Abstract
Epigenetic regulation of gene expression is critical to phenotypic maintenance and transition of human breast cancer cells. HOX antisense intergenic RNA (HOTAIR) is a long intergenic non-coding RNA that epigenetically represses gene expression via recruitment of enhancer of zeste homolog 2 (EZH2), a histone methyltransferase. Elevated expression of HOTAIR promotes progression of breast cancer. In the current study we examined the expression and function of HOTAIR in MCF-7-TNR cells, a derivative of the luminal-like breast cancer cell line MCF-7 that acquired resistance to TNF-α-induced cell death. The expression of HOTAIR, markers of the luminal-like and basal-like subtypes, and growth were compared between MCF-7 and MCF-7-TNR cells. These variables were further assessed upon inhibition of HOTAIR, EZH2, p38 MAPK, and SRC kinase in MCF-7-TNR cells. When compared with MCF-7 cells, MCF-7-TNR cells exhibited an increase in the expression of HOTAIR, which correlated with characteristics of a luminal-like to basal-like transition as evidenced by dysregulated gene expression and accelerated growth. MCF-7-TNR cells exhibited reduced suppressive histone H3 lysine27 trimethylation on the HOTAIR promoter. Inhibition of HOTAIR and EZH2 attenuated the luminal-like to basal-like transition in terms of gene expression and growth in MCF-7-TNR cells. Inhibition of p38 and SRC diminished HOTAIR expression and the basal-like phenotype in MCF-7-TNR cells. HOTAIR was robustly expressed in the native basal-like breast cancer cells and inhibition of HOTAIR reduced the basal-like gene expression and growth. Our findings suggest HOTAIR-mediated regulation of gene expression and growth associated with the basal-like phenotype of breast cancer cells.
Keywords: HOTAIR, lincRNA, EZH2, breast cancer
INTRODUCTION
Altered death receptor signaling and resistance to subsequent apoptosis is a critical determinant in response to chemotherapy in patients with breast cancer. Laboratory derived chemo-resistant variants of breast cancer cell lines provide a platform to explore the molecular mechanisms of drug resistance. We have generated and investigated a MCF-7 variant, MCF-7-TNR cells that survived progressive exposure to tumor necrosis factor-α (TNF-α) and acquired resistance to cell death induced by TNF-α and several chemotherapeutic agents [1–4]. As revealed by gene expression arrays, MCF-7-TNR cells exhibit profound changes in three major signaling pathways that are intrinsically linked to multidrug resistance phenotype: 1) Attenuated estrogen receptor; 2) Diminished death receptor; and 3) Activated epithelial to mesenchymal transition (EMT) [4]. When implanted orthotopically in mice MCF-7-TNR cells exhibit greater tumorigenic activity than MCF-7 cells [4]. The MAPK pathway mediates dysregulated gene expression and aggressive behaviors in MCF-7-TNR cells. MCF-7-TNR cells exhibit greater basal activity of p38 MAPK than MCF-7 cells [1]. As a consequence, MCF-7-TNR displays hyperactive NF-κB pathway, a pro-survival pathway that is downstream of the p38 MAPK pathway [5]. Activation of another MAPK kinase, MEK5-ERK5 mediates the TNF-α resistance and EMT phenotype in MCF-7-TNR cells [6].
A large number of long intergenic non-coding RNAs (lincRNA) regulate gene expression via their interaction with chromatin modification complexes [7,8]. One of the lincRNA-regulated chromatin modification complexes is polycomb repressive complex2 (PRC2) that catalyzes tri-methylation of histone H3 lysine 27 (H3K27me3), a histone code for transcriptional repression [9,10]. Among the PRC2-interacting lincRNAs, HOX antisense intergenic RNA (HOTAIR) is a HOXC cluster-derived lincRNA that recruits PRC2 to its target genes [11,12]. HOTAIR is a novel oncogene in breast cancer because elevated expression of HOTAIR promotes metastasis of breast cancer cells and correlates with poor prognosis in patients with breast cancer [13]. How the expression of HOTAIR is up-regulated in breast cancer remains unclear.
In the current study we investigate HOTAIR expression and function in MCF-7-TNR cells, the multidrug resistant variant of MCF-7 breast cancer cells, and in the basal-like breast cancer cells.
MATERIALS AND METHODS
Reagents and Plasmids
3-Deazaneplanocin A (DZNEP), a pharmacological inhibitor of enhancer of zeste homolog 2 (EZH2), the catalytic subunit of PRC2, was purchased from Cayman Chemical (Cat #13828, Ann Arbor, MI) [14]. Bosutinib, an inhibitor of SRC kinase, was purchased from LC Laboratories (Cat #B1788, Woburn MA) [15]. SB203580, a p38-specific inhibitor was purchased from InvivoGen (Cat #tlrl-sb20, San Diego CA) [16]. Recombinant human TNF-α was purchased from R&D Systems (Cat #210-TA-005, Minneapolis MN). Paclitaxel, a therapeutic inhibitor of mitosis, was purchased from Sigma (Cat #T1912, St. Louis MO).
Cell Culture
The luminal-like human breast cancer cell line MCF-7 cells (N variant) and its multidrug resistant derivative MCF-7-TNR cells were cultured as we previously described [17]. MCF-7-TNR cells are a variant of the parental MCF-7 cells that survived progressive exposure to TNF-α and acquired resistance to multiple chemotherapeutic agents, such as taxol [1,4–6,18]. The basal-like human breast cancer cell lines BT-549 and MDA-MB-157 were purchased from ATCC (Manassas VA) and cultured in DMEM supplemented with 10% FBS.
Transient Transfection
The human HOTAIR-specific Mission siRNA (HOTAIRsiRNA) and the control Mission siRNA were purchased from Sigma (St. Louis MO). Most of the HOTAIR knockdown was achieved using the HOTAIRsiRNA with a Sigma ID SASI_Hs02_00380445. The salient findings were confirmed using a second HOTAIRsiRNA with a Sigma ID SASI_Hs02_00380446. The HOTAIRsiRNA or a control siRNA (CTLsiRNA, Sigma) was transfected at 60 nM into MCF-7-TNR cells using the reverse transfection protocol with RNAiMAX per the provider’s instructions (Invitrogen, Carlsbad CA) [19]. Cell culture was terminated at 72 hrs after transfection for cell counting, RNA extraction, and protein extraction.
Colony Formation in Soft Agar Assay
Colony formation in soft agar culture was used to measure anchorage independent growth of MCF-7 and MCF-7-TNR cells as described elsewhere [20]. Briefly, MCF-7 and MCF-7-TNR cells were seeded at 4,000 cells/well in soft agar in a 6-well culture plate. Colony formation was monitored for three weeks. Cell culture was stained using crystal violet and the visible colonies were enumerated. The number of colonies from three independent experiments was averaged and compared between MCF-7 and MCF-7-TNR cells.
Cell Proliferation Assay
MCF-7 and MCF-7-TNR cells were seeded at 3 × 105 cells/well in 6-well culture plates and cultured for 48 hrs. After trypsinization live cells were counted using a hematocytometer coupled with trypan blue exclusion method. A fold change in cell count was determined by calculating the ratios of the cell number at 48 hrs after seeding over the cell number at seeding. In the selected experiments, the indicated inhibitors or DMSO was added into the culture at 24 hrs after seeding and the treated cells were counted at 72 hrs after seeding. When MCF-7-TNR cells were transfected with either HOTAIRsiRNA or CTLsiRNA the transfected cells were counted at 72 hrs after seeding. Each comparison of the treated groups and their corresponding controls consisted of three independent experiments.
Cell Viability Assay
Cell viability post exposure to TNF-α or pacilitaxel was compared between MCF-7-TNR cells transfected with CTLsiRNA or HOTAIRsiRNA using XTT based In Vitro Toxicology Assays (Sigma, St Louis MO) as previously described [21]. At 24 hrs after transfection MCF-7-TNR cells were exposed TNF-α (10 ng/ml) for 24 hrs or pacilitaxel (100 nM) for 48 hrs according to our previous observations [4]. The number of live cells was determined by measuring absorbance at the wavelength of 450 nm. Three independent transfections were carried out in each treated group.
RNA extraction and quantitative RT-PCR
Total cell RNA was extracted using Trizol (Invitrogen Cat #15596026, Carlsbad CA) as we previously described [22]. The mRNA levels of each gene of interest were measured using reverse transcription coupled with quantitative real-time PCR (qRT-PCR) on an iCycler (BIO-RAD, Hercules CA) as we previously described [23]. The sequences of the primers were provided in Supplementary Materials Table 1. A fold change of each transcript was obtained by normalizing each transcript to the values of 36B4, a house keep gene and by setting the values from the control group to one [24].
Immunoblot
Total cell lysates were extracted from MCF-7 and MCF-7-TNR cells exposed to the indicated treatments using 1 × Laemmli buffer. The protein levels of the following gene products were measured using immunoblots as we previously described [25,26]. The antibodies specific for the following proteins were used: EZH2 (BD Biosciences Cat #612666, San Jose CA); p38 MAPK (Cell Signaling Cat # 4511, Danvers MA); p38 MAPK phosphorylated at Thr180/Tyr182 (Cell Signaling Cat #9212); heat shock protein 27 (HSP27, Santa Cruz Cat #sc-1049, Dallas TX); HSP27 phosphorylated at Ser82 (Cell Signaling Cat #2401); HOXC11 (Sigma, Cat #AV32642); Keratin 8 (Millipore Cat #MAB3414, Billerica MA); Versican (VCAN, Thermo Fisher Cat #PA1-1748A, Waltham MA); Stratifin (SFN, Sigma Cat #SAB2500001); β-actin (Cell Signaling Cat #4967). The bound primary antibody was detected using an IRDye800-conjugated anti-rabbit IgG on an Odyssey Infrared Imaging System (Licor Biosciences, Lincoln NE) as we previously described [27]. The immunoblots for KRT8 in Figure 1E and for HOXC11 in Figure 2B shared the same loading control β-actin. Each immunoblot was quantified by densitometry using NIH Image J. A fold change of each protein of interest was obtained by normalizing to its corresponding loading control and setting the values from the control groups to one. The image of each immunoblot was a representative of three independent experiments.
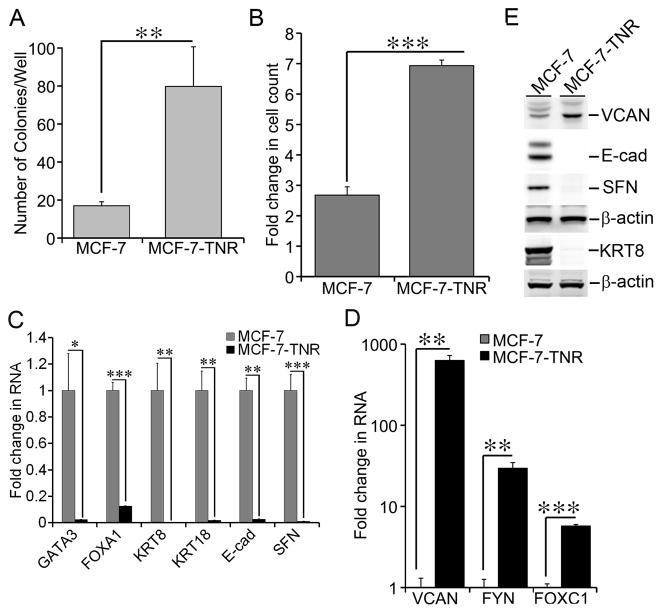
Figure 1. Dysregulated growth and gene expression in MCF-7-TNR cells.
A) Growth of MCF-7 and MCF-7-TNR cells was compared using soft agar colony formation assays. After three weeks of culture in soft agar, the colonies formed by each cell line were enumerated and compared. B) Proliferation of MCF-7 and MCF-7-TNR cells was assessed by counting live cells after 48 hrs in culture. A fold increase in the number of cells was obtained by calculating the ratios of the number of cells at 48 hrs post-seeding over the initial number of cells seeded. C) Total cell RNA was extracted from MCF-7 and MCF-7-TNR cells. The mRNA levels of SFN and the selected markers for the luminal-like subtype were compared between MCF-7 and MCF-7-TNR cells. A fold change of each transcript in MCF-7-TNR cells over that in MCF-7 cells was obtained by normalizing to the house keeping gene 36B4 and setting the values from MCF-7 cells to one. D) Similar to part C except that the mRNA levels of the selected markers for the basal-like subtype were compared between MCF-7 and MCF-7-TNR cells. E) Total cell lysates were extracted from MCF-7 and MCF-7-TNR cells. The protein levels of the indicated genes were measured using immunoblots. When presented, means and standard deviations were obtained from three independent experiments. *, **, and *** indicate a P value < 0.05, 0.01, 0.001, respectively.
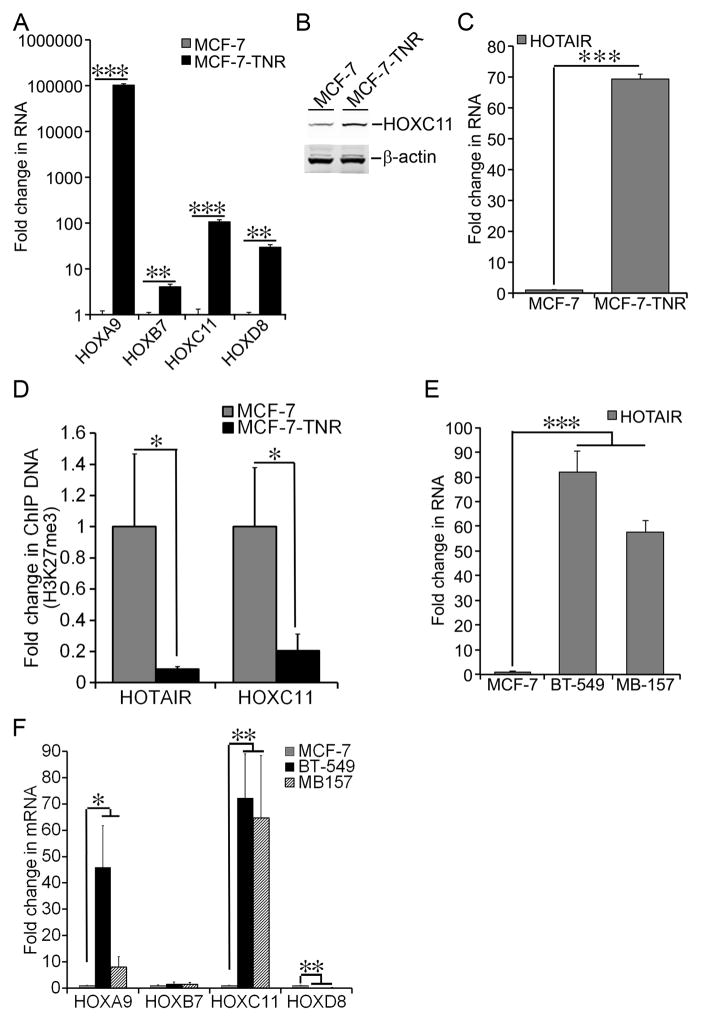
Figure 2. Elevated Expression of HOTAIR in MCF-7-TNR cells.
A) Total cell RNA was extracted from MCF-7 and MCF-7-TNR cells. The mRNA levels of the selected HOX genes were determined using qRT-PCR. A fold change of each transcript in MCF-7-TNR cells over that in MCF-7 cells was obtained by normalizing to the house keeping gene 36B4 and setting the values from MCF-7 cells to one. B) Total cell lysates were extracted from MCF-7 and MCF-7-TNR cells. The protein levels of HOXC11 were measured using immunoblots. The same loading control β-actin was shared between HOXC11 in Figure 2B and KRT8 in Figure 1E. C) Similar to part A except that the RNA levels of HOTAIR were compared between MCF-7 and MCF-7-TNR cells. D) ChIP assays were carried out using an H3K27me3-specific antibody in MCF-7 and MCF-7-TNR cells. The amount of the H3K27me3 bound HOTAIR and HOXC11 promoters was measured using qPCR and normalized to their corresponding input. A fold change of each promoter was obtained by setting the values from MCF-7 cells to one. E) Similar to part A except that the RNA levels of HOTAIR were measured and compared among MCF-7, MDA-MB-157 (MB-157), and BT-549 cells. F) Similar to part A except that the mRNA levels of the selected HOX genes were measured and compared among MCF-7, MDA-MB-157 (MB-157), and BT-549 cells. When presented, means and standard deviations were obtained from three independent experiments. *, **, and *** indicate a P value < 0.05, 0.01, 0.001, respectively.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation was performed as we previously described with minor modifications [21,28]. EZ ChIP Kit (Cat #17-371) and a ChIP grade H3K27me3-specific antibody (Cat #17-622) were purchased from Millipore (Darmstadt, Germany). MCF-7 and MCF-7-TNR cells were cross-linked in 1% formaldehyde at room temperature for 10 min. Sheared chromatin prepared from roughly 107 cells was immunoprecipitated with either the H3K27me3-specific antibody or a negative control antibody. The immunoprecipitated DNA was then recovered, purified, and dissolved in 40 μl of nuclease-free water. Two μl of each sample was used for quantitative PCR (qPCR). The sequences of the primers specific for the human HOTAIR and HOXC11 promoters were provided in Supplementary Materials Table 1. The input of each gene promoter was also measured using qPCR. The ratios of the immunoprecipitated promoters versus their corresponding input were compared between MCF-7 and MCF-7-TNR cells. A fold change of each promoter was established by setting the values from MCF-7 cells to one.
Statistical Analysis
When presented, means and standard deviations were obtained from at least 3 independent experiments. A P value between any two compared groups was determined using unpaired two-tailed Student’s T-test (GraphPad Prism, Version 5).
RESULTS
Dysregulated growth and gene expression in MCF-7-TNR cells
MCF-7-TNR cells are a MCF-7 variant that survived progressive exposure to TNF-α and acquired resistance to cell death induced by TNF-α and several chemotherapeutic reagents [1,4,5,29]. In congruence to their striking phenotypic difference 3404 genes are significantly differentially expressed between MCF-7 and MCF-7-TNR cells (P value<0.05, fold change>2) as revealed by gene expression arrays [4]. Those genes can be clustered into functional signaling categories using the Kyoto Encyclopedia of Genes and Genomes database (KEGG) and Gene Ontology algorithms as described in Table 1 in our previous report [4]. The clustered signaling categories revealed alterations in three major signaling pathways: 1) Attenuated estrogen receptor signaling; 2) Diminished death receptor signaling; and 3) Activated epithelial to mesenchymal transition (EMT) signaling [4]. The KEGG analysis also revealed enrichment of two growth related signaling pathways, i.e., p53 Signaling and Cell Cycle (see Table 1 in the referred article) [4]. Twenty-eight differentially expressed genes were clustered into the KEGG Cell Cycle pathway and nineteen differentially expressed genes were clustered into the KEGG p53 Signaling pathway (Supplementary Tables 2 & 3). These findings prompted us to examine growth of MCF-7-TNR cells in vitro. In soft agar colony formation assays, MCF-7-TNR cells exhibited a 4.7-fold increase in colony formation (79.8±20.86/well) over that of MCF-7 cells (17±2.12/well) (Figure 1A, P<0.01). In cell proliferation assays, MCF-7-TNR cells exhibited a remarkable 6.9-fold increase in cell count after 48 hrs in culture, whereas MCF-7 exhibited a modest 2.7-fold increase (Figure 1B, P<0.001). Dysregulated expression of the KEGG p53 Signaling and Cell Cycle genes were highlighted with reduced expression of the mediators of cell cycle arrest and apoptosis, e.g., p21waf1/cip1 (CDKN1A), caspase 8 (CASP8), and Growth arrest and DNA-damage-inducible protein GADD45 gamma (GADD45G) in MCF-7-TNR cells (Supplementary Tables 2 & 3). We chose to focus on Stratifin (SFN, also named 14-3-3σ) because 1) SFN was one of the most repressed genes in both signaling pathways (Supplementary Tables 2 & 3); 2) SFN arrests cell proliferation and functions as a tumor suppressor in breast cancer [30]; 3) Expression of SFN is repressed in breast carcinoma cells through epigenetically hypermethylation of the SFN promoter [31]. Suppression of SFN expression was confirmed by qRT-PCR as the mRNA levels of the SFN gene in MCF-7-TNR cells were reduced to 1% of that in MCF-7 cells (Figure 1C, P<0.001). This observation led us to investigate whether silencing of SFN in MCF-7-TNR cells required HOTAIR in the latter part of the current study. Although not addressed in the current manuscript it is noteworthy that p21waf1/cip1 is recently reported as a target gene of HOTAIR and EZH2 [32,33]. Thus simultaneous repression of multiple growth inhibitory genes may contribute to accelerated growth of MCF-7-TNR cells.
Accelerated proliferation generally impairs maintenance of cell differentiation [34]. In accordance, the tumors formed by the grafted MCF-7-TNR cells exhibit a loss of glandular histology that is characteristic of the grafted MCF-7 cells [4]. To search for the dysregulated gene expression programs underlying the phenotypic change in MCF-7-TNR cells we analyzed the differentially expressed genes between MCF-7 and MCF-7-TNR cells using Gene Set Enrichment Analysis (GSEA) coupled with C2 the Curated Gene Sets in the GSEA’s MSigDB collection [35]. We used a cutoff FDR value of 25% as recommended by GSEA’s user guide. Thirty-seven gene sets were negatively enriched in MCF-7-TNR cells relative to MCF-7 cells (Supplementary Table 4). Forty-three gene sets were positively enriched in MCF-7-TNR cells relative to MCF-7 cells (Supplementary Table 5). The enriched gene sets featured two gene sets that distinguish the luminal-like breast cancer cells from the basal-like cancer cells as reported in a comprehensive gene expression profiling of human breast cancer cell lines (source of the gene sets: Table 2S in the referred article) [36]. MCF-7-TNR cells were negatively enriched in the gene set CHARAFE_BREAST_CANCER_LUMINAL_VS_BASAL_UP that consists of the genes of higher expression in the luminal-like breast cancer cell lines than the basal-like ones (Supplementary Table 6). In contrast MCF-7-TNR cells were positively enriched in the gene set CHARAFE_BREAST_CANCER_LUMINAL_VS_BASAL_DN that consists of the genes of higher expression in the basal-like breast cancer cell lines than the luminal-like ones (Supplementary Table 7). We inferred from these findings that MCF-7-TNR cells lost expression of the luminal-like markers and simultaneously acquired expression of the basal-like markers. This luminal to basal transition was solidified by dysregulated expression of the selected luminal-like/basal-like markers in MCF-7-TNR cells. The mRNA levels of the selected luminal-like markers GATA3, FOXA1, keratin 8 (KRT8), keratin (KRT18), and E-cadherin (E-cad) in MCF-7-TNR cells were substantially reduced when compared to MCF-7 cells (2% in GATA3, P < 0.05; 12% in FOXA1, P < 0.001; 0.4% in KRT8, P < 0.01; 1.7% in KRT18, P < 0.01; 2.7% in E-cad, P < 0.01) (Figure 1C). In contrast the mRNA levels of the selected basal-like markers FOXC1, FYN, and versican (VCAN) displayed a substantial increase in MCF-7-TNR cells over that in MCF-7 cells (629–fold in VCAN, P < 0.01; 6–fold in FOXC1, P < 0.001; 30–fold in FYN, P < 0.01) (Figure 1D). We further confirmed the dysregulated expression of the growth regulators and luminal-like/basal-like markers using immunoblots. The protein levels of E-cad, KRT8, and SFN were nearly undetectable in MCF-7-TNR cells when compared with that in MCF-7 cells (Figure 1E). In contrast, the protein levels of the basal-like marker VCAN exhibited a 2.5±0.3-fold increase (P < 0.01) in MCF-7-TNR cells over that in MCF-7 cells (Figure 1E). These results correlated accelerated growth with dysregulated expression of the luminal-like/basal-like markers in MCF-7-TNR cells.
Elevated expression of HOTAIR in MCF-7-TNR cells
In our GSEA analysis we noticed that 4 enriched gene sets were related to PRC2. Those gene sets were 1) BENPORATH_SUZ12_TARGETS; 2) BENPORATH_EED_TARGETS; 3) BENPORATH_PRC2_TARGETS; 4) BENPORATH_ES_WITH_H3K27ME3 (Supplementary Table 5) (source of the gene set: Table 1S in the referred article) [37]. Out of the PRC2 target genes in the BENPORATH_PRC2_TARGETS gene set, the expression of 145 genes were altered in MCF-7-TNR cells with ~60% of them being up-regulated and ~40% of them being down-regulated when compared with MCF-7 cells (Supplementary Table 8). Interestingly this gene set featured the HOX gene family that is a well-characterized transcriptional target of PRC2 [38]. Indeed 27 HOX genes were substantially up-regulated in MCF-7-TNR cells when compared with MCF-7 cells (Supplementary Table 9). To confirm a broadly dysregulated expression of the HOX genes we encompassed all four HOX gene clusters by choosing HOXA9, HOXB7, HOXC11, and HOXD8. HOXA9 was chosen because it regulates the expression of BRCA1, a master regulator of breast cancer [39]. HOXB7 was chosen because its overexpression in breast cancer mediates drug resistance and EMT [40]. HOXC11 was chosen because it mediates resistance to endocrine therapy via interaction with steroid receptor coactivator-1 and its expression correlates with poor disease-free survival in breast cancer [41]. Moreover the transcription start site of HOXC11 is roughly 3 kb away from the transcription start site of HOTAIR and HOTAIR is transcribed in an antisense fashion relative to HOXC11 [11]. Therefore those two genes potentially share a bi-directional promoter and are intertwined with respect to their transcriptional regulation. HOXD8 was chosen because it resides in the HOXD cluster that is repressed by HOTAIR in normal human fibroblasts [11]. The aberrant expression of those 4 HOX genes from each cluster was confirmed as the four selected HOX genes exhibited substantial increase in their mRNA levels in MCF-7-TNR over MCF-7 (102597–fold in HOXA9, P < 0.001; 4.1–fold in HOXB7, P < 0.01; 106–fold in HOXC11, P < 0.001; 30–fold in HOXD8, P < 0.01) (Figure 2A). Moreover the protein levels of HOXC11 exhibited a 2.7±0.6-fold increase (P < 0.01) in MCF-7-TNR cells over that in MCF-7 cells (Figure 2B). Elevated expression of HOXC11 prompted us to examine the expression of its neighbor, the tumor-promoting lincRNA HOTAIR gene in MCF-7-TNR cells [11,13]. Similar to HOXC11, the RNA levels of HOTAIR in MCF-7-TNR cells exhibited a 69-fold increase over that in MCF-7 cells (Figure 2C, P < 0.001). Because the HOX genes are tightly regulated by PRC2-mediated H3K27me3, we performed H3K27me3-specific ChIP assays to examine H3K27me3 occupancy on the HOTAIR and HOXC11 promoters. Consistent with induction of HOXC11 and HOTAIR, the HOXC11 and HOTAIR promoters exhibited a 90% and 80% decrease in association with H3K27me3, respectively (Figure 2D, P < 0.05).
Because MCF-7-TNR cells featured gene expression characteristic of the basal-like subtype we examined the RNA levels of HOTAIR in two native basal-like subtype cell lines, BT-549 and MDA-MB-157 [36,42–49]. Indeed the RNA levels of HOTAIR in BT-549 and MDA-MB-157 cells were 80- and 60-fold higher than that in MCF-7 cells, respectively (Figure 2E, P < 0.001). We questioned whether the protein-coding HOX genes were also differentially expressed between the luminal-like and the basal-like cells. We examined the mRNA levels of HOXA9, HOXB7, HOXC11, and HOXD8 using qRT-PCR. Similar to their expression in MCF-7-TNR cells HOXA9 and HOXC11 exhibited higher expression in BT-549 and MDA-MB-157 than that in MCF-7 (HOXA9: 46-fold and 8-fold higher, respectively; HOXC11: 72-fold and 65-fold higher, respectively) (Figure 2F). On the other hand BT-549 and MB-157 exhibited a modestly higher expression of HOXB7 and a significantly lower expression of HOXD8 than MCF-7 cells (Figure 2F). These results revealed largely similar, but not identical expression profiles of the HOX genes in the native basal-like BT-549 and MDA-MB-157 cells and the basal-like MCF-TNR cells transitioned from the luminal-like MCF-7 cells.
Requirement of HOTAIR for dysregulated growth and gene expression in MCF-7-TNR cells
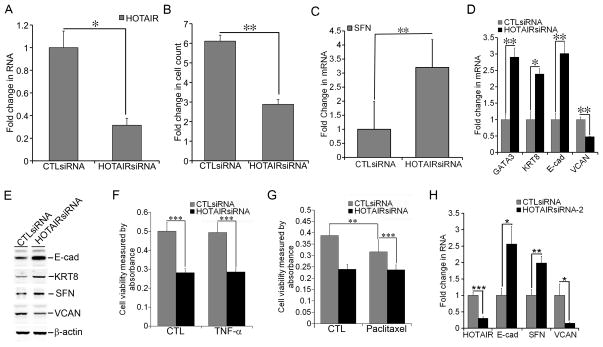
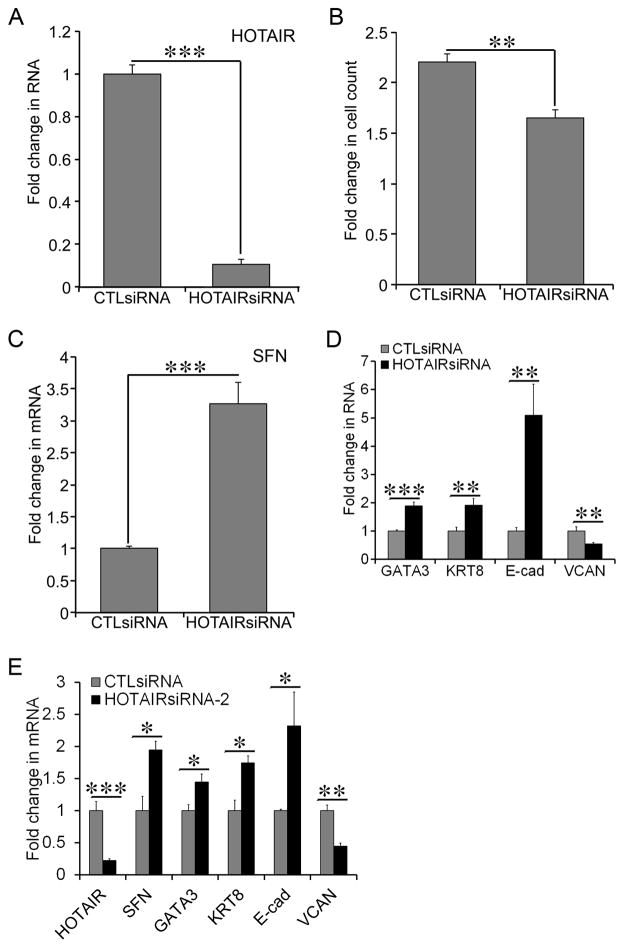
Elevated expression of HOTAIR prompted us to examine the role of HOTAIR in dysregulated growth and gene expression in MCF-7-TNR cells. We transfected MCF-7-TNR cells with either HOTAIRsiRNA or CTLsiRNA. RNAi-mediated knockdown of HOTAIR was confirmed by a 70% reduction of the RNA levels of HOTAIR in the HOTAIRsiRNA transfected group when compared with that in the CTLsiRNA transfected group (Figure 3A, P < 0.05). HOTAIRsiRNA diminished proliferation of MCF-7-TNR cells as the HOTAIRsiRNA group exhibited a 2.9-fold increase in cell count at 72 hrs after transfection, whereas the CTLsiRNA group exhibited a 6.1-fold increase (Figure 3B, P < 0.01). Consequently we examined the expression of the growth inhibitor SFN upon transfection of the HOTAIRsiRNA. HOTAIRsiRNA induced a 3.2-fold increase in the mRNA levels of SFN over that in the CTLsiRNA group (Figure 3C, P < 0.01). Moreover HOTAIRsiRNA increased the mRNA levels of the luminal markers GATA3 (2.9-fold, P < 0.01), KRT8 (2.4-fold, P < 0.05), and E-cad (3-fold, P < 0.01) (Figure 3D). In contrast HOTAIRsiRNA reduced the mRNA levels of the basal marker VCAN (50%, P < 0.01) (Figure 3D). To confirm the HOTAIRsiRNA-altered gene expression at the protein levels we immunoblotted the cell lysates for the luminal-like markers E-cad and KRT8, the basal-like marker VCAN, and the growth inhibitor SFN. HOTAIRsiRNA increased the protein levels of E-cad (2.7±0.3-fold, P < 0.01), KRT8 (2.3±0.3-fold, P < 0.001), and SFN (1.8±0.2-fold, P < 0.05) when compared with the CTLsiRNA (Figure 3E). In contrast HOTAIRsiRNA reduced the protein levels of VCAN by 52±9% (P < 0.01) (Figure 3E). Because MCF-7-TNR cells exhibit decreaded cell death signaling and resistance to cell death induced by TNF-α and chemo reagents we compared cell viability using XTT assays when MCF-7-TNR cells were exposed to various combinations of HOTAIRsiRNA, TNF-α, and paclitaxel [4]. As expected, TNF-α at 10 ng/ml did not reduce cell viability in MCF-7-TNR cells (Figure 3F). In contrast HOTAIRsiRNA alone caused a >40% decrease in cell viability regardless of TNF-α (Figure 3F, P < 0.001). Paclitaxel (100 nM) alone caused a modest ~20% decrease (P < 0.01) in cell viability, whereas HOTAIRsiRNA caused ~40% decrease (P < 0.001) regardless of paclitaxel (Figure 3G). Although addition of HOTAIRsiRNA to TNF-α or paclitaxel did not result in any synergistic or additive cell death, HOTAIRsiRNA abolished MCF-7-TNR cells’ resistance to TNF-α or paclitaxel (Figure 3, F & G). Similar to the first HOTAIRsiRNA employed in our study a second HOTAIR-specific siRNA HOTAIRsiRNA-2 substantially increased the expression of E-cadherin (2.6-fold, P <0.05) and SFN (2-fold, P < 0.01), and decreased the expression of VCAN (85%, P < 0.05) (Figure 3H). In summary these findings indicated requirement of HOTAIR for dysregulated growth and gene expression in MCF-7-TNR cells.
Figure 3. Requirement of HOTAIR for growth and gene expression in MCF-7-TNR cells.
A) MCF-7-TNR cells were transfected with either a control siRNA or a HOTAIR-specific siRNA (HOTAIRsiRNA). The RNA levels of HOTAIR were measured using qRT-PCR. A fold change was obtained by normalizing to the house keeping gene 36B4 and setting the values from the control siRNA (CTLsiRNA) group to one. B) Proliferation of MCF-7-TNR cells was assessed in the presence of CTLsiRNA or HOTAIRsiRNA. A fold increase in the number of cells was obtained by calculating the ratios of the number of cells at 72 hrs post-seeding over the number of cells seeded. C) Similar to part A except that the mRNA levels of SFN were compared between the CTLsiRNA and HOTAIRsiRNA transfected cells using qRT-PCR. D) Similar to part A except that the mRNA levels of the selected markers for the luminal-like and the basal-like subtypes were compared using qRT-PCR. E) Similar to part A except that the proteins levels of SFN and the selected markers for the luminal-like and the basal-like subtypes were measured using immunoblots. F) MCF-7-TNR cells were transfected with either CTLsiRNA or HOTAIRsiRNA. Cell viability of the transfected MCF-7-TNR cells was examined after 48 hrs exposure to TNF-α (10 ng/ml) using XTT assays. G) Similar to part F except that the transfected cells were exposed to paclitaxel (100 nM) for 24 hrs. H) The mRNA levels of the selected markers for the luminal-like and basal-like subtypes were examined in MCF-7-TNR cells transfected with a second HOTAIRsiRNA (HOTAIRsiRNA-2) or CTLsiRNA using qRT-PCR. When presented, means and standard deviations were obtained from three independent experiments. *, **, *** indicate a P value < 0.05, 0.01, 0.001, respectively.
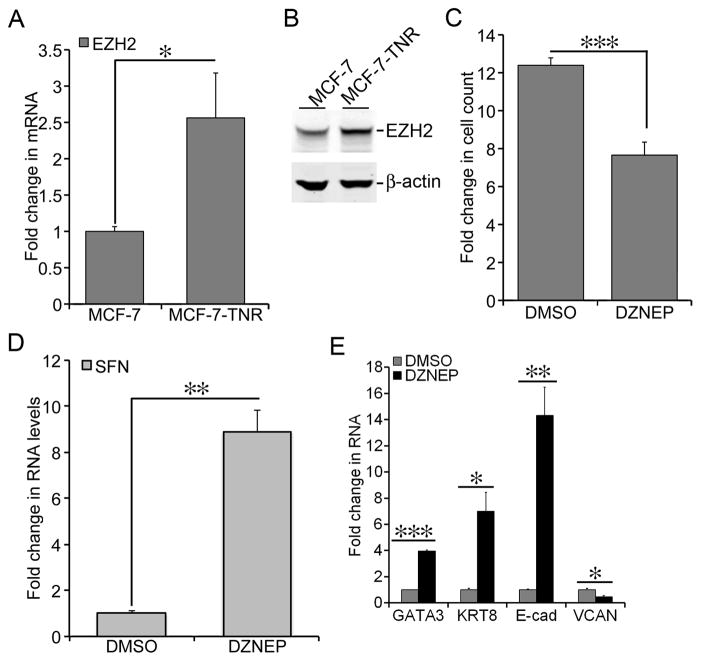
HOTAIR binds to EZH2, the histone methyltransferase in the PRC2 complex, and thereby recruits PRC2 to its target genes [11,13]. Therefore, we examined the expression of EZH2 in MCF-7-TNR cells. The mRNA levels of EZH2 in MCF-7-TNR cells were 2.6-fold higher than that in MCF-7 cells (Figure 4A, P < 0.05). In congruence, the protein levels of EZH2 exhibited a 1.9-fold increase over that in MCF-7 cells (Figure 4B). To test whether EZH2 was required for dysregulated growth and gene expression in MCF-7-TNR we exposed MCF-7-TNR cells to DZNEP, an EZH2 inhibitor (5 μM) for 72 hrs. DZNEP reduced cell count of MCF-7-TNR cells to a modest 7.6-fold increase vs a 12.3-fold increase in the control group (Figure 4C, P < 0.001). In concert DZNEP up-regulated the mRNA levels of SFN to a 8.9-fold increase over the control group (Figure 4D, P < 0.01). Moreover, DZNEP increased the mRNA levels of the luminal-like markers GATA3 (4-fold, P < 0.001), KRT8 (7-fold, P < 0.05), and E-cad (14-fold, P < 0.01) over the control group (Figure 4E). In contrast the mRNA levels of the basal-like marker VCAN were reduced to 48% by DZNEP (Figure 4E, P < 0.05). These results suggested requirement of EZH2 in dysregulated growth and gene expression in MCF-7-TNR cells.
Figure 4. Elevated expression of EZH2 in MCF-7-TNR cells.
A) Total cell RNA was extracted from MCF-7 and MCF-7-TNR cells. The mRNA levels of EZH2 were measured using qRT-PCR. A fold change of the EZH2 mRNA in MCF-7-TNR cells over that in MCF-7 cells was obtained by normalizing to the house keeping gene 36B4 and setting the values from MCF-7 cells to one. B) Similar to part A except that the protein levels of EZH2 were determined using immunoblots with an EZH2-specific antibody. C) MCF-7-TNR cells were cultured for 72 hrs with or without a 48-hr exposure to DZNEP (1 μM). A fold increase in the number of cells was obtained by calculating the ratios of the number of cells at 72 hrs post-seeding over the initial number of cells seeded. D) The culture condition was similar to part C. Total cell RNA was extracted from MCF-7-TNR cells. The mRNA levels of SFN were compared between the control and the DZNEP treated cells using qRT-PCR. A fold change of the SFN mRNA in the DZNEP-treated group over that in the DMSO vehicle group was obtained by normalizing to the house keeping gene 36B4 and setting the values from the DMSO group to one. E) Similar to part C except that the mRNA levels of the selected markers for the luminal-like and basal-like subtypes were measured and compared. When presented, means and standard deviations were obtained from three independent experiments. *, **, and *** indicate a P value < 0.05, 0.01, 0.001, respectively.
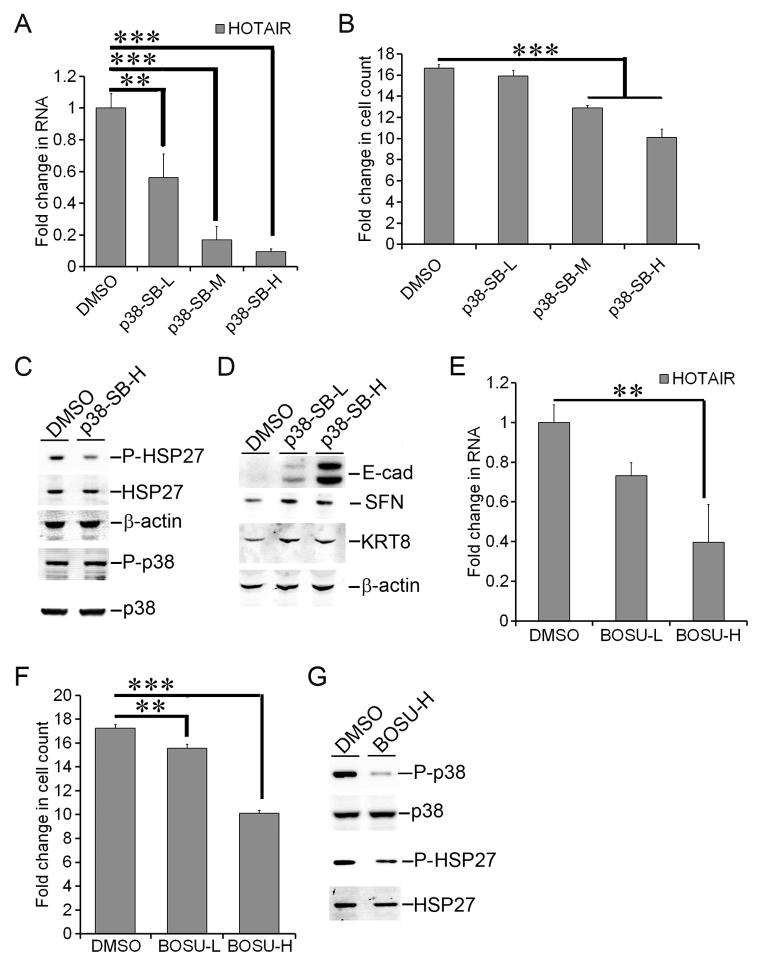
Suppression of HOTAIR expression by inhibition of SRC and p38
SRC and p38 MAPK kinases mediate cell responses to TNF-α [50–52]. Moreover SRC mediates activation of p38 [53,54]. As a consequence of their prolonged exposure to TNF-α, MCF-7-TNR cells exhibit hyperactive MAPK signaling, particularly p38 [1–3,55]. These observations prompted us to investigate the role of SRC and p38 in elevated expression of HOTAIR in MCF-7-TNR cells. We exposed MCF-7-TNR cells to increasing doses of SB203580, a p38-specific inhibitor (10, 25, and 50 μM) for 72 hrs. The RNA levels of HOTAIR were reduced by SB203580 in a dose dependent fashion as HOTAIR declined to a range from 56% to 10% of that in the control group (Figure 5A, P < 0.01 & 0.001). In concert SB203580 reduced cell proliferation to a 13-fold increase in cell count at 25 μM and a 10-fold increase at 50 μM, whereas the control group exhibited a 16.7-fold increase (Figure 5B, P < 0.001). To confirm SB203580-mediated inhibition of p38 MAPK, we examined phosphorylation of HSP27 at Ser82, a phosphorylation event that is dependent on active p38 MAPK and inhibited by SB203580 [56,57]. As expected one hour exposure of MCF-7-TNR cells to SB203580 resulted in a substantial decrease in phosphorylation of HSP27 at Ser82 (56±5%, P < 0.01) (Figure 5C). Consistent with the previous reports SB203580 did not inhibit activating phosphorylation of p38 at Thr180/Tyr182 (Figure 5C) [56,57]. Moreover SB203580 (10 and 50 μM) increased the protein levels of E-cad (9.3±1.3-fold by 50 μM, P < 0.001), SFN (1.8±0.2-fold by 50 μM, P < 0.01), and KRT8 (1.5±0.2-fold by 50 μM, P < 0.001) (Figure 5D). We then exposed MCF-7-TNR cells to increasing doses of Bosutinib, an SRC inhibitor (1 & 3 μM) for 72 hrs. The RNA levels of HOTAIR were reduced to 73% by 1 μM Bosutinib and 40% by 3 μM Bosutinib (P < 0.01) over that in the control group (Figure 5E). In concert Bosutinib reduced cell count to a 15.6-fold increase at 1 μM and a 10.1-fold increase at 3 μM in (P < 0.05) versus a 17.1-fold increase in the control group (Figure 5F). Moreover 1 hr exposure to Bosutinib (3 μM) reduced phosphorylation of p38 at Thr180/Tyr182 (23±4%, P < 0.05) and phosphorylation of HSP27 at Ser82 (63±6%, P < 0.05) (Figure 5G). These findings suggested requirement of p38 and SRC kinases in up-regulation of HOTAIR in MCF-7-TNR cells.
Figure 5. Suppression of HOTAIR expression by inhibition of p38 and SRC.
A) MCF-7-TNR cells were cultured for 96 hrs with or without exposure to SB203580, a p38 inhibitor for 72 hrs (p38-SB-L, -M, and -H indicate a dose at 10 μM, 25 μM, 50 μM, respectively). The RNA levels of HOTAIR were measured using qRT-PCR. A fold change was obtained by normalizing to the house keeping gene 36B4 and setting the values from the DMSO control group to one. B) The culture condition was identical to part A. Cell proliferation was assessed by counting the cells at seeding and harvest. A fold increase in the number of cells was obtained by calculating the ratios of the number of cells at 96 hrs post-seeding over the initial number of cells seeded. C) MCF-7-TNR cells were exposed to DMSO or SB203580 (p38-SB-H indicates a dose at 50 μM) for 1 hr. Immunoblots were carried out to examine the proteins levels of total p38, p38 with Thr180/Tyr182 phosphorylated (P-p38), total HSP27, and HSP27 with Ser82 phosphorylated (P-HSP27). D) MCF-7-TNR cells were exposed to SB203580 for 72 hrs (p38-SB-L indicates a dose at 10 μM and p38-SB-H indicates a dose at 50 μM). The protein levels of the indicated genes were measured using immunoblots. E) Similar to part A except that MCF-7-TNR cells were exposed to the SRC inhibitor Bosutinib (BOSU-L indicates a dose at 1 μM and BOSU-H indicates a dose at 3 μM). The RNA levels of HOTAIR were assessed as in part A. F) The culture condition was identical to part E. Cell proliferation was assessed as described in part B. G) Similar to party C except that MCF-7-TNR cells were exposed to Bosutinib (BOSU-H, 3 μM) for 1 hr. When presented, means and standard deviations were obtained from three independent experiments. ** and *** indicate a P value < 0.01 and 0.001, respectively.
Requirement of HOTAIR for the basal-like gene expression and growth in MDA-MB-157 cells
Robust expression of HOTAIR in the basal-like breast cancer cells prompted us to examine the role of HOTAIR in growth and gene expression in the native basal-like breast cancer cells. We transfected MDA-MB-157 cells with either HOTAIRsiRNA or CTLsiRNA. RNAi-mediated knockdown of HOTAIR was confirmed as the RNA levels of HOTAIR in the HOTAIRsiRNA transfected group were reduced to 10% of that in the CTLsiRNA transfected group (Figure 6A, P < 0.001). HOTAIRsiRNA reduced proliferation of MDA-MB-157 cells because the HOTAIRsiRNA group exhibited a 1.6-fold increase in cell count at 72 hrs after transfection, whereas the CTLsiRNA group exhibited a 2.3-fold increase (Figure 6B, P < 0.01). Consistent with its inhibition of growth, HOTAIRsiRNA induced a 3.3-fold increase in the mRNA levels of SFN over that in the CTLsiRNA group (Figure 6C, P < 0.001). Moreover the HOTAIRsiRNA group exhibited an increase in the mRNA levels of the luminal markers GATA3 (1.9-fold, P < 0.001), KRT8 (1.9-fold, P < 0.01), and E-cad (5.1-fold, P < 0.01), as well as a ~50% decrease in the mRNA levels of the basal marker VCAN (P < 0.01) (Figure 6D). To confirm our salient findings we transfected MDA-MB-157 cells with a second HOTAIR-specific siRNA HOTAIRsiRNA-2. HOTAIRsiRNA-2 also caused an increase in SFN (1.9-fold, P < 0.05), GATA3 (1.4-fold, P < 0.05), KRT8 (1.7-fold, P < 0.05), and E-cad (2.3-fold, P < 0.05), as well as a decrease in VCAN (56%, P < 0.01) (Figure 6E). These findings indicated requirement of HOTAIR for gene expression and growth associated with the basal-like breast cancer cells.
Figure 6. Requirement of HOTAIR for the basal-like growth and gene expression in MDA-MB-157 cells.
A) MDA-MB-157 cells were transfected with either control siRNA (CTLsiRNA) or HOTAIR-specific siRNA (HOTAIRsiRNA). The RNA levels of HOTAIR were determined using qRT-PCR. A fold change of the HOTAIR transcript was obtained by normalizing to the house keeping gene 36B4 and setting the values from the control siRNA group to one. B) Proliferation of MDA-MB-157 cells was assessed in the culture conditions as in part A. A fold increase in the number of cells was obtained by calculating the ratios of the number of cells at 72 hrs post-seeding over the initial number of cells seeded. C) Similar to part A except that the mRNA levels of SFN were compared between the CTLsiRNA and HOTAIRsiRNA transfected groups using qRT-PCR. D) Similar to part A except that the mRNA levels of GATA3, KRT8, Ecad, and VCAN were compared between the CTLsiRNA and HOTAIRsiRNA transfected groups. E) Similar to part D except that a second HOTAIRsiRNA (HOTAIRsiRNA-2) was transfected into MDA-MB-157 cells. When presented, means and standard deviations were obtained from three independent experiments. *, **, and *** indicate a P value < 0.01 and 0.001, respectively.
DISCUSSION
Human breast tumors are heterogeneous in their origin, histology, course of progression, and responsiveness to therapy. Genome wide expression profiling clusters human breast tumors into 4 “intrinsic subtypes”: normal-like, erbB2+, basal-like, and luminal-like [49]. Clinical presentation of each molecular subtype is tightly linked to its gene expression signature. This gene expression based clustering is further validated in the established human breast cancer cell lines [36,42]. Plasticity of breast cancer cells may underlie generation and transition of the molecular subtypes [58]. In the current study we investigate the expression and function of HOTAIR in MCF-7-TNR cells, a basal-like variant that originated from the luminal-like MCF-7 cells.
Epigenetic regulation of gene expression, particularly chromatin modifications, plays a critical role in lineage determination and maintenance of molecular subtypes of breast cancer [43,44,46]. Our findings suggest HOTAIR as a novel epigenetic determinant of the molecular subtypes of breast cancer. HOTAIR and its partner EZH2 are up-regulated in MCF-7-TNR cells, the basal-like derivative of the luminal-like MCF-7 cells (Figures 2 & 4). The HOTAIR-EZH2 complex plays a critical role in maintenance of the basal-like phenotype because inhibition of either HOTAIR or EZH2 attenuates dysregulated expression of luminal-like and basal-like markers as well as growth in MCF-7-TNR cells (Figures 3 & 4). This notion is strengthened by higher expression of HOTAIR in the native basal-like BT-549 and MDA-MB157 cells than the luminal-like MCF-7 cells and requirement of HOTAIR for the basal-like gene expression and growth in MDA-MB-157 cells (Figure 6). It is noteworthy that HOTAIR does not necessarily suppress the HOXD cluster in breast cancer cells because expression of HOTAIR and HOXD8 are concurrently higher in MCF-7-TNR cells, but in opposite direction in MB-157 and BT-549 cells (Figure 2, A, C, & E).
Increased expression of EZH2 and HOTAIR is linked to aggressive progression and poor prognosis in breast cancer [13,59]. Moreover simultaneous up-regulation of HOTAIR and EZH2 might be clinically significant because a recent study of a breast cancer archive correlates simultaneous high expression of EZH2 and HOTAIR with aggressive growth and metastasis [60]. Concurrent increase in HOTAIR and EZH2 in MCF-TNR cells is consistent with the above observations in patients with breast cancer. The expression of EZH2 can be up-regulated through the ERK pathway in breast cancer [61]. The ERK pathway is also hyperactive in MCF-7-TNR cells [2]. It is plausible that in MCF-7-TNR cells the MAPK pathway coordinates the epigenetic program of gene expression via the p38 MAPK mediated up-regulation of HOTAIR (as discussed below) and the ERK MAPK mediated up-regulation of EZH2. EZH2 can confer constitutive activation of the NF-κB target genes in the basal-like breast cancer cells, but not in the luminal-like breast cancer cells [62]. Moreover EZH2 acts as a transcriptional co-activator for NF-κB independent of its histone methyltransferase activity [62]. Because HOTAIR expression is higher in the basal-like breast cells (MCF-7-TNR, BT-549, and MDA-MB-157) than the luminal-like breast cancer cells (MCF-7) (Figures 2–4), it is worth exploring the role of the EZH2-HOTAIR complex in NF-κB-mediated gene activation in the basal-like breast cancer cells.
Despite mounting evidence of elevated expression of HOTAIR in several types of cancer, regulation of HOTAIR expression in cancer cells remains largely unknown. A recent study indicates that HOTAIR is transcriptionally activated by estradiol in MCF-7 cells [63]. Moreover the expression of HOTAIR can be up-regulated by the tumor modulating cues from the tumor microenvironment. For instance, type I collagen, a tumor-promoting extracellular matrix is aberrantly enriched in the tumor microenvironment and up-regulates the expression of HOTAIR in lung cancer cells [64]. The current study links up-regulated expression of HOTAIR in MCF-7-TNR cells to prolonged and progressive exposure to TNF-α, a cytokine that is enriched in the tumor microenvironment. As a consequence of its prolonged exposure to TNF-α, MCF-7-TNR cells exhibit hyperactive kinase activity of p38 [1–3,55]. In accordance inhibition of p38 and SRC kinases, two mediators of the cell responses to TNF-α, can diminish the expression of HOTAIR and restore the expression of SFN, E-cad, and KRT8 in MCF-7-TNR cells (Figure 5) [51,52]. Moreover inhibition of p38 and SRC exerts a growth inhibitory effect similar to that by inhibition of HOTAIR or EZH2 (Figures 3–5). Inhibition of either p38 or SRC results in decreased phosphorylation of HSP27 at Ser82, a signaling event essential for the molecular chaperone function of HSP27 in response to stress (Figure 5, C & G) [65]. SRC inhibition also results in decreased phosphorylation of p38 at Thr180/Tyr182, which is consistent with SRC-mediated activation of p38 in response to diverse stimuli (Figure 5G) [53,54]. These findings implicate an SRC-p38-HSP27 signaling cascade that activates the expression of HOTAIR in response to cellular stress, such as TNF-α.
The magnitude of induction of HOTAIR expression suggests that the HOTAIR gene is switched from a transcriptionally silent stage in MCF-7 cells to a transcriptionally active stage in MCF-7-TNR cells. Our findings suggest a relief of gene suppression because the HOTAIR and HOXC11 promoters exhibit a decreased occupancy of H3K27me3, a histone code for gene suppression, in MCF-7-TNR cells when compared with that in MCF-7 cells (Figure 2D). On the other hand, estradiol activates HOTAIR expression via recruitment of histone methyltransferases mixed lineage leukemia proteins (MLL) to the HOTAIR promoter, and the consequent trimethylation of histone H3 lysine 4 (H3K4me3), a histone code for gene activation [63]. Our results warrant further investigation to determine whether the histone codes for gene activation, such as MLL-mediated H3K4me3, concur with reduced H3K27me3 on the HOTAIR promoter. However it is likely that H3K4me3 on the HOTAIR promoter, if present in MCF-7-TNR cells, is achieved through an ER-independent mechanism because the expression of ER is silenced in MCF-7-TNR cells [4].
In summary we report an induction of HOTAIR expression during transition from the luminal-like MCF-7 to the basal-like MCF-7-TNR cells. Our results further suggest a critical role of HOTAIR in growth and gene expression in the basal-like breast cancer cells.
Supplementary Material
Acknowledgments
This work is supported in party by Washington State University Startup Fund awarded to BS and by NIH R01GM097571 to JJ.
Abbreviations
- TNF-α
tumor necrosis factor-α
- lincRNA
long intergenic non-coding RNA
- HOTAIR
HOX transcript antisense RNA
- PRC2
polycomb repressive complex 2
- EZH2
enhancer of zeste homolog 2
- qRT-PCR
quantitative Reverse Transcription-Polymerase Chain Reaction
- qPCR
quantitative Polymerase Chain Reaction
- H3K27me3
histone H3 lysine 27 tri-methylation
- H3K4me3
histone H3 lysine 4 tri-methylation
- MLL
mixed lineage leukemia
Contributor Information
Yan Zhuang, Email: yzhuang1@tulane.edu.
Hong T. Nguyen, Email: hnguyen9@tulane.edu.
Matthew E. Burow, Email: mburow@tulane.edu.
Ying Zhuo, Email: zhuoying1@yahoo.com.
Samir S. El-Dahr, Email: seldahr@tulane.edu.
Xiao Yao, Email: xyao@tulane.edu.
Subing Cao, Email: scao1@tulane.edu.
Erik K. Flemington, Email: eflemin@tulane.edu.
Kenneth P. Nephew, Email: knephew@indiana.edu.
Fang Fang, Email: ffang@indiana.edu.
Bridgette Collins-Burow, Email: bcollin1@tulane.edu.
Lyndsay V. Rhodes, Email: lvanhoy@tulane.edu.
Qiang Yu, Email: yuq@gis.a-star.edu.sg.
Janarthanan Jayawickramarajah, Email: jananj@tulane.edu.
Bin Shan, Email: bin.shan@wsu.edu.
References
- 1.Weldon CB, Parker AP, Patten D, et al. Sensitization of apoptotically-resistant breast carcinoma cells to TNF and TRAIL by inhibition of p38 mitogen-activated protein kinase signaling. International journal of oncology. 2004;24(6):1473–1480. [PubMed] [Google Scholar]
- 2.Weldon CB, Scandurro AB, Rolfe KW, et al. Identification of mitogen-activated protein kinase kinase as a chemoresistant pathway in MCF-7 cells by using gene expression microarray. Surgery. 2002;132(2):293–301. doi: 10.1067/msy.2002.125389. [DOI] [PubMed] [Google Scholar]
- 3.Weldon CB, Burow ME, Rolfe KW, Clayton JL, Jaffe BM, Beckman BS. NF-kappa B-mediated chemoresistance in breast cancer cells. Surgery. 2001;130(2):143–150. doi: 10.1067/msy.2001.115512. [DOI] [PubMed] [Google Scholar]
- 4.Antoon JW, Lai R, Struckhoff AP, et al. Altered death receptor signaling promotes epithelial-to-mesenchymal transition and acquired chemoresistance. Sci Rep. 2012;2:539. doi: 10.1038/srep00539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antoon JW, White MD, Slaughter EM, et al. Targeting NFkB mediated breast cancer chemoresistance through selective inhibition of sphingosine kinase-2. Cancer Biol Ther. 2011;11(7):678–689. doi: 10.4161/cbt.11.7.14903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou C, Nitschke AM, Xiong W, et al. Proteomic analysis of tumor necrosis factor-alpha resistant human breast cancer cells reveals a MEK5/Erk5-mediated epithelial-mesenchymal transition phenotype. Breast Cancer Res. 2008;10(6):R105. doi: 10.1186/bcr2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spitale RC, Tsai MC, Chang HY. RNA templating the epigenome: long noncoding RNAs as molecular scaffolds. Epigenetics : official journal of the DNA Methylation Society. 2011;6(5):539–543. doi: 10.4161/epi.6.5.15221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pauler FM, Barlow DP, Hudson QJ. Mechanisms of long range silencing by imprinted macro non-coding RNAs. Current opinion in genetics & development. 2012 doi: 10.1016/j.gde.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao J, Ohsumi TK, Kung JT, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40(6):939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khalil AM, Guttman M, Huarte M, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106(28):11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44(4):667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan J, Yang X, Zhuang L, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21(9):1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Golas JM, Arndt K, Etienne C, et al. SKI-606, a 4-anilino-3-quinolinecarbonitrile dual inhibitor of Src and Abl kinases, is a potent antiproliferative agent against chronic myelogenous leukemia cells in culture and causes regression of K562 xenografts in nude mice. Cancer Res. 2003;63(2):375–381. [PubMed] [Google Scholar]
- 16.Barancik M, Bohacova V, Kvackajova J, Hudecova S, Krizanova O, Breier A. SB203580, a specific inhibitor of p38-MAPK pathway, is a new reversal agent of P-glycoprotein-mediated multidrug resistance. Eur J Pharm Sci. 2001;14(1):29–36. doi: 10.1016/s0928-0987(01)00139-7. [DOI] [PubMed] [Google Scholar]
- 17.Zhuang Y, Nguyen HT, Lasky JA, et al. Requirement of a novel splicing variant of human histone deacetylase 6 for TGF-beta1-mediated gene activation. Biochemical and biophysical research communications. 2010;392(4):608–613. doi: 10.1016/j.bbrc.2010.01.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antoon JW, White MD, Burow ME, Beckman BS. Dual inhibition of sphingosine kinase isoforms ablates TNF-induced drug resistance. Oncol Rep. 2012;27(6):1779–1786. doi: 10.3892/or.2012.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shan B, Yao TP, Nguyen HT, et al. Requirement of HDAC6 for transforming growth factor-beta1-induced epithelial-mesenchymal transition. The Journal of biological chemistry. 2008;283(30):21065–21073. doi: 10.1074/jbc.M802786200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brugge JS, Butel JS. Role of simian virus 40 gene A function in maintenance of transformation. Journal of virology. 1975;15(3):619–635. doi: 10.1128/jvi.15.3.619-635.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shan B, Zhuo Y, Chin D, Morris CA, Morris GF, Lasky JA. Cyclin-dependent kinase 9 is required for tumor necrosis factor-alpha-stimulated matrix metalloproteinase-9 expression in human lung adenocarcinoma cells. The Journal of biological chemistry. 2005;280(2):1103–1111. doi: 10.1074/jbc.M406293200. [DOI] [PubMed] [Google Scholar]
- 22.Nguyen HT, Zhuang Y, Sun L, et al. Src-mediated morphology transition of lung cancer cells in three-dimensional organotypic culture. Cancer cell international. 2013;13(1):16. doi: 10.1186/1475-2867-13-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shan B, Hagood JS, Zhuo Y, et al. Thy-1 attenuates TNF-alpha-activated gene expression in mouse embryonic fibroblasts via Src family kinase. PloS one. 2010;5(7):e11662. doi: 10.1371/journal.pone.0011662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shan B, Morris CA, Zhuo Y, Shelby BD, Levy DR, Lasky JA. Activation of proMMP-2 and Src by HHV8 vGPCR in human pulmonary arterial endothelial cells. Journal of molecular and cellular cardiology. 2007;42(3):517–525. doi: 10.1016/j.yjmcc.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 25.Shan B, Morris GF. Binding sequence-dependent regulation of the human proliferating cell nuclear antigen promoter by p53. Experimental cell research. 2005;305(1):10–22. doi: 10.1016/j.yexcr.2004.09.033. [DOI] [PubMed] [Google Scholar]
- 26.Saito S, Lasky JA, Guo W, et al. Pharmacological inhibition of HDAC6 attenuates endothelial barrier dysfunction induced by thrombin. Biochemical and biophysical research communications. 2011;408(4):630–634. doi: 10.1016/j.bbrc.2011.04.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li C, Nguyen HT, Zhuang Y, et al. Post-transcriptional up-regulation of miR-21 by type I collagen. Molecular carcinogenesis. 2011;50(7):563–570. doi: 10.1002/mc.20742. [DOI] [PubMed] [Google Scholar]
- 28.Shan B, Xu J, Zhuo Y, Morris CA, Morris GF. Induction of p53-dependent activation of the human proliferating cell nuclear antigen gene in chromatin by ionizing radiation. The Journal of biological chemistry. 2003;278(45):44009–44017. doi: 10.1074/jbc.M302671200. [DOI] [PubMed] [Google Scholar]
- 29.Antoon JW, Meacham WD, Bratton MR, et al. Pharmacological inhibition of sphingosine kinase isoforms alters estrogen receptor signaling in human breast cancer. Journal of molecular endocrinology. 2011;46(3):205–216. doi: 10.1530/JME-10-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang H, Zhang Y, Zhao R, et al. Negative cell cycle regulator 14-3-3sigma stabilizes p27 Kip1 by inhibiting the activity of PKB/Akt. Oncogene. 2006;25(33):4585–4594. doi: 10.1038/sj.onc.1209481. [DOI] [PubMed] [Google Scholar]
- 31.Ferguson AT, Evron E, Umbricht CB, et al. High frequency of hypermethylation at the 14-3-3 sigma locus leads to gene silencing in breast cancer. Proc Natl Acad Sci U S A. 2000;97(11):6049–6054. doi: 10.1073/pnas.100566997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Z, Sun M, Lu K, et al. The long noncoding RNA HOTAIR contributes to cisplatin resistance of human lung adenocarcinoma cells via downregualtion of p21(WAF1/CIP1) expression. PloS one. 2013;8(10):e77293. doi: 10.1371/journal.pone.0077293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu Z, Lee ST, Qiao Y, et al. Polycomb protein EZH2 regulates cancer cell fate decision in response to DNA damage. Cell Death Differ. 2011;18(11):1771–1779. doi: 10.1038/cdd.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 35.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Charafe-Jauffret E, Ginestier C, Monville F, et al. Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene. 2006;25(15):2273–2284. doi: 10.1038/sj.onc.1209254. [DOI] [PubMed] [Google Scholar]
- 37.Ben-Porath I, Thomson MW, Carey VJ, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nature genetics. 2008;40(5):499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nature reviews Cancer. 2010;10(5):361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 39.Gilbert PM, Mow JK, Unger MA, et al. HOXA9 regulates BRCA1 expression to modulate human breast tumor phenotype. The Journal of clinical investigation. 2010;120(5):1535–1550. doi: 10.1172/JCI39534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu X, Chen H, Parker B, et al. HOXB7, a homeodomain protein, is overexpressed in breast cancer and confers epithelial-mesenchymal transition. Cancer Res. 2006;66(19):9527–9534. doi: 10.1158/0008-5472.CAN-05-4470. [DOI] [PubMed] [Google Scholar]
- 41.McIlroy M, McCartan D, Early S, et al. Interaction of developmental transcription factor HOXC11 with steroid receptor coactivator SRC-1 mediates resistance to endocrine therapy in breast cancer [corrected] Cancer Res. 2010;70(4):1585–1594. doi: 10.1158/0008-5472.CAN-09-3713. [DOI] [PubMed] [Google Scholar]
- 42.Kao J, Salari K, Bocanegra M, et al. Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery. PloS one. 2009;4(7):e6146. doi: 10.1371/journal.pone.0006146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maruyama R, Choudhury S, Kowalczyk A, et al. Epigenetic regulation of cell type-specific expression patterns in the human mammary epithelium. PLoS genetics. 2011;7(4):e1001369. doi: 10.1371/journal.pgen.1001369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bloushtain-Qimron N, Yao J, Snyder EL, et al. Cell type-specific DNA methylation patterns in the human breast. Proc Natl Acad Sci U S A. 2008;105(37):14076–14081. doi: 10.1073/pnas.0805206105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park SY, Lee HE, Li H, Shipitsin M, Gelman R, Polyak K. Heterogeneity for stem cell-related markers according to tumor subtype and histologic stage in breast cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010;16(3):876–887. doi: 10.1158/1078-0432.CCR-09-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shipitsin M, Campbell LL, Argani P, et al. Molecular definition of breast tumor heterogeneity. Cancer cell. 2007;11(3):259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 47.Marotta LL, Almendro V, Marusyk A, et al. The JAK2/STAT3 signaling pathway is required for growth of CD44(+)CD24(−) stem cell-like breast cancer cells in human tumors. The Journal of clinical investigation. 2011;121(7):2723–2735. doi: 10.1172/JCI44745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marotta LL, Polyak K. Unraveling the complexity of basal-like breast cancer. Oncotarget. 2011;2(8):588–589. doi: 10.18632/oncotarget.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 50.Raingeaud J, Gupta S, Rogers JS, et al. Pro-inflammatory cytokines and environmental stress cause p38 mitogen-activated protein kinase activation by dual phosphorylation on tyrosine and threonine. The Journal of biological chemistry. 1995;270(13):7420–7426. doi: 10.1074/jbc.270.13.7420. [DOI] [PubMed] [Google Scholar]
- 51.Paul A, Wilson S, Belham CM, et al. Stress-activated protein kinases: activation, regulation and function. Cell Signal. 1997;9(6):403–410. doi: 10.1016/s0898-6568(97)00042-9. [DOI] [PubMed] [Google Scholar]
- 52.Page TH, Smolinska M, Gillespie J, Urbaniak AM, Foxwell BM. Tyrosine kinases and inflammatory signalling. Curr Mol Med. 2009;9(1):69–85. doi: 10.2174/156652409787314507. [DOI] [PubMed] [Google Scholar]
- 53.van Vliet C, Bukczynska PE, Puryer MA, et al. Selective regulation of tumor necrosis factor-induced Erk signaling by Src family kinases and the T cell protein tyrosine phosphatase. Nature immunology. 2005;6(3):253–260. doi: 10.1038/ni1169. [DOI] [PubMed] [Google Scholar]
- 54.Kant S, Swat W, Zhang S, et al. TNF-stimulated MAP kinase activation mediated by a Rho family GTPase signaling pathway. Genes Dev. 2011;25(19):2069–2078. doi: 10.1101/gad.17224711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Antoon JW, Bratton MR, Guillot LM, et al. Pharmacology and anti-tumor activity of RWJ67657, a novel inhibitor of p38 mitogen activated protein kinase. Am J Cancer Res. 2012;2(4):446–458. [PMC free article] [PubMed] [Google Scholar]
- 56.Hedges JC, Dechert MA, Yamboliev IA, et al. A role for p38(MAPK)/HSP27 pathway in smooth muscle cell migration. The Journal of biological chemistry. 1999;274(34):24211–24219. doi: 10.1074/jbc.274.34.24211. [DOI] [PubMed] [Google Scholar]
- 57.Cuenda A, Rouse J, Doza YN, et al. SB 203580 is a specific inhibitor of a MAP kinase homologue which is stimulated by cellular stresses and interleukin-1. FEBS letters. 1995;364(2):229–233. doi: 10.1016/0014-5793(95)00357-f. [DOI] [PubMed] [Google Scholar]
- 58.Eroles P, Bosch A, Perez-Fidalgo JA, Lluch A. Molecular biology in breast cancer: intrinsic subtypes and signaling pathways. Cancer Treat Rev. 2012;38(6):698–707. doi: 10.1016/j.ctrv.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 59.Hussein YR, Sood AK, Bandyopadhyay S, et al. Clinical and biological relevance of enhancer of zeste homolog 2 in triple-negative breast cancer. Human pathology. 2012;43(10):1638–1644. doi: 10.1016/j.humpath.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chisholm KM, Wan Y, Li R, Montgomery KD, Chang HY, West RB. Detection of long non-coding RNA in archival tissue: correlation with polycomb protein expression in primary and metastatic breast carcinoma. PloS one. 2012;7(10):e47998. doi: 10.1371/journal.pone.0047998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fujii S, Tokita K, Wada N, et al. MEK-ERK pathway regulates EZH2 overexpression in association with aggressive breast cancer subtypes. Oncogene. 2011;30(39):4118–4128. doi: 10.1038/onc.2011.118. [DOI] [PubMed] [Google Scholar]
- 62.Lee ST, Li Z, Wu Z, et al. Context-specific regulation of NF-kappaB target gene expression by EZH2 in breast cancers. Mol Cell. 2011;43(5):798–810. doi: 10.1016/j.molcel.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 63.Bhan A, Hussain I, Ansari KI, Kasiri S, Bashyal A, Mandal SS. Antisense Transcript Long Noncoding RNA (lncRNA) HOTAIR is Transcriptionally Induced by Estradiol. J Mol Biol. 2013 doi: 10.1016/j.jmb.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhuang Y, Wang X, Nguyen HT, et al. Induction of long intergenic non-coding RNA HOTAIR in lung cancer cells by type I collagen. J Hematol Oncol. 2013;6:35. doi: 10.1186/1756-8722-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rogalla T, Ehrnsperger M, Preville X, et al. Regulation of Hsp27 oligomerization, chaperone function, and protective activity against oxidative stress/tumor necrosis factor alpha by phosphorylation. The Journal of biological chemistry. 1999;274(27):18947–18956. doi: 10.1074/jbc.274.27.18947. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.