SUMMARY
The ER Sec61 translocon is a large macromolecular machine responsible for partitioning secretory and membrane polypeptides into the lumen, cytosol, and lipid bilayer. Because the Sec61 protein-conducting channel has been isolated in multiple membrane-derived complexes, we determined how the nascent polypeptide modulates translocon component associations during defined cotranslational translocation events. The model substrate preprolactin (pPL) was isolated principally with Sec61αβγ upon membrane targeting, whereas higher-order complexes containing OST, TRAP, and TRAM were stabilized following substrate translocation. Blocking pPL translocation by passenger domain folding favored stabilization of an alternate complex that contained Sec61, Sec62 and Sec63. Moreover, Sec62/63 stabilization within the translocon occurred for native endogenous substrates, such as the prion protein, and correlated with a delay in translocation initiation. This data shows that cotranslational translocon contacts are ultimately controlled by the engaged nascent chain and the resultant substrate-driven translocation events.
INTRODUCTION
Approximately 30% of the cellular proteome enters the endoplasmic reticulum (ER) through the Sec61αβγ protein-conducting channel (PCC) and its associated proteins, collectively referred to here as the translocon. The translocon accepts nascent secretory and membrane proteins from the signal recognition particle (SRP) and, together with the translating ribosome, cotranslationally directs their topology by partitioning the elongating polypeptide into the ER lumen, cytosol, and lipid bilayer (Nyathi et al., 2013; Park and Rapoport, 2012; Shao and Hegde, 2011; Skach, 2009). The 10-spanning transmembrane (TM) protein, Sec61α, comprises the PCC core. A central constricted pore accommodates nascent chain during translocation, whereas a site between TM2b and TM7 opens laterally to allow TM segment integration into the adjacent lipid bilayer (du Plessis et al., 2009; Martoglio et al., 1995;; Van den Berg et al., 2004).
Translocon accessory factors adjacent to the PCC are responsible for a variety of activities, such as removal of N-terminal signal peptides by the signal peptidase complex and covalent attachment of N-linked carbohydrates by oligosaccharyltransferase (OST) (Auclair et al., 2012; Kelleher and Gilmore, 2006). Translocation and TM segment integration are facilitated by the translocon-associated protein complex (TRAPαβδγ) (Fons et al., 2003; Sommer et al., 2013). The translocating chain-associated membrane protein (TRAM) resides directly adjacent to signal sequences upon ER targeting as well as to TM segments during membrane insertion (Martoglio et al., 1995). Although TRAM has not been visualized or isolated in translocon complexes directly, it is essential or stimulatory for translocation (Gorlich et al., 1992;; Voigt et al., 1996).
Sec61 also associates with Sec62 and Sec63 in mammalian organisms (Meyer et al., 2000). Similar complexes in S. Cerevisiae mediate post-translational translocation of cytosolic substrates and serve to recruit the ATP-driven ER HSP70, Kar2p (BiP in mammals) that binds incoming nascent chain to facilitate forward movement into the ER lumen (Matlack et al., 1999; Panzner et al., 1995). In mammals, Sec63 has been implicated in the cotranslational mode of ER import for select substrates such as the prion protein (PrP) and ERj3 (Lang et al., 2012). Although Sec63 may associate with ribosome-translocon complexes (RTCs) indirectly through Sec62 (Muller et al., 2010), how it participates in cotranslational translocation remains to be shown.
Analyses of the RTC have generally portrayed the translocon’s organization as rather static apart from conformational shifts in Sec61 itself to accommodate the translocating polypeptide (Menetret et al., 2005; Pfeffer et al., 2014; Potter and Nicchitta, 2000; Snapp et al., 2004; Voorhees et al., 2014). For instance, TRAP and TRAM remain adjacent to Sec61 regardless of the nascent chain’s presence, and accordingly, both crosslink targeted polypeptides (Martoglio et al., 1995; Menetret et al., 2005; Snapp et al., 2004; Wiedmann et al., 1987). Sec61, TRAP, and OST complexes are observed during translocation events but also remain intact after translational termination (Pfeffer et al., 2014; Potter and Nicchitta, 2002). In addition, all cryo-EM structures to date have visualized a large 12–17Å gap between targeted ribosomes and the ER membrane, thereby, failing to identify any obvious structural features that might control directional polypeptide movement into the lumen versus cytosol (Frauenfeld et al., 2011; Pfeffer et al., 2014). However, analyses of nascent chain accessibility using fluorescence quenching indicate that the RTC undergoes specific architectural transitions during the translocation process (Crowley et al., 1994; Liao et al., 1997; Lin et al., 2011).
Here we address whether the nascent chain impacts translocon component associations by isolating Sec61-bound ribosomes during progressive stages of substrate translocation that contain precisely defined nascent chains. The model substrate preprolactin (pPL) was isolated principally with Sec61αβγ upon membrane targeting. pPL translocation occurred by chain lengths of 163 aa and stabilized high-order complexes containing Sec61, OST, TRAP and TRAM. However, an alternate substrate, the prion protein (PrP), that did not initiate translocation until the longer lengths of 188–207 aa, was captured in a 500 kD complex containing Sec61, Sec62, and Sec63. This same complex was stabilized when pPL translocation was blocked by stable passenger domain folding within the RTC. We now demonstrate the presence of Sec62/63 in the translocon during mammalian cotranslational translocation. Taken together our results indicate that translocon associations are modulated by distinct, substrate-driven translocation events.
RESULTS
Strategy to Characterize Functionally-Engaged ER Translocons
ER microsomes have provided a rich source of information on RTC structure and composition. However, even in fresh ER microsome preparations, most RTCs are not engaged in translation and the minor population of active RTCs is producing a diverse array of substrates at variable stages of translocation (Voorhees et al., 2014). To overcome these limitations, in vitro translation in rabbit reticulocyte lysate (RRL) was used to populate canine rough ER microsomes (CRMs) with defined radiolabeled nascent chains that were synchronously captured at distinct stages of biosynthesis. This approach recapitulates translocation events that include signal peptide recognition and cleavage, N-linked glycosylation, nascent chain translocation, and insertion and orientation of TM segments into the lipid bilayer (Devaraneni et al., 2011; Sadlish et al., 2005; Shao and Hegde, 2011; Skach, 2009). The detergent digitonin was employed for membrane solubilization due to its ability to maintain translocon integrity (Devaraneni et al., 2011; Matlack and Walter, 1995; Potter and Nicchitta, 2000; Shibatani et al., 2005).
Solubilized RTCs from total membrane extracts were collected by ultracentrifugation, and ribosome-associated membrane proteins (RAMPs) were released with puromycin and 1M NaCl to obtain a highly enriched population of ER-derived membrane protein complexes that were analyzed by Blue Native Polyacrylamide Gel Electrophoresis (BN-PAGE) (Figures 1A, B and S1A, B). RAMPs were also subjected to SDS-PAGE in the second dimension (2D-BN/SDS-PAGE) to visualize distinct protein complexes as identified in our previous studies by Liquid Chromatography – Tandem Mass Spectrometry (LC-MS/MS) and immunoblotting (Figure 1C) (Shibatani et al., 2005). In total, six different complexes were identified including: Sec61αβγ (~130 kD), TRAPαβγδ (~140 kD), OST (~525 kD; STT3A, ribophorins I & II, OST48, DAD1, KCP2, and DC2), Sec61-TRAP (~300 kD), Sec61-OST (~650 kD), and Sec61-OST-TRAP (~750 kD) (Kelleher and Gilmore, 2006; Shibatani et al., 2005).
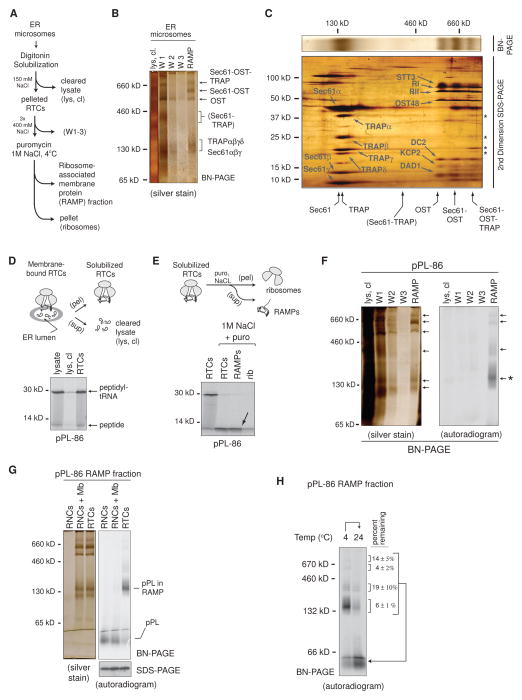
Figure 1. Nascent pPL-86 Binds to a Subset of ER-Derived RAMPs.
(A) Schematic of protocol used to isolate ribosome-associated membrane proteins (RAMPs). (B) Blue native (BN) - PAGE analysis of RAMPs labeled with translocon complexes. (C) 2nd dimension SDS-PAGE analysis of a BN-PAGE gel slice (silver stain) containing translocon components and RAMP complexes as labeled. Asterisks denote TRAP components in the 750 kD RAMP complex. (D) pPL-86 ribosome-translocon complexes (RTCs) were solubilized and pelleted (pel) by ultracentrifugation and analyzed by SDS-PAGE (autoradiogram). Cleared lysate (lys, cl) shows little pPL-86 remaining in the supernatant (sup). (E) SDS-PAGE (autoradiogram) of pPL-86 RTCs before and after NaCl and puromycin (puro) treatment followed by fractionation using ultracentrifugation into RAMP fractions (arrow) and ribosome pellets (rib). (F) BN-PAGE of cleared lysate, wash (W), and RAMP fractions obtained from pPL-86 in vitro translation reactions. (G) pPL-86 ribosome-nascent chain complexes with or without addition of solubilized membranes (RNCs or RNCs + Mb) did not associate with RAMP complexes on BN-PAGE. (H) Incubation of pPL-86 RAMP complexes at 24°C prior to BN-PAGE. Percent nascent chain remaining associated with each RAMP complex after incubation is shown (± SEM, n=3). See also Figure S1.
RAMP Complexes Stably Associate with a Nascent Secretory Substrate
To identify translocon components specifically associated with a substrate, CRMs were programmed by in vitro translation with the model secretory protein preprolactin (pPL). mRNA was initially truncated at codon 86 to generate translationally-arrested radiolabeled polypeptides (pPL-86) that remained stably bound to the ribosome via a covalent peptidyl-tRNA bond and therefore represent physiological translocation intermediates. pPL-86 peptidyl-tRNA pelleted with RTCs, whereas full-length pPL was released into the ER lumen after translational termination (Figures 1D and S1C). Treatment of RTCs with puromycin and 1M NaCl, but not NaCl or puromycin alone, released pPL-86 from ribosomes and into the soluble RAMP fraction where it migrated primarily in a protein complex of ~140 kD on BN-PAGE, similar in size to Sec61αβγ (Figures 1E, F and S1D). Controls show that pPL-86 complexes were specifically formed during the translocation process and were highly stable at 4°C but not 24°C (Figure 1G, H). Surprisingly, only a minor fraction of nascent pPL-86 was found in the three large complexes prominent in silver-stained gels that contained combinations of Sec61, OST, and TRAP.
Nascent Chain Elongation Stabilizes Higher-Order RAMP Complexes
To determine whether different isolated translocon complexes could be stabilized by substrate at different biosynthetic stages, pPL was serially truncated to generate nascent chains with lengths ranging from 86 to 223 aa (near full length). Signal sequence cleavage was first observed at truncation 137 and was completed by a length of 163 aa (Figure 2A). As the nascent chain was elongated, radiolabeled polypeptides isolated in the RAMP fractions were redistributed between five separate complexes on BN-PAGE, labeled A–E (Figure 2B). Prior to signal cleavage, nascent chains migrated principally with complex E. As chain length was extended to 137 aa, a minor RAMP complex C, with an apparent mass of ~500 kD, was observed, whose intensity decreased to ~2% at 163 and 188 aa, but rose to ~5% at 223 aa (Figure 2B and C). At longer lengths, pPL was progressively captured in larger RAMP complexes with a 7-fold increase in the fraction of nascent chains migrating in complexes A and B at truncation 163 (44 ± 5%)(Figure 2C). Thus, pPL elongation results in stabilization of larger 650 and 750 kD RAMP complexes that correlate with signal peptide removal.
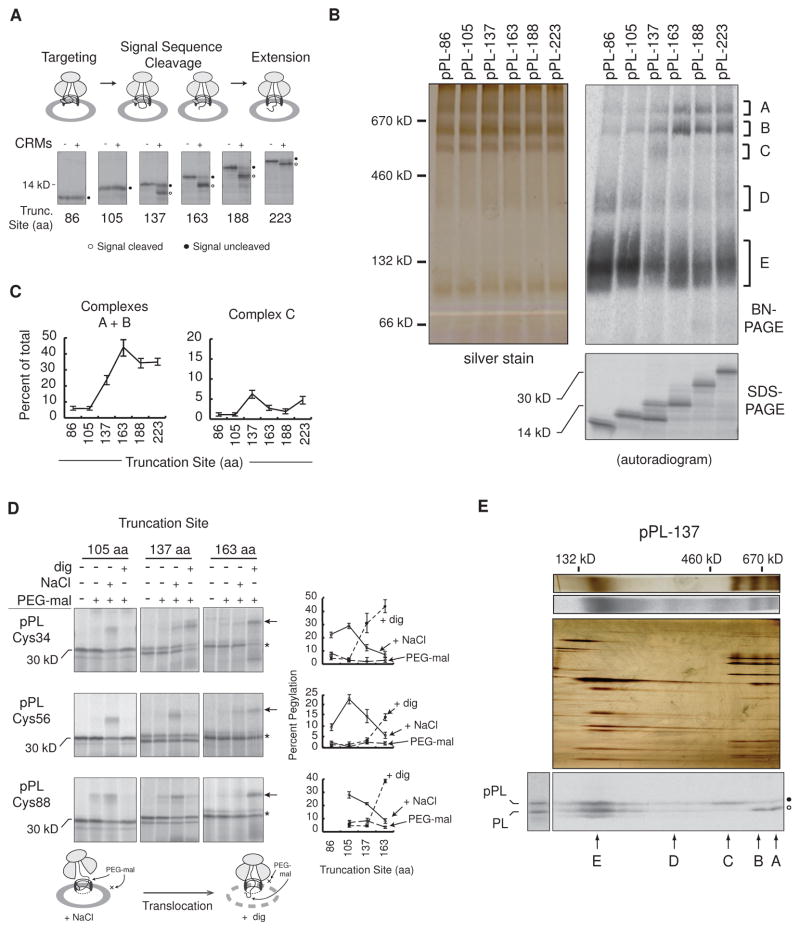
Figure 2. Nascent Chain Translocation Stabilizes Higher-Order RAMP Complexes.
(A) SDS-PAGE of translations ± CRMs at indicated pPL truncations. tRNA was removed with RNase. (B) BN- and SDS-PAGE of RAMPs, corresponding to translations (+CRMs) in panel A. Major complexes A–E are indicated. (C) Mean percent of RAMPs recovered in complexes A and B or C at specified chain lengths ± SEM (n=6). (D) SDS-PAGE of truncated single-cysteine pPL mutants (Cys34, Cys56, Cys88) expressed in CRMs and treated with 0.5M NaCl or digitonin (dig) prior to pegylation (PEG-mal). “*” and “←” indicate unmodified and pegylated peptidyl-tRNA respectively. Schematic diagram illustrates effects of salt and digitonin on assembled RTC intermediates. Mean percent pegylation is shown ± SEM (n ≥ 3). (E) BN-PAGE of pPL-137 RAMPs (top) aligned with corresponding 2D-BN/SDS-PAGE (bottom) labeled with complexes A–E. Silver stains, color; autoradiograms, grayscale. See also Figure S2.
Higher-Order RAMP Complexes Coincide with Substrate Translocation
Because translocation, signal cleavage and recognition of glycosylation motifs often occur coincidentally, we determined which of these events correlated with higher-order RAMP complex formation. The signal sequence consensus site was mutated to eliminate signal peptide removal, whereas non-cognate glycosylation motifs (Q-X-S/T) (Nilsson et al., 2003) were either eliminated or changed into consensus N-X-S/T sites (Figure S2). Despite these changes, a similar proportion of nascent chain was found in higher-order complexes by truncation 163 aa compared to WT pPL (Figure S2). In addition, N-linked glycosylation stabilized one or more additional components in the 650 kD complex.
To determine when the nascent chain entered the ER lumen, endogenous cysteines were removed, and a single cysteine was placed at pPL residue 34, 56, or 88. Microsomes containing truncated translocation intermediates were then incubated in PolyEthyleneGlycol maleimide-5,000 Da (PEG-mal) under conditions that perturb either ribosome binding (high salt) or membrane integrity (digitonin) (Devaraneni et al., 2011). Nascent chains 105 aa and shorter were protected from modification in both intact and permeabilized microsomes but underwent pegylation when the ribosome-translocon junction was disrupted (Figure 2D, left column). The pPL passenger domain, therefore, resided in a cytosolically-restricted space within the RTC at these lengths.
For pPL-137 (Figure 2D, middle column), Cys34 was pegylated after high salt treatment and more effectively following membrane permeabilization, whereas Cys56 and Cys88 were accessible primarily after high salt treatment. Potentially, OST and TRAP could have blocked lumenal Cys56 pegylation at truncation 137, although partial signal cleavage and salt sensitivity indicated that this residue is not fully translocated in all nascent chains. With continued translation to 163 aa, the passenger domain between residues 34 and 88 was pegylated principally in the presence of digitonin (Figure 2D, right panels). Although observed pegylation efficiencies reported on a population average of nascent chain conformations, pegylation at all three probe sites were salt-sensitive at truncation 105 and became digitonin-sensitive at truncation 163. Thus, signal cleavage and pegylation data taken together indicate that pPL translocation has been fully initiated by chain lengths of 163 aa. The 650 and 750 kD complexes of pPL-137 RAMPs contained predominantly signal cleaved polypeptide on 2D-BN/SDS-PAGE (Figure 2E). Translocated as well as signal uncleaved nascent chains were also associated with the 140 kD complex E, whereas the 500 kD complex C only bound signal uncleaved nascent chain.
pPL-Associated RAMPs Contain Sec61, TRAP, OST, and TRAM
Although pPL-associated RAMP complexes migrated at similar sizes as those derived from bulk CRMs, they represented only a minor fraction of total proteins isolated (Figures 2B, compare pPL-86 silver stain and autoradiographic signals). We, therefore, identified components in the pPL-RAMP complexes. Nascent pPL-163 containing a Nε-5-azido-2-nitrobenzoyl-Lys (ANB-Lys) photocrosslinking probe at residue 114 generated a 55 kD Sec61α adduct following UV irradiation that was visualized on 2D-BN/SDS-PAGE in complexes A, B, and E (Figure 3A and B). Thus, these pPL-associated RAMP complexes minimally contained Sec61α. Photoadducts comprised nearly 50% of bands A & B, suggesting either that crosslinking is more favorable in these larger complexes or photoadduct formation may contribute to their stabilization.
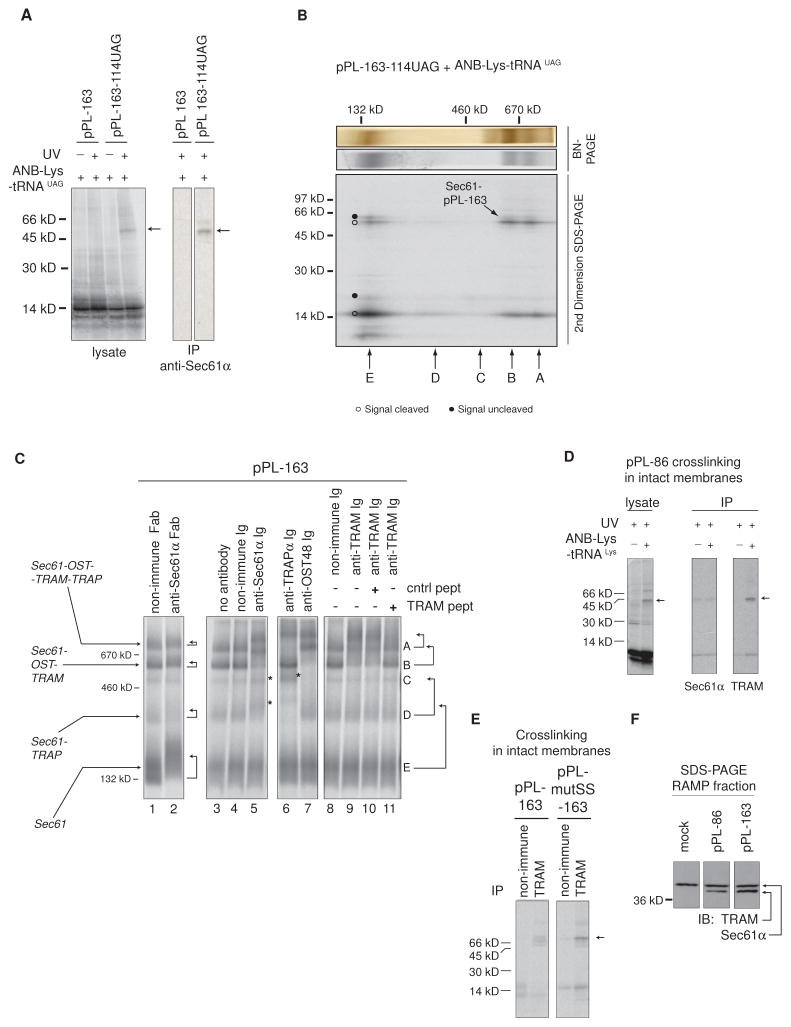
Figure 3. Nascent pPL-Associated RAMPs Contain Sec61, OST, TRAM, and TRAP.
(A) Translation of pPL-163-114UAG in presence of ANB-Lys-tRNAUAG incorporated ANB-Lys into position 114. UV light exposure as indicated (left) induced a 55 kD adduct (arrows) identified as Sec61α by immunoprecipitation (IP) (right). Peptidyl-tRNA was removed with RNase. SDS-PAGE autoradiograms are shown. (B) 2D-BN/SDS-PAGE analysis of photocrosslinked RAMPs showing location of Sec61α photoadduct (autoradiogram) and migration of complexes A–E. (C) Antibody (Ig) and Fab gel-shifts ± indicated peptide as performed on pPL-163-derived RAMPs and analyzed by BN-PAGE (autoradiogram). Arrows indicate changes in band A-E migration. Asterisks denote new autoradiographic signal intensity on top of band C or D resulting from gel-shifts of lower MW bands. Gel-shift subpanels were cropped from separate experiments performed identically. (D) ANB-Lys incorporation into pPL-86 lysine residues by translation in the presence of ANB-Lys-tRNALys produced a 45 kD photo-adduct (arrow) with TRAM as identified by immunoprecipitation (IP). SDS-PAGE autoradiograms are shown. (E) Preventing signal cleavage by pPL mutation (pPL-mutSS as in Figure S2) retained TRAM photocrosslinking (arrow) at truncation 163 as performed in panel D and as revealed by IP of UV-exposed lysates. SDS-PAGE autoradiograms are shown. (F) Western blots (IB) on SDS-PAGE of indicated RAMP fractions for TRAM and Sec61α simultaneously. IPs and IBs within panels were cropped from non-adjacent lanes on the same gel..
To identify additional components, we employed a gel-shift assay by incubating RAMP complexes with purified antibodies (Ig) prior to BN-PAGE. For pPL-163, anti-Sec61α Ig (~150 kD) and/or Fab antibody fragments (~50 kD) slowed migration of bands A, B, D, and E according to the immune protein’s size (Figure 3C, lanes 1–5). Notably, bands E and D were shifted by Ig near or directly on top of bands D and C, respectively (Figure 3C, asterisks). Antibodies against the OST subunit, OST48, shifted only bands A and B, (Figures 3C lane 7) as expected (Shibatani et al., 2005). Anti-TRAPα Ig primarily altered migration of bands A and D, but not band B (Figure 3C lane 6), again in good agreement with previous studies (Shibatani et al., 2005). Thus, we can assign with some certainty the identity of pPL-associated RAMPs, although we cannot rule out the possibility that a small proportion of other RAMP components might also co-migrate with complexes A-E on BN-PAGE.
Surprisingly, anti-TRAM Ig also shifted bands A and B (Figure 3C, lane 8 vs. 9), even though TRAM was not previously reported in bulk RAMP complexes (Figure 1C and (Potter and Nicchitta, 2000; Shibatani et al., 2005)). Binding specificity was confirmed by peptide competition (Figure 3C, lane 10 vs. 11). As previously reported, pPL’s signal sequence photocrosslinked TRAM in intact membranes at truncation 86 (Martoglio et al., 1995) as well as at truncation 163 after pPL mutations were introduced to eliminate signal cleavage (pPL-mutSS) (Figure 3D, E, arrow, and S2A). This indicates that TRAM resides near the nascent chain at all lengths. Accordingly, TRAM was detected by western blot in pPL-86 and pPL-163 RAMP fractions (Figure 3F). Thus, results in Figure 2 and 3 provide evidence that pPL translocon stabilizes interactions between Sec61, OST, TRAP, and TRAM as the nascent chain is targeted, extended, and translocated into the ER lumen.
Blocking pPL Translocation Stabilizes RAMP Complex C
To test our hypothesis that these events are triggered specifically by translocation, we performed “loss-of-function” experiments by preventing pPL lumenal entry. pPL residues 45–75 or 55–85 were replaced with a small zinc finger domain (ZnF) (pPL45-Zn and pPL55-Zn, respectively) that stably folds in the presence of Zn+2 and sterically blocks translocation (Figure 4A) (Conti et al., 2014). Thus, in the presence of Zn+2, ~70% of pPL45-Zn remained signal uncleaved and was susceptible to proteinase K digestion (Figure 4B). Under these conditions, pPL45-Zn began transitioning into BN-PAGE complex C at truncation 137, where it remained as synthesis continued (Figure 4C, right panel). Ultimately ~41% of nascent chains resided in RAMP complex C at truncation 188 (Figure 4D). pPL55-Zn polypeptides also failed to undergo efficient signal cleavage in the presence of Zn+2, trapping the uncleaved population in complex C (Figures 3E and S3A,B). In the absence of Zn+2, RAMPs were distributed between complexes A-E, similar to WT pPL (Figure 4C, left panel). Band D, although present, did not resolve well in some BN-PAGE gel preparations (Figure 4C, right panel). Zn+2 treatment had no effect on WT pPL RAMPs (Figure S3C). Surprisingly, approximately 88% and 95% of pPL-163 and pPL45-Zn-163 (produced in the presence of Zn+2) nascent chains respectively were retained within complex C after a 1 hour incubation at 24°C (Figure 4F and G). In contrast, pPL’s association with Sec61 (complex E) was stabilized at chain lengths of 163 aa, potentially because the translocating nascent chain was passing directly through the PCC channel.
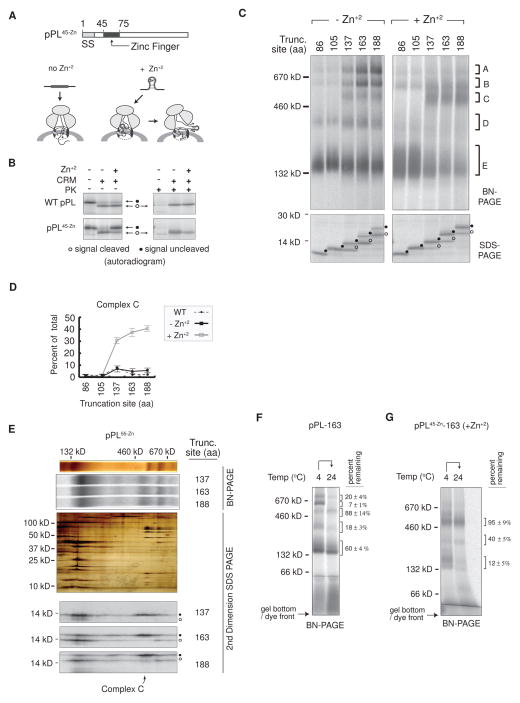
Figure 4. Blocking pPL Translocation Stabilizes RAMP Complex C.
(A) Schematic of pPL45-Zn construct showing effect of Zn+2 on passenger translocation. (B) SDS-PAGE of pPL and pPL45-Zn translated ± CRMS, ± Zn+2 both before and after proteinase K digestion (PK). (C) BN- and SDS-PAGE of RAMPs derived from the indicated pPL45-Zn truncations that were produced in the presence (right) and absence (left) of Zn+2. (D) Mean percent of RAMPs recovered in complex C at specified chain lengths ± SEM (n=3). (E) From top to bottom. Silver stain and corresponding autoradiogram of 1D BN-PAGE gel from pPL55-Zn construct truncated at indicated residues and translated in presence of Zn+2. Middle panel is a silver-stained representative 2D-BN/SDS-PAGE gel aligned with upper gel slices showing composition of RAMP complexes. Bottom panels show autoradiograms of 2D-BN/SDS-PAGE gels derived from indicated translation reactions. (F),(G) Percent of nascent chains remaining associated with each RAMP complex after 24°C incubation for pPL-163 and pPL45-Zn-163 (produced in the presence of Zn+2) ± SEM, n=3. See also Figure S3.
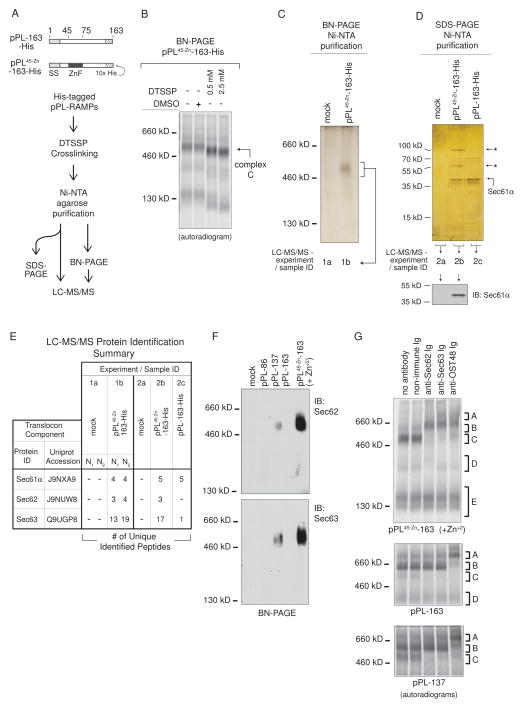
Complex C Contains Sec61, Sec62, and Sec63
To identify components within complex C, we developed a His-tag-based purification strategy (Figure 5A). Translations were carried out in the presence of Zn+2 using pPL-163 and pPL45-Zn-163 containing a 10x C-terminal His-tag (pPL-163-His and pPL45-Zn-163-His respectively) to allow isolation of the corresponding RAMPs. Complex C integrity was maintained during purification by treatment with the reversible, bifunctional crosslinker 3,3′-dithiobis[sulfosuccinimidylpropionate] (DTSSP). Crosslinked complex C migrated slightly faster on BN-PAGE with no detectable additional intercomplex associations (Figure 5B). Analysis of samples on reduced versus non-reduced SDS-PAGE showed crosslinks were indeed reversible with DTT treatment (Figure S4A and B).
Figure 5. RAMP Complex C Contains Sec61, Sec62, and Sec63.
(A) Purification scheme used to isolate and identify pPL-bound RAMP components. “-His” refers to truncations encoding a 10x His-tag. In panels (B)–(E), all RAMPs were derived from translations in the presence of Zn+2. (B) BN-PAGE of pPL45-Zn-163-His RAMPs treated with DMSO or DTSSP. (C) BN-PAGE (silver stain) of pPL45-Zn-163-His RAMP purifications confirmed complex C isolation. (D) SDS-PAGE of purified pPL45-Zn-163-His RAMPs, compared to those of mock control and pPL-163-His, indicated isolation of 55 and 100 kD proteins (top, silver stain, “←*”). Western blot (bottom) identified the 38 kD band as Sec61α. (E) Samples from experiment 1 and 2, as prepared and designated in panels C and D, were analyzed by LC-MS/MS. Translocon components identified in pPL45-Zn-163-containing samples (1b and 2b) are listed along with numbers of peptide matches, as compared to those identified in mock control (1a and 2a) or pPL-163-His (2c) purified RAMP samples. N1 and N2 are analyses of repeated experiments performed on different days. (F) Western blots confirmed complex C generated from pPL-137 and pPL45-Zn-163(+Zn+2) (ie pPL45-Zn-163 translated in the presence of Zn+2) contained Sec62 and Sec63. (G) Gel-shifts of complex C derived from pPL45-Zn-163(+Zn+2), pPL-163, and pPL-137 translations with indicated antibodies. See also Figure S4 – 6.
Ni-NTA purification of pPL45-Zn-163-His RAMPs followed by BN-PAGE analysis confirmed complex C isolation (Figure 5C), and SDS-PAGE revealed 55 and 100 kD proteins that were absent in those derived from mock control or pPL-163-His translations (Figure 5D). The prominent 38 kD band in experimental samples was identified as Sec61α by western blot (Figure 5D). Other translocon antibodies failed to react against their corresponding component after DTSSP modification (data not shown).
Therefore LC-MS/MS was performed on samples derived directly from complex C cut from BN-PAGE gels (Figure 5C, 1a–b) from two separate experiments (N1 and N2) and on samples eluted directly from Ni-NTA beads from an additional experiment (Figure 5D, 2a–c). Resultant LC-MS/MS spectra were compared to a canine protein sequence database supplemented with the human Sec63 due to the absence of the full-length canine homologue. Protein identification required two or more high probability peptide matches. Surprisingly, Sec62 (55 kD) and Sec63 (100 kD) were identified in pPL45-Zn-derived samples (Figure 5E). A single peptide for Sec63 was found in the pPL-163-His sample and likely originated from small amounts of complex C observed at this truncation (Figure 2B and 3B). Sec61α was present in all His-tagged samples, consistent with western blot results. Sequences corresponding to Sec61, Sec62, and Sec63, as well as additional peptides identified, were compiled in Figure S5 and S6.
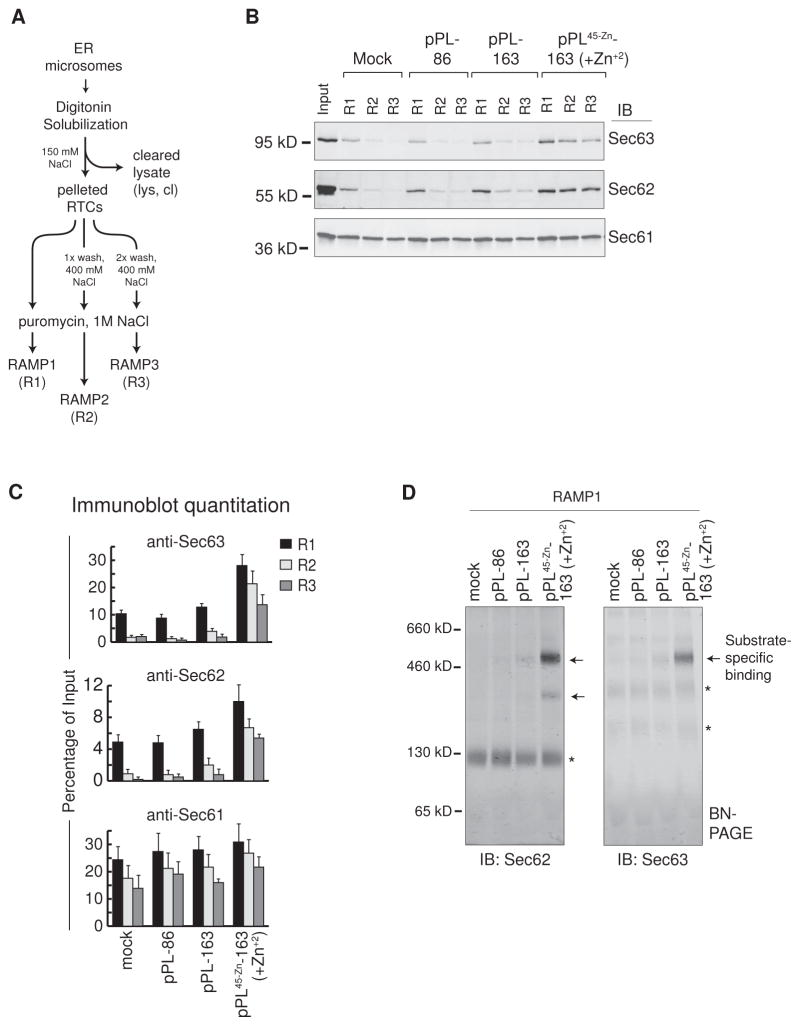
BN-PAGE western blots confirmed that Sec62 and Sec63 were contained within the 500 kD complex C in RAMPs prepared from pPL137 and pPL45-Zn-163 (+Zn+2) (i.e., pPL45-Zn -163 translations in the presence of Zn+2) (Figure 5F). TRAM was not abundant in pPL45-Zn -163 (+Zn+2) RAMPs compared to those derived from WT pPL (Figure S4C). Gel-shift experiments further demonstrated the presence of Sec62/63 in complex C (Figure 5G). Complex C also contained OST48. Anti-Sec62 also shifted complex D but only from pPL45-Zn -163 (+Zn+2) RAMPs. Taken together, results from Figure 5 indicate that complex C contains Sec61, Sec62, Sec63 and potentially a subset of other translocon components. Interestingly, pPL45-Zn -163 (+Zn+2)-driven stabilization of Sec62/63 binding was apparent even in RAMP fractions that were released after only one or two rounds of RTC pelleting and resuspension (ie RAMP1 or RAMP2 fractions), when compared to mock, pPL-86, and pPL-163 (Figure 6A–C). Western blots confirmed that the 500 kD complex C was present in RAMP1 fractions of pPL45-Zn -163 (+Zn+2), while being a minor component or absent in comparable mock, pPL-86 and pPL-163 samples (Figure 6D, arrows). Sec62 was also bound to pPL45-Zn -163 (+Zn+2) at ~ 300 kD on BN-PAGE, consistent with gel-shift experiments (Figure 5G).
Figure 6. Sec62/63 is Associated with Complex C upon Initial Membrane Solubilization.
(A) Schematic of RAMP1, 2, and 3 (R1, R2, and R3) isolation procedure consisting of one, two, or three rounds of RTC pelleting and resuspension before RAMP release. (B) Immunoblots of Sec63, Sec62, and Sec61 in RAMP fractions 1–3 that were generated by translation of the indicated nascent chains as compared to input (left lane). (C) Quantitation of immunoblots as in panel B. Percent of Sec63, Seec62, or Sec61 remaining in RAMP fractions 1–3 as compared to input is shown for indicated nascent chain translations ± SEM (n=3).(D) Immunoblots of RAMP1 fractions show location of Sec62 and Sec63 complexes on BN-PAGE with and without translated nascent chains (arrows and asterisks respectively).
These results indicate that altering translocation events, independent of the primary nascent chain sequence, stabilizes RTC associations of different translocon components. In particular, if pPL nascent chain was extended beyond ~130–160 residues and failed to initiate translocation, the Sec62/63 complex C becomes the major RAMP component. Interestingly, translocation of an alternate endogenous substrate, the prion protein (PrP), was reported to require Sec63 for translocation (Lang et al., 2012). PrP contains a large intrinsically disordered N-terminal domain and its translocation is dependent upon synthesis of helical domains located at residues 172–226 (Pfeiffer et al., 2013). Thus, we wondered whether PrP translocation initiation might be delayed in vitro and whether it might also stabilize Sec61/62/63 complexes.
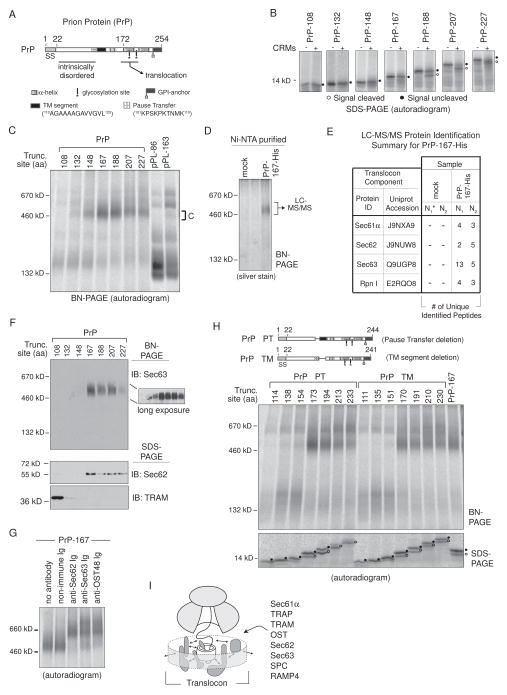
PrP Stabilizes Sec62 and Sec63 RAMP Complexes
PrP is a topologically complex protein that contains an N-terminal signal sequence, a hydrophilic pause-transfer sequence that immediately precedes a weak TM segment, and two C-terminal helical domains that are reportedly required for translocation due to the aforementioned intrinsically disordered N-terminal domain (Figure 7A) (Miesbauer et al., 2009; Nakahara et al., 1994; Pfeiffer et al., 2013). A glycophosphatidylinositol (GPI) -anchor site causes C-terminal cleavage and a S231 linkage to membrane lipids in cells. As a result, PrP can take three different topologies but predominantly acquires a signal cleaved and secreted form when synthesized in RRL (Lopez et al., 1990).
Figure 7. PrP Stabilizes Sec62 and Sec63 RAMP Complexes.
(A) A schematic of the prion protein (PrP) indicates location of signal sequence (SS), an N-terminal intrinsically disordered region, glycosylation sites, a GPI-anchor site, two helical domains, a pause-transfer sequence, and a TM segment. (B) SDS-PAGE analysis of translated PrP RTC intermediates ± CRMs. Peptidyl-tRNA was removed with RNase. (C) BN-PAGE of RAMPs prepared from indicated PrP and pPL translations. (D) Complex C isolation from PrP-167-His RAMP fractions by Ni-NTA purification after DTSSP crosslinking. (E) LC-MS/MS identified proteins from excised ~500 kD gel bands generated from mock or PrP-167-His Ni-NTA purified RAMP complexes as in (D). The asterisk denotes mock sample N1 that served as the control in a single experiment containing both pPL45-Zn-163-His (Figure 5E) and PrP-167-His samples. (F) Western blots of PrP RAMP fractions analyzed on BN- and SDS-PAGE. (G) Gel-shifts of PrP-167 RAMP complex C with indicated antibodies. (H) BN-PAGE of RAMPs prepared from indicated PrP truncations which lack the pause-transfer (PrP ΔPT) or TM segment (PrP ΔTM). (I) General model of dynamic translocon contacts (arrows) driven by nascent chain that could reflect repositioning of components within the RTC, recruitment of new factors, or affinity changes between bound components and/or polypeptide. See also Figure S5 and S7.
PrP signal cleavage was not completed until chain lengths of 207–227 aa (Figure 7B), roughly corresponding to helical domain emergence from the ribosomal exit tunnel. We therefore tested whether the Sec62/63 RAMP complex C was stabilized at similar lengths compared to pPL45-Zn -163 (+Zn+2). Indeed, PrP association with complex C first occurred at truncations 132 and 148, peaked at truncations 167 and 188, and then weakened upon signal cleavage (Figure 7C). At longer chain lengths, 207 and 227 aa, nascent chains partially transitioned into larger complexes. To confirm the observed 500 kD complex contained Sec61/62/63, the His-tag-based purification strategy was used to isolate complex C derived from PrP-167-His RAMP samples (Figure 7D), and the band was processed for LC-MS/MS analysis in two separate experiments. As expected, Sec61, Sec62, and Sec63 were predominant proteins within complex C (Figure 7E). The OST subunit ribophorin I (Rpn I) was also identified. Translocation protein sequences identified are listed in Figure S5. Immunoblotting confirmed that Sec63 was contained in PrP complex C and Sec62 was also detected in PrP-167, 188, 207, and 227 RAMP fractions (Figure 7F), which correlated with complex C association. TRAM associated with PrP-108 RTCs, although it did not bind efficiently at longer chain lengths (Figure 7F). Full or partial gel-shifts further confirmed the presence of Sec61, Sec62, and Sec63 as well as OST48 in complex C (Figure 7G). Finally, deletion of either the pause-transfer (PrP ΔPT) or TM segment (PrP ΔTM), which reduced the nascent chain’s hydrophilicity or hydrophobicity respectively, had little effect on stabilization of complex C (Figure 7H). However, TM segment deletion delayed or inhibited signal cleavage. Thus, Sec62/63 interactions with a native substrate are strengthened for prolonged durations throughout cotranslational translocation.
DISCUSSION
We examined solubilized translocon complexes formed by programming membrane-bound RTCs with nascent chains synchronously arrested at specific biosynthetic stages. Because the only variable in these experiments is nascent chain length, the dramatic changes observed here must reflect changes in translocon contacts driven by the nascent chain itself (Figure 7I). Upon ER targeting, nascent chains remained associated principally with Sec61αβγ heterotrimers in RAMP fractions and relatively weakly with other translocon components. As the nascent chain was extended, a greater proportion of polypeptide was captured in higher-order RAMP complexes dependent upon the ensuing translocation event. For pPL, efficient translocation initiation at lengths of ~163 aa stabilized association of additional components that minimally included OST, TRAM, and TRAP. In contrast, 500 kD Sec61/62/63 complexes were stabilized if the nascent chain failed to initiate translocation (as judged by signal cleavage) due to either experimentally-induced passenger folding within the RTC (pPL45-Zn) or the natural biosynthesis of an endogenous substrate (PrP). These findings demonstrate that the Sec61 translocon is not static in the ER membrane but undergoes dynamic transitions during specific translocation events that are driven by topogenic behavior of the nascent substrate.
A number of different mechanisms could be responsible for the observed variations in RAMP complex distribution. On one hand, the translocon’s composition could remain unaltered regardless of bound nascent chain, and therefore, different RAMP complexes would either reflect component repositioning within the translocon, new contacts with the nascent chain itself, or affinity changes between components due to conformational shifts. Upon isolation, the translocon complex would therefore disassemble differently, or to varying degrees, dependent upon the nascent chain. On the other hand, components within the ER membrane could be recruited more favorably immediately preceding or during translocation events, which would directly lead to different RAMP complex composition. Importantly, both mechanisms could be utilized to varying degrees dependent upon the translocon component. The former model would more easily explain several observations in the current study including the presence of Sec62/63 in all RAMP1 fractions (Figure 6), the association of translocated nascent chain (signal cleaved and glycosylated) with Sec61 alone (Figure 2E and S2H, complex E), some pPL-86 association with higher-order RAMP complexes A and B (Figure 2B and C), and the presence of OST and TRAP in RAMP fractions without any association to Sec61 (Figure 1B and C). Most studies suggest the translocon is static and components, such as TRAM, TRAP and OST, are contained within the complex continuously (Menetret et al., 2005; Muller et al., 2010; Pfeffer et al., 2012; Potter and Nicchitta, 2002; Snapp et al., 2004). Currently, less evidence exists for different translocon populations in intact membranes before solubilization (Pfeffer et al., 2014). Although distinguishing between these models remains challenging in the membrane environment, current results clearly indicate translocon contacts can be altered significantly, dependent upon nascent chain synthesis and the ensuing translocation events.
Attesting to both the validity of the experimental approach and the translocon’s dynamic state, inefficient translocation initiation as nascent chain extends beyond ~130–160 aa markedly reconfigures translocon contacts and results in strong stabilization of Sec61/62/63 RAMP complexes. Interestingly, this can be accomplished either by artificially blocking translocation with a folded domain (pPL45-Zn) or by the lack of structured domains in the passenger region altogether (PrP). In S. Cerevisiae, Sec61/62/63 homologues, as well as Sec71p and Sec72p which are absent in mammalian organisms, constitute an SRP-independent, post-translational translocation complex (Panzner et al., 1995). In this system, Sec63p (Sec63 in mammals) provides a J-domain, which recruits the ATP-driven ER HSP70 chaperone Kar2p (BiP in mammals) to ratchet the nascent chain into the lumen (Matlack et al., 1999; Misselwitz et al., 1998). Mammalian Sec62 has gained the additional ability to bind ribosomes (Muller et al., 2010), which may allow Sec62/63 to participate in a similar mechanism to import substrate cotranslationally.
An attractive theory is that the stabilization of Sec62/63-containing complexes by nascent chain correlates with its dependence on Sec62/63 for translocation. PrP was recently reported to be partially dependent upon Sec63 for translocation (Lang et al., 2012). In fact, an additional substrate ERj3, whose translocation depends completely on Sec63 (Lang et al., 2012), also strongly binds Sec61/62/63 RAMP complexes (Figure S7). Thus, Sec62/63 association with the translocon may be strengthened at critical stages of translocation. In this role, Sec62/63 may actually assist in the translocation of select substrates (e.g. PrP and ERj3) and not others (Lang et al., 2012), similar to OST’s utilization only when a glycosylation consensus site exists. Minimally, stable Sec63 association with the translocon provides a direct mechanism for recruitment of BiP, which is critical in mediating cotranslational ER import and possibly translocon gating (Brodsky et al., 1995; Haigh and Johnson, 2002; Young et al., 2001).
Therefore, a primary question for future investigations is what translocon contacts are introduced or altered that cause Sec62/63 stabilization? Because of limited space within the RTC, nascent chain may begin to sample or reside in the cytosol if translocation initiation is delayed. For proteins with N-terminal signal sequences this would therefore appear to occur at lengths of 130–60 aa. Nascent chain accumulation on the cytosolic side of the membrane could weaken the RTC junction, thereby altering translocon contacts and leading to strengthened Sec62/63 binding (Devaraneni et al., 2011; Voigt et al., 1996). Alternatively, Sec62/63 could exist as a peripheral sensor, engaging substrates that sample the cytosolic environment. Indeed, peripheral proteins have been visualized in RTC structures (Pfeffer et al., 2012). Regardless, Sec62/63 appears to participate in deciding whether nascent chain on the cytosolic side of the membrane will be translocated, positioned in the cytosol, or alternatively removed by ER-associated degradation machinery (Jung et al., 2014; Reithinger et al., 2013; Rubenstein et al., 2012).
Taken together, our results now indicate that transitions in Sec61 translocon organization and/or stability are triggered by distinct translocation events. These results open up multiple questions to be addressed in future studies. Structurally, how do TRAM, Sec62, and Sec63 interact with the RTC? Because membrane protein biogenesis requires both ER and cytosolic delivery of substrate, how do these events influence translocon component interactions? How do results here reflect complexes formed as translation proceeds in vivo with faster kinetic rates? Understanding how dynamic changes in the ER translocon architecture mediate these complex events remains a fundamental challenge in the cell biology of protein trafficking.
EXPERIMENTAL PROCEDURES
In Vitro Transcription and Translation
All bovine preprolactin and hamster prion protein plasmids were constructed in the pSP64T vector (Promega). pPL45-Zn and pPL55-Zn replaced 29 pPL amino acids starting at the designated residue with the S. Cervisiae ADR1a zinc finger sequence. RNA transcripts were synthesized using SP6 polymerase from PCR- generated cDNA templates that were full-length or truncated at the indicated open reading frame codon. Unless otherwise indicated translations were carried out at 24°C in 40% (v/v) rabbit reticulocyte lysate containing 35S-methionine and CRMs to generate radiolabeled nascent chains.
RAMP Isolation and BN-PAGE Analysis
Pelleted CRMs from translation reactions were lysed in buffer containing 1.2% (w/v) digitonin. Solubilized RTCs were collected and washed by repeated rounds of ultracentrifugation and resuspension. RAMPs were eluted with 1mM puromycin and 1M NaCl for BN-PAGE analysis. Samples were kept at 4°C throughout. Proteins were visualized by silver staining or phosphorimaging of 35S-containing polypeptides. Chemical crosslinking was performed with 2 mM DTSSP at 4°C. For antibody gel-shifts, purified Igs or Fab fragments were incubated with RAMP complexes (and competing peptide where indicated) immediately before BN-PAGE. For 2D- BN/SDS-PAGE, excised BN-PAGE gel strips were applied directly on top of SDS-PAGE gels (Shibatani et al., 2005). Where indicated RAMP samples were analyzed by SDS- or BN-PAGE, transferred to PVDF membrane, and immunoblotted with indicated antibodies.
Pegylation Assay
CRMs containing specified translation products were pelleted and resuspended, treated with 0.5 M NaCl or 1% (w/v) digitonin, and incubated with 2 mM PEG-mal. Samples were incubated at 4°C for 1 hour and quenched with 200 mM DTT prior to SDS-PAGE and phosphorimaging.
Photocrosslinking
Nε-5-azido-2-nitrobenzoyl-Lys (ANB-Lys) was incorporated into nascent chains at lysines or the engineered UAG codon by supplementing translation reactions with ANB-Lys-tRNALys or ANB-Lys-tRNAUAG respectively. Translation products were irradiated with UV light to activate the ANB moiety and photocrosslink adjacent proteins prior to denaturation, RNase treatment, and SDS-PAGE.
Ni-NTA Purification and Immunoprecipitation
Translation reactions or DTSSP crosslinked RAMPs were denatured in 1% (w/v) SDS, diluted 20-fold in 1% (v/v) Triton X-100 in phosphate buffer, and incubated with Ni-NTA-agarose or with Protein A-agarose beads and antibody as indicated. Beads were washed extensively preceding analysis by PAGE or LC-MS/MS.
LC – MS/MS
Reduced, alkylated, and trypsinized samples were analyzed by nanospray LC-MS/MS on Thermo Scientific LTQ Velos or Orbitrap Fusion mass spectrometers. Spectra were matched to canine protein sequences with the Proteome Discoverer 1.4 software package, using the Sequest HT and Percolator modules. Identified peptides assigned with a false discovery rate q-value of 0.01 or less were considered high confidence matches.
Supplementary Material
Acknowledgments
We thank R. Zimmermann, R. Gilmore, S. High, and K.E. Matlack for providing antibody reagents. This work was supported by US National Institutes of Health (NIH) grants GM53457 (W.R.S.), DK51818 (W.R.S.), F32 GM083568 (B.J.C.), and T32 HL083808 (B.J.C.), by NIH core grants P30EY010572 and P30CA069533, by the NIH shared Instrument grant S10OD012246, and by the Cystic Fibrosis Foundation Therapeutics grant SKACH05XX0 (W.R.S.).
Footnotes
AUTHOR CONTRIBUTIONS
B.J.C. conceived the project, designed and executed experiments, analyzed results, and wrote the manuscript. P.K.D. contributed to the conceptual development of the project. Z.Y. designed and generated molecular biology reagents. L.L.D. performed the LC-MS/MS analysis. W.R.S. designed experiments, analyzed results, and assisted in writing the manuscript.
Supplemental information includes seven figures and Supplemental Experimental Procedures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Auclair SM, Bhanu MK, Kendall DA. Signal peptidase I: cleaving the way to mature proteins. Protein Sci. 2012;21:13–25. doi: 10.1002/pro.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL, Goeckeler J, Schekman R. BiP and Sec63p are required for both co- and posttranslational protein translocation into the yeast endoplasmic reticulum. Proc Natl Acad Sci U S A. 1995;92:9643–9646. doi: 10.1073/pnas.92.21.9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti BJ, Elferich J, Yang Z, Shinde U, Skach WR. Cotranslational folding inhibits translocation from within the ribosome-Sec61 translocon complex. Nat Struct Mol Biol. 2014;21:228–235. doi: 10.1038/nsmb.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley KS, Liao S, Worrell VE, Reinhart GD, Johnson AE. Secretory proteins move through the endoplasmic reticulum membrane via an aqueous, gated pore. Cell. 1994;78:461–471. doi: 10.1016/0092-8674(94)90424-3. [DOI] [PubMed] [Google Scholar]
- Devaraneni PK, Conti B, Matsumura Y, Yang Z, Johnson AE, Skach WR. Stepwise insertion and inversion of a type II signal anchor sequence in the ribosome-Sec61 translocon complex. Cell. 2011;146:134–147. doi: 10.1016/j.cell.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Plessis DJ, Berrelkamp G, Nouwen N, Driessen AJ. The lateral gate of SecYEG opens during protein translocation. J Biol Chem. 2009;284:15805–15814. doi: 10.1074/jbc.M901855200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fons RD, Bogert BA, Hegde RS. Substrate-specific function of the translocon-associated protein complex during translocation across the ER membrane. J Cell Biol. 2003;160:529–539. doi: 10.1083/jcb.200210095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frauenfeld J, Gumbart J, Sluis EO, Funes S, Gartmann M, Beatrix B, Mielke T, Berninghausen O, Becker T, Schulten K, et al. Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nat Struct Mol Biol. 2011;18:614–621. doi: 10.1038/nsmb.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D, Hartmann E, Prehn S, Rapoport TA. A protein of the endoplasmic reticulum involved early in polypeptide translocation. Nature. 1992;357:47–52. doi: 10.1038/357047a0. [DOI] [PubMed] [Google Scholar]
- Haigh NG, Johnson AE. A new role for BiP: closing the aqueous translocon pore during protein integration into the ER membrane. J Cell Biol. 2002;156:261–270. doi: 10.1083/jcb.200110074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung SJ, Kim JE, Reithinger JH, Kim H. The Sec62-Sec63 translocon facilitates translocation of the C-terminus of membrane proteins. J Cell Sci. 2014;127:4270–4278. doi: 10.1242/jcs.153650. [DOI] [PubMed] [Google Scholar]
- Kelleher DJ, Gilmore R. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology. 2006;16:47R–62R. doi: 10.1093/glycob/cwj066. [DOI] [PubMed] [Google Scholar]
- Lang S, Benedix J, Fedeles SV, Schorr S, Schirra C, Schauble N, Jalal C, Greiner M, Hassdenteufel S, Tatzelt J, et al. Different effects of Sec61alpha, Sec62 and Sec63 depletion on transport of polypeptides into the endoplasmic reticulum of mammalian cells. J Cell Sci. 2012;125:1958–1969. doi: 10.1242/jcs.096727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S, Lin J, Do H, Johnson AE. Both lumenal and cytosolic gating of the aqueous ER translocon pore are regulated from inside the ribosome during membrane protein integration. Cell. 1997;90:31–41. doi: 10.1016/s0092-8674(00)80311-6. [DOI] [PubMed] [Google Scholar]
- Lin PJ, Jongsma CG, Liao S, Johnson AE. Transmembrane segments of nascent polytopic membrane proteins control cytosol/ER targeting during membrane integration. J Cell Biol. 2011;195:41–54. doi: 10.1083/jcb.201103117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez CD, Yost CS, Prusiner SB, Myers RM, Lingappa VR. Unusual topogenic sequence directs prion protein biogenesis. Science. 1990;248:226–229. doi: 10.1126/science.1970195. [DOI] [PubMed] [Google Scholar]
- Martoglio B, Hofmann MW, Brunner J, Dobberstein B. The protein-conducting channel in the membrane of the endoplasmic reticulum is open laterally toward the lipid bilayer. Cell. 1995;81:207–214. doi: 10.1016/0092-8674(95)90330-5. [DOI] [PubMed] [Google Scholar]
- Matlack KE, Walter P. The 70 carboxyl-terminal amino acids of nascent secretory proteins are protected from proteolysis by the ribosome and the protein translocation apparatus of the endoplasmic reticulum membrane. J Biol Chem. 1995;270:6170–6180. doi: 10.1074/jbc.270.11.6170. [DOI] [PubMed] [Google Scholar]
- Matlack KES, Misselwitz B, Plath K, Rapoport TA. BiP Acts as a Molecular Ratchet during Posttranslational Transport of Prepro-α Factor across the ER Membrane. Cell. 1999;97:553–564. doi: 10.1016/s0092-8674(00)80767-9. [DOI] [PubMed] [Google Scholar]
- Matsumura Y, Rooney L, Skach WR. In vitro methods for CFTR biogenesis. Methods Mol Biol. 2011;741:233–253. doi: 10.1007/978-1-61779-117-8_16. [DOI] [PubMed] [Google Scholar]
- Menetret JF, Hegde RS, Heinrich SU, Chandramouli P, Ludtke SJ, Rapoport TA, Akey CW. Architecture of the ribosome-channel complex derived from native membranes. J Mol Biol. 2005;348:445–457. doi: 10.1016/j.jmb.2005.02.053. [DOI] [PubMed] [Google Scholar]
- Meyer HA, Grau H, Kraft R, Kostka S, Prehn S, Kalies KU, Hartmann E. Mammalian Sec61 Is Associated with Sec62 and Sec63. Journal of Biological Chemistry. 2000;275:14550–14557. doi: 10.1074/jbc.275.19.14550. [DOI] [PubMed] [Google Scholar]
- Miesbauer M, Pfeiffer NV, Rambold AS, Muller V, Kiachopoulos S, Winklhofer KF, Tatzelt J. alpha-Helical domains promote translocation of intrinsically disordered polypeptides into the endoplasmic reticulum. J Biol Chem. 2009;284:24384–24393. doi: 10.1074/jbc.M109.023135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misselwitz B, Staeck O, Rapoport TA. J proteins catalytically activate Hsp70 molecules to trap a wide range of peptide sequences. Mol Cell. 1998;2:593–603. doi: 10.1016/s1097-2765(00)80158-6. [DOI] [PubMed] [Google Scholar]
- Muller L, de Escauriaza MD, Lajoie P, Theis M, Jung M, Muller A, Burgard C, Greiner M, Snapp EL, Dudek J, et al. Evolutionary gain of function for the ER membrane protein Sec62 from yeast to humans. Mol Biol Cell. 2010;21:691–703. doi: 10.1091/mbc.E09-08-0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara DH, Lingappa VR, Chuck SL. Translocational pausing is a common step in the biogenesis of unconventional integral membrane and secretory proteins. J Biol Chem. 1994;269:7617–7622. [PubMed] [Google Scholar]
- Nilsson I, Kelleher DJ, Miao Y, Shao Y, Kreibich G, Gilmore R, von Heijne G, Johnson AE. Photocross-linking of nascent chains to the STT3 subunit of the oligosaccharyltransferase complex. J Cell Biol. 2003;161:715–725. doi: 10.1083/jcb.200301043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyathi Y, Wilkinson BM, Pool MR. Co-translational targeting and translocation of proteins to the endoplasmic reticulum. Biochimica et biophysica acta. 2013;1833:2392–2402. doi: 10.1016/j.bbamcr.2013.02.021. [DOI] [PubMed] [Google Scholar]
- Panzner S, Dreier L, Hartmann E, Kostka S, Rapoport TA. Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell. 1995;81:561–570. doi: 10.1016/0092-8674(95)90077-2. [DOI] [PubMed] [Google Scholar]
- Park E, Rapoport TA. Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annu Rev Biophys. 2012;41:21–40. doi: 10.1146/annurev-biophys-050511-102312. [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Brandt F, Hrabe T, Lang S, Eibauer M, Zimmermann R, Forster F. Structure and 3D arrangement of endoplasmic reticulum membrane-associated ribosomes. Structure. 2012;20:1508–1518. doi: 10.1016/j.str.2012.06.010. [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Dudek J, Gogala M, Schorr S, Linxweiler J, Lang S, Becker T, Beckmann R, Zimmermann R, Forster F. Structure of the mammalian oligosaccharyl-transferase complex in the native ER protein translocon. Nature communications. 2014;5:3072. doi: 10.1038/ncomms4072. [DOI] [PubMed] [Google Scholar]
- Pfeiffer NV, Dirndorfer D, Lang S, Resenberger UK, Restelli LM, Hemion C, Miesbauer M, Frank S, Neutzner A, Zimmermann R, et al. Structural features within the nascent chain regulate alternative targeting of secretory proteins to mitochondria. Embo J. 2013;32:1036–1051. doi: 10.1038/emboj.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter MD, Nicchitta CV. Ribosome-independent regulation of translocon composition and Sec61alpha conformation. J Biol Chem. 2000;275:2037–2045. doi: 10.1074/jbc.275.3.2037. [DOI] [PubMed] [Google Scholar]
- Potter MD, Nicchitta CV. Endoplasmic reticulum-bound ribosomes reside in stable association with the translocon following termination of protein synthesis. J Biol Chem. 2002;277:23314–23320. doi: 10.1074/jbc.M202559200. [DOI] [PubMed] [Google Scholar]
- Reithinger JH, Kim JE, Kim H. Sec62 protein mediates membrane insertion and orientation of moderately hydrophobic signal anchor proteins in the endoplasmic reticulum (ER) J Biol Chem. 2013;288:18058–18067. doi: 10.1074/jbc.M113.473009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein EM, Kreft SG, Greenblatt W, Swanson R, Hochstrasser M. Aberrant substrate engagement of the ER translocon triggers degradation by the Hrd1 ubiquitin ligase. J Cell Biol. 2012;197:761–773. doi: 10.1083/jcb.201203061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadlish H, Pitonzo D, Johnson AE, Skach WR. Sequential triage of transmembrane segments by Sec61alpha during biogenesis of a native multispanning membrane protein. Nat Struct Mol Biol. 2005;12:870–878. doi: 10.1038/nsmb994. [DOI] [PubMed] [Google Scholar]
- Shao S, Hegde RS. Membrane protein insertion at the endoplasmic reticulum. Annu Rev Cell Dev Biol. 2011;27:25–56. doi: 10.1146/annurev-cellbio-092910-154125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibatani T, David LL, McCormack AL, Frueh K, Skach WR. Proteomic analysis of mammalian oligosaccharyltransferase reveals multiple subcomplexes that contain Sec61, TRAP, and two potential new subunits. Biochemistry. 2005;44:5982–5992. doi: 10.1021/bi047328f. [DOI] [PubMed] [Google Scholar]
- Skach WR. Cellular mechanisms of membrane protein folding. Nat Struct Mol Biol. 2009;16:606–612. doi: 10.1038/nsmb.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapp EL, Reinhart GA, Bogert BA, Lippincott-Schwartz J, Hegde RS. The organization of engaged and quiescent translocons in the endoplasmic reticulum of mammalian cells. J Cell Biol. 2004;164:997–1007. doi: 10.1083/jcb.200312079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer N, Junne T, Kalies KU, Spiess M, Hartmann E. TRAP assists membrane protein topogenesis at the mammalian ER membrane. Biochimica et biophysica acta. 2013;1833:3104–3111. doi: 10.1016/j.bbamcr.2013.08.018. [DOI] [PubMed] [Google Scholar]
- Van den Berg B, Clemons WM, Jr, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- Voigt S, Jungnickel B, Hartmann E, Rapoport TA. Signal sequence-dependent function of the TRAM protein during early phases of protein transport across the endoplasmic reticulum membrane. J Cell Biol. 1996;134:25–35. doi: 10.1083/jcb.134.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees RM, Fernandez IS, Scheres SH, Hegde RS. Structure of the Mammalian ribosome-sec61 complex to 3.4 a resolution. Cell. 2014;157:1632–1643. doi: 10.1016/j.cell.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedmann M, Kurzchalia TV, Hartmann E, Rapoport TA. A signal sequence receptor in the endoplasmic reticulum membrane. Nature. 1987;328:830–833. doi: 10.1038/328830a0. [DOI] [PubMed] [Google Scholar]
- Young BP, Craven RA, Reid PJ, Willer M, Stirling CJ. Sec63p and Kar2p are required for the translocation of SRP-dependent precursors into the yeast endoplasmic reticulum in vivo. EMBO J. 2001;20:262–271. doi: 10.1093/emboj/20.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.