Abstract
Purpose
To evaluate the sensitivity and specificity of classification of pathomimetically-degraded bovine nasal cartilage (BNC) at 3 T and 37°C using univariate MRI measurements of both pure parameter values and intensities of parameter-weighted images.
Materials and Methods
Pre- and post-trypsin degradation values of T1, T2, T2*, magnetization transfer ratio (MTR), and apparent diffusion coefficient (ADC), and corresponding weighted images, were analyzed. Classification based on the Euclidean distance was performed and the quality of classification was assessed through sensitivity, specificity and accuracy (ACC).
Results
The classifiers with the highest accuracy values were ADC (ACC = 0.82 ± 0.06), MTR (ACC = 0.78 ± 0.06), T1 (ACC = 0.99 ± 0.01), T2 derived from a 3D spin-echo sequence (ACC = 0.74 ± 0.05), and T2 derived from a 2D spin-echo sequence (ACC = 0.77 ± 0.06), along with two of the diffusion-weighted signal intensities (b = 333 s/mm2: ACC = 0.80 ± 0.05; b = 666 s/mm2: ACC = 0.85 ± 0.04). In particular, T1 values differed substantially between the groups, resulting in atypically high classification accuracy. The second-best classifier, diffusion weighting with b = 666 s/mm2, as well as all other parameters evaluated, exhibited substantial overlap between pre- and post-degradation groups, resulting in decreased accuracies.
Conclusion
Classification according to T1 values showed excellent test characteristics (ACC = 0.99), with several other parameters also showing reasonable performance (ACC > 0.70). Of these, diffusion weighting is particularly promising as a potentially practical clinical modality. As in previous work, we again find that highly statistically significant group mean differences do not necessarily translate into accurate clinical classification rules.
Keywords: osteoarthritis, bovine nasal cartilage, quantitative MRI, classification, sensitivity, specificity
INTRODUCTION
MRI is widely used to assess cartilage matrix status in a variety of experimental and clinical settings. Matrix-sensitive quantitative MRI measurements include the relaxation time values T1, T2, T1ρ, and dGEMRIC index (T1,Gd), as well as apparent diffusion coefficient (ADC), magnetization transfer ratio (MTR), and magnetization transfer rate (km). Numerous in vivo and ex vivo studies have demonstrated the dependence of group mean values of these MRI parameters on biochemical content (1–4) and location within the joint (5). Means have also been found to differ between clinical subject groups in many contexts, including OA status (6), knee injury (7) and physical activity (8). Described trends include increased T1, T2, T1ρ, and ADC values, and decreased MTR, km, and T1,Gd, as cartilage degradation progresses. Further, a degree of macromolecular specificity is exhibited by some of these parameters (9). The results from these MRI studies have been interpreted in terms of cartilage macromolecular characteristics with a primary focus on the content, distribution, and orientation of proteoglycan (PG) and collagen in addition to water content (10,11).
In spite of the these results, the extent to which matrix-sensitive MRI measurements can define the status of cartilage in a clinical setting remains an open question (12). As an example, it is unclear whether a statistically significant difference in group means between patients with differing osteoarthritis (OA) status can be translated into a useful clinical test (12); classification accuracy requires that parameter distributions for two groups exhibit limited overlap, which may or may not correspond to a statistically significant difference in group means.
Previous work has explored the sensitivity (SE) and specificity (SP) of classifying pathomimetically-degraded bovine nasal cartilage (BNC) using cartilage matrix-sensitive MRI measurements (13). The classification algorithm used a decision rule defined by group arithmetic means for T1, T2, km, ADC, and fixed charge density (FCD) as determined by dGEMRIC. For both18-h trypsin and 20-h collagenase degradation protocols, T1 values were found to be the best univariate classifier, exhibiting a SE and SP in the range of 0.9 or better. ADC also exhibited favorable classification properties, while other MRI parameters performed substantially worse.
This work established a framework for viewing MRI measurements in terms of SE and SP, rather than solely in terms of the statistical significance of differences between means (12). Nevertheless, these previous results were limited to experimental conditions in which data were acquired at 4°C at a preclinical field strength of 9.4 T. However, temperature is known to affect virtually all imaging parameters (14), while field strength has a known substantial effect on T1 as well as less well-documented effects on certain other MR parameters. Finally, in our previous work classification was performed only according to pure quantitative MRI parameters, rather than incorporating clinically-relevant weighted image signal intensities (SI). In particular, results of classification according to the T1-weighted fast low angle shot (FLASH) sequence, proton density (PD)-, intermediate (ID)- and T2-weighted images obtained using the Osteoarthritis Initiative (OAI) acquisition protocol would be of particular interest (15).
In accordance with the above, the purpose of this work is to investigate the accuracy of univariate MR classification of degraded cartilage under clinical research conditions, incorporating i) less severe degradation, ii) parameter–weighted imaging data, iii) acquisition at physiological temperature, and iv) a clinical field strength of 3T. This lays the foundation for further clinical studies to address the additional complexities of articular cartilage.
MATERIALS & METHODS
Sample Preparation
The nasal septum was obtained from two freshly slaughtered mature beef cattle (Green Village Packing, Green Village, NJ). The septa were placed in gauze soaked in Dulbecco's phosphate-buffered saline at 4°C (DPBS; Invitrogen, Grand Island, NY) for three days, at which time 30 cylindrical plugs of 8 mm length and 3 mm diameter were punched from each septum, resulting in a total sample size of n = 60. Thirty of these samples were imaged in one imaging session, with plugs skewered securely onto hollow polyethylene tubes with Teflon spacers to separate the 10 plugs per tube. The three skewers of plugs were placed respectively into three of the wells of a four-well susceptibility-matched polyetherimide (ULTEM) sample holder (each well 9 mm in diameter, 8.5 cm deep). Each of these three wells was filled with Fluorinert® FC-77 (Sigma-Aldrich, St. Louis, MO), which served as a hydrophobic, susceptibility-matched bath surrounding the plugs during imaging. The fourth well was filled with DPBS at pH 7.5 ± 0.1 and served as a standard for normalizing the SI of the contrast-weighted images obtained over the four separate imaging sessions. This permitted pre-degradation images to be obtained from all 60 control samples over two imaging sessions. After this was completed, samples were retained on their skewers and digested for 2.5 hours with 1 mg/ml trypsin (Sigma-Aldrich, St. Louis, MO) in DPBS at 37°C in a 5% CO2 atmosphere, resulting in a matched set of 60 degraded samples. Following this, the samples were rinsed thoroughly to remove the trypsin solution and then treated with a protease inhibitor prepared according to the manufacturer's protocol (Sigma-Aldrich, St. Louis, MO) and Ilomastat matrix metalloproteinase inhibitor (16 µL/ml; Chemicon International, Temecula, CA) in DPBS at pH 7.0 ± 0.1 to arrest further digestion; this resulted in the cartilage maintaining structural integrity but sustaining an sulfated glycosaminoglycan (sGAG) loss of 84% by wet weight as quantified by the dimethyl methylene blue (DMMB) assay (16–18). With this protocol, a total of 60 samples were imaged pre-degradation and then again post-degradation..
MRI Measurements
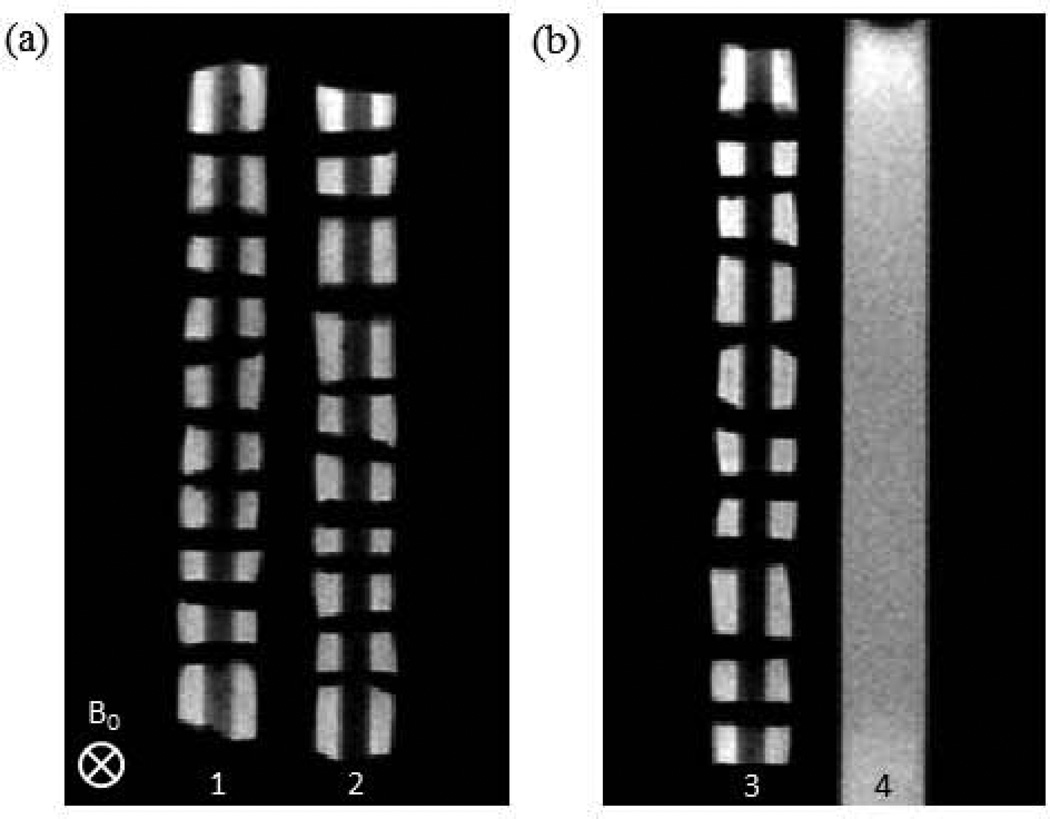
Imaging was performed using a 3 T Philips Achieva MRI scanner (Philips Healthcare, Best, The Netherlands) equipped with an 8-channel SENSE knee coil. As shown in Figure 1, the wells of the sample holder were positioned perpendicular to the B0 static magnetic field. Two slices were obtained, with the first bisecting the cartilage samples in wells 1 and 2 lengthwise, and the second bisecting the cartilage in well 3 and the DPBS in well 4, again lengthwise. The slice thickness was dependent upon available radio frequency bandwidth and gradient strength for the selected image acquisition sequence.
FIGURE 1.
Proton density weighted images (2D spin echo with TE = 10 ms) of the ULTEM sample holder containing ten BNC plugs per well (wells 1, 2, and 3), along with a DPBS standard (well 4) for intensity normalization of weighted images. BNC plugs were threaded onto hollow polyethylene tubes and separated by Teflon spacers. A single ROI consisted of two BNC regions, on either side of a polyethylene tube, from a single plug. These images indicate the slice orientation for each contrast modality. Note that the first slice bisected wells 1 and 2 (a), while the second bisected wells 3 and 4 (b).
All samples were studied at physiological temperature (37 ± 0.1°C), maintained in the magnet using an MRI-compatible warm air heating module (SA Instruments, Stony Brook, NY) connected to flexible tubing. Temperature was monitored using a MRI-compatible thermistor attached to the sample holder.
Quantitative and contrast-weighted MRI measurements were acquired using protocols readily available on clinical scanners, including two of the weighted scans used in the OAI Clinical Protocol (15). The parameters measured were:
Diffusion
A 2D diffusion-prepared sequence with echo planar imaging (EPI) readout and with echo time (TE) = 62 ms, repetition time (TR) = 2 s, and EPI factor = 3 was used to acquire two 4 mm thick imaging slices with b-values of 0, 333, 666, 1000, 1333, 1666, and 2000 s/mm2 applied in three orthogonal gradient directions, with Δ = 25.3 ms, δ = 12.4 ms, acquisition bandwidth (BW) = 12 kHz, field of view (FOV) = 75 × 43.75 mm2 (vertical × horizontal), acquisition matrix/in-plane voxel dimensions (MTX) = 96 × 43/0.78 × 0.95 mm2, and number of signals averaged (NSA) = 1. Total scan time = 15.5 minutes. Both ADC and diffusion weighted (DW) image intensity values were derived from this dataset.
Magnetization Transfer (MT)
A 2D gradient echo sequence with TE = 2.4 ms, TR = 517 ms, and flip angle (FA) = 25° preceded by rectangular saturation pulses with B1 = 2.15 µT, 1 kHz resonance offset and saturation time durations (Tsat) = 50 ms, 100 ms, 150 ms, and 200 ms was used to acquire two 5 mm thick slices. Other acquisition parameters included BW = 98.9 kHz, FOV = 75 × 45 mm2, MTX = 152 × 73/0.49 × 0.62 mm2, and NSA = 2. Total scan time = 10 minutes. Both MTR and MT weighted (MTW) image intensities were derived from this sequence.
T1
A 2D Look-Locker sequence with EPI readout and TE = 5 ms, TR = 6 s, 55 inversion times (TI) ranging from 18 ms to 2757 ms, FA = 14°, and EPI factor = 3 was used to acquire two 4 mm thick imaging slices with BW = 17.5 kHz, FOV = 75 × 44.5 mm2, MTX = 120 × 66/0.63 × 0.67 mm2, and NSA = 2. Total scan time = 7.7 minutes.
T1-weighted (T1W) Acquisition (OAI protocol)
A T1W 3D FLASH gradient echo sequence (TE = 7.57 ms, TR = 20 ms, FA = 13°, partial Fourier factor = 0.75) was used with BW = 31.1 kHz, FOV = 75 × 45 × 24 mm3, MTX = 240 × 144 × 16/0.31 × 0.31 × 1.50 mm3, and NSA = 1. Total scan time = 14 s.
T2
A 3D multi-echo spin echo sequence with TE = 12 ms, TR = 767 ms, and echo train length (ETL) = 30 was used with BW = 28.2 kHz, FOV = 75 × 45 × 23 mm3, MTX = 188 × 78 × 7/0.40 × 0.58 × 3.30 mm3, and NSA = 1. Total scan time = 13.0 minutes. All 30 images with TE ranging from 12 ms to 360 ms with echo spacing of 12 ms were used for T2 calculation.
T2-weighted (T2W) Acquisition (OAI protocol)
A 2D multi-slice multi-echo spin echo sequence with TE = 10 ms, TR = 2700 ms, and ETL = 7 was used to acquire two 3 mm thick slices with BW = 59.8 kHz, FOV = 75 × 45 mm2, MTX = 240 × 101/0.31 × 0.45 mm2, and NSA = 1. Total scan time = 4.5 minutes. Although all seven echo times from this T2W sequence were analyzed, only three weightings, corresponding to a proton density-weighted image (PDW; TE = 10ms), an intermediate-weighted (IDW, TE = 30ms) image, and a T2-weighted (T2W; TE = 60ms) image, are reported (15). All seven images obtained with TE ranging from 20 ms to 70 ms with echo spacing of 10 ms were used for T2 calculation.
T2*
A 2D gradient echo sequence with TE = 1.5 ms, ΔTE = 4.2 ms, TR = 2 s, FA = 25°, and ETL = 30 was used to acquire two 3.5 mm thick slices with BW = 98.9 kHz, FOV = 75×45 mm2, MTX = 152 × 73/0.49 × 0.62 mm2, and NSA = 2. Scan time = 4.9 minutes.
MRI Data Analysis
Each BNC plug image was bisected by the skewer, and, for each plug, a single region of interest (ROI) was selected excluding the region defined by the skewer, with SI defined as the average value across the ROI. ADC, T1, T2, and T2* values were obtained through a conventional three-parameter, monoexponential fit including baseline offset (19,20). For 3D spin echo T2 acquisitions, all echoes were fit to calculate T2 values. Due to stimulated echo effects, the 10 ms echo was excluded from the calculation of T2 values based on the 2D spin echo acquisitions. MTR was calculated as 1 − (Msat / M0) with Msat taken as SI for Tsat = 200 ms and M0 taken as the SI in the absence of off-resonance saturation. Weighted image SIs were calculated by normalizing the observed SI of the plug by the SI of the DPBS standard, resulting in DW, MTW, T1W, PDW, IDW, and T2W intensity values.
Inter-session variability was assessed based on measurements of a single control BNC plug collected in one day over five independent sessions, with the sample removed after each imaging session and re-inserted prior to the next measurement, with orientation preserved. These data were used to calculate the coefficient of variation (CV) for each measured parameter.
Statistical Analysis
Sample and MRI parameter data are reported as mean ± standard deviation. Parameter changes due to degradation were calculated using a paired, two-tailed t-test with statistical significance denoted as p < 0.05 (*) or p < 0.01 (**).
Euclidean distance classification analysis was performed as follows. For each MRI measurement, SE, SP, and accuracy (ACC) of classification into the control group (Ctl, nCtl = = 60) or trypsin degraded group (Deg, nDeg = 60) was assessed using a Euclidean distance metric as previously described (13). Briefly, a training set was constructed from of a random selection of two-thirds of the set of Ctl and Deg samples (nTr = 80), with the remaining samples forming a validation set (nVal = 40). For each MRI measurement, the arithmetic means of the control (μCtl) and degraded (μDeg) samples in the training set were calculated and then used to assign each of the validation set samples (Snew) based on its parameter value (pnew). Assignment was performed according to:
| [1] |
and
| [2] |
The number of correct and incorrect classifications for control and degraded samples was recorded over 100 independent iterations of the above procedure, that is, over 100 independent random selections of training and validation sets, to account for possible training set selection bias. This randomization procedure resulted in an overall equal selection of Ctl and Deg samples in the training sets to within 0.15%. Results for the training and validation sets are reported separately as the means ± standard deviations, taken over these 100 random selections, of SE, that is, the rate of true positives, SP, that is, the rate of true negatives, and ACC. For a balanced dataset such as this, with equal numbers of control and degraded samples, ACC is the average of SE and SP. All calculations were performed using a MATLAB script (The MathWorks, Inc. Natick, MA) developed in-house.
RESULTS
Table 1 shows group mean MRI parameter values indicating, in most cases, a statistically significant difference between the control and trypsin degraded BNC. In particular, changes in all pure parameter values, as opposed to changes in several normalized parameter-weighted SIs, were statistically significant. As expected, ADC, T1, T2, and T2* values increased, and MTR decreased, with degradation. However, these group means do not clearly indicate the degree of overlap between groups. Table 1 also shows the CV for each MRI parameter obtained from repeated experiments on a single sample, as described above. Pure parameter values were stable with CVs of 2.8%, 1.7%, 2.9%, 2.5%, and 5.7% for ADC, MTR, T1, T2-3D, and T2*, respectively. The T2 derived from six echoes from the 2D spin echo sequence had a higher CV of 9.0%. Certain contrast-weighted measurements had a wider range of variation, with DW measurements exhibiting a CV of up to ~15% and T1W measurements showing a CV of ~7%.
TABLE 1.
MRI parameters means in control and 2.5-h trypsin degraded BNC samples
| MRI Measurement |
Control (n = 60) |
2.5-h Trypsin (n = 60) |
Coefficient of Variation (%) |
|---|---|---|---|
| ADC (× 10−3 mm2/s) | 1.90 ± 0.10 | 2.08 ± 0.09 ** | 2.8 |
| DW (b = 333 s/mm2) | 1.12 ± 0.11 | 1.01 ± 0.06 ** | 2.3 |
| DW (b = 666 s/mm2) | 1.79 ± 0.20 | 1.52 ± 0.11 ** | 4.6 |
| DW (b = 1000 s/mm2) | 2.60 ± 0.41 | 2.39 ± 0.36 ** | 12.5 |
| DW (b = 1333 s/mm2) | 1.89 ± 0.61 | 1.85 ± 0.55 | 6.3 |
| DW (b = 1666 s/mm2) | 1.14 ± 0.20 | 1.17 ± 0.17 | 13.5 |
| DW (b = 2000 s/mm2) | 0.97 ± 0.13 | 1.09 ± 0.14 ** | 14.2 |
| MTR | 0.46 ± 0.02 | 0.43 ± 0.02 ** | 1.7 |
| MTW (Tsat = 50 ms) | 0.69 ± 0.04 | 0.69 ± 0.03 | 1.5 |
| MTW (Tsat = 100 ms) | 0.68 ± 0.04 | 0.69 ± 0.03 | 2.0 |
| MTW (Tsat = 150 ms) | 0.58 ± 0.06 | 0.60 ± 0.03 ** | 1.5 |
| MTW (Tsat = 200 ms) | 0.53 ± 0.03 | 0.54 ± 0.03 * | 1.4 |
| T1 (ms) | 866.8 ± 31.4 | 1000.7 ± 31.7 ** | 2.9 |
| T1W | 1.31 ± 0.15 | 1.32 ± 0.09 | 7.2 |
| T2 - 3D (ms) | 78.5 ± 6.3 | 88.3 ± 7.9 ** | 2.5 |
| T2 - 2D (ms) | 50.2 ± 6.1 | 63.7 ± 9.9 ** | 9.0 |
| PDW (TE = 10 ms) | 1.20 ± 0.13 | 1.16 ± 0.11 ** | 0.9 |
| IDW (TE = 30 ms) | 0.97 ± 0.11 | 0.97 ± 0.09 | 0.9 |
| T2W (TE = 60 ms) | 0.72 ± 0.09 | 0.75 ± 0.08 ** | 0.7 |
| T2* (ms) | 21 ± 7 | 25 ± 6 ** | 5.7 |
p < 0.05 control vs. 2.5-h trypsin,
p < 0.01 control vs. 2.5-h trypsin
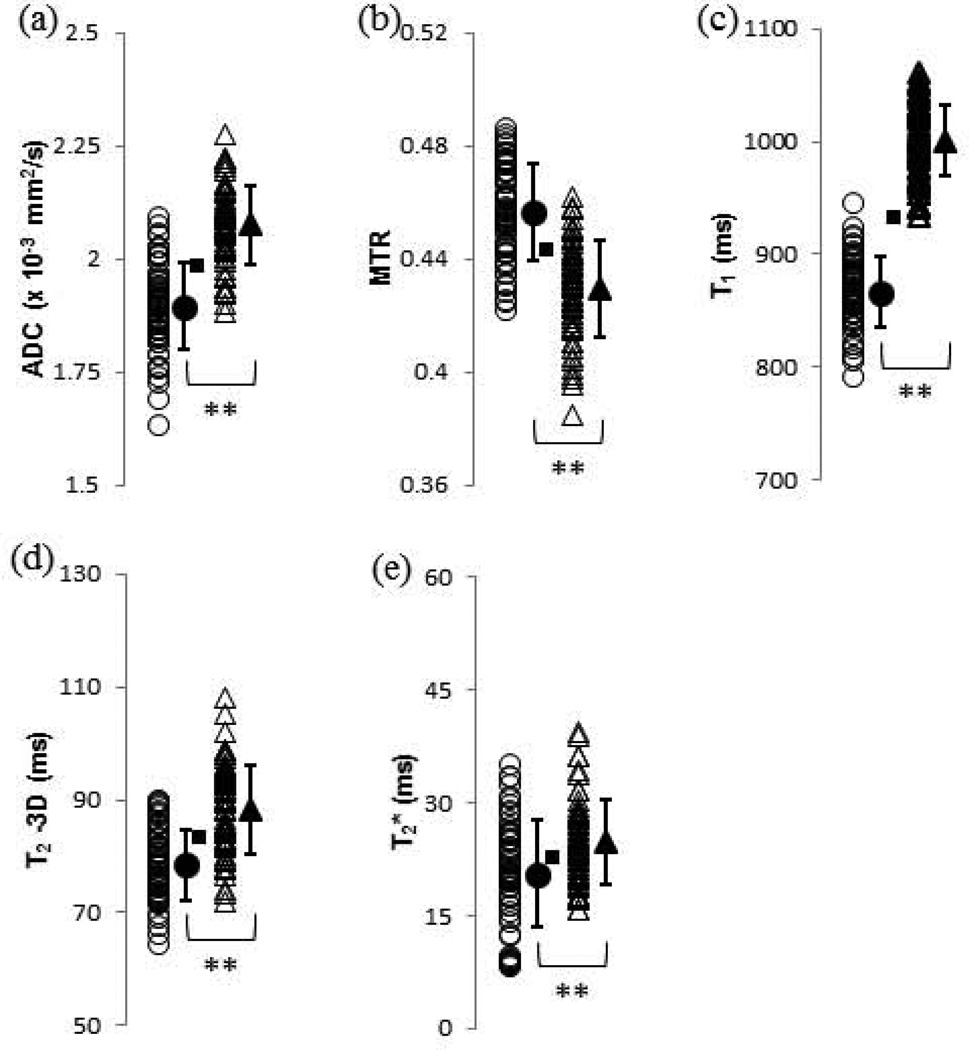
Figure 2 shows individual measured values for each of the pure parameters, each of which exhibited a statistically significant change (p<0.01) upon degradation. The pulse sequences used for ADC (2a), T1 (2c) and T2 (2d), using a 3D spin echo acquisition, are relatively time-consuming, while MTR (2b), using a single 200 ms saturation pulse, and T2* (2e) acquisitions are relatively rapid. With the exception of T1 values, all parameters showed considerable overlap in the values obtained before and after degradation.
FIGURE 2.
Values of pure MR parameters for control (open circles) and 2.5-h trypsin-degraded (open triangles) BNC samples. Group means and standard deviations are shown as adjacent solid shapes and error bars. The classification boundary based on the Euclidean distance metric is indicated with a square to indicate which samples which would have been misclassified based on the group means of the total set of control (n = 60) and 2.5-h trypsin degraded (n = 60) datasets. The actual classification results were obtained using a cross-validation procedure as described in the text. (a) ADC, with 12 control and 9 degraded misclassified samples; (b) MTR, with 15 control and 12 degraded misclassified samples; (c) T1, with 1 control and 0 degraded misclassified samples; (d) T2 – 3D, with 13 control and 17 degraded misclassified samples; and (e) T2*, with 22 control and 23 degraded misclassified samples. * p < 0.05 control vs. 2.5-h trypsin, ** p < 0.01 control vs. 2.5-h trypsin
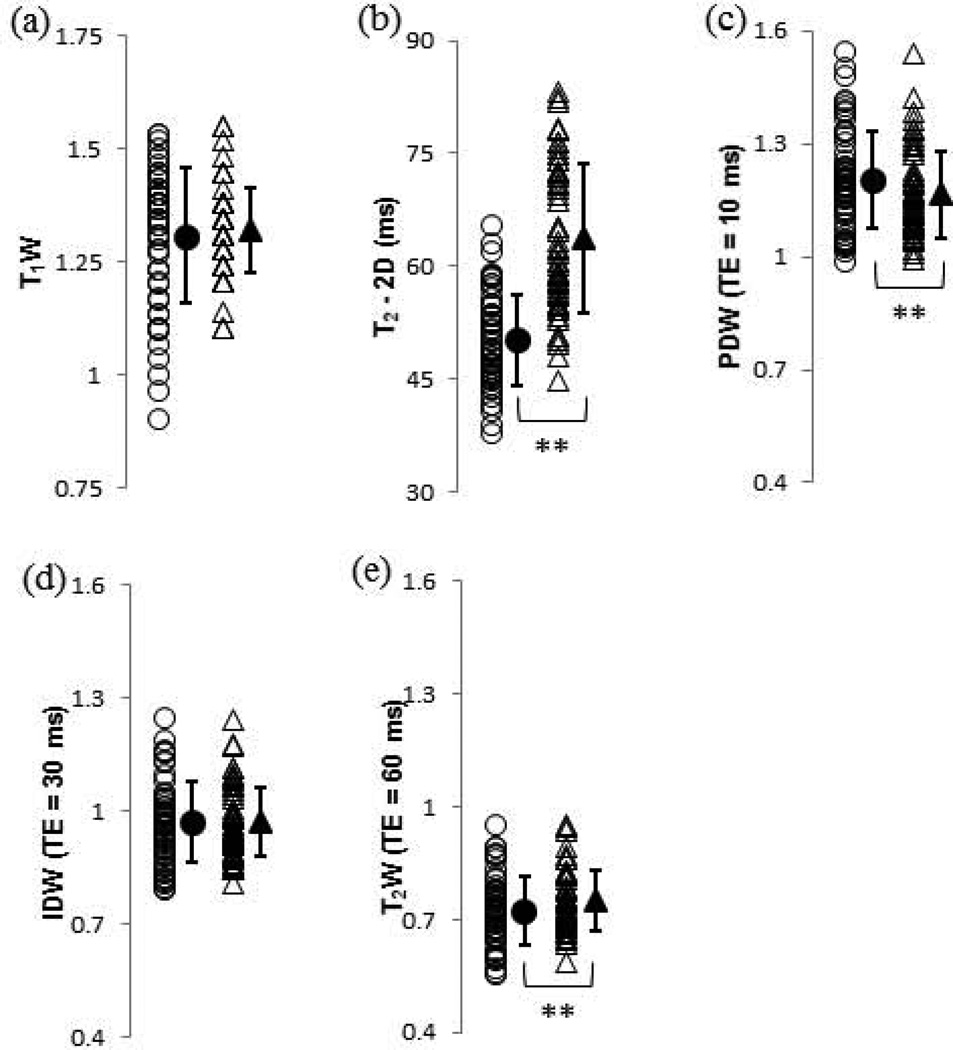
Figure 3 shows the DPBS-normalized T1W results from the FLASH sequence and the T2 relaxation time and DPBS-normalized PDW, IDW, and T2W results from the 2D spin echo sequence corresponding to the OAI protocols. Statistically significant (p<0.01) changes (Table 1) were seen in the T2 value and the PDW and T2W images. However, all DPBS-normalized SI parameters, including those that changed with a high degree of statistical significance upon degradation, exhibited a substantial overlap in the values obtained before and after degradation.
FIGURE 3.
Values of the DPBS-normalized T1 weighted MRI measurements, T2 relaxation time, and T2 weighted MRI measurements obtained using the OAI protocol for control (open circles) and 2.5-h trypsin degraded (open triangles) BNC samples. Group means and standard deviations are shown as adjacent solid shapes and error bars. (a) T1W; (b) T2 – 2D (ms); (c) PDW (TE = 10 ms); (d) IDW (TE = 30 ms) and (e) T2W (TE = 60 ms). * p < 0.05 control vs. 2.5-h trypsin, ** p < 0.01 control vs. 2.5-h trypsin
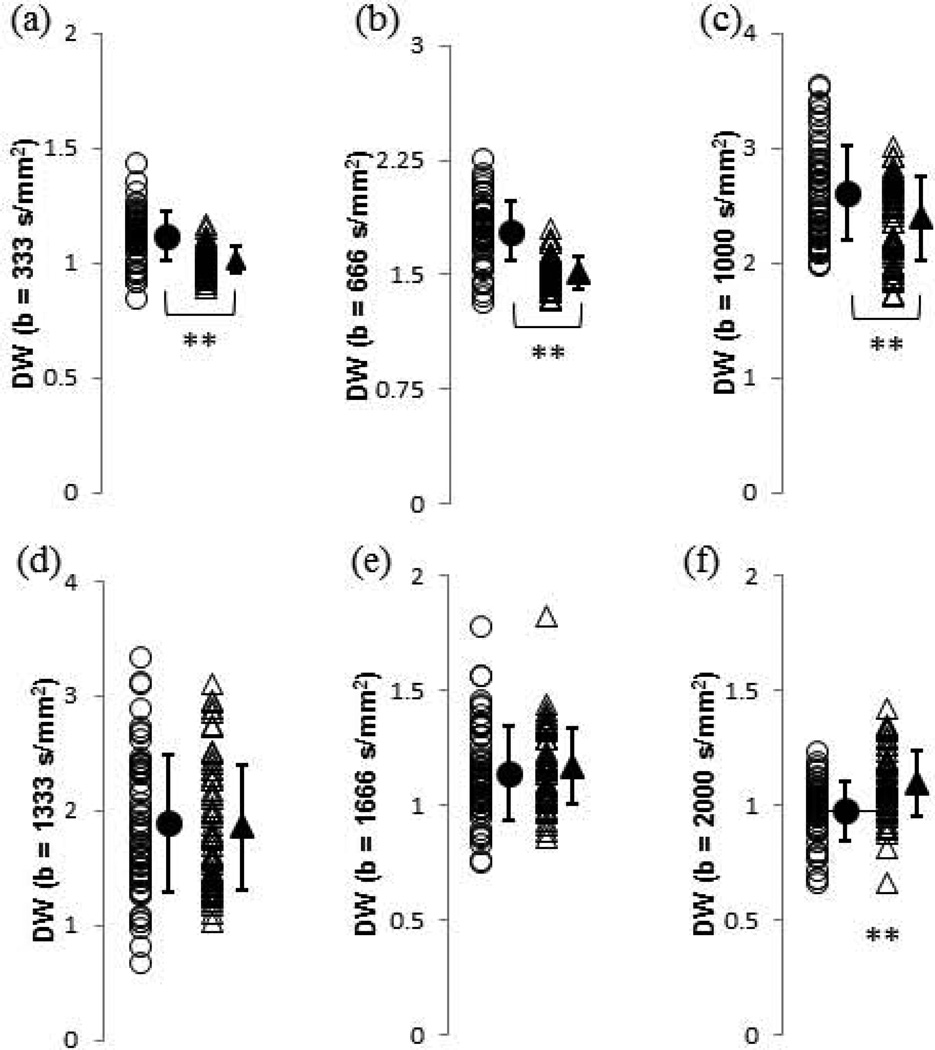
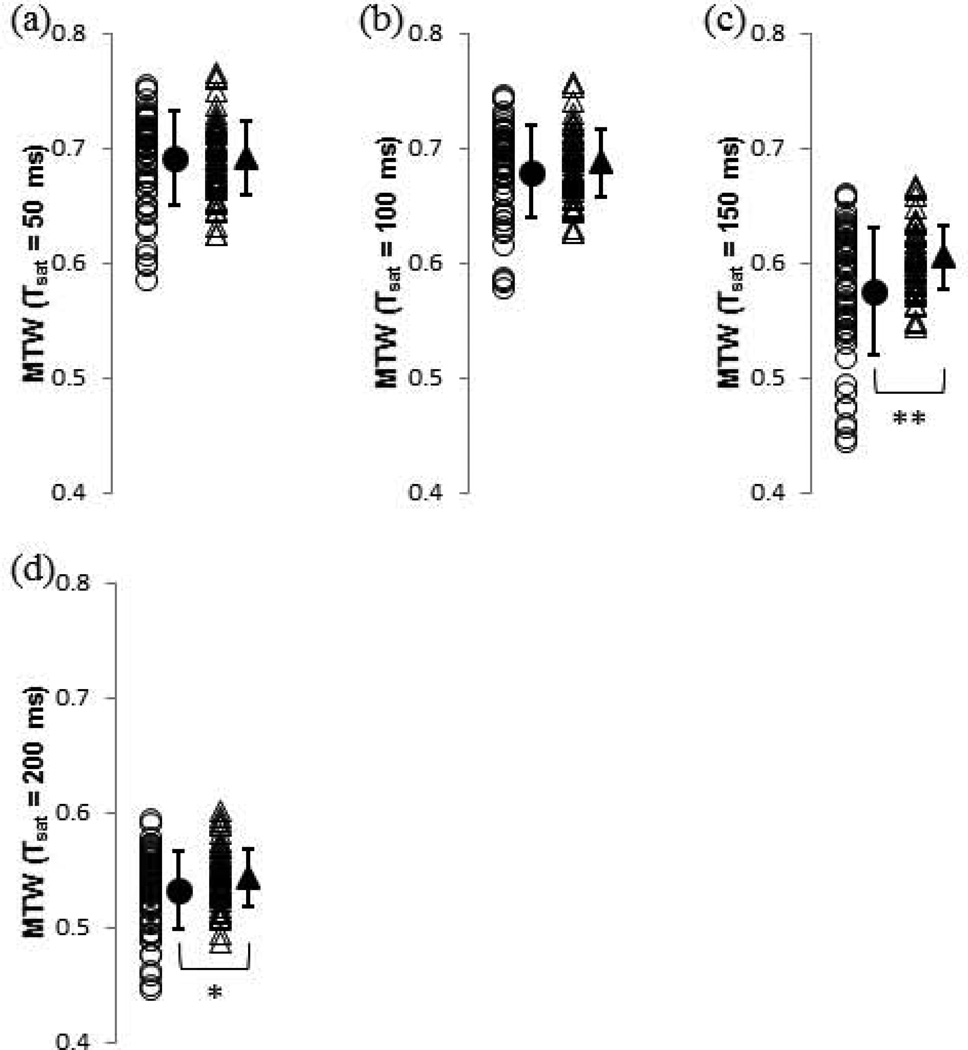
Figure 4 shows results for each of the DW image SIs, while Figure 5 shows results for the SI of each of the MTW images. Again, even for statistically significant differences in group means, there is a great deal of overlap between pre- and post-degradation values.
FIGURE 4.
Values of DPBS-normalized diffusion-weighted MRI measurements for control (open circles) and 2.5-h trypsin degraded (open triangles) BNC samples. Group means and standard deviations are shown as adjacent solid shapes and error bars. (a) DW (b = 333 s/mm2); (b) DW (b = 666 s/mm2); (c) DW (b = 1000 s/mm2); (d) DW (b = 1333 s/mm2); (e) DW (b = 1666 s/mm2); and (f) DW (b = 2000 s/mm2). * p < 0.05 control vs. 2.5-h trypsin, ** p < 0.01 control vs. 2.5-h trypsin
FIGURE 5.
Values of DPBS-normalized magnetization transfer-weighted MRI measurements for control (open circles) and 2.5-h trypsin degraded (open triangles) BNC samples. Group means and standard deviations are shown as adjacent solid shapes and error bars. (a) MTW (Tsat = 50 ms); (b) MTW (Tsat = 100 ms); (c) MTW (Tsat = 150 ms); and (d) MTW (Tsat = 200 ms). * p < 0.05 control vs. 2.5-h trypsin, ** p < 0.01 control vs. 2.5-h trypsin
As noted, the goal of the present work is to provide SE and SP values for classification of BNC samples via parameter measurements made under clinical MRI acquisition conditions. These test characteristics, as derived from our experimental results, are shown in Table 2. Results shown for the training and validation sets indicated, as expected, somewhat lower SE, SP, and ACC for the latter. However, the differences between results for the training and validation sets are small, indicating that the iterated simple-split classification analysis performed here did not result in a significant degree of overfitting (13). Results described below are those for the validation sets.
TABLE 2.
Univariate classification results for MRI parameters in control and 2.5-h trypsin degraded BNC samples
| Training Set | Validation Set | |||||
|---|---|---|---|---|---|---|
| MRI Measurement | Sensitivity | Specificity | Accuracy | Sensitivity | Specificity | Accuracy |
| ADC | 0.85 ± 0.03 | 0.80 ± 0.04 | 0.83 ± 0.03 | 0.85 ± 0.07 | 0.80 ± 0.07 | 0.82 ± 0.06 ** |
|
DW (b = 333 s/mm2) |
0.82 ± 0.04 | 0.79 ± 0.04 | 0.80 ± 0.03 | 0.82 ± 0.08 | 0.77 ± 0.08 | 0.80 ± 0.05 ** |
|
DW (b = 666 s/mm2) |
0.91 ± 0.03 | 0.82 ± 0.03 | 0.87 ± 0.02 | 0.90 ± 0.07 | 0.80 ± 0.07 | 0.85 ± 0.04 ** |
| DW (b = 1000 s/mm2) |
0.54 ± 0.04 | 0.54 ± 0.04 | 0.54 ± 0.03 | 0.53 ± 0.11 | 0.53 ± 0.11 | 0.53 ± 0.06 ** |
| DW (b = 1333 s/mm2) |
0.56 ± 0.08 | 0.51 ± 0.05 | 0.54 ± 0.03 | 0.50 ± 0.13 | 0.44 ± 0.09 | 0.47 ± 0.06 |
| DW (b = 1666 s/mm2) |
0.53 ± 0.04 | 0.61 ± 0.06 | 0.57 ± 0.04 | 0.53 ± 0.11 | 0.58 ± 0.12 | 0.55 ± 0.08 |
| DW (b = 2000 s/mm2) |
0.69 ± 0.04 | 0.69 ± 0.05 | 0.69 ± 0.03 | 0.70 ± 0.11 | 0.68 ± 0.13 | 0.69 ± 0.07 ** |
| MTR | 0.80 ± 0.03 | 0.78 ± 0.03 | 0.79 ± 0.03 | 0.79 ± 0.09 | 0.77 ± 0.10 | 0.78 ± 0.06 ** |
| MTW (Tsat = 50 ms) |
0.52 ± 0.05 | 0.48 ± 0.12 | 0.50 ± 0.08 | 0.46 ± 0.13 | 0.42 ± 0.13 | 0.44 ± 0.08 |
| MTW (Tsat = 100 ms) |
0.52 ± 0.05 | 0.46 ± 0.04 | 0.49 ± 0.04 | 0.50 ± 0.11 | 0.45 ± 0.11 | 0.48 ± 0.07 |
| MTW (Tsat = 150 ms) |
0.71 ± 0.06 | 0.52 ± 0.04 | 0.62 ± 0.04 | 0.70 ± 0.11 | 0.52 ± 0.11 | 0.61 ± 0.06 ** |
| MTW (Tsat = 200 ms) |
0.58 ± 0.04 | 0.47 ± 0.04 | 0.52 ± 0.03 | 0.58 ± 0.11 | 0.48 ± 0.11 | 0.53 ± 0.07 * |
| T1 | 0.99 ± 0.01 | 0.98 ± 0.01 | 0.99 ± 0.01 | 0.99 ± 0.02 | 0.99 ± 0.02 | 0.99 ± 0.01 ** |
| T1W | 0.52 ± 0.08 | 0.46 ± 0.10 | 0.49 ± 0.06 | 0.49 ± 0.15 | 0.42 ± 0.11 | 0.45 ± 0.07 |
| T2 - 3D | 0.72 ± 0.03 | 0.77 ± 0.04 | 0.75 ± 0.03 | 0.72 ± 0.09 | 0.76 ± 0.09 | 0.74 ± 0.05 ** |
| T2 - 2D | 0.68 ± 0.03 | 0.82 ± 0.03 | 0.75 ± 0.03 | 0.70 ± 0.10 | 0.83 ± 0.07 | 0.77 ± 0.06 ** |
| PDW (TE = 10 ms) |
0.65 ± 0.06 | 0.53 ± 0.04 | 0.59 ± 0.04 | 0.66 ± 0.14 | 0.53 ± 0.12 | 0.60 ± 0.09 |
| IDW (TE = 30 ms) |
0.54 ± 0.08 | 0.52 ± 0.04 | 0.53 ± 0.04 | 0.46 ± 0.12 | 0.46 ± 0.12 | 0.46 ± 0.06 |
| T2W (TE = 60 ms) |
0.48 ± 0.04 | 0.56 ± 0.04 | 0.52 ± 0.04 | 0.48 ± 0.10 | 0.58 ± 0.13 | 0.53 ± 0.07 ** |
| T2* | 0.62 ± 0.04 | 0.63 ± 0.04 | 0.62 ± 0.03 | 0.62 ± 0.11 | 0.62 ± 0.11 | 0.62 ± 0.06 ** |
Best-performing MRI measurements are indicated in bold.
p < 0.05 control vs. 2.5-h trypsin,
p < 0.01 control vs. 2.5-h trypsin (indicating statistical significance from Table 1)
The best classifiers (Table 2) were the pure MRI parameter values of ADC, MTR, T1, and T2 from the 3D and 2D spin echo sequences, along with two of the relatively modestly-weighted DW image SIs. Classification cutoffs for pure parameters, as defined by group means according to Eqs. (1) and (2), are indicated in Figure 2. Samples in a given group that exhibit parameter values closer to the mean of the other group are those that are misclassified.
As expected, the limited overlap in experimental T1 values between the two groups resulted in a statistically significant difference in group means, and nearly-perfect classification results. However, results from the other parameters show that this is not generalizable. For example, the mean T2-3D value increased by over 10% upon degradation, from 78.5 ms to 88.3 ms (P < 0.01). This is both a highly statistically significant difference and a substantial increase in absolute terms (CV = 2.5%). However, the classification based on this result is quite poor, with a SE of 0.72 ± 0.09, SP of 0.76 ± 0.09, and ACC of 0.74 ± 0.05. Similar comments apply to MTR and ADC, the other highly statistically significant outcomes, with SE of 0.79 ± 0.09, SP of 0.77 ± 0.10, and ACC of 0.78 ± 0.06, and SE of 0.85 ± 0.07, SP of 0.80 ± 0.07, and ACC of 0.82 ± 0.06, respectively.
In addition to classification by these pure parameters, classification according to normalized DW SIs acquired with b-values of 333 and 666 s/mm2 was comparable to classification according to ADC, with SE of 0.82 ± 0.08, SP of 0.77 ± 0.08, and ACC of 0.80 ± 0.05, and SE of 0.90 ± 0.07, SP of 0.80 ± 0.07, and ACC of 0.85 ± 0.04, respectively.
DISCUSSION
There is a substantial literature relating MRI measurements to both cartilage degradation, as assessed by histology and biochemistry, and to clinical or other status of research subjects (1–8). In spite of this, no range of normal values has been established, or even proposed, for any MRI parameter. One difficulty in moving towards this goal is the variability in the values obtained through use of pulse sequences that differ subtly between vendors, through different analysis methods, or even between different similarly-named sequences on a given instrument. However, a substantial overlap in parameter values between control and degraded cartilage is seen even under highly controlled experimental conditions. The resulting difficulties in classification and diagnosis have been pointed out previously (13).
The present work extends this analysis to clinically relevant magnetic field and sample temperature. In addition, we incorporated normalized contrast-weighted images, rather than focusing only on parameter values, due to their ubiquity in the clinical and clinical research literature. We elected to retain the same type of experimental samples, that is, digested BNC, in order to isolate the effect of imaging conditions on the results, although we applied a less severe degradation protocol as a further move towards a more clinically-relevant analysis. Of course, the numerical results obtained on native and pathomimetically-degraded BNC are not expected to correspond to those obtained from human subjects. Nevertheless, it is of interest to investigate the results of the specific contrast weightings available in the OAI data in the context of the present experimental design.
Of the best-performing univariate classifiers, six were pure parameters: ADC, MTR, T2 derived from 2D and 3D spin-echo sequences, and T1. ADC and DW images may be of particular interest because they can reflect in a fairly direct fashion underlying tissue architecture (2,21). In contrast, the interpretation of T2, as a correlation-function dependent relaxation time, is substantially more complicated (22), with sensitivity to multiple details of macromolecular content and architecture. In the case of BNC, with relatively limited anisotropy as compared to articular cartilage, changes in ADC are most readily interpretable in terms of net water molecule displacement through tissue. We found that ADC, as well as the DW images with b-values of 333 s/mm2 and 666 s/mm2, performed well as classifiers. Although these relatively small b-values are expected to result in less sensitivity to small differences in diffusivity than would be obtained with stronger diffusion weighting, this disadvantage is compensated for by their higher SNR (21). The ability to classify according to these weighted images, which can be acquired much more rapidly than an ADC map, is potentially of great clinical utility. It is of note that diffusion was the only parameter for which the pure parameter and weighted images achieved comparably high classification accuracy.
In contrast, MTW images did not perform as well as MTR. MTR showed a much greater difference in pre- and post-degradation means, and exhibited much more favorable classification characteristics. This again indicates the lack of close correspondence between statistical significance and test characteristics. MTR has been explored as a biomarker for changes in macromolecular content in cartilage (23,24). Yao et al. reported no significant differences in MTR between normal subjects and patients diagnosed with OA, but as demonstrated here, statistical significance may be a limited indicator of the SE and SP of group assignment (25). We should emphasize that due to specific absorption of radiation (SAR) and hardware duty cycle limitations, the available MT weightings presented are insufficient to reach steady state.
The near-perfect classification ACC using T1 relaxation time values was an unexpected result, although the fact that T1 was the best classifier was consistent with earlier findings at 9.4 T (13). The usefulness of T1 as a classifier is limited by its relatively lengthy acquisition time. In addition, the SI for T1W FLASH, using the parameters indicated, resulted in very poor classification ACC. Although T1 values may be related primarily to PG content and tissue hydration status (26), interpretation in terms of tissue properties remains incomplete.
The T2 relaxation time of cartilage is dependent upon water and collagen content, and collagen orientation (27–29). In contrast, the direct effect of PG content on T2 value is relatively modest (11,22,30–32). Trypsin acts primarily to deplete the PG component of cartilage, as is also seen in early OA (16). Nevertheless, changes in water content resulting from PG loss lead indirectly to an influence of PG content on T2 (10). We found the differences between the mean values of T2 in the pre- and post-degradation groups to be of the same order of magnitude as in previous work, as well as classification results that were comparable (13). While T2 did show some classification capability, it was more modest than that obtained from ADC, MTR, or T1 values. In addition, classification according to any of the normalized T2W image SI was quite poor. There was no appreciable difference in classification performance based on T2's derived from the 2D and the 3D spin echo sequences.
Although Mamisch et al. showed that T2* relaxation times was correlated with T2 values in measurements made on healthy volunteers and on subjects after microfracture cartilage repair (33), T2* resulted in the worst classification ACC of any of the pure parameters. Indeed, as seen in Fig. 2, T2* exhibited the greatest overlap in values between the control and 2.5-h trypsin samples, with the group mean values falling within one standard deviation of each other. Interpretation of tissue T2* measurements presents substantial difficulties due to the influence of both intrinsic tissue susceptibility and local field inhomogeneities. However, we incorporated T2* values into our analysis because of the rapidity of gradient-echo imaging as compared to spin-echo imaging. The modest efficacy of T2* relaxation times for classification may indicate limited clinical utility for diagnosis based on parameter values, as this study was performed under optimized experimental conditions using a small well-shimmed sample.
Overall, pure MRI parameter values were more effective for classification than their weighted counterparts. This may be due in part to the effective averaging of noise that results from fitting noisy data to multiple points of a defined curve. However, weighted images are more readily available than are pure component maps, so that this issue, including the potential utility of diffusion-weighted images, merits further investigation.
Measurement of the pure parameter ADC, MTR, and T1 values are all relatively time-consuming. However, these were the three most effective classifiers. In contrast, T2-2D and T2* relaxation times can be measured relatively rapidly, but neither displayed favorable classification characteristics. Of the contrast-weighted images, with relatively favorable acquisition times, DW contrast showed the greatest potential as a useful classifier.
Our study has several important limitations. The analysis was performed on BNC, permitting a relatively direct assessment of parameter classification based on cartilage matrix characteristics. In fact, earlier work of fundamental importance (34,35), as well as more recent work (36–38), also used BNC for this reason. In actual clinical practice, however, classification must be performed on human articular cartilage, which is highly structured, especially in terms of its collagen network, and which also displays certain biochemical differences as compared to BNC, such as a lower degree of hydration (16,39). In addition, although we evaluated classification under clinical imaging conditions, the study was performed on cartilage explants, eliminating such considerations as scanning time and motion. We further note that an enzymatic degradation paradigm was used to explore classification characteristics. While enzymatic degradation is referred to as pathomimetic, in fact enzymatic degradation differs substantially from the degradation process that occurs in human OA (40).
The present work investigated binary classification of cartilage samples. However, OA is a progressive disease, and effective characterization with MRI or any other modality requires the ability to provide graded evaluation. The extension from binary to continuous grading can be performed in a number of ways; several examples are provided in the recent literature (3,13,18). The approaches described in these papers all provide a probabilistic measure of degradation status; the approach of Lin et al. (13) can be applied without difficulty to the present results. We have not reproduced this analysis in the present work.
In conclusion, we have extended previous work on pathomimetically-degraded BNC by evaluating classification under clinical imaging conditions, and incorporating weighted images, as opposed to restricting attention to pure parameter maps. The results of this analysis identified pure parameter maps as, overall, the best classifiers, but DW contrast as a promising and potentially more practical modality. Further, we again find that highly statistically significant group differences do not necessarily translate into highly accurate decision rules.
ACKNOWLEDGEMENTS
This work was supported by the National Institutes of Health, National Institute on Aging, Intramural Research Program. The authors are grateful to Michael Schär for his assistance.
REFERENCES
- 1.Regatte RR, Akella SV, Reddy R. Depth-dependent proton magnetization transfer in articular cartilage. J Magn Reson Imaging. 2005;22:318–323. doi: 10.1002/jmri.20377. [DOI] [PubMed] [Google Scholar]
- 2.Mlynarik V, Sulzbacher I, Bittsansky M, Fuiko R, Trattnig S. Investigation of apparent diffusion constant as an indicator of early degenerative disease in articular cartilage. Journal of Magnetic Resonance Imaging. 2003;17:440–444. doi: 10.1002/jmri.10276. [DOI] [PubMed] [Google Scholar]
- 3.Lin PC, Reiter DA, Spencer RG. Classification of degraded cartilage through multiparametric MRI analysis. Journal Of Magnetic Resonance. 2009;201:61–71. doi: 10.1016/j.jmr.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nissi MJ, Toyras J, Laasanen MS, et al. Proteoglycan and collagen sensitive MRI evaluation of normal and degenerated articular cartilage. Journal of Orthopaedic Research. 2004;22:557–564. doi: 10.1016/j.orthres.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 5.Wiener E, Pfirrmann CW, Hodler J. Spatial variation in T1 of healthy human articular cartilage of the knee joint. Br J Radiol. 2010;83:476–485. doi: 10.1259/bjr/62779246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L, Regatte RR. Quantitative mapping of human cartilage at 3.0T: parallel changes in T2, T1rho, and dGEMRIC. Acad Radiol. 2014;21:463–471. doi: 10.1016/j.acra.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedrich KM, Shepard T, de Oliveira VS, et al. T2 measurements of cartilage in osteoarthritis patients with meniscal tears. AJR Am J Roentgenol. 2009;193:W411–W415. doi: 10.2214/AJR.08.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stehling C, Liebl H, Krug R, et al. Patellar cartilage: T2 values and morphologic abnormalities at 3.0-T MR imaging in relation to physical activity in asymptomatic subjects from the osteoarthritis initiative. Radiology. 2010;254:509–520. doi: 10.1148/radiol.09090596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burstein D, Gray M. New MRI techniques for imaging cartilage. J Bone Joint Surg Am. 2003;85-A(Suppl 2):70–77. doi: 10.2106/00004623-200300002-00009. [DOI] [PubMed] [Google Scholar]
- 10.Lusse S, Claassen H, Gehrke T, et al. Evaluation of water content by spatially resolved transverse relaxation times of human articular cartilage. Magn Reson Imaging. 2000;18:423–430. doi: 10.1016/s0730-725x(99)00144-7. [DOI] [PubMed] [Google Scholar]
- 11.Shapiro EM, Borthakur A, Kaufman JH, Leigh JS, Reddy R. Water distribution patterns inside bovine articular cartilage as visualized by 1H magnetic resonance imaging. Osteoarthritis Cartilage. 2001;9:533–538. doi: 10.1053/joca.2001.0428. [DOI] [PubMed] [Google Scholar]
- 12.Spencer RG, Pleshko N. How do statistical differences in matrix-sensitive magnetic resonance outcomes translate into clinical assignment rules? J Am Acad Orthop Surg. 2013;21:438–439. doi: 10.5435/JAAOS-21-07-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin PC, Reiter DA, Spencer RG. Sensitivity and specificity of univariate MRI analysis of experimentally degraded cartilage. Magnetic Resonance in Medicine. 2009;62:1311–1318. doi: 10.1002/mrm.22110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rieke V, Butts Pauly K. MR thermometry. J Magn Reson Imaging. 2008;27:376–390. doi: 10.1002/jmri.21265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peterfy CG, Schneider E, Nevitt M. The osteoarthritis initiative: report on the design rationale for the magnetic resonance imaging protocol for the knee. Osteoarthritis Cartilage. 2008;16:1433–1441. doi: 10.1016/j.joca.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buckwalter JA, Mankin HJ. Articular cartilage: tissue design and chondrocyte-matrix interactions. Instr Course Lect. 1998;47:477–486. [PubMed] [Google Scholar]
- 17.Farndale RW, Buttle DJ, Barrett AJ. Improved quantitation and discrimination of sulfated glycosaminoglycans by use of dimethylmethylene blue. Biochimica et Biophysica Acta. 1986;883:173–177. doi: 10.1016/0304-4165(86)90306-5. [DOI] [PubMed] [Google Scholar]
- 18.Lin PC, Irrechukwu O, Roque R, Hancock B, Fishbein KW, Spencer RG. Multivariate analysis of cartilage degradation using the support vector machine algorithm. Magn Reson Med. 2012;67:1815–1826. doi: 10.1002/mrm.23189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bydder GM, Rutherford MA, Hajnal JV. How to perform diffusion-weighted imaging. Childs Nerv Syst. 2001;17:195–201. doi: 10.1007/s003810000281. [DOI] [PubMed] [Google Scholar]
- 20.Liu J, Nieminen AOK, Koenig JL. Calculation of T1, T2, and proton spin density images in nuclear magnetic resonance imaging. Journal of Magnetic Resonance. 1969;85:95–110. 1989. [Google Scholar]
- 21.Burstein D, Gray ML, Hartman AL, Gipe R, Foy BD. Diffusion Of Small Solutes In Cartilage As Measured By Nuclear-Magnetic-Resonance (NMR) Spectroscopy And Imaging. Journal Of Orthopaedic Research. 1993;11:465–478. doi: 10.1002/jor.1100110402. [DOI] [PubMed] [Google Scholar]
- 22.David-Vaudey E, Ghosh S, Ries M, Majumdar S. T2 relaxation time measurements in osteoarthritis. Magn Reson Imaging. 2004;22:673–682. doi: 10.1016/j.mri.2004.01.071. [DOI] [PubMed] [Google Scholar]
- 23.Gray ML, Burstein D, Lesperance LM, Gehrke L. Magnetization transfer in cartilage and its constituent macromolecules. Magn Reson Med. 1995;34:319–325. doi: 10.1002/mrm.1910340307. [DOI] [PubMed] [Google Scholar]
- 24.Wachsmuth L, Juretschke HP, Raiss RX. Can magnetization transfer magnetic resonance imaging follow proteoglycan depletion in articular cartilage? MAGMA. 1997;5:71–78. doi: 10.1007/BF02592269. [DOI] [PubMed] [Google Scholar]
- 25.Yao W, Qu N, Lu Z, Yang S. The application of T1 and T2 relaxation time and magnetization transfer ratios to the early diagnosis of patellar cartilage osteoarthritis. Skeletal Radiol. 2009;38:1055–1062. doi: 10.1007/s00256-009-0769-8. [DOI] [PubMed] [Google Scholar]
- 26.Julkunen P, Korhonen RK, Nissi MJ, Jurvelin JS. Mechanical characterization of articular cartilage by combining magnetic resonance imaging and finite-element analysis: a potential functional imaging technique. Phys Med Biol. 2008;53:2425–2438. doi: 10.1088/0031-9155/53/9/014. [DOI] [PubMed] [Google Scholar]
- 27.Menezes NM, Gray ML, Hartke JR, Burstein D. T2 and T1rho MRI in articular cartilage systems. Magnetic Resonance in Medicine. 2004;51:503–509. doi: 10.1002/mrm.10710. [DOI] [PubMed] [Google Scholar]
- 28.Mosher TJ, Dardzinski BJ. Cartilage MRI T2 relaxation time mapping: overview and applications. Seminars in Musculoskeletal Radiology. 2004;8:355–368. doi: 10.1055/s-2004-861764. [DOI] [PubMed] [Google Scholar]
- 29.Xia Y, Moody JB, Alhadlaq H. Orientational dependence of T2 relaxation in articular cartilage: a microscopic MRI (micro MRI) study. Magnetic Resonance in Medicine. 2002;48:460–469. doi: 10.1002/mrm.10216. [DOI] [PubMed] [Google Scholar]
- 30.Mlynarik V, Trattnig S, Huber M, Zembsch A, Imhof H. The role of relaxation times in monitoring proteoglycan depletion in articular cartilage. J Magn Reson Imaging. 1999;10:497–502. doi: 10.1002/(sici)1522-2586(199910)10:4<497::aid-jmri1>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 31.Reiter DA, Roque RA, Lin P-C, et al. Mapping Proteoglycan-Bound Water in Cartilage: Improved Specificity of Matrix Assessment Using Multiexponential Transverse Relaxation Analysis. Magnetic Resonance in Medicine. 2011;65:377–384. doi: 10.1002/mrm.22673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toffanin R, Mlynarik V, Russo S, Szomolanyi P, Piras A, Vittur F. Proteoglycan depletion and magnetic resonance parameters of articular cartilage. Arch Biochem Biophys. 2001;390:235–242. doi: 10.1006/abbi.2001.2338. [DOI] [PubMed] [Google Scholar]
- 33.Mamisch TC, Hughes T, Mosher TJ, et al. T2* relaxation times for assessment of articular cartilage at 3 T: a feasibility study. Skeletal Radiol. 2012;41:287–292. doi: 10.1007/s00256-011-1171-x. [DOI] [PubMed] [Google Scholar]
- 34.Bashir A, Gray ML, Burstein D. Gd-DTPA2- as a measure of cartilage degradation. Magn Reson Med. 1996;36:665–673. doi: 10.1002/mrm.1910360504. [DOI] [PubMed] [Google Scholar]
- 35.Allen RG, Burstein D, Gray ML. Monitoring glycosaminoglycan replenishment in cartilage explants with gadolinium-enhanced magnetic resonance imaging. J Orthop Res. 1999;17:430–436. doi: 10.1002/jor.1100170320. [DOI] [PubMed] [Google Scholar]
- 36.Magin RL, Li W, Velasco MP, et al. Anomalous NMR relaxation in cartilage matrix components and native cartilage: Fractional-order models. Journal of Magnetic Resonance. 2011 doi: 10.1016/j.jmr.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng S, Xia Y. On the measurement of multi-component T2 relaxation in cartilage by MR spectroscopy and imaging. Magnetic Resonance Imaging. 2010;28:537–545. doi: 10.1016/j.mri.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng S, Xia Y. The impact of the relaxivity definition on the quantitative measurement of glycosaminoglycans in cartilage by the MRI dGEMRIC method. Magn Reson Med. 2010;63:25–32. doi: 10.1002/mrm.22169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reiter DA, Roque RA, Lin P-C, Doty SB, Pleshko N, Spencer RG. Improved specificity of cartilage matrix evaluation using multexponential transverse relaxation analysis applied to pathomimetically degraded cartilage. NMR in Biomedicine. 2011 doi: 10.1002/nbm.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bay-Jensen AC, Hoegh-Madsen S, Dam E, et al. Which elements are involved in reversible and irreversible cartilage degradation in osteoarthritis? Rheumatol Int. 2010;30:435–442. doi: 10.1007/s00296-009-1183-1. [DOI] [PubMed] [Google Scholar]