Abstract
Mycobacterium bovis is the major causative agent of bovine tuberculosis, one of the most relevant zoonoses in the world, and affects a wide range of wild and domesticated animals. Development of screening panels in mycobacterial genotyping, according to specific geographical regions, is strongly needed. The aim of this study is to select a panel, constituted by highly polymorphic MIRU-VNTR loci, to discriminate clinical isolates of M. bovis in Mexico. In this study, 65 isolates of M. bovis obtained from clinical bovine samples proceeding from different geographic regions of Mexico were identified by phenotypic and genotypic tests and subsequently genotyped by a 24-locus MIRU-VNTR panel. The most polymorphic loci were selected to build a panel with a high discriminatory power similar to the 24-locus panel results. A panel of seven elements (QUB 11a, MIRU 26, ETR-A, QUB 26, MIRU 16, MIRU 27, and MIRU 39) with the highest allelic diversity showed an appropriate differentiation. The selected MIRU-VNTR elements, according to the regional allelic variability, may be used in the preliminary genotyping of Mycobacterium bovis isolates in Mexico.
1. Introduction
Mycobacterium bovis is the major causative agent of bovine tuberculosis (BTB), one of the most relevant zoonoses in the world, showing a wide range of effects on wild and domesticated animals [1–3]. BTB is a major worldwide animal health problem, which negatively affects animal production as well as national and international trade of livestock [4]. In developed countries, which have a tradition of cattle farming, the prevalence of BTB has reached very low levels because of strict control policies [5], but even in the United States of America M. bovis continues infecting cattle at a low level despite control and eradication efforts [6]. Currently, only three countries, Canada, Mexico, and Australia, are authorized to trade live cattle with the United States [6]. However, BTB precludes animal-related trade and production, causing financial losses to farming worldwide, and remains a public health hazard [1].
There is limited evidence of human-to-human transmission of BTB. In fact, recent research findings suggest that transmission occurs mainly within cattle populations and less frequently between cattle and humans or between humans [7]. The geographical distribution of M. bovis cases in more-rural areas is consistent with zoonotic transmission [8]. However, the accurate incidence of BTB may be underestimated given that the infection is clinically indistinguishable from that caused by M. tuberculosis and because it is difficult to differentiate M. tuberculosis from M. bovis with traditional diagnostic methods [9, 10].
Furthermore, for BTB control it is required to accurately know how many and which M. bovis strains are located in each geographic region, and classical bacteriological methods are unable to distinguish strains among the same species [2]. Strain genotyping may contribute to a better understanding of BTB transmission and could lead to identification of outbreaks and tracking dissemination of particular strains, to explore relations between domestic and wild BTB, to improve bovine TB control strategies, or even to reconstruct the evolution of a certain group of bacteria [3, 5, 11, 12].
There is a growing research showing that mycobacterial interspersed repetitive unit variable number tandem repeat (MIRU-VNTR) is the most discriminatory technique to genotype M. bovis isolates [1, 13, 14]. This method can therefore be used to confirm conventional epidemiological links, trace transmission routes, determine the source of infection and outbreaks, understand the relationship between different outbreaks, and identify wild animal reservoir of M. bovis [5, 8].
In Europe, a network of laboratories has agreed on a consensus of six VNTR loci to genotype M. bovis [12]; however, these loci are still not widely used, which can hamper interlaboratory exchange [11, 13]. In Mexico, where resources for diagnosis and molecular typing are scarce, development of screening panels in mycobacterial genotyping is strongly needed. The aim of this study was to select a minimal MIRU-VNTR loci panel to discriminate locally prevalent M. bovis population.
2. Materials and Methods
2.1. Clinical Isolates
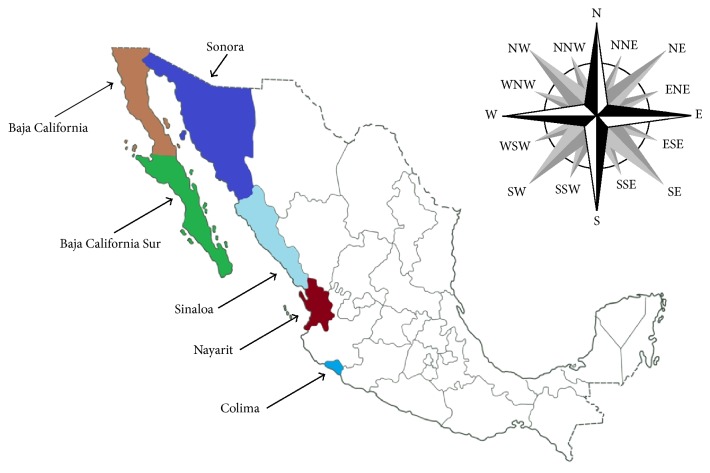
Sixty-five M. bovis isolates, originally cultured from bovine tuberculosis lesions, were included in this study. The isolates were obtained from five states of Mexico (Figure 1), Baja California (11 isolates), Baja California Sur (5), Colima (12), Nayarit (12), Sinaloa (12), and Sonora (13), from 2010 to 2013 and sent to a reference laboratory (Laboratorio Estatal de Salud Pública) in Sonora, México. All isolates were identified as M. bovis by phenotypic (standard biochemical assays) [15] and genotypic (gyrB-RFLP and RD1) tests [16, 17]. All isolates were frozen in skim milk and kept at −20°C until use. Mycobacterium tuberculosis H37Rv strain was used as control.
Figure 1.
Map of Mexico and geographical origin of the 65 Mycobacterium bovis isolates included in this study.
2.2. DNA Preparation
M. bovis isolates and Mycobacterium tuberculosis H37Rv strain frozen in skim milk were thawed and cultured on Stonebrink or Löwenstein-Jensen media, respectively, prepared as solid slants in screw-cap tubes, for 3 to 4 weeks at 37°C. For each isolate, four to five colonies were transferred into 500 μL of TE buffer (0.01 M Tris-HCl, 0.01 M EDTA [pH 8.0]). The suspended colonies were washed twice with TE and boiled for 30 min in Chelex-100 (10% in MilliQ water), and the bacterial lysate was used directly in PCRs (modified from [18, 19]).
2.3. MIRU-VNTR
Twenty-four genomic loci were amplified in separate PCR reactions with the primers previously described [20, 21]. PCRs were performed on 2.5 μL of DNA sample in a final volume of 25 μL. All PCR reactions were carried out according to the method of Supply et al. [22] with the following modifications: the final MgCl2 concentration was 1.5 mM, and 0.5 U of TaqDNA polymerase (Promega) was used in each reaction. Additionally, QUB 26 loci required 0.05% (v/v) DMSO for optimal results. All MIRU-VNTR loci were amplified in individual reactions. The amplification program consisted of 2 min at 95°C, followed by 40 cycles of 60 s at 94°C, 60 s at 59°C, and 90 s at 72°C and a final extension at 72°C for 10 min. The number of tandem repeats (alleles) was estimated after electrophoresis on 2% agarose (Sigma) gels at 90 V with a 100 bp ladder (Promega) according to the allele calling table. Mycobacterium tuberculosis H37Rv strain was used as positive control in every PCR reaction and electrophoresis procedure.
2.4. Data Analysis
The Hunter-Gaston index (HGI) was calculated to determine the discriminatory power for individual VNTR loci [23]. MIRU-VNTR profiles were recorded as character data and analyzed using Bionumerics software v 6.6 (Applied Maths, St-Martin-Latem, Belgium). Dendrograms were generated by using the categorical character option and the UPGMA (for unweighted pair-group method with arithmetic averages) clustering method.
3. Results
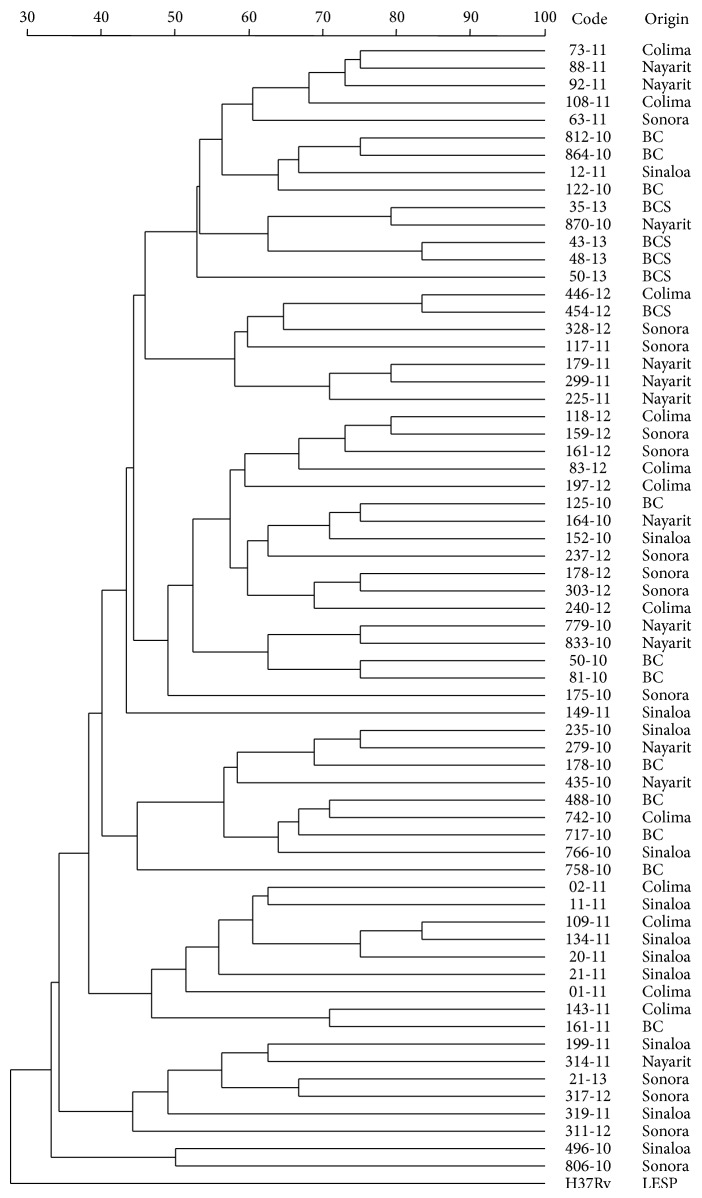
As expected, the 24-locus MIRU-VNTR panel allowed differentiation of 65 isolates. Figure 2 shows that each M. bovis isolate has a unique MIRU-VNTR profile and H37Rv M. tuberculosis strain is quite different from the rest of the mycobacteria. Despite achieving individual differentiation of each microorganism evaluated, some kind of association, regarding the geographical origin of each isolate, was not possible to obtain.
Figure 2.
Dendrogram generated by using categorical character and the UPGMA clustering method with 24-locus MIRU-VNTR panel results of 65 isolates of M. bovis and M. tuberculosis strain H37Rv.
The Hunter-Gaston index differed for individual loci, ranging from 0.34 to 0.86 (Table 1). Loci QUB3232 and QUB11a were the most discriminant, whereas QUB 1895 and MIRU 20 had the lowest diversity index. Locus QUB3232 showed several problems and was subsequently excluded from the final selection of highly discriminative MIRU-VNTR loci.
Table 1.
Allelic diversity of 24-locus MIRU-VNTR in 65 Mycobacterium bovis Mexican isolates.
| Locus | Number of copies | HGI | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | ||
| QUB 3232 | 4 | 3 | 6 | 15 | 14 | 9 | 6 | 4 | 4 | 0.86 | |||||
| QUB 11a | 2 | 4 | 1 | 4 | 2 | 11 | 20 | 16 | 5 | 0.81 | |||||
| MIRU 26 | 1 | 1 | 7 | 14 | 21 | 14 | 5 | 2 | 0.80 | ||||||
| ETR A | 1 | 4 | 2 | 14 | 22 | 18 | 4 | 0.77 | |||||||
| QUB 26 | 1 | 23 | 27 | 10 | 4 | 0.69 | |||||||||
| MIRU 16 | 24 | 28 | 10 | 3 | 0.66 | ||||||||||
| MIRU 27 | 4 | 37 | 10 | 10 | 1 | 3 | 0.63 | ||||||||
| MIRU 39 | 2 | 36 | 14 | 10 | 1 | 2 | 0.63 | ||||||||
| MIRU 2 | 10 | 34 | 20 | 1 | 0.62 | ||||||||||
| MIRU 31 | 1 | 23 | 33 | 8 | 0.61 | ||||||||||
| QUB 3336 | 1 | 40 | 4 | 3 | 6 | 8 | 1 | 2 | 0.60 | ||||||
| QUB 23 | 16 | 39 | 7 | 2 | 1 | 0.58 | |||||||||
| ETR C | 3 | 8 | 12 | 41 | 1 | 0.56 | |||||||||
| QUB 11b | 2 | 4 | 6 | 41 | 12 | 0.56 | |||||||||
| ETR B | 1 | 2 | 40 | 21 | 1 | 0.52 | |||||||||
| MIRU 40 | 18 | 42 | 5 | 0.51 | |||||||||||
| MIRU 23 | 1 | 24 | 40 | 0.49 | |||||||||||
| QUB 18 | 9 | 45 | 9 | 2 | 0.49 | ||||||||||
| MIRU 10 | 17 | 44 | 4 | 0.48 | |||||||||||
| MIRU 4 | 43 | 22 | 0.45 | ||||||||||||
| MIRU 24 | 22 | 43 | 0.45 | ||||||||||||
| QUB 15 | 3 | 48 | 13 | 1 | 0.42 | ||||||||||
| QUB 1895 | 12 | 50 | 3 | 0.38 | |||||||||||
| MIRU 20 | 8 | 52 | 5 | 0.34 | |||||||||||
HGI: Hunter-Gaston index.
Once the most polymorphic MIRU-VNTR loci were identified, additional analysis was performed to identify combinations for improved differentiation of mycobacteria included in the present study.
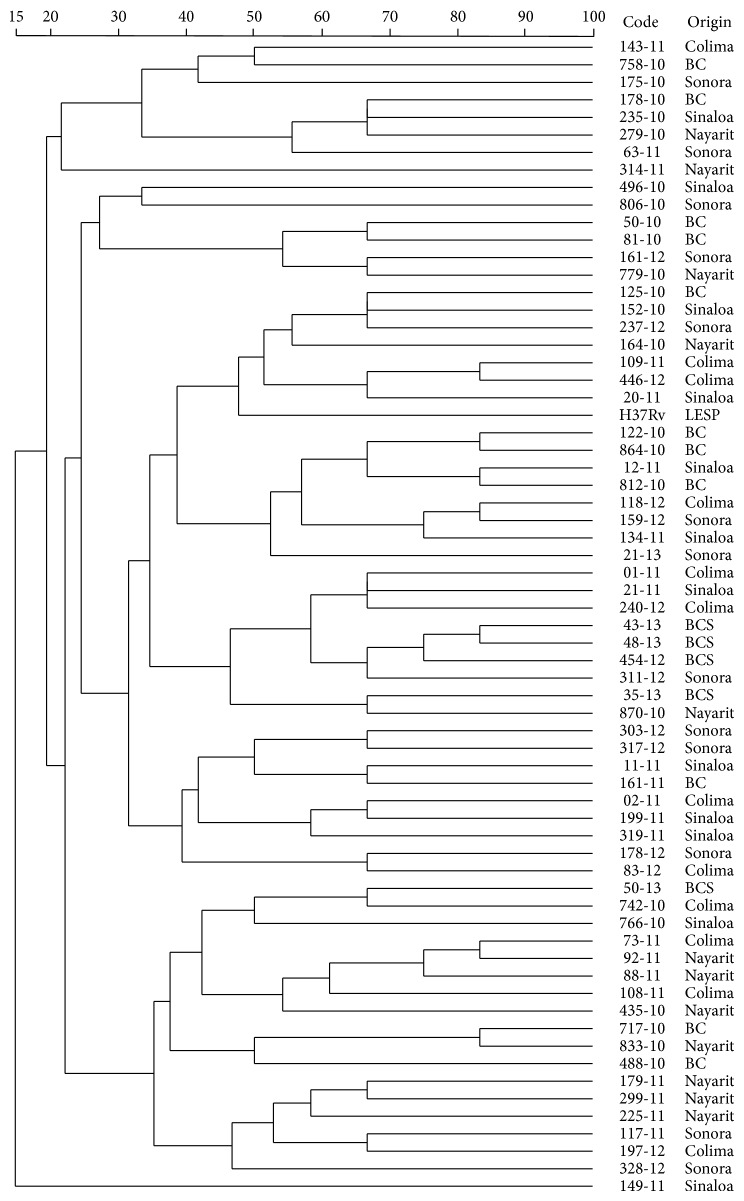
In the first approach, five MIRU-VNTR typing loci (excluding QUB 3232) showed a very high discriminatory index for our set of isolates and were particularly useful to differentiate almost all the mycobacterial isolates, except 73-11 and 92-11 isolates that displayed an identical profile (data not shown). By including MIRU 27, differentiation of each of the isolations was achieved; however, when we incorporated MIRU 39 into the panel (seven-locus MIRU-VNTR panel), the differentiation was kept and additionally it was possible to observe some clusters, according to the geographical region of origin (Figure 3).
Figure 3.
Dendrogram generated by using categorical character and the UPGMA clustering method with 7-locus MIRU-VNTR panel results of 65 isolates of M. bovis and M. tuberculosis strain H37Rv.
4. Discussion
The value of mycobacterial interspersed repetitive units variable number tandem repeats (MIRU-VNTR) as a genotyping technique for Mycobacterium bovis has previously been confirmed [1]. However, a standardized panel of MIRU-VNTR loci has not yet been adopted for M. bovis, since allelic diversity of each locus differs among countries [1, 4, 11]. Thus, it is important to standardize a panel of loci for future interlaboratory comparisons [4].
The first study in Mexico where genetic diversity of mycobacterial strains was evaluated using MIRU-VNTR revealed seven distinct patterns in nine M. bovis strains [10]. Recently Laniado-Laborín and colleagues found 26 strains of M. bovis in 600 clinical mycobacterial isolates [9]. In both studies, however, the mycobacteria were recovered from human clinical samples. In this study we have identified 65 M. bovis isolates with unique MIRU-VNTR patterns, but all of them were recovered from bovine sources.
Although all the 24 loci MIRU-VNTR showed good discriminatory power, the loci with the highest diversity were QUB 3232, QUB 11a, MIRU 26, ETR A, QUB 26, MIRU 16, MIRU 27, MIRU 39, MIRU 2, MIRU 31, and QUB 3336 (HGI 0.85–0.60). Meanwhile, the loci with intermediate diversity were QUB 23, ETR C, QUB 11b, ETR B, MIRU 40, MIRU 23, QUB 18, MIRU 10, MIRU 4, MIRU 24, and QUB 15 (HGI 0.58–0.42). Finally loci QUB 1895 (HGI: 0.38) and MIRU 20 (HGI: 0.34) were considered with low discriminatory power.
Our findings are consistent with data reported by previous researchers that consider QUB 3232 as the most variable locus [1, 4, 11, 14, 24]. QUB 11a, the second most variable locus in this study, has been reported with high [1, 7, 11, 13, 14, 20], intermediate [24], or low diversity index [4]. This situation was detected for MIRU 26 (high [1, 3, 7, 9, 20], intermediate [14, 24], and low diversity index [2]) and QUB 26 (high [7, 11, 20] and intermediate diversity index [2, 14, 24]).
Our results are also consistent with previous studies where ETR A was considered as a highly polymorphic locus [1, 4, 7, 11, 14, 20, 24]. However we observed considerable differences in utility of the loci MIRU 2, 27, 31 and 39, which in previous studies were considered as having a discriminatory power of intermediate or scarce value [3, 24]. Interestingly, in a previous research, MIRU 31 (HGI = 0.61, in this study) showed null discriminatory power in the genotyping of M. bovis recovered from human samples in Mexican population [9].
Regarding the loci that showed an intermediate or low value of HGI, our results correlate more appropriately with previous studies [1–4, 11, 13, 14, 20, 24].
By testing combinations of the most variable elements, within the set of 24 loci, we found that a satisfactory degree of discrimination could be achieved using a minimal group of six loci; however, we recommend extending the set of markers to include MIRU 39 (7-locus MIRU-VNTR panel), since it is possible to observe some tendency to clustering, according to the geographical region of origin (Figure 3). Several attempts to reduce the number of repetitive elements and maintain the most discriminative power have been done, and that includes six [4], eight [1], nine [20], or even thirteen MIRU-VNTR loci panels [24]. However, the optimal procedure to use for strain typing of M. bovis will depend on the strains present in a region, the number of isolates to be typed, and availability of resources.
As already mentioned, even though QUB 3232 was the most discriminative locus, difficulties with the reproducibility of amplification and gel band previously reported [9, 20, 24] lead us to not recommending this locus for routine use.
This is a preliminary screening investigation to establish a minimal panel for the large-scale genotyping analysis of M. bovis in Mexico. Our findings suggest the MIRU-VNTR markers that have high discriminative power and may be used to identify M. bovis isolates from the northwest and probably all of the Mexican territory. However, further research is recommended, to determine MIRU-VNTR loci that would be sufficiently discriminating in different settings/profiles of circulating strains, as previously proposed [8, 12].
5. Conclusions
The present study demonstrated the usefulness of 24-locus MIRU-VNTR panel for discriminating 65 Mycobacterium bovis isolates from six different regions of Mexico. This study also presents seven-locus MIRU-VNTR panel with high discriminatory power. Additional studies are needed to validate the proposed scheme.
Acknowledgments
This study was supported by “Apoyo a la Incorporación de Nuevos PTC,” convocatoria 2010, and “Fortalecimiento de los Cuerpos Académicos,” convocatoria 2011 del Programa para el mejoramiento del Profesorado (PROMEP).
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Duarte E. L., Domingos M., Amado A., Cunha M. V., Botelho A. MIRU-VNTR typing adds discriminatory value to groups of Mycobacterium bovis and Mycobacterium caprae strains defined by spoligotyping. Veterinary Microbiology. 2010;143(2–4):299–306. doi: 10.1016/j.vetmic.2009.11.027. [DOI] [PubMed] [Google Scholar]
- 2.Jeon B.-Y., Je S., Park J., et al. Variable number tandem repeat analysis of Mycobacterium bovis isolates from Gyeonggi-do, Korea. Journal of Veterinary Science. 2008;9(2):145–153. doi: 10.4142/jvs.2008.9.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parreiras P. M., Andrade G. I., do Nascimento T. D. F., et al. Spoligotyping and variable number tandem repeat analysis of Mycobacterium bovis isolates from cattle in Brazil. Memórias do Instituto Oswaldo Cruz. 2012;107(1):64–73. doi: 10.1590/s0074-02762012000100009. [DOI] [PubMed] [Google Scholar]
- 4.Lamine-Khemiri H., Martínez R., García-Jiménez W. L., et al. Genotypic characterization by spoligotyping and VNTR typing of Mycobacterium bovis and Mycobacterium caprae isolates from cattle of Tunisia. Tropical Animal Health and Production. 2014;46(2):305–311. doi: 10.1007/s11250-013-0488-y. [DOI] [PubMed] [Google Scholar]
- 5.Ramos D. F., Tavares L., Silva P. E. A., Dellagostin O. A. Molecular typing of Mycobacterium bovis isolates: a review. Brazilian Journal of Microbiology. 2014;45(2):365–372. doi: 10.1590/s1517-83822014005000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsao K., Robbe-Austerman S., Miller R. S., Portacci K., Grear D. A., Webb C. Sources of bovine tuberculosis in the United States. Infection, Genetics and Evolution. 2014;28:137–143. doi: 10.1016/j.meegid.2014.09.025. [DOI] [PubMed] [Google Scholar]
- 7.Sanou A., Tarnagda Z., Kanyala E., et al. Mycobacterium bovis in Burkina Faso: epidemiologic and genetic links between human and cattle isolates. PLoS Neglected Tropical Diseases. 2014;8(10) doi: 10.1371/journal.pntd.0003142.e3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mandal S., Bradshaw L., Anderson L. F., et al. Investigating transmission of Mycobacterium bovis in the United Kingdom in 2005 to 2008. Journal of Clinical Microbiology. 2011;49(5):1943–1950. doi: 10.1128/jcm.02299-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laniado-Laborín R., Muñiz-Salazar R., García-Ortiz R. A., Vargas-Ojeda A. C., Villa-Rosas C., Oceguera-Palao L. Molecular characterization of Mycobacterium bovis isolates from patients with tuberculosis in Baja California, Mexico. Infection, Genetics and Evolution. 2014;27:1–5. doi: 10.1016/j.meegid.2014.06.020. [DOI] [PubMed] [Google Scholar]
- 10.Lopez-Alvarez R., Badillo-Lopez C., Cerna-Cortes J. F., et al. First insights into the genetic diversity of Mycobacterium tuberculosis isolates from HIV-infected Mexican patients and mutations causing multidrug resistance. BMC Microbiology. 2010;10, article 82 doi: 10.1186/1471-2180-10-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gormley E., Corner L. A. L., Costello E., Rodriguez-Campos S. Bacteriological diagnosis and molecular strain typing of Mycobacterium bovis and Mycobacterium caprae . Research in Veterinary Science. 2014;97(supplement):S30–S43. doi: 10.1016/j.rvsc.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 12.Navarro Y., Herranz M., Romero B., et al. High-throughput multiplex MIRU-VNTR typing of Mycobacterium bovis . Research in Veterinary Science. 2014;96(3):422–425. doi: 10.1016/j.rvsc.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 13.McLernon J., Costello E., Flynn O., Madigan G., Ryan F. Evaluation of mycobacterial interspersed repetitive-unit-variable-number tandem-repeat analysis and spoligotyping for genotyping of Mycobacterium bovis isolates and a comparison with restriction fragment length polymorphism typing. Journal of Clinical Microbiology. 2010;48(12):4541–4545. doi: 10.1128/jcm.01175-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez-Campos S., Navarro Y., Romero B., et al. Splitting of a prevalent Mycobacterium bovis spoligotype by variable-number tandem-repeat typing reveals high heterogeneity in an evolving clonal group. Journal of Clinical Microbiology. 2013;51(11):3658–3665. doi: 10.1128/jcm.01271-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vestal A. L. Procedures of isolation and identification of mycobacteria. U.S. Department of Health, Education, and Welfare publication (CDC), 8230, 1977.
- 16.Niemann S., Harmsen D., Rusch-Gerdes S., Richter E. Differentiation of clinical Mycobacterium tuberculosis complex isolates by gyrB DNA sequence polymorphism analysis. Journal of Clinical Microbiology. 2000;38(9):3231–3234. doi: 10.1128/jcm.38.9.3231-3234.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huard R. C., Lazzarini L. C. O. O., Butler W. R., van Soolingen D., Ho J. L. PCR-based method to differentiate the subspecies of the Mycobacterium tuberculosis complex on the basis of genomic deletions. Journal of Clinical Microbiology. 2003;41(4):1637–1650. doi: 10.1128/jcm.41.4.1637-1650.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iralu J. V., Sritharan V. K., Pieciak W. S., Wirth D. F., Maguire J. H., Barker R. H., Jr. Diagnosis of Mycobacterium avium bacteremia by polymerase chain reaction. Journal of Clinical Microbiology. 1993;31(7):1811–1814. doi: 10.1128/jcm.31.7.1811-1814.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sampaio J. L. M., Chimara E., Ferrazoli L., et al. Application of four molecular typing methods for analysis of Mycobacterium fortuitum group strains causing post-mammaplasty infections. Clinical Microbiology and Infection. 2006;12(2):142–149. doi: 10.1111/j.1469-0691.2005.01312.x. [DOI] [PubMed] [Google Scholar]
- 20.Sun Z., Cao R., Tian M., et al. Evaluation of spoligotyping and MIRU-VNTR for Mycobacterium bovis in Xinjiang, China. Research in Veterinary Science. 2012;92(2):236–239. doi: 10.1016/j.rvsc.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 21.Kam K. M., Yip C. W., Tse L. W., et al. Optimization of variable number tandem repeat typing set for differentiating Mycobacterium tuberculosis strains in the Beijing family. FEMS Microbiology Letters. 2006;256(2):258–265. doi: 10.1111/j.1574-6968.2006.00126.x. [DOI] [PubMed] [Google Scholar]
- 22.Supply P., Lesjean S., Savine E., Kremer K., Van Soolingen D., Locht C. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. Journal of Clinical Microbiology. 2001;39(10):3563–3571. doi: 10.1128/jcm.39.10.3563-3571.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hunter P. R., Gaston M. A. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. Journal of Clinical Microbiology. 1988;26(11):2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boniotti M. B., Goria M., Loda D., et al. Molecular typing of Mycobacterium bovis strains isolated in Italy from 2000 to 2006 and evaluation of variable-number tandem repeats for geographically optimized genotyping. Journal of Clinical Microbiology. 2009;47(3):636–644. doi: 10.1128/jcm.01192-08. [DOI] [PMC free article] [PubMed] [Google Scholar]