Abstract
Cystic fibrosis transmembrane conductance regulator (CFTR)-mediated chloride secretion is critical to maintaining airway surface hydration and efficient mucociliary clearance in the upper airways. Mutations in CFTR in cystic fibrosis lead to reduced expression of functional CFTR channels at the apical plasma membrane of the airway epithelium, leading to dehydration of the airway surface liquid and diminished mucociliary clearance. Cell surface CFTR is modulated by changes in CFTR endocytosis and recycling, effectively altering the cell surface abundance of the channel. This chapter examines current methods employed to measure the cell surface expression of CFTR, as well as methods to monitor CFTR movement through the endocytic pathway.
Keywords: Cystic fibrosis, cystic fibrosis transmembrane conductance regulator, endocytosis, recycling, intracellular trafficking
1. Introduction
The number of CFTR channels in the apical plasma membrane of polarized epithelial cells – and thus, the rate of transepithelial Cl secretion – is determined in part by regulating the endocytic trafficking of CFTR (1). Endocytic trafficking is the process of sequestration and internalization of cargo proteins from the plasma membrane into endocytic vesicles, called endosomes, trafficking of endosomes among intracellular organelles, and then recycling of endosomes back to the plasma membrane, or delivery of endosomes to lysosomes where cargo proteins are degraded (2). In general, endocytosed proteins are delivered to early endosomes and either recycle back to the plasma membrane via recycling endosomes or move progressively from multivesicular bodies to late endosomes and to lysosomes where cargo is degraded. A fascinating variety of proteins regulate the endocytic trafficking of cargo proteins, including Rab GTPases, SNAREs, PDZ domain-containing proteins, myosin motors, numerous lipids including phosphoinositide-P (3) and phosphoinositide-P2 (3, 5), and a host of other regulatory proteins (1, 2).
In this chapter, we will discuss methods to detect and quantify the apical membrane abundance of CFTR and follow the intracellular trafficking of CFTR through the endocytic pathway. Cell surface biotinylation methods will be discussed as an effective measure of apical membrane CFTR abundance (3, 4). This chapter will also present an OptiPrep gradient fractionation method to separate subcellular compartments and a cell surface biotinylation-based endocytosis and recycling assay (4). These methods provide the ability to quantify the trafficking of CFTR through the endocytic pathway, as well as the ability to measure the effect of a particular treatment on CFTR trafficking through this pathway.
2. Materials
All reagents were purchased from Sigma-Aldrich, unless noted otherwise. Ezrin antibody was purchased from BD Biosciences (San Jose, CA), CFTR antibody (clone 596) from University of North Carolina Cystic Fibrosis Center (funded by the Cystic Fibrosis Foundation), NHS-LC-biotin (Cat. # 21335) from Thermo Scientific (Waltham, MA), and streptavidin agarose beads (Cat. # 20349) from Thermo Scientific (Waltham, MA). Phosphate buffered saline (PBS, 500 ml bottle) was purchased pre-made from Invitrogen (Carlsbad, CA). Permeable membrane supports were purchased from Corning (Costar 24-mm Transwell support, 0.4 μm pore, Cat. # 3412; Corning, Inc., Corning, NY).
2.1. Buffers
-
Dulbecco’s PBS/Mg/Ca, pH 7.0 (PBS/Mg/Ca):
1 ml of 0.5 M MgCl2
0.1 ml of 0.5 M CaCl2
-
Lysis buffer (LB), pH 8.2:
25 mM HEPES, pH 8.0
1% (v/v) Triton X-100
10% (v/v) glycerol
-
LB + protease inhibitor (LB[C]):
One complete protease inhibitor tablet (Roche, Cat. # 1697498)
50 ml lysis buffer
-
Loading sample buffer (LSB):
Laemmli buffer (Bio-Rad Laboratories, Hercules, CA)
90 mM DTT
-
Homogenization buffer
0.25 M sucrose
78 mM KCl
4 mM MgCl2
8.4 mM CaCl2
10 mM EGTA
50 mM HEPES–NaOH, pH 7.0
One complete protease inhibitor tablet (Roche, Cat. # 1697498)/50 ml of buffer
-
Working solution diluent
78 mM KCl
4 mM MgCl2
8.4 mM CaCl2
10 mM EGTA
50 mM HEPES–NaOH, pH 7.0
-
Glutathione solution, pH 8.6 (GSH; Sigma, Cat. # G6529), in deionized water:
50 mM GSH
75 mM NaCl
1 mM MgCl2
0.1 mM CaCl2
Add just before use:
80 mM NaOH
10% FBS
*The NaOH neutralizes the carboxyl group and deprotonates half the cysteine residues in glutathione. It is strongly buffered at the pKa of this cysteine, which is pH 8.6.
3. Methods
3.1. Cell Surface CFTR
-
Culture human airway epithelial cells on permeable membrane supports at air–liquid interface to polarize cells and perform desired treatments.
24-mm Transwell permeable membrane support/treatment.
Remove all fluid by placing the filters at an angle to allow draining of the fluid to one side.
Wash wells with cold PBS/Mg/Ca, pH 8.2, three times, each wash for 2 min (see Note 1).
Incubate with 1 ml of NHS-LC-biotin (in PBS/Mg/Ca, pH 8.2; 1 mg/ml) on the apical side of the membrane support in the dark for 60 min at 4°C (see Note 2).
Wash wells with cold PBS/Mg/Ca, pH 8.2, three times, each wash for 2 min.
Add 300 μl of LB[C] to each well.
Incubate in the dark for 15 min shaking vigorously on an orbital shaker at 4°C.
Scrape cells from filter with a rubber policeman and collect in a 1.5-ml Eppendorf tube.
Homogenize each sample with a pipette tip and spin at 14,000×g for 10 min at 4°C.
Add 50 μl of sample buffer (LSB) to 30 μl of supernatant and place sample at 85°C for 5 min (whole cell lysate (WCL) sample).
Prepare 100 μl aliquots of 50% streptavidin agarose beads and aspirate all fluid.
Wash with 1 ml of PBS/Mg/Ca twice, pelleting the beads with a pulse spin between the washes.
Wash with 1 ml LB[C] once (to equilibrate the beads before adding the cell lysates); suction dry the beads (see Note 3).
Incubate the remaining supernatants with washed streptavidin beads (bring the volume up to 1.0 ml with LB[C]) rotating in the dark for at least 2 h or overnight at 4°C.
Wash beads with 1 ml LB three times, each wash for 2 min, pulse spin the beads between washes, and suction dry the fluid after the last wash.
Incubate beads with 80 μl LSB at 85°C for 5 min.
Pulse spin the beads.
Collect the supernatant in 1.5-ml Eppendorf tubes and place sample at −20°C until ready to perform Western blot analysis.
Load samples on 7.5% Tris–HCl ready gels (Bio-Rad Laboratories, Hercules, CA) and run at 120 mV.
Transfer onto polyvinylidene fluoride (PVDF) membrane in cold room for 1.5–2 h at 95 mV.
Incubate membranes in 5% milk in TBS+0.1% Tween 20 blocking buffer at room temperature for 1 h.
Perform Western blot analysis with both WCL and biotinylation reaction samples, probing blots overnight with 1:1000 CFTR monoclonal antibody (clone 596) and 1:1000 ezrin monoclonal antibody (loading control) in 5% bovine serum albumin (BSA) in PBS+0.1% Tween 20 at 4°C (see Note 4). Goat anti-mouse secondary antibody conjugated to horseradish peroxidase (1:3000) in 5% milk in TBS+0.1% Tween 20 is used for detection.
3.2. CFTR Trafficking Through Intracellular Compartments Utilizing OptiPrep Gradients
Protocol adapted from Biemesderfer D, et al. (2002) AJP Renal 282:F785-94(6). Please see Note 5:
-
Culture human airway epithelial cells on permeable membrane supports at air–liquid interface to polarize cells and perform desired treatments:
24-mm or (1) 100-mm Transwell permeable membrane support/treatment.
-
Prepare OptiPrep gradient:
Add 1.9 ml volume of 20% OptiPrep in working solution diluent to the bottom of a 4.4-ml ultracentrifuge tube.
Add 1.9 ml of 5% OptiPrep in working solution diluent on top of the 20% layer (with peristaltic pump).
Allow continuous gradient to form over 3–4 h at room temperature (or overnight at 4°C) or use gradient maker to generate continuous gradient.
Remove all fluid by placing the filters at an angle to allow draining of the fluid to one side.
Wash filters with cold PBS/Mg/Ca, pH 8.2, three times, each wash for 2 min (see Note 1).
Incubate with 1 ml of NHS-LC-biotin (in PBS/Mg/Ca, pH 8.2; 1 mg/ml) in the dark for 60 min at 4°C.
Wash wells with cold PBS/Mg/Ca, pH 8.2, three times, each wash for 2 min (see Note 2).
Lyse cells on filters with 200 μl/well of 1% Triton X-100 in homogenization buffer.
Scrape the filters and pass through a 22-gauge, 3–in. needle 20 times.
Spin lysates at 1,500×g for 8 min.
Transfer 0.5 ml of the supernatant to a TH-660 ultracentrifuge tube (polyallomer), overlaying continuous OptiPrep gradient (with p200, 100 μl at a time very gently!!).
After overlaying prepared lysates, centrifuge gradients for 18 h at 100,000×g in TH-660 swing-bucket rotor in Sorvall Ultra Pro 80 ultracentrifuge.
Analyze the gradient in 0.25 ml fractions by WB analysis (18 fractions total; last fraction contains plasma membrane pellet).
Add 50 μl of sample buffer to 30 μl of supernatant and place at 85°C for 5 min (whole cell lysate (WCL) sample).
Prepare 100 μl aliquots of 50% streptavidin agarose beads for each fraction and aspirate all fluid.
Wash with 1 ml of PBS/Mg/Ca twice, pelleting the beads with a pulse spin between the washes.
Wash with 1 ml LB[C] once [to equilibrate the beads before adding the cell lysates]; suction dry the beads (see Note 3).
Incubate the remaining supernatants from each fraction of the gradient with washed streptavidin beads (bring the volume up to 1.0 ml with LB[C]) rotating in the dark for at least 2 h or overnight at 4°C.
Wash beads with 1 ml LB three times, each wash for 2 min, pulse spin the beads between washes, and suction dry the fluid after the last wash.
Incubate beads with 80 μl LSB at 85°C for 5 min.
Pulse spin the beads.
Collect the supernatant in 1.5-ml Eppendorf tubes and place at −20°C until ready to perform Western blot analysis.
Load samples on 7.5% Tris–HCl ready gels (Bio-Rad) and run at 120 mV.
Transfer onto PVDF membrane in cold room for 1.5–2 h at 95 mV.
Incubate membranes in 5% milk in TBS+0.1% Tween 20 blocking buffer at room temperature for 1 h.
Perform Western blot analysis with both WCL and biotinylation reaction samples, probing blots overnight with 1:1000 CFTR monoclonal antibody (clone 596) and 1:1000 ezrin monoclonal antibody (loading control) in 5% bovine serum albumin (BSA) in PBS+0.1% Tween 20 at 4°C (see Notes 4 and 6). Goat anti-mouse secondary antibody conjugated to horseradish peroxidase (1:3000) in 5% milk in TBS+0.1% Tween 20 is used for detection.
-
Intracellular compartment location in the gradient is detected by Western blot analysis, probing with antibodies specific for each compartment.
Plasma membrane (fraction 18): Na/K ATPase, gp135 (BD Biosciences, San Jose, CA);
Endosome (fractions 9–13): Rab5a (Santa Cruz Biotechnology, Santa Cruz, CA);
Lysosome (fractions 5–9): LAMP-1 (BD Biosciences, San Jose, CA).
3.3. CFTR Endocytosis and Recycling Assays
Culture human airway epithelial cells on permeable membrane supports at air–liquid interface to polarize cells. Plates A–D (see below) are needed for a recycling assay, but only plates A–C are needed if endocytosis is being measured.
*(1) 24-mm Transwell permeable membrane support/treatment.
3.3.1. Prepare the Previous Night
PBS/Mg/Ca, pH 8.2, at 37°C.
PBS/Mg/Ca, pH 8.2, at 4°C.
PBS/Mg/Ca, pH 8.6, at 4°C.
LB[C] at 4°C.
-
Deionized water at 4°C.
*see Note 7.
3.3.2. Day of the Experiment
Use warm PBS/Mg/Ca at 37°C to fill wells of six-well plates as needed for endocytosis and recycling assays. Place these plates in the 37°C incubator that will be used for the warming during the assay.
3.3.3. Biotinylation Reaction
-
1
Perform desired treatments on cultured human airway epithelial cells.
-
2
Place filters on ice (see Note 8).
-
3
Wash the apical and basolateral sides of filters with ice-cold PBS/Mg/Ca, pH 8.2, three times, each wash for 2 min (see Notes 9 and 10).
-
4
Incubate with 1.0 ml of 1 mg/ml NHS-S-S-biotin in PBS/Mg/Ca, pH 8.2, buffer on the apical side, 1.5 ml PBS/Mg/Ca, pH 8.2, on the basolateral side in the dark in the cold room for 60 min (see Note 9).
-
5
Wash with PBS/Mg/Ca, pH 8.2, three times, each wash for 2 min.
-
6
Leave PBS/Mg/Ca, pH 8.2, on all filters.
-
7
Arrange filters into plates that will receive the same treatment in the endocytosis/recycling assays as indicated in Table 18.1 (see Note 11).
Table 18.1.
Arrangement of filters in plates for the endocytosis/recycling assays
| Plate A | Plate B | Plate C | Plate D | |
|---|---|---|---|---|
| Number of filters | 2–3 | 2–3 | 2–3 | 2–3 |
| Biotinylation | + | + | + | + |
| 1st Warming | + 5 min | + 5 min | ||
| 1st GSH | + | + | + | |
| 2nd Warming | +5 min | |||
| 2nd GSH | + |
3.3.4. Endocytic Assay
-
8
Leave plates A and B in the cold room.
-
9
Place plates C and D on ice and bring the plates on ice close to the 37°C incubator (used for endocytosis and recycling time points).
-
10
Pour off cold PBS/Mg/Ca and put filters in wells of warm PBS/Mg/Ca, pH 8.2, at 30 s intervals. Incubate at 37°C for 5 min. Raising the temperature to 37°C will induce endocytosis.
-
11
At 5 min, put filters in six-well plates containing ice-cold PBS/Mg/Ca, pH 8.2, on ice (keep the same timing as in the initiation of endocytosis, this will mean staggering by 30 s). This step will terminate endocytosis (see Note 12).
-
12
Bring plates C and D on ice back to the cold room.
-
13
Remove all PBS/Mg/Ca from plates A to D.
-
14
Incubate plates B–D with 1.0 ml of glutathione (GSH) solution, pH 8.6, on the apical side and 1.5 ml PBS/Mg/Ca, pH 8.6, on the basolateral side, in the cold room in the dark five times, each time for 15 min (don’t need to change the PBS on the basolateral side) (make enough GSH solution for just endocytosis washes, see Note 13).
-
15
At the same time, incubate plate A with PBS/Mg/Ca, pH 8.6 (see Note 14).
-
16
After GSH incubation is finished, wash plates B and C with PBS/Mg/Ca, pH 8.6, three times, each wash for 2 min.
-
17
Wash plate D with PBS/Mg/Ca, pH 8.2, three times, each wash for 2 min.
-
18
Leave plates A–C in the cold room.
3.3.5. Recycling Assay
-
19
Subject the remaining plate (D) to a second warming for 5 min (plate D), staggering each filter by 30 s, as above for endocytosis protocol (see Note 15). This incubation at 37°C allows endocytosed CFTR to recycle.
-
20
Put the filters in ice-cold PBS/Mg/Ca, pH 8.2, and wash with PBS/Mg/Ca, pH 8.2, three times, each wash for 2 min. This step terminates recycling.
-
21
Incubate plate D with freshly made glutathione (GSH) solution, pH 8.6, in the dark three times, each GSH treatment for 15 min (see Note 13) at 4°C.
-
22
At the same time, incubate plates A–C with PBS/Mg/Ca, pH 8.6.
-
23
Remove all fluid by placing the plates at an angle to allow draining of the fluid to one side.
-
24
Add 300 μl LB[C] to each filter.
-
25
Incubate in the dark at 4°C for 15 min by shaking vigorously on an orbital shaker.
-
26
Scrape the cells from filter with a rubber policeman and collect in a 1.5-ml Eppendorf tube.
-
27
Homogenize each sample with a pipette tip and spin at 14,000×g for 10 min at 4°C.
-
28
Add 50 μl of sample buffer to 30 μl of supernatant and put at 85°C for 5 min (whole cell lysate (WCL) sample).
-
29
Prepare 100 μl aliquots of 50% streptavidin agarose beads and aspirate all fluid.
-
30
Wash with 1 ml of PBS/Mg/Ca twice, pelleting the beads with a pulse spin between the washes.
-
31
Wash with 1 ml LB[C] once (to equilibrate the beads before adding the cell lysates); suction dry the beads (see Note 3).
-
32
Incubate the remaining supernatants with washed streptavidin agarose beads (bring the volume up to 1.0 ml with LB[C]) rotating in the dark for at least 2 h or overnight at 4°C.
-
33
Wash beads with 1 ml LB three times, each wash for 2 min, pulse spin the beads between washes, and suction dry the fluid after the last wash.
-
34
Incubate beads with 80 μl LSB at 85°C for 5 min.
-
35
Pulse spin the beads.
-
36
Collect the supernatant in 1.5-ml Eppendorf tubes and place at −20°C until ready to perform Western blot analysis.
-
37
Load samples on 7.5% Tris–HCl ready gels (Bio-Rad Laboratories, Hercules, CA) and run at 120 mV.
-
38
Transfer onto PVDF membrane in cold room for 1.5–2 h at 95 mV.
-
39
Incubate membranes in 5% milk in TBS+0.1% Tween 20 blocking buffer at room temperature for 1 h.
-
40
Perform Western blot analysis with both WCL and biotinylation reaction samples, probing blots overnight with 1:1000 CFTR monoclonal antibody (clone 596) and 1:1000 ezrin monoclonal antibody (loading control) in 5% bovine serum albumin (BSA) in PBS+0.1% Tween 20 at 4°C (see Note 4). Goat anti-mouse secondary antibody conjugated to horseradish peroxidase (1:3000) in 5% milk in TBS+0.1% Tween 20 is used for detection.
3.3.6. Analysis of Data
-
41
Biotinylated CFTR is quantitated and normalized for WCL CFTR or ezrin abundance (see Note 4).
-
42
Samples from plate A are set as the control biotinylated signal (set to 100%) (Fig. 18.1).
-
43
GSH cleavage efficiency is examined by percentage of plate A biotinylated CFTR signal found in plate B (see Note 16, Fig. 18.1):
GSH cleavage efficiency: ((100–5)/100)100 = 95%.
-
44
Endocytosis of CFTR is calculated by subtracting the biotinylated CFTR signal from plate C from the signal of GSH-cleaved biotinylated CFTR in plate B. This number is expressed as the percentage of total biotinylated CFTR in plate A:
Endocytosis of CFTR: 80–5 = 75;
Percentage of total CFTR endocytosed: (75/100)100 =75%.
-
45
Recycled CFTR is the fraction of the biotinylated CFTR signal from plate D of the endocytosed CFTR calculation from step 42. This number is then expressed as the percentage of endocytosed CFTR that recycles:
Recycling of CFTR: (20/75)100 = 26.67;
Percentage of endocytosed CFTR recycled: 100–26.67 = 73.33.
-
46
The results of the experiment depicted in Fig. 18.1 are as follows:
GSH cleavage efficiency is 95%.
The percentage of total biotinylated CFTR that endocytosed in 5 min is 75%.
The percentage of endocytosed CFTR that recycled in 5 min is 73.33%.
Fig. 18.1.
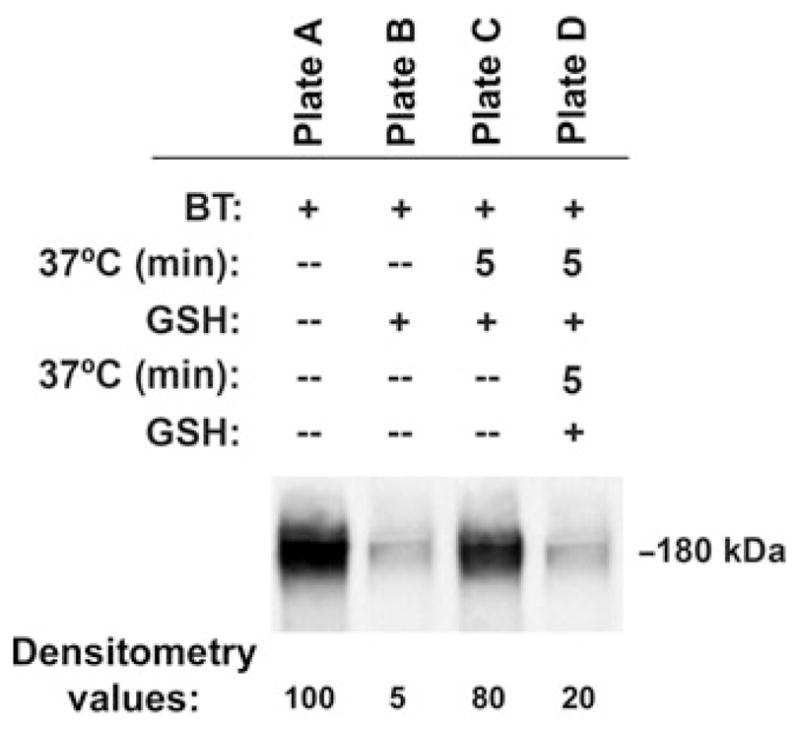
The endocytic recycling of CFTR. Polarized CFBE4lo- cells were grown on 24-mm Transwell permeable membrane supports for 7 days. A, Endocytosis and recycling experiments were performed as described in text and detailed in figure. B, Representative blots from endocytic and recycling assays. Cells from all four lanes were cooled to 4°C and biotinylated. The amount of biotinylated CFTR remaining in the plasma membrane after GSH treatment at 4°C (lane 2) was subtracted from the amount of CFTR remaining biotinylated after warming to 37°C and GSH treatment (lane 3) to determine the amount of endocytosed CFTR. Cells from lane 3 were then warmed at 37°C to allow recycling of endocytosed CFTR before a second GSH treatment. CFTR recycling was calculated as the difference between the amount of biotinylated CFTR after the first (lane 3) and second (lane 4) GSH treatments.
Footnotes
The biotinylation labeling efficiency has been optimized for pH 8.2 of the PBS/Mg/Ca used in the labeling reaction. Other pH of PBS/Mg/Ca will work, but pH 8.2 is the most effective at labeling cell surface proteins.
Gently washing cells before and after biotinylation procedure, as well as strict adherence to temperatures called for in protocol, will yield most reliable cell surface CFTR measures. This will also limit inappropriate biotinylation of intracellular CFTR proteins.
When aspirating the streptavidin agarose beads dry, use a 1-in., 27-gauge syringe needle with suction. The 27-gauge needle will remove all liquid, but not aspirate the beads.
Cell surface CFTR abundance can be normalized to total CFTR expressed in the WCL or to ezrin in the WCL sample. Both can be effective loading controls, given that the treatments being assessed do not change the expression of either WCL CFTR or ezrin. In our experience, many treatments alter actin expression in the cell lysate, while ezrin abundance remains quite constant. Therefore, we chose to normalize protein abundance in our samples to ezrin abundance.
This method enables the tracking of a cell surface protein through the endocytic pathway. By biotinylating cell surface proteins before treatments are applied, you can track the movement of a particular protein (i.e., CFTR) that begins the time course at the cell surface and analyze how different treatments alter the endocytic trafficking of that protein.
CFTR abundance in each fraction is represented as a percentage of total CFTR in all fractions.
All steps are done on ice or in the cold room, unless noted otherwise. Maintaining cold temperature, except during endocytosis or recycling time points, is absolutely critical in obtaining accurate measures of CFTR trafficking in this assay.
All treatments within the experiment are performed in duplicate or triplicate.
Check the pH of the various PBS/Mg/Ca solutions on the day of the experiment to make sure they are correct. An incorrect pH of the PBS/Mg/Ca will reduce the efficiency of the biotin labeling reaction, thus increasing the noise in the experiment.
Gently washing cells before and after biotinylation procedure, as well as strict adherence to temperatures called for in protocol, will yield most reliable cell surface CFTR measures. This will also limit inappropriate biotinylation of intracellular CFTR proteins. Be sure to use NHS-S-S biotin in this protocol to allow cleavage by GSH.
Arranging the filters in six-well plates (plates A–D), according to the condition during the endocytosis/recycling assay, is incredibly helpful during the endocytosis and recycling time points of the assay. Be sure to label plates carefully, detailing which well received which treatment.
The endocytosis and recycling time points are the most critical steps to strictly adhere to the temperatures called for in the protocol. Using large volumes of PBS/Mg/Ca for the washes ensures rapid temperature changes during time points. Previous experiments have been performed in CFBE41o- cells to determine that CFTR endocytosis is linear between 1 and 5 min. Performing this assay in a new cell type would require a time course to determine at which time points the endocytosis of CFTR is linear and suitable for the assay. Because endocytosed CFTR recycles rapidly, time points longer than ~5 min measure a composite of endocytosis and recycling back to the plasma membrane.
For best results with GSH cleavage of biotin, strictly adhere to the pH recommended in the protocol and make GSH just before use in each step.
Sequester plate A during the GSH washes, so GSH is not inadvertently added to plate A. This is the control that all samples will be compared to during analysis. If the biotin is cleaved from this plate, there is no control and the experiment is lost.
Previous experiments have been performed in CFBE4lo-cells to determine that CFTR recycling is linear at 2.5 and 5 min. Performing this assay in a new cell type would require a time course to determine at which time points the recycling of CFTR is linear and suitable for the assay.
GSH treatment (plate B) must achieve 85% cleavage of NHS-S-S-biotin from control filters (plate A) to be considered a successful experiment. Lesser cleavage makes interpretation and quantitation of results quite difficult (5).
References
- 1.Guggino WB, Stanton BA. New insights into cystic fibrosis: Molecular switches that regulate CFTR. Nat Rev Mol Cell Biol. 2006;7:426–436. doi: 10.1038/nrm1949. [DOI] [PubMed] [Google Scholar]
- 2.Gruenberg J, van der Goot FG. Mechanisms of pathogen entry through the endosomal compartments. Nat Rev Mol Cell Biol. 2006;7:495–504. doi: 10.1038/nrm1959. [DOI] [PubMed] [Google Scholar]
- 3.Swiatecka-Urban A, Boyd C, Coutermarsh B, Karlson KH, Barnaby R, Aschenbrenner L, et al. Myosin VI regulates endocytosis of the cystic fibrosis transmembrane conductance regulator. J Biol Chem. 2004;279:38025–38031. doi: 10.1074/jbc.M403141200. [DOI] [PubMed] [Google Scholar]
- 4.Bomberger JM, Barnaby RL, Stanton BA. The deubiquitinating enzyme USP10 regulates the postendocytic sorting of cystic fibrosis transmembrane conductance regulator in airway epithelial cells. J Biol Chem. 2009;284:18778–18789. doi: 10.1074/jbc.M109.001685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swiatecka-Urban A, Talebian L, Kanno E, Moreau-Marquis S, Coutermarsh B, Hansen K, et al. Myosin VB is required for trafficking of CFTR in RAB11A-specific apical recycling endosomes in polarized human airway epithelial cells. J Biol Chem. 2007;282:23725–23736. doi: 10.1074/jbc.M608531200. [DOI] [PubMed] [Google Scholar]
- 6.Biemesderfer D, Mentone SA, Mooseker M, Hasson T. Expression of myosin VI within the early endocytic pathway in adult and developing proximal tubules. Am J Physiol Renal Physiol. 2002;282:F785–F794. doi: 10.1152/ajprenal.00287.2001. [DOI] [PubMed] [Google Scholar]


