SUMMARY
Chromosomes of metazoan organisms are partitioned in the interphase nucleus into discrete topologically associating domains (TADs). Borders between TADs are formed in regions containing active genes and clusters of architectural protein binding sites. Transcription of most genes is repressed after temperature stress in Drosophila. Here we show that temperature stress induces relocalization of architectural proteins from TAD borders to inside TADs, and this is accompanied by a dramatic rearrangement in the 3D organization of the nucleus. TAD border strength declines, allowing for an increase in long-distance inter-TAD interactions. Similar but quantitatively weaker effects are observed upon inhibition of transcription or depletion of individual architectural proteins. Heat shock-induced inter-TAD interactions result in increased contacts among enhancers and promoters of silenced genes, which recruit Pc and form Pc bodies in the nucleolus. These results suggest that the TAD organization of metazoan genomes is plastic and can be quickly reconfigured.
Keywords: epigenetics, transcription, chromatin, CTCF, Hi-C
INTRODUCTION
Architectural proteins (also known as insulator proteins) facilitate contacts among enhancers and promoters and are also responsible for bringing Pc-repressed genes together to form Pc bodies (Bonora et al., 2014; Li et al., 2013). The complex pattern of these interactions in the nucleus of eukaryotic cells can be visualized using Hi-C, and is manifested in the form of Topologically Associating Domains (TADs) (Nora et al., 2013). TADs are defined as regions of the genome undergoing high frequency of local contacts while interacting infrequently with sequences present in adjacent TADs (Nora et al., 2012). Borders between TADs are located in regions densely populated by actively transcribed genes, including housekeeping genes, and architectural proteins such as CTCF and Rad21 {Dixon, 2012 #51;Hou, 2012 #52;Sexton, 2012 #59}.
Although TADs are partially conserved, a subset of TADs differs among cell types (Dixon et al., 2012). Thus, TAD organization must change to some degree as cells implement different patterns of gene expression during differentiation. To address the dynamics of TAD organization during the establishment of new transcription programs, we examined possible changes in chromatin organization during the heat shock response. Drosophila cells respond to temperature stress by undergoing dramatic changes in their transcription profile. Expression of most active genes is quickly repressed while a few previously silenced genes, the heat shock genes, are up-regulated (Gonsalves et al., 2011; Guertin et al., 2012). The mechanisms underlying the silencing of most of the genome during temperature stress has not been explored in detail. The distribution of the CP190 architectural protein is altered after heat shock (Wood et al., 2011), suggesting that alterations in the genome-wide distribution of architectural proteins as a consequence of temperature stress may lead to changes in TAD organization, which in turn participate in the genome-wide silencing of gene expression.
Several architectural proteins have been characterized in Drosophila, including the DNA binding proteins CCCTC-binding factor (CTCF), Suppressor of Hairy-wing [Su(Hw)], Boundary Element Associated Factor 32 (BEAF-32), DNA Replication Related Element Binding Factor (DREF), Z4 (also known as Putzig), and Transcription Factor IIIC (TFIIIC). These proteins bind to specific sequences in the DNA and recruit a series of accessory factors essential for the establishment of interactions between distant elements, including Centrosomal Protein 190 (CP190), Modifier of mdg4 [Mod(mdg4)], Rad21 (cohesin), Cap-H2 (condensin II), the long isoform of Female sterile homeotic on chromosome 1 [Fs(1)h-L], L(3)mbt, Z4, and Chromator (also known as Chriz) (Van Bortle et al., 2014). These architectural proteins are distributed in the genome in an intricate pattern that correlates with their function. Architectural protein binding sites (APBSs) occur in clusters of variable complexity, resulting in genomic sites with different numbers of architectural proteins that display distinct functional properties. APBSs containing 9–13 architectural proteins are enriched at TAD borders and show classical insulator activity in functional reporter assays. APBSs with 5–8 architectural proteins are present inside TADs or at weak TAD borders, and show position-dependent weak insulator activity. Finally, sites in the genome with 4 or fewer architectural proteins are enriched inside TADs and show no insulator activity. These observations suggest that changes in the distribution of architectural proteins in the genome could result in APBSs with increased or decreased numbers of these proteins, altering their functional properties and their ability to form TAD borders (Van Bortle et al., 2014).
Here we use Hi-C to examine changes in the TAD organization of Drosophila chromosomes after heat shock. Results suggest that cells can regulate the distribution of architectural proteins, which become relocated from TAD borders to inside TADs to facilitate long-range interactions among enhancers and promoters after heat shock. The heat shock-specific enhancer-promoter interactions in turn become sequestered by the Pc silencing machinery to specific nuclear compartments to repress transcription genome-wide during temperature stress.
RESULTS
Chromatin 3D organization is dynamic and changes in response to temperature stress
Kc167 cells were exposed to heat shock for 20 min, fixed, and DNA was digested with HindIII to generate Hi-C genomic libraries. Sequencing of Hi-C libraries from two biological replicates resulted in a total of 503 million read pairs. After removal of duplicates, read pairs from ligation between adjacent fragments, and further normalization (Hou et al., 2012), we obtained 151 million read pairs uniquely aligned to the D. melanogaster dm3 reference genome (Table S1).
To investigate global changes in 3D chromatin architecture, we analyzed chromatin interaction frequencies across the entire genome for cells grown at normal temperature (NT) and subjected to heat shock (HS). We subtracted the NT contact matrix from that of HS (Figures S1A and S1B). Chromatin interactions within each chromosome arm increase dramatically after heat shock and interactions between telomeres increase, particularly on chromosome 3 (Figure S1A). Interestingly, interactions between the two chromosome arms increase for chromosome 3, producing a Rabl conformation, but do not change for chromosome 2, indicating that the changes observed are not a consequence of a non-specific, thermodynamically driven increase in interactions (Figures S1A and S1B). Furthermore, heat shock also results in the reduction of some chromatin interactions, specifically between centromeres and interactions of the predominantly heterochromatic chromosome 4 (Figure S1A).
We next compared intra-chromosomal contacts between NT and HS samples at single fragment resolution. For comparison, two-dimensional chromatin contact matrices were normalized by the total number of intra-chromosomal arm read pairs. We observe that the heat shock response increases long range chromatin interactions, often spanning large genomic distances, which contrasts with the distinct triangular TAD structures observed at normal temperature (Figure 1A and S1C). Subtracting chromatin contacts observed in NT from those observed in HS samples confirms the increase in long range chromatin contact frequencies and highlights a reciprocal decrease in contact frequency between neighboring loci (Figure 1B and S1D). Analysis of the decay in contact frequency with increasing genomic distance quantified the changes observed between NT and HS samples (Figure 1C). Chromatin interactions less than 140 kb apart were more abundant in NT (regression exponents NT=−0.926 and HS=−0.751), while contact frequencies spanning 140 kb to 1 Mb were more abundant in HS (regression exponents NT=−1.524 and HS=−1.095) (Figure 1C). Since the average TAD size in Drosophila cells is ~140 kb, this observation suggests that intra-TAD interactions are higher under normal temperature, while inter-TAD interactions are more prominent after heat shock (Figures 1D and S1F). Heat shock-specific interactions are mostly inter-TAD, while normal temperature-specific interactions are predominantly intra-TAD (Figures 1E and 1F). Together, these observations suggest that TAD structure is dynamic and can rapidly reorganize, within 20 min, in response to temperature stress.
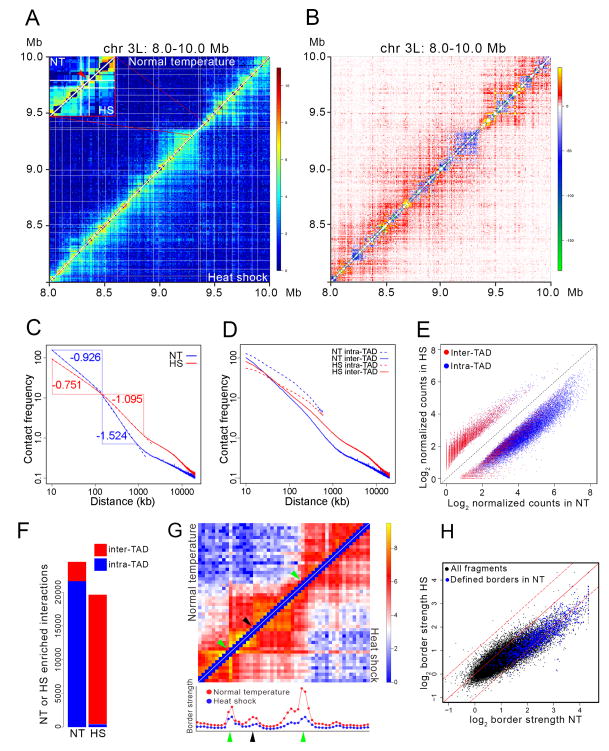
Figure 1. Changes in TAD organization after heat shock.
(A) Hi-C contact maps at single fragment resolution for control (upper left) and heat shock (lower right) Kc167 cells plotted with normalized intra-chromosomal arm read pairs in a 2 Mb region of chromosome 3L. White lines denote TAD borders in control cells. Inset shows an enlargement of a 34-HindIII fragment region of chromosome 3L containing several heat shock genes within a 16 kb interval; red arrow indicates the location of these genes.
(B) Subtraction of normal temperature control from heat shock contact matrices shows changes in contact frequencies between the two Hi-C samples. Region of chromosome 3L depicted is the same as in Figure 1A.
(C) Intra-chromosome arm chromatin contact frequency decay curves of control (NT, blue) and heat shock (HS, red) Hi-C data. Point of intersection between the vertical blue and horizontal red lines is located at 140 kb.
(D) Intra-chromosome arm chromatin contact frequency decay curves of control (NT, blue) and heat shock (HS, red) for intra- and inter-TAD interactions.
(E) Scatter plot of intra- and inter-TAD contact frequencies of control-specific (NT) and heat shock-specific (HS) interactions. Interactions were calculated based on counts present in 5 kb bins. Diagonal dashed black line shows the result assuming there is no difference in intra- or inter-TAD contacts between NT and HS.
(F) Number of intra- and inter-TAD interactions specific for the control (NT) and heat shock (HS) Hi-C data sets.
(G) Contact matrices of a ~200 kb region from cells grown at normal temperature (upper left) or heat shocked (lower right) at single fragment resolution. Green arrows indicate the location of TAD borders determined computationally. Black arrow indicates the location of a sub-TAD border determined visually. Graphs below the heat maps indicate the border strength for each restriction fragment shown in the heat maps above. Red and blue dots indicate border strength in arbitrary units in control and heat shocked cells, respectively.
(H) Scatter plot comparing TAD border strength in control (NT) and heat shock (HS) Hi-C samples. Blue dots indicate border strength for TAD borders determined using a probability-based model. Black dots indicate border strength for all HindIII restriction fragments in the genome. Dashed lines above and below the diagonal indicate + or − one standard deviation, respectively.
See also Figure S1.
Temperature stress alters TAD border strength
Changes in TAD borders could explain the redistribution of interactions from within TADs to between TADs observed after heat shock. To describe TAD borders in a quantitative manner, we and others have established the concept of TAD border strength: the ratio between inter-TAD and intra-TAD interactions on either side of the border (Sofueva et al., 2013; Van Bortle et al., 2014). Using this definition, every restriction site in the genome can be assigned a quantitative border strength and any specific fragment above a certain threshold will be defined as a TAD border by previously published methods. This strategy allows us to quantitatively evaluate the ability of every restriction fragment in the genome to form a border, independent of whether it is defined as a TAD border by standard methods. To illustrate this concept, we evaluated border strength after heat shock for a ~200 kb region (Figure 1G). Border strength decreases after heat shock at every restriction fragment present in this region, including both standard TAD borders (green arrows) and a non-borders (black arrow). We then extended this analysis of border strength to the whole Drosophila genome in NT and HS samples (Figure 1H). Border strength at most standard TAD borders (blue dots) decreases after heat shock. This is also observed for most HindIII fragments in the genome (black dots), although a subset of fragments show an increase in border strength in cells subjected to temperature stress (Figure 1H). Thus, heat shock reduces border strength at a majority of HindIII fragments and traditional TAD borders, which agrees with the observed increase in inter-TAD interactions.
Inhibition of transcription has a weak effect on TAD border strength and 3D organization
Since TAD borders are present in regions containing highly transcribed genes, the decrease in TAD border strength after heat shock may be a consequence of the absence of transcription. In addition, the decrease in intra-TAD interactions, may represent lost contacts between promoters and regulatory sequences after transcriptional silencing. This is suggested by ChIP-seq analysis of RNA polymerase II (RNAPII) distribution, which shows a dramatic decrease in RNAPII at gene promoters and coding regions in heat shocked cells (Figure 2A). To explore the possibility that transcriptional silencing contributes to the changes in 3D organization observed after heat shock, we inhibited transcription by treating cells with flavopiridol or triptolide. Treatment with flavopiridol inhibits elongation and pauses RNAPII (Jonkers et al., 2014), resulting in lower levels of RNAPII phosphorylated in Ser5 and loss of RNAPII phosphorylated in Ser2 (Figure 2B). Triptolide inhibits transcription initiation and results in degradation of RNAPII (Jonkers et al., 2014), resulting in a further decrease of RNAPII phosphorylated in Ser5 and absence of RNAPII phosphorylated in Ser2 (Figure 2B). We then compared RNAPII occupancy after treatment with transcription inhibitors or heat shock using ChIP-seq (Figure 2A). Flavopiridol depletes RNAPII throughout the coding regions of genes, resulting in an accumulation of total RNAPII at promoters. In contrast, triptolide results in a decrease in RNAPII at promoters and coding regions of genes similar to that observed after heat shock (Figure 2A). Heat shock dramatically decreases RNAPII at most promoters, with most TSSs containing low levels of RNAPII (Figure 2C). Cells treated with flavopiridol show a modest decrease in the number of enhancers and promoters containing high levels of RNAPII, while triptolide treatment exhibits a more pronounced decrease (Figure 2C).
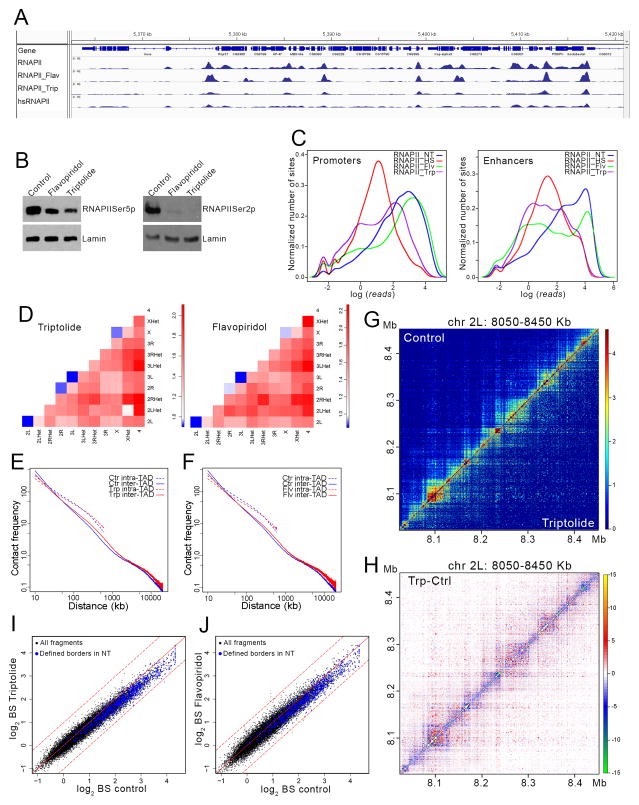
Figure 2. Effect of transcription inhibition on 3D organization.
(A) Screenshot of a region of the Drosophila genome showing ChIP-seq signal for RNAPII in control, flavopiridol-treated, triptolide-treated, and heat shocked cells.
(B) Western analysis of control, flavopiridol-treated and triptolide-treated cells using antibodies to the CTCD domain of RNAPII phosphorylated in serine 5 (left) or serine 2 (right). Antibodies to lamin were used for the loading control (bottom).
(C) Distribution of ChIP-seq normalized reads with respect to sites in the genome at enhancers and promoters in cells grown at normal temperature, heat shocked, and treated with tripdolide or flavopiridol.
(D) Changes in frequency of intra- and inter-arm interactions obtained from Hi-C data in cells treated with triptolide or flavopiridol relative to untreated cells.
(E) Decay curves of intra- and inter-TAD interactions with respect to distance for control and triptolide-treated cells.
(F) Decay curves of intra- and inter-TAD interactions with respect to distance for control and flavopiridol-treated cells.
(G) Two-dimensional contact matrices of Hi-C data obtained with control (upper left) and triptolide-treated (bottom right) cells.
(H) Subtraction heat map in which the contact matrix for a 400 kb of chromosome 2L obtained from Hi-C data in control cells has been subtracted from that obtained in triptolide-treated cells.
(I) Scatter plot comparing border strength (BS) in Hi-C samples from control and triptolide-treated cells. Blue dots indicate TAD borders determined using a probability-based model. Black dots indicate all DpnII restriction fragments in the genome. Dashed lines above and below the diagonal indicate + or − one standard deviation, respectively.
(J) Scatter plot comparing border strength (BS) in Hi-C samples from control and flavopiridol-treated cells. Blue dots indicate TAD borders determined using a probability-based model. Black dots indicate all DpnII restriction fragments in the genome. Dashed lines above and below the diagonal indicate + or − one standard deviation, respectively.
To examine the effects of inhibiting transcription initiation or elongation on the 3D nuclear interactome, we generated Hi-C libraries, using DpnII instead of HindIII, with Kc167 cells treated with triptolide or flavopiridol. We obtained 169 million processed read pairs for the untreated control, 65.8 million read pairs for triptolide-treated and 37.8 million for flavopiridol-treated cells from two biological replicates for each condition (Table S1). The changes in chromatin contact frequencies are very similar for cells treated with either triptolide or flavopiridol. Interactions between centromeres increase after transcription inhibition, contrasting with the marked decrease observed after heat shock (Figure 2D). Decay curves of intra-arm interactions follow a similar but less pronounced pattern as that observed after temperature stress (Figure 2E and 2F), and inter-chromosomal interactions increase with respect to untreated cells (Figure 2D). Intra-TAD interactions decrease slightly whereas inter-TAD interactions increase, most notably at distances above 100 kb. These changes can be visually observed in two-dimensional contact matrices (Figure 2G), and in heat maps in which the contact matrix of control cells has been subtracted from that of treated cells (Figure 2H). Similar to heat shocked cells, chromatin interactions are redistributed, resulting in fewer intra-TAD interactions and more inter-TAD interactions compared to control. In addition, TAD border strength also decreases after treatment with triptolide or flavopiridol (Figure 2I and 2J). These findings suggest that the genome-wide inhibition of transcription that occurs after temperature stress may contribute to the observed changes in TAD organization but that additional factors are required to account for all of the changes observed after heat shock.
The genome-wide distribution of architectural proteins is altered after temperature stress
Since architectural proteins are enriched at TAD borders, it is possible that they also contribute to the changes in 3D organization observed after temperature stress. To determine whether the distribution of architectural proteins is affected by heat shock, we conducted ChIP-seq with antibodies to Su(Hw), CTCF, BEAF-32, TFIIIC, DREF, Rad21, Cap-H2, L(3)mbt, Fs(1)h-L, Mod(mdg4), CP190, Z4, and Chromator in NT and HS cells. The number of sites in the genome containing architectural proteins increases for all proteins except for Cap-H2 and Rad21 (Figure 3A). We next evaluated TAD border strength in relation to architectural protein occupancy. We divided all restriction fragments in the genome into ten groups based on border strength and assigned each group a border strength between 1 and 10. Each group was then subdivided into 12 subgroups based on the number of architectural proteins present (0–2, 3, 4, through 13) and, for each subgroup, we determined the amount of RNAPII present based on the number of normalized ChIP-seq reads. Most TAD borders with the highest strengths contain high amounts of RNAPII and more APBSs highly occupied by architectural proteins, whereas the weakest borders are depleted of RNAPII and contain APBSs with low occupancy (Figure 3B). This relationship is maintained in HS cells (Figure 3C), in which case the border strength is lower for the same number of architectural proteins present at specific borders (Figure 3D).
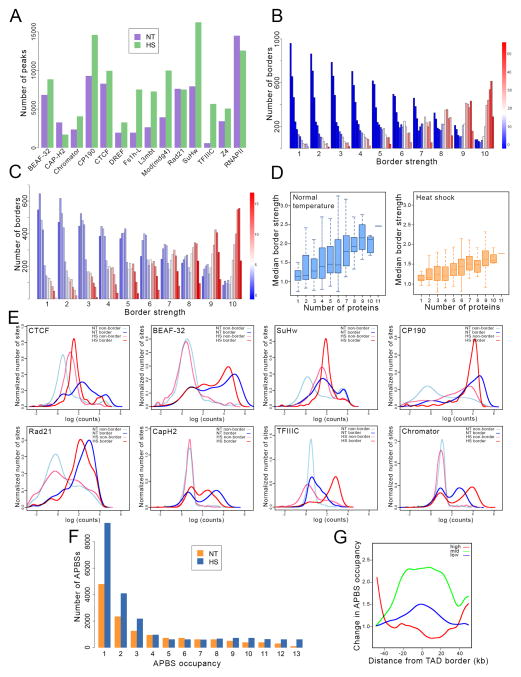
Figure 3. Genome-wide redistribution of architectural proteins during heat shock.
(A) Number of ChIP-seq peaks for different architectural proteins and RNAPII in control (NT) and heat shocked (HS) cells.
(B) Number of borders found in Hi-C data from control cells for each border strength, which ranges from 1 to 10 in arbitrary units. For each border strength, the number of borders containing different amounts of RNAPII (red to blue) and APBSs occupied with different numbers of architectural proteins (12 columns for each border strength ranging in occupancy from 1 to 12 from left to right). Color bar for RNAPII indicates number of mapped ChIP-seq reads in 50 bp bins normalized relative to the total number of reads.
(C) Number of borders found in Hi-C data from heat shocked cells for each border strength, which ranges from 1 to 10 in arbitrary units. For each border strength, the number of borders containing different amounts of RNAPII (red to blue) and APBSs occupied with different numbers of architectural proteins (12 columns for each border strength ranging in occupancy from 1 to 12 from left to right).
(D) Box plots indicating the relationship between median border strength and the number of architectural proteins present at each border for control (left) and heat shocked (right) cells.
(E) Relationship between the normalized number of sites in the genome and the number of normalized ChIP-seq reads for different architectural proteins at TAD borders and non-borders in control and heat shocked cells.
(F) Change in the number of architectural protein binding sites (APBSs) with different numbers of architectural proteins (APBS occupancy) in heat shocked (HS) with respect to control (NT) cells.
(G) Changes in APBS occupancy with respect to the distance from TAD borders for TAD borders with low (1–4) mid (5–8) and high (9–13) number of architectural proteins in heat shocked cells with respect to control.
We then compared changes in architectural proteins at TAD borders and non-border regions. In all cases, with the exception of Cap-H2, the number of sites containing high amounts of bound architectural proteins increases at non-borders with respect to those at TAD borders after temperature stress (Figure 3E). This reorganization results in a higher number of low and high occupancy APBSs, whereas the number of medium occupancy APBSs is unchanged (Figure 3F). Furthermore, the numbers of low and medium occupancy APBS sites increases at TAD borders after heat shock. However, the number of highly occupied APBSs increases in regions located away from TAD borders (Figure 3G). The decrease in APBS occupancy at TAD borders and its increase inside TADs after heat shock may contribute to the observed decrease in TAD border strength and increase in inter-TAD interactions.
Highly occupied APBSs established after heat shock are enriched at enhancers and promoters
To gain insight into the function of high occupancy APBSs located inside TADs in HS cells, we examined the location of architectural proteins before and after heat shock with respect to enhancers and promoters. We used enhancer sequences identified by two different methods (see supplemental information). The results are very similar for chromatin-based and functionally defined enhancers, so only the results for the latter are shown (Figure 4A). After heat shock, the amount of architectural proteins, with the exception of Cap-H2, bound at enhancers, and the number of enhancer sites bound by architectural proteins, increases. Cap-H2 follows the opposite pattern, decreasing in both the number of enhancers bound and the amount of CAP-H2 bound to enhancers. A similar pattern is observed at gene promoters (Figure 4A), suggesting that the changes in architectural protein distribution observed after heat shock may serve to mediate novel interactions among these sequences.
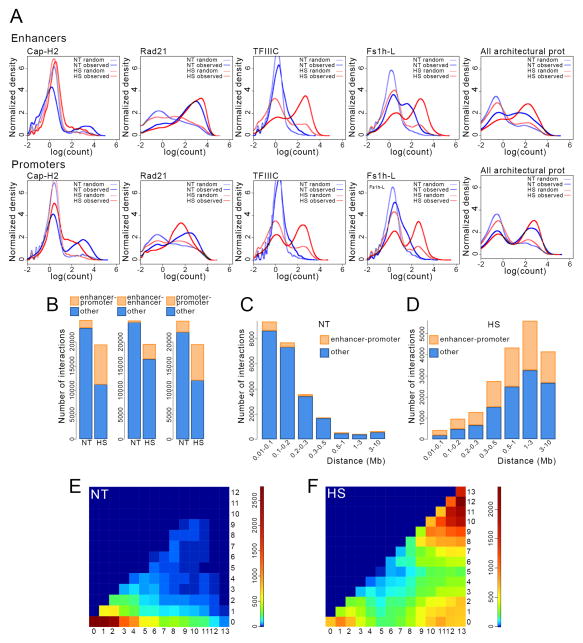
Figure 4. Changes in enhancer-promoter interactions in heat shocked cells.
(A) Relationship between the number of sites and the ChIP-seq read count for various architectural proteins in control and heat shocked cells at enhancers and promoters.
(B) Changes in the number of interactions among enhancers and promoters in heat shocked (HS) with respect to control (NT) cells.
(C) Number of interactions at different distances for enhancer-promoter contacts in cells grown at normal temperature (NT).
(D) Number of interactions at different distances for enhancer-promoter contacts in heat shocked cells (HS).
(E) Frequency of interactions between enhancers and promoters containing different numbers (1–13) of architectural proteins in cells grown at normal temperature (NT).
(F) Frequency of interactions between enhancers and promoters containing different numbers (1–13) of architectural proteins in cells subjected to temperature stress (HS).
See also Figure S2.
To determine whether increased architectural protein occupancy at enhancers and promoters changes the pattern of their chromatin contacts, we compared the chromatin interaction frequency data from Hi-C in NT and HS samples. Enhancer-promoter, enhancer-enhancer, and promoter-promoter interactions increase dramatically after heat shock, comprising >90% of all the heat shock-specific interactions (Figure 4B). Interactions between enhancers and promoters predominantly occur within relatively short distances (< 300 kb) at normal temperature (Figure 4C), while enhancer-promoter contacts span up to 10 Mb after heat shock (Figure 4D). In agreement with these observations, most enhancer-promoter interactions occur intra-TAD in control cells and inter-TAD after heat shock (Figure S2A). In general, most enhancer-promoter contacts in control cells involve APBSs with an occupancy of 0 to 4 architectural proteins (Figure 4E) and involve sequences located in the same TAD (Figure S2B). However, enhancer-promoter interactions that occur between TADs are mediated by medium occupancy APBSs (Figure S2B). In contrast, heat shock results in an enrichment of APBSs containing 9–13 architectural proteins at sites of enhancer-promoter interactions (Figure 4F) with most interactions occurring between TADs (Figure S2B). These results suggest that redistribution of architectural proteins after heat shock results in the formation of new highly occupied APBSs, which then mediate long-rage inter-TAD enhancer-promoter interactions and contribute to the global 3D chromatin changes observed after temperature stress.
Cohesin and Condensin II have opposite effects on enhancer-promoter interactions
Contrary to other architectural proteins, redistribution of Cap-H2 after heat shock leads to fewer sites and lower levels at enhancers and promoters (Figure 4A). Cap-H2 has been shown to disrupt transvection by antagonizing inter-chromosome contacts required for enhancer-promoter interactions in trans {Bauer, 2012 #270;Hartl, 2008 #276}. Thus, the normal role of Cap-H2 may include the negative regulation of enhancer-promoter contacts in cis. Downregulation of Cap-H2 after heat shock may then be required for disruption of short-range intra-TAD interactions. To determine the contribution of Cap-H2 downregulation to dynamic 3D chromatin remodeling after heat shock, we examined the effect of Cap-H2 depletion on chromatin interactions (Figure S2C). ChIP-seq analysis demonstrates that RNAi treatment results in depletion of Cap-H2 from chromatin (Figure S3A). We then generated Hi-C libraries from Cap-H2 depleted cells using DpnII and obtained a total of 139 million processed read pairs from two biological replicates (Table S1) and we subtracted two dimensional contact matrices of control cells from Cap-H2-depleted cells. Contrary to observations in heat shocked cells, intra-chromosome arm interactions are reduced with respect to control (Figure 5A). However, as is the case in cells subjected to temperature stress, interactions between pericentromeric regions, telomeres and chromosome arms increase, such that chromosomes 2 and 3 appear to acquire a Rabl configuration (Figure 5A and S3B). These data suggest that Cap-H2 downregulation reproduces some aspects of the global changes in chromatin architecture observed after heat shock.
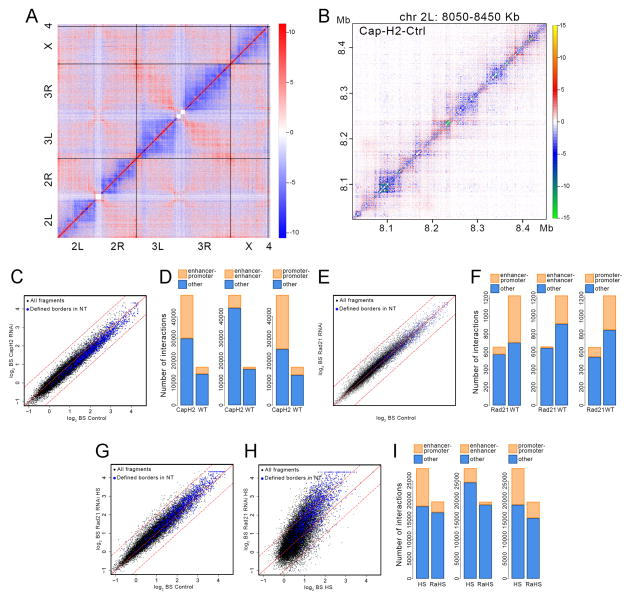
Figure 5. Effect of Cap-H2 and Rad21 depletion on 3D chromatin organization.
(A) Whole genome heat map obtained by subtracting the two dimensional contact matrix of Hi-C data from control cells from that of Cap-H2 depleted cells. Black vertical and horizontal lines indicate chromosome boundaries.
(B) Heat map for a 400 kb region of chromosome 2L obtained by subtracting the two dimensional contact matrix of Hi-C data from control cells from that of Cap-H2 depleted cells.
(C) Scatter plot comparing border strength (BS) in Hi-C samples from control and Cap-H2 depleted cells. Blue dots indicate TAD borders determined using a probability-based model. Black dots indicate all DpnII restriction fragments in the genome. Dashed lines above and below the diagonal indicate + or − one standard deviation, respectively.
(D) Number of interactions among enhancers and promoters specific for normal (WT) and Cap-H2-depleted (CapH2) cells.
(E) Scatter plot comparing border strength (BS) in Hi-C samples from control and Rad21 depleted cells. Symbols are as in panel C.
(F) Number of interactions among enhancers and promoters specific for normal (WT) and Rad21-depleted (Rad21) cells.
(G) Scatter plot comparing border strength in Hi-C samples from control and Rad21 depleted cells subjected to heat shock. Symbols are as in panel C.
(H) Scatter plot comparing border strength in Hi-C samples from normal control and Rad21 depleted cells, both subjected to heat shock. Symbols are as in panel C.
(I) Number of interactions among enhancers and promoters specific for heat shocked (HS) and Rad21-depleted cells subjected to heat shock (RaHS).
See also Figure S3.
At a higher resolution, changes in 3D interactions are similar but less pronounced after Cap-H2 knockdown than after heat shock (Figure 5B and S3C). Intra-TAD interactions decrease after Cap-H2 depletion (Figure 5B), but regions containing Cap-H2 in control cells show an increase of short-range intra-TAD interactions after heat shock (Figure S3D). The redistribution of chromatin interactions after Cap-H2 depletion can be illustrated by changes in border strength, which is slightly reduced, both at TAD borders determined in control cells and at most DpnII fragments in the genome (Figure 5C). Interestingly, interactions among enhancers and promoters increase in Cap-H2 depleted cells when compared to control, suggesting that Cap-H2 antagonizes enhancer-promoter interactions in normal cells (Figure 5D).
The antagonizing effects of Cap-H2 on enhancer-promoter interactions contrast with the function of another architectural protein, Rad21. In mammals, Rad21 interacts with CTCF or Mediator to facilitate interactions between regulatory sequences (DeMare et al., 2013; Dowen et al., 2014; Kagey et al., 2010; Phillips-Cremins et al., 2013). To investigate the possible contribution of Rad21 to heat shock-induced changes in enhancer-promoter interaction, we depleted Rad21 in Kc167 cells using dsRNAs (Figure S2D) and generated Hi-C libraries using DpnII. We obtained a total of 103 million processed read pairs from two biological replicates (Table S1) and compared the chromatin interactions to controls. Downregulation of Rad21 causes a small but detectable reduction in border strength at TAD borders defined in control cells and at most DpnII fragments in the genome (Figure 5E). Contrary to Cap-H2, depletion of Rad21 results in a decrease in enhancer-promoter, enhancer-enhancer and promoter-promoter interactions, in agreement with studies in mammals suggesting that Rad21 facilitates interactions between regulatory regions (Figure 5F).
To further test the role of Rad21 during heat shock, we subjected Rad21-depleted cells to temperature stress and we generated Hi-C libraries. We obtained a total of 83 million processed read pairs (Table S1). Contrary to the effects observed after heat shock alone, Rad21-depleted cells subjected to temperature stress show a slight increase in border strength with respect to cells grown at normal temperature (Figure 5G). This increase in TAD border strength is more dramatic when compared to normal cells subjected to heat shock (Figure 5H). Furthermore, Rad21-depleted cells subjected to heat shock show a decrease in interactions among enhancers and promoters with respect to normal heat shocked cells (Figure 5I). These results suggest that Rad21 plays an important role in mediating changes in enhancer-promoter interactions during the heat shock response.
Changes in chromatin structure in heat shocked cells
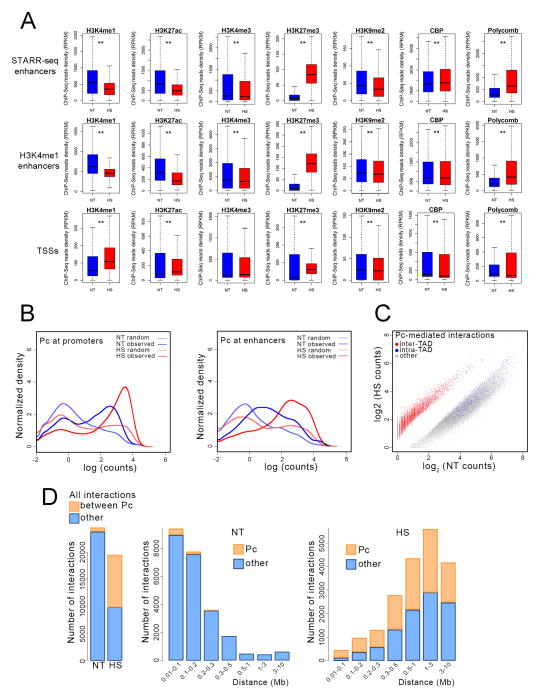
It is possible that the widespread silencing of most of the genome in heat shocked cells involves not only changes in the 3D enhancer-promoter interactomes but also alterations in covalent histone modifications. To investigate this possibility we conducted ChIP-seq in control and heat shocked cells with antibodies to H3K4me1, H3K27ac, H3K4me3, H3K27me3, H3K9me2, CBP, and Polycomb (Pc). Consistent with global downregulation of transcription, histone modifications characteristic of actively transcribed genes decrease at enhancers and promoters in heat shocked cells, H3K27me3 increases at these two types of regulatory sequences, and H3K9me2 remains the same (Figure 6A, S4A and S4B). Interestingly, CBP levels do not change at enhancers and promoters after heat shock, in spite of the decrease in H3K27ac, whereas levels of Pc increase dramatically at these sequences (Figure 6A, S4A and S4B). Furthermore, there is an increase in both the number of sites and the amount of protein present at Pc sites in both enhancers and promoters after heat shock (Figure 6B).
Figure 6. Changes in histone modifications and chromatin proteins in heat shocked cells.
(A) Box plots indicating changes in the levels of various histone modifications and chromatin proteins at enhancers and promoters of control (blue) versus heat shocked (red) cells. ** indicates p<0.001
(B) Changes in the number of Pc sites at enhancers and promoters and the read number at each site in control (NT) versus heat shocked (HS) cells.
(C) Scatter plot showing changes in intra- and inter-TAD interactions between DNA fragments containing Pc in control (NT) versus heat shocked (HS) cells.
(D) Changes in the number of interactions and the distance between the interacting Pc-containing DNA fragments in control (NT) versus heat shocked (HS) cells.
See also Figure S4
To further analyze the possible involvement of Pc in the heat shock-induced silencing of most previously active genes, we examined contacts between fragments containing Pc at both ends of the interacting sites. In normal control cells, most contacts between Pc-containing fragments take place within TADs, with a few inter-TAD contacts, whereas most Pc-containing interactions in heat shocked cells occur between fragments located in different TADs (Figure 6C). Furthermore, cells subjected to temperature stress display a dramatic increase in interactions between Pc-containing fragments (Figure 6D). In addition, whereas Pc-containing interactions are short-range in control cells, these interactions take place between fragments located at long distances from each other in cells undergoing heat shock (Figure 6D).
Pc-containing enhancers and promoters form long-distance interactomes in heat shocked cells
It is possible that architectural proteins are involved in the establishment of new Pc-containing interactions in heat shocked cells, which in turn may play a role in bringing enhancers and promoters of silenced genes together for efficient silencing. To test this hypothesis, we examined interactions between fragments with Pc-containing enhancers or promoters. In control cells, most interactions between enhancers and promoters lack Pc, whereas most interactions among enhancers and promoters in heat shocked cells also contain Pc (Figure 7A). We then used the HiC data to examine the effect of depleting architectural proteins on Pc-containing enhancer-promoter interactions. Depletion of Rad21 results in a decrease of Pc-containing interactions among enhancers and promoters (Figure 7B), whereas depletion of Cap-H2 has the opposite effect (Figure 7C). Furthermore, depletion of Rad-21 reverses the effect of heat shock on Pc-containing interactions among enhancers and promoters, causing a decrease in the frequency of these contacts (Figure 7D).
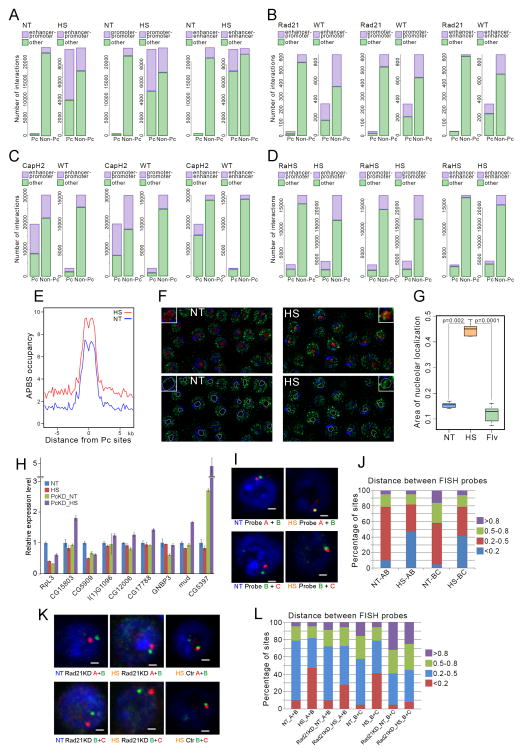
Figure 7. Analysis of Pc-mediated interactions by Hi-C and FISH.
(A) Number of interactions among Pc-containing enhancers and promoters in control (NT) and heat shocked (HS) cells.
(B) Number of interactions among enhancers and promoters specific for normal (WT) and Rad21-depleted (Rad21) cells.
(C) Number of interactions among enhancers and promoters specific for normal (WT) and Cap-H2-depleted (CapH2) cells.
(D) Number of interactions among enhancers and promoters specific for normal (HS) and Rad21-depleted (RaHS) cells, both subjected to heat shock.
(E) Distribution of APBSs with different occupancies around Pc sites in control and heat shocked cells
(F) Immunofluorescence microscopy of control (NT) and heat shocked (HS) cells stained with antibodies to Pc (green) and fibrillarin (red, nucleolus); DAPI is indicated in blue. The location of the nucleolus has been traced with a white line in the lower panels based on the location of fibrillarin signal. Insets show an enlarged region containing the nucleolus from one of the cells.
(G) Box plots showing levels of Pc protein in the nucleolar region of control (NT), heat shocked (HS) and flavopiridol-treated (Flv) cells.
(H) RNA expression levels of nine different genes under different experimental conditions. NT denotes control cells grown at normal temperature and HS indicates control cells subjected to heat shock at 36.5 C for 20 min. PcKD denotes cells depleted of Pc by RNAi and grown at normal temperature (PcKD_NT) or subjected to heat shock (PcKD_HS).
(I) FISH using three Pc-containing DNA fragments, A, B and C, shown to interact by Hi-C. NT denotes cells grown at normal temperature and HS indicates cells heat shocked at 36.5 °C. White bar indicates 1 μm.
(J) Analysis of distances between FISH probes using cells grown at normal temperature (NT) or subjected to heat shock (HS). Distances are indicated in μm.
(K) FISH using three Pc-containing DNA fragments, A, B and C, shown to interact by Hi-C. NT denotes cells grown at normal temperature and HS indicates cells heat shocked at 36.5 °C. White bar indicates 1 μm. Rad21KD denotes cells depleted of Rad21 by RNAi and Ctr indicates untreated control cells.
(L) Analysis of distances between FISH probes using control or Rad21 depleted (Rad21KD) cells grown at normal temperature (NT) or subjected to heat shock (HS). Distances are indicated in μm.
See also Figure S3.
Pc-mediated interactions normally result in the formation of Pc bodies, which are thought to be the manifestation of multiple Pc sites interacting in the 3D nuclear space (Grimaud et al., 2006). To test whether the pattern of Pc bodies is altered in nuclei from cells exposed to temperature stress, we carried out immunofluorescence microscopy with antibodies to Pc. Results show that the distribution of Pc bodies is different in heat shocked versus control cells (Figure 7E). More specifically, Pc is excluded from the nucleolar region in cells grown at normal temperature, but Pc bodies are clearly detected within the nucleolus after heat shock (Figures 7F and 7G). This result is not simply a consequence of inhibition of transcription, since treatment of cells with flavopiridol has no effect on Pc body distribution (Figure 7G). Furthermore, the levels of Pc at rDNA loci do not increase in cells exposed to temperature stress, indicating that the formation of nucleolar Pc bodies is not due to silencing of rDNA genes during heat shock (Figure S3E). This was confirmed by measuring uridine incorporation into the nucleoli of cells grown at normal temperature or heat shocked, showing no difference in incorporation between the two conditions (Figure S2F). Therefore, the observed changes in Pc body distribution support the model that Pc-containing enhancers and promoters interact throughout the nucleus after heat shock, and may contribute to the generalized silencing of most of the genome observed in heat shocked cells. To test this hypothesis, we chose 9 genes previously found to undergo a decrease in RNA levels after heat shock (Gonsalves et al., 2011; Guertin et al., 2012) and used qPCR to measure the levels of transcripts encoded by these genes in normal cells and in cells depleted of Pc using RNAi (Figure S2D). With the exception of CG5397, most genes do not become de-repressed in cells depleted of Pc and grown at normal temperature, probably due to the fact that Pc does not normally silence these genes (Figure 7H). However, expression of all genes fails to be silenced in heat shocked cells depleted of Pc. These results support the hypothesis that Pc contributes to the silencing of most genes during the heat shock response.
To further test the role of architectural proteins in the long range interactions among Pc-containing enhancers and promoters observed in heat shocked cells, we chose three regions, A, B, and C, spanning 10 kb each. These three regions contain Pc sites after heat shock and were found by Hi-C to interact together in temperature-stressed but not control cells. We then carried out fluorescence in situ hybridization (FISH) with pair-wise combinations of these three regions. Results from these experiments indicate that the three regions co-localize at low frequency in the nucleus in control cells but co-localize very often in cells exposed to temperature stress (Figure 7I). Quantification of the distance between the two probes in each pairwise combination indicates that the loci co-localize (less than 0.2 μm distance) in 5–10% of control cells, but this number increases to 40–45% in heat shocked cells (Figure 7I and 7J). These results validate observations obtained from Hi-C experiments suggesting that these three sequences interact in the nucleus of heat shocked but not control cells. We then tested the possible role of Rad21 in the maintenance of interactions among the three loci after heat shock using FISH. Depletion of Rad21 using RNAi (Figure S2E) does not cause a measurable alteration in interactions between loci A, B and C in cells grown at normal temperature (Figure 7K). This is expected, since these three loci do not interact in control cells. However, depletion of Rad21 interferes with the ability of loci A, B and C to interact in cells subjected to heat shock (Figure 7K). Quantification of the results indicates that depletion of Rad21 results in a decrease of overlapping frequency (<0.2 μm) in heat shocked cells from 47% to 28% for loci A and B, and from 41 % to 8 % for loci B and C (Figure 7L). These results support observations obtained by Hi-C and suggest that Rad21 participates in the establishment and/or maintenance of long range interactions in heat shocked cells.
DISCUSSION
To try to gain insights into whether TAD organization is a cause or a consequence of changes in gene expression we examined differences in 3D organization during the heat shock response in Drosophila. We find that transcription changes taking place after temperature stress are accompanied by dramatic alterations in border strength throughout the genome, including standard TAD borders. These changes can be interpreted to suggest that TADs do not actually form de novo or disappear. Instead, cells are able to alter the strength of TAD borders, thus biasing intra- versus inter-TAD interactions. Alterations in TAD border strength observed in heat shocked cells could be a consequence, rather than an effector, of transcription changes. However, the decrease in TAD border strength observed after inhibition of transcription by treatment with triptolide or flavopiridol is modest and much smaller than that observed after heat shock, suggesting that absence of transcription is not sufficient to account for the decline in TAD border strength observed in heat shocked cells. Instead, architectural proteins, which undergo a dramatic rearrangement during heat shock, appear to play a more critical role in the observed alterations in TAD border strength. We suggest a model whereby, in response to temperature stress, transcription is paused and elongation ceases due to dephosphorylation of H3S10 {Karam, 2010 #119}. Architectural proteins are then re-distributed from TAD borders to sites located inside TADs, probably by mechanisms that include covalent modification of some of these proteins The newly formed high occupancy APBSs located inside TADs can now mediate long-range interactions with sequences present in other TADs. According to this model, TAD-based organization is not a consequence of transcription. Instead, the redistribution of architectural proteins during heat shock establishes new interaction patterns in the genome, which in turn determine TAD organization.
Specific architectural proteins may have distinct roles in the establishment of 3D genome organization. However, results presented here suggest that border strength directly correlates with the number and levels of all architectural proteins present at the border, rather than the presence of specific proteins. Nevertheless, knock down of either Rad21 or Cap-H2 has important consequences on long-range interactions among enhancers and promoters. Furthermore, depletion of Rad21 protein followed by heat shock reverses the effect of temperature stress on enhancer-promoter contacts, underscoring its role in mediating long-range interactions among these regulatory sequences after heat shock. The function of Cap-H2 in chromatin biology during interphase is less understood than that of Rad21. The observation that interactions among promoters and enhancers increase in cells depleted of Cap-H2 is in line with previous findings suggesting a role for this protein in transvection (Bauer et al., 2012; Hartl et al., 2008; Joyce et al., 2012). The opposite roles of Rad21 and Cap-H2 on enhancer-promoter interactions is surprising, and points to very specific mechanisms by which the cohesin and condensin complexes seem to operate to ensure proper enhancer-promoter contacts. We suggest that the apparent antagonistic roles of these two proteins may be aimed at ensuring the non-promiscuity of enhancer-promoter communication.
During the heat shock response intra- and inter-chromosomal interactions are redistributed from short-range intra-TAD to long-range inter-TAD. This redistribution may be critical in the chain of events leading to the silencing of most of the Drosophila genome that follows temperature stress. Enhancers and promoters of previously active genes become populated by Pc during the heat shock response. This increase in Pc could be sufficient for the observed repression of transcription after heat shock. However, Pc silencing requires trans interactions among several Pc-containing loci to increase the local concentration of this and other PRC1 components for efficient repression (Cheutin and Cavalli, 2014; Delest et al., 2012; Pirrotta and Li, 2012; Schwartz and Pirrotta, 2007). These long range contacts among Pc-containing sites in HS cells are possible by the decrease in TAD border strength and the ensuing switch from intra-TAD to inter-TAD interactions observed after temperature stress. How do Pc-containing sequences come together in the 3D nuclear space? In Drosophila, most Pc response elements (PREs) are adjacent to binding sites for architectural proteins, and it has been suggested that these proteins play an important role in bringing together distant Pc-containing sequences (Li et al., 2011; Li et al., 2013). Therefore, the redistribution of architectural proteins after temperature stress may have two related goals: to decrease border strength in order to allow inter-TAD interactions, and to assemble new highly occupied APBSs at Pc-containing enhancers and promoters to facilitate their contact for efficient silencing. This conclusion is supported by the fact that depletion of Rad21 results in a decrease in contacts between Pc-containing sites. The assembly of new Pc bodies containing Pc-interacting loci appears to take place, at least in part, at the nucleolus. The mechanisms by which these bodies are formed in this specific region of the nucleus are unclear. It is possible that TFIIIC, which is highly induced and present at a large number of sites in the genome of heat shocked versus control cells, plays a role in this recruitment, since this protein and tRNA genes have been shown to be present in the perinucleolar region (D’Ambrosio et al., 2008; Haeusler et al., 2008; Thompson et al., 2003). If so, a specific role for TFIIIC in the heat shock response, together with those found for Rad21 and Cap-H2, would suggest a division of labor among the different architectural proteins in order to elicit changes in 3D organization that, together with alterations in the structure of the 10 nm chromatin fiber, cooperate to silence most of the Drosophila genome in a rapid and efficient manner.
EXPERIMENTAL PROCEDURES
Cell culture
Kc167 cells were grown to confluence in SFX medium at 25°C and collected for Hi-C experiments. For heat shock, cells were cultured at 36.5°C for 20 min. For transcription inhibition studies, cells were treated with 10 μM triptolide or 1 μM flavopiridol for 3 h.
Hi-C, sequencing, mapping and normalization
Hi-C experiments were performed using cells subjected to the various treatments described in the Results. Hi-C libraries of two biological replicates were generated as described (Hou et al., 2012; Lieberman-Aiden et al., 2009). Paired reads were aligned to the dm3 reference genome using Bowtie 0.12.7. GC content, mappability, and fragment length effects were normalized as described (Hou et al., 2012). A summary of the results and the quality control steps is shown in Table S1. Additional details on the computational analyses can be found in the Extended Experimental Procedures.
ChIP-seq and data analysis
Chromatin immunoprecipitation on control or heat shocked Kc167 cells was performed as described with slight modifications (Van Bortle et al., 2012). Antibodies to architectural proteins have been previously described (Van Bortle et al., 2014). ChIPed and input DNA was amplified following standard protocols using indexed primers and sequenced as above. Paired reads were aligned using Bowtie and used for peak calling using MACS (Langmead, 2010; Zhang et al., 2008). Peaks with p values lower than 1×10−10 were used for further analysis.
Immunofluorescence analysis and FISH
Kc167 cells were fixed with 1% formaldehyde and permeabilized with 0.3% TritonX-100. After blocking, cells were incubated with primary antibodies, washed, and incubated with goat anti-rabbit Alexa fluor 488 and anti-mouse Alexa fluor 594. For FISH analyses, cells were attached to slides, incubated in RNAse A for 1h, and washed in PBS. Probes were denatured at 80°C for 4 min and then added to cells. Coverslips were sealed with rubber cement, and slides were incubated overnight at 37°C. Images were taken on an Olympus FV1000 laser scanning confocal microscope. Distances between the centers of FISH signals were measured using Fiji Software as previously described (Phillips-Cremins et al., 2013).
Supplementary Material
Acknowledgments
We thank Drs. John T. Lis for providing antibodies against RNAPII, and Vincent Pirrotta and Richard Jones for providing antibodies against Pc. We would like to thank the Genomic Services Lab at the HudsonAlpha Institute for Biotechnology, and specially Braden Boone, Angela Jones and Terri Pointer, for their help in performing Illumina sequencing of ChIP-Seq samples. This work was supported by U.S. Public Health Service Awards R01 GM035463 to V.G.C., R01 HG005119 to Z.S.Q., R01 GM069462 to G.B, and funds from the Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases DK015602 to E.P.L.
Footnotes
ACCESSION NUMBERS
Raw Hi-C and ChIP-seq paired reads and processed datasets have been deposited in GEO under accession number GSE63518. For cells grown at normal temperature, additional replicates of ChIP-seq experiments for BEAF-32, CTCF, CP190, Mod(mdg4) and Su(Hw) are deposited under accession numbers GSE36944 and GSE54529. Additional replicates of ChIP-seq experiments for H3K4me1, H3K4me3 and H3K4ac are deposited under accession number GSE36374.
Supplemental information, including Extended Experimental Procedures, four figures, and one table are provided with this article.
AUTHOR CONTRIBUTIONS
E.P.L., G.B., Z.S.Q., and V.G.C. designed the experiments. X.L., C.H., N.T., H.Q.N., and C.-T. O. conducted the experiments. L.L., C.H. and M.H. analyzed the data. C. C.-P. and V.G.C. wrote the manuscript.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bauer CR, Hartl TA, Bosco G. Condensin II promotes the formation of chromosome territories by inducing axial compaction of polyploid interphase chromosomes. PLoS genetics. 2012;8:e1002873. doi: 10.1371/journal.pgen.1002873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonora G, Plath K, Denholtz M. A mechanistic link between gene regulation and genome architecture in mammalian development. Current opinion in genetics & development. 2014;27:92–101. doi: 10.1016/j.gde.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheutin T, Cavalli G. Polycomb silencing: from linear chromatin domains to 3D chromosome folding. Current opinion in genetics & development. 2014;25:30–37. doi: 10.1016/j.gde.2013.11.016. [DOI] [PubMed] [Google Scholar]
- D’Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, Shirahige K, Uhlmann F. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes & development. 2008;22:2215–2227. doi: 10.1101/gad.1675708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delest A, Sexton T, Cavalli G. Polycomb: a paradigm for genome organization from one to three dimensions. Current opinion in cell biology. 2012;24:405–414. doi: 10.1016/j.ceb.2012.01.008. [DOI] [PubMed] [Google Scholar]
- DeMare LE, Leng J, Cotney J, Reilly SK, Yin J, Sarro R, Noonan JP. The genomic landscape of cohesin-associated chromatin interactions. Genome research. 2013;23:1224–1234. doi: 10.1101/gr.156570.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, Weintraub AS, Schuijers J, Lee TI, Zhao K, et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014;159:374–387. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonsalves SE, Moses AM, Razak Z, Robert F, Westwood JT. Whole-genome analysis reveals that active heat shock factor binding sites are mostly associated with non-heat shock genes in Drosophila melanogaster. PloS one. 2011;6:e15934. doi: 10.1371/journal.pone.0015934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaud C, Bantignies F, Pal-Bhadra M, Ghana P, Bhadra U, Cavalli G. RNAi components are required for nuclear clustering of Polycomb group response elements. Cell. 2006;124:957–971. doi: 10.1016/j.cell.2006.01.036. [DOI] [PubMed] [Google Scholar]
- Guertin MJ, Martins AL, Siepel A, Lis JT. Accurate prediction of inducible transcription factor binding intensities in vivo. PLoS genetics. 2012;8:e1002610. doi: 10.1371/journal.pgen.1002610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler RA, Pratt-Hyatt M, Good PD, Gipson TA, Engelke DR. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes & development. 2008;22:2204–2214. doi: 10.1101/gad.1675908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl TA, Smith HF, Bosco G. Chromosome alignment and transvection are antagonized by condensin II. Science. 2008;322:1384–1387. doi: 10.1126/science.1164216. [DOI] [PubMed] [Google Scholar]
- Hou C, Li L, Qin ZS, Corces VG. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Molecular cell. 2012;48:471–484. doi: 10.1016/j.molcel.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers I, Kwak H, Lis JT. Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. eLife. 2014;3:e02407. doi: 10.7554/eLife.02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce EF, Williams BR, Xie T, Wu CT. Identification of genes that promote or antagonize somatic homolog pairing using a high-throughput FISH-based screen. PLoS genetics. 2012;8:e1002667. doi: 10.1371/journal.pgen.1002667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B. Aligning short sequencing reads with Bowtie. In: Baxevanis Andreas D, et al., editors. Current protocols in bioinformatics. Unit 11. Chapter 11. 2010. p. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HB, Muller M, Bahechar IA, Kyrchanova O, Ohno K, Georgiev P, Pirrotta V. Insulators, not Polycomb response elements, are required for long-range interactions between Polycomb targets in Drosophila melanogaster. Molecular and cellular biology. 2011;31:616–625. doi: 10.1128/MCB.00849-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HB, Ohno K, Gui H, Pirrotta V. Insulators target active genes to transcription factories and polycomb-repressed genes to polycomb bodies. PLoS genetics. 2013;9:e1003436. doi: 10.1371/journal.pgen.1003436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Dekker J, Heard E. Segmental folding of chromosomes: a basis for structural and regulatory chromosomal neighborhoods? BioEssays : news and reviews in molecular, cellular and developmental biology. 2013;35:818–828. doi: 10.1002/bies.201300040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, Ong CT, Hookway TA, Guo C, Sun Y, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V, Li HB. A view of nuclear Polycomb bodies. Current opinion in genetics & development. 2012;22:101–109. doi: 10.1016/j.gde.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nature reviews Genetics. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- Sofueva S, Yaffe E, Chan WC, Georgopoulou D, Vietri Rudan M, Mira-Bontenbal H, Pollard SM, Schroth GP, Tanay A, Hadjur S. Cohesin-mediated interactions organize chromosomal domain architecture. The EMBO journal. 2013;32:3119–3129. doi: 10.1038/emboj.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M, Haeusler RA, Good PD, Engelke DR. Nucleolar clustering of dispersed tRNA genes. Science. 2003;302:1399–1401. doi: 10.1126/science.1089814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bortle K, Nichols MH, Li L, Ong CT, Takenaka N, Qin ZS, Corces VG. Insulator function and topological domain border strength scale with architectural protein occupancy. Genome biology. 2014;15:R82. doi: 10.1186/gb-2014-15-5-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bortle K, Ramos E, Takenaka N, Yang J, Wahi JE, Corces VG. Drosophila CTCF tandemly aligns with other insulator proteins at the borders of H3K27me3 domains. Genome research. 2012;22:2176–2187. doi: 10.1101/gr.136788.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AM, Van Bortle K, Ramos E, Takenaka N, Rohrbaugh M, Jones BC, Jones KC, Corces VG. Regulation of chromatin organization and inducible gene expression by a Drosophila insulator. Molecular cell. 2011;44:29–38. doi: 10.1016/j.molcel.2011.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.