SUMMARY
Super-enhancers and stretch enhancers (SEs) drive expression of genes that play prominent roles in normal and disease cells, but the functional importance of these clustered enhancer elements is poorly understood, so it is not clear why genes key to cell identity have evolved regulation by such elements. Here we show that SEs consist of functional constituent units that concentrate multiple developmental signaling pathways at key pluripotency genes in embryonic stem cells and confer enhanced responsiveness to signaling of their associated genes. Cancer cells frequently acquire SEs at genes that promote tumorigenesis, and we show that these genes are especially sensitive to perturbation of oncogenic signaling pathways. Super-enhancers thus provide a platform for signaling pathways to regulate genes that control cell identity during development and tumorigenesis.
INTRODUCTION
Mammalian cells contain tens of thousands of transcriptional enhancers that control their specific gene expression programs (Bulger and Groudine, 2011; ENCODE Project Consortium et al., 2012). Clusters of enhancers, called super-enhancers or stretch enhancers (SEs), control expression of genes that have especially prominent roles in cell type-specific processes (Hnisz et al., 2013; Parker et al., 2013; Whyte et al., 2013). Cancer cells acquire super-enhancers to drive high-level transcription of oncogenes (Chapuy et al., 2013; Groschel et al., 2014; Hnisz et al., 2013; Loven et al., 2013; Mansour et al., 2014; Northcott et al., 2014), and sequence variation associated with other diseases is especially enriched in super-enhancers (Farh et al., 2014; Hnisz et al., 2013; Parker et al., 2013; Pasquali et al., 2014). While it is clear that super-enhancers play important roles in control of cell identity, there is limited understanding of the functions of super-enhancers and thus the reasons why most genes that control cell identity have evolved regulation by these elements.
To gain insights into the functions of embryonic stem cell (ESC) super-enhancers, we characterized their constituent enhancers with a combination of luciferase reporter assays, CRISPR/Cas9-mediated genetic perturbation, and analysis of transcription factor occupancy and function. The results revealed that super-enhancer constituents generally function as active enhancer elements that have cell type-specific, OCT4-dependent functions. Importantly, SEs are more frequently occupied by terminal transcription factors of the Wnt, TGF-β and LIF signaling pathways than typical enhancers, and manipulation of these three developmentally important signaling pathways preferentially affected expression of SE-associated genes in ESCs. We also found that tumor cells dependent on oncogenic Wnt signaling acquire SEs at key genes associated with tumorigenesis and that perturbation of the Wnt pathway had especially profound effects on these genes.
RESULTS
Enhancer activity of super-enhancer constituents
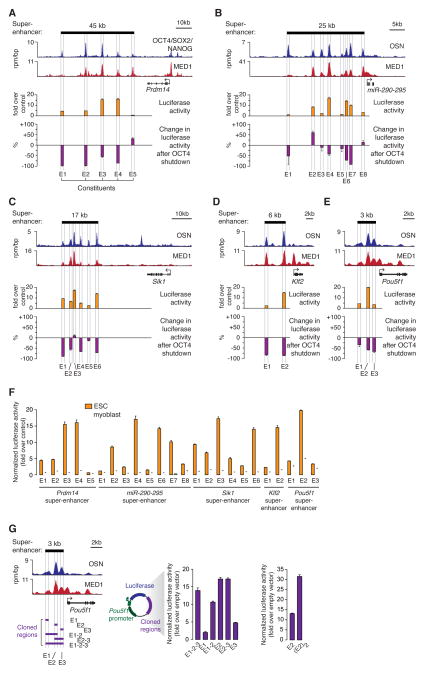
We used murine embryonic stem cells (ESCs) as a model to investigate the functional properties of super-enhancers. In ESCs, co-occupancy of genomic sites by the pluripotency transcription factors (TFs) OCT4, SOX2 and NANOG is highly predictive of enhancer activity (Chen et al., 2008), and ESC super-enhancers have been defined as clusters of sites occupied by OCT4, SOX2 and NANOG, together with exceptionally high levels of Mediator and other components of the transcription apparatus (Whyte et al., 2013). We first investigated whether each individual constituent of super-enhancers has enhancer activity and if that activity is cell type-specific and dependent on the pluripotency TFs. We selected five super-enhancers that control genes key to ESC identity and that broadly represent the 231 ESC super-enhancers in terms of size, occupancy by Mediator and the presence of other enhancer-associated features such as H3K27Ac enrichment and DNase hypersensitivity (Figure 1A–E, Figure S1A–B). These five super-enhancers control transcription of Prdm14, miR-290-295, Sik1, Klf2 and Pou5f1 (Oct4), which play important roles in ESC self-renewal, pluripotency and differentiation (Hall et al., 2009; Ma et al., 2011; Medeiros et al., 2011; Nichols et al., 1998; Romito et al., 2010). The five super-enhancers show physical interactions with their respective associated genes and are located within insulated neighborhoods in the ESC genome (Figure S1C), (Dowen et al., 2014), suggesting that these genes represent the bona fide physiological targets of the five SEs.
Figure 1. Activities of super-enhancer constituents.
ChIP-Seq binding profiles for OCT4, SOX2 and NANOG (merged) and Mediator (MED1) at the (A) Prdm14, (B) miR-290-295, (C) Sik1, (D) Klf2 and (E) Pou5f1 (Oct4) loci in ESCs. Enhancer activity measured in luciferase reporter assays in wild type cells and the change in enhancer activity after OCT4 shutdown is plotted for each constituent enhancer within the super-enhancer. The super-enhancer is depicted as a black bar above the binding profiles. The difference in values after OCT4 shutdown is statistically significant for all constituents, except from miR-290-295 M1, M3 and M5 (P<0.05, Student’s t-test). (F) Enhancer activity of the SE constituents measured in ESCs and myoblasts. The difference between the two values is statistically significant for each active constituent (P<0.01, Student’s t-test). (G) Enhancer activities of indicated fragments (purple) of the Pou5f1 SE. Throughout the figure, values correspond to mean +SD from three biological replicate experiments.
See also Figure S1, S2, Table S1.
We cloned individual constituent enhancers of the five super-enhancers into enhancer-reporter vectors, and found that the majority (21/24) of super-enhancer constituents were active in luciferase reporter assays (>1.5 fold over control, P<0.01 Student’s t-test) in ESCs (Figure 1A–E, Table S1). The enhancer activity was specific to ESCs, since all constituents active in ESCs showed decreased enhancer activity when transfected into murine myoblasts, murine embryonic fibroblasts, or several human cell lines (P<0.01 versus the activity measured in ESCs in each pair-wise comparison, Student’s t-test) (Figure 1F, Figure S2A). Furthermore, depletion of the pluripotency TF OCT4 in ESCs led to a reduction of the enhancer activity of 20/24 (83%) of the super-enhancer constituents (Figure 1A–E, Figure S2B–F). These results demonstrate that ESC super-enhancers generally consist of clusters of active enhancers that have OCT4-dependent and ESC-specific functions.
It is possible that SE constituents function additively, synergistically or exert a more complex influence on one another’s activity. We used the reporter system to investigate how various combinations of the constituents of the Pou5f1 super-enhancer behaved in this assay. The Pou5f1 super-enhancer was selected for this experiment because its SE was small enough to be fully accommodated by the reporter vector. The results revealed that the three constituent enhancers produced slightly less activity than E2 alone, which produced the largest signal (Figure 1G). The combination of E1 and E2 produced a signal intermediate between E1 and E2 alone. The combination of E2 and E3 had the same activity as E2 alone. It was possible to obtain an additive effect with two enhancer constituents in this assay, because two tandem copies of E2 produced approximately twice the activity of a single copy of E2 (Figure 1G). These results indicate that the multiple enhancer constituents in the Pou5f1 super-enhancer do not have an additive or synergistic function when each is present in single copy, and the results are consistent with the constituents having a complex influence on one another’s activity.
Perturbation of super-enhancer constituents
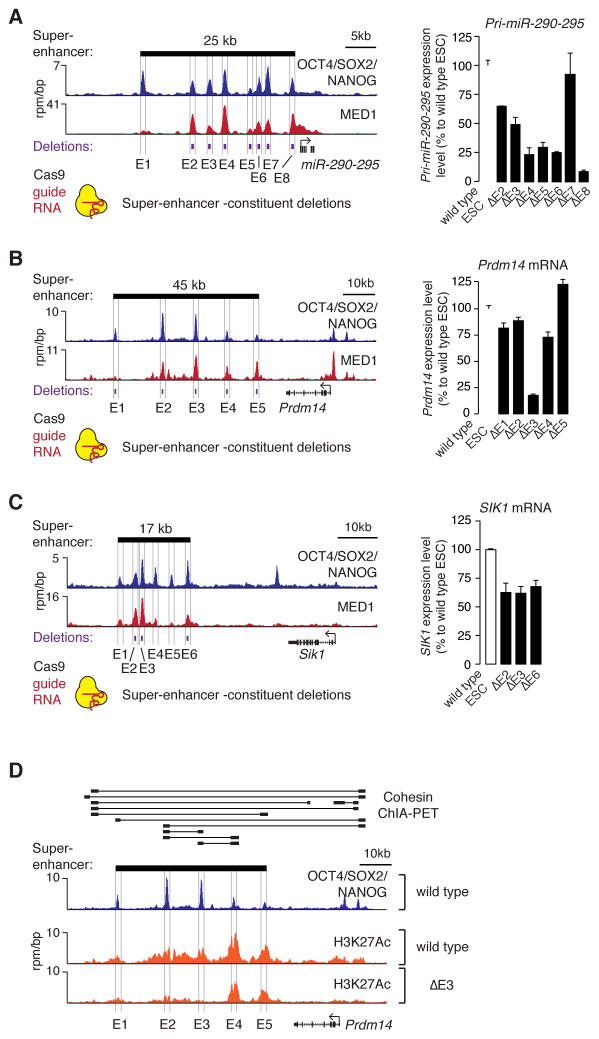
Previous studies have described how multiple enhancers can contribute to regulation of a single gene (Frankel et al., 2010; Hong et al., 2008; Perry et al., 2011); in some cases the individual enhancers can appear to be redundant but contribute to robustness (Hong et al., 2008), while in others the individual enhancers each make a demonstrable contribution to gene transcription (Bender et al., 2012). We investigated whether the individual ESC super-enhancer constituents have redundant or non-redundant functions in vivo by deleting individual constituents for three super-enhancers using a CRISPR/Cas9-based approach (Figure 2A–C). If the constituents have redundant functions, we would expect that loss of any single constituent might have little effect on gene expression. In contrast, if most of the constituents contribute to normal levels of gene expression, then we would expect that loss of any constituent would impact gene expression. The results showed that deletion of most (12/14) super-enhancer constituents led to reduced expression of the associated gene, while in one case (Prdm14 E5) the deletion caused a small increase in expression of the associated gene (Figure 2A–C). The effects on expression ranged from modest (<50% difference vs wild type; Prdm14 E1, E2, E4, E5; miR-290-295 E2, E3, and Sik1 E2, E3, E6, P<0.01 Student’s t-test) to substantial (>50% difference vs wild type; Prdm14 E3 and miR-290-295 E4, E5, E6, E8, P<0.01 Student’s t-test). These results suggest that most super-enhancer constituents make a positive contribution to transcriptional activity, but some constituents may have the opposite effect and contribute to a more complex SE environment.
Figure 2. Contributions of super-enhancer constituents to gene expression in vivo.
(A) (left) ChIP-Seq binding profiles for OCT4, SOX2 and NANOG (merged) and Mediator (MED1) at the miR-290-295 locus in ESCs. (right) miR-290-295 expression level in ESCs in which the indicated super-enhancer constituents were deleted. Values correspond to mean +SD from three biological replicate experiments. (B) Gene expression analysis at the Prdm14 locus after deletion of super-enhancer constituents. (C) Gene expression analysis at the Sik1 locus after deletion of SE constituents. All values correspond to mean +SD from three biological replicate experiments. The difference of all values measured in deletion lines, except for miR-290-295 E7, are statistically significant compared to wild type (P<0.05, Student’s t-test). (D) ChIP-Seq binding profiles for H3K27Ac in wild type and Prdm14 E3 deleted ESCs. Cohesin (SMC1) ChIA-PET data for ESCs is shown above the binding profiles, where thick black bars connected by lines indicate regions that show high-confidence interactions (Dowen et al., 2014).
See also Table S2.
The expression of some SE-associated genes was found to be especially dependent on particular SE constituents; deletion of Prdm14 E3 led to the most substantial loss of gene expression in these experiments (Figure 2B). Chromatin interaction (cohesin ChIA-PET) data shows that Prdm14 E3 interacts with both E2 and E4 (Figure 2D), so we postulated that the large effect of E3 deletion might be a consequence of a functional interdependence between E3 and the other super-enhancer constituents. To test this idea, we investigated the effect of the E3 deletion on the active enhancer mark H3K27Ac across the Prdm14 super-enhancer. The results show that deletion of E3 caused a substantial loss of active enhancer signal at the other SE constituents (reduction of 43% at E1, 56% at E2, 28% at E4, and 21% at E5 based on ChIP-Seq density) (Figure 2D). These results suggest that the constituent enhancers of the Prdm14 super-enhancer may be functionally interdependent. Taken together with the chromatin interaction data, these results indicate that there are physical and functional interactions among multiple super-enhancer constituents at the Prdm14 locus.
Signaling modules at super-enhancers
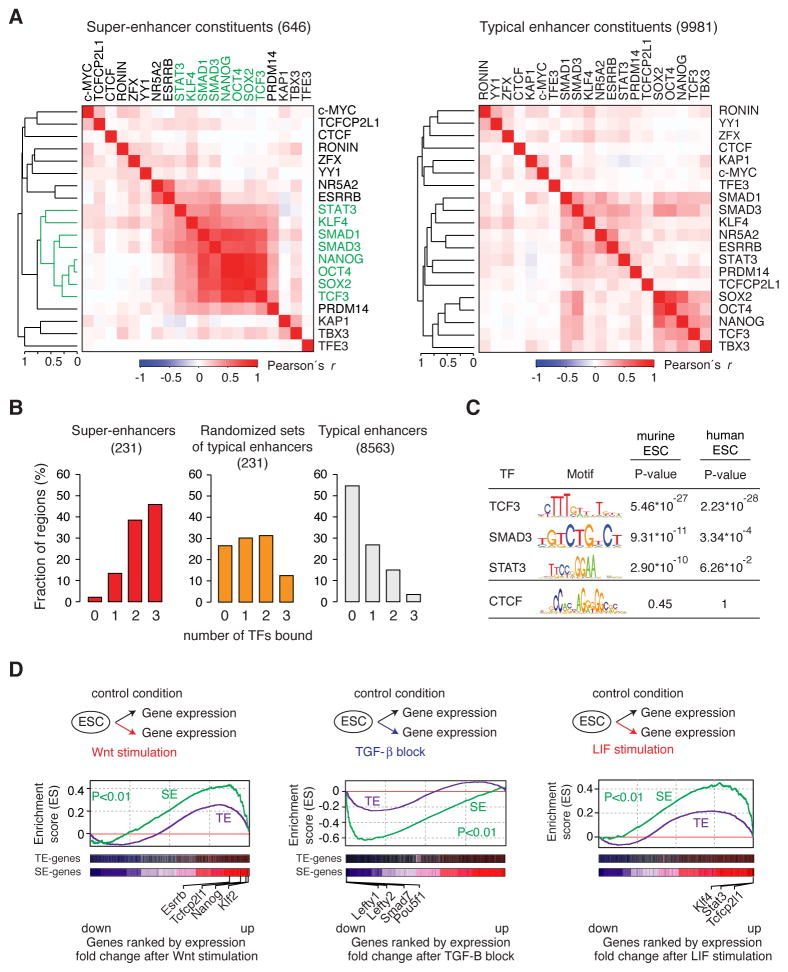
The evidence that many individual super-enhancer constituents contribute to the full transcriptional activity of their target gene (Figures 1 and 2), are occupied by high levels of cofactors relative to typical enhancers (Hnisz et al., 2013; Whyte et al., 2013), and make physical contacts with one another (Dowen et al., 2014) is consistent with the view that these clusters may facilitate high levels of transcription (Siersbaek et al., 2014; Whyte et al., 2013). However, these observations do not explain why most key cell identity genes have evolved this clustered enhancer structure because many housekeeping genes are expressed at high levels, yet are not driven by super-enhancers. An examination of the pattern of transcription factor binding to super-enhancer constituents provided a hypothesis to resolve this conundrum (Figure 3A). The terminal TFs of the Wnt (TCF3), TGF-β (SMAD3) and LIF (STAT3) signaling pathways, which play essential roles in transcriptional control of the stem cell state (Ng and Surani, 2011; Young, 2011), were among the TFs whose binding pattern to SE constituents was most similar to that of OCT4, SOX2 and NANOG at SE-constituents (Figure 3A). Most SE constituents (75%) were occupied by at least one of these three TFs, whereas only 43% of typical enhancer constituents were bound by one of the three (Figure S3A). More importantly, 98% of super-enhancers were bound by at least one, 86% were bound by at least two, and 46% were bound by all three signaling TFs, whereas a much smaller fraction of typical enhancers were bound by these TFs (Figure 3B, Figure S3B–F). Clusters of randomly selected typical enhancers, constructed to mimic the potential enrichment effects of clustering, failed to show this enrichment of signaling TFs noted for the super-enhancers (Figure 3B). If the clustered enhancer structure of SEs has evolved to provide a means to respond to the developmental signaling pathways, then one might expect that the enrichment of signaling TF motifs in SEs would be evolutionarily conserved. Indeed, we found enrichment of the cognate binding motifs for TCF3, SMAD3 and STAT3 in both murine and human ESC super-enhancer constituents (Figure 3C). These results indicate that signaling TFs are enriched in embryonic stem cell super-enhancers and suggest that these super-enhancers provide their target genes with a much higher probability of responding to signaling.
Figure 3. Signaling modules at super-enhancers.
(A) Hierarchical clustering of 20 transcription factor ChIP-Seq binding profiles at super-enhancer and typical enhancer constituents. A set of factors with binding profiles similar to Oct4, Sox2 and Nanog is highlighted in green. (B) Percentage of super-enhancers and typical enhancers bound by the indicated number of signaling TFs (TCF3, SMAD3, STAT3). Randomized sets of typical enhancers indicate sets of typical enhancers where the numbers in the sets correspond to the number of constituents within super-enhancers. (C) Binding motifs for TCF3, SMAD3 and STAT3 and the P-values for their enrichment in super-enhancer constituent enhancers in murine and human ESCs. The motif of CTCF is not found enriched, and serves as a negative control. The P-values in mESCs are re-analysis from (Whyte et al., 2013) (D) Gene set enrichment analysis (GSEA) of gene expression changes after manipulation of the Wnt, TGF-β and LIF pathways. “SE-genes” indicate genes associated with SEs, “TE-genes” with typical enhancers, respectively.
If super-enhancers confer responsiveness to the Wnt, TGF-β and LIF pathways more frequently than typical enhancers, then stimulation or perturbation of these pathways should have a more profound effect on super-enhancer–associated genes than typical enhancer–associated genes. The results of transcriptional profiling and gene set enrichment analysis in ESCs confirm this prediction (Figure 3D); super-enhancer associated genes were found enriched among the genes whose expression exhibited the most profound changes after pathway stimulation or perturbation (Wnt: P<0.01; TGF-β: P<0.01; LIF: P<0.01). In contrast, the enrichment for genes associated with typical enhancers was more moderate (Figure 3D). The super-enhancer–associated genes that showed a profound response to signaling included previously reported targets of these pathways that play key roles in ESC self-renewal and differentiation (Figure 3D, Figure SG). A subset of the Prdm14 SE-constituents that are bound by signaling TFs were found to be responsive to perturbation of these signaling pathways in reporter assays (Figure S3H). These results lead us to propose that key cell identity genes have evolved a clustered enhancer structure to provide a means to respond directly to these developmentally important signaling pathways.
Signaling to super-enhancers acquired in tumor cells
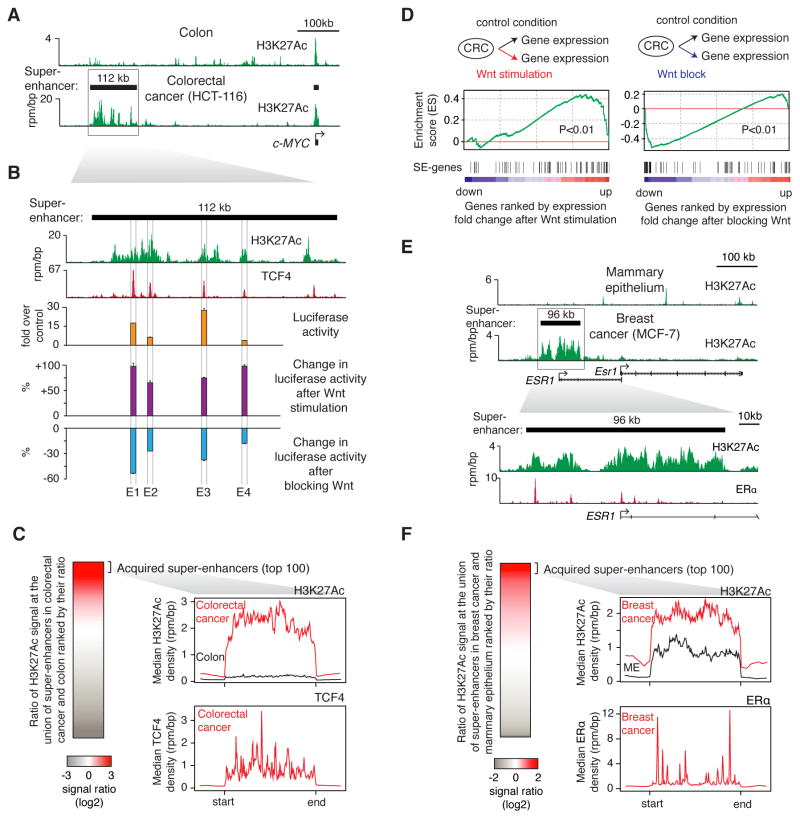
Cancer cells frequently acquire super-enhancers at oncogenes (Chapuy et al., 2013; Hnisz et al., 2013; Mansour et al., 2014; Northcott et al., 2014) and dysregulation of signaling pathways is a hallmark of cancer (Hanahan and Weinberg, 2011), leading us to investigate whether these two features may be linked. We used a colorectal cancer model, where tumorigenesis depends on hyperactivation of the Wnt pathway (Clevers and Nusse, 2012), to investigate whether the acquisition of super-enhancers provides for TCF binding and Wnt responsiveness at associated genes. The high density of H3K27Ac signal that is characteristic of super-enhancers was used to identify super-enhancers in normal human colon tissue and in the colorectal cancer cell (CRC) line HCT-116 (Figure 4A, Figure S4A), as described in (Hnisz et al., 2013). Oncogene-associated super-enhancers, not present in normal colon cells, including those at the c-MYC locus, were found in the colorectal cancer cell line (Figure 4A, Figure S4B), indicating that they are acquired in the tumor cells. Analysis of ChIP-seq binding profiles for multiple TFs in this cell line revealed that TCF4 (Tcf7l2), the terminal TF of the Wnt pathway in colorectal cancer cells, indeed occupies the super-enhancer at the c-MYC locus (Figure 4B, Figure S4C), which is a well-established target of Wnt signaling (He et al., 1998). Four of the TCF4-bound sites within the c-MYC super-enhancer region were found to have enhancer activity when introduced into reporter vectors in CRC cells, and this activity was responsive to stimulation or inhibition of the Wnt pathway (Figure 4B).
Figure 4. TCF4 occupancy and Wnt responsiveness of super-enhancers acquired in colorectal cancer.
(A) ChIP-Seq binding profiles for H3K27Ac at the c-MYC locus in colon and colorectal cancer cells (HCT-116). (B) A blow-up of the region indicated by a box on panel (A). TCF4 binding profile in the HCT-116 is displayed, along with the enhancer activity of TCF4-bound constituents of the acquired super-enhancer at MYC locus. Luciferase reporter activity in HCT-116 cells and the change in enhancer activity after Wnt stimulation or blockage are plotted. Values correspond to mean +SD from three biological replicate experiments. (C) (left) Ratio of H3K27Ac in CRC (HCT-116) vs. normal colon tissue used densities at the union of SEs identified in the two samples. (right) Metagene representation of H3K27Ac and TCF4 ChIP-Seq densities at the regions corresponding to the top 100 acquired super-enhancers. (D) Gene set enrichment analysis (GSEA) of gene expression changes after manipulation of the Wnt pathway. “SE-genes” indicate genes that are associated with acquired SEs. (E) (top) ChIP-Seq binding profiles for H3K27Ac at the ESR1 locus in mammary epithelium and breast cancer cells (MCF-7). (bottom) A blow-up of the region indicated on top. ChIP-Seq binding profile for ERα is displayed. (F) (left) Ratio of H3K27Ac in breast cancer (MCF-7) vs. mammary epithelium (ME) at the union of SEs identified in the two samples. (right) Metagene representation of H3K27Ac and ERα ChIP-Seq densities at the regions corresponding to the top 100 acquired super-enhancers.
We next investigated whether TCF4 occupied other acquired CRC super-enhancers and if this made this set of genes associated with the acquired SEs especially responsive to the manipulation of the Wnt pathway. Acquired super-enhancers, which showed the highest levels of H3K27Ac signal in CRC relative to normal cells, showed strong evidence of TCF4 binding (Figure 4C). Genes associated with these acquired super-enhancers were enriched for expression changes after stimulation or blockage of the Wnt pathway (stimulation: P<0.01; blockage: P<0.01), although not all super-enhancer genes showed this response (Figure 4D). These results indicate that acquired super-enhancers in colorectal cancer can be occupied by TCF4 and tend to be especially responsive to perturbation of the oncogenic Wnt pathway.
Oncogenic signaling plays important roles in many different cancers, so we sought evidence that acquired super-enhancers in other cancers are occupied by oncogenic signaling factors. Some breast cancer cells are dependent on estrogen signaling, so we investigated whether acquired super-enhancers in ER-positive breast cancer cells are bound by high amounts of the estrogen receptor (Figure 4E, 4F). Examination of H3K27Ac ChIP-seq data in normal breast epithelium and in the ER-positive cell line MCF-7 revealed a super-enhancer at the ESR1 gene, which encodes estrogen receptor alpha (ERα), only in the tumor cells (Figure 4E). This acquired super-enhancer is bound at multiple sites by ERα, indicating that estrogen signaling is brought to ESR1 via an acquired super-enhancer (Figure 4E). Furthermore, binding of ERα was observed at additional SEs acquired in this tumor line (Figure 4F). These results are consistent with the idea that tumor cells evolve SEs at key oncogenes, at least in part, to enhance the connection to oncogenic signaling pathways.
DISCUSSION
Super-enhancers control genes that play especially prominent roles in cellular physiology and disease (Brown et al., 2014; Chapuy et al., 2013; Groschel et al., 2014; Herranz et al., 2014; Hnisz et al., 2013; Loven et al., 2013; Mansour et al., 2014; Northcott et al., 2014; Parker et al., 2013; Siersbaek et al., 2014; Whyte et al., 2013), but there is a limited understanding of the functions of these clustered elements, and thus why they have evolved to drive genes that play key roles in cell type-specific biology. Our results reveal that SEs can provide a platform for signaling pathways to regulate genes that control cell identity during development and tumorigenesis.
Previous studies in metazoans have shown that multiple enhancers can contribute to gene regulation in many different ways (Frankel et al., 2010; Hong et al., 2008; Perry et al., 2011; Spitz and Furlong, 2012). For example, Locus Control Regions (LCRs) contain multiple enhancers that are active at different developmental stages and can regulate different genes within a locus (Grosveld et al., 1987; Li et al., 2002). Multiple enhancers for a single gene can have apparently redundant functions in gene activation but contribute to phenotypic robustness, as has been proposed for some “shadow enhancers” in Drosophila and for elements of the “regulatory archipelago” at the mammalian HoxD locus (Hong et al., 2008; Montavon et al., 2011). Multiple enhancers can ensure precise spatial fine-tuning of gene expression in response to a morphogen gradient in Drosophila (Perry et al., 2011). It is also possible that multiple enhancers produce additive or synergistic interactions to drive high levels of gene expression (Bender et al., 2012; Prazak et al., 2010). Some mammalian super-enhancers may incorporate features and functions previously described for multiple enhancer loci, but the results described here suggest that super-enhancers have additional features that help explain why they have evolved to be associated with genes that have important roles in cell identity.
Several lines of evidence argue that the constituent enhancers of at least some super-enhancers can act as an interdependent structural and functional unit to control their associated genes. Our results show that ESC SEs generally consist of clusters of active enhancers that have OCT4-dependent and ESC-specific functions (Figure 1) and demonstrate that optimal transcriptional activity of target genes is dependent on the presence of most of the constituent enhancers (Figure 2). Chromatin interaction data indicates that constituent enhancers physically interact within the SEs; indeed, the interactions among SE constituents in ESCs appear to be more frequent than interactions between the SE-constituents and their associated gene promoters, and interactions between typical enhancers (Dowen et al., 2014). We previously noted that enhancer clusters can be gained or lost as a unit during development or oncogenesis (Hnisz et al., 2013) and have shown that large tumor SEs can collapse as a unit when depleted of the enhancer cofactor BRD4 (Loven et al., 2013) or when a constituent is deleted (Mansour et al., 2014). In some T cell acute lymphoblastic leukemia (T-ALL) cells, a small mono-allelic insertion that creates a binding site for a master transcription factor can nucleate the formation of an oncogenic super-enhancer that involves establishment of additional transcriptional components in adjacent sites (Mansour et al., 2014). Super-enhancers produce relatively high levels of enhancer RNAs (Hnisz et al., 2013) and a recent study showed that inflammation-dependent super-enhancers form domains of coordinately regulated enhancer RNAs (Hah et al., 2015). These results, taken together, suggest that the constituent enhancers of super-enhancers can interact physically and functionally to coordinate transcriptional activity.
Our results reveal that SEs are occupied more frequently by terminal transcription factors of the Wnt, TGF-β and LIF signaling pathways than typical enhancers in ESCs, and genes driven by SEs show a more pronounced response to the manipulation of these pathways than genes driven by typical enhancers (Figure 3). Thus, the clustered enhancer architecture of SEs may have evolved, at least in part, to provide a conduit for these signaling pathways to signal maintenance or change at genes that are key to control of cell identity. Our results also suggest that one reason that tumor cells evolve SEs at key oncogenes is to enhance the connection to oncogenic signaling pathways. The recent report of NOTCH1-driven SEs in T-ALL likely represents another example of this phenomenon (Herranz et al., 2014; Wang et al., 2014). An implication of this model is that therapies that target both oncogenic signaling pathways and super-enhancer components may be especially effective in tumor cells that have signaling and transcriptional dependencies.
EXPERIMENTAL PROCEDURES
Cell culture
V6.5 and Zhbtc4 murine ESCs, C2C12 murine myoblasts, primary murine embryonic fibroblasts (MEFs), HEK-293T cells and HCT-116 human colorectal cancer cells were cultured under previously described conditions detailed in the supplemental information.
Luciferase reporter assays
Super-enhancer constituent enhancers (~400bp) were cloned into Firefly luciferase reporters driven by a minimal Oct4 promoter (for murine cells), or a minimal c-Myc promoter (for human cells). Zhbtc4 cells were transfected using Lipofectamine 2000 (Invitrogen), C2C12 cells and MEFs were transfected using Lipofectamine 3000 (Invitrogen), HEK-293T and HCT-116 cells were transfected using pPEI (Sigma). A Renilla luciferase control plasmid was co-transfected as a normalization control. Luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega). In Zhbtc4 cells OCT4-expression was shut down by addition of doxycycline to the culture media during transfection. In HCT-116 cells, the Wnt pathway was stimulated by co-transfection of a S33Y-β-catenin expressing vector (Addgene: 19286), and was blocked by co-transfection of a ΔNTCF4 expressing vector (generous gift from Hans Clevers).
Genome editing
Super-enhancer constituent enhancers (~400bp) were deleted in V6.5 murine ESCs using the CRISPR/Cas9 system. sgRNAs were cloned into the pX330 vector (Addgene: 42230) containing Cas9. Cells were transfected with two plasmids expressing Cas9 and an sgRNA complementary to each end of the targeted super-enhancer constituent using X-fect reagent (Clontech). A plasmid expressing PGK-puroR was co-transfected for selection. One day after transfection, cells were re-plated on DR4 MEF feeder layers. One day after re-plating, puromycin (2ug/ml) was added for three days. Subsequently, puromycin was withdrawn for three to four days. Individual colonies were picked and genotyped by PCR. Deletion alleles were verified by sequencing.
Supplementary Material
Acknowledgments
We thank Nicholas Kwiatkowski and Chikdu Shivalila for experimental assistance, Rudolf Jaenisch for sharing reagents, Tom Volkert, Jennifer Love, Sumeet Gupta, and Jeong-Ah Kwon at the Whitehead Genome Technologies Core for Solexa sequencing; and members of the Young lab for helpful discussions. This work was supported by the National Institutes of Health grant HG002668 (R.A.Y.), an Erwin Schrödinger Fellowship (J3490) from the Austrian Science Fund (FWF) (D.H.), a Rubicon Fellowship for the Life Sciences from the Netherlands Organization for Scientific Research (NWO) (J.S.), and a grant from the U.S. Department of Defense CDMRP CA120184 (C.Y.L). The authors declare competing financial interests: R.A.Y. and J.E.B. are founders of Syros Pharmaceuticals.
Footnotes
Accession numbers
Sequencing data have been deposited at the Gene Expression Omnibus (GEO) under the accession number GSE64188.
AUTHOR CONTRIBUTIONS
D.H. and R.A.Y. conceived the project. D.H., J.S. and A.S.W. designed and performed experiments with input from T.I.L. D.H., J.S. and A.S.W. analyzed experimental data. C.Y.L. and B.J.A. designed and performed genomics data analyses. D.H. and R.A.Y co-wrote the paper with input from J.E.B. All authors edited the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bender MA, Ragoczy T, Lee J, Byron R, Telling A, Dean A, Groudine M. The hypersensitive sites of the murine beta-globin locus control region act independently to affect nuclear localization and transcriptional elongation. Blood. 2012;119:3820–3827. doi: 10.1182/blood-2011-09-380485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JD, Lin CY, Duan Q, Griffin G, Federation AJ, Paranal RM, Bair S, Newton G, Lichtman AH, Kung AL, et al. NF-kappaB Directs Dynamic Super Enhancer Formation in Inflammation and Atherogenesis. Molecular cell. 2014 doi: 10.1016/j.molcel.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MG, Qi J, Rahl PB, Sun HH, Yeda KT, Doench JG, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer cell. 2013;24:777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, Weintraub AS, Schuijers J, Lee TI, Zhao K, et al. Control of cell identity genes occurs in insulated neighborhoods in Mammalian chromosomes. Cell. 2014;159:374–387. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh KK, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, Shoresh N, Whitton H, Ryan RJ, Shishkin AA, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2014 doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel N, Davis GK, Vargas D, Wang S, Payre F, Stern DL. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature. 2010;466:490–493. doi: 10.1038/nature09158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groschel S, Sanders MA, Hoogenboezem R, de Wit E, Bouwman BA, Erpelinck C, van der Velden VH, Havermans M, Avellino R, van Lom K, et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell. 2014;157:369–381. doi: 10.1016/j.cell.2014.02.019. [DOI] [PubMed] [Google Scholar]
- Grosveld F, van Assendelft GB, Greaves DR, Kollias G. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- Hah N, Benner C, Chong LW, Yu RT, Downes M, Evans RM. Inflammation-sensitive super enhancers form domains of coordinately regulated enhancer RNAs. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E297–302. doi: 10.1073/pnas.1424028112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J, Guo G, Wray J, Eyres I, Nichols J, Grotewold L, Morfopoulou S, Humphreys P, Mansfield W, Walker R, et al. Oct4 and LIF/Stat3 additively induce Kruppel factors to sustain embryonic stem cell self-renewal. Cell stem cell. 2009;5:597–609. doi: 10.1016/j.stem.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- Herranz D, Ambesi-Impiombato A, Palomero T, Schnell SA, Belver L, Wendorff AA, Xu L, Castillo-Martin M, Llobet-Navas D, Cordon-Cardo C, et al. A NOTCH1-driven MYC enhancer promotes T cell development, transformation and acute lymphoblastic leukemia. Nature medicine. 2014;20:1130–1137. doi: 10.1038/nm.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JW, Hendrix DA, Levine MS. Shadow enhancers as a source of evolutionary novelty. Science. 2008;321:1314. doi: 10.1126/science.1160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Peterson KR, Fang X, Stamatoyannopoulos G. Locus control regions. Blood. 2002;100:3077–3086. doi: 10.1182/blood-2002-04-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Swigut T, Valouev A, Rada-Iglesias A, Wysocka J. Sequence-specific regulator Prdm14 safeguards mouse ESCs from entering extraembryonic endoderm fates. Nature structural & molecular biology. 2011;18:120–127. doi: 10.1038/nsmb.2000. [DOI] [PubMed] [Google Scholar]
- Mansour MR, Abraham BJ, Anders L, Berezovskaya A, Gutierrez A, Durbin AD, Etchin J, Lawton L, Sallan SE, Silverman LB, et al. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014 doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros LA, Dennis LM, Gill ME, Houbaviy H, Markoulaki S, Fu D, White AC, Kirak O, Sharp PA, Page DC, et al. Mir-290-295 deficiency in mice results in partially penetrant embryonic lethality and germ cell defects. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:14163–14168. doi: 10.1073/pnas.1111241108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, Splinter E, de Laat W, Spitz F, Duboule D. A regulatory archipelago controls Hox genes transcription in digits. Cell. 2011;147:1132–1145. doi: 10.1016/j.cell.2011.10.023. [DOI] [PubMed] [Google Scholar]
- Ng HH, Surani MA. The transcriptional and signalling networks of pluripotency. Nature cell biology. 2011;13:490–496. doi: 10.1038/ncb0511-490. [DOI] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Northcott PA, Lee C, Zichner T, Stutz AM, Erkek S, Kawauchi D, Shih DJ, Hovestadt V, Zapatka M, Sturm D, et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature. 2014;511:428–434. doi: 10.1038/nature13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker SC, Stitzel ML, Taylor DL, Orozco JM, Erdos MR, Akiyama JA, van Bueren KL, Chines PS, Narisu N, Program NCS, et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:17921–17926. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquali L, Gaulton KJ, Rodriguez-Segui SA, Mularoni L, Miguel-Escalada I, Akerman I, Tena JJ, Moran I, Gomez-Marin C, van de Bunt M, et al. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nature genetics. 2014;46:136–143. doi: 10.1038/ng.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry MW, Boettiger AN, Levine M. Multiple enhancers ensure precision of gap gene-expression patterns in the Drosophila embryo. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:13570–13575. doi: 10.1073/pnas.1109873108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prazak L, Fujioka M, Gergen JP. Non-additive interactions involving two distinct elements mediate sloppy-paired regulation by pair-rule transcription factors. Developmental biology. 2010;344:1048–1059. doi: 10.1016/j.ydbio.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romito A, Lonardo E, Roma G, Minchiotti G, Ballabio A, Cobellis G. Lack of sik1 in mouse embryonic stem cells impairs cardiomyogenesis by down-regulating the cyclin-dependent kinase inhibitor p57kip2. PloS one. 2010;5:e9029. doi: 10.1371/journal.pone.0009029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siersbaek R, Rabiee A, Nielsen R, Sidoli S, Traynor S, Loft A, Poulsen LL, Rogowska-Wrzesinska A, Jensen ON, Mandrup S. Transcription factor cooperativity in early adipogenic hotspots and super-enhancers. Cell reports. 2014;7:1443–1455. doi: 10.1016/j.celrep.2014.04.042. [DOI] [PubMed] [Google Scholar]
- Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nature reviews Genetics. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- Wang H, Zang C, Taing L, Arnett KL, Wong YJ, Pear WS, Blacklow SC, Liu XS, Aster JC. NOTCH1-RBPJ complexes drive target gene expression through dynamic interactions with superenhancers. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:705–710. doi: 10.1073/pnas.1315023111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.