SUMMARY
The remarkable capacity for pluripotency and self-renewal in embryonic stem cells (ESCs) requires a finely-tuned transcriptional circuitry wherein the pathways and genes that initiate differentiation are suppressed, but poised to respond rapidly to developmental signals. To elucidate transcriptional control in mouse ESCs in the naïve, ground state, we defined the distribution of engaged RNA polymerase II (Pol II) at high-resolution. We find that promoter-proximal pausing of Pol II is most enriched at genes regulating cell cycle and signal transduction, and not, as expected, at developmental or bivalent genes. Accordingly, ablation of the primary pause-inducing factor NELF does not increase expression of lineage markers, but instead causes proliferation defects, embryonic lethality and dysregulation of ESC signaling pathways. Indeed, ESCs lacking NELF have dramatically attenuated FGF/ERK activity, rendering them resistant to differentiation. This work thus uncovers a key role for NELF-mediated pausing in establishing the responsiveness of stem cells to developmental cues.
INTRODUCTION
Elucidating the molecular mechanisms underlying pluripotency and self-renewal in ESCs is fundamental to understanding mammalian development and critical for the growing field of regenerative medicine (Hackett and Surani, 2014; Young, 2011). Clearly, establishment and maintenance of the ESC state entails a sophisticated transcriptional network that involves a core set of DNA-binding transcription factors and distinct epigenetic features (Bernstein et al., 2006; Marks et al., 2012; Wray et al., 2010; Young, 2011). In particular, the expression of cell lineage-specific regulators must be repressed in ESCs, but potentiated for efficient activation by developmental signals. Several mechanisms have been proposed to facilitate this plasticity, including pausing of Pol II during early transcription elongation (Amleh et al., 2009; Marks et al., 2012) and the presence of bivalent chromatin domains that contain both active and repressive histone modifications (Bernstein et al., 2006; Brookes et al., 2012; Ku et al., 2008). However, the role of Pol II pausing or bivalency in shaping the gene expression program in ESCs has remained enigmatic, as has the relationship between these regulatory strategies (Bernstein et al., 2006; Brookes et al., 2012; Ku et al., 2008; Marks et al., 2012; Min et al., 2011; Tee et al., 2014).
Pol II pausing is a conceptually appealing way to generate a poised state, wherein developmentally-regulated promoters could be loaded with Pol II in anticipation of future activation (Adelman and Lis, 2012; Levine, 2011). Pausing occurs when the early transcription elongation complex, associated with a short nascent RNA, comes under control of the pause-inducing factors NELF and DSIF. The association of NELF (comprised of four subunits: NELF-A, -B, -C/D and -E; Yamaguchi et al., 1999) with engaged polymerase inhibits further elongation, stably holding Pol II within the promoter-proximal region (Cheng and Price, 2007; Henriques et al., 2013; Li et al., 2013). Release of paused Pol II into the gene body is triggered by recruitment of the kinase P-TEFb, which phosphorylates Pol II and NELF, dissociating NELF from the polymerase and enabling productive elongation (Adelman and Lis, 2012; Cheng and Price, 2007; Peterlin and Price, 2006).
Widespread pausing of Pol II was first noted in Drosophila embryos, embryo-derived S2 cells and human ESCs (Guenther et al., 2007; Muse et al., 2007; Zeitlinger et al., 2007), suggesting a role in metazoan development. Indeed, pausing is strongly enriched at Drosophila genes within developmental gene ontology (GO) categories (Muse et al., 2007; Zeitlinger et al., 2007). Accordingly, the presence of pre-loaded and paused Pol II at lineage-specifying genes in the early Drosophila embryo is thought to enable their rapid, synchronous activation upon receipt of developmental cues (Lagha et al., 2013). Moreover, recent kinetic analyses of Pol II distribution during Drosophila morphogenesis revealed Pol II recruitment to developmental promoters in advance of gene activation, implying that the establishment of paused Pol II could render a promoter permissive for future gene expression (Gaertner et al., 2012).
The role of pausing in mammalian development, however, remains unclear. Genetic knock-out of the pause-inducing factor NELF causes deficiencies in the inner cell mass and peri-implantation lethality (Amleh et al., 2009), indicating an essential function; but the targets and mechanisms underlying this are poorly understood. A detailed study of engaged Pol II in mouse ESCs using the highly-sensitive global run-on sequencing (GRO-seq) assay reported that pausing was prevalent at genes involved in metabolism/catabolism, cell cycle and translation but not at genes under developmental control (Min et al., 2011). Moreover, developmental genes that displayed Pol II occupancy were generally found to be active, rather than repressed. In fact, fewer than 2% of inactive genes in ESCs were found to be associated with paused Pol II (Min et al., 2011), suggesting that pausing is not a common mechanism for gene repression in ESCs and that pausing plays a different role in pluripotent stem cells than in Drosophila embryos.
Notably, prior GRO-seq experiments were performed on ESCs grown in serum-containing culture conditions (Jonkers et al., 2014; Min et al., 2011) that allow for the low-level, heterogeneous expression of a number of lineage markers: so-called ‘lineage priming’. Thus, the observed expression of developmental genes and lack of paused Pol II (Min et al., 2011) may reflect ESCs grown under these differentiation-permissive conditions. In contrast, ESCs cultured in a defined medium containing inhibitors of kinases in the β-catenin and FGF/ERK (fibroblast growth factor/ extracellular-signal-regulated-kinase) pathways display greatly reduced expression of many lineage markers (Marks et al., 2012). The two inhibitor (or 2i) media effectively blunts differentiation-inducing signals transmitted through these key developmental cascades, promoting homogeneity in the ESC population and enriching for ESCs in the naïve, ground state of pluripotency (Kunath et al., 2007; Stavridis et al., 2007; Wray et al., 2010; Ying et al., 2008).
Studies of Pol II occupancy using Pol II ChIP-seq in ESCs grown under serum-containing vs. 2i conditions have indicated that ESCs cultured in 2i display a global increase in Pol II promoter signal, including at developmental regulators and repressed bivalent genes (Marks et al., 2012). These data imply that ESCs in the naïve, ground state might employ Pol II pausing more broadly than ESCs grown under permissive serum-containing conditions. Nonetheless, to date, no comprehensive analysis has been performed of the targets and consequences of pausing in ground state ESCs.
To definitively identify genes that harbor paused Pol II in naïve ESCs, we performed GRO-seq under 2i conditions. An assay such as GRO-seq that monitors actively engaged Pol II is essential to confirm that Pol II detected by ChIP-seq is indeed paused during early elongation. Further, the high-resolution and strand-specific nature of GRO-seq enables rigorous analysis of Pol II distribution at individual genes. Using this strategy, we report that pausing is not enriched at lineage-specifying, developmental-control or bivalent genes in ESCs grown in 2i, but is prevalent at genes involved in cell cycle control and signaling pathways. Importantly, evaluation of previously published total Pol II ChIP-seq and GRO-seq data sets demonstrates that this conclusion is valid regardless of cell culture conditions or methodology for mapping Pol II distribution.
In agreement with a role for pausing in cell cycle and developmental signaling in ESCs, genetic ablation of NELF causes profound proliferation defects and dysregulation of genes controlling cell growth, metabolism and kinase activity. Critically, loss of NELF does not lead to spontaneous differentiation or up-regulation of lineage-specifying genes in ESCs grown in 2i or serum-conditions. To the contrary, we find that ESCs lacking NELF become refractory to differentiation cues. In particular, FGF/ERK activity is markedly attenuated upon loss of NELF, blocking the response to differentiation-inducing cues through this pathway. Thus, we conclude that Pol II pausing regulates developmental potential by controlling the basal expression and activity of the signal transduction machineries that govern and maintain the ESC state.
RESULTS
Defining the Distribution of Pol II in ESCs Grown in 2i Conditions
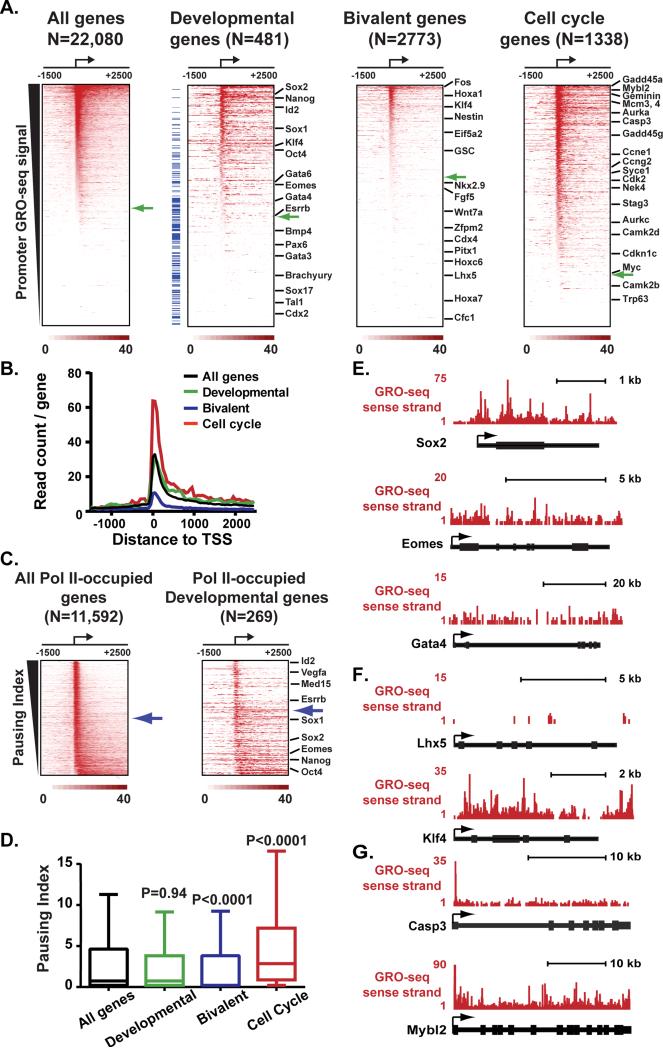
We used GRO-seq (Core et al., 2008) to define the distribution of engaged Pol II in mouse ESCs grown in 2i (Figures S1A and S1B). More than half of all RefSeq genes displayed significant sense-strand GRO-seq signal near their promoters (Figure 1A, All genes; 52% above green arrow denoting significant promoter GRO-seq signal), consistent with earlier work (Core et al., 2008; Jonkers et al., 2014; Min et al., 2011). Developmental genes, categorized by Gene Ontology (GO) as regulators of cell fate and stem cell development, displayed comparable levels of Pol II occupancy to genes on average (Figure 1A; 56% of Developmental genes have significant promoter GRO-seq signal). The similarity in Pol II levels at developmental regulators vs. all genes was also borne out by evaluation of the composite Pol II signal around promoters in each gene group (Figure 1B, compare black and green lines).
Figure 1. Pol II Pausing is Enriched at ESC Genes Involved in Cell Cycle Regulation and Proliferation.
(A) Heatmap depiction of sense-strand GRO-seq reads around mouse RefSeq TSSs, ranked by decreasing promoter GRO-seq signal (±150nt). The number of genes used for each heatmap is noted at top: Developmental genes include GO terms:0045165, 0048864, 0007498; Bivalent genes were defined in (Ku et al., 2008), Cell Cycle genes are from GO term:0007049. Green arrows depict the position in each heatmap where promoter GRO-seq signal fails to be statistically significant (P<0.001), and the position of selected pluripotency and lineage markers is shown. Blue tick marks at left of Developmental heatmap denote Developmental genes characterized as bivalent.
(B) Average sense-strand GRO-seq signal around all genes as compared to the signal derived from genes in each group described in (A).
(C) Pol II distribution at genes with significant promoter Pol II signal is shown as heatmaps ranked by decreasing Pausing Index. Blue arrows depict the point at which the Pausing Index drops below 4.
(D) Comparison of Pausing Indices across gene groups reveals significantly lower levels of pausing among Bivalent genes and higher levels among Cell Cycle genes (as compared to all genes, P-values are from Mann-Whitney U-test).
(E-G) Examples of GRO-seq signal at (E) the pluripotency gene Sox2 and lineage-markers Eomes and Gata4, (F) Bivalent genes Lhx5 and Klf4, or (G) cell cycle genes Caspase3 and C-Myb (Mybl2). Gene models are shown below with TSSs designated by arrows.
See also Figure S1.
To probe in detail the level of pausing among different gene classes, we calculated the Pausing Index for all genes. The Pausing Index is the ratio of Pol II signal density near a gene promoter to signal density within the gene body (Adelman and Lis, 2012), such that higher Pausing Indices reflect a greater enrichment of promoter-paused Pol II. Ranking all Pol II-occupied genes in ESCs (i.e. those with statistically significant promoter Pol II GRO-seq signal) by decreasing Pausing Index showed that 51% exhibit a Pausing Index greater than four, indicative of strong promoter bias of Pol II signal (Figure 1C, left, shown by blue arrow). By comparison, only 42% of Pol II-occupied developmental genes have a Pausing Index greater than four (Figure 1C, right). Statistical analysis demonstrated no difference in the Pausing Indices between developmental genes and genes on average (Figure 1D). We conclude that, although some developmental genes may be controlled at the level of pausing, this mechanism of regulation is no more common at developmental genes than expected by chance. Accordingly, genes encoding master regulators of pluripotency exhibited Pol II signal across the gene, indicative of productive transcription elongation (e.g., Figure 1E, Sox2). Likewise, cell-lineage markers tended to have a low, uniform distribution of Pol II across the gene with little evidence of pausing (e.g., Figure 1E, Eomes and Gata4) or lacked significant Pol II signal (Figure 1A, Developmental; below green arrow).
We further noted that Developmental genes characterized as having bivalent chromatin modifications typically exhibited low occupancy by engaged Pol II (shown in Figure 1A as blue tick marks at left of Developmental heat map; (Ku et al., 2008). This was of interest, because previous work had indicated that genes defined as bivalent in serum conditions gained Pol II occupancy in 2i, leading to a model wherein pausing plays a role in repressing bivalent gene expression under naïve conditions (Marks et al., 2012). Therefore, we investigated the distribution of engaged Pol II at all genes characterized by bivalent histone modifications in serum (Ku et al., 2008). This revealed generally low GRO-seq signals near the promoters in 2i media, with a majority of bivalent genes lacking transcriptionally engaged Pol II (Figure 1A, Bivalent, green arrow, 39% Pol II-occupied; e.g. Figure 1F, Lhx5). In addition, bivalent genes displayed significantly lower Pausing Indices vs. genes on average (Figure 1D). Bivalent genes with detectable Pol II, such as the actively transcribed Klf4 and Goosecoid genes (Figure 1F and S1C), typically displayed Pol II occupancy across the gene body, without promoter-enrichment of signal. Thus, our data from ESCs grown in 2i agree fully with GRO-seq from serum-grown ESCs (Figures S1C-S1E; Jonkers et al., 2014; Min et al., 2011) and do not support models that involve pausing of Pol II or control of transcription elongation as a central regulator of bivalent gene expression (Brookes et al., 2012; Marks et al., 2012). Consistent with this, recent evidence from ESCs grown in 2i indicates that Pol II detected near bivalent promoters has likely not undergone transcription initiation (Tee et al., 2014).
To identify gene classes most likely to be regulated by pausing, we performed gene ontology (GO) analysis of the decile of Pol II-occupied genes with the highest Pausing Indices (n=1,159). This revealed significant enrichment in GO terms related to cell cycle, response to DNA damage stimulus, metabolism and regulation of protein kinase activity (P<0.005). The GO category most enriched among highly paused genes is Cell cycle (GO:0007049; Figures 1A and 1B), with a number of cyclin-dependent kinases, their inhibitors, and other cell cycle regulatory factors displaying paused Pol II in ESCs (e.g. Figure 1G). Comparative analyses of Pausing Indices within each gene group confirmed a significant enrichment of pausing at cell cycle genes (Figure 1D). This finding is intriguing, given the extremely rapid cell cycle progression and absence of checkpoint control in ESCs, and suggests that pausing might impact these unique aspects of stem cell biology (Knoepfler, 2009).
Importantly, the GRO-seq assay for engaged Pol II yields consistent conclusions for ESCs grown in either 2i (this work) or serum-containing media (Jonkers et al., 2014; Min et al., 2011): paused Pol II is not enriched at promoters of lineage-specifying, developmental control or bivalent genes, but is prevalent at cell cycle regulators and genes involved in signaling (Figures S1C-1E). Hence, these findings suggest that Pol II pausing plays a different role in ESCs than predicted previously.
Pol II Promoter Profiles are Broadly Conserved in ESCs
The strong agreement between GRO-seq data sets derived from ESCs grown under different culture conditions prompted us to investigate the concordance of Pol II distribution between GRO-seq and Pol II ChIP-seq data sets. We compared Pol II signal near all RefSeq promoters (±150bp) among data sets derived from 2i vs. serum-containing culture conditions, GRO-seq and ChIP-seq using various Pol II antibodies (Figure S2A). As described above, the correlation between GRO-seq data sets from ESCs in 2i and serum was very good (Spearman's rho=0.88). Notably, Pol II ChIP-seq data from ESCs grown in serum (Rahl et al., 2010) that used an antibody recognizing all Pol II species (regardless of the phosphorylation state of the Pol II C-terminal domain) exhibited promoter Pol II levels that were highly correlated with both GRO-seq data sets (Spearman's rho=0.85 for GRO-seq in 2i, =0.86 for GRO-seq in serum). Thus, this “total” Pol II ChIP-seq data set accurately captures the majority of engaged Pol II (Rahl et al., 2010). By comparison, Pol II ChIP-seq performed using antibodies that recognize specific phosphorylation states of the Pol II C-terminal domain (CTD), which correspond to specific stages in the transcription cycle, showed globally lower concordance with GRO-seq data (Spearman's rho=0.77 to 0.82, see Figure S2A). Nevertheless, all data sets displayed generally strong agreement.
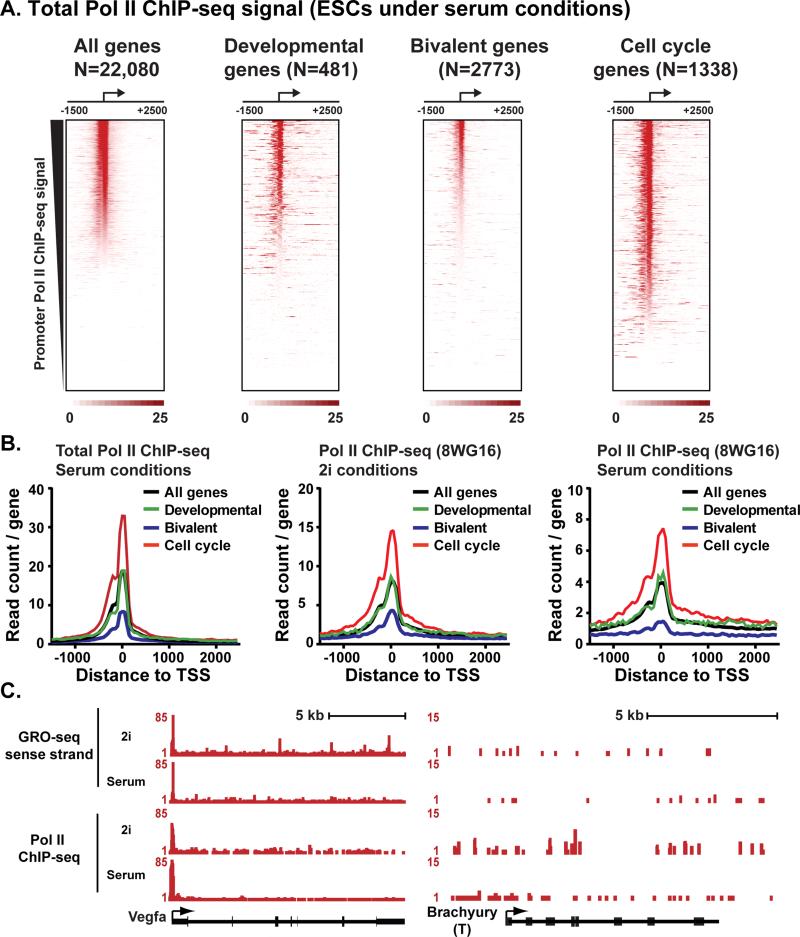
To determine whether the observed similarity in Pol II distribution between GRO-seq and ChIP-seq data extends from an analysis of all promoters to particular gene classes, we generated heat maps of Pol II ChIP-seq signal from ESCs grown under serum (Figure 2A) and 2i conditions (Figure S2B) at the gene groups analyzed in Figure 1. These heat maps reveal Pol II profiles across the gene classes that are very similar to GRO-seq data and to each other. Accordingly, composite metagene analysis of Pol II ChIP-seq signal exhibited equivalent profiles for various data sets (Figure 2B): Pol II signal was indistinguishable between genes on average (all genes, black lines) and developmental genes (green lines). In contrast, Pol II occupancy was markedly lower at bivalent genes (blue lines) and higher at cell cycle regulators (red lines). Thus, the relative distribution of Pol II at different gene classes is nearly identical among different cell culture conditions and techniques for mapping Pol II occupancy.
Figure 2. GRO-seq and Pol II ChIP-seq Data Sets Show Similar Pol II Profiles.
(A) Heatmap depiction of Pol II ChIP-seq reads (total Pol II antibody; data from (Rahl et al., 2010) from ESCs grown in serum-containing conditions. Genes are aligned around mouse RefSeq TSSs and ranked by decreasing promoter Pol II ChIP-seq signal (±150bp). Gene groups are shown as in Figure 1A.
(B) Composite Pol II ChIP-seq signal around all genes (black) as compared to the signal derived from Developmental genes (green), Bivalent genes (blue) and Cell cycle regulators (red). Data sets represent: Pol II ChIP-seq using a total Pol II antibody (Rahl et al., 2010) in serum-grown ESCs (as in Figure 2A) and Pol II ChIP-seq from 2i or serum conditions using the 8WG16 antibody that enriches in Pol II with a hypophosphorylated CTD (Marks et al., 2012).
(C) Examples of a gene that broadly display pausing across data sets, Vegfa (left), and a gene with little signal in any data set, Brachyury (right). Data are: GRO-seq in 2i (this work) or serum (Jonkers et al., 2014), Pol II ChIP-seq in 2i (Marks et al., 2012) or serum conditions (Rahl et al., 2010).
See also Figure S2.
We note that our analyses of comparable Pol II ChIP-seq data sets from ESCs grown in 2i vs. serum (Marks et al., 2012) recapitulate the previously reported broad increase in promoter Pol II levels in 2i media (compare peak levels in Figure 2B, center with 2B, right panel; Marks et al., 2012). But, we find that this phenomenon affects all gene groups equivalently, with no apparent specificity for developmental regulators or bivalent genes. Examination of individual developmental genes confirmed that Pol II distribution was similar among conditions and methodologies (Figure 2C and Figure S2C).
Ablation of NELF-B Results in Loss of the NELF complex and Proliferation Defects
To further define the functional targets of pausing during mammalian development, we generated mice with inducible knock-out of the pause-inducing factor, NELF. We used a targeting strategy in which LoxP sites were inserted flanking the NELF-B promoter and first four exons (Figures S3A-S3C). The expression of Cre recombinase in cells bearing the floxed NELF-B allele triggers recombination between the LoxP sites, excising this region of NELF-B and abrogating expression. Importantly, by targeting the central NELF-B subunit for genetic ablation, we can achieve depletion of all NELF complex members, since it is known that NELF subunits are inter-dependent for their stability (Narita et al., 2007).
Deletion of one NELF-B allele by crossing homozygous floxed mice (NELF-B Fl/Fl) with those expressing a ubiquitous Cre driver early in development (Zp3-Cre, see Supplemental Experimental Procedures) did not generate a phenotype. Heterozygous NELF-B Δ/Wt offspring were viable, fertile, and displayed no gross or histological abnormalities, in agreement with an earlier report (Amleh et al., 2009). In contrast, the absence of NELF-B is lethal during early development: crossing heterozygous NELF-BΔ/Wt animals failed to yield homozygous NELF-B Δ/Δ mice (Figure S3D).
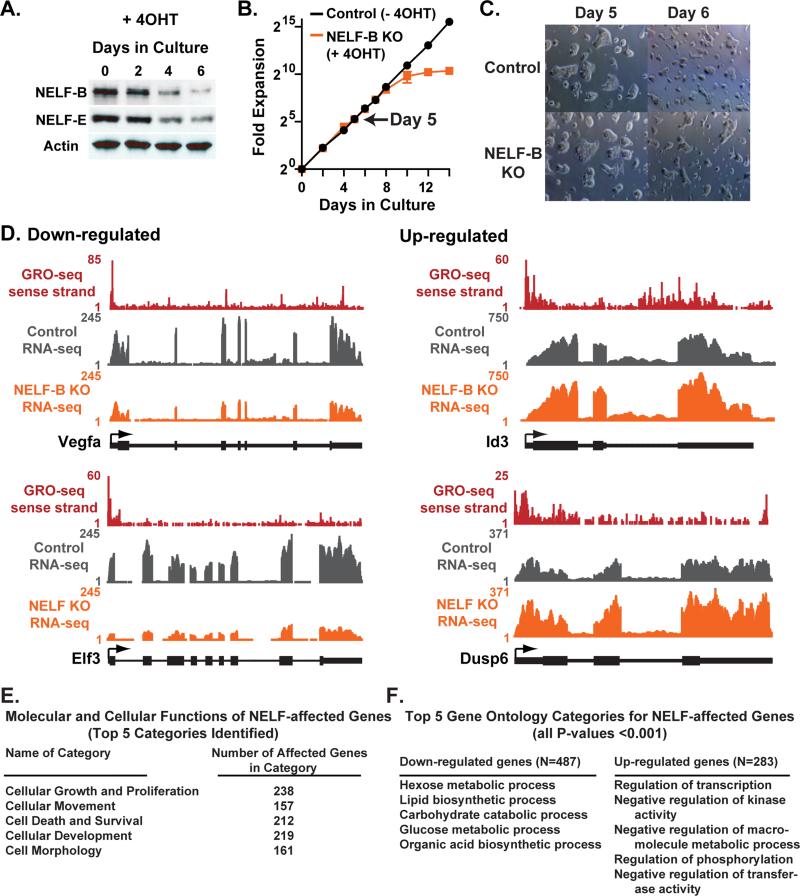
To probe the mechanisms that underlie an essential role for NELF-B in development, we generated ESCs in which NELF-B could be conditionally deleted by treatment with 4-hydroxy-Tamoxifen (4OHT) using an inducible Cre-Estrogen Receptor fusion protein (Cre-ER; Figure S3E). Upon treatment with 4OHT, ESCs derived from these mice underwent recombination of the NELF-B locus, and NELF-B knockout (KO) led to loss of other NELF complex subunits (e.g. Figure 3A). Consistent with a high level of pausing at genes associated with cell cycle, NELF-B KO in ESCs caused a dramatic decline in proliferation over time (Figure 3B). However, there was a delay between marked depletion of NELF-B and the onset of this defect, providing a window of opportunity (Figure 3B, Day 5, arrow) for analysis of the proximate targets of NELF-mediated pausing in ESCs with normal morphology (Figure 3C) and cell cycle progression (Figure S3F).
Figure 3. NELF-B Knockout in 2i Media Dysregulates Expression of Genes Involved in Proliferation, Signal Transduction and Metabolism.
(A) Western analysis shows that NELF-B and NELF-E proteins are lost with similar kinetics after conditional NELF-B KO ESCs are treated with 4OHT. Actin serves as a loading control.
(B) Viable ESCs were counted at the time points shown to determine the effects of NELF-B KO on cell proliferation.
(C) Images depict typical morphology of conditional NELF-B ESCs following vehicle or 4OHT treatment. Cells were split between Day 5 and Day 6, reflecting continued potential for self-renewal at this time point.
(D) Examples of genes significantly affected by NELF-B KO in RNA-seq data. The distributions of sense-strand GRO-seq reads and RNA-seq reads for Control and NELF-B KO ESCs are shown. Gene models are depicted with arrows indicating TSSs.
(E) A list of 771 NELF-affected genes was examined using Ingenuity Pathway Analysis, and the top Molecular and Cellular Functions categories were identified as shown.
(F) Lists of Entrez gene IDs corresponding to 487 genes significantly down-regulated (left) or 283 genes up-regulated (right) upon loss of NELF-B was analyzed for enriched GO categories (P<0.001). The top five GO categories for each class are shown.
NELF-B KO Causes Broad Dysregulation of Genes in Signaling and Metabolic Pathways
We performed RNA-seq on ESCs harboring the conditional NELF-B allele, grown in 2i media with and without 4OHT treatment. Within 5 days of recombination, loss of NELF-B significantly affected the expression of 771 genes (Table S1; P<0.01). As observed in other systems (Gilchrist et al., 2008; Narita et al., 2007), perturbation of NELF led to both up- and down-regulation of gene expression (e.g., Figure 3D). Loss of NELF-B did not significantly alter expression of pluripotency or lineage-specifying genes under these conditions (Table S1). Instead, pathway analysis of RNA-seq data identified cellular growth and proliferation as the molecular and cellular function most affected by loss of NELF-B (Figure 3E). Notably, a number of regulators of cell growth and cell cycle that display Pol II pausing, such as B-myb, Caspase3 (both shown in Figure 1G), Aurora kinase C, Gadd45g, Ccng2, Stag3 and Ndrg1 were dysregulated in NELF-B KO ESCs. GO analysis of genes down-regulated by NELF-B KO revealed enrichment in metabolic and biosynthetic processes (Figure 3F, left), whereas genes up-regulated in NELF-B KO ESCs were enriched in transcription regulation and negative regulation of kinase activity (Figure 3F, right). Interestingly, up-regulated genes include a number of transcription factors and proteins such as Id3 that suppress ESC differentiation, and several repressors of FGF/ERK signaling (e.g. Dusp6, Spry4, Spry2, Spred3; Villegas et al., 2010; Ying et al., 2003).
Loss of NELF-B Affects Promoter Pol II Distribution
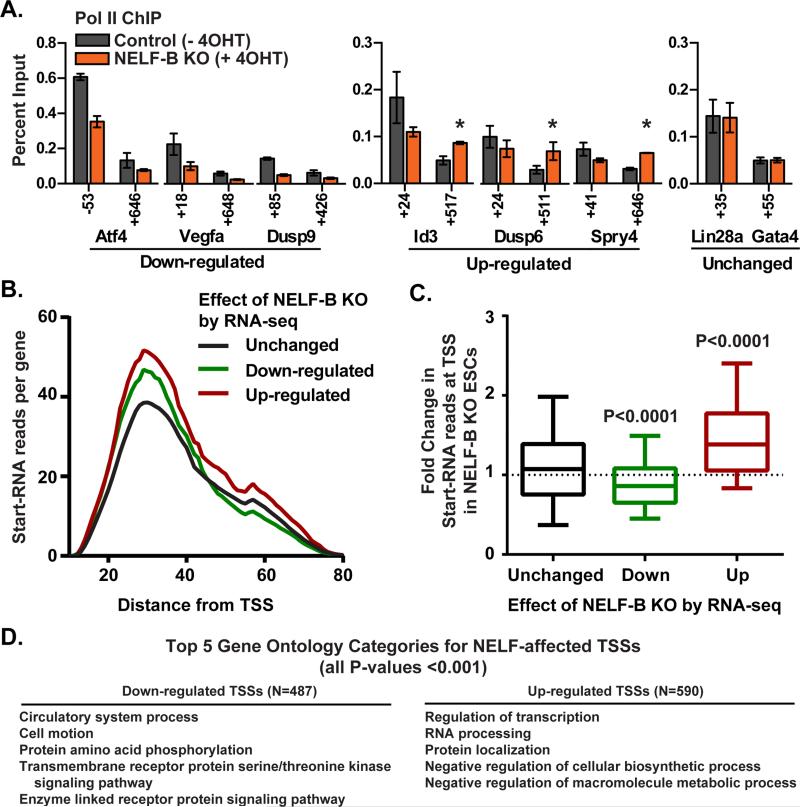
To define the effects of NELF-B KO under 2i conditions on the distribution of Pol II and the NELF complex, ChIP was performed at individual target genes. ChIP signal for the NELF-E protein was reduced to near background levels in NELF-B KO ESCs, confirming loss of the pause-inducing activity of the NELF complex as a whole (Figures S4A and S4B). Pol II occupancy was reduced at both promoter and downstream regions of genes down-regulated upon ablation of NELF-B (Figure 4A), consistent with work in Drosophila demonstrating a broad loss of Pol II at such genes (Gilchrist et al., 2010). Also as expected, genes up-regulated by disruption of NELF-mediated pausing showed an increase in elongating Pol II released into the gene body (Figure 4A, asterisks). By comparison, no changes in Pol II ChIP signal were detected at genes unaffected by NELF-B KO such as Gata4, an early marker of cell differentiation that does not display pausing (e.g. Figure 4A, Unchanged).
Figure 4. Effects of NELF-B KO on Gene Expression Reflect Changes in Promoter Pol II.
(A) ChIP was performed to verify changes in Pol II distribution upon NELF-B KO. Percent input is shown for N=3 clones ± SEM. Positions noted on the x-axis denote the center point of each primer pair with respect to the TSS. Significant increases in Pol II signal within up-regulated genes are noted by asterisks (P<0.05).
(B) Shown are the number and length of Start-RNA reads observed in control ESCs (N=3) for genes unchanged, down-regulated, or up-regulated in RNA-seq analysis of NELF-B KO ESCs. The distribution of distances between the TSS (defined by the 5’-end of the Start-RNA) and the position of Pol II within the promoter region (defined by the Start-RNA 3’-end) is similar for all gene groups (no significant differences). But, both down- and up-regulated genes display significantly more Start-RNA reads mapping near their promoters than genes unchanged by loss of NELF-B (Down-regulated vs. Unchanged P=0.05; Up-regulated vs. Unchanged, P<0.0001; Mann-Whitney U test).
(C) Fold change in Start-RNA reads upon deletion of NELF-B at genes unchanged, down- and up-regulated by NELF-B KO in RNA-seq experiments. For this analysis, Start-RNA reads with 5’-ends mapping within 100 nt of RefSeq TSSs were counted using normalized (N=3) data sets from control vs. NELF-B KO ESCs.
(D) TSSs with significantly fewer (left) or more (right) Start-RNAs upon loss of NELF-B were analyzed for enriched GO categories (P<0.001), based on the Entrez gene ID associated with each affected TSS. The top five GO categories for each class are shown.
See also Figure S4.
To broadly confirm that the effects of NELF-B KO on gene expression occur through altered behavior of promoter-proximal Pol II, we employed a sensitive technique developed to measure the level and position of early elongation complexes genome-wide (Nechaev et al., 2010). Importantly, this method captures the nascent transcription start site-associated RNAs (hereafter referred to as Start-RNAs) associated with both paused and actively elongating Pol II, revealing the precise position and total amount of engaged polymerase located within the initially transcribed sequence (approximately the first 100 nt, Nechaev et al., 2010).
First, we analyzed Start-RNAs from control ESCs grown in 2i conditions. We found similar distributions of Start-RNA 3-prime ends for all genes: elongation complexes were focused between 25-40 nt downstream of the transcription start site (TSS), precisely within the region where NELF-mediated pausing occurs (Nechaev et al., 2010). As shown in Figure 4B, the 3-prime end positions were comparable regardless of whether a gene was affected by NELF-B KO in RNA-seq (Figure 4B, Unchanged, Down-regulated, and Up-regulated). These data provide broad evidence of promoter-proximal pausing in ESCs, even at genes whose expression is unchanged by loss of NELF-B.
Whereas the position of pausing is similar for all gene groups, the level of promoter-proximally paused Pol II in control ESCs is greater at genes that are affected by NELF-B KO: we observed significantly more reads mapping near the promoters of genes that are either down- or up-regulated upon NELF-B KO than at genes whose expression is unchanged (Figure 4B). This indicates that genes with higher levels of Pol II paused within their promoter-proximal regions are more likely to be impacted by dysregulation of NELF.
Next, we determined the change in Start-RNA levels observed in Control vs. NELF-B KO ESCs grown under the same 2i conditions as used for RNA-seq, to validate that the changes in steady-state RNA levels resulted from differential pausing in the absence of NELF. We found that genes down-regulated in RNA-seq exhibited significantly reduced Start-RNA reads upon NELF-B KO (Figure 4C), confirming a decrease in promoter Pol II occupancy at these genes in the absence of NELF. Conversely, genes up-regulated in RNA-seq experiments showed significantly increased levels of Start-RNAs upon loss of NELF-B (Figure 4C), signifying an increase in Pol II transiting through the promoter region of these genes. This finding is consistent with a direct reduction in pause duration in the absence of NELF, enabling more frequent initiation and promoter escape by Pol II at up-regulated genes, as exemplified by the Hsp70 gene in Drosophila (Gilchrist et al., 2008).
Importantly, there is significant overlap in the genes defined as altered by NELF-B KO in RNA-seq and the TSSs affected in Start-RNA-seq (Overlap among Down-regulated genes and TSSs: P= 6×10−4; among Up-regulated genes and TSSs: P < 1×10−11). Further, analysis of transcripts whose TSSs show significant decreases or increases in Start-RNA levels in NELF-B KO ESCs reveals enrichment in GO categories very similar to those identified by RNA-seq, including regulation of transcription, metabolic processes, and signaling pathways (Figures 4D). Thus, evaluation of Start-RNA reads validates that defects in steady state RNA levels upon NELF-B KO are directly related to changes in promoter Pol II and implicates pausing in control of key facets of the ESC state.
NELF-B KO ESCs Resist Spontaneous Differentiation in Serum Conditions
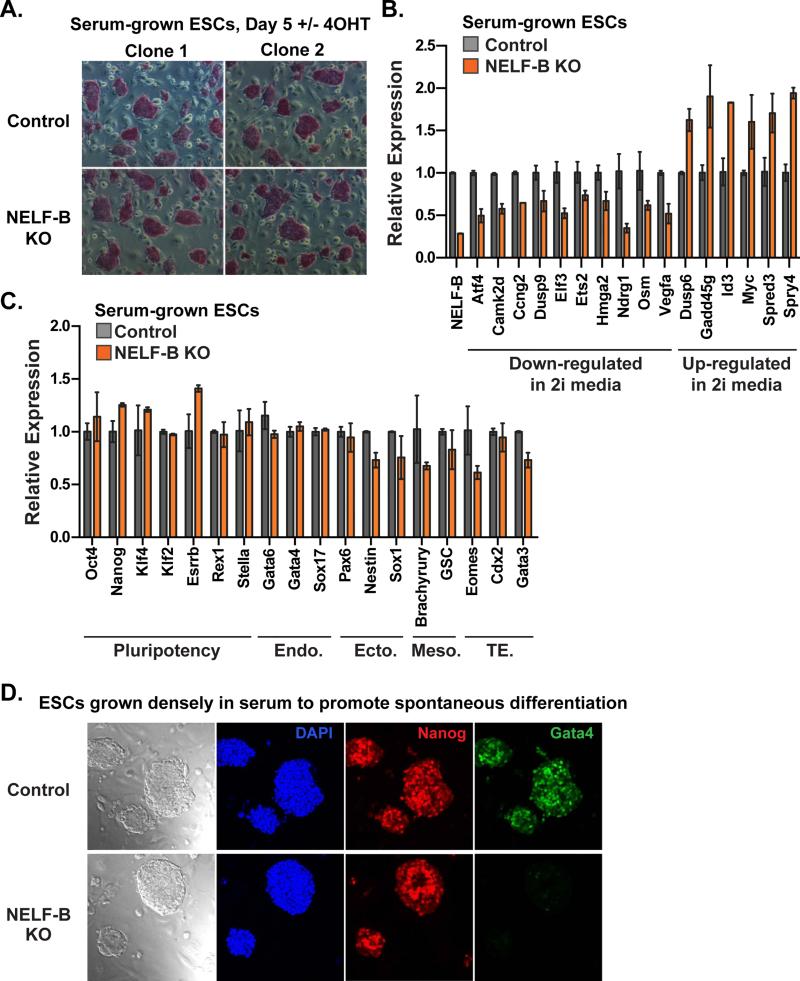
To further evaluate gene expression and developmental signaling in NELF-B KO ESCs, cells were maintained in conventional non-restrictive (without 2i) media containing serum. Five days after recombination of NELF-B, cells in serum exhibited colony morphology and alkaline phosphatase staining characteristic of pluripotent ESCs (Figure 5A). Gene expression analysis of these serum-grown ESCs by qRT-PCR confirmed that regulators of cell cycle affected by NELF-B KO in 2i media were similarly altered in serum (Figure 5B; e.g. Camk2d, Ccng2, Ndrg1, Osm, Gadd45g). Moreover, developmental signaling genes dysregulated upon loss of NELF-B in both media conditions include several components of the FGF/ERK pathway (Figure 5B and S4C), with increased expression of repressors like Spry4, Spred3, and Dusp6. Critically, the expression of pluripotency genes was not altered, and no cell-lineage marker tested displayed increased expression in NELF-B KO ESCs grown in serum (Figures 5C and S5A). Thus, these data confirm our findings in 2i conditions that NELF-mediated pausing is not a primary means of repressing lineage-specific gene expression.
Figure 5. Ablation of NELF-B Renders ESCs Resistant to Differentiation in Serum.
(A) Alkaline phosphatase staining of Control and NELF-B KO ESCs grown in serum on feeders (Day 5 after treatment +/− 4OHT). NELF-B KO ESCs display similar colony morphology and size, as well as equal staining intensity with alkaline phosphatase.
(B) Quantitative RT-PCR analysis of mRNA in NELF-B KO ESCs grown in serum on feeders. Genes involved in proliferation and FGF/ERK signaling were similarly dysregulated in serum-exposed NELF-B KO ESCs as compared to ESCs grown in 2i.
(C) Pluripotency marker expression is similar between Control and NELF-B KO ESCs, while lineage-specific genes are unchanged, or even down-regulated in NELF-B KO cells grown in serum conditions. Markers represent: endoderm (Endo.), ectoderm (Ecto.), mesoderm (Meso.) and trophectoderm (TE) lineages. Values shown represent N=2 independent clones ± range, with control levels set as 1.
(D) Bright field images depict typical differences in morphology between Control and NELF-B KO ESCs grown to high density in serum on feeders to induce spontaneous differentiation (Day 5 after treatment +/− 4OHT). Immunofluorescence is shown for DAPI, Nanog, and Gata4. Representative images are merged z-stack sections, to show the absence of Gata4 within the entire field of view.
See also Figure S5.
To probe the sensitivity of NELF-B KO ESCs to differentiation cues, ESCs were plated at a high-density in serum-containing media and grown for three days without passage to induce spontaneous differentiation. Indeed, ruffled morphology indicative of differentiation was observed on the edges of control ESC colonies (Figure 5D). In contrast, NELF-B KO colonies maintained the smooth external appearance typical of pluripotent cells. Immunostaining was then performed for the pluripotency marker Nanog and an early marker of primitive endoderm formation, Gata4. Importantly, Gata4 is used here as a marker of cell differentiation since Gata4 expression is not directly affected by NELF-B KO (Figure 5C). As anticipated, large colonies of control ESCs exhibited clear staining for Gata4, revealing spontaneous differentiation towards endoderm within the colony population (Figure 5D). However, Gata4-staining was absent in cells depleted of NELF-B (Figure 5D). This finding suggests that NELF-B KO ESCs resist intrinsic differentiation signals due to up-regulation of factors that antagonize pro-differentiation signaling cascades (e.g. Figure S4C). This idea was further supported by alkaline phosphatase staining (Figure S5B) over an extended time course of ESC growth in serum without passage. Control ESCs underwent spontaneous differentiation, as determined by expanded, altered colony morphology and reduced alkaline phosphatase staining (Figure S5B). In contrast, robust alkaline phosphatase staining persisted with no changes in NELF-B KO colony morphology over a 6-day time period. We conclude that NELF-B KO ESCs are refractory to spontaneous differentiation.
NELF Activity is Required for Signal Transduction through the FGF/ERK Pathway
To better understand the mechanisms underlying the resistance of NELF-B KO ESCs to differentiation, we identified genes affected by NELF-B KO within developmental signaling cascades. As noted above, this analysis revealed that a number of negative regulators of ERK activity were up-regulated upon NELF-B deletion (Figure S4C), implying that NELF-B KO might attenuate FGF/ERK signaling. Given the pivotal role of the FGF/ERK cascade as a trigger for cell fate commitment in ESCs (Kunath et al., 2007; Stavridis et al., 2007), these findings suggested that repression of ERK activity in NELF-B KO ESCs could underlie their blunted response to differentiation cues.
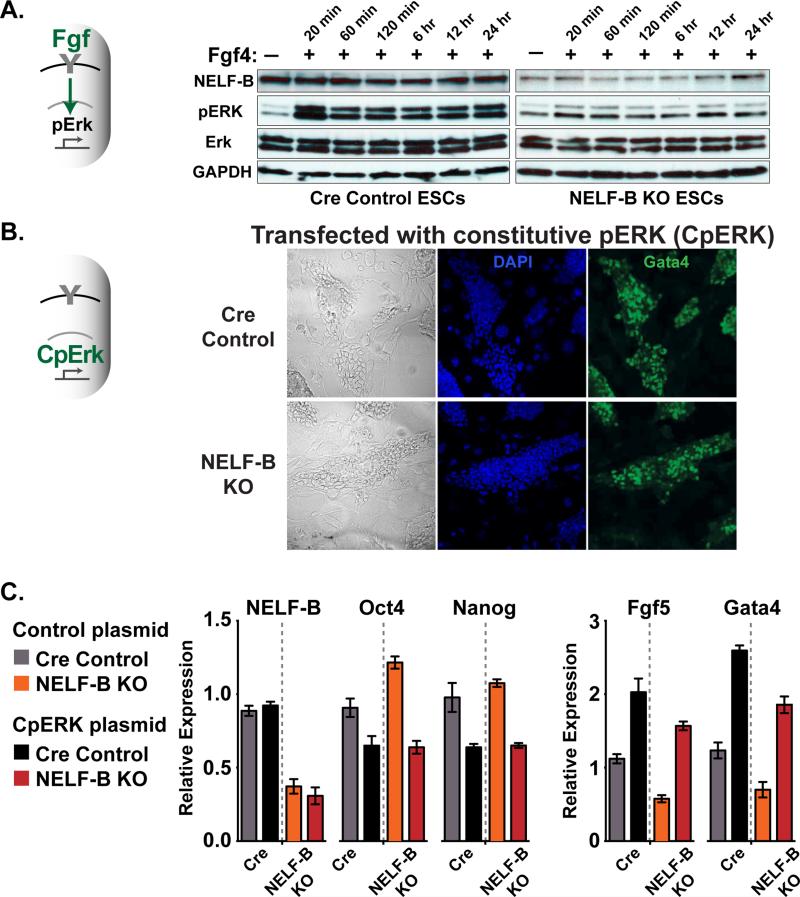
To directly test activity of the FGF/ERK pathway, Cre control and NELF-B KO ESCs were treated with Fgf4 and the resulting levels of activated, phosphorylated ERK (pERK) determined by western blotting. As expected, stimulation of control ESCs with Fgf4 led to marked increases in pERK that were sustained over a 24 hour time course (Figure 6A). In contrast, NELF-B KO ESCs exhibited little increase in pERK at any time point following the addition of Fgf4 (Figures 6A and S6A show this effect in distinct ESC clones). Thus, loss of NELF-B causes a strong, persistent attenuation of FGF/ERK signaling.
Figure 6. NELF-B KO ESCs are Refractory to FGF/ERK Signaling Upon Treatment with Fgf4, But are Induced to Differentiate with Constitutively Active ERK.
(A) Fgf4 stimulation of NELF-B KO ESCs fails to trigger phosphorylation of ERK (pERK). Cre control and NELF KO ESCs were cultured in serum for 4 days after 4OHT treatment, and Fgf4 added to the media (10 ng/ml) for indicated times. Western analysis of phosphorylated and total ERK is shown, with NELF-B and GAPDH as loading controls. ERK antibodies recognize ERK1 (upper) and ERK2 (lower band).
(B) Constitutively active ERK protein (CpERK) triggers differentiation and Gata4 expression in NELF-B KO ESCs. Cre control and conditional NELF-B KO ESCs were treated with 4OHT prior to transfection with CpERK. ESCs plated on feeders in serum-containing media were analyzed by immunofluorescence for DAPI and Gata4 on day 5 after 4OHT treatment. Images are merged z-stack sections.
(C) Analysis of mRNA levels for pluripotency (Oct4 and Nanog) and early differentiation (Fgf5 and Gata4) markers. Cre Control and NELF-B KO ESCs were transfected with plasmids expressing GFP (Control), or GFP and CpERK (CpERK). Transfected, GFP-positive cells were isolated by FACS and RNA analyzed by qRT-PCR. Values represent N=3 clones ± SEM.
See also Figure S6.
We next asked whether ESCs lacking NELF-B could successfully differentiate if ERK activity were rescued. To accomplish this, control and NELF-B KO ESCs were transfected with a constitutively active ERK protein (CpERK, described in Robinson et al., 1998) or a control vector for comparison. As visualized by bright field imaging and immunofluorescence (Figure 6B), the expression of CpERK in NELF-B KO ESCs resulted in differentiated cell morphology and expression of Gata4. Further, RT-PCR analysis of transfected ESCs showed that expression of CpERK led to reduced expression of pluripotency genes Oct4/Pou5f1 and Nanog in both control and NELF-B KO ESCs, and increased mRNA levels of differentiation markers Fgf5 and Gata4 (Figure 6C). Thus, bypass of the defective FGF/ERK pathway in NELF-B KO ESCs enabled differentiation, pinpointing this pathway as a key functional target of NELF-mediated pausing in pluripotent cells.
DISCUSSION
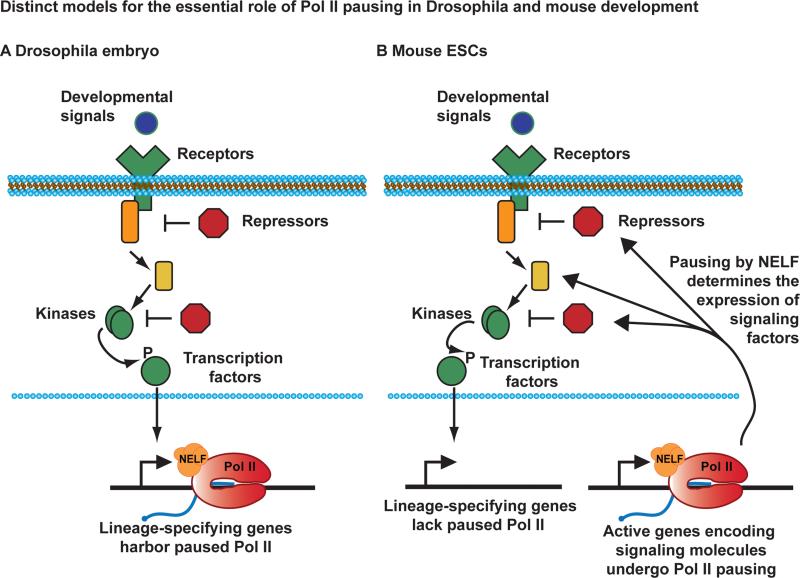
By combining our high-resolution genomic analysis of engaged Pol II in naïve ESCs with a genetic dissection of NELF activity in both naïve and serum-exposed ESCs, we define a broad, essential function for Pol II pausing in regulating stem cell differentiation potential. This work greatly increases our understanding of the transcriptional control of pluripotency and cell fate specification. We propose a paradigm for the role of Pol II pausing in mammalian ESCs that does not involve active suppression of developmental regulators and lineage-specifying genes (Figure 7). In contrast to the situation in Drosophila embryos, where pausing at such genes is common (Figure 7A), in naïve mouse ESCs promoters targeted by developmental signals exhibit Pol II pausing less frequently than expected by chance (Figure 1C). Instead, we find that NELF activity facilitates plasticity and cell fate commitment in mouse ESCs by establishing the appropriate expression level of signaling molecules. Pausing would thus control the responsiveness of signaling cascades to differentiation cues through direct regulation of signal transduction machineries (Figure 7B).
Figure 7. NELF-mediated Pausing of Pol II Regulates Developmental Signaling Cascades in ESCs.
Developmental signaling pathways converge on key transcription factors that enter the nucleus to activate genes involved in differentiation and cell fate specification.
(A) In the Drosophila embryo, many of the genes involved in morphogenesis and cell fate harbor paused Pol II (Red, shown associated with NELF in orange and a short, capped RNA transcript in blue). Pausing enables these promoters to be rapidly and synchronously induced by developmental cues.
(B) In mouse ESCs, genes involved in lineage commitment do not frequently display Pol II pausing, perhaps allowing them to be more tightly repressed. Pausing is instead prevalent at active genes encoding signaling molecules, where NELF activity governs the expression and responsiveness of signal transduction machineries.
Both the NELF complex and Pol II pausing are absent from lower organisms, such as yeast and C. elegans (Narita et al., 2003), suggesting that pausing could have originated to enable fine-tuned transcriptional profiles required for establishment of complex body plans. We suggest that the differences in the function of pausing in flies and mouse may reflect fundamental differences in their developmental programs: whereas synchronous cell cycling and extremely rapid gene activation are essential for Drosophila embryos (Lagha et al., 2013; Levine, 2011), cell cycle progression and cell fate specification are inherently less coordinated and more probabilistic in mammals.
In support of a connection between Pol II pausing and the FGF/ERK pathway, previous investigation of Pol II ChIP-seq data from ESCs grown in serum found an enrichment of pausing at genes in the ERK network (Gilchrist et al., 2012). This enrichment likely reflects the importance of establishing stable, yet flexible ERK signaling activity. Pluripotent cells are highly vulnerable to dysregulation of ERK signaling and must balance a requirement to suppress ERK activity in the early blastocyst with the necessity for effective signaling within the inner cell mass during the development of the primitive endoderm (Chazaud et al., 2006; Villegas et al., 2010). Moreover, recent results indicate that ERK can phosphorylate the Pol II C-terminal domain, raising the intriguing possibility of a feedback loop between Pol II transcriptional status and the ERK pathway (Tee et al., 2014). Furthermore, ESCs must tightly control their cell cycle progression and metabolic profile, reflecting other pathways enriched in paused Pol II. Thus, we propose that pausing provides ESCs a mechanism for coordinating gene expression in order to achieve and maintain the equilibrium required for pluripotency and self-renewal.
Notably, not all developmental signaling pathways are equally susceptible to perturbations in pausing; for example, most targets of the β-catenin/Wnt pathway were unchanged in NELF-B KO ESCs. This finding appears to contrast with a previous study suggesting that deletion of NELF-B causes up-regulation of Lef1, a transcriptional mediator of Wnt signaling (Amleh et al., 2009). However, we find no evidence of Pol II pausing at Lef1 in our GRO-seq data, nor does loss of NELF-B affect Lef1 expression by RNA-seq in 2i (Figure S7A). Moreover, despite an increase in Lef1 mRNA in NELF-B KO ESCs grown in serum, our data (Figure S7B) and prior microarray analysis (Figure S7C) indicates that Lef1 target genes are broadly unaffected by depletion of NELF-B, arguing against activation of canonical Wnt signaling in NELF-B KO ESCs.
In summary, this study reveals that control of signal transduction activity in ESCs can be accomplished at the level of Pol II pausing. By regulating many genes within a given signaling network, NELF exerts potent influence over pathway activity, as exemplified by the attenuation of FGF/ERK signaling in ESCs lacking NELF-B. Further, we find that pausing governs multiple aspects of the ESC state, including proliferative capacity and metabolic characteristics. This suggests that further studies of pausing could provide new insights into the absence of checkpoint control in ESCs and their inherent tumorigenicity (Knoepfler, 2009). Likewise, from a translational perspective, future investigation of NELF targets in human ESCs and different stem cell compartments might identify strategies for improving cellular reprogramming efficiency and could facilitate resetting of human ESCs to the naïve ground state.
EXPERIMENTAL PROCEDURES
Cell culture and ESC clones
GRO-seq was performed in the C2 ESC line. Conditional NELF-B and Control ESCs were derived from NELF-Bwt/wt, CreER+/− and NELF-BFl/Fl, CreER+/− mice on a C57Bl/6 background, as per standard protocols. Where indicated, cells were treated with 100 nM 4OHT (Sigma) to recombine out the floxed NELF-B alleles. ESCs were maintained in 2i media (Ying et al., 2008) with 1 μM PD0325901 (Stegment), 3 μM CHIR99021 (Stegment), and 1000 U/mL ESGRO, or non-restrictive M15 media (15% ES-grade serum in DMEM plus 1000 U/mL ESGRO) on feeders.
GRO-seq, RNA-seq, Start-seq and ChIP
Detailed methods for are available in Extended Experimental Procedures. GRO-seq assays were performed as previously described (Core et al., 2008) and libraries prepared using the Illumina small RNA TruSeq kit. For RNA-seq, total RNA was extracted from whole cells using Trizol reagent (Invitrogen) and exogenous RNAs were spiked-in to each sample for use as internal normalization controls. Libraries were prepared according to the TruSeq Stranded Total RNA Gold Kit (Illumina). Isolation and sequencing of Start-RNAs was performed as described (Nechaev et al., 2010). To ensure normalization between samples, 15 synthetic capped RNAs were spiked into the Trizol preparation as in (Henriques et al., 2013). ChIP was performed as previously described (Gilchrist et al., 2010) using antibodies targeting the Rpb3 subunit of Pol II and NELF-E (#HPA007187; Sigma).
Data Analysis
Two independent samples had mappable read depths of 33,123,684 and 25,976,028. Agreement between replicates was strong (R2=0.96), thus the data was combined for further analysis. Pausing indices were calculated as the ratio of read density (read pairs/kb) in the promoter window (TSS±150 nt) over the gene body density (+250 to +2250nt). For RNA-seq, a total of 113,817,860 and 108,809,117 read pairs were successfully aligned for the Control and NELF-B KO samples, respectively. Differentially expressed genes were identified with DESeq at an adjusted p-value of <0.01. Start-RNA reads from 3 replicates were in good agreement, with a mappable read depth of 82,685,356 for Control and 77,635,525 for NELF-B KO ESCs. After normalizing for sequencing depth, the return of spike-in RNAs was very similar for all samples, so no additional normalization was performed. Pathway analysis employed Ingenuity software. GO analysis was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID ) v6.7 (http:david.abcc.ncifcrf.gov/home.jsp). The top 5 most enriched (P<0.001) GO categories are shown.
Immunofluorescence and Alkaline Phosphatase Staining
For immunostaining, fixed and permeabilized cells were incubated with the following antibodies: Gata4 (Santa Cruz Biotechnology, #sc25310, 1:50) and Nanog (Cosmo Bio #RCAB0002P-F, 1:1000). Alkaline phosphatase staining was performed using the Stemgent AP Staining kit II (#00-0055).
FGF Stimulation and Constitutive ERK Activation
To stimulate the FGF/ERK pathway, cells were cultured in M15 in the presence of 10 ng/mL Fgf4 with 1 ug/mL heparin. Transfections were performed using previously described ERK2-MEK1-LA expression vector (CpERK) (Robinson et al., 1998) and a GFP expressing plasmid, to assist in the isolation of transfected cells for qRT-PCR.
Supplementary Material
Highlights.
- Pausing of Pol II does not repress expression of bivalent or developmental genes
- Disruption of pausing attenuates proliferative and developmental signaling pathways
- ESCs lacking NELF are refractory to FGF signaling and lineage commitment
- Regulated Pol II pausing tunes the differentiation potential of stem cells
ACKNOWLEDGEMENTS
We thank Dr. Lars Pedersen (NIEHS) for generation of the NELF-B protein, Paul Wade (NIEHS) for the Rpb3 antibody and Dr. Melanie Cobb (University of Texas Southwestern) for providing the constitutively active ERK expression vector. This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences to K.A. (Z01 ES101987).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
L.H.W., G.F. and K.A. designed the study. L.H.W. and G.F. performed the majority of experiments described. G.W.M did mouse breeding and phenotyping. G.H provided technical guidance. N.G. and T.H. carried out GRO-seq and Start-seq experiments, respectively. A.B., D.C.F and K.A. analyzed data. K.A. wrote the paper with the help of L.H.W.
Accession Number
All genomic data described herein are deposited in Gene Expression Omnibus with accession #GSE43390.
REFERENCES
- Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nature reviews. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amleh A, Nair SJ, Sun J, Sutherland A, Hasty P, Li R. Mouse cofactor of BRCA1 (Cobra1) is required for early embryogenesis. PloS one. 2009;4:e5034. doi: 10.1371/journal.pone.0005034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Brookes E, de Santiago I, Hebenstreit D, Morris KJ, Carroll T, Xie SQ, Stock JK, Heidemann M, Eick D, Nozaki N, et al. Polycomb associates genome-wide with a specific RNA polymerase II variant, and regulates metabolic genes in ESCs. Cell stem cell. 2012;10:157–170. doi: 10.1016/j.stem.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazaud C, Yamanaka Y, Pawson T, Rossant J. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Developmental cell. 2006;10:615–624. doi: 10.1016/j.devcel.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Cheng B, Price DH. Properties of RNA Polymerase II Elongation Complexes Before and After the P-TEFb-mediated Transition into Productive Elongation. The Journal of biological chemistry. 2007;282:21901–21912. doi: 10.1074/jbc.M702936200. [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science (New York, N.Y. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertner B, Johnston J, Chen K, Wallaschek N, Paulson A, Garruss AS, Gaudenz K, De Kumar B, Krumlauf R, Zeitlinger J. Poised RNA polymerase II changes over developmental time and prepares genes for future expression. Cell reports. 2012;2:1670–1683. doi: 10.1016/j.celrep.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Fromm G, dos Santos G, Pham LN, McDaniel IE, Burkholder A, Fargo DC, Adelman K. Regulating the regulators: the pervasive effects of Pol II pausing on stimulus-responsive gene networks. Genes & development. 2012;26:933–944. doi: 10.1101/gad.187781.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Nechaev S, Lee C, Ghosh SK, Collins JB, Li L, Gilmour DS, Adelman K. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes & development. 2008;22:1921–1933. doi: 10.1101/gad.1643208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JA, Surani MA. Regulatory principles of pluripotency: from the ground state up. Cell stem cell. 2014;15:416–430. doi: 10.1016/j.stem.2014.09.015. [DOI] [PubMed] [Google Scholar]
- Henriques T, Gilchrist DA, Nechaev S, Bern M, Muse GW, Burkholder A, Fargo DC, Adelman K. Stable Pausing by RNA Polymerase II Provides an Opportunity to Target and Integrate Regulatory Signals. Molecular cell. 2013;52:517–528. doi: 10.1016/j.molcel.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers I, Kwak H, Lis JT. Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. eLife. 2014;3:e02407. doi: 10.7554/eLife.02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS. Deconstructing stem cell tumorigenicity: a roadmap to safe regenerative medicine. Stem cells (Dayton, Ohio) 2009;27:1050–1056. doi: 10.1002/stem.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS genetics. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunath T, Saba-El-Leil MK, Almousailleakh M, Wray J, Meloche S, Smith A. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development (Cambridge, England) 2007;134:2895–2902. doi: 10.1242/dev.02880. [DOI] [PubMed] [Google Scholar]
- Lagha M, Bothma JP, Esposito E, Ng S, Stefanik L, Tsui C, Johnston J, Chen K, Gilmour DS, Zeitlinger J, Levine MS. Paused Pol II coordinates tissue morphogenesis in the Drosophila embryo. Cell. 2013;153:976–987. doi: 10.1016/j.cell.2013.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M. Paused RNA polymerase II as a developmental checkpoint. Cell. 2011;145:502–511. doi: 10.1016/j.cell.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Liu Y, Rhee HS, Ghosh SK, Bai L, Pugh BF, Gilmour DS. Kinetic competition between elongation rate and binding of NELF controls promoter-proximal pausing. Molecular cell. 2013;50:711–722. doi: 10.1016/j.molcel.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Stewart AF, Smith A, Stunnenberg HG. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes & development. 2011;25:742–754. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nature genetics. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita T, Yamaguchi Y, Yano K, Sugimoto S, Chanarat S, Wada T, Kim DK, Hasegawa J, Omori M, Inukai N, et al. Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex. Molecular and cellular biology. 2003;23:1863–1873. doi: 10.1128/MCB.23.6.1863-1873.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita T, Yung TM, Yamamoto J, Tsuboi Y, Tanabe H, Tanaka K, Yamaguchi Y, Handa H. NELF interacts with CBC and participates in 3' end processing of replication-dependent histone mRNAs. Molecular cell. 2007;26:349–365. doi: 10.1016/j.molcel.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science (New York, N.Y. 2010;327:335–338. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Molecular cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MJ, Stippec SA, Goldsmith E, White MA, Cobb MH. A constitutively active and nuclear form of the MAP kinase ERK2 is sufficient for neurite outgrowth and cell transformation. Curr Biol. 1998;8:1141–1150. doi: 10.1016/s0960-9822(07)00485-x. [DOI] [PubMed] [Google Scholar]
- Stavridis MP, Lunn JS, Collins BJ, Storey KG. A discrete period of FGF-induced Erk1/2 signalling is required for vertebrate neural specification. Development (Cambridge, England) 2007;134:2889–2894. doi: 10.1242/dev.02858. [DOI] [PubMed] [Google Scholar]
- Tee WW, Shen SS, Oksuz O, Narendra V, Reinberg D. Erk1/2 activity promotes chromatin features and RNAPII phosphorylation at developmental promoters in mouse ESCs. Cell. 2014;156:678–690. doi: 10.1016/j.cell.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villegas SN, Canham M, Brickman JM. FGF signalling as a mediator of lineage transitions--evidence from embryonic stem cell differentiation. Journal of cellular biochemistry. 2010;110:10–20. doi: 10.1002/jcb.22536. [DOI] [PubMed] [Google Scholar]
- Wray J, Kalkan T, Smith AG. The ground state of pluripotency. Biochemical Society transactions. 2010;38:1027–1032. doi: 10.1042/BST0381027. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nature genetics. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.