneous injection of Pentylenetetrazole, a widely used convulsant in studies of epilepsy.64 Additionally, numerous rodent studies have also demonstrated that blockade of TrkB receptors leads to increased neuroprotection from electrical and chemoconvulsant-induced seizure activity, and in TrkB knockout mice prevents epileptogenesis.49,65 In agreement with these prior studies, we demonstrate that BDNF is abnormally regulated in the epileptic hippocampus in association with increased spiking activity. Nevertheless, while our findings in the present study provide basic insights into the epigenetic regulation of memory-permissive genes such as Bdnf during memory formation with epilepsy, future electrophysiological studies of epilepsy are necessary to further explore how BDNF-TrkB signaling couples with DNA methylation mechanisms to influence spiking activity and theta rhythms in epilepsy.
In additional studies, we found that these effects were indeed DNA methylation-dependent, as DNMT inhibition with 5-AZA attenuated the Met-enhancing effect on Bdnf DNA methylation levels and the normalization of Bdnf gene transcription in the epileptic hippocampus during memory consolidation. Furthermore, DNMT inhibition prevented the memory rescue effect of methyl supplementation with Met. Collectively, these data support the hypothesis that epilepsy induces memory deficits via a DNA methylation-regulatory mechanism, which can be overcome by methyl supplementation with Met. However, it is possible that methyl supplementation with Met could have initiated other metabolic pathways involved in the rescue of the epilepsy-related memory deficits (Fig.6f). Indeed, we observed a marked decrease in glycine levels in the hippocampus of epileptic animals (Table1 and Fig.6f). Glycine is converted to sarcosine, which may account for a large amount of Met consumption.66 Therefore, deficiencies in hippocampal glycine may contribute to the observed increases in baseline DNA methylation levels in the epileptic hippocampus compared to a non-epileptic hippocampus following Met administration.
In summary, we conclude that methyl supplementation with Met has profound effects on hippocampal Bdnf DNA methylation, gene transcription, and cognition in a rodent model of TLE. The present findings provide novel insight into a powerful transcriptional regulator of gene expression that may contribute to memory impairments associated with epilepsy disorders. Specifically, we expand on the role of Bdnf transcriptional regulation in hippocampus-dependent memory formation and how abnormal activity of this gene may affect memory formation with TLE. In silico analysis of the Bdnf gene (Fig.2f), along with previous studies17,67 reveal several transcription factor-binding sites at Bdnf promoter 4 that underscores the importance of altering DNA methylation-mediated gene transcription in the epileptic hippocampus. Importantly, we provide a novel therapeutic approach for treatment of memory impairments that often accompany epilepsies and other neurodegenerative disorders as well. Our data show that a single, acute administration of Met has a positive effect on hippocampus-dependent memory formation by altering DNA methylation-mediated transcriptional events and reducing cellular excitability in a rodent model of TLE. We caution, however, that it has been reported that chronic administration of Met at high doses can lead to hypermethioninemia, a condition that occurs when there is a high amount of methionine in the body.68 High doses of Met have been shown to result in memory deficits in zebrafish.69 Therefore, future studies should focus on determining the long-term effects of Met supplementation in epilepsy. Additionally, while our results are promising, here we tested the effect of Met treatment in one experimental rodent model of TLE. Thus, further work is necessary to determine its effectiveness in other models of epilepsy. Indeed, intriguing questions still remain as to whether acute or continuous treatment with Met can impact seizure frequency, other epileptiform activity patterns, progression of epilepsy, or sustain the associated rescue of memory deficits with TLE.
Acknowledgments
We thank members of the Lubin laboratory including Tim Jarome and Meribeth Parrish for their thoughtful comments regarding the manuscript. We also thank M. Crowley and M. Han and the University of Alabama at Birmingham Heflin Genomics Core Facility for assistance in DNA sequencing. This work was supported by the National Institute of Mental Health (MH082106, MH097909), the UAB Intellectual and Developmental Disabilities Research Center (P30-HD38985), the Epilepsy Foundation, the Civitan International, and the Evelyn F. McKnight Brain Research Foundation. The authors declare no competing financial interests.
Conflict of Interest
None declared.
References
- Kleen JK, Scott RC, Lenck-Santini PP, Holmes GL. Cognitive and behavioral co-morbidities of epilepsy. In: Noebels JL, Avoli M, Rogawski MA, Olsen RW, Delgado-Escueta AV, editors. Jasper's basic mechanisms of the epilepsies. 4th ed. Bethesda, MD: Oxford University Press; 2012. pp. 915–929. [Google Scholar]
- Levenson JM, Roth TL, Lubin FD, et al. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J Biol Chem. 2006;281:15763–15773. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson ED, Kavalali ET, Monteggia LM. Activity-dependent suppression of miniature neurotransmission through the regulation of DNA methylation. J Neurosci. 2008;28:395–406. doi: 10.1523/JNEUROSCI.3796-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Ma DK, Mo H, et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 2011;14:1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Fan G. The role of DNA methylation in the central nervous system and neuropsychiatric disorders. Int Rev Neurobiol. 2009;89:67–84. doi: 10.1016/S0074-7742(09)89004-1. [DOI] [PubMed] [Google Scholar]
- Kobow K, Jeske I, Hildebrandt M, et al. Increased reelin promoter methylation is associated with granule cell dispersion in human temporal lobe epilepsy. J Neuropathol Exp Neurol. 2009;68:356–364. doi: 10.1097/NEN.0b013e31819ba737. [DOI] [PubMed] [Google Scholar]
- Miller-Delaney SF, Das S, Sano T, et al. Differential DNA methylation patterns define status epilepticus and epileptic tolerance. J Neurosci. 2012;32:1577–1588. doi: 10.1523/JNEUROSCI.5180-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobow K, Kaspi A, Harikrishnan KN, et al. Deep sequencing reveals increased DNA methylation in chronic rat epilepsy. Acta Neuropathol. 2013;126:741–756. doi: 10.1007/s00401-013-1168-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryley Parrish R, Albertson AJ, Buckingham SC, et al. Status epilepticus triggers early and late alterations in brain-derived neurotrophic factor and NMDA glutamate receptor Grin2b DNA methylation levels in the hippocampus. Neuroscience. 2013;248C:602–619. doi: 10.1016/j.neuroscience.2013.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams-Karnesky RL, Sandau US, Lusardi TA, et al. Epigenetic changes induced by adenosine augmentation therapy prevent epileptogenesis. J Clin Invest. 2013;123:3552–3563. doi: 10.1172/JCI65636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharfman HE, Goodman JH, Sollas AL, Croll SD. Spontaneous limbic seizures after intrahippocampal infusion of brain-derived neurotrophic factor. Exp Neurol. 2002;174:201–214. doi: 10.1006/exnr.2002.7869. [DOI] [PubMed] [Google Scholar]
- Binder DK. The role of BDNF in epilepsy and other diseases of the mature nervous system. Adv Exp Med Biol. 2004;548:34–56. doi: 10.1007/978-1-4757-6376-8_3. [DOI] [PubMed] [Google Scholar]
- McNamara JO, Huang YZ, Leonard AS. Molecular signaling mechanisms underlying epileptogenesis. Science's STKE: signal transduction knowledge environment. 2006;2006:re12. doi: 10.1126/stke.3562006re12. [DOI] [PubMed] [Google Scholar]
- Liu IY, Lyons WE, Mamounas LA, Thompson RF. Brain-derived neurotrophic factor plays a critical role in contextual fear conditioning. J Neurosci. 2004;24:7958–7963. doi: 10.1523/JNEUROSCI.1948-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha C, Angelucci A, D'Antoni A, et al. Brain-derived neurotrophic factor (BDNF) overexpression in the forebrain results in learning and memory impairments. Neurobiol Dis. 2009;33:358–368. doi: 10.1016/j.nbd.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, et al. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- Bekinschtein P, Cammarota M, Izquierdo I, Medina JH. BDNF and memory formation and storage. Neuroscientist. 2008;14:147–156. doi: 10.1177/1073858407305850. [DOI] [PubMed] [Google Scholar]
- Racine RJ. Modification of seizure activity by electrical stimulation. II. Motor seizure. Electroencephalogr Clin Neurophysiol. 1972;32:281–294. doi: 10.1016/0013-4694(72)90177-0. [DOI] [PubMed] [Google Scholar]
- Rubin RA, Ordonez LA, Wurtman RJ. Physiological dependence of brain methionine and S-adenosylmethionine concentrations on serum amino acid pattern. J Neurochem. 1974;23:227–231. doi: 10.1111/j.1471-4159.1974.tb06938.x. [DOI] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol Psychiatry. 2009;65:760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish RR, Day JJ, Lubin FD. Direct bisulfite sequencing for examination of DNA methylation with gene and nucleotide resolution from brain tissues. Curr Protoc Neurosci. 2012 doi: 10.1002/0471142301.ns0724s60. ;Chapter 7:Unit 7 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Zhang Y, Fei J, et al. Rapid quantification of DNA methylation by measuring relative peak heights in direct bisulfite-PCR sequencing traces. Lab Invest. 2010;90:282–290. doi: 10.1038/labinvest.2009.132. [DOI] [PubMed] [Google Scholar]
- Assini FL, Duzzioni M, Takahashi RN. Object location memory in mice: pharmacological validation and further evidence of hippocampal CA1 participation. Behav Brain Res. 2009;204:206–211. doi: 10.1016/j.bbr.2009.06.005. [DOI] [PubMed] [Google Scholar]
- Ennaceur A, Neave N, Aggleton JP. Spontaneous object recognition and object location memory in rats: the effects of lesions in the cingulate cortices, the medial prefrontal cortex, the cingulum bundle and the fornix. Exp Brain Res. 1997;113:509–519. doi: 10.1007/pl00005603. [DOI] [PubMed] [Google Scholar]
- Kim JJ, Rison RA, Fanselow MS. Effects of amygdala, hippocampus, and periaqueductal gray lesions on short- and long-term contextual fear. Behav Neurosci. 1993;107:1093–1098. doi: 10.1037//0735-7044.107.6.1093. [DOI] [PubMed] [Google Scholar]
- Phillips RG, LeDoux JE. Differential contribution of amygdala and hippocampus to cued and contextual fear conditioning. Behav Neurosci. 1992;106:274–285. doi: 10.1037//0735-7044.106.2.274. [DOI] [PubMed] [Google Scholar]
- Binder DK, Scharfman HE. Brain-derived neurotrophic factor. Growth Factors. 2004;22:123–131. doi: 10.1080/08977190410001723308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso M, Vianna MR, Izquierdo I, Medina JH. Signaling mechanisms mediating BDNF modulation of memory formation in vivo in the hippocampus. Cell Mol Neurobiol. 2002;22:663–674. doi: 10.1023/A:1021848706159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao LR, Dudek FE. Increased excitatory synaptic activity and local connectivity of hippocampal CA1 pyramidal cells in rats with kainate-induced epilepsy. J Neurophysiol. 2004;92:1366–1373. doi: 10.1152/jn.00131.2004. [DOI] [PubMed] [Google Scholar]
- Smith BN, Dudek FE. Short- and long-term changes in CA1 network excitability after kainate treatment in rats. J Neurophysiol. 2001;85:1–9. doi: 10.1152/jn.2001.85.1.1. [DOI] [PubMed] [Google Scholar]
- Sakata K, Martinowich K, Woo NH, et al. Role of activity-dependent BDNF expression in hippocampal-prefrontal cortical regulation of behavioral perseverance. Proc Natl Acad Sci USA. 2013;110:15103–15108. doi: 10.1073/pnas.1222872110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinowich K, Schloesser RJ, Jimenez DV, et al. Activity-dependent brain-derived neurotrophic factor expression regulates cortistatin-interneurons and sleep behavior. Mol Brain. 2011;4:11. doi: 10.1186/1756-6606-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G, Feng C, Wu F, et al. Methionine choline reverses lead-induced cognitive and N-methyl-d-aspartate receptor subunit 1 deficits. Toxicology. 2010;272:23–31. doi: 10.1016/j.tox.2010.03.018. [DOI] [PubMed] [Google Scholar]
- LaPlant Q, Vialou V, Covington HE, III, et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci. 2010;13:1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W, Zhao M, Li M, et al. Reversal of cocaine-conditioned place preference through methyl supplementation in mice: altering global DNA methylation in the prefrontal cortex. PLoS One. 2012;7:e33435. doi: 10.1371/journal.pone.0033435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, Champagne FA, Brown SE, et al. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: altering epigenetic marking later in life. J Neurosci. 2005;25:11045–11054. doi: 10.1523/JNEUROSCI.3652-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, Meaney MJ, Szyf M. Maternal care effects on the hippocampal transcriptome and anxiety-mediated behaviors in the offspring that are reversible in adulthood. Proc Natl Acad Sci USA. 2006;103:3480–3485. doi: 10.1073/pnas.0507526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan OI, Zhao Q, Miller F, Holmes GL. Interictal spikes in developing rats cause long-standing cognitive deficits. Neurobiol Dis. 2010;39:362–371. doi: 10.1016/j.nbd.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleen JK, Scott RC, Holmes GL, Lenck-Santini PP. Hippocampal interictal spikes disrupt cognition in rats. Ann Neurol. 2010;67:250–257. doi: 10.1002/ana.21896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauviere L, Rafrafi N, Thinus-Blanc C, et al. Early deficits in spatial memory and theta rhythm in experimental temporal lobe epilepsy. J Neurosci. 2009;29:5402–5410. doi: 10.1523/JNEUROSCI.4699-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge M, Wang D, Dong G, et al. Transient impact of spike on theta rhythm in temporal lobe epilepsy. Exp Neurol. 2013;250:136–142. doi: 10.1016/j.expneurol.2013.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto JY, Stead M, Kucewicz MT, et al. Network oscillations modulate interictal epileptiform spike rate during human memory. Brain. 2013;136:2444–2456. doi: 10.1093/brain/awt159. (Pt 8): [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duzel E, Penny WD, Burgess N. Brain oscillations and memory. Curr Opin Neurobiol. 2010;20:143–149. doi: 10.1016/j.conb.2010.01.004. [DOI] [PubMed] [Google Scholar]
- Saletu B, Anderer P, Di Padova C, et al. Electrophysiological neuroimaging of the central effects of S-adenosyl-l-methionine by mapping of electroencephalograms and event-related potentials and low-resolution brain electromagnetic tomography. Am J Clin Nutr. 2002;76:1162S–1171S. doi: 10.1093/ajcn/76.5.1162S. [DOI] [PubMed] [Google Scholar]
- Sales AJ, Biojone C, Terceti MS, et al. Antidepressant-like effect induced by systemic and intra-hippocampal administration of DNA methylation inhibitors. Br J Pharmacol. 2011;164:1711–1721. doi: 10.1111/j.1476-5381.2011.01489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao X, Finkbeiner S, Arnold DB, et al. Ca2+ influx regulates BDNF transcription by a CREB family transcription factor-dependent mechanism. Neuron. 1998;20:709–726. doi: 10.1016/s0896-6273(00)81010-7. [DOI] [PubMed] [Google Scholar]
- Binder DK, Croll SD, Gall CM, Scharfman HE. BDNF and epilepsy: too much of a good thing? Trends Neurosci. 2001;24:47–53. doi: 10.1016/s0166-2236(00)01682-9. [DOI] [PubMed] [Google Scholar]
- Heinrich C, Lahteinen S, Suzuki F, et al. Increase in BDNF-mediated TrkB signaling promotes epileptogenesis in a mouse model of mesial temporal lobe epilepsy. Neurobiol Dis. 2011;42:35–47. doi: 10.1016/j.nbd.2011.01.001. [DOI] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Papaleo F, Silverman JL, Aney J, et al. Working memory deficits, increased anxiety-like traits, and seizure susceptibility in BDNF overexpressing mice. Learn Mem. 2011;18:534–544. doi: 10.1101/lm.2213711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Benedek DM, Fullerton CS, et al. PTSD risk is associated with BDNF Val66Met and BDNF overexpression. Mol Psychiatry. 2014;19:8–10. doi: 10.1038/mp.2012.180. [DOI] [PubMed] [Google Scholar]
- Cao XJ, Huang SH, Wang M, et al. S-adenosyl-l-methionine improves impaired hippocampal long-term potentiation and water maze performance induced by developmental lead exposure in rats. Eur J Pharmacol. 2008;595:30–34. doi: 10.1016/j.ejphar.2008.07.061. [DOI] [PubMed] [Google Scholar]
- Perry S, Levasseur J, Chan A, Shea TB. Dietary supplementation with S-adenosyl methionine was associated with protracted reduction of seizures in a line of transgenic mice. Comp Med. 2008;58:604–606. [PMC free article] [PubMed] [Google Scholar]
- Tremolizzo L, Carboni G, Ruzicka WB, et al. An epigenetic mouse model for molecular and behavioral neuropathologies related to schizophrenia vulnerability. Proc Natl Acad Sci USA. 2002;99:17095–17100. doi: 10.1073/pnas.262658999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Wang L, Zhang Y, et al. Increased expression of DNA methyltransferase 1 and 3a in human temporal lobe epilepsy. J Mol Neurosci. 2012;46:420–426. doi: 10.1007/s12031-011-9602-7. [DOI] [PubMed] [Google Scholar]
- Cao XJ, Wang M, Chen JT, Ruan DY. Protection effects of S-adenosyl-l-methionine on lead-exposed rats during development and its mechanism of long-term potentiation. Zhonghua Yu Fang Yi Xue Za Zhi. 2008;42:151–155. [PubMed] [Google Scholar]
- Holmes GL, Lenck-Santini PP. Role of interictal epileptiform abnormalities in cognitive impairment. Epilepsy Behav. 2006;8:504–515. doi: 10.1016/j.yebeh.2005.11.014. [DOI] [PubMed] [Google Scholar]
- Buzsaki G. Theta rhythm of navigation: link between path integration and landmark navigation, episodic and semantic memory. Hippocampus. 2005;15:827–840. doi: 10.1002/hipo.20113. [DOI] [PubMed] [Google Scholar]
- Hyman JM, Wyble BP, Goyal V, et al. Stimulation in hippocampal region CA1 in behaving rats yields long-term potentiation when delivered to the peak of theta and long-term depression when delivered to the trough. J Neurosci. 2003;23:11725–11731. doi: 10.1523/JNEUROSCI.23-37-11725.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard GR, Titiz A, Tyler A, et al. Speed modulation of hippocampal theta frequency correlates with spatial memory performance. Hippocampus. 2013;23:1269–1279. doi: 10.1002/hipo.22164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimesch W, Sauseng P, Hanslmayr S. EEG alpha oscillations: the inhibition-timing hypothesis. Brain Res Rev. 2007;53:63–88. doi: 10.1016/j.brainresrev.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Lahteinen S, Pitkanen A, Saarelainen T, et al. Decreased BDNF signalling in transgenic mice reduces epileptogenesis. Eur J Neurosci. 2002;15:721–734. doi: 10.1046/j.1460-9568.2002.01897.x. [DOI] [PubMed] [Google Scholar]
- Liu X, Liu J, Liu J, et al. BDNF-TrkB signaling pathway is involved in pentylenetetrazole-evoked progression of epileptiform activity in hippocampal neurons in anesthetized rats. Neurosci Bull. 2013;29:565–575. doi: 10.1007/s12264-013-1326-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara JO, Scharfman HE. Temporal lobe epilepsy and the BDNF receptor, TrkB. In: Noebels JL, Avoli M, Rogawski MA, Olsen RW, Delgado-Escueta AV, editors. Jasper's basic mechanisms of the epilepsies. 4th ed. Bethesda, MD: Oxford University Press; 2012. pp. 514–531. [Google Scholar]
- Mudd SH, Brosnan JT, Brosnan ME, et al. Methyl balance and transmethylation fluxes in humans. Am J Clin Nutr. 2007;85:19–25. doi: 10.1093/ajcn/85.1.19. [DOI] [PubMed] [Google Scholar]
- Takeuchi Y, Miyamoto E, Fukunaga K. Analysis on the promoter region of exon IV brain-derived neurotrophic factor in NG108-15 cells. J Neurochem. 2002;83:67–79. doi: 10.1046/j.1471-4159.2002.01096.x. [DOI] [PubMed] [Google Scholar]
- Stefanello FM, Matte C, Scherer EB, et al. Chemically induced model of hypermethioninemia in rats. J Neurosci Methods. 2007;160:1–4. doi: 10.1016/j.jneumeth.2006.07.029. [DOI] [PubMed] [Google Scholar]
- Vuaden FC, Savio LE, Piato AL, et al. Long-term methionine exposure induces memory impairment on inhibitory avoidance task and alters acetylcholinesterase activity and expression in zebrafish (Danio rerio. Neurochem Res. 2012;37:1545–1553. doi: 10.1007/s11064-012-0749-6. [DOI] [PubMed] [Google Scholar]