Abstract
The DEAD-box RNA helicase DDX3X is frequently mutated in pediatric medulloblastoma. We dissect how these mutants affect DDX3X function with structural, biochemical, and genetic experiments. We identify an N-terminal extension (“ATP-binding loop”, ABL) that is critical for the stimulation of ATP hydrolysis by RNA. We present crystal structures that suggest the ABL interacts dynamically with ATP and confirm the interaction occurs in solution by NMR chemical shift perturbation (CSP) and isothermal calorimetry (ITC). DEAD-box helicases require interaction between two conserved RecA-like helicase domains, D1 and D2 for function. We use NMR CSP to show that DDX3X interacts specifically with double-stranded RNA (dsRNA) through its D1 domain, with contact mediated by residues G302 and G325. Mutants of these residues, G302V and G325E, are associated with pediatric medulloblastoma. These mutants are defective in RNA-stimulated ATP hydrolysis. We show that DDX3X complements the growth defect in a ded1 temperature-sensitive strain of S. pombe, but the cancer-associated mutants G302V and G325E do not complement and exhibit protein expression defects. Taken together, our results suggest that impaired translation of important mRNA targets by mutant DDX3X represents a key step in the development of medulloblastoma.
Keywords: crystallography, NMR, biochemistry, genetics
Introduction
Medulloblastoma is the most common malignant childhood brain tumor. It has been divided into four subgroups based upon transcriptional analysis of primary tumors: WNT, SHH, Group 3, and Group 4 [1]. Although the molecular basis of each subgroup differs, all patients with medulloblastoma nevertheless receive the same combination therapy of surgery, radiation, and chemotherapy [2]. Approximately 70% of patients are cured, but their quality of life is severely compromised. Subgroup-specific treatments could potentially increase cure rates and/or increase the quality of life for medulloblastoma survivors by decreasing side effects that include long-term neurocognitive deficits [3]. The St. Jude-Washington University Pediatric Cancer Genome Project [4] (PCGP) performed whole genome sequencing of several medulloblastoma tumors in each subgroup [2] and demonstrated a strong association between mutation of the protein DDX3X and specific subgroups of medulloblastoma, particularly the WNT subgroup [2]. Other studies have also linked DDX3X mutation to medulloblastoma [5-7]. Of 9 DDX3X mutants identified by the PCGP, 8 were associated with medulloblastoma [2], and one (R351W) was identified in an infant acute lymphoblastic leukemia (James Downing and Jinghui Zhang, personal communication).
DDX3X is a member of a large family of RNA helicases known as “DEAD-box” helicases [8, 9]. This name is derived from the conserved Asp-Glu-Ala-Asp sequence motif [10] in “helicase motif” II [11] that contains the catalytic base for ATP hydrolysis. The DEAD-box helicases belong to a larger group of helicases known as “Super Family 2” (SF2), which is similar to the “Super Family 1” (SF1) group of helicases. Both of these super families (reviewed in [12-14] are defined by conserved “helicase motifs” [11] that reside on adjacent RecA-like domains (D1 and D2) connected by a linker (Fig. 1a). Interdomain movements generated by ATP-binding and hydrolysis at the D1-D2 interface are expected to drive manipulation of substrate nucleic acid [12]. For DNA helicases, such as the SF1 helicases PcrA [15] and Dda [16] or the SF2 helicase RecQ [17], a robust in vitro strand displacement activity is observed with synthetic DNA substrates [18-21]. In contrast, DEAD-box helicases often do not display an in vitro unwinding activity [22], and when an unwinding activity is observed, the duplex region is generally not longer than 10 base-pairs [23], suggesting DEAD-box helicases might serve more of a remodeling function [22, 24] than a strand-displacement function.
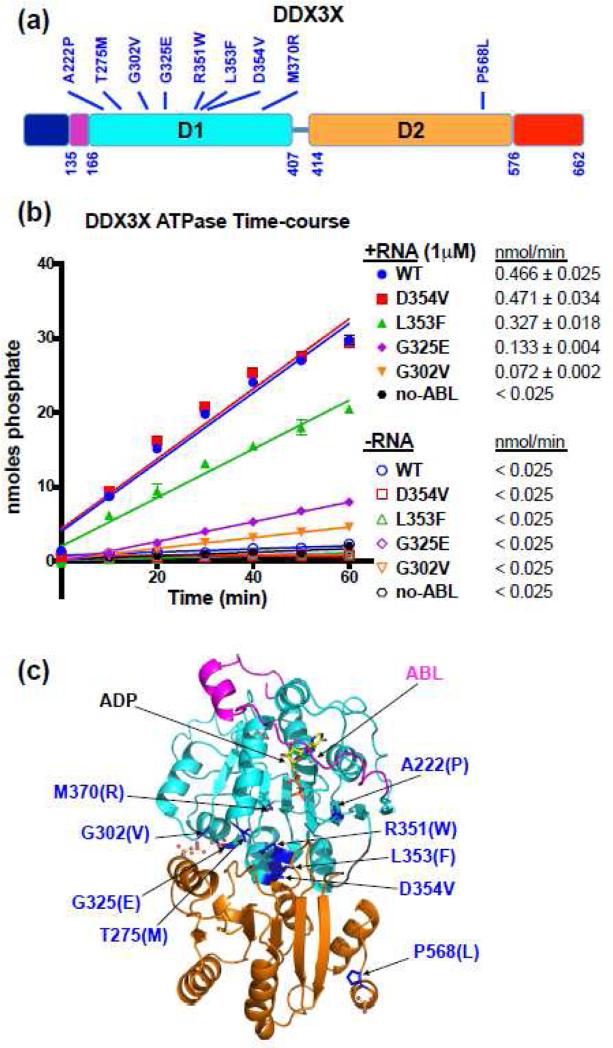
Figure 1. The domain architecture of DDX3X.
(a) Two RecA-like domains (cyan and light orange) flanked by N-and C-terminal extensions (dark blue and red, respectively). An N-terminal “ATP-binding loop” (ABL) described in this study is shown in magenta. The positions of cancer-associated mutations are noted above.
(b) Time-dependent RNA-stimulated ATPase activity of 3 μM DDX3X and its cancer-associated mutants in the presence of 2 mM ATP. For DDX3X constructs with the ABL (DDX3X 135-582), wild-type, D354V, and L353F all showed significant ATPase activity in the presence of 1 μg 5’-tailed duplex RNA (1 μM duplex). The same proteins had negligible ATPase activity in the absence of RNA or in the presence of ssRNA or blunt duplex RNA (Fig. S2a). In contrast, the G302V and G325E mutants are defective in ATPase activity. The mean and standard deviation of two independent experiments for each protein are shown. The enzymatic activity of each protein was determined as the slope (nmol phosphate released/min) following linear regression (right).
(c) Crystal structure of DDX3X (135-582) D354V. The bound ADP nucleotide is shown in stick. The ATP-binding loop (ABL) that sits on top of the nucleotide is shown in magenta. Domain D1 is shown in cyan, and domain D2 is depicted in orange. All the PCGP-identified mutation positions are shown in blue with the side-chains of the D354V structure shown in stick. Co-crystallized phosphate ions are shown as ball-and-stick.
The DEAD-box helicase family is characterized by additional specialized sequence motifs [8]. These include the Q-motif [25], a conserved phenylalanine (“F-motif”) [25], motif-Ib (“GG motif”) [26], and motif-IVa (“QxxR motif”) [27]. Some of the helicase motif residues are involved in ATP-binding or hydrolysis, while others bind substrate nucleic acid [8]. DDX3X is highly similar to the Ded1 proteins of S. cerevisiae (Sc) and S. pombe (Sp), and to Drosophila melanogaster (Dm) Belle and Vasa in sequence (see Fig. S1b) [28], and the DDX3X D1-D2 structure closely resembles the Vasa D1-D2 structure [29, 30]. The likely roles of the conserved sequence motif residues in DDX3X are illustrated in the crystal structure of DmVasa in complex with single-stranded RNA (ssRNA) and an ATP analog [30]. The structure demonstrates how Vasa binds RNA and ATP in a conformation that appears poised for ATP hydrolysis [30], termed the “closed conformation” [31].
Of the 9 DDX3X mutations identified by the PCGP, 8 are in D1 (Fig. 1a, Fig. S1a-b), suggesting that DDX3X function is more sensitive to mutation in D1 than in D2. Many of the mutations are located in or adjacent to the helicase sequence motifs, suggesting that the mutations may disrupt fundamental activities. Three (T275M, G302V, and G325E) are in helicase motifs implicated in binding RNA [8]. These motifs presumably interact with one strand of RNA in the manner observed in the crystal structure of Vasa [30], which shows that main-chain amides in each of the three corresponding regions directly contact the backbone of one ssRNA strand. Although side-chain mutagenesis does not directly alter interactions with the main-chain, these mutations could disrupt binding to the RNA strand by altering the main-chain structure itself or by sterically preventing the proximity needed for interaction. In order to remodel RNA secondary structure, a DEAD-box helicase may also interact with an opposing RNA strand. A good candidate for such interactions is the region described as Motif IIa [32]. Although this motif does not have a well-conserved sequence, the structural element is often observed to interact with the complementary strand of nucleic acid [32]. Interestingly, three of the DDX3X mutations cluster directly after Motif II (R351W, L353F, and D354V). Thus, if DDX3X binds dsRNA, three of the mutations (T275M, G302V, and G325E) potentially alter binding to one RNA strand, and three of the mutations (R351W, L353F, and D354V) potentially alter binding to the complementary RNA strand.
DDX3X has been implicated in many aspects of RNA processing [33-35], including the positive regulation of translation of transcripts with complex UTR sequences [36-38]. DDX3X has been implicated in binding mRNAs containing the TAR motif (trans-activation response element) of HIV and in facilitating their translation [37]. The Ded1 proteins of S. cerevisiae [39] and S. pombe [40] are essential proteins that share strong sequence homology with DDX3X. In fission yeast, Ded1 is required for translation, particularly for mRNAs with complex UTRs [41]. Thus, in addition to strong sequence homology (Fig. S1b), SpDed1 appears to share functional homology with DDX3X and therefore may be a useful model to elucidate the biological and biochemical functions of DDX3X and the cancer-associated mutants.
In this study, we characterize the properties of DDX3X and DDX3X mutants identified in pediatric cancer. We show that the ATPase activity of DDX3X is specifically stimulated by a hybrid double-stranded/single-stranded RNA substrate, but is not stimulated by blunt dsRNA or by ssRNA. Stimulation of the ATPase activity relies on the presence of a DDX3X structural loop N-terminal to the first RecA domain that we define as the “ATP-binding loop” (ABL). We show that the ABL is a flexible loop that sits on top of the bound nucleotide in the crystal structure of medulloblastoma mutant D354V and confirm the interaction between the ABL and ATP for wild-type DDX3X in solution by NMR chemical shift perturbation and by isothermal calorimetry (ITC). We show that DDX3X residues G302 and G325 are involved in double-stranded RNA (dsRNA) binding, and that the pediatric medulloblastoma mutants G302V and G325E are severely defective for RNA-stimulated ATPase activity. In contrast, the pediatric medulloblastoma mutant D354V exhibits normal RNA-stimulated ATPase activity, and L353F shows only a minor decrease in this ATPase activity. We show that DDX3X and medulloblastoma mutants L353F and D354V can complement temperature-sensitive ded1 alleles in fission yeast. In contrast, the medulloblastoma mutants G302V and G325E cannot complement ded1, and exhibit protein expression defects. We suggest that some of the DDX3X mutants identified in pediatric medulloblastoma have a defective RNA-stimulated ATPase activity that impairs translation of critical cellular regulators and thus contributes to medulloblastoma.
Results
We first sought to identify a minimal ATPase active construct of wild-type DDX3X that would be amenable to crystallographic and biochemical investigation. A crystal structure of a DDX3X construct comprising the minimal RecA domains D1 and D2 (168-582) has previously been reported [29]. However, this protein was reported to lack RNA-stimulated ATPase activity [29]. We noted that the N-terminus of the construct corresponds to the minimal N-terminus of the first RecA fold, similar to the native N-terminus of the DEAD-box helicase eIF4A (Fig. S1b) [27]. In contrast, the crystal structure of the more closely related Vasa helicase included additional N-terminal residues that interact with a bound ATP analog [30], and this protein construct had ATPase activity [30]. We found that a DDX3X construct with a similar N-terminal extension (135-582) had an ATPase activity that was stimulated by RNA (Fig. 1b, Fig. S2a-b) while DDX3X (168-582) did not exhibit significant RNA-stimulated ATPase activity, as reported previously [29]. Following this identification, we generated DDX3X (135-582) expression reagents for each of the PCGP-identified mutants. Several of the mutant DDX3X (135-582) proteins were isolated in good yield, namely: wild-type, G302V, G325E, L353F, and D354V. The other DDX3X mutants appeared to generate exclusively insoluble protein during bacterial expression (data not shown).
ATPase activity is specifically stimulated by hybrid ds/ssRNA
While none of the DDX3X (135-582) proteins showed significant basal ATPase activity, wild-type, L353F, and D354V had substantial ATPase activity in the presence of a palindromic RNA substrate consisting of 10-mer dsRNA with 20-mer single-stranded 5’-overhangs (Fig. 1b, Fig. S2a-b). The ATPase activity is time-dependent (Fig. 1b) and protein concentration-dependent (Fig. S2b). All mutants, except D354V, showed a significant decrease in ATP hydrolysis rate. The G302V and G325E mutants were especially defective, exhibiting extremely low RNA-stimulated ATPase activities (Fig. 1b, Fig. S2a-b). For the proteins that show ATPase activity, this activity was not stimulated by 14-mer blunt duplex RNA or by 10-mer oligo-rU ssRNA (Fig. S2a), suggesting that DDX3X is specialized for a ds/ssRNA hybrid substrate. The requirement for a ssRNA tail to stimulate ATPase activity is consistent with several other DEAD-box helicases [42, 43].
DDX3X D354V crystal structure reveals an “ATP-binding loop” (ABL)
To structurally characterize the role of the N-terminal extension in enabling ATPase activity, we extensively screened each DDX3X protein construct for crystallization and obtained crystals for mutant D354V (135-582). Crystals grew in the presence of ADP and sodium/potassium phosphate, and the structure was solved to 3.2 Å resolution (Fig. 1c; Fig. S3-S4; Table S1). The D354V mutation is at the center of the D1-D2 interface possibly stabilizing the two domains to facilitate crystallization of this mutant. The individual RecA domains are very similar in structure to those in the previously reported structure of DDX3X (168-582) [29] (D1 168-246, 263-404 Cα RMSD: 0.52 Å; D2 414-576 RMSD: 1.46 Å). Hence, the D354V mutation does not significantly alter the structure of D1 or D2. The D1 and D2 domains in the D354V structure have a different relative orientation than the previously published structure of DDX3X (168-582) [29]. In a D1-based alignment of the two structures, the D2 center-of-mass positions are separated by 44 Å, and the D2 domains are rotated by approximately 180°. Both DDX3X structures represent “open conformations” rather than the “closed conformation” observed in the Dm Vasa structure [30] (Fig. S3). We expect DDX3X to adopt a “closed conformation” highly similar to that observed in Vasa in catalyzing ATP hydrolysis. Thus, although the D354V mutation appears to stabilize a specific interdomain conformation for crystallization, this conformation is not locked because D354V is able to catalyze ATP hydrolysis (Fig. 1b).
The structure reveals that part (residues 152-160) of the N-terminal extension that is necessary for RNA-stimulated ATP hydrolysis (see Fig. 1b) appears to have a role in binding ATP, and we shall refer to it as the “ATP-binding loop” (ABL). Most of the ABL forms a similar structure to the N-terminal extension of Vasa, which has been implicated in ATP binding [30], but the DDX3X residues N-terminal to L146 are directed differently (Fig. S3a). The extension consists of a short α-helix with a conserved phenylalanine (F151) that projects into a hydrophobic pocket formed by L197, Y291, L150, and the aliphatic portion of the K288 and E147 side chains (Fig. S5a). Vasa F215 similarly projects into a hydrophobic pocket formed by the aliphatic portion of K262, F343, I214, and the aliphatic portion of K340, and A211 (Fig. S5b). This phenylalanine and the hydrophobic pocket residues are also conserved in the sequence of fission yeast Ded1 (Fig. S1b, yellow arrow), suggesting that this interaction is an important feature of this subgroup of DEAD-box helicases. Although the loop between this helix and the start of the first RecA-like domain (corresponding to DDX3X amino acids 152-168) is well-ordered in the Vasa structure with observed interactions with the AMP-PNP molecule [30], this loop is more weakly ordered in DDX3X with elevated B-factors. Due to limited side-chain density, part of the region has been modeled as poly-alanine. The main chain appears to interact with the sugar of the ADP molecule, but specific residue interactions cannot be assigned from the present crystal structure.
The ABL enhances DDX3X affinity for ATP
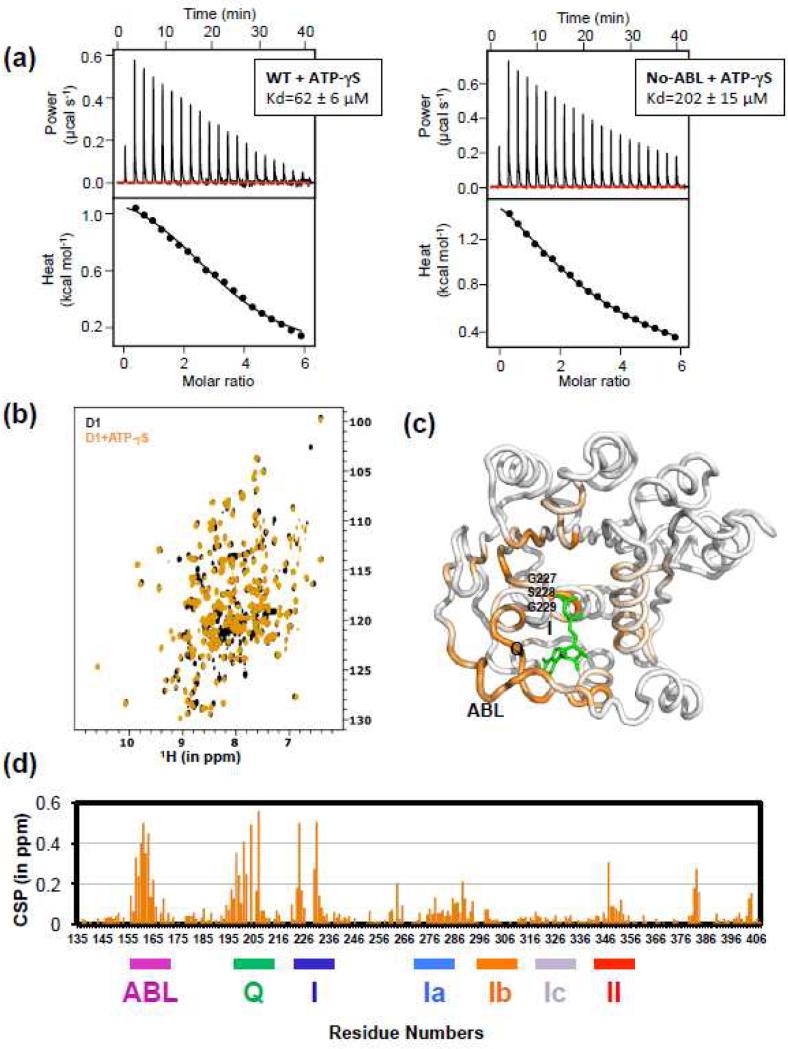
To determine the role of the ABL in binding ATP, we determined the equilibrium dissociation constants (Kd) for the binding of ATP-γS to DDX3X (135-582) and to DDX3X-no-ABL (168-582) by isothermal titration calorimetry. The dissociation constants reveal that the protein lacking the ABL (Kd = 202 ± 15 μM) binds with a 3-fold weaker affinity than the protein with the ABL (Kd = 62 ± 6 μM) (Fig. 2a), suggesting a significant and direct role for the ABL in binding ATP.
Figure 2. DDX3X D1 interaction with ATP analog in solution.
(a) The ABL enhances DDX3X affinity for ATP. Isotherms were measured by titrating 2.5 μM ATP-γS into 90 μM solutions of DDX3X either with the ABL (135-582; left) or lacking the ABL (168-582; right). The determined dissociation constants revealed that the construct with the ABL (62 ± 6 μM) binds 3-fold tighter than the construct lacking the ABL (202 ± 15 μM).
(b) [1H,15N] TROSY spectrum of the DDX3X D1 (in black) superimposed with the same in the presence of ATP-γS in the molar ratio of 1:5 (in orange).
(c) CSP values for the D1:ATP-γS complex are mapped onto the crystal structure of D1 shown in loop representation in shades of orange for values from 0.06 to 0.6 ppm. The bound AMP-PNP molecule is shown in stick representation. The ABL, Q-motif, and Motif-I residue positions are labeled.
(d) CSP values for the D1:ATP-γS complex versus residue number. The largest shifts are in the ATP-binding loop (ABL), the Q-motif, and Motif-I.
NMR Chemical Shift Perturbation confirms ABL:ATP interaction
We used NMR chemical shift perturbation to confirm direct interaction between the ABL and ATP suggested by the crystal structure. As the D1-D2 protein (53 kDa) is too large to analyze by NMR chemical shift perturbation, we chose to analyze D1 in isolation (33 kDa) because the interactions between DDX3X and ADP exclusively reside in D1 in our crystal structure of DDX3X mutant D354V. First, we confirmed that wild-type D1 (135-407) in isolation is similarly structured to the D1 portion of D354V (135-582) and that it interacts equivalently with ATP-analog as with ADP by determining a crystal structure of wild-type DDX3X D1 in complex with AMP-PNP to 2.3 Å resolution (Fig. S6-S7; Table S1). The structure consists of 3 monomers in the asymmetric unit, all very similar to the D1 portion of the D354V D1-D2 structure (RMSD: 0.71, 0.72, and 0.76 Å for the 3 crystallographically unique monomer C-alpha positions). Each monomer is bound to a nucleotide modeled as ADP because the γ-phosphate of the AMP-PNP molecule is not observed in the electron density. Similar to the D1-D2 D354V crystal structure, the ABL is weakly ordered and has only been assigned for one of the monomers. For this monomer, the ABL is positioned consistently as in the D354V structure. Thus, this crystal structure indicates that the D1 structure and its interaction with nucleotide are not affected by removal of D2. Next, we measured a TROSY spectrum for this D1 construct (Fig. 2b). We assigned the backbone resonances in the well-dispersed 1H-15N TROSY spectrum of DDX3X (135-407) with 90% of the backbone resonances assigned without ambiguity by conventional triple resonance TROSY based techniques for large biomolecules (Table S2). The C-alpha deviations from random coil values that reflect the secondary structure (Fig. S6b) confirm that the solution conformation of D1 is very similar to the crystal structure.
The binding of D1 to an ATP-analog was then monitored by residue specific chemical shift perturbation (CSP) in the spectra. We recorded the 1H-15N TROSY spectrum of D1 in the presence of the ATP analog, ATP-γS (Fig. 2b, orange spectrum). Several backbone amides in the binding pocket were in slow exchange in the NMR spectrum, and so this spectrum was reassigned. These experiments confirmed a key role for the ABL in ATP interaction by showing strong CSPs in the ABL upon binding ATP-γS. The CSP magnitudes in the ABL were comparable to those in well-established ATP-binding regions, namely the Q motif and motif I (Fig. 2c and d). When mapped onto the structure of D1, the chemical shift perturbations correlate with the binding region of nucleotide in the crystal structures (Fig. 2c). The backbone amide resonances of three residues, G227, S228 and G229, part of motif-I, close to the ATP-γS binding site (Fig. 2c), were not observed in the free spectrum or in the ATP-γS bound form, suggesting that this loop region in helicase motif I undergoes motion in the millisecond to second time scale. Even ATP-γS binding to this region does not stabilize the loop, and therefore the dynamics of this loop could be functionally important in ATP binding and/or hydrolysis. Additional, smaller shifts were observed in motifs Ia, II, and III upon binding ATP-γS. None of the DDX3X D1 residues associated with medulloblastoma by the PCGP [2] or by other studies [6, 7] were significantly shifted upon binding ATP-γS, indicating that the residues associated with cancer mutants are not directly involved in ATP-binding.
NMR CSP shows DDX3X D1 binds dsRNA differently than ssRNA and DNA
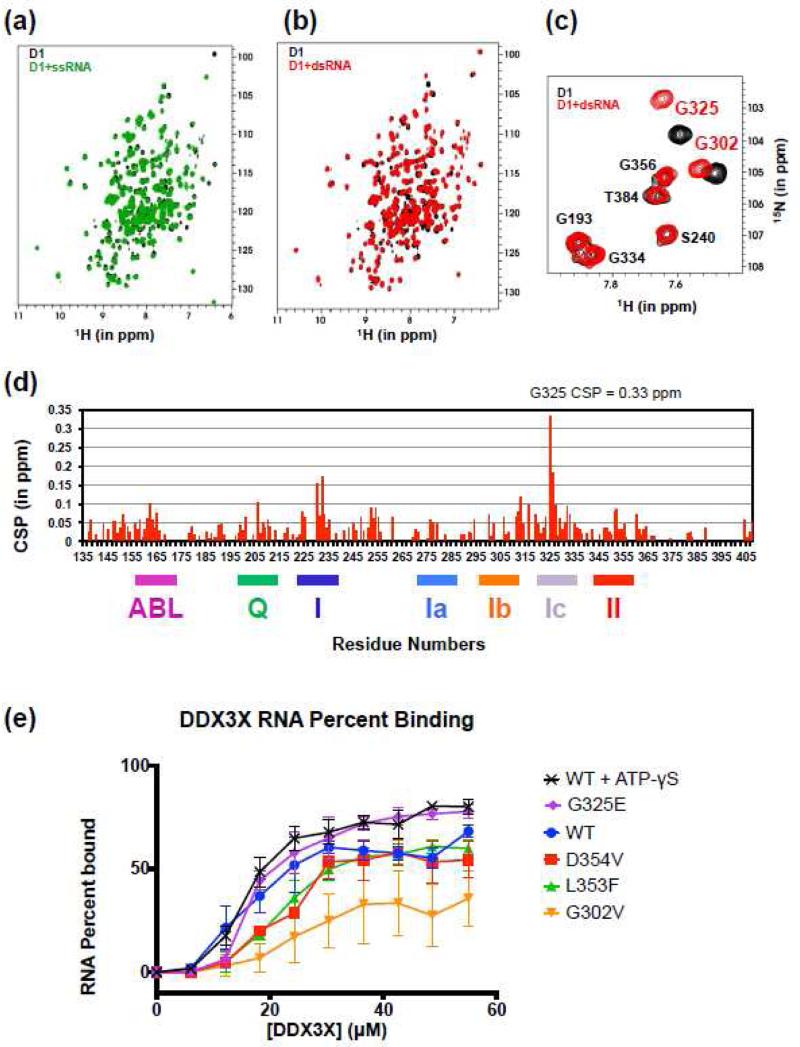
Because ATP-hydrolysis was exclusively stimulated by a dsRNA/ssRNA hybrid substrate, we used NMR CSP experiments to deconvolute and localize the interactions with each form of nucleic acid. For these experiments, we used the same labeled D1 construct as in the ATP-γS experiments. Unfortunately, we found that D2 could not be concentrated to the level needed for the NMR experiments without including high salt concentrations, consistent with a previous report [44]. High salt concentrations are incompatible with nucleic-acid binding experiments by NMR CSP, and as a result, the experiments were carried out exclusively with D1. NMR spectra were measured for D1 in combination with single- and double-stranded RNA or DNA substrates (Fig. 3, Fig. S8). Large chemical shift perturbations were observed uniquely upon addition of dsRNA to D1 (Fig. 3b-c). In contrast, only minimal chemical shift perturbations or peak broadening were observed on addition of dsDNA (Fig. S8a). This observation is consistent with previous reports that DEAD-box helicases specifically bind A-form substrates and do not bind to the B-form structure of dsDNA [9]. Upon addition of ssDNA (Fig. S8b) or ssRNA (Fig. 3a) to D1, the chemical shift perturbations observed were smaller than the CSPs after addition of dsRNA, and were not confined to specific regions of the protein, suggesting a less specific mode of interaction, such as an overall electrostatic interaction between D1 and these substrates. Taken together, the chemical shift perturbation experiments indicate that DDX3X D1 is specialized for binding dsRNA. When ATP-γS was added to the D1:dsRNA complex, the chemical shift perturbations were confined to residues that shifted upon binding ATP-γS in the absence of RNA (Fig. S8e), suggesting that the mode of dsRNA-binding was not significantly different after ATP-binding, consistent with a requirement for both D1 and D2 to generate ATP-dependent changes in RNA interaction.
Figure 3. DDX3X D1 interaction with nucleic acids in solution.
(a) Superposition of the [1H,15N] TROSY spectra of DDX3X D1 (in black) with the same in the presence of ss-RNA (in green) in the molar ratio of 1:1.2.
(b) Superposition of the [1H,15N] TROSY spectra of DDX3X D1 (in black) with the same in the presence of ds-RNA (in red) in the molar ratio of 1:1.2.
(c) Expansion of the glycine region of (b) highlighting the shifts for G325, which showed the largest chemical shift perturbation, and G302.
(d) CSP values for the D1:dsRNA complex versus residue number. Residue G325 shows the largest CSP, 2-fold greater than the next highest CSP.
(e) Electrophoretic mobility shift assays show that the G302V mutant that is severely defective in RNA-stimulated ATPase activity is also defective in binding double-stranded RNA. The mean and standard deviation of three independent binding experiments with 40 nM blunt duplex RNA are shown for each protein. Successive mean data points are connected by linear segments. Representative EMSA gel images are provided in Fig. S9.
Comparison of the 1H-15N TROSY spectrum of D1 with that of the two-domain construct, 52.6 kDa, DDX3X (135-582) showed that the conformation of D1 is similar in both constructs (Fig. S8f). The D1 resonances that shift upon binding nucleic acid remain unshifted in the spectra of free-D1 and free D1-D2, suggesting that the observed binding for D1 in isolation is also possible for the 2-domain protein. None of the surface residues of D1 showed significant CSP in the D1-D2 protein, suggesting that the orientation of the two domains is flexible in solution, perhaps contributing to the difficulties in crystallizing most of the D1-D2 DDX3X constructs. Such flexibility is not surprising because interdomain motion is expected to be necessary to generate unwinding or remodeling activities.
The strongest peak shift in D1 upon binding dsRNA corresponds to G325 (in helicase motif Ic) (Fig. 3b-d). Residue G302 (motif Ib) is also clearly shifted (Fig. 3c). These shifts suggest that G302 and G325 have key roles in the interaction with dsRNA. Strikingly, the ATP-binding region of D1 was also a prominent region of CSP in the presence of dsRNA (Fig. 3d). Although the protein is not expected to catalyze ATP hydrolysis in the absence of critical residues on D2 such as motif VI [8], the CSP in the ATP-binding region of D1 in the presence of dsRNA is consistent with dsRNA inducing conformational changes at the ATP-binding site and could contribute in part to the stimulation of ATPase activity by tailed dsRNA.
The cancer-associated mutants of DDX3X (135-582) were investigated for binding to the same dsRNA substrate of the NMR experiments by electrophoresis mobility shift assays (EMSAs) to evaluate the identified interactions in the D1-D2 protein (Fig. 3e, Fig. S9). The L353F and D354V mutants showed very minor defects in binding the dsRNA substrate. In contrast, the G302V mutant was significantly defective in binding the blunt dsRNA probe. Interestingly, G325E showed no defect in binding this blunt RNA substrate despite the strong conservation of glycine at this position in DEAD-box helicase motif Ic (Fig. S1b). Although the strong CSP for G325 indicates that this residue intimately associates with dsRNA, the glycine itself is not needed to bind dsRNA. The glycine instead appears to play a much stronger role in generating RNA-stimulated ATP-hydrolysis as shown by the severely defective G325E mutant (Fig. 1b). The conservation of the glycine is not likely specialized for a backbone structural requirement because the crystal structure of D354V shows that G325 is at the start of an α-helix in a conformation allowable for any residue (phi/psi = −54/−52).
DDX3X:dsRNA model
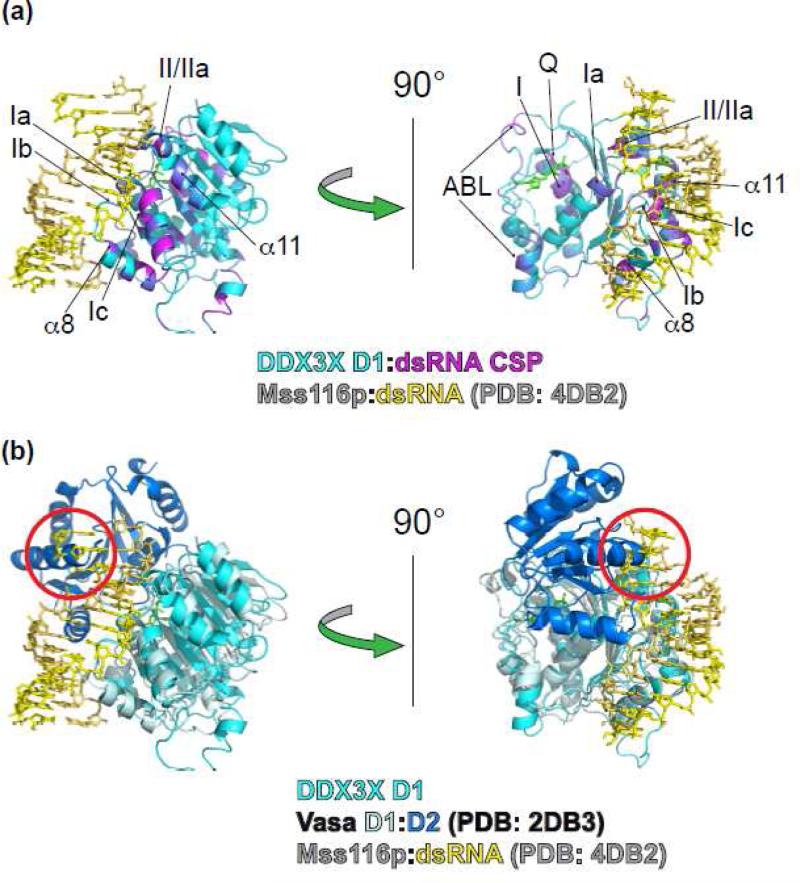
The chemical shift perturbations that resulted from binding dsRNA were evaluated for compatibility with the dsRNA molecules of existing RNA helicase:dsRNA co-crystal structures. No crystal structures are available to show how D1 of a DEAD-box helicase binds dsRNA. In the D354V crystal structure, residues G302 and G325, in RNA-binding motifs Ib and Ic [8], are observed to interact with co-crystallized phosphate ions that are positioned similar to phosphates in the RNA-backbone of the aligned structure of DmVasa [30] (Fig. S10), reinforcing the likelihood that DDX3X interacts with RNA similar to Vasa. We investigated three related scenarios for their fit to the NMR CSP data: 1) D1 of the DEAD-box helicase Vasa bound to ssRNA [30]; 2) D1 of the DECH-box helicase RIG-I bound to dsRNA [45, 46]; and 3) D2 of the DEAD-box helicase Mss116p bound to dsRNA [9]. Mss116p D2 was used based on the previously identified relationships of D1 and D2 for nucleic acid binding [47] and also because the dsRNA substrate of the CSP experiment was identical to that present in the Mss116p D2:dsRNA crystal structure. Specifically, DDX3X residues 274-277 (Motif Ia), 301-304 (Motif Ib), and 323-326 (Motif Ic) were used as a reference to align the RNA-binding regions of the other structures followed by inspection of the interactions between each protein and RNA and how the positioned RNA agreed with the DDX3X D1:dsRNA CSP data. We especially focused on multiple RNA-binding interactions that involve main-chain amides that are likely to be consistent among all the proteins and not subject to side-chain differences. We first aligned residues 326-329, 353-356, and 375-378 of DmVasa bound to ssRNA [30] (Fig. S11a) to provide a benchmark for binding one strand. RIG-I belongs to the DECH-box family of RNA helicases that is closely related to the DEAD-box helicase family [48]. The RIG-I:dsRNA structure [45] was aligned based on residues 298-301, 325-328, and 347-350 (Fig. S11b). To align Mss116p D2 with DDX3X, an overall alignment of the RecA-folds was first performed to identify the structurally equivalent residues, which were then used to optimize the alignment of the RNA-binding regions. Mss116p D2 residues 381-384 (Motif IV), 407-410 (Motif IVa), and 433-436 (Motif V) were aligned with the DDX3X residues described above (Fig. S11c).
The residues that align with DDX3X R276 (motif Ia) and G302 (Motif Ib) show consistent interactions between main-chain amides and sequential RNA phosphates for all structures (Fig. S11a-c). In contrast, the residues that align with Motif Ic (including DDX3X G325) showed significant differences in the structure of Vasa bound to ssRNA versus the dsRNA-bound structures. In Vasa, the residues that align with DDX3X 323-326 form significant interactions with the RNA backbone, while the corresponding atoms in the dsRNA-bound structures are too distant to interact with the same RNA nucleotide. As noted previously [30], this region of the RNA strand deviates from an idealized A-form trajectory and precludes an idealized A-form dsRNA structure. Thus, in order to interact with dsRNA, the residues of Motif Ic, such as G325, would need to move more extensively than the RNA-binding residues in Motif Ia and Ib. Our CSP data are fully consistent with this scenario because the largest shifts that occur upon binding dsRNA and are located in Motif Ic at residues G325 and G326.
Among the candidate alignments, we selected the dsRNA of the Mss116p D2:dsRNA complex [9] as the best qualitative fit to the CSP data (Fig. 4a). The resulting dsRNA position was found to align well with the regions of DDX3X that exhibited the largest chemical shift perturbations for dsRNA: motifs Ia, Ib, Ic, II, IIa, and helices α9, α10 and the start of α11 (Fig. 4a). The superimposed Vasa D1-D2 model showed significant steric clashes between D2 and the modeled dsRNA (Fig. 4b), consistent with the previous mechanistic suggestion that D1 and D2 cannot simultaneously bind the same dsRNA molecule in the closed conformation [9]. The binding model suggests that most of the DDX3X D1:dsRNA interactions would involve the sugar-phosphate backbone of one strand of RNA, but the region around Motif IIa might interact directly with RNA bases in the dsRNA minor groove (Fig. 4a) or with the complementary strand.
Figure 4. DDX3X D1:dsRNA binding model.
(a) Orthogonal views of the DDX3X D1-dsRNA model based on the chemical shift perturbations and alignment of Mss116p D2:dsRNA [9]. The superposition was by least-squares alignment of DDX3X 274-277 (Motif Ia), 301-304 (Motif Ib), and 323-326 (Motif Ic) with Mss116p residues 381-384 (Motif IV), 407-410 (Motif IVa), and 433-436 (Motif V). DDX3X is shown in cyan with CSP in the presence of dsRNA in shades of red. The Mss116p dsRNA is shown in yellow/yellow-orange. The dsRNA model aligns well with the regions of CSP at the right side of the molecule (right panel). Other prominent regions of CSP in the presence of dsRNA are at the left side of the molecule in the ATP-binding site. The ADP molecule of DDX3X is shown in green stick to highlight the region. These shifts are consistent with dsRNA inducing a change in the ATP-binding site and could contribute to RNA-stimulated ATP hydrolysis.
(b) The Vasa:ssRNA structure (PDB: 2DB3) [30] superimposed on the DDX3X D1:dsRNA model in (a) with Vasa D1 shown in light blue and Vasa D2 shown in dark blue. D2 shows significant steric clashes with the modeled dsRNA (red circles), suggesting that the two domains are not mutually compatible with binding the same dsRNA, consistent with Mss116p [9].
DDX3X complements a temperature-sensitive ded1 allele in S. pombe
We screened the functional consequences of the PCGP mutations in a fission yeast complementation assay. Ded1, the fission yeast ortholog of DDX3X, is encoded by an essential gene [40]. We first tested whether expression of DDX3X could complement for viability of the fission yeast ded1-1D5 thermosensitive mutant [41] at restrictive temperature. We found that episomal expression of either wild-type SpDed1 or HsDDX3X rescued the temperature-sensitive growth defect of ded1-1D5 cells (Fig. S12a). The level of vector-expressed Ded1 was similar to endogenous Ded1 based upon immunoblotting with a Ded1 antibody [49], and expression levels of the ectopic DDX3X and Ded1 were also similar (Fig. S12b).
The DDX3X mutations associated with pediatric cancer [2] (and personal communication, Jinghui Zhang and James Downing) were tested for complementation of the ded1-1D5 temperature-sensitive growth defect (Fig. S13a). An ATPase defective Walker-A DDX3X mutant, K230A, was also tested as an internal control. The mutants A222P, K230A (Walker-A), G302V, G325E, and P568L all exhibited growth defects at restrictive temperature. The severity of the growth defect was assessed by use of serial dilution assays and by plating cells at an intermediate temperature of 33°C (Fig. S13b). This analysis revealed that K230A (Walker-A) was the most defective mutant, and negatively impacted growth of ded1-1D5 cells even at the permissive temperature of 25°C. G302V, G325E, and A222P exhibited intermedia te defects, and P568L was also somewhat impaired.
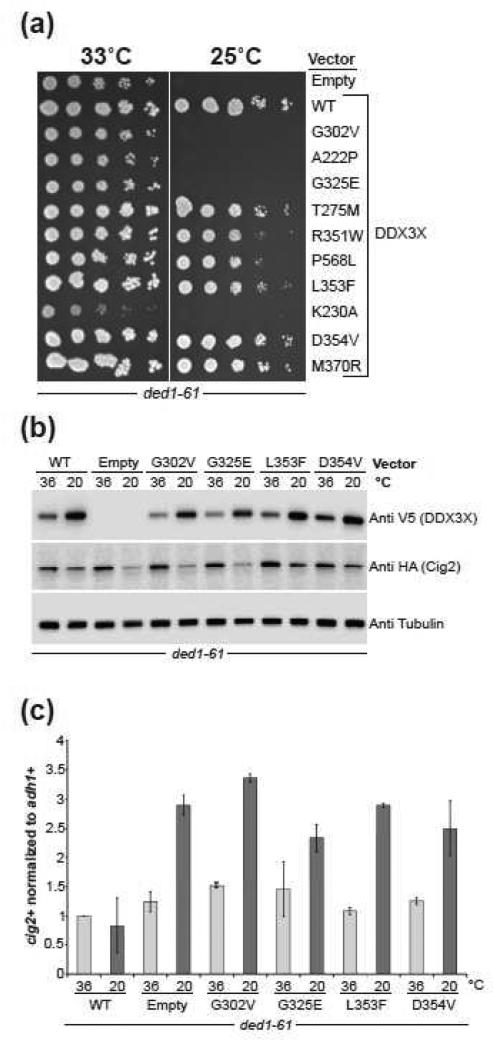
All DDX3X proteins were expressed at slightly lower levels at the restrictive temperature, with a more marked reduction in expression of the non-complementing mutants (Fig. S13c). To address the possibility that reduced protein level contributed to the phenotype, we transformed a cold-sensitive ded1 mutant strain, ded1-61 [41], and found that the same set of mutants failed to complement for growth at restrictive cold temperature (25°C) (Fig. 5a). Under these condition s, however, protein levels for all DDX3X proteins were elevated, indicating that cell growth differences are not caused by altered DDX3X expression levels (Fig. 5b). Thus, we differentiate the cancer-associated DDX3X mutants into two classes. Those that can functionally replace Ded1 are referred to as “functional mutants” (T275M, R351W, L353F, D354V, and M370R), and “non-complementing mutants” are those that cannot functionally replace Ded1 in ded1 thermosensitive mutants at restrictive temperature (A222P, G302V, G325E, P568L).
Figure 5. DDX3X functionally complements SpDed1.
(a) Complementation of the cold-sensitive ded1-61 strain by expression of the DDX3X mutant plasmids was examined in a serial dilution assay. The Walker-A mutant, K230A, was the most defective and impacted growth at the permissive temperature of 33°C. Growth of strains expressing the mutants A222P, G302V, and G325E was significantly impaired at the restrictive temperature (25°C).
(b) Growth defects of affected strains correlate with lower expression of Cig2 protein following incubation at low temperature (20°C) acco rding to immunoblotting of the HA epitope tagged Cig2.
(c) The decrease in expression is not due to lower mRNA levels as indicated by qRT-PCR of cig2+ transcripts normalized to adh1+.
Functional DDX3X facilitates expression of cyclin Cig2 protein
The ded1-1D5 mutation is associated with decreased overall protein translation at the non-permissive temperature [41], and translation of the cyclins Cig2 and Cdc13 is especially sensitive [41]. We monitored levels of Cig2-3xHA protein in the different strains. Cig2 expression levels were impaired after 2.5 hours of growth at 36°C in ded1-1D5 mutant cells, but were unaffected in cells expressing wild-type Ded1 or DDX3X (Fig. S12c). The non-complementing mutants A222P, G302V, and G325E all showed reduced Cig2 levels at 36°C when compared to wild-t ype DDX3X (Fig. S13c). The P568L mutant, which exhibited only very minor defects at the semi-permissive temperature, did not show significantly different Cig2 expression levels at 25°C versus 36°C. Similar results were observed in ded1-61 cells following shift to 20°C for 3 hours (Fig. 5b). The decreased Cig2 protein level in non-complementing mutants is not due to a transcriptional defect because the defective mutants exhibited high cig2+ transcript levels (Fig. 5c and Fig. S13d) when measured by quantitative real time PCR after 2.5 hours growth at restrictive temperature. The elevation in cig2+ transcripts may be due to arrest of the non-complementing mutant cells at G1 phase of the cell cycle when cig2+ is maximally transcribed.
Discussion
DDX3X belongs to the DEAD-box family of helicases, which hydrolyze ATP and remodel RNA secondary structure through the combined activities of several conserved helicase motif residues on two tandem RecA-like domains. Here we show that, in addition to the canonical DEAD-box helicase motifs, DDX3X also uses an N-terminal ABL (residues 135 to 168) to interact with ATP. While Motif-I, Motif-II, and the Q-motif interact with Mg/ATP with well-defined characteristic interactions [8], the ABL does not seem to use a single set of specific interactions. The dynamic nature of ABL:ATP interaction is shown by the high B-factors in the ABL and the difficulty in assigning the sequence registry in the ABL region. Although dynamic, the ABL clearly interacts with ATP as shown by the CSP and ITC experiments, and the ABL plays a decisive role in RNA-stimulated ATPase activity. This role is specific to DDX3X and its close relatives because most DEAD-box helicases do not possess this ABL. The ABL is therefore not intrinsically required to catalyze ATP hydrolysis. Instead, we suggest that the ABL is involved in stimulating ATP hydrolysis by preferred RNA substrates. Consistent with this role, the ABL shows CSP upon addition of dsRNA, suggesting that the ABL can sense the presence of bound dsRNA. An intriguing possibility is that the ABL is normally dynamic, but becomes well-structured when DDX3X binds a specific form of RNA, thus facilitating ATP hydrolysis. Consistent with this hypothesis, the crystal structure of DmVasa has RNA bound [30] and shows a well-ordered ABL.
The ATPase activity of DDX3X is stimulated by a hybrid ds/ssRNA substrate, but not by blunt dsRNA or ssRNA, suggesting that DDX3X is specialized for specific RNA structures. The specific biological RNA substrate(s) of DDX3X are not known, and it is possible that the synthetic ds/ssRNA substrate of our assay is not the optimal substrate for DDX3X ATPase activity. Nevertheless, our CSP experiments show that D1 has specificity for dsRNA, and that binding involves residues G302 and G325 in canonical RNA-binding sequence motifs [8]. The CSP experiments also indicate that D1 interacts nonspecifically with ssRNA. We therefore suggest that the RNA-bound species that leads to ATPase stimulation has double-stranded RNA bound at G302 and G325 (motifs Ib and Ic) as shown in our dsRNA binding model (Fig. 4) and ssRNA interacting at a region that is yet to be identified. Possible candidates for ssRNA interaction include Motifs IV, IVa, and V in D2, canonical RNA-binding motifs [8] in a domain that we were unable to examine by NMR.
Consistent with a strong role in acting on cellular RNA targets, the medulloblastoma mutants G302V and G325E are severely defective for RNA-stimulated ATPase activity and cannot functionally complement Ded1 in fission yeast. Although ATP hydrolysis is clearly required for the functional complementation based upon the profound growth defect of the DDX3X K230A (Walker-A/Motif-I) ATPase defective mutant, none of the residues within D1 that are mutated in medulloblastoma show significant interaction with ATP-γS in our NMR experiments. Instead, the ATPase defects of the G302V and the G325E mutants likely arise from severely compromised RNA-stimulation of the ATPase activity. Consistent with this model, both residues are clearly shifted upon binding dsRNA (Fig. 3), and both are in canonical RNA-binding DEAD-box helicase motifs (Motifs Ib and Ic). The unperturbed affinity of G325E for dsRNA in EMSAs indicates that RNA-binding alone does not dictate ATPase stimulation and that other critical features must be elucidated. Our dsRNA binding model (Fig. 4) suggests that G302 and G325 bind one strand of dsRNA, and the region described as Motif IIa [32] binds in the minor groove, perhaps also binding the complementary strand. In this model, the medulloblastoma mutants R351W, L353F, and D354V could have potential roles in sequence specificity. Based on this model, DDX3X interaction with the RNA minor groove by Motif IIa is likely not as essential as the set of interactions with the first strand by Motifs Ib and Ic. The motif IIa mutants L353F and D354V exhibit only limited RNA-binding and ATPase defects and can functionally complement Ded1 while the G302V and G325E mutants have severe RNA-stimulated ATPase defects and cannot functionally complement SpDed1.
DDX3X, like Ded1, has been implicated in the positive regulation of translation of transcripts with complex UTR sequences, including G1/S cyclins. We propose that a subset of the DDX3X mutants identified in pediatric medulloblastomas have reduced RNA-dependent ATPase activity that impairs translation of critical cellular regulators and contributes to the development of the disease. The Ded1 complementation experiments clearly show that expression of Cig2 is disrupted by mutants of DDX3X with highly reduced RNA-stimulated ATPase activity. It has been demonstrated that Ded1 is required for the effective translation of Cig2, perhaps playing a role in remodeling the long structured 5’-UTR of the transcript [41]. If the 5’-UTR is shortened, this requirement is reduced [41]. Complementation by wild-type and mutant DDX3X produces phenotypes that are fully consistent with Cig2 5’-UTR processing in a ded1 temperature sensitive strain. Similarly, in human cells, cyclin E1 levels are greatly reduced upon knockdown of DDX3X, but are restored when the 5’-UTR is exchanged for that of cyclin D1, which is unaffected by DDX3X knockdown [38]. The 5’-UTR of human cyclin E1 is 82.5% GC and is also predicted to have a highly stable secondary structure [38]. Several retroviruses also depend on cellular DDX3XDed1 for the translation of viral transcripts. HIV-1 is dependent on DDX3X for translation, and hence DDX3X has been studied as a potential anti-HIV drug target [50, 51]. Similarities between some of the viral RNAs and human transcripts that are regulated by DDX3X are the presence of long UTR sequences with secondary structure, and a very high GC-content that stabilizes this secondary structure. DDX3X has been reported to reduce the secondary structure of retroviral UTRs, allowing the 43S ribosome to bind for translation [37]. We suggest that DDX3X is needed to process long structured 5’-UTR sequences, and that this activity is lost in the G302V and G325E mutants. An intriguing possibility is that DDX3X acts upon the 5’-UTRs of multiple mRNAs to correctly balance levels of growth promoting proteins with tumor suppressors. A mutation that decreases DDX3X activity could promote tumorigenesis by preventing translation of tumor suppressor(s) while a mutation that enhances DDX3X activity could promote tumorigenesis through excessive translation of growth promoting protein(s). It will be interesting to determine whether select DDX3X mutants contribute to human medulloblastoma through translational control of such cell cycle regulators.
Experimental Procedures
Media and Chemicals
Fission yeast were maintained on rich media (YES), unless nutritional selection was required for maintenance of the LEU2 marked plasmid (pREP41-3xV5 series), when cells were grown on PMG media with appropriate supplements [52]. All chemicals were purchased from Sigma unless otherwise indicated.
Cloning and Mutagenesis for Plasmid Construction
E.coli expression plasmids
The wild-type hsDDX3X gene was obtained as an IMAGE clone from openbiosystems. The full gene was amplified by PCR using primers flanked by EcoRI and XhoI restriction sites. The PCR product was digested with EcoRI/XhoI and ligated into an EcoRI/XhoI-digested, phosphatase-treated pET-28a vector. The T7 tag was removed from the construct by mutagenesis to generate an expression construct with a thrombin-cleavable His6-tag. Expression plasmids for N- or C-terminal deletions were prepared by PCR of the entire plasmid, excluding the region to be deleted, with phosphorylated primers followed by circularization with ligase, producing constructs for: His6-DDX3X (135-582) (pLE193), His6-DDX3X (168-582) (LE194), His6-DDX3X (135-407) (pLE231), and His6-DDX3X (408-582) (pLE235). Mutations were similarly introduced with phosphorylated mutagenic primers to generate His6-DDX3X (135-582) G302V (pLE210), His6-DDX3X (135-582) L353F (pLE224), His6-DDX3X (135-582) G325E (pLE228), and His6-DDX3X (135-582) D354V (pLE230). Sequences were verified by the Hartwell Center DNA Sequencing Facility (St. Jude Children's Research Hospital).
S.pombe expression plasmids
ded1+ was amplified from S. pombe genomic DNA using primers bearing SalI sites and 15 bases of homology with the vector at the 5’ and 3’ ends of the gene. This PCR fragment was cloned into a LEU2 marked fission yeast expression vector carrying an N terminal 3xV5 epitope tag under control of a mid-strength nmt1 promoter (pREP41-V5 (JP1340), kind gift from P. Bjerling) which was linearized with SalI downstream of the epitope tag. Cloning was performed using InFusion HD (Clontech): 50 ng each of vector and insert were incubated with InFusion mix at 50°C for 15 minutes and on ice for 15 minute s, prior to transformation of Stellar competent cells (Clontech). Clones were screened by PCR, and full sequencing of the ded1 insert was performed. Construction of plasmids expressing full length wild-type DDX3X was performed similarly, using a cloned DDX3X cDNA (kind gift from JP. Taylor) as template. Vectors containing mutant DDX3X were produced through PCR mutagenesis of the JP2104 DDX3X construct using Phusion high fidelity DNA polymerase (Thermo F-553). Mutations made were: A222P, K230A, T275M, G302V, G325E, R351W, L353F, D354V, M370R, and P568L. Sequencing was performed on the entire DDX3X gene of these plasmids to verify that only the required mutation was created.
Protein Expression and Purification
Each construct was freshly transformed into BL21(DE3)-RIPL (Stratagene) chemically competent cells and grown overnight in a 100 mL starter culture containing 30 mg/L kanamyacin and 8 g/L glucose. Large-scale cultures were prepared by inoculating 10 mL of starter culture per liter of fresh LB media containing 30 mg/L kanamyacin and 8 g/L glucose. The cultures were shaken at 200 rpm at 37°C and induced at 18°C by 0.5 mM IPTG once the O.D. reached 0.7-0.8. After growing overnight, cells were harvested by centrifugation for 15 minutes in an SLC-6000 rotor at 3200 rcf. The cells were resuspended in lysis buffer (50mM Tris 8.3; 250 mM NaCl; 10% glycerol; 2 mM BME; Roche Complete EDTA-free protease inhibitor tablets) and lysed using a microfluidizer. The soluble fraction was isolated after centrifugation in an SLC-1500 rotor (13,000 rpm in a Sorvall RC-6+ centrifuge) for 45 minutes. For samples used in biochemical assays, sodium chloride was added to a final concentration of 1M, and then nucleic acids were precipitated by adding 10% polyethylenimine/10% hydrochloric acid to a final concentration of 0.3%. This solution was centrifuged at 2,900 rcf for 15 minutes in a benchtop centrifuge, and the supernatant was collected. Ammonium sulfate was added to 70% saturation, and the precipitate was isolated by centrifugation for one hour at 13,000 rpm in an SLC-1500 rotor in a Sorvall RC-6+ centrifuge. The pellet was resuspended in buffer NBB (50 mM Tris 8.3; 500 mM NaCl; 25 mM Imidazole; 10% glycerol; 2mM BME) and bound batchwise to Qiagen Ni-NTA agarose. After 8 washings of the Ni-NTA agarose with NBB, the protein was eluted in a single step with 50mM Tris 8.3; 500 mM NaCl; 10% glycerol; 2mM BME; 250 mM imidazole. The proteins were further purified by size-exclusion chromatography with a Superdex 26/60 column. For crystallization samples, the final gel-filtration buffer was 20mM Hepes 7.6; 150 mM KCl; 5 mM DTT; 5 mM MgCl2. Other samples of DDX3X (135-582) were purified in 20 mM Hepes 7.6; 200 mM NaCl; 2mM DTT. The DDX3X (168-582) sample was purified in 20 mM Hepes 7.6; 500 mM NaCl; 10% glycerol; 2 mM TCEP. Protein concentrations were determined by absorbance at 280 nm (ε135-582 = 38,850; ε135-407 = 20,400, ε168−582=31,860). Extinction coefficients were calculated using the ProtParam tool on the ExPasy server (http://web.expasy.org/protparam/).
Crystallization, Data-collection, Structure-solution, Model-building, and Refinement
For DDX3X (135-582) D354V, the protein was co-concentrated with 2.5 mM ADP and 25 mM MgCl2 to 15 mg/ml. Crystals grew when mixed 1:1 with 1.66 M NaH2PO4/0.240 M K2HPO4 at 18°C. Crystals were quickly passed through a 1 :3 solution of ethylene glycol in the crystallization well solution and flash frozen in liquid nitrogen. Data were collected at SER-CAT beamline 22-ID at the Advanced Photon Source at Argonne National Lab at 1.0 Å wavelength. Data were integrated and scaled with the HKL-2000 package [53] to 3.2 Å resolution. Initial phases were determined by molecular replacement by the program Phaser [54] that placed one copy of the individual D1 and D2 domains of HsDDX3X (PDB: 2I4I) [29]. The model was manually improved with Coot [55] and refined at various stages with CNS [56, 57] and refmac5 [58]. The final refinement was carried out with CNS. Figures were prepared with the program PyMol [59] and Coot [55]. Although DDX3X (135-582) D354V was crystallized in the presence of Mg2+ and ADP, electron density was not observed for Mg2+ ions.
For DDX3X (135-407), the protein was initially concentrated to 5 mg/mL when it started precipitating. Addition of MgCl2 and either AMP-PNP, AMP-PNP + U10 RNA, or ATP-γS improved the solubility and permitted higher concentrations. Each sample was screened for crystallization on an Art Robbins Phoenix Robot with commercial kits from Qiagen and Hampton Research, which produced lead crystals for the AMP-PNP-treated samples. A fresh sample was co-concentrated with 10 mM AMP-PNP and 20 mM DTT to a final concentration of 15 mg/mL and filtered. Single crystals grew in a few days when mixed 1:1 with 50 mM Hepes 7.6; 7% PEG 3350 and incubated at 18°C. Crystals were quickly passed through a 1:3 solution of ethylene glycol in the crystallization well solution and flash frozen in liquid nitrogen. Data were collected at SER-CAT beamline 22-ID at the Advanced Photon Source at Argonne National Lab. Data were collected at 1.0 Å wavelength in 0.5° oscillations for a total o f 125 degrees of crystal rotation. Data were integrated and scaled with the HKL-2000 package [53] to 2.3 Å resolution. Initial phases were determined by molecular replacement by the program Phaser [54] that placed 3 copies of the N-terminal domain of HsDDX3X (PDB: 2I4I) [29]. The model was refined at various stages with CNS [56, 57] and refmac5 [58]. The final refinement was carried out with CNS using 3 NCS restraint groups for the 3 copies. Figures were prepared with the program PyMol [59] and Coot [55]. Although DDX3X D1 was crystallized in the presence of Mg2+ and AMP-PNP, electron density was not observed for Mg2+ ions or for the γ-phosphate. The nucleotide has been modeled as ADP in the final refinement.
ATPase Assay
ATPase experiments were all performed at 30 °C in 5 0 μL reactions containing 40 mM Hepes pH 7.6, 35 mM KCl, 2 mM DTT, 5 mM MgCl2, and 2 mM ATP. Reactions with added RNA included 1 μg of one of the following sequences: UUUUUUUUUU (U10), GGGCGGGCCCGCCC (blunt dsRNA; palindromic 14-mer), UUUUUUUUUUUUUUUUUUUUGGCGGCCGCC (palindromic 10-mer dsRNA with 20-mer 5’-overhangs). Initial experiments to determine an RNA substrate that would stimulate ATPase activity were performed for 30 minutes with 3 μM protein in triplicate. Time-course and concentration-dependence experiments were performed with the palindromic 10-mer dsRNA with 20-mer 5’-overhangs. Time-course experiments were performed in duplicate with 3 μM final concentration of the indicated protein. Protein concentration dependence experiments were performed in duplicate, by incubating protein concentrations ranging from 0 to 3 μM for 30 minutes. Released phosphate was quantified with the Biomol Green (enzolifesciences) detection kit. Standard curve reactions were prepared with NaH2PO4 as a phosphate standard in identical buffer conditions. GraphPad Prism was used to calculate released phosphate by linear regression and for plotting of results.
Electromobility Shift Assay
A palindromic 14-mer RNA, GGGCGGGCCCGCCC, previously shown to bind to the DEAD-box helicase Mss116p, was obtained commercially (Sigma Aldrich) with a 3’-fluorescein modification. Protein samples were dialyzed into 50 mM Tris 7.5; 125 mM KCl; 5 mM DTT; 10 mM MgCl2; 10% glycerol; 0.05% Tween-20 and adjusted to 3.2 mg/mL as determined by absorbance at 280 nm on a NanoDrop spectrophotometer. Binding reactions contained a total volume of 18 μl of protein/dialysis buffer, ranging from 0 to 55 μM final protein concentration, and 2 μl of fluorescently-labeled RNA (40 nM final concentration). Wild-type DDX3X (135-582) was also titrated in the presence of 1 mM ATP-γS. After incubating at 25°C for 30 minutes, 5 μl of 40% sucrose was added, and 20 μl were loaded in a 4-20% gradient native TBE polyacrylamide gel (Biorad) and run for one hour at 150 V in 1X TBE buffer. The gel was then imaged with a SybrGreen filter for 2 minutes on a FujiFilm LAS4000. Three independent titrations were performed for each sample.
Isothermal Calorimetry
Thermodynamic parameters for the interaction of DDX3X-with-ABL (135-582) and DDX3X-No-ABL (168-582) with ATP-analog were measured with a MicroCal auto-iTC 200 (GE Healthcare). Protein samples were exchanged into 20 mM HEPES (pH 7.6), 150 mM KCl, 2 mM Tris (2-carboxyethyl)phosphine, and 5 or 10% glycerol prior to the experiments. Titrations were performed by first injecting 0.5 μl of 2.5 mM ATP-analog (ATP-γS) into a solution of 90 μM DDX3X-with-ABL or DDX3X-No-ABL, followed by additional 2 μl injections. Experiments were carried out at 25 °C. Results were analyzed using Origin software (OriginLab) provided by MicroCal. Binding constants (Kd) were calculated from the average of three individual experiments by fitting the data to a single site binding model using a nonlinear least-squares fitting algorithm.
Strains
Ded1 mutant strains are described in [41]. S. pombe: The ded1-1D5 mutant allele was sequenced and found to contain two point mutations, S282F, and R285C. The ded1-61 mutant allele contained a P263L mutation. The ded1-61 cig2-3XHA strain (PY7803) was used for all ded1-61 analyses. The leu1-32 mutation was introduced into the ded1-1D5 cig2-3xHA strain (PY 7885) prior to transformation.
Plating Assays
For the simple temperature sensitivity plating assay, ded1-1D5 strains were streaked onto PMG agar lacking leucine to maintain the plasmid, and incubated for 2 to 3 days at either 25°C or 36°C before being photographed. For the serial dilution growth assay, ded1-1D5 cells were grown at 25°C and ded1-61 cells at 32°C to a density of 5 × 10 6 cells/ml in PMG –Leu media. Five-fold serial dilutions were made, and were spotted onto PMG –Leu agar plates such that 1.2 × 104 cells were contained in the first spot. Plates were incubated at 25°C or 33°C for 5-7 days.
Protein Extracts and Western Blotting
Cells were grown overnight at 25°C in PMG –Leu to a density of 5.0 × 106 cells per ml. Protein extracts were made by bead beating cells in 2X SDS sample buffer with chilled glass beads for 3 minutes and heating to 90°C for 2 minutes. The samples were spun out at 13K RPM for 10 minutes and the supernatant was collected. Extracts were run on NuPage 4-12% Bis-Tris minigels (Invitrogen) in NuPage MOPS buffer (Invitrogen) and transferred to Protran membrane (GE Life Science). Western blotting was done using the following mouse monoclonal antibodies: anti-V5 antibody (AbD Serotec MCA1360) and anti-Tubulin antibody (kind gift from K. Gull) at dilutions of 1:2000 in PBS + 0.2% tween (PBST) for 1 hour at room temperature. Rat monoclonal anti-HA 3F10 antibody (Roche 1867423) was used at a dilution of 1:500 overnight at 4°C. Rabbit polyclonal anti-Ded1p antibody (kind gift from N. Walworth) was used at a dilution of 1:50,000 overnight at 4°C. Incubations were performed with s econdary antibodies (anti-mouse IgG peroxidase (Sigma A4416), anti-rabbit IgG peroxidase (Sigma A6154), and anti-rat IgG peroxidase (Sigma SAB3700595)) at dilutions of 1:5000 in 3% milk PBST for 1 hour at room temperature. Detection was performed using Pierce ECL substrate and exposure to Hyperfilm (GE Life Sciences).
Temperature shift assays on ded1-1D5 cultures were performed by growing cells in PMG –Leu to a density of ~4.0 × 106 cells/ml at 25°C. The culture was split, and one culture was kept at 25°C while the other was shifte d to 36°C for 3 hours. For ded1-61 cultures, cells were grown to a density of ~4.0 × 106 cells/ml at 32°C, split and cultured at 20°C or 36°C for 3 hours. Total cell extracts we re made as described above.
Transcript Analyses
For ded1-1D5, cells were grown to a density of ~4.0 × 106 cells/ml at 25°C in PMG – Leu, split and grown at 25°C or 36°C for 2.5 hours. For ded1-61 cultures, cells were grown at 32°C prior to splitting and growing at 20° C or 36°C for 3 hours. Random priming of total cellular RNA was used to prepare cDNA as described previously [60]. Quantitative real time PCR was performed to measure transcript levels of cig2+ (JPO 2990 TTTGTTTAATGCCCGAAACC and JPO 2991 TGCTAGCGATGAGAAGAGCA) and adh1+ as the euchromatic control (JPO 793 AACGTCAAGTTCGAGGAAGTCC and JPO 794 AGAGCGTGTAAATCGGTGTGG). Real time PCR was performed using an Eppendorf Mastercycler ep Realplex machine and Quantifast Sybr green (Qiagen). cig2 transcript levels were normalized to adh1, and results represent the mean of two biological replicates. The linear range of amplification for each set of primers was verified, and experiments were performed within this range. The ΔΔCt method was used for the analysis of transcript levels.
Supplementary Material
Acknowledgements
This work was supported by Comprehensive Cancer Center Support Grant Developmental Funds [5 P30 CA021765-32 to EJE]; by Cancer Center support grant [CCSG 2 P30 CA21765 to St. Jude]; and by ALSAC of St. Jude Children's Research Hospital. We are grateful to Beata Grallert for the ded1 strains, to Nancy Walworth for the Ded1 antibody. We thank Stephen White for insightful discussions and support; Richard Gilbertson for enthusiastic encouragement and for sharing DDX3X mutant data prior to publication; and the St. Jude DDX3X working group and Partridge and Enemark lab members for helpful discussions. We thank Cristina Guibao and Jie Zheng for advice on NMR sample preparation, Weixing Zhang for technical assistance with NMR experiments, and Judith Hyle and Jill Lahti for initial ATPase activity measurements. We thank Charles Rock for advice on ATPase experiments and Sivaraja Vaithiyalingam (Molecular Interaction Analysis Shared Resource, St. Jude Children's Research Hospital) for performing and analyzing ITC experiments. Thanks also to Martine Roussel and Stephen White for critical reading of the manuscript. Data were collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found at www.ser-cat.org/members.html. We are grateful to SER-CAT staff for experimental support. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38.
Biography
 Leslie B. Epling
Leslie B. Epling
 Christy R. Grace
Christy R. Grace
 Brandon R. Lowe
Brandon R. Lowe
 Janet F. Partridge
Janet F. Partridge
 Eric J. Enemark
Eric J. Enemark
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
L.B.E. prepared and purified recombinant protein; performed crystallographic, NMR, and biochemical experiments; and analyzed results. C.R.G. designed, performed, and analyzed NMR experiments. B.R.L. performed and analyzed S. pombe complementation and biochemical experiments. All authors contributed to writing and revising the manuscript.
ACCESSION NUMBERS: Coordinates and structure factors have been deposited in the Protein Data Bank with accession numbers 4PX9 and 4PXA.
References
- 1.Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123:465–72. doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488:43–8. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mulhern RK, Palmer SL, Merchant TE, Wallace D, Kocak M, Brouwers P, et al. Neurocognitive consequences of risk-adapted therapy for childhood medulloblastoma. J Clin Oncol. 2005;23:5511–9. doi: 10.1200/JCO.2005.00.703. [DOI] [PubMed] [Google Scholar]
- 4.Downing JR, Wilson RK, Zhang J, Mardis ER, Pui CH, Ding L, et al. The Pediatric Cancer Genome Project. Nat Genet. 2012;44:619–22. doi: 10.1038/ng.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–9. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones DT, Jager N, Kool M, Zichner T, Hutter B, Sultan M, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–5. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pugh TJ, Weeraratne SD, Archer TC, Pomeranz Krummel DA, Auclair D, Bochicchio J, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106–10. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12:505–16. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- 9.Mallam AL, Del Campo M, Gilman B, Sidote DJ, Lambowitz AM. Structural basis for RNA-duplex recognition and unwinding by the DEAD-box helicase Mss116p. Nature. 2012;490:121–5. doi: 10.1038/nature11402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linder P, Lasko PF, Ashburner M, Leroy P, Nielsen PJ, Nishi K, et al. Birth of the D E-A-D box. Nature. 1989;337:121–2. doi: 10.1038/337121a0. [DOI] [PubMed] [Google Scholar]
- 11.Gorbalenya AE, Koonin EV. Helicases - Amino-Acid-Sequence Comparisons and Structure-Function-Relationships. Current Opinion in Structural Biology. 1993;3:419–29. [Google Scholar]
- 12.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 13.Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys. 2008;37:317–36. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 14.Byrd AK, Raney KD. Superfamily 2 helicases. Front Biosci (Landmark Ed) 2012;17:2070–88. doi: 10.2741/4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 16.He X, Byrd AK, Yun MK, Pemble CWt, Harrison D, Yeruva L, et al. The T4 phage SF1B helicase Dda is structurally optimized to perform DNA strand separation. Structure. 2012;20:1189–200. doi: 10.1016/j.str.2012.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernstein DA, Zittel MC, Keck JL. High-resolution structure of the E.coli RecQ helicase catalytic core. EMBO J. 2003;22:4910–21. doi: 10.1093/emboj/cdg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bird LE, Brannigan JA, Subramanya HS, Wigley DB. Characterisation of Bacillus stearothermophilus PcrA helicase: evidence against an active rolling mechanism. Nucleic Acids Res. 1998;26:2686–93. doi: 10.1093/nar/26.11.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raney KD, Benkovic SJ. Bacteriophage T4 Dda helicase translocates in a unidirectional fashion on single-stranded DNA. J Biol Chem. 1995;270:22236–42. doi: 10.1074/jbc.270.38.22236. [DOI] [PubMed] [Google Scholar]
- 20.Harmon FG, Kowalczykowski SC. Biochemical characterization of the DNA helicase activity of the escherichia coli RecQ helicase. J Biol Chem. 2001;276:232–43. doi: 10.1074/jbc.M006555200. [DOI] [PubMed] [Google Scholar]
- 21.Bachrati CZ, Hickson ID. Analysis of the DNA unwinding activity of RecQ family helicases. Methods Enzymol. 2006;409:86–100. doi: 10.1016/S0076-6879(05)09005-1. [DOI] [PubMed] [Google Scholar]
- 22.Banroques J, Cordin O, Doere M, Linder P, Tanner NK. A conserved phenylalanine of motif IV in superfamily 2 helicases is required for cooperative, ATP-dependent binding of RNA substrates in DEAD-box proteins. Mol Cell Biol. 2008;28:3359–71. doi: 10.1128/MCB.01555-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanner NK, Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol Cell. 2001;8:251–62. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- 24.Jankowsky E, Bowers H. Remodeling of ribonucleoprotein complexes with DExH/D RNA helicases. Nucleic Acids Res. 2006;34:4181–8. doi: 10.1093/nar/gkl410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanner NK, Cordin O, Banroques J, Doere M, Linder P. The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol Cell. 2003;11:127–38. doi: 10.1016/s1097-2765(03)00006-6. [DOI] [PubMed] [Google Scholar]
- 26.Schmid SR, Linder P. D-E-A-D protein family of putative RNA helicases. Mol Microbiol. 1992;6:283–91. doi: 10.1111/j.1365-2958.1992.tb01470.x. [DOI] [PubMed] [Google Scholar]
- 27.Caruthers JM, Johnson ER, McKay DB. Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc Natl Acad Sci U S A. 2000;97:13080–5. doi: 10.1073/pnas.97.24.13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tarn WY, Chang TH. The current understanding of Ded1p/DDX3 homologs from yeast to human. RNA Biol. 2009;6:17–20. doi: 10.4161/rna.6.1.7440. [DOI] [PubMed] [Google Scholar]
- 29.Hogbom M, Collins R, van den Berg S, Jenvert RM, Karlberg T, Kotenyova T, et al. Crystal structure of conserved domains 1 and 2 of the human DEAD-box helicase DDX3X in complex with the mononucleotide AMP. J Mol Biol. 2007;372:150–9. doi: 10.1016/j.jmb.2007.06.050. [DOI] [PubMed] [Google Scholar]
- 30.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 31.Korolev S, Hsieh J, Gauss GH, Lohman TM, Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–47. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- 32.Luo D, Kohlway A, Pyle AM. Duplex RNA activated ATPases (DRAs): platforms for RNA sensing, signaling and processing. RNA Biol. 2013;10:111–20. doi: 10.4161/rna.22706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soto-Rifo R, Ohlmann T. The role of the DEAD-box RNA helicase DDX3 in mRNA metabolism. Wiley Interdiscip Rev RNA. 2013;4:369–85. doi: 10.1002/wrna.1165. [DOI] [PubMed] [Google Scholar]
- 34.Rosner A, Rinkevich B. The DDX3 subfamily of the DEAD box helicases: divergent roles as unveiled by studying different organisms and in vitro assays. Curr Med Chem. 2007;14:2517–25. doi: 10.2174/092986707782023677. [DOI] [PubMed] [Google Scholar]
- 35.Soto-Rifo R, Rubilar PS, Ohlmann T. The DEAD-box helicase DDX3 substitutes for the cap-binding protein eIF4E to promote compartmentalized translation initiation of the HIV-1 genomic RNA. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lai MC, Lee YH, Tarn WY. The DEAD-box RNA helicase DDX3 associates with export messenger ribonucleoproteins as well as tip-associated protein and participates in translational control. Mol Biol Cell. 2008;19:3847–58. doi: 10.1091/mbc.E07-12-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soto-Rifo R, Rubilar PS, Limousin T, de Breyne S, Decimo D, Ohlmann T. DEAD- box protein DDX3 associates with eIF4F to promote translation of selected mRNAs. EMBO J. 2012;31:3745–56. doi: 10.1038/emboj.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lai MC, Chang WC, Shieh SY, Tarn WY. DDX3 regulates cell growth through translational control of cyclin E1. Mol Cell Biol. 2010;30:5444–53. doi: 10.1128/MCB.00560-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Struhl K. Nucleotide sequence and transcriptional mapping of the yeast pet56-his3- ded1 gene region. Nucleic Acids Res. 1985;13:8587–601. doi: 10.1093/nar/13.23.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawamukai M. Isolation of a novel gene, moc2, encoding a putative RNA helicase as a suppressor of sterile strains in Schizosaccharomyces pombe. Biochim Biophys Acta. 1999;1446:93–101. doi: 10.1016/s0167-4781(99)00071-8. [DOI] [PubMed] [Google Scholar]
- 41.Grallert B, Kearsey SE, Lenhard M, Carlson CR, Nurse P, Boye E, et al. A fission yeast general translation factor reveals links between protein synthesis and cell cycle controls. J Cell Sci. 2000;113(Pt 8):1447–58. doi: 10.1242/jcs.113.8.1447. [DOI] [PubMed] [Google Scholar]
- 42.Bizebard T, Ferlenghi I, Iost I, Dreyfus M. Studies on three E. coli DEAD-box helicases point to an unwinding mechanism different from that of model DNA helicases. Biochemistry. 2004;43:7857–66. doi: 10.1021/bi049852s. [DOI] [PubMed] [Google Scholar]
- 43.Yang Q, Jankowsky E. The DEAD-box protein Ded1 unwinds RNA duplexes by a mode distinct from translocating helicases. Nat Struct Mol Biol. 2006;13:981–6. doi: 10.1038/nsmb1165. [DOI] [PubMed] [Google Scholar]
- 44.Rodamilans B, Montoya G. Expression, purification, crystallization and preliminary X-ray diffraction analysis of the DDX3 RNA helicase domain. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007;63:283–6. doi: 10.1107/S1744309107006434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luo D, Ding SC, Vela A, Kohlway A, Lindenbach BD, Pyle AM. Structural insights into RNA recognition by RIG-I. Cell. 2011;147:409–22. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, et al. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–35. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 47.Milner-White EJ, Pietras Z, Luisi BF. An ancient anion-binding structural module in RNA and DNA helicases. Proteins. 2010;78:1900–8. doi: 10.1002/prot.22704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20:313–24. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu HY, Nefsky BS, Walworth NC. The Ded1 DEAD box helicase interacts with Chk1 and Cdc2. J Biol Chem. 2002;277:2637–43. doi: 10.1074/jbc.M109016200. [DOI] [PubMed] [Google Scholar]
- 50.Maga G, Falchi F, Radi M, Botta L, Casaluce G, Bernardini M, et al. Toward the discovery of novel anti-HIV drugs. Second-generation inhibitors of the cellular ATPase DDX3 with improved anti-HIV activity: synthesis, structure-activity relationship analysis, cytotoxicity studies, and target validation. ChemMedChem. 2011;6:1371–89. doi: 10.1002/cmdc.201100166. [DOI] [PubMed] [Google Scholar]
- 51.Radi M, Falchi F, Garbelli A, Samuele A, Bernardo V, Paolucci S, et al. Discovery of the first small molecule inhibitor of human DDX3 specifically designed to target the RNA binding site: towards the next generation HIV-1 inhibitors. Bioorg Med Chem Lett. 2012;22:2094–8. doi: 10.1016/j.bmcl.2011.12.135. [DOI] [PubMed] [Google Scholar]
- 52.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 53.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–26. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 54.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–74. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 56.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 57.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2:2728–33. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 58.Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, et al. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr. 2004;60:2184–95. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 59.Schrodinger, LLC. The PyMOL Molecular Graphics System, Version 1.3r1. 2010 [Google Scholar]
- 60.Partridge JF, DeBeauchamp JL, Kosinski AM, Ulrich DL, Hadler MJ, Noffsinger VJ. Functional separation of the requirements for establishment and maintenance of centromeric heterochromatin. Mol Cell. 2007;26:593–602. doi: 10.1016/j.molcel.2007.05.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.