Abstract
Directly modulating the activity of genome-editing proteins has the potential to increase their specificity by reducing activity following target locus modification. We developed Cas9 nucleases that are activated by the presence of a cell-permeable small molecule by inserting an evolved 4-hydroxytamoxifen (4-HT)-responsive intein at specific positions in Cas9. In human cells, conditionally active Cas9s modify target genomic sites with up to 25-fold higher specificity than wild-type Cas9.
The RNA-guided endonuclease Cas9 from the type II CRISPR-Cas system enables simple and efficient genome editing in a wide variety of organisms. Virtually any target DNA locus can be cleaved by programming Cas9 with a single guide RNA (sgRNA) that contains a stretch of ~20 nucleotides complementary to the target sequence1–3. Due to its simplicity and robustness, the Cas9 system has been widely adopted for biological research and therapeutic development. The DNA cleavage specificity of Cas9 is imperfect4–8, however, raising concerns over off-target genome modification that may limit its usefulness in therapeutic or research applications. Researchers have reduced Cas9 off-target activity through protein9–12 and sgRNA13 engineering, and by direct delivery of Cas9:sgRNA protein:RNA complexes into cells14–16.
A complementary, underexplored strategy to improve Cas9 specificity is to reduce its activity once it has had sufficient opportunity to modify the target DNA locus. Indeed, higher concentrations of Cas9 in cells have been observed to degrade specificity4–6 (defined as the ratio of on-target:off-target DNA cleavage activity), presumably because any Cas9 protein present after the target locus has been modified can only process off-target substrates. Unfortunately, wild-type Cas9 nucleases are not known to be regulated by other molecules and therefore are used in constitutively active form. While Cas9 can be regulated at the transcriptional level through the use of inducible promoters17,18, transcriptional control cannot limit activity to the short temporal windows that may be necessary to maximize genome-editing specificity16,19, in contrast with the high temporal resolution of post-translational strategies that directly control protein activity.
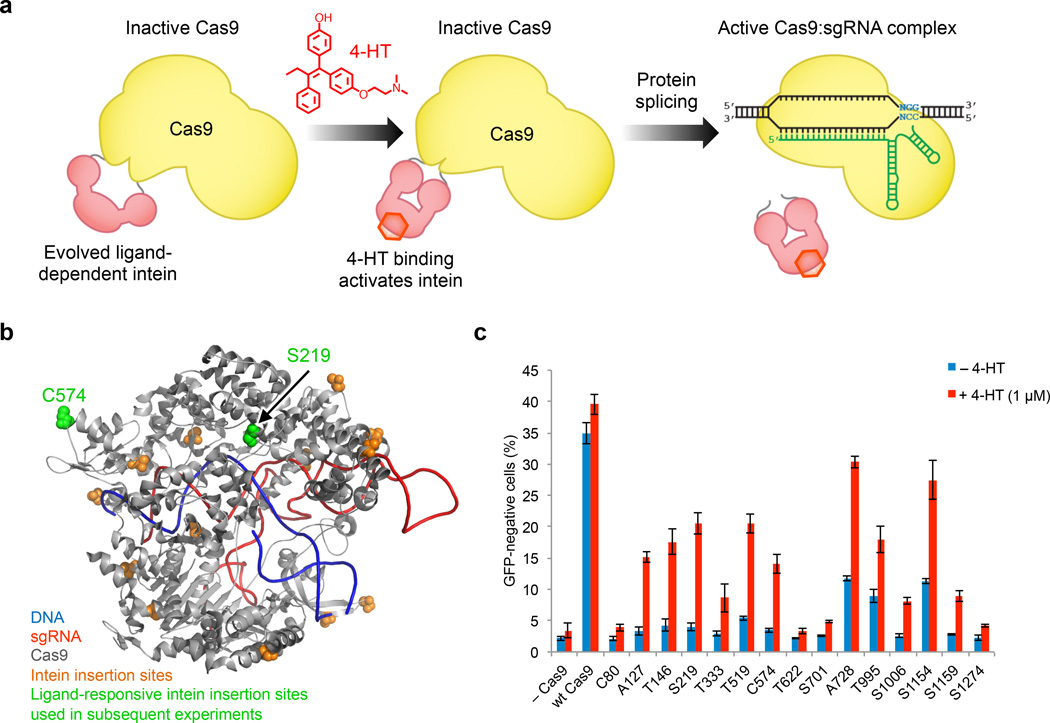
To enable post-translational control over Cas9 in cells, we sought to engineer variants of Cas9 that can be controlled with a readily available, cell-permeable small molecule. We previously evolved inteins that undergo protein splicing only in the presence of 4-hydroxytamoxifen (4-HT)20. These inteins were developed by inserting the human estrogen receptor ligand-binding domain into the M. tuberculosis RecA intein and evolving the resulting inactive fusion protein into a conditionally active intein that requires the presence of 4-HT20–22. Subsequent evolution at 37 °C yielded a second-generation intein, 37R3-2, with improved splicing properties in mammalian cells22. We envisioned that inserting the 37R3-2 intein into Cas9 at a location that disrupts Cas9 activity until protein splicing has taken place could result in conditionally active Cas9 nucleases that are active only in the presence of 4-HT (Fig. 1a).
Figure 1.
Insertion of an evolved ligand-dependent intein enables small-molecule control of Cas9. (a) Intein insertion renders Cas9 inactive. Upon 4-HT binding, the intein undergoes conformational changes that trigger protein splicing and restore Cas9 activity. (b) The evolved intein was inserted to replace each of the colored residues. Intein-inserted Cas9 variants at S219 and C574 (green) were used in subsequent experiments. (c) Genomic EGFP disruption activity of wild-type Cas9 and intein-Cas9 variants in the absence or presence of 4-HT. Intein-Cas9 variants are identified by the residue replaced by the intein. Error bars reflect the standard deviation of three biological replicates.
We genetically inserted the 4-HT-dependent intein at each of fifteen positions in Cas9 (Cys80, Ala127, Thr146, Ser219, Thr333, Thr519, Cys574, Thr622, Ser701, Ala728, Thr995, Ser1006, Ser1154, Ser1159, and Ser1274), chosen to distribute the location of the intein across the structural domains of Cas923 (Fig. 1b). Because intein splicing leaves behind a single Cys residue, the intein was inserted in place of one Cas9 amino acid in each of the 15 candidate constructs. In addition to replacing natural Cys amino acids, we also favored replacing Ala, Ser, or Thr residues to minimize the likelihood that the resulting Cys point mutation resulting from protein splicing would disrupt Cas9 activity. The 15 intein-Cas9 candidates were expressed in HEK293-GFP cells together with a sgRNA that targets the genomic EGFP locus in these cells. Twelve hours post-transfection, cells were treated with or without 1 µM 4-HT. Five days post-transfection, cells were analyzed on a flow cytometer for loss of GFP expression from Cas9-mediated EGFP cleavage and subsequent non-homologous end joining.
Eight of the candidates, corresponding to intein insertion at A127, T146, S219, T333, T519, C574, S1006, and S1159, demonstrated 4-HT-dependent loss of GFP expression consistent with 4-HT-triggered Cas9 activity (Fig. 1c). Interestingly, three intein-Cas9 proteins (insertion at A728, T995, and S1154) showed high DNA modification rates both in the presence and absence of 4-HT, suggesting that large protein insertions at these positions do not significantly inhibit nuclease activity, or that the intein lost its 4-HT dependence due to context-dependent conformational perturbations. We speculate that it may be possible to engineer split Cas9 variants by dividing the protein at these locations, given their tolerance of a 413-residue insertion. The lack of nuclease activity of the remaining four Cas9-inteins (insertion at C80, T622, S701, and S1274) in the presence or absence of 4-HT could result from the inability of the intein to splice in those contexts, the inability of Cas9 to refold properly following splicing, or intolerance of replacement of native Thr or Ser residues with Cys. We pursued two intein-Cas9 variants corresponding to insertion at S219 and C574 (Fig. 1b). These two variants combined high activity in the presence of 4-HT and low activity in the absence of 4-HT.
To evaluate the genome modification specificity of conditionally active Cas9 variants, we expressed intein-Cas9(S219), intein-Cas9(C574), and wild-type Cas9 in HEK293-GFP cells together with each of three previously described11 sgRNAs that target the well-studied EMX, VEGF, and CLTA genomic loci. We assayed these Cas9:sgRNA combinations in human cells for their ability to modify the three on-target loci as well as 11 known off-target genomic sites (Supplementary Results, Supplementary Table 1)4,5,10,13. Cells were treated with or without 1 µM 4-HT during transfection, and after 12 h the media was replaced with fresh media lacking 4-HT. We observed no cellular toxicity arising from 12 or 60 h of treatment with 1 µM 4-HT in untransfected or transfected HEK293 cells (Supplementary Fig. 1). Genomic DNA was isolated 60 h post-transfection and analyzed by high-throughput DNA sequencing
Overall on-target genome modification frequency of intein-Cas9(S219) and intein-Cas9 (C574) in the presence of 1 µM 4-HT was similar to that of wild-type Cas9 (Fig. 2a, Supplementary Tables 2 and 3, and Supplementary Data Set 1). On-target modification frequency in the presence of 4-HT was 3.4- to 7.3-fold higher for intein-Cas9(S219), and 3.6- to 9.6-fold higher for intein-Cas9(C574), than in the absence of 4-HT, whereas modification efficiency for wild-type Cas9 was 1.2- to 1.8-fold lower in the presence of 4-HT (Fig. 2a). Both intein-Cas9 variants exhibited a low level of background activity in the absence of 4-HT, consistent with previous reports20–22. Western blot analysis of intein-Cas9(S219) from transfected HEK293 cells confirmed the presence of spliced product at the earliest assayed time point (4 h) following 4-HT treatment; no spliced product was detected in the absence of 4-HT (Supplementary Fig. 2). Together, these results indicate that intein-Cas9(S219) and intein-Cas9(C574) are slightly less active than wild-type Cas9 in the presence of 4-HT, likely due to incomplete splicing (Supplementary Fig. 2), but much less active in the absence of 4-HT.
Figure 2.
Genomic DNA modification by intein-Cas9(S219), intein-Cas9(C574), and wild-type Cas9. (a) Indel frequency from high-throughput DNA sequencing of amplified genomic on-target sites in the absence or presence of 4-HT. Note that a significant number of indels were observed at the CLTA on-target site even in the absence of a targeting sgRNA (Supplementary Table 7). (b–d) DNA modification specificity, defined as on-target:off-target indel frequency ratio4–6, normalized to wild-type Cas9. Cells were transfected with 500 ng of the Cas9 expression plasmid. P-values are < 10−15 for the Fisher exact test (one-sided up) on comparisons of indel modification frequency in the presence versus the absence of 4-HT for intein-Cas9(S219) and intein-Cas9(C574). P-values were adjusted for multiple comparisons using the Benjamini-Hochberg method, and are listed in Supplementary Table 3. Error bars reflect the range of two independent experiments conducted on different days.
High-throughput sequencing of 11 previously described off-target sites that are modified by wild-type Cas9:sgRNA complexes targeting the EMX, VEGF, and CLTA loci revealed that both intein-Cas9 variants when treated with 4-HT for 12 h exhibit substantially improved specificity compared to that of wild-type Cas9 (Supplementary Fig. 3, Supplementary Tables 2, 4, and 5, and Supplementary Data Sets 1 and 2). On-target:off-target indel modification ratios for both intein-Cas9 variants were on average 6-fold higher, and as much as 25-fold higher, than that of wild-type Cas9 (Fig. 2b–d). In the absence of 4-HT, the genome modification specificity of both intein-Cas9 variants was on average 14-fold higher than that of wild-type Cas9 in the absence of 4-HT (Supplementary Fig. 4), presumably resulting from the much lower activity of the intein-Cas9 variants in the absence of 4-HT4–6.
Since intein-Cas9s can result in slightly lower on-target modification rates compared to wild-type Cas9 (Fig. 2a), we sought to verify that the improvements in specificity among the intein-Cas9s were not simply a result of reduced activity. Both on- and off-target activity of Cas9 has been shown to be dependent on the amount of Cas9 expression plasmid transfected4–6. By transfecting lower amounts of the wild-type Cas9 expression plasmid, we compared intein-Cas9s with wild-type Cas9 under conditions that result in very similar levels of on-target modification. To minimize potential differences in transfection efficiency, we supplemented with a plasmid that does not express Cas9 so that the same total amount of plasmid DNA was transfected into each sample. High-throughput sequencing revealed that wild-type Cas9 shows slightly improved specificity, as expected, as the on-target cleavage rate is reduced. The intein-Cas9 variants, however, remain substantially more specific than wild-type Cas9 at similar on-target DNA cleavage rates (Supplementary Figs. 5–7, Supplementary Tables 4 and 6, and Supplementary Data set 2). For example, intein-Cas9(C574) and wild-type Cas9 (80 ng) have virtually identical on-target DNA cleavage rates (both 6.4%) at the EMX locus but all four off-target sites are modified at an average of 4-fold lower frequencies (P < 1 × 10−13) by intein-Cas9(C574) than by wild-type Cas9. These findings indicate that specificity improvements of intein-Cas9 variants do not simply arise from differences in overall genome editing activity.
Intein 37R3-2 can be activated by other estrogen receptor modulators. To enable intein-Cas9 applications in which endogenous β-estradiol is present, we inserted into the estrogen receptor ligand-binding domain a point mutation (G521R) that renders the domain more specific for 4-HT24. This mutation slightly reduces affinity for 4-HT but almost abolishes affinity for β-estradiol. The addition of this mutation to intein-Cas9(S219) eliminates the ability of β-estradiol to trigger Cas9 activity (Supplementary Fig. 8).
The intein-Cas9 variants developed here enable small-molecule control of Cas9 function, thereby enhancing genome-modification specificity. Together with our recent observations that direct delivery of transient Cas9 protein into cells can also improve its specificity16, these findings strongly suggest that limiting the opportunity of genome-editing agents to modify off-target loci following a period of activity sufficient to induce desired levels of on-target modification results in substantial specificity improvements. Our findings complement those of a recent study25 describing small molecule-dimerized split Cas9 halves with improved specificity, published while this study was in review. Since the intein-Cas9s, unlike split Cas9 proteins, are expressed as single polypeptides that, upon splicing, generate Cas9 proteins with identical (or nearly identical) amino acid sequence as wild-type Cas9, these two approaches offer complementary strengths. We anticipate that the use of ligand-dependent Cas9 variants will provide greater control over genomic modification efficiencies and specificities than is currently achievable with constitutively active or transcriptionally regulated genome editing. In principle, this approach could synergize with other specificity-augmenting strategies such as using truncated guide RNAs13, paired Cas9 nickases9,10, or FokI-dCas9 fusions11,12. This approach could also be applied to other genome engineering proteins to enable, for example, small-molecule control of TALE-based or Cas9-mediated transcriptional regulators.
ONLINE METHODS
Cas9, intein-Cas9, and sgRNA expression plasmids
A plasmid encoding the human codon-optimized Streptococcus pyogenes Cas9 nuclease with an NLS and 3×FLAG tag (Addgene plasmid 43861)5 was used as the wild-type Cas9 expression plasmid. Intein 37R3-2 was subcloned at the described positions into the wild-type Cas9 expression plasmid using USER (NEB M5505) cloning. sgRNA expression plasmids used in this study have been described previously11. Plasmid constructs generated in this work will be deposited with Addgene.
Modification of genomic GFP
HEK293-GFP stable cells (GenTarget), which constitutively express Emerald GFP, served as the reporter cell line. Cells were maintained in “full serum media”: Dulbecco’s Modified Eagle’s Media plus GlutaMax (Life Technologies) with 10% (vol/vol) FBS and penicillin/streptomycin (1×, Amresco). 5 × 104 cells were plated on 48-well collagen-coated Biocoat plates (Becton Dickinson). 16–18 h after plating, cells were transfected with Lipofectamine 2000 (Life Technologies) according to the manufacturer’s protocol. Briefly, 1.5 µL of Lipofectamine 2000 was used to transfect 650 ng of total plasmid: 500 ng Cas9 expression plasmid, 125 ng sgRNA expression plasmid, and 25 ng near-infrared iRFP670 expressing plasmid (Addgene plasmid 45457)26. 12 h after transfection, the media was replaced with full serum media, with or without 4-HT (1 µM, Sigma-Aldrich T176). The media was replaced again 3–4 days after transfection. Five days after transfection, cells were trypsinized and resuspended in full serum media and analyzed on a C6 flow cytometer (Accuri) with a 488-nm laser excitation and 520-nm filter with a 20-nm band pass. Transfections and flow cytometry measurements were performed in triplicate.
High-throughput DNA sequencing of genome modifications
HEK293-GFP stable cells were transfected with plasmids expressing Cas9 (500 ng) and sgRNA (125 ng) as described above. For treatments in which a reduced amount of wild-type Cas9 expression plasmid was transfected, pUC19 plasmid was used to bring the total amount of plasmid to 500 ng. 4-HT (1 µM final), where appropriate, was added during transfection. 12 h after transfection, the media was replaced with full serum media without 4-HT. Genomic DNA was isolated and pooled from three biological replicates 60 h after transfection using a previously reported11 protocol with a DNAdvance Kit (Agencourt). 150 ng or 200 ng of genomic DNA was used as a template to amplify by PCR the on-target and off-target genomic sites with flanking HTS primer pairs described previously11. PCR products were purified using RapidTips (Diffinity Genomics) and quantified using the PicoGreen dsDNA Assay Kit (Invitrogen). Purified DNA was PCR amplified with primers containing sequencing adaptors, purified with the MinElute PCR Purification Kit (Qiagen) and AMPure XP PCR Purification (Agencourt). Samples were sequenced on a MiSeq high-throughput DNA sequencer (Illumina), and sequencing data was analyzed as described previously4.
Western blot analysis of intein splicing
HEK293-GFP stable cells were transfected with 500 ng Cas9 expression plasmid and 125 ng sgRNA expression plasmid. 12 h after transfection, the media was replaced with full serum media, with or without 4-HT (1 µM). Cells were lysed and pooled from three technical replicates 4, 8, 12, or 24 h after 4-HT treatment. Samples were run on a Bolt 4–12% Bis-Tris gel (Life Technologies). An anti-FLAG antibody (Sigma-Aldrich F1804) and an anti-mouse 800CW IRDye (LI-COR) were used to visualize the gel on an Odyssey IR imager.
Statistcal analysis
Statistical tests were performed as described in the figure captions. All p-values were calculated with the R software package. p-values for the Fisher exact test were calculated using the fisher.test function, with a one-sided alternative hypothesis (alternative = "greater" or alternative = "less", as appropriate). Upper bounds on p-values that are close to zero were determined manually. The Benjamini-Hochberg adjustment was performed using the R function p.adjust (method = "fdr").
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health R01 GM095501, Defense Advanced Research Projects Agency HR0011-11-2-0003 and N66001-12-C-4207, and the Howard Hughes Medical Institute (HHMI). K.M.D. acknowledges the Natural Sciences and Engineering Research Council (NSERC) of Canada for a PGS D scholarship. J.A.Z. acknowledges NIH fellowship support (F32GM106601). We are grateful to Jennifer Doudna, Sam Sternberg, David Taylor, Martin Jinek, and Fuguo Jiang for providing the structural coordinates of Cas9, and to John Guilinger, Yongjoo Kim, Margie Li, and Ahmed Badran for helpful discussions.
Footnotes
Author contributions
K.M.D., V.P., D.B.T., and D.R.L. designed the research. K.M.D. performed the experiments, and J.A.Z. assisted with high-throughput sequencing. K.M.D., V.P., D.B.T., and J.A.Z. analyzed the data. D.R.L. supervised the research. All authors wrote the manuscript.
Competing financial interests
The authors declare competing financial interests: the co-authors have filed a provisional patent application related to this work. D.R.L is a consultant for Editas Medicine, a company that applies genome-editing technologies.
References
- 1.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hwang WY, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nature biotechnology. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pattanayak V, et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nature biotechnology. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nature biotechnology. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nature biotechnology. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho SW, et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome research. 2014;24:132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsai SQ, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nature biotechnology. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mali P, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nature biotechnology. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ran FA, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nature biotechnology. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai S Q, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nature biotechnology. 2014;32 doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nature biotechnology. 2014;32 doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim S, Kim D, Cho SW, Kim J, Kim JS. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome research. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramakrishna S, et al. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome research. 2014;24:1020–1027. doi: 10.1101/gr.171264.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuris JA, et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nature biotechnology. 2015;33:73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez F, et al. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell stem cell. 2014;15:215–226. doi: 10.1016/j.stem.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pruett-Miller SM, Reading DW, Porter SN, Porteus MH. Attenuation of zinc finger nuclease toxicity by small-molecule regulation of protein levels. PLoS genetics. 2009;5:e1000376. doi: 10.1371/journal.pgen.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buskirk AR, Ong YC, Gartner ZJ, Liu DR. Directed evolution of ligand dependence: small-molecule-activated protein splicing. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:10505–10510. doi: 10.1073/pnas.0402762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuen CM, Rodda SJ, Vokes SA, McMahon AP, Liu DR. Control of transcription factor activity and osteoblast differentiation in mammalian cells using an evolved small-molecule-dependent intein. Journal of the American Chemical Society. 2006;128:8939–8946. doi: 10.1021/ja062980e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peck SH, Chen I, Liu DR. Directed evolution of a small-molecule-triggered intein with improved splicing properties in mammalian cells. Chemistry & biology. 2011;18:619–630. doi: 10.1016/j.chembiol.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jinek M, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343:1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Danielian PS, White R, Hoare SA, Fawell SE, Parker MG. Identification of residues in the estrogen receptor that confer differential sensitivity to estrogen and hydroxytamoxifen. Molecular endocrinology. 1993;7:232–240. doi: 10.1210/mend.7.2.8469236. [DOI] [PubMed] [Google Scholar]
- 25.Zetsche B, Volz SE, Zhang F. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nature biotechnology. 2015;33:139–142. doi: 10.1038/nbt.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shcherbakova DM, Verkhusha VV. Near-infrared fluorescent proteins for multicolor in vivo imaging. Nature methods. 2013;10:751–754. doi: 10.1038/nmeth.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.