Abstract
During development cells interact mechanically with their microenvironment through cell-cell and cell-matrix adhesions. Many proteins involved in these adhesions serve both mechanical and signaling roles. In this review we will focus on the mechanical roles of these proteins and their complexes in transmitting force or stress from cell to cell or from cell to the extracellular matrix. As forces operate against tissues they establish tissue architecture, extracellular matrix assembly, and pattern cell shapes. As tissues become more established, adhesions play a major role integrating cells with the mechanics of their local environment. Adhesions may serve as both a molecular-specific glue, holding defined populations of cells together, and as a lubricant, allowing tissues to slide past one another. We review the biophysical principles and experimental tools used to study adhesion so that we may aid efforts to understand how adhesions guide these movements and integrate their signaling functions with mechanical function. As we conclude we review efforts to develop predictive models of adhesion that can be used to interpret experiments and guide future efforts to control and direct the process of tissue self-assembly during development.
Keywords: morphogenesis, elastic modulus, adhesion energy, surface tension, stiffness, binding energy
Introduction
Over several summers at the Beaufort Laboratory in North Carolina, H.V. Wilson conducted a remarkable series of experiments with cells isolated from sponges (Wilson, 1907); these cells, in isolation and when aggregated exhibited a range of phenomena including distinctive cell motility, adhesion, differentiation, development, and tissue homeostasis. Wilson dissociated cells from adult and larval sponges and by combining them he observed their remarkable ability to regenerate the structure and form of adult sponges. Wilson and others observed sorting, protrusive behaviors, tissue self-assembly, and regeneration based on the cell type origin, stage, individual, and species. These observations inspired later workers such as Holtfreter, Steinberg, Trinkaus, and others to consider the role of adhesion and cell motility in driving development and tissue-self assembly. Ultimately, this work led to the discovery of the molecules regulating cell-cell and cell-substrate adhesion, the founding of the field of cell mechanics, and a resurgent interest in the physical principles of early development, morphogenesis, organogenesis, stem cell biology, regeneration, wound healing, and disease. We focus in this review on recent efforts to understand the physical role of adhesion during development and how molecular mechanisms of adhesion generate biological form. In the following sections we introduce biophysical methods of investigating cell adhesion and its contribution to the mechanical properties and force production within developing embryos. In addition to biophysical studies on embryos we include studies with cultured cells and cells isolated from adult tissues to demonstrate how cells coordinate biochemical and mechanical signaling through cell-cell and cell-substrate adhesions.
Adhesion couples cell populations that establish mechanical support in tissues, allowing cells to be “fixed” with varying degrees of freedom to certain structures. For instance, epithelial cells can be constrained by their apical adhesive junctions to a two-dimensional plane, similarly, mesenchymal cells may form a monolayer as they bind a distinctive layer of extracellular matrix. Different cell types and diverse cell substrates can restrict cell movements along interfaces where adhesion receptors or ligands are present or direct the force they generate at these interfaces along specific directions. Context- or stage-dependent changes in adhesion may occur as cells contact new neighbors or as cells change their expression or activity of their adhesions and contacts. As tissues assemble into more complex structures adhesion can serve to couple forces produced by cytoskeletal dynamics in one cell to drive deformation and movement of a field of cells (Gardel et al., 2010; Kasza and Zallen, 2011; Parsons et al., 2010).
Adhesion is also thought to contribute to cell sorting during tissue assembly. Sorting refers to the rearrangement of scattered mixtures of two or more cell types into homogeneous clusters. Cell sorting has been observed in aggregates of different cell types (Steinberg, 1963), in aggregates of cells expressing different levels of adhesion molecules (Foty and Steinberg, 2005), and in aggregates of cells from different germ layers of the early embryo (Townes and Holtfreter, 1955). This later observation suggested that cell sorting might contribute to the mechanical processes that drive gastrulation. After germ layer determination in embryos, adhesions alone or adhesion-dependent cell behaviors may drive sorting to specific locations based on the type and density of adhesion proteins (Steinberg and Takeichi, 1994). For instance, assembly of extracellular matrix (ECM) at an interface between prospective notochord and paraxial mesoderm cells could attract other notochord and paraxial cells to each face of the boundary. Mixtures of notochord and paraxial cells might then sort at this boundary and then adopt specific behaviors along the interface, maintaining or strengthening that critical structure. The theories that adhesion alone is capable of driving cell rearrangement and tissue morphogenesis have been contentious (Harris, 1976) but have inspired alternative theories in which cell adhesions regulate cell behaviors, or that adhesions might be coordinated with apico-basal or planar polarity cues to create asymmetric patterns of actomyosin contractility.
Formal definitions of the mechanics of adhesion
Adhesion can be defined as the bonding of two distinct entities in a manner that resists their subsequent separation. In the context of cell biology, these entities can be held together through either homotypic or heterotypic protein-protein interactions. Cohesion is the specific adhesion formed via homotypic interactions. To understand the role of adhesion in development we must understand how cell and tissue adhesion resists or enables separation of these adhesions. Both heterotypic and homotypic adhesions resist detachment in the direction normal to the surface, e.g. tension, of the adhesive interface and can resist movement parallel to the surface, e.g. shear. In the case of shear, resistance to movement can be linear or can exhibit complex non-linear responses such as stiction when increasing force causes a tissue to "slip" along a boundary after a critical level of applied force is reached. To understand the biophysical response of cell- and tissue-level adhesion to mechanical loads found in embryos we adopt standard terminology from physics and engineering such as stress. Stress is defined in terms of the force applied over a surface (units of force/area; Newton/meter2 or Pascal). Stress that is uniform in all three directions is pressure; a surface is under tension when the forces are applied in a direction that would cause separation and an interface is under compression when forces are applied that would bring the objects on either sides of the interface closer together. Once a force, or load, is applied the tissues can change shape or deform. Since the geometry of interfaces can take many forms, the term strain is a more useful description. Strain describes the amount of deformation per the scale of the object being deformed. The degree of strain a material exhibits when a stress is applied is formally defined as the modulus. A material with a high modulus deforms less under a fixed load compared to a material with a lower modulus. The compressive modulus describes the degree a material resists compressive loads whereas the shear modulus represents how a material will change shape if a shear stress, e.g. a load applied parallel to a surface, is applied. In the practice of mechanical engineering a material may have different modulus in each of the three cardinal directions and along the six shearing surfaces. Mathematically, the modulus is a 3D tensor. When considering a material that slips at a surface we can define a yield stress which is the stress at which a material slips at the interface; such a material is referred to as a plastic and is permanently deformed once the shear stress is removed. The yield stress can be defined from the stiction force needed to overcome static friction when stationary objects are in contact.
Adhesions and biological materials can behave very differently under mechanical loads. A material is elastic if it returns to its original shape after deforming stresses are removed. The degree to which stress produces strain in a material defines the material's elastic modulus. By contrast, a material is viscous if it deforms over time to a new shape that is not restored after stress is removed. The degree to which stress produces a time-dependent strain-rate in a material defines the material's viscosity. In practice, biological materials and adhesive structures fall between the two extremes of elastic and viscous behavior and may simultaneously exhibit both viscous and elastic behavior. These intermediate behaviors can be viscoelastic or viscoplastic. The behavior of these types of materials depend critically on the rate at which a force or stress is applied. A tissue may behave as an elastic material if the force is applied rapidly but may deform more like a liquid if the force is applied over a longer time-scale. Tissues can even exhibit superplasticity - a term borrowed from descriptions of solid crystalline materials (Valiev et al., 1991); superplastic materials can deform well beyond the usual breaking point of an elastic material through rearrangements of grains at specific temperatures or strain rates. Mechanical engineers formulate theory describing such complex material behaviors from the behaviors of submicroscopic components of the materials, however, the elemental components of cells, e.g. multiple classes of interacting polarized polymers, motors, dynamic cross-linkers and their regulators may exhibit new "physics" (e.g. soft condensed matter physics) that are not well represented by the "orderly" behaviors of simple elastic or viscous materials.
Biophysical descriptions of adhesion
The dynamics of adhesions must be considered when considering their mechanical function. Biophysical models of cell adhesion can be based on kinetic, thermodynamic and mechanical descriptions of adhesion (Zhu et al., 2000). Kinetic models represent adhesion by the rates in which adhesion receptors bind and dissociate and on their differential binding affinities. Thermodynamic models seek to explain adhesion through the differential chemical potentials of the receptors, ligands and bonds. Mechanical models approach the problems of adhesion through adhesion energy density, γ, defined as the mechanical work required to separate a unit area of the adherent surface. Each of these models can be used to predict experimental properties of adhesions. For instance, mechanical engineers can measure γ using a peel-test. A peel test is a method used in materials and mechanical engineering to test adhesiveness of two materials bonded along a planar adhesive interface. In brief, one of the materials is attached to a force transducer positioned perpendicular to the planar adhesive interface and pulled away from the interface at predetermined velocities. For example, consider the removal of a piece of adhesive tape from a table top. The forces required to pull the one material from the other is measured and can be used to calculate the adhesion energy density of the interface. Peel tests have limited utility in measuring the adhesion between two tissues when adhesion strength between the tissues is greater than their cohesion strength. Analogous biophysical techniques have been used to measure cohesion using atomic force microscopy between single cells.
Another aspect of adhesion which makes it difficult to study is that the cytoskeleton and the adhesive machinery in the cell are not only coupled physically but are also coupled through intracellular signaling pathways. Changes in adhesion can alter the cytoskeleton (Kovacs et al., 2011), which in turn can alter cell mechanics (Zhou et al., 2009). Integrins and cadherins have parallel roles in their respective adhesion complexes and involve several common scaffolding and cytoskeletal proteins such as α-actinin and vinculin and engage similar signaling pathways both independently and through cross-talk (Weber et al., 2011). The overlap between integrin and cadherin function and signaling suggest mechanosensing pathways in the embryo could be activated by cell-matrix adhesion and cytoskeletal tension, e.g. activation of ROCK by RhoA (Bhadriraju et al., 2007). Similar signaling pathways mediate both the formation and maturation of integrin based contacts (Huveneers and Danen, 2009) and the accumulation of E-cadherin at cell-cell contacts (Shewan et al., 2005). The direct physical contact between these adhesion systems plays a critical role transmitting force across cells (Maruthamuthu et al., 2010). Intracellular feedback through signaling has been proposed to drive the formation of the notochord-somite boundary during gastrulation in the frog Xenopus laevis (Fagotto et al., 2013). This study observed that changes in cadherin clustering at the prospective notochord-somite drives the actomyosin cortex to detach from the plasma membrane. Destabilized actomyosin contractions then drive distinctive cell blebbing which captures and traps cells at the newly formed interface .
The structural and mechanical function of adhesions during development
Adhesion plays a vital role in the establishment of tissue structure by shaping and maintaining the polarity of cell associations with their neighbors and surrounding ECM. Thus, the biological form of embryos and organs is driven in part by expression patterns and activity of adhesion proteins and the matrix elements they assemble. In this review we focus on classical cadherin and integrin family proteins since they are the most extensively characterized families of adhesion proteins. Cadherins mediate cell-cell adhesion and are transmembrane proteins consisting of an N-terminal extracellular region, a single-pass transmembrane segment and a C-terminal cytoplasmic tail (for a full review on cadherin structure and function, (Leckband and Prakasam, 2006)). The extracellular region consists of repeated sequences that are sites necessary for Ca2+ binding and cell adhesion. The intracellular portion mediates linkage to the actin cytoskeleton via catenin-family proteins, including p120 and beta-catenin. Integrins mediate cell-extracellular matrix adhesion and are heterodimeric transmembrane proteins consisting of alpha and beta subunits (for a full review on integrin structure and function, (Campbell and Humphries, 2011)). Most of the receptor dimer is extracellular, but both subunits traverse the plasma membrane and terminate in short cytoplasmic domains. The cytoplasmic portion initiates the assembly of large signaling complexes to bridge the extracellular matrix to the cell cytoskeleton. Molecular-level changes in adhesion regulate cell- and tissue-level processes, both biochemically and mechanically. Much attention has been given to the cell- and tissue- scale mechanics of cell cohesion and adhesion in the past two decades in an effort to answer long-standing questions in biology on the mechanism of cell sorting and cancer metastasis. Understanding the mechanics governing cell cohesion and cell adhesion during development and in adult tissues is critical not only to further basic science, but also for the development of cellular therapies and tissue engineering applications.
Tissues are dynamic composite materials and thus have complex material properties. These material properties can be represented by mathematical models adapted from mechanical engineering. Engineering models of embryonic tissues range from elastic solid-like to viscous liquid-like, depending on the time-scale of measurement. Some of these models have likened tissues to granular powders or colloids. These models are derived from mechanical interactions between smaller structural components and can include cell-cell adhesion forces needed to maintain the integrity of the tissue and cell-ECM adhesion forces to maintain contacts between cells and an external substrate.
Modulating Adhesions and Mechanics at Different Scales
Adhesion has been studied at many different scales; from single molecule to cellular to whole tissue. Molecular scale experiments isolate adhesion proteins of interest to elucidate the strength of adhesions in an acellular environment, removing variables such as cytoskeletal linkages and intracellular signaling. Adhesion studies at these scales typically use immortalized cell lines or dissociated embryonic primary cells to test adhesion strengths. These experiments cannot reveal the role of tissue mechanical and biochemical cues from neighboring cells that might influence adhesion dynamics in vivo. Cell aggregate and cell sheet experiments are more difficult to control during adhesion measurements but attempt to retain some of the native cues cells experience.
Experiments at each scale bring valuable insights but also come with unique caveats. In simple engineered materials, larger scale adhesion can be inferred by the sum of its parts, e.g. the total adhesion strength of a strip of Velcro could be calculated by summing the individual adhesion strength of its constituent ‘hooks’ and ‘loops’. In live cells, it is much more difficult to transition between scales because adhesion systems operate within the dynamic biology of motile cells in tissues. Single molecule experiments are very well controlled but lack the dynamic cellular microenvironment while larger scale aggregate and tissue scale adhesion tests are excellent in vivo models of adhesion but retain many uncontrollable variables. Tissue scale tests provide bulk measurements but such bulk mechanical responses may hide other cellular processes that produce the same apparent adhesion (David et al., 2014). For instance, spatial and temporal changes in cell-cell or cell-ECM strength within cell sheets may be coordinated or may be involved in regulatory feedback networks. Such examples might be found during cadherin contact inhibition at the notochord-somite boundary (Fagotto et al., 2013) and in cell junction remodeling by oscillation and asymmetric polarity of E-cadherin (Levayer and Lecuit, 2013). By coordinating experiments across multiple scales, we gain further insight into the mechanics of cellular adhesion.
At the molecular scale
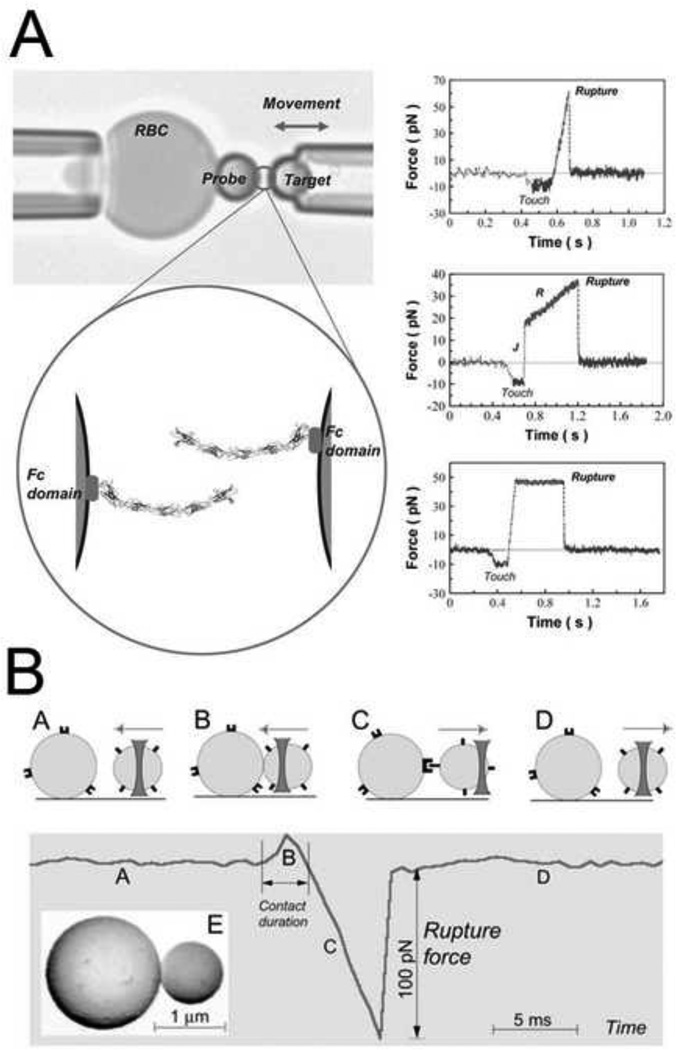
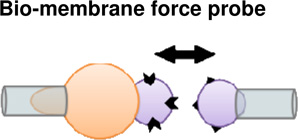
Cohesion and adhesion in animal tissues can be determined by the specificity and general structure of cell surface adhesion proteins (e.g. (Bayas et al., 2006); Figure 1A), and their adhesive strengths (e.g. (Perret et al., 2004)). A range of different experimental approaches have been developed to probe the single molecule level adhesive properties of homophilic cadherin and integrin pairs. Glass beads coated with cell surface receptor proteins such as cadherins and integrins have been used to test interactions when their respective ligands are brought into contact. Using a bio-membrane force probe (BFP), rupture strength between reconstituted E-cadherin pairs is tested by loading with precise forces over time from 0.001 to > 1 sec (Perret et al., 2004). The BFP is similar to atomic force microscopy, but the dynamic range of loading and the force sensitivity are greater. There are 3 modes for the BMP; steady-ramp, jump/ramp, and constant force (Figure 1A). Each modality begins by bringing beads into contact with a force of ~10pN and held for 0.1 s. In the steady-ramp mode, beads were separated at a constant loading rate (pN/s) until failure. In the jump/ramp mode, the beads were first pulled apart at a constant loading rate until the force reached a preset value. Surviving bonds were then pulled apart at a lower rate until failure. Jump/ramp mode was used to distinguish between bound states with different rupture strengths. Finally, in the constant force mode, beads were pulled apart at a constant force until failure. Constant force mode measurements verified the multi-state dynamics of homophilic cadherin adhesion. Using the BFP, the interaction between different recombinant fragments of extracelluar fragments were tested along with the contribution of individual fragment domains to adhesion strength. The cadherin extracellular segment is organized in five tandem repeats, numbered from the outermost N-terminal domain (EC1-EC5). These experiments revealed a rupture strength hierarchy for bonds between the full five-domain cadherin fragments, which represented different sub-states of cadherin binding depending on domain engagement (Perret et al., 2004). Similar results were found in studies using reconstituted C-cadherin (Bayas et al., 2006; Sivasankar et al., 1999). It is important to note that the original model of cadherin homophilic binding is that binding is mediated by tandem repeats of 5 EC domains. A newer model is that cadherin adhesion is a cis dimer formed by binding of the EC domains of 2 cadherins on the same cell surface. Cell-cell adhesion is initiated by the formation of trans adhesive complexes between the EC1 domains of cadherin cis dimers on opposing cell surfaces (Zhang et al., 2009). Additionally, different levels of Ca2+ can modulate cadherin interactions with increasing Ca leading to more rigid ectodomains and further increases in Ca leading to complex rings between dimers (Leckband and Sivasankar, 2000).
Figure 1. Biophysical analysis of adhesion at the molecular scale.
(A) Biomembrane force probe (BFP) to measure the strengths and lifetimes of homophilic cadherin bonds. Latex beads immobilized with cadherins are brought into contact with one another. After adhesions are established, the forces required to pull the beads apart are measured. The upper panel shows an aspirated RBC with an attached bead and the probe bead held by the second micropipette. The lower panel is a cartoon of the bound cadherins at the bead-bead junction. On the right, representative force versus time profiles obtained with the steady-ramp (upper), jump/ramp (middle), and constant force (lower) modes of the BFP (Bayas et al., 2006). (B) Laser tweezer system to measure receptor–ligand bond strengths. Integrin molecules are attached to polyacrylamide-coated, spherical silica pedestals and fibrinogen molecules are attached to smaller, laser tweezer controlled, latex beads. The upper panel shows the various steps before and after pedestal-bead contact. The lower panel shows the force profile during the time course of the experiment. Receptor-ligand rupture force is the force required to separate the pedestal and bead after adhesions are made (Litvinov et al., 2002).
Another approach to test single molecule adhesion strength involves laser tweezers. In these experiments, either purified proteins attached to beads or living cells were brought into contact with their ligand and pulled apart. Rupture forces required to separate single ligand-receptor pairs (fibrinogen-αIIbβ3 integrin) exhibited peak yield strengths of 80–100 pN (Litvinov et al., 2002) (Figure 1B).
Other studies sought to quantify adhesion by calculating binding probabilities using a dual micropipette system. In this system, live cells with surface-bound receptors and ligands are aspirated into opposite glass micropipettes, and brought into contact for a defined period. In this extension of the classical dual pipette assay, an ultrasensitive red blood cell picoforce transducer is used to detect adhesion strength. CHO cells expressing full length, wild-type C-cadherin were brought into contact with red blood cells modified to bind soluble hexahistidine-tagged C-cadherin ectodomain fragments. Cell pairs were subjected to 50 to 100 contact-retraction cycles which were assigned a 1 or 0 depending on whether adhesion was observed. Binding probability is the average of all cycles. Cadherin binding curves exhibited a fast, low probability binding state and a second, high probability binding state, which forms more slowly and required the full extracellular segment. In the first two seconds the binding probability rapidly increases to a plateau of 0.2, followed by a two to five second lag phase and transition to a high probability binding state after 20 seconds. Disrupting coordinate binding to a single site drove binding probability curves to adopt a simple monophasic form, as predicted for a single site binding mechanism (Chien et al., 2008).
Controlled molecular-scale studies are informative, for instance demonstrating that complex adhesion dynamics can be derived from receptor-scale phenomena, however, single molecule scale experiments cannot capture the role of cell signaling or the role of feedback loops that may occur in vivo.
Scaling-up: from single-cells to multiple cells and aggregates
Studies of cell aggregates or cohesion between identical cell types highlight the interplay between cell-cell and cell-ECM adhesion and the role of the cytoskeleton on adhesion strength and cortical tension. Cells within embryos are presented with complex mechanical and biochemical cues which may alter their behavior. Just as it is difficult to extrapolate the role of single molecular adhesion to single cells it is further challenging to extrapolate the in vivo dynamics of adhesion mechanics in embryos from single cell experiments. To understand adhesion mechanics on this scale requires experiments at the scale of cell sheets and aggregates with physiologically relevant microenvironments. These experiments come with many uncontrollable variables but provide details on the role of adhesion during collective mechanical processes such as tissue spreading and establishment of surface tension.
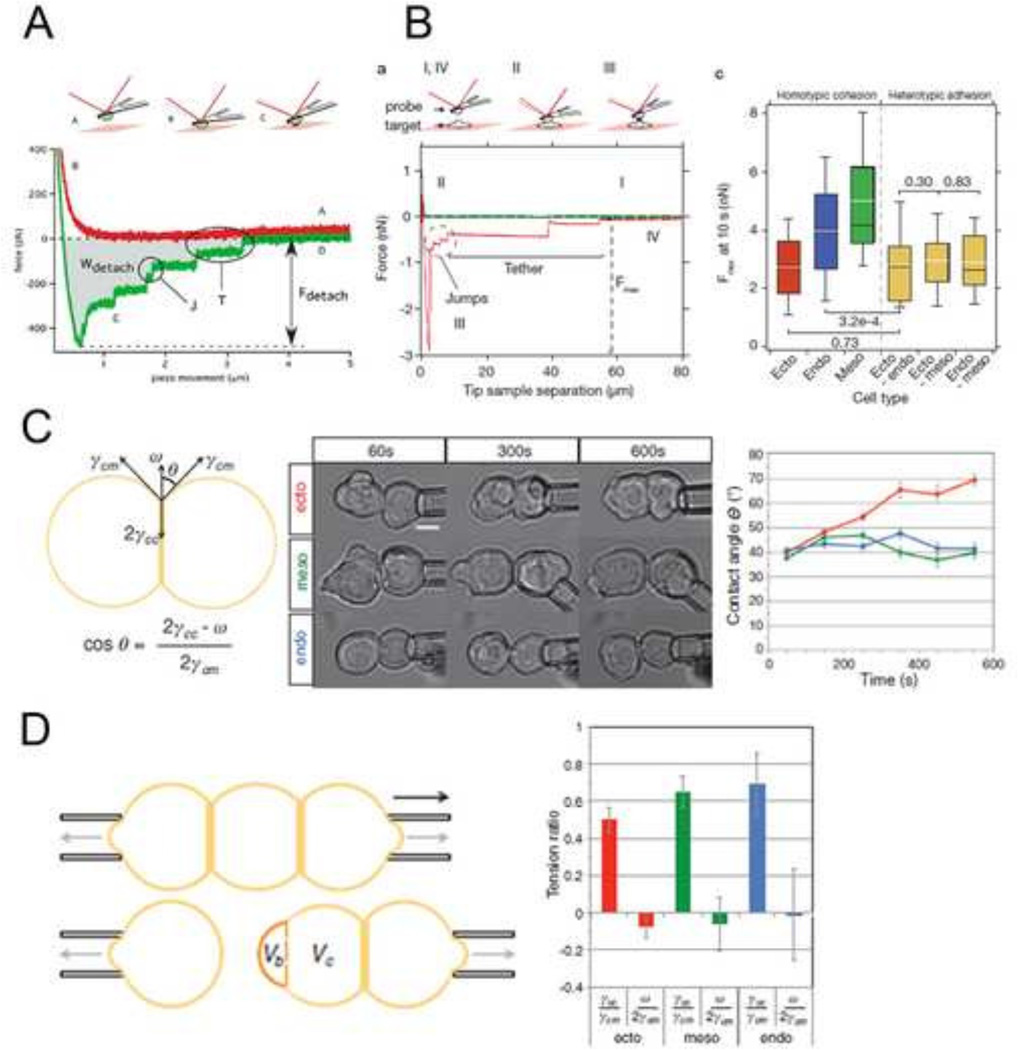
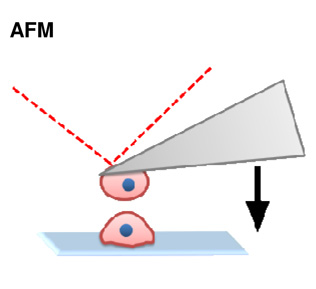
During development, cell surface adhesion proteins are integrated with the cytoskeleton to hold cells to their environment. Much of the motivation driving these experiments has been aimed to understand how cells sort from one another during animal development and tissue self-assembly, specifically how germ layers segregate and compartmentalize as development progresses. A leading theory is that cells exhibit differential adhesion for one another and that cells with strongest cohesion end up in the interior and cells with weakest adhesion sort to the exterior (DAH; Differential Adhesion Hypothesis, (Foty and Steinberg, 2005)). Recent efforts to test these hypotheses have used atomic force microscopy (Krieg et al., 2008; Puech et al., 2005) (Figure 2A–B) and micropipette aspiration assays (Maitre et al., 2012) (Figure 2C–D). In these approaches, cells from different germ layers are attached to AFM cantilevers or aspirated into micropipettes and brought into contact with another cell or substrate. The force required to break the adhesion from the cell-cell or cell-substrate interaction is a function of contact time and can be measured using either technique. AFM force traces reveal that bonds break sequentially as the micropipettes are pulled apart, suggesting differential bond strength across the adhesion site (Figure 2A–B). In homotypic cell-cell experiments, ectoderm cell pairs exhibited significantly less cohesion compared with mesoderm and endoderm counterparts. Additionally, heterotypic adhesion forces were similar to homotypic ectoderm cohesion. While the DAH predicts that protein-level discrimination between different cadherins drives sorting, studies have shown that cadherins can cross react and heterophilic cadherin adhesion is not substantially weaker than homophilic adhesion (Prakasam et al., 2006).
Figure 2. Strategies for measuring adhesion at the cell scale.
(A) Atomic force microscopy (AFM) to measure cell-ECM adhesion forces. Mesendodermal progenitor cells from zebrafish embryos are attached to AFM cantilevers and brought into contact with a fibronectin coated substrate. After adhesions are established, the cantilever/cell is withdrawn from the substrate and the bonds that have been formed sequentially break. The force of detachment is the force required to pull the cell off the substrate after adhesions are established. Numerous distinct ‘detachment events’ are measured during the time-course of cell retraction (Puech et al., 2005). (B) Atomic force microscopy (AFM) to measure cell-cell adhesion forces. This system is identical to (A) except instead of bring cells into contact with an ECM-coated substrate; cells are brought into contact with other cells. Homotypic cohesion and heterotypic adhesion is measured using endo-, meso- and ectoderm primary zebrafish cells (Krieg et al., 2008). (C) Cell doublet shape analysis to measure cell cortical tension. The left panel illustrates how contact angle θ results from the balance between adhesion tension CO and the cortex tensions at the cell medium γ and cell-cell interfaces γ. The middle panel shows aspirated homotypic cell doublets during the time course of experimentation. The right panel shows contact angles of cell doublets plotted over time (Maitre et al., 2012). (D) Cell triplet shape analysis to measure cell cortical tension. The left panel illustrates how cell triplets are manipulated with dual microaspirators. Bulge volume Vb is measured at the former cell-cell contact after separation and normalized to the cell body volume Vc. Vb/Vc is a measure of cortex tension at the cell-cell interface. The right panel shows tension ratios computed from homotypic triplet and doublet shapes for each germ layer (Maitre et al., 2012).
The theory of differential cell contractility (DCC) has also been suggested to account for the mechanics of cell sorting (Brodland, 2002; Harris, 1976). According to this theory, cells develop different levels of contractility at different locations such as the apical versus the basolateral cortex. Differential contractility of cells within a cohesive tissue may drive sorting independent of differences in adhesion strength. High levels of contraction generated parallel to the surface of the apical cell would appear as a surface tension. Both DAH and DCC have been formally tested with aggregates and single cells isolated from Zebrafish embryos to estimate surface tension. In these studies ectoderm cells had the highest actomyosin-dependent cell-cortex tension followed by mesoderm and then endoderm progenitors (Krieg et al., 2008).
Other approaches have focused on quantifying adhesion by analyzing traction forces between cell pairs. If cells in pairs are considered in mechanical equilibrium, the unbalanced traction force on an ECM substrate reflects the force exerted via cell-cell adhesion (Maruthamuthu et al., 2011). Endogenous cell-cell forces were estimated at approximately 100 nN and interestingly, were independent of cell-cell junction length. In a similar study using microneedles as force sensors rather than traction force gels, VE-cadherin mutants resulted in reduced intercellular forces, while traction forces remained similar (Liu et al., 2010). Additionally, junction size was directly correlated with intercellular force, unlike findings from Maruthamuthu et al. These studies describe a direct relationship between total cellular traction force and cell-cell force generation which suggests functional cross-talk between cell-ECM adhesion proteins and cell-cell adhesion proteins.
Sheets, aggregates, and assembly of large multi-tissue structures
Molecular specificity of adhesion serves to integrate a group of like cells into single mechanical units (e.g. sheets, aggregates, tubes, keels, masses, etc). Such units can then respond to applied forces by distributing strain over the structure and avoid "tears" or “rips". Units can remain together, moving over other tissues through active traction or by passively sliding. Examples of "mechanical units" include epithelial sheets that remain cohesive during epiboly and neurulation. Mesenchymal tissues can also form well defined structures such as the neural keel in teleosts (Papan and Camposortega, 1994) or the notochord in vertebrates (Stemple, 2005). Mechanically integrated tissues may shear or slide over one another. Such shearing movements could occur when clusters of cells actively migrate along interfaces, for example the movement of streaming neural crest (Loring and Erickson, 1987) may be driven passively in response to forces generated elsewhere. Many mesenchymal tissues shear with respect to adjacent layers, such as head mesoderm or mesendoderm tissues move during gastrulation (Davidson et al., 2002; Winklbauer, 1990).
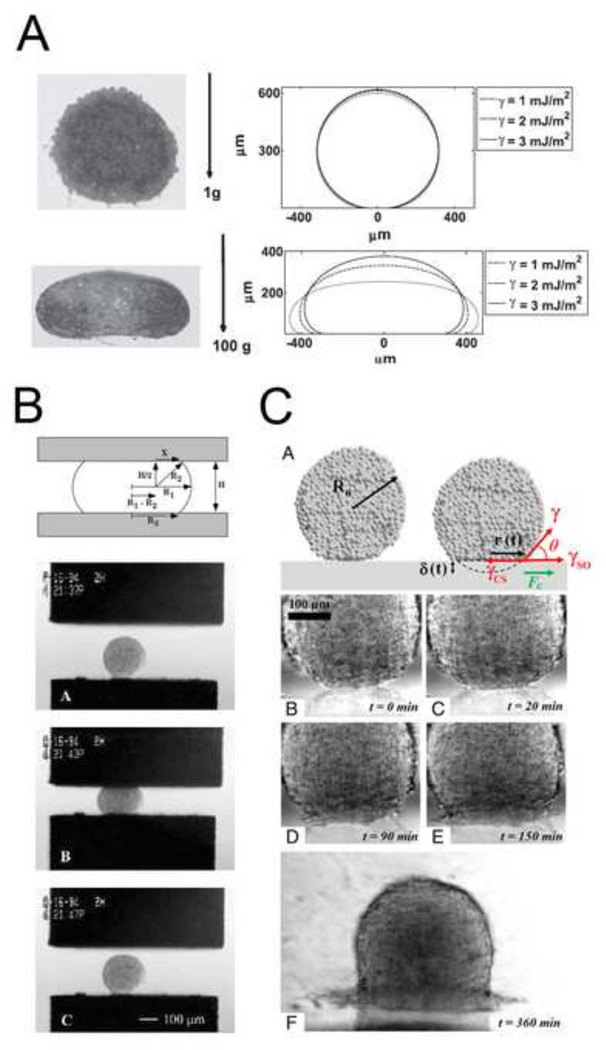
Experimental approaches to study tissue cohesiveness on this scale, like efforts to understand single cell adhesion, draw physical analogies to surface tension. Considered as an analog to surface tension between immiscible fluids, adhesion forces are thought to act parallel to the cell membrane and seek to minimize the exposed area of the aggregate (Kalantarian et al., 2009) (Figure 3A). Surface-tension forces driven by differential adhesion can be offset by cortical tension (Manning et al., 2010). Approaches to measure surface tension are similar to those used to estimate elastic and viscoelastic properties and include parallel plate compression and drop-shape analysis. Parallel plate compression of spheroidal aggregates between two parallel plates requires measurement of the force of compression and the shape of the deformed aggregate. By analogy to simple theoretical models of surface tension between two immiscible fluids, the applied force, and geometry of the deformed aggregate allow calculation of an equivalent surface tension for the multi-cellular aggregate. Compression-, sessile- or drop-shape analysis of the aggregate can provides an estimate of the surface tension within the aggregate after forces have been applied. Using these approaches, the contribution of cadherin levels to surface tension have been estimated. Steinberg and co-workers generated several cell lines expressing varying degrees of various cadherin proteins (Foty and Steinberg, 2005) and estimated the surface tension of multi-cellular aggregates using parallel plate compression. Similar experiments have been carried out using cells isolated from different regions of the chick embryo where differences in cohesion parallel the capacity of the tissue to sort out (Foty et al., 1994b; Foty et al., 1996) (Figure 3B). Using drop shape analysis, EP/C-cadherin knockdown, by cytoplasmically truncated EP/C-cadherin expression (Kalantarian et al., 2009) and EP/C-cadherin morpholinos (Ninomiya et al., 2012), has been shown to decrease surface tension by at least two fold.
Figure 3. Investigating the biophysics of adhesion at the tissue scale.
(A) Drop shape analysis for estimating surface tension of cell aggregates. Tissue aggregates under variable loading conditions (left) with complementary drop shape analyses and surface tension prediction (right) (Kalantarian et al., 2009). (B) Upper panel is a schematic of a liquid droplet compressed between parallel plates. In brief, radii, degree of compression, and force of compression measurements are combined to calculate surface tension. Bottom panel shows a spherical heart aggregate before, during, and after compression (Foty et al., 1996). (C) Aggregate spreading analysis for characterizing liquid-gas transition behavior. Upper panel is a schematic of an aggregate spreading and parameters used in analyses. All subsequent panels show the progression of aggregate spreading. Spreading dynamics were altered when changing levels of cohesion via cadherins (Douezan et al., 2011).
The process of tissue spreading depends on many of the same adhesion-dependent mechanical processes as sorting and engulfment. Spreading of an aggregate can be used to challenge the forces of cohesion that keep the aggregate together with forces of adhesion which drive spreading onto an ECM substrate. For instance, spheroidal aggregates spreading uniformly on ECM can be modeled by the spreading of liquid on a surface with spreading rates being proportional to cohesiveness of the cell aggregate (Beaune et al., 2014). In highly cohesive aggregates, tissues flow like a liquid, however when cohesiveness is reduced, by perturbing E-cadherin expression, the cells in the tissue act independently. Like molecules in a two dimensional gas, cells detach from the monolayer and move independently (Douezan et al., 2011) (Figure 3C).
Efforts to integrate the physics of tissue adhesion at the single cell scale with the properties of embryonic tissues or aggregates often have multiple plausible explanations and these can be further confounded by complex cell signaling and motility within dense tissues. Cells in tissues are always in contact with neighbors or extracellular substrates which can provide cues for asymmetric contractility, cytoskeletal dynamics, as well as adhesion. For instance, perturbing contacts between cells may alter conventional signaling pathways. However, new biophysical approaches combining fluorescence reporters of molecular strain with systems biology tools such as computational models discussed in the next section can resolve some of these issues.
New efforts to extend mechanics of molecular adhesion to tissue scales: molecular strain and tension sensors
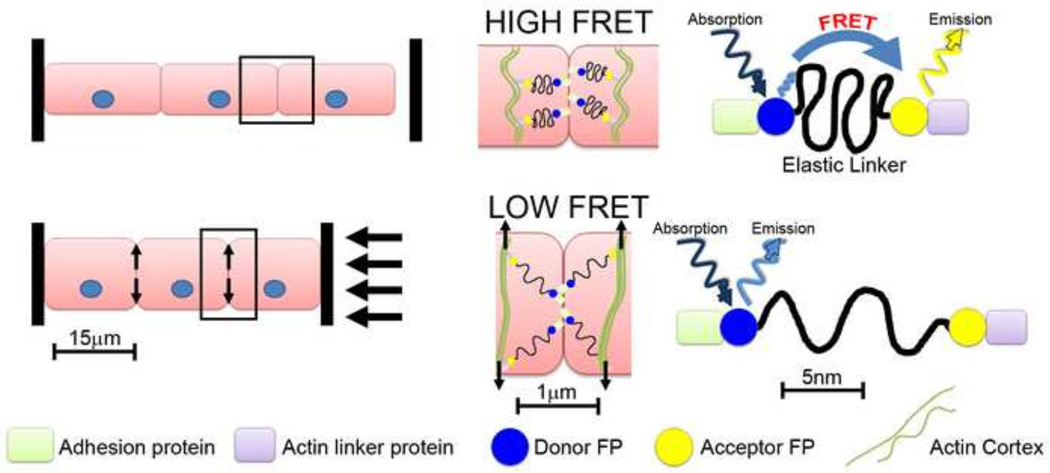
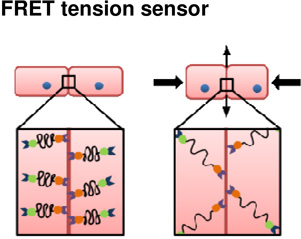
Combined advances in light microscopy and protein-encoded fluorescent probes have made possible the construction of fluorescent reporters of molecular strain at adhesive junctions (Borghi et al., 2012; Grashoff et al., 2010). Molecular strain sensors, also referred to as intramolecular tension sensors use Förster resonant energy transfer (FRET) to report the magnitude of tension at cell-cell or cell-ECM junctions within cultured cells (Figure 5). These sensors are based on a variety of protein modules that unfold or stretch with increasing force. Strain sensors are constructed with a FRET pair of donor and acceptor fluorescent proteins positioned at either end of the unfolding protein module. Under conditions of low or no tension the donor and acceptor are in close proximity and a virtual photon can be transferred directly from the donor to the acceptor. The missing emission by the donor and extra emission by the acceptor can be observed due to the difference in emission wavelengths of the FRET pair of fluorescent proteins. As force is applied to the adhesion complex, the FRET pair is separated and photon transfer ceases. Early applications of these molecular strain sensors have been promising. One question as these probes are applied is whether the molecular strain reported within the adhesion complex is relevant to the cell and tissue-scale tensions that drive morphogenesis. Another question concerns the dynamic range of these tension sensors, e.g. is the force sensing protein module stiff enough to report intermediate strains? Furthermore , if the protein module is too stiff the fluorescent proteins themselves can be denatured yielding a false indication of reduced FRET(Saeger et al., 2012). One significant limitation of these sensors is that they do not detect the direction of strain only its absolute magnitude. For instance, the reported FRET signal may indicate strain along the apical-basal axis of the cell and not the strain directed within the plane of the epithelium. However, even with these caveats, these probes are already raising new questions regarding our current views of strain within molecular complexes that mediate adhesion (Conway et al., 2013) and may reveal novel feedback between adhesion and cell signaling pathways.
Figure 5. How molecular FRET tension sensors might probe the mechanics of adhesion at cell junctions in tissues under compression.
Several tension sensors have been developed to interrogate the mechanical status of sub-cellular structures. This theoretical example shows such a tension sensor that includes an actin-binding protein on one end and an adhesion protein on the other end. Under no compressive load (top row), cortical tensions will be at a baseline level and therefore the FRET sensor will be in the ‘off position, indicating high FRET signal. After compression of the tissue (bottom row) the load will be transferred perpendicular to the applied force to stretch the cell cortices and cortical F-actin networks. As the F-actin networks are stretched, the tension sensor will turn on, causing low FRET signal. In this example, a compressive load at the tissue level is converted to tensile loads at the cell level.
Mechanics and adhesion in the embryo
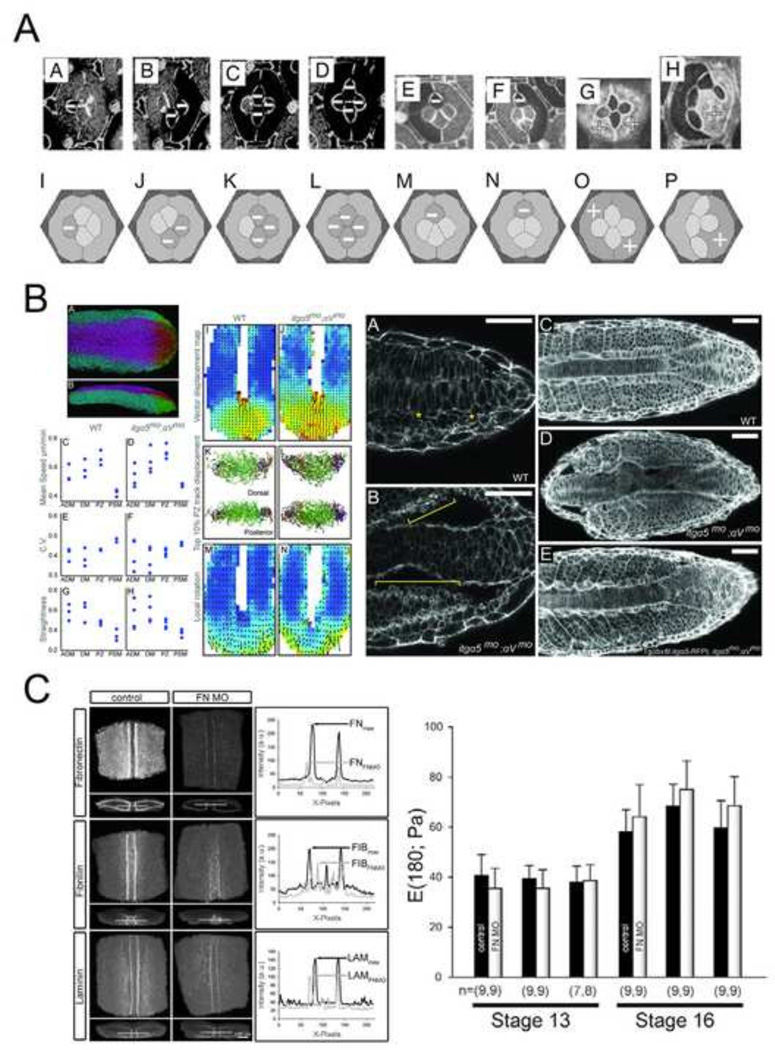
To better understand the role of adhesion mechanics in the embryo, animal models with controllable adhesion levels have been developed. An elegant study investigating the role of adhesion mechanics in vivo focused on patterning within stereotyped cell clusters in the Drosophila retina (Hayashi and Carthew, 2004). During pattern formation, cell types within the retina become determined, and adopt identity-specific cell shapes. DN- and DE- cadherins mediate apical adhesion between retina epithelial cells and establish the shape of cone cells. Interestingly, wild type cone cell configurations are nearly identical to soap bubbles confined within similar clusters. In soap bubble clusters these configurations are governed by surface energy minimization. Even mutants, with variable numbers of cone cells, exhibit soap bubblelike configurations (Hilgenfeldt et al., 2008; Kafer et al., 2007) (Figure 4A). With mosaic analysis of adhesion mutant clones, these studies showed that differential DN-cadherin between cone cells and primary pigment cells altered cell shapes into soap bubble idealized configurations. When DN-cadherin is misexpressed in both primary pigment cells and in cone cells, the cone cells did not assume soap bubble, surface area minimized, configurations. While the physical processes control cell shape in the Drosophila retina may be more complex than surface-tension mechanics, this study demonstrates the interplay between genetic control of adhesion and physical mechanical processes; regulated levels of cadherin expression adopt shapes analogous to predicted configurations. The success of physical analog models in predicting cell shapes in the retina demonstrates the plausibility of these mechanisms for coordinating local cell shape and geometric order within cell clusters; the question of whether these mechanisms are universal or can produce sufficient stress to shape larger tissues remains unanswered.
Figure 4. Roles of adhesion at the embryo scale.
(A) Drosophila retina ommatidium shapes are governed by cell adhesion and cortex contractility. This panel illustrates various N-cadherin mutants resulting in abnormal cell shapes. Using computer simulations and knock down experiments, Kafer et al. show that a simple surface minimization model is insufficient to model the experimentally observed shapes and packing patterns of ommatidium. The authors develop a model in which adhesion leads to surface increase, balanced by cell contraction, which successfully predicts experimental observations (Kafer et al., 2007). (B) Cell-fibronectin interactions determine tissue fluidity in zebrafish tail bud. Left panel shows the quantitative analysis of cell motion in wildtype and integrin mutant embryos. Right panel is a series of confocal images of phalloidin stained wildtype, integrin mutant, and rescued embryos showing shortened body elongation and increased blebbing (yellow brackets) in integrin mutants (Dray et al., 2013). (C) Fibrillar fibronectin does not contribute to dorsal tissue stiffness in Xenopus laevis embryos. The left panel shows representative confocal images of dorsal tissues stained for fibronectin, fibrillin, and laminin in control and ECM knock-down explants. The right panel shows that the stiffness of explants injected with fibronectin morpholinos (FNMO) does not differ from uninjected explants at either stage 13 or at stage 16 (Zhou et al., 2009).
Another whole embryo model focused on the role of ECM adhesion mechanics during zebrafish tailbud trunk elongation. Paraxial cell movements in these stages appear fluid-like and suggest that cells reduce their elastic coupling. Tissue fluidity during trunk elongation is estimated noninvasively by combining whole embryo imaging and cell tracking analysis. The discovery of highly ordered collective migration and its dependence on cell-substrate interactions (Lawton et al., 2013) has been interpreted to support the role of ECM in maintaining tissue fluidity in the tailbud. To test the role of cell-fibronectin (FN) interactions in vertebrate body elongation, embryos whose integrin receptors for ECM have been reduced generate a body truncation (Dray et al., 2013) (Figure 4B). Surprisingly, characteristic features of migrating cells were unaltered after abrogation of cell-FN interactions. Knock-down embryos exhibited abnormally anisotropic FN along the mesoderm-notochord boundary with high rates of cell blebbing and detachment from the notochord in paraxial mesoderm surface cells. The low rates of blebbing in WT cells suggested that FN matrix constrains blebbing and accompanying changes in cytosolic pressure. Expression of integrin α5 specifically expressed in the paraxial mesoderm rescued paraxial mesoderm FN assembly, paraxial mesoderm-notochord adhesion, and normal blebbing. This study revealed that FN is not only a substrate for cell migration but is also integrated mechanically with cells during tissue morphogenesis.
Cell-cell and cell-matrix adhesions may play direct roles in establishing tissue stiffness during morphogenesis. Dorsal tissues within Xenopus laevis embryos increase in stiffness six-fold as from gastrula through neurula stages (Zhou et al., 2009). Over these same stages the embryo assembles a complex fibrillar ECM at all major tissue interfaces (Davidson et al., 2008). Such a correlation between histology and physical mechanics suggested that ECM could be a major contributor to increasing levels of stiffness. Testing the hypothesis that FN matrix mediated the observed increase in tissue stiffness measurements were collected from embryos injected with FN morpholinos. Interestingly, even though FN, laminin, and fibrillin fibrils were reduced in FN-knockdown embryos, dorsal tissues showed no difference in stiffness from controls(Zhou et al., 2009) (Figure 4C). FN does not contribute to bulk tissue stiffness, however, FN and other matrix components may still contribute to other mechanical aspects of tissue development, for instance, serving as a lubricant for shear at tissue interfaces, as a medium for force transmission through tissue types, or as a permissive signal for cell motility (Rozario et al., 2009).
In silico analyses of adhesion
Theoretical and computational models of adhesion serve to extend intuition to physical mechanics of the cell and tissue scale that are not familiar to the researcher. Models are also used to establish the plausibility of physical hypotheses on the role of adhesion mechanics and how adhesion may be integrated with cell signaling pathways. Such plausibility tests may be combined with experiment to narrow the range of permissive conditions or identify physical parameters of adhesion and tissue mechanics that might be used as a further test of the model. However, modeling alone cannot "prove" a hypothesis. For a review of these approaches we direct readers to two theoretical frameworks that have been used extensively to demonstrate how adhesion coordinates cell mechanics and signaling during morphogenesis. Cellular Potts models have been used extensively to study signaling and movement in multicellular tissues (Glazier and Graner, 1993; Zajac et al., 2000). For instance, the process of somitogenesis has been extensively explored with models that integrate signaling with cell rearrangement (Cooke and Zeeman, 1976; Dias et al., 2014; Dubrulle et al., 2001). Analysis of epithelial morphogenesis, especially in the fly wing disk has been significantly aided by vertex network models based on cell-shaped arrays of elastic springs, viscous dashpots, and adhesive surface tensions(Staple et al., 2010). Vertex network models have been used to understand the origin and patterning of segment boundaries in the wing and the maintenance of geometric order (Farhadifar et al., 2007) and domain size (Aliee et al., 2012). Both Cellular Potts and Vertex network models represent tissues as arrays of discretely shaped cells and allow explicit representation of adhesion- and elastic-energy as well as intercellular signaling and gene regulatory networks. Theoretical analyses and computational models of adhesion and mechanics are important experimental tools to define the role of adhesion and mechanics within multi-cellular tissues and provide testable predictions for new lines of biomechanical studies.
Conclusion
Throughout this review we have described biophysical approaches to measuring adhesion and mechanical properties of embryonic tissues. The fundamental role of adhesion is to connect cells to their microenvironment allowing force-transmission across a tissue. While this is a mechanical process, the forces and stresses originate from the cytoskeleton of individual cells pulling on neighboring cells and matrix. Thus, when considering potential cases of mechanical feedback, it is critical to distinguish between the force-transmission and mechano-transduction roles of these multifunctional systems. Biomechanical tests using controlled strains and strain-rates within physiological levels are critical to distinguishing the mechanical roles played by adhesion.
Mechano-transduction occurs when a cell converts a mechanical stimulus into a chemical cue that can be relayed across the cell or to other cells via more conventional biochemical signaling pathways. To understand the mechanochemical circuits that use adhesion to drive and control development we will need to consider how adhesion operates at the molecular level and how forces and signals are transmitted throughout the tissue. New molecular strain sensors will be powerful tools to probe how forces are transmitted through space and time and across length scales (Fig. 5). Using these tools we will need to answer several questions: How does the action at the level of molecular adhesions alter tissues 10,000-fold larger? How are the direction and magnitude of stresses and strains transmitted over the intervening length scales?
Molecular level mechanics and kinetics provide the most fundamental descriptions of adhesion. The work of many molecules on the surface of cells governs the potential mechanical roles of adhesion and cohesion. Cell dynamics and cues from the environment must be integrated with developmental processes that guide cells in their fate and differentiation. How integrin or cadherin activation drives and organizes the cytoskeleton within the embryo and how their expression and activity are regulated spatially and temporally remain open questions.
| TECHNIQUE | SCALE | ADVANTAGES | LIMITATIONS | REFERENCES |
|---|---|---|---|---|
 |
Molecular | + Ability to measure forces at molecular level in intact tissues | − Difficult validation process − Unable to obtain directional information − Molecular scale may not reflect cell- or tissue- scale tensions |
(Conway et al., 2013; Saeger et al., 2012) |
 |
Molecular | + Ability to tether proteins and cells at nanometer resolution and under physiologica conditions. | − Receptor- ligand bond strengths may differ from in
vivo due to complex biological interactions. − Difficult to run experiments at high velocities due to viscous drag from cantilevers. |
(Puech et al., 2005) |
 |
Molecular | + Detection of bond rupture forces with picoNewton sensitivity. + Micron sized trapped beads allow for more |
− Receptor- ligand bond strengths may differ from in
vivo due to complex biological interactions. − Limited rangel of detectable forces (large forces yield non-linear force- deformation Irelationships |
(Perret et al., 2004) (Bayas et al., 2006) |
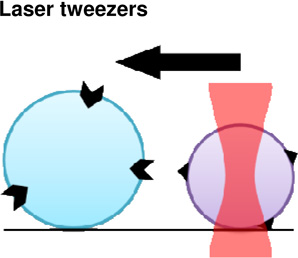 |
Molecular- Cell | + Isolation of single molecules + Precise control of force application. |
− Receptor- ligand bond strengths may differ from in
vivo due to complex biological interactions. − Requires optically homogeneous and highly purified samples. − Local heating of sample, potentially perturbing mechanics |
(Litvinov et al., 2002) |
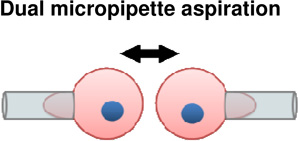 |
Cell | + Ability to grasp and move cells into contact with other cells or substrate | − Requires substantial deformation of cells. − Cells removed from native environment |
(Maitre et al., 2012) |
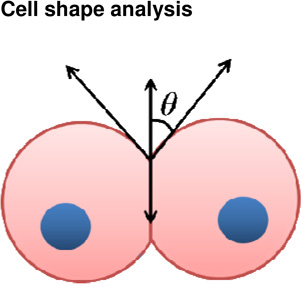 |
Cell | + Obtain cell surface tensions | − Dependent on contact time between cell pairs − Cells removed from native environment |
(Maitre et al., 2012) (Manning et al., 2010) |
 |
Cell-tissue | + Can be done in vivo + Genotype to phenotype relationship |
− Not mechanically quantitative − May change cell identity over time course of experiment |
(Kafer et al., 2007) (Dray et al., 2013) |
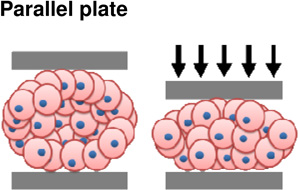 |
Tissue | + Directly measure tissue relaxation after application of known loads | − Requires spherical aggregates − Risk of changing cell identities over time course of experiment |
(Foty et al., 1994a) |
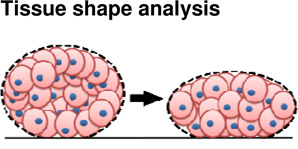 |
Tissue | + Simple prep + Obtain whole tissue surface tension measureme nts |
− Requires spherical aggregates − Risk of changing cell identities over time course of experiment |
(Kalantarian et al., 2009) |
Highlights.
Important role of adhesion in the mechanics of development.
Engineering terms that are useful when considering the contribution of cell adhesion to mechanics.
Experimental approaches to measuring mechanics of adhesion at multiple spatial scales, from single molecule to whole embryo.
Overview of methods at each scale with their advantages and limitations.
Acknowledgements
This material is based upon work supported by the National Science Foundation (IOS-0845775 and CMMI-1100515) and the National Institutes of Health (HD044750). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aliee M, Röper J-C, Landsberg KP, Pentzold C, Widmann TJ, Jülicher F, Dahmann C. Physical Mechanisms Shaping the Drosophila Dorsoventral Compartment Boundary. Current Biology. 2012;22:967–976. doi: 10.1016/j.cub.2012.03.070. [DOI] [PubMed] [Google Scholar]
- Bayas MV, Leung A, Evans E, Leckband D. Lifetime measurements reveal kinetic differences between homophilic cadherin bonds. Biophys J. 2006;90:1385–1395. doi: 10.1529/biophysj.105.069583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaune G, Stirbat TV, Khalifat N, Cochet-Escartin O, Garcia S, Gurchenkov VV, Murrell MP, Dufour S, Cuvelier D, Brochard-Wyart F. How cells flow in the spreading of cellular aggregates. Proceedings of the National Academy of Sciences. 2014;111:8055–8060. doi: 10.1073/pnas.1323788111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadriraju K, Yang M, Alom Ruiz S, Pirone D, Tan J, Chen CS. Activation of ROCK by RhoA is regulated by cell adhesion, shape, and cytoskeletal tension. Exp Cell Res. 2007;313:3616–3623. doi: 10.1016/j.yexcr.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghi N, Sorokina M, Shcherbakova OG, Weis WI, Pruitt BL, Nelson WJ, Dunn AR. E-cadherin is under constitutive actomyosin-generated tension that is increased at cell-cell contacts upon externally applied stretch. Proceedings of the National Academy of Sciences. 2012;109:12568–12573. doi: 10.1073/pnas.1204390109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodland GW. The Differential Interfacial Tension Hypothesis (DITH): a comprehensive theory for the self-rearrangement of embryonic cells and tissues. J Biomech Eng. 2002;124:188–197. doi: 10.1115/1.1449491. [DOI] [PubMed] [Google Scholar]
- Campbell ID, Humphries MJ. Integrin structure, activation, and interactions. Cold Spring Harbor perspectives in biology. 2011:3. doi: 10.1101/cshperspect.a004994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien YH, Jiang N, Li F, Zhang F, Zhu C, Leckband D. Two stage cadherin kinetics require multiple extracellular domains but not the cytoplasmic region. J Biol Chem. 2008;283:1848–1856. doi: 10.1074/jbc.M708044200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway DE, Breckenridge MT, Hinde E, Gratton E, Chen CS, Schwartz MA. Fluid shear stress on endothelial cells modulates mechanical tension across VE-cadherin and PECAM-1. Current Biology. 2013;23:1024–1030. doi: 10.1016/j.cub.2013.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke J, Zeeman EC. A clock and wavefront model for control of the number of repeated structures during animal morphogenesis. J Theor Biol. 1976;58:455–476. doi: 10.1016/s0022-5193(76)80131-2. [DOI] [PubMed] [Google Scholar]
- David R, Luu O, Damm EW, Wen JW, Nagel M, Winklbauer R. Tissue cohesion and the mechanics of cell rearrangement. Development. 2014;141:3672–3682. doi: 10.1242/dev.104315. [DOI] [PubMed] [Google Scholar]
- Davidson LA, Dzamba BD, Keller R, Desimone DW. Live imaging of cell protrusive activity, and extracellular matrix assembly and remodeling during morphogenesis in the frog, Xenopus laevis. Dev Dyn. 2008;237:2684–2692. doi: 10.1002/dvdy.21600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson LA, Hoffstrom BG, Keller R, DeSimone DW. Mesendoderm extension and mantle closure in Xenopus laevis gastrulation: combined roles for integrin alpha5beta1, fibronectin, and tissue geometry. Dev Biol. 2002;242:109–129. doi: 10.1006/dbio.2002.0537. [DOI] [PubMed] [Google Scholar]
- Dias AS, de Almeida I, Belmonte JM, Glazier JA, Stern CD. Somites without a clock. Science. 2014;343:791–795. doi: 10.1126/science.1247575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douezan S, Guevorkian K, Naouar R, Dufour S, Cuvelier D, Brochard-Wyart F. Spreading dynamics and wetting transition of cellular aggregates. Proc Natl Acad Sci U S A. 2011;108:7315–7320. doi: 10.1073/pnas.1018057108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dray N, Lawton A, Nandi A, Julich D, Emonet T, Holley SA. Cell-fibronectin interactions propel vertebrate trunk elongation via tissue mechanics. Current biology : CB. 2013;23:1335–1341. doi: 10.1016/j.cub.2013.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrulle J, McGrew MJ, Pourquie O. FGF signaling controls somite boundary position and regulates segmentation clock control of spatiotemporal Hox gene activation. Cell. 2001;106:219–232. doi: 10.1016/s0092-8674(01)00437-8. [DOI] [PubMed] [Google Scholar]
- Fagotto F, Rohani N, Touret AS, Li R. A molecular base for cell sorting at embryonic boundaries: contact inhibition of cadherin adhesion by ephrin/ Eph-dependent contractility. Dev Cell. 2013;27:72–87. doi: 10.1016/j.devcel.2013.09.004. [DOI] [PubMed] [Google Scholar]
- Farhadifar R, Roper JC, Aigouy B, Eaton S, Julicher F. The influence of cell mechanics, cell-cell interactions, and proliferation on epithelial packing. Current biology : CB. 2007;17:2095–2104. doi: 10.1016/j.cub.2007.11.049. [DOI] [PubMed] [Google Scholar]
- Foty RA, Forgacs G, Pfleger CM, Steinberg MS. Liquid properties of embryonic tissues: Measurement of interfacial tensions. Phys Rev Lett. 1994a;72:2298–2301. doi: 10.1103/PhysRevLett.72.2298. [DOI] [PubMed] [Google Scholar]
- Foty RA, Forgacs G, Pfleger CM, Steinberg MS. Liquid properties of embryonic tissues: measurement of interfacial tensions. Physical Review Letters. 1994b;72:2298–2301. doi: 10.1103/PhysRevLett.72.2298. [DOI] [PubMed] [Google Scholar]
- Foty RA, Pfleger CM, Forgacs G, Steinberg MS. Surface tensions of embryonic tissues predict their mutual envelopment behavior. Development. 1996;122:1611–1620. doi: 10.1242/dev.122.5.1611. [DOI] [PubMed] [Google Scholar]
- Foty RA, Steinberg MS. The differential adhesion hypothesis: a direct evaluation. Dev Biol. 2005;278:255–263. doi: 10.1016/j.ydbio.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Gardel ML, Schneider IC, Aratyn-Schaus Y, Waterman CM. Mechanical integration of actin and adhesion dynamics in cell migration. Annual review of cell and developmental biology. 2010;26:315–333. doi: 10.1146/annurev.cellbio.011209.122036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazier JA, Graner F. Simulation of the differential adhesion driven rearrangement of biological cells. Physical review. E, Statistical physics, plasmas, fluids, and related interdisciplinary topics. 1993;47:2128–2154. doi: 10.1103/physreve.47.2128. [DOI] [PubMed] [Google Scholar]
- Grashoff C, Hoffman BD, Brenner MD, Zhou R, Parsons M, Yang MT, McLean MA, Sligar SG, Chen CS, Ha T. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466:263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AK. Is Cell sorting caused by differences in the work of intercellular adhesion? A critique of the Steinberg hypothesis. J Theor Biol. 1976;61:267–285. doi: 10.1016/0022-5193(76)90019-9. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Carthew RW. Surface mechanics mediate pattern formation in the developing retina. Nature. 2004;431:647–652. doi: 10.1038/nature02952. [DOI] [PubMed] [Google Scholar]
- Hilgenfeldt S, Erisken S, Carthew RW. Physical modeling of cell geometric order in an epithelial tissue. Proc Natl Acad Sci U S A. 2008;105:907–911. doi: 10.1073/pnas.0711077105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huveneers S, Danen EH. Adhesion signaling - crosstalk between integrins, Src and Rho. J Cell Sci. 2009;122:1059–1069. doi: 10.1242/jcs.039446. [DOI] [PubMed] [Google Scholar]
- Kafer J, Hayashi T, Maree AF, Carthew RW, Graner F. Cell adhesion and cortex contractility determine cell patterning in the Drosophila retina. Proc Natl Acad Sci U S A. 2007;104:18549–18554. doi: 10.1073/pnas.0704235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantarian A, Ninomiya H, Saad SM, David R, Winklbauer R, Neumann AW. Axisymmetric drop shape analysis for estimating the surface tension of cell aggregates by centrifugation. Biophys J. 2009;96:1606–1616. doi: 10.1016/j.bpj.2008.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasza KE, Zallen JA. Dynamics and regulation of contractile actin-myosin networks in morphogenesis. Current opinion in cell biology. 2011;23:30–38. doi: 10.1016/j.ceb.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs EM, Verma S, Ali RG, Ratheesh A, Hamilton NA, Akhmanova A, Yap AS. N-WASP regulates the epithelial junctional actin cytoskeleton through a non-canonical post-nucleation pathway. Nat Cell Biol. 2011;13:934–943. doi: 10.1038/ncb2290. [DOI] [PubMed] [Google Scholar]
- Krieg M, Arboleda-Estudillo Y, Puech PH, Kafer J, Graner F, Muller DJ, Heisenberg CP. Tensile forces govern germ-layer organization in zebrafish. Nat Cell Biol. 2008;10:429–436. doi: 10.1038/ncb1705. [DOI] [PubMed] [Google Scholar]
- Lawton AK, Nandi A, Stulberg MJ, Dray N, Sneddon MW, Pontius W, Emonet T, Holley SA. Regulated tissue fluidity steers zebrafish body elongation. Development. 2013;140:573–582. doi: 10.1242/dev.090381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leckband D, Prakasam A. Mechanism and dynamics of cadherin adhesion. Annual review of biomedical engineering. 2006;8:259–287. doi: 10.1146/annurev.bioeng.8.061505.095753. [DOI] [PubMed] [Google Scholar]
- Leckband D, Sivasankar S. Mechanism of homophilic cadherin adhesion. Current opinion in cell biology. 2000;12:587–592. doi: 10.1016/s0955-0674(00)00136-8. [DOI] [PubMed] [Google Scholar]
- Levayer R, Lecuit T. Oscillation and polarity of E-cadherin asymmetries control actomyosin flow patterns during morphogenesis. Dev Cell. 2013;26:162–175. doi: 10.1016/j.devcel.2013.06.020. [DOI] [PubMed] [Google Scholar]
- Litvinov RI, Shuman H, Bennett JS, Weisel JW. Binding strength and activation state of single fibrinogen-integrin pairs on living cells. Proc Natl Acad Sci U S A. 2002;99:7426–7431. doi: 10.1073/pnas.112194999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Tan JL, Cohen DM, Yang MT, Sniadecki NJ, Ruiz SA, Nelson CM, Chen CS. Mechanical tugging force regulates the size of cell-cell junctions. Proc Natl Acad Sci U S A. 2010;107:9944–9949. doi: 10.1073/pnas.0914547107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loring JF, Erickson CA. Neural crest cell migratory pathways in the trunk of the chick embryo. Dev Biol. 1987;121:220–236. doi: 10.1016/0012-1606(87)90154-0. [DOI] [PubMed] [Google Scholar]
- Maitre JL, Berthoumieux H, Krens SF, Salbreux G, Julicher F, Paluch E, Heisenberg CP. Adhesion functions in cell sorting by mechanically coupling the cortices of adhering cells. Science. 2012;338:253–256. doi: 10.1126/science.1225399. [DOI] [PubMed] [Google Scholar]
- Manning ML, Foty RA, Steinberg MS, Schoetz EM. Coaction of intercellular adhesion and cortical tension specifies tissue surface tension. Proc Natl Acad Sci U S A. 2010;107:12517–12522. doi: 10.1073/pnas.1003743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruthamuthu V, Aratyn-Schaus Y, Gardel ML. Conserved F-actin dynamics and force transmission at cell adhesions. Current opinion in cell biology. 2010;22:583–588. doi: 10.1016/j.ceb.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruthamuthu V, Sabass B, Schwarz US, Gardel ML. Cell-ECM traction force modulates endogenous tension at cell-cell contacts. Proc Natl Acad Sci U S A. 2011;108:4708–4713. doi: 10.1073/pnas.1011123108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninomiya H, David R, Damm EW, Fagotto F, Niessen CM, Winklbauer R. Cadherin-dependent differential cell adhesion in Xenopus causes cell sorting in vitro but not in the embryo. Journal of Cell Science. 2012;125:1877–1883. doi: 10.1242/jcs.095315. [DOI] [PubMed] [Google Scholar]
- Papan C, Camposortega JA. On the Formation of the Neural Keel and Neural-Tube in the Zebrafish Danio (Brachydanio) Rerio. Roux Arch Dev Biol. 1994;203:178–186. doi: 10.1007/BF00636333. [DOI] [PubMed] [Google Scholar]
- Parsons JT, Horwitz AR, Schwartz MA. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat Rev Mol Cell Biol. 2010;11:633–643. doi: 10.1038/nrm2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perret E, Leung A, Feracci H, Evans E. Trans-bonded pairs of E-cadherin exhibit a remarkable hierarchy of mechanical strengths. Proc Natl Acad Sci U S A. 2004;101:16472–16477. doi: 10.1073/pnas.0402085101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakasam AK, Maruthamuthu V, Leckband DE. Similarities between heterophilic and homophilic cadherin adhesion. P Natl Acad Sci USA. 2006;103:15434–15439. doi: 10.1073/pnas.0606701103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puech PH, Taubenberger A, Ulrich F, Krieg M, Muller DJ, Heisenberg CP. Measuring cell adhesion forces of primary gastrulating cells from zebrafish using atomic force microscopy. J Cell Sci. 2005;118:4199–4206. doi: 10.1242/jcs.02547. [DOI] [PubMed] [Google Scholar]
- Rozario T, Dzamba B, Weber GF, Davidson LA, DeSimone DW. The physical state of fibronectin matrix differentially regulates morphogenetic movements in vivo. Developmental Biology. 2009;327:386–398. doi: 10.1016/j.ydbio.2008.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeger J, Hytonen VP, Klotzsch E, Vogel V. GFP's mechanical intermediate states. PLoS One. 2012;7:e46962. doi: 10.1371/journal.pone.0046962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewan AM, Maddugoda M, Kraemer A, Stehbens SJ, Verma S, Kovacs EM, Yap AS. Myosin 2 is a key Rho kinase target necessary for the local concentration of E-cadherin at cell-cell contacts. Mol Biol Cell. 2005;16:4531–4542. doi: 10.1091/mbc.E05-04-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasankar S, Brieher W, Lavrik N, Gumbiner B, Leckband D. Direct molecular force measurements of multiple adhesive interactions between cadherin ectodomains. Proc Natl Acad Sci U S A. 1999;96:11820–11824. doi: 10.1073/pnas.96.21.11820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staple D, Farhadifar R, Röper J-C, Aigouy B, Eaton S, Jülicher F. Mechanics and remodelling of cell packings in epithelia. The European Physical Journal E: Soft Matter and Biological Physics. 2010;33:117–127. doi: 10.1140/epje/i2010-10677-0. [DOI] [PubMed] [Google Scholar]
- Steinberg MS. Reconstruction of tissues by dissociated cells. Some morphogenetic tissue movements and the sorting out of embryonic cells may have a common explanation. Science. 1963;141:401–408. doi: 10.1126/science.141.3579.401. [DOI] [PubMed] [Google Scholar]
- Steinberg MS, Takeichi M. Experimental specification of cell sorting, tissue spreading, and specific spatial patterning by quantitative differences in cadherin expression. Proc Natl Acad Sci U S A. 1994;91:206–209. doi: 10.1073/pnas.91.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemple DL. Structure and function of the notochord: an essential organ for chordate development. Development. 2005;132:2503–2512. doi: 10.1242/dev.01812. [DOI] [PubMed] [Google Scholar]
- Townes PL, Holtfreter J. Directed movements and selective adhesion of embryonic amphibian cells. Journal of Experimental Zoology. 1955;128:53–120. doi: 10.1002/jez.a.114. [DOI] [PubMed] [Google Scholar]
- Valiev RZ, Krasilnikov N, Tsenev N. Plastic deformation of alloys with submicron-grained structure. Materials Science and Engineering: A. 1991;137:35–40. [Google Scholar]
- Weber GF, Bjerke MA, DeSimone DW. Integrins and cadherins join forces to form adhesive networks. J Cell Sci. 2011;124:1183–1193. doi: 10.1242/jcs.064618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson HV. On some phenomena of coalescence and regeneration in sponges. Journal of Experimental Zoology. 1907;5:245–258. [Google Scholar]
- Winklbauer R. Mesodermal cell migration during Xenopus gastrulation. Dev Biol. 1990;142:155–168. doi: 10.1016/0012-1606(90)90159-g. [DOI] [PubMed] [Google Scholar]
- Zajac M, Jones GL, Glazier JA. Model of convergent extension in animal morphogenesis. Phys Rev Lett. 2000;85:2022–2025. doi: 10.1103/PhysRevLett.85.2022. [DOI] [PubMed] [Google Scholar]
- Zhang YX, Sivasankar S, Nelson WJ, Chu S. Resolving cadherin interactions and binding cooperativity at the single-molecule level. P Natl Acad Sci USA. 2009;106:109–114. doi: 10.1073/pnas.0811350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Kim HY, Davidson LA. Actomyosin stiffens the vertebrate embryo during critical stages of elongation and neural tube closure. Development. 2009;136:677–688. doi: 10.1242/dev.026211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Bao G, Wang N. Cell mechanics: mechanical response, cell adhesion, and molecular deformation. Annual review of biomedical engineering. 2000;2:189–226. doi: 10.1146/annurev.bioeng.2.1.189. [DOI] [PubMed] [Google Scholar]