Abstract
Genome-wide analyses have revolutionized our ability to study the transcriptional regulation of circadian rhythms. The advent of next-generation sequencing methods has facilitated the use of two such technologies, ChIP-seq and RNA-seq. In this chapter, we describe detailed methods and protocols for these two techniques, with emphasis on their usage in circadian rhythm experiments in the mouse liver, a major target organ of the circadian clock system. Critical factors for these methods are highlighted and issues arising with time series samples for ChIP-seq and RNA-seq are discussed. Finally detailed protocols for library preparation suitable for Illumina sequencing platforms are presented.
Eukaryotic genomes, the substrate of much of modern biology, are packaged into dynamically regulated units, chromatin, a macromolecular structure consisting of DNA, protein, and RNA. DNA is wrapped around a histone octamer forming a nucleosome, the basic unit of chromatin. Each nucleosome consists of two copies of core histones (H2A, H2B, H3, H4) that assemble when two dimers of H3/H4 form a tetramer and complex with two H2A/H2B dimers. 147 base pairs of DNA are wrapped around each histone octamer and constitute a nucleosome. Histone amino-end tails protrude from the nucleosome core and are extensively and dynamically modified. Arrays of nucleosomes referred to as “beads on a string” are further organized into a 30 nM fiber that are packaged in to higher order structures within the nucleus. Transcription, replication, repair and recombination of DNA have to occur in the context of chromatin. Chromatin is generally thought to act as a physical barrier that must be overcome in order to access DNA for basic function, and is known to be highly dynamic, with open and closed states. The constituency of chromatin and its interaction, DNA binding factors and cofactors are critical for transcriptional regulation and techniques to dissect these regulatory roles are important for understanding of any biological processes including circadian rhythms.
Mammalian circadian rhythms are regulated by a transcription-translation feedback loop in which the bHLH-PAS transcription factors, CLOCK (and its paralog NPAS2) and BMAL1 (ARNTL) dimerize and activate transcription of the Period (Per1, Per2) and Cryptochrome (Cry1, Cry2) genes (Bunger et al., 2000; Gekakis et al., 1998; King et al., 1997; Kume et al., 1999). As the PER proteins accumulate, they form complexes with the CRY proteins, translocate into the nucleus, and interact with the CLOCK/BMAL1 complex to inhibit their own transcription (Chen et al., 2009; Lee et al., 2001). As the inhibitory complex turns over and declines, the repression phase ends, and the cycle starts again with a new round of CLOCK/BMAL1-activated transcription.
In many ways the circadian system is ideally suited to study the many facets of transcription. There exists a well-defined cohort of central regulators with strong genetic and biochemical validity of the system (Lowrey and Takahashi, 2004, 2011). Many mutant alleles of the core components exist that can be exploited. The ~24 hour pace of the transcriptional oscillation allows for the study of a naturally occurring endogenous system that is conserved from behavior at the organismal level to the single cells. Large amounts of starting material can be obtained from homogeneous tissues such as liver, at specified times from entrained mice for complex biochemical analysis. At the same time, the circadian field can benefit from the application of modern molecular biology approaches developed by the transcription field. One such approach is Chip-seq which we detail here, as well as, RNA-seq, which we describe next. Such genome-wide analyses have recently been published by a number of labs (Hatanaka et al., 2010; Koike et al., 2012; Le Martelot et al., 2012; Menet et al., 2012; Rey et al., 2011; Vollmers et al., 2012).
Chromatin Immunoprecipitation (ChIP) is a powerful technique for detection of protein-DNA interactions and combined with modern next generation sequencing (NGS), Chip-Seq has revolutionized modern systems-level understanding of transcriptional landscape. First developed in the late 1970 and early 1980s, ChIP was used to understand the organization of nucleosomes on DNA in its native state (Jackson, 1978; Solomon and Varshavsky, 1985). A variety of reagents were used to crosslink DNA to proteins including formaldehyde, dimethyl sulfate and UV (Gilmour and Lis, 1984; Karpov et al., 1984; Welsh, 1984). The key insight from these pioneering studies was that in vivo crosslinking with formaldehyde preserves chromatin structure and the process of crosslinking does not radically alter DNA-histone interactions (Jackson and Chalkley, 1981). Thus these early biochemical and biophysical studies reliably established the technique of crosslinking critical for ChIP. The methods used currently for ChIP were developed in the mid 1990s when antibodies to individual histones and to various modified histones were first made available (Kuo and Allis, 1999). Genome wide analysis through ChIP was attempted using microarrays in (Chip-Chip) however this technique was not widely adapted due to several factors (Buck and Lieb, 2004; Ren et al., 2000).
The principles of ChIP are fairly straightforward but its successful execution relies on many critical factors that need optimization based on cell type and antibody used and antigen probed. We will attempt to outline the basic steps in ChIP followed by library construction for NGS that has worked well in our laboratory with the caveat that this protocol must be optimized for individual antibodies, factors and tissue used.
The basic steps in ChIP are outlined in (see accompanying Chapter by Zhou et al.). DNA is covalently bound to surrounding proteins, presumably in its native state by using the chemical crosslinker, formaldehyde. The cells are then lysed and the nucleoprotein complex is sheared using either sonication or nuclease, the target crosslinked protein is then immunoprecipitated, and after extensive washing to remove background contaminants, the crosslinks are reversed, the DNA isolated, libraries are made for next generation sequencing studies.
Critical factors
Antibody
The antibody is the most critical factor in ChIP experiments. An antibody that functions in western or immunohistochemistry may not always perform in ChIP. There is considerable batch to batch variability in polyclonal and monoclonal antibodies from commercial suppliers. We routinely purchase large batches of a particular “working” antibody from commercial vendors. When obtaining a new antibody it is critical to confirm its usefulness with a known positive and negative target. In this protocol, we detail the use of the following antibodies.
Antibodies against PER1, PER2, CLOCK, and BMAL1 were made as described previously (Lee et al., 2001). CRY1 antibody was made as described (Lee et al., 2004). CRY2 (epitope: residues 514–592) and p300 (epitope; residues 60–242 of human p300) antibodies were generated using guinea pigs (Cocalico Biological) and serum was affinity purified using the same protein used to raise antibody. NPAS2 antibody (Reick et al., 2001) was a kind gift from Dr. Steven McKnight (UT Southwestern Medical Center). RNAPII-8WG16 (MMS-126R) antibody (Jones et al., 2004) was purchased from Covance. RNAPII-Ser5P (clone 3E8, 04-1572) antibody (Chapman et al., 2007) was purchased from Millipore and RNAPII-Ser5P (ab5131) antibody (Rahl et al., 2010) was purchased from Abcam. H3K4me1 (ab8895), H3K4me3 (ab1012), H3K9ac (ab4441), H3K27ac (ac4729), H3K36me3 (ab9050) and H3K79me2 (ab3594) antibodies were purchased from Abcam. CBP antibody was monoclonal AC238 culture supernatant (Eckner et al., 1996).
Crosslinking/fixation
Formaldehyde covalently links peptide side-chain nitrogens of lysines, arginine, histidine as well as the α-amino groups of all amino acids to exocyclic amino groups and the endocyclic imino groups of DNA bases (Chaw et al., 1980; McGhee and von Hippel, 1975a, b). Because of its ease of use, fast-acting nature and crosslink reversibility it is the most commonly used crosslinker for ChIP. Formaldehyde, which crosslinks reactive groups within a 2Å distance, is best suited for studying direct protein-DNA interactions. Formaldehyde crosslinking can be optimized by varying the time of fixation, temperature, and concentration. Typically short times are required for immunoprecipitating with core histones and DNA binding factors, however extended times are required for cofactors that indirectly bind DNA. When fixation is too short, stable DNA-protein complexes that can be pulled down with the antibody will not form. When samples are over fixed, sonication, pulldown and de-crosslinking will be inefficient, leading to lowered yield. In order to study cofactors that bind several layers away in the sandwich, dual cross-linkers or increased length of crosslinking with formaldehyde can be used.
We have used two crosslinking methods, depending on antigen targeted. If the protein of interest is a DNA binding protein, 1% formaldehyde works well in most cases. However formaldehyde has a short cross-linking spacer arm and is not efficient to examine the proteins indirectly associated with DNA, such as PERs and CRYs. Dual crosslinking using a protein-protein crosslinker and formaldehyde works better in these cases (Koike et al., 2012; Nowak et al., 2005; Zeng et al., 2006).
Sonication
Too much sonication can disrupt the protein-DNA complex or cause damage to DNA and lead to low levels of immunoprecipitated DNA. Low levels of sonication will lead to large DNA fragment length and low resolution of the genomic region that is immunoprecipitated. Sonication is strongly affected by the type and concentration of detergent used and length of fixation.
Detergents
Detergents such as SDS or sarcosyl have multiple functions, they lyse crosslinked cells, expose and solubilize the antigenic complex, are important for proper sonication, and decrease background binding. But they can also denature the antigen and disrupt the antigen-antibody interaction surface of some antibodies, lowering yield. In many cases, gentler detergents such as Triton-X100 must be used, but this will lead to decreases in sonication efficiency. Thus it is important to characterize each antibody with a range of detergent concentrations and types.
Bioinformatics
Computational analysis of ChIP-seq data varies between labs and can be a source of irreproducibility. Even when the software that is used is consistent, parameters used should be properly documented. Circadian data is further complicated by the cyclical nature of interactions that we are interested in detecting. We use analyze cycling using three independent programs, COSOPT (Panda et al., 2002), JTK cycle (Hughes et al., 2010) and ARSER (Yang and Su, 2010). For example in one study (Koike et al., 2012), in order for a gene to be considered cycling, it was scored as cycling if 2 out of the 3 software programs detected it.
ChIP-seq Method for Mouse Liver
Tissue Sampling
Male C57BL/6J at 8–12 weeks of age are housed in light-tight boxes and entrained to LD 12:12 conditions for minimum of 7 days. Thirty-six hours after mice are transferred to constant darkness, liver samples are collected every 4 hours. We dissect the entire liver.
ChIP-seq
- 1% formaldehyde crosslinking
- Homogenize mouse livers immediately in 4 ml per liver of 1× PBS containing 1% formaldehyde.
- Wash liver with PBS by soaking
- Mince liver with a razor blade into small pieces.
- Add liver pieces to 4 ml (per liver) of PBS containing 1% formaldehyde.
- Homogenize with a Dounce homogenizer (7 strokes each with A(loose) and B(tight) pestle)
- Incubate for 8 min at room temperature.
- Add 250µl of 2.5 M glycine to stop the reaction on ice.
- Dual crosslinking
- Homogenize mouse livers immediately in 4 ml per liver of 1× PBS containing 2 mM EGS (Ethylene glycol bis[succinimidylsuccinate]).
- Incubate for 20 min at room temperature.
- Add formaldehyde to final concentration of 1%.
- Incubate for 8 min at room temperature.
- Add 250 µl of 2.5 M glycine to stop the reaction on ice.
- Nuclei isolation
- Add 10ml of ice-cold 2.3M sucrose containing 150mM glycine, 10mM HEPES pH 7.6, 15mM KCl, 2mM EDTA, 0.15mM spermine, 0.5mM spermidine, 0.5mM DTT and 0.5mM PMSF to the homogenate.
- Layer the homogenate on the top of a 3 ml cushion of 1.85M sucrose (containing the same ingredients and including 10% glycerol).
- Centrifuge for 1hr at 24,000 rpm at 4°C in a Beckman SW32.1 rotor.
- Wash the precipitated nuclei with 1 ml of 10 mM Tris pH 7.5, 150 mM NaCl, 2 mM EDTA and transfer to a 1.5 ml microfuge tube.
- Centrifuge for 3 min at 3000 rpm at 4°C and washed again.
- Stored at −80°C until use.
-
Chromatin sonication
We used two different sonicators (Covaris S2 and Misonix S-4000) for chromatin shearing and four different buffers (1% SDS, 0.5% Sarkosyl, 1% Triton-X100, or MNase digestion buffers) depending on the antibody. As previously stated this should be optimized- BMAL1 and RNAPII-8WG16 antibodies
- Resuspend the formaldehyde-crosslinked nuclei in 0.8 ml per liver of lysis buffer (50 mM Tris pH 7.5, 10 mM EDTA, 1% SDS, 1 mM PMSF and Roche complete EDTA free protease inhibitor cocktail).
- Sonicate 10 times for 30 sec at 4°C using a Covaris S2 ultrasonicator.
- Dilute tenfold with IP buffer (10 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 1 mM PMSF and protease inhibitor cocktail).
- CLOCK and NPAS2 antibodies
- Resuspend the dual crosslinked nuclei in 0.8 ml per liver of Sarkosyl lysis buffer (50 mM Tris pH 7.5, 10 mM EDTA, 0.5% N-lauroylsarcosine, 1 mM PMSF and Roche complete EDTA free protease inhibitor cocktail).
- Sonicate 6 times for 30 sec at 4°C using a Covaris S2 ultrasonicator.
- Dilute tenfold with IP buffer.
- PER1, PER2, CRY1, CRY2, CBP and p300 antibodies
- Resuspend the dual-crosslinked nuclei in 3 ml per liver of IP buffer.
- Sonicate 48 times for 5 sec on ice using a Misonix S-4000 sonicator.
- RNAPII-Ser5P antibody
- Resuspend formaldehyde-crosslinked nuclei in 0.8 ml per liver of Sarkosyl lysis buffer.
- Sonicate 6 times for 30 sec at 4°C using a Covaris S2 ultrasonicator
- Dilute 10-fold with IP buffer.
- H3K4me1, H3K4me3, H3K9ac, H3K27ac, H3K36me3 and H3K79me2 antibodies
- Resuspend the formaldehyde-crosslinked nuclei in 0.7 ml per liver of MNase Buffer (10mM Tris pH 7.5 and 100 mM NaCl).
- Sonicate 5 times for 30 sec at 4°C using a Covaris S2 ultrasonicator.
- Incubate for 40 min at 37°C with 200 kunitz units of Micrococcal Nuclease and 2 mM CaCl2.
- Stop the reaction with 10 mM EGTA and 1% SDS.
- Dilute 10-fold with IP buffer.
-
ChIP
We used approximately 120 µg (for transcription factors) or 80 µg (for histones) of fragmented chromatin for ChIP-seq.- Pre-clear sonicated nuclear lysates. Add 40 µl (final bed volume) of protein A beads (pre-blocked with PBS containing 5mg/ml BSA) to each lysate and incubate for 2 hrs in rotation at 4 °C.
- Centrifuge the beads at 14000 rpm for 10 min at 4 °C.
- Carefully take out supernatant. Remove 1/10 to 1/20 of the lysate and save as INPUT (go to Step F for reverse crosslinking).
- The amount of antibody per IP should be determined by careful titration.
- Add antibody to the pre-cleared chromatin and incubate overnight at 4°C on a rotating wheel.
- Add 10 µl (final bed volume) of Protein A/G Plus-agarose (Santa Cruz, sc-2003) and incubated for 1.5 hr at 4°C.
- Centrifuge the beads at 5000 rpm for 1 min at 4°C.
- Remove the supernatant.
- Wash twice with IP buffer
- Wash twice with high salt wash buffer (20 mM Tris pH 7.5, 500 mM NaCl, 2 mM EDTA, 1% Triton X-100, 1 mM PMSF).
- Wash twice with LiCl wash buffer (20 mM Tris pH 7.5, 250 mM LiCl, 2 mM EDTA, 0.5% Igepal CA-630, 1% sodium deoxycholate, 1 mM PMSF).
- Wash once with TE.
- Carefully remove residual TE.
- Add 50µl of Elution Buffer (20 mM Tris pH 7.5, 5 mM EDTA, 0.5% SDS).
- Place tubes in 65°C heat block for 10min. Gently vortex.
- Centrifuge the tube at 5000 rpm for 1min.
- Transfer the supernatant to new tube.
- Repeat the elution one more time. Final elution volume is 100µl.
- Reverse crosslinking
- Incubate the eluted chromatin at 65°C for 5–8 hrs or up to 12 hrs to reverse the crosslinking.
- Add 10 µg of RNaseA and incubate for 30 min at 37°C.
- Add 160µg of proteinase K and incubate for 30 min at 55°C.
- Purify DNA using a Qiaquick PCR purification Kit (Qiagen).
Library preparation for ChIP-seq
This protocol uses previously isolated ChIP DNA and converts it into DNA libraries suitable for subsequent cluster generation and sequencing. The protocol is based on the Illumina workflow and is comparable to the Illumina® TruSeq® ChIP Sample Preparation Kit which has been used as a reference.
Equipment and reagents needed
1.5 ml nuclease free tubes
96 well PCR plate, non-skirted
Adhesive PCR plate seal
2, 10, 20, 200 and 1000 µl pipettes and 200 µl multichannel pipette
PCR machine
Magnetic stand-96
0.2 ml and 1.5 ml nuclease free tubes
Agencourt Ampure XP beads (Beckman Coulter)
Bioanalyzer
Kapa Library Quantification kit (Kapa Biosystems)
qPCR machine
Buffers and enzyme mixes recipes
| Ligase storage buffer | |||
|---|---|---|---|
| Component | Final concentration | Stock Solution | 10 ml |
| Water | 4.39 ml | ||
| Tris-HCl, pH 7.4 | 10 mM | 1 M pH 7.4 at RT | 100 µl |
| EDTA, pH 8.0 | 0.1 mM | 500 mM pH 8.0 at RT | 2 µl |
| DTT | 1mM | 1M | 10 µl |
| KCl | 50 mM | 1 M | 500 µl |
| Glycerol | 50% | 100% | 5 ml |
Store at −20°C
| End repair buffer | |||
|---|---|---|---|
| Component | Volume/reaction (µl) | Vendor | Catalog number |
| 10 mM dNTPs | 2.5 | Enzymatics | N2050-10-L |
| 10× End repair buffer | 4.5 | Enzymatics | B9140 |
Store at −20°C
| End repair enzyme | |||
|---|---|---|---|
| Component | Volume/reaction (µl) | Vendor | Catalog number |
| End-repair mix (Low concentration) | 3 | Enzymatics | Y9140-LC-L |
Store at −20°C
| A-tailing mix | |||
|---|---|---|---|
| Component | Volume/reaction (µl) | Vendor | Catalog number |
| 10 mM dATP | 1 | Enzymatics | N2010-A-L |
| 10× Blue buffer | 2 | Enzymatics | B0110 |
| Klenow (3'–5' exo-) (Low concentration) | 0.5 | Enzymatics | P7010-LC-L |
Store at −20°C
| Ligation mix | |||
|---|---|---|---|
| Component | Volume/reaction (µl) | Vendor | Catalog number |
| 2× Ligase buffer | 25 | Enzymatics | B1010L |
| Ligase storage buffer | 2 | - | - |
| T4 DNA ligase (Rapid) | 1 | Enzymatics | L6030-HC-L |
Store at −20°C
| PCR amplification mix | |||
|---|---|---|---|
| Component | Volume/reaction (µl) | Vendor | Catalog number |
| Kapa dNTP Mix | 1 | Kapa Biosystems | KK2101 |
| KAPA HiFi Fidelity Buffer (5×) | 10 | Kapa Biosystems | KK2101 |
| KAPA HiFi DNA Polymerase (1 U/µL) | 1 | Kapa Biosystems | KK2101 |
Store at −20°C
| Library dilution buffer | |||
|---|---|---|---|
| Component | Final concentration | Stock Solution | 100 ml |
| Tris-HCl, pH 8.0 | 10 mM | 1M | 1 ml |
| Tween-20 | 0.05% | 100% | 50 µL |
Store at room temperature
| Library normalization buffer | |||
|---|---|---|---|
| Component | Final concentration | Stock Solution | 100 ml |
| Tris-HCl, pH 8.5 | 10 mM | 1M | 1 ml |
| Tween-20 | 0.1% | 100% | 100 µL |
Adapters and Primers
The barcoded Y-shaped adapters are ordered from Bioo Scientific (Catalog # 514123). They are stored at −20°C.
- The PCR primers are ordered from Integrated DNA Systems and subsequently reconstituted at 100 µM and then diluted to 25 µM each and mixed in equal volume to make a 12.5µM PCR primer mix. Store at −20°C.
- PCR primer 1: 5'AATGATACGGCGACCACCGAGATCTACAC
- PCR primer 2: 5'CAAGCAGAAGACGGCATACGAGAT
Detailed Protocol
Step 1: End-repair
The end repair step converts DNA with overhangs to 5’ phosphorylated, blunt-ended DNA that can be subsequently used for adapter ligation. The conversion of fragmented DNA to blunt-ended is carried out by the 3’ to 5’ and 5’ to 3’ exonuclease activities of T4 DNA polymerase and the 5’ phosphorylation is carried out by the T4 Polynucleotide Kinase in the enzyme mix.
- Perform the following reaction in a 96 well plate. Mix,
40 µl ChIP DNA 7 µl End repair buffer 3 µl End repair enzyme Incubate at 25°C for 30 minutes.
Step 2: Bead based size selection
Bead based size selection is based on removing large DNA fragments first by binding them on the beads and doing a supernatant transfer and then subsequently binding all the DNA on the beads except small fragments (less than 100 bp) and then eluting the DNA of interest from the beads.
For performing a 150 bp size selection, add 60 µL of well-mixed AMPure XP Beads to each sample and mix thoroughly by pipetting.
Incubate the plate for 5 minutes at room temperature.
Place the plate on the magnetic stand for 5 minutes at room temperature or until the liquid appears completely clear.
DO NOT discard the supernatant. Gently transfer 108 µL of the supernatant to a fresh well from the plate without disturbing the beads.
Add 40 µL of well-mixed AMPure XP Beads to each sample and mix thoroughly by pipetting.
Incubate the plate for 5 minutes at room temperature.
Place the plate on the magnetic stand for 5 minutes at room temperature or until the liquid appears completely clear.
Remove and discard all of the supernatant from the plate taking care not to disturb the beads.
With plate on stand, add 200 µL of freshly prepared 80% ethanol to each well without disturbing the beads and incubate the plate for at least 30 seconds at room temperature. Carefully remove and discard all the supernatant.
Repeat step 9, for a total of two ethanol washes. Ensure the ethanol has been removed.
Remove the plate from the magnetic stand and let it dry at room temperature for 2 minutes.
Resuspend dried beads in 18 µL Resuspension Buffer. Gently, pipette the entire volume up and down to mix thoroughly. Ensure that the beads are completely rehydrated and re-suspended.
Incubate resuspended beads at room temperature for 2 minutes.
Place the plate on the magnetic stand for 5 minutes at room temperature or until the supernatant appears completely clear.
Gently transfer 17 µL of the clear supernatant to a fresh well.
The procedure may be stopped at this point and the reactions stored at −20°C.
Tip
Use multichannel pipette for performing bead cleanups to ensure consistency in processing across samples. Ensure beads don’t crack when drying. Complete resuspension of beads will maximize recovery.
Step 3: A-tailing
A-tailing is performed by utilizing the polyermase activity of Klenow (3'–5' exo-) in presence of dATP to add a single ‘A’ to the 3' end of a blunt, double-stranded DNA. A-tailing prevents the blunt fragments from self ligating during the adapter ligation step.
- For each reaction, mix:
17 µl end repaired DNA 3.5 µl A-tailing mix Mix well by pipetting and then incubate at 37°C for 30 minutes followed by 70° for 5 minutes. Immediately proceed to adapter ligation
Step 4: Y-shaped adapter ligation
The ligation step ligates barcoded Y-shaped adapters to the ends of A-tailed DNA fragments. The adapters have a ‘T’ overhang, which is complementary to the adenylated DNA. The ligation step prepares the DNA fragments for subsequent hybridization onto the flow cells.
- For each reaction, mix:
20.5 µl Adenylated DNA 2 µl NEXTflex™ Barcoded Adapter (0.6 µM) 28 µl Ligation mix Mix well by pipetting and then incubate at 22°C for 15 minutes.
STEP 5: Double bead cleanup
Double-bead cleanup is performed at the end of ligation to remove any excess adapters that might have been self-ligated or be free floating and prevent them from getting amplified during PCR.
Add 50.5 µL of well-mixed AMPure XP Beads to each sample and mix thoroughly by pipetting.
Incubate the plate for 5 minutes at room temperature.
Place the plate on the magnetic stand for 5 minutes at room temperature or until the liquid appears completely clear.
Remove and discard all of the supernatant from the plate taking care not to disturb the beads.
With plate on stand, add 200 µL of freshly prepared 80% ethanol to each well without disturbing the beads and incubate the plate for at least 30 seconds at room temperature. Carefully, remove and discard the supernatant.
Repeat step 5, for a total of two ethanol washes. Ensure the ethanol has been removed.
Remove the plate from the magnetic stand and let dry at room temperature for 2 minutes.
Resuspend dried beads in 51 µL Resuspension Buffer. Gently, pipette the entire volume up and down to mix thoroughly. Ensure that the beads are completely rehydrated and re-suspended.
Incubate resuspended beads at room temperature for 2 minutes.
Place the plate on the magnetic stand for 5 minutes at room temperature or until the supernatant appears completely clear.
Gently transfer 50 µL of the clear supernatant to a fresh well.
Add 50 µL of well-mixed AMPure XP Beads to each well containing sample and mix thoroughly by pipetting.
Incubate the plate for 5 minutes at room temperature.
Place the plate on the magnetic stand for 5 minutes at room temperature or until the liquid appears completely clear.
Remove and discard all of the supernatant from the plate taking care not to disturb the beads.
With plate on stand, add 200 µL of freshly prepared 80% ethanol to each well without disturbing the beads and incubate the plate for at least 30 seconds at room temperature. Carefully, remove and discard the supernatant.
Repeat step 16, for a total of two ethanol washes. Ensure the ethanol has been removed.
Remove the plate from the magnetic stand and let dry at room temperature for 2 minutes.
Resuspend dried beads in 36 µL Resuspension Buffer. Gently, pipette the entire volume up and down to mix thoroughly. Ensure that the beads are completely rehydrated and re-suspended.
Incubate resuspended beads at room temperature for 2 minutes.
Place the plate on the magnetic stand for 5 minutes at room temperature or until the supernatant appears completely clear.
Gently transfer 35 µL of the clear supernatant to a fresh well.
The procedure may be stopped at this point and the reactions stored at −20°C.
STEP 6: PCR amplification
PCR amplification is performed to selectively amplify the DNA fragments that have adapters bound to them. The PCR primers anneal in part to the adapter sequences.
- For each reaction, mix:
35 µl Adapter ligated DNA 12 µl PCR amplification mix 2 µl PCR primer mix (12.5 µM) Mix well by pipetting.
- PCR cycling:
98°C 2 minutes 98°C 30 seconds 65°C 30 seconds repeat for 12–20 cycles 72°C 60 seconds 72°C 4 minutes
Tip
Always do the minimum number of PCR cycles possible.
STEP 7: Double bead cleanup
Post PCR amplification a double bead cleanup is performed to get rid of excess primer and primer dimers.
Add 50 µL of well-mixed AMPure XP Beads to each sample and mix thoroughly by pipetting.
Incubate the plate for 5 minutes at room temperature.
Place the plate on the magnetic stand for 5 minutes at room temperature or until the liquid appears completely clear.
Remove and discard all of the supernatant from the plate taking care not to disturb the beads.
With plate on stand, add 200 µL of freshly prepared 80% ethanol to each well without disturbing the beads and incubate the plate for at least 30 seconds at room temperature. Carefully, remove and discard the supernatant.
Repeat step 5, for a total of two ethanol washes. Ensure the ethanol has been removed.
Remove the plate from the magnetic stand and let dry at room temperature for 2 minutes.
Resuspend dried beads in 51 µL Resuspension Buffer. Gently, pipette the entire volume up and down to mix thoroughly. Ensure that the beads are completely rehydrated and re-suspended.
Incubate resuspended beads at room temperature for 2 minutes.
Place the plate on the magnetic stand for 5 minutes at room temperature or until the supernatant appears completely clear.
Gently transfer 50 µL of the clear supernatant to a fresh well.
Add 50 µL of well-mixed AMPure XP Beads to each well containing sample and mix thoroughly by pipetting.
Incubate the plate for 5 minutes at room temperature.
Place the plate on the magnetic stand for 5 minutes at room temperature or until the liquid appears completely clear.
Remove and discard all of the supernatant from the plate taking care not to disturb the beads.
With plate on stand, add 200 µL of freshly prepared 80% ethanol to each well without disturbing the beads and incubate the plate for at least 30 seconds at room temperature. Carefully, remove and discard the supernatant.
Repeat step 16, for a total of two ethanol washes. Ensure the ethanol has been removed.
Remove the plate from the magnetic stand and let dry at room temperature for 2 minutes.
Resuspend dried beads in 32 µL Resuspension Buffer. Gently, pipette the entire volume up and down to mix thoroughly. Ensure that the beads are completely rehydrated and re-suspended.
Incubate resuspended beads at room temperature for 2 minutes.
Place the plate on the magnetic stand for 5 minutes at room temperature or until the supernatant appears completely clear.
Gently transfer 30 µL of the clear supernatant to a fresh well.
The procedure may be stopped at this point and the libraries stored at −20°C until they are validated for quality and quantified for sequencing.
Quality control
Check the size and quality of the library by running it on a Bioanalyzer using the High Sensitivity DNA assay. If on the Bioanalyzer trace there are two bands, one of expected size and one of higher molecular weight, it’s indicative of a bubble product. This double product will not affect the outcome to the sequencing run as double stranded product is denatured prior to sequencing. As an extra verification step, a portion of this product (1–2 µL) can be denatured manually by heating the sample to 95°C for 5 minutes and then placing it on ice and subsequently be run on a Bioanalyzer RNA Pico 6000 Chip Kit. The denatured product should appear as a single band on a Pico 6000 chip.
Quantification of libraries
In order to get consistent number of reads across different samples it is important to accurately quantify the DNA library templates and then normalize all the samples before sequencing. To get the best sequencing results, its important to get optimum cluster densities across every lane on every flow cell and this also makes quantification an important step. qPCR-based quantification is the most accurate method for quantifying library templates. Use the right Kapa library quantification kit by Kapa Biosystems for your qPCR instrument.
Setting up qPCR
Make a 1:1000 dilution of each of your library template for quantification in library dilution buffer.
Subsequently, use the 1:1000 dilution to make a 1:10,000 dilution.
Set up 10 µl qPCR reactions as described in the Technical Data Sheet for the Kapa kit
Analyze the qPCR data as described in the data sheet and then calculate the concentration of each of the library template.
Normalizing and pooling libraries for sequencing
If you have barcoded libraries, follow Bioo Scientific’s guidelines in the barcode manual for pooling normalized samples for sequencing.
Normalize the concentration of each library to 20 nM using Library normalization buffer and then pool samples samples at equimolar concentration.
Based on the coverage you want you can determine how many samples to pool per lane as 50 bp single end sequencing.
Data Analysis for ChIP-seq
Data Analysis
Reads were trimmed using fastq-mcf (https://code.google.com/p/ea-utils/wiki/FastqMcf, reads less than 35bp after trimming were discarded.
Bowtie2 was used with the program default values for these two relevant parameters –end-to-end & –sensitive (Langmead and Salzberg, 2012). After mapping, we removed reads of quality less than Q10 using SAMtools (Li et al., 2009).
Remove duplicates using Picard MarkDuplicates (http://picard.sourceforge.net).
Perform random sampling. In order to normalized for differences in sequencing depth among timed ChIP-seq samples, the sequence reads are “down sampled” to the lowest number of the uniquely mapped reads with duplicates among the time points for each ChIP factor.
The peaks are identified from uniquely mapped reads without duplicates using MACS with following parameters: genomic size = mm (1.87 Gb), shift = 60 and input chromatin samples as control data (Zhang et al., 2008). We use a p-value threshold of 10−5 (default) and a ratio between the ChIP-seq tag count and λlocal of 10 (fold_enrichment threshold). The false peaks called by MACS that repeatedly emerged from low complexity sequence are removed from further analysis.
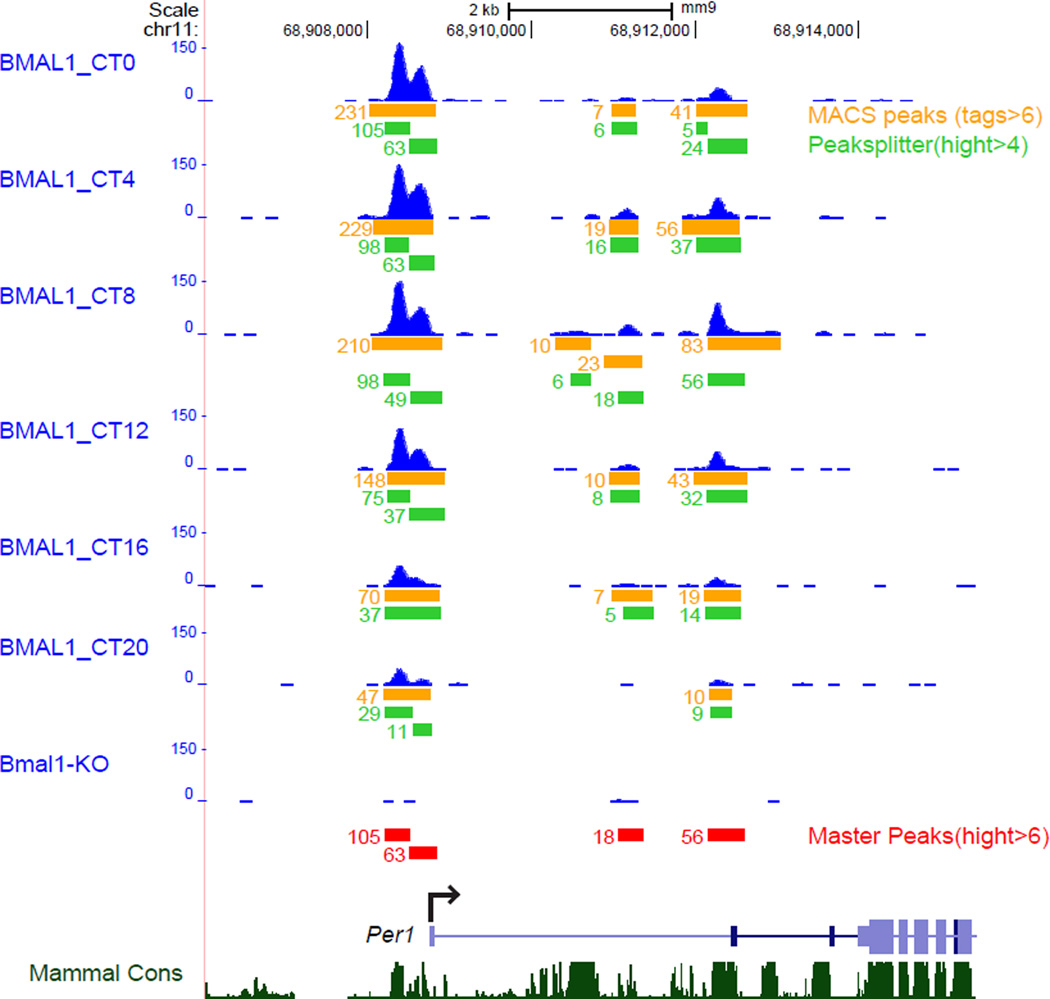
The peaks are then subdivided by PeakSplitter (Salmon-Divon et al., 2010) with options of –valley 0.7 and –cutoff 7. To construct a master peak list from the six time points, the peaks with summit height more than 6 obtained after PeakSplitter are merged, compare for overlaps and the peak with the highest summit value is chosen if the summit coordinates were within 120 bp. Figure 1 illustrates the master peak process in which MACS peaks are called, then subdivided with PeakSplitter and then compared for overlap and summit height (Koike et al., 2012).The ChIP-seq peak overlaps (peak summit +/− 120bp) from the master peak lists were determined using HOMER (Heinz et al., 2010).
Results were analyzed using HOMER (Heinz et al., 2010). A tool, “makeTagDirectory”, creates Tag Directories for each samples. The numbers of mapped reads in each peak can be quantified by HOMER using a Perl script, “annotatePeaks.pl”. HOMER also provides a tool, “makeUCSCfile”, to create UCSC visualization file, which can be uploaded as a custom track to UCSC genome browser. We normalized genome browser views to display uniquely mapped reads per 10 million uniquely mapped reads with duplicates.
Figure 1.
UCSC genome browser view of MACS peak calls for six timed BMAL1 ChIP-seq occupancy at the Per1 gene. Orange bars indicate the MACS peak calls and green bars indicate the peak after using Peaksplitter. The numbers to the left of each bar refer to the peak height. Red bars at the bottom show the final consolidated peaks used to construct the master peak list. The Peaksplitter peak with the largest peak height in the region of overlap of peaks is chosen to represent this peak in the master peak list. Data adapted from Koike et al. 2012.
RNA-Seq Method for Mouse Liver
Overview of RNA-seq strategy
Isolate total RNAs from mouse livers using Trizol reagent (Life Technologies).
Determine the quality of isolated total RNAs by Agilent 2011 Bioanalyzer. We usually use total RNA with RIN values of more than 8.
For whole transcriptome (WT) RNA-seq, deplete ribosomal RNAs in 10 µg of total RNA pooled from three mice using Ribo-Zero Gold kit for Human/Mouse/Rat (Illumina) using the manufacturer’s instructions. For mRNA-seq, follow the detailed protocol below.
Construct strand specific RNA-seq libraries using the detailed protocol below to make sequencing libraries for the Illumina HiSeq 2500 platform.
For Illumina platforms, we use 50 bp single end reads or 100bp × 100 bp paired-end reads for WT RNA-seq. The samples can be multiplexed using barcode primers (below). The ability to detect low copy number transcripts depends on the depth of sequencing. A minimum depth of 100 million reads is currently recommended for a typical mammalian tissue, according to Standards, Guidelines and Best Practices for RNA-Seq from the ENCODE consortium (https://genome.ucsc.edu/ENCODE/protocols/dataStandards/RNA_standards_v1_2011_May.pdf). We recommend at least 100 million reads for WT RNA-seq and at least 30 million reads for mRNA-seq.
Library Preparation for RNA-Seq
This protocol uses total RNA and provides instructions on enriching mRNA that can be subsequently converted into DNA libraries retaining strand origin information. This library can then be used for cluster generation and DNA sequencing. The protocol is based on the Illumina workflow and is comparable to the Illumina® TruSeq® Stranded mRNA Sample Preparation Kit which has been used as a reference.
Equipment and reagents needed
NEXTflex™ Poly(A) Beads
1.5 ml nuclease free tubes
96 well PCR plate, non-skirted
Adhesive PCR plate seal
2, 10, 20, 200 and 1000 µl pipettes and 200 µl multichannel pipette
PCR machine
Magnetic stand-96
0.2 ml and 1.5 ml nuclease free tubes
Agencourt Ampure XP beads (Beckman Coulter)
Bioanalyzer
Kapa Library Quantification kit (Kapa Biosystems)
qPCR machine
Buffers and enzyme mixes recipes
| Actinomycin D (1 mg/ml) | |||
|---|---|---|---|
| Component | Vendor | Catalog number | 5 ml |
| Actinomycin D | Sigma Aldrich | A1410 | 5 mg |
| 100% ethanol | 5 ml | ||
Actinomycin D stock is stored at −80°C as aliquots in lightsafe microcentrifuge tubes
| 120 ng/µl Actinomycin D | |||
|---|---|---|---|
| Component | Final concentration | Stock Solution | 1 ml |
| Actinomycin D | 120 ng/µl | 1 mg/ml | 120 µl |
| 100% ethanol | 880 µl | ||
Actinomycin D stock is stored at −80°C as aliquots in lightsafe microcentrifuge tubes
Store at −20°C
| Ligase storage buffer | |||
|---|---|---|---|
| Component | Final concentration | Stock Solution | 10 ml |
| Water | 4.39 ml | ||
| Tris-HCl, pH 7.4 | 10 mM | 1 M pH 7.4 at RT | 100 µl |
| EDTA, pH 8.0 | 0.1 mM | 500 mM pH 8.0 at RT | 2 µl |
| DTT | 1mM | 1M | 10 µl |
| KCl | 50 mMa | 1 M | 500 µl |
| Glycerol | 50% | 100% | 5 ml |
Store at −20°C
| RNA fragmentation buffer | |||
|---|---|---|---|
| Component | Final concentration | Stock Solution | 10 ml |
| Tris-HCl, pH 8.3 | 250 mM | 1 M | 2.5 ml |
| KCl | 375 mM | 1 M | 3.75 ml |
| MgCl2 | 10 mM | 1 M | 100 µl |
Store at −20°C
| First strand synthesis buffer (stranded) | |||
|---|---|---|---|
| Component | Volume/reaction (µl) | Vendor | Catalog number |
| 100 mM DTT | 2 | Enzymatics | Supplied with EnzScript™ |
| 10 mM dNTPs | 1 | Enzymatics | N2050-10-L |
| 120 ng/µl Antinomycin D | 0.5 | - | - |
| Rnase Inhibitor | 0.5 | Enzymatics | Y9240L |
Store at −20°C
| EnzScript™ | |||
|---|---|---|---|
| Component | Volume/reaction (µl) | Vendor | Catalog number |
| EnzScript™ (M-MLV Reverse Transcriptase RNase H minus) | 0.5 | Enzymatics | P7600L |
Store at −20°C
| Second strand synthesis mix (stranded) | |||
|---|---|---|---|
| Component | Volume/reaction (µl) | Vendor | Catalog number |
| 10× Blue buffer | 3 | Enzymatics | Supplied with DNA Polymerase I |
| dNTP/dUTP mix(1:1:1:2) | 1 | Enzymatics | - |
| Rnase H | 0.5 | Enzymatics | Y9220L |
| DNA Polymerase I | 1 | Enzymatics | P7050L |
Store at −20°C
| A-tailing mix | |||
|---|---|---|---|
| Component | Volume/reaction (µl) | Vendor | Catalog number |
| 10 mM dATP | 1 | Enzymatics | N2010-A-L |
| 10× Blue buffer | 2 | Enzymatics | B0110 |
| Klenow (3'–5' exo-) (Low concentration) | 0.5 | Enzymatics | P7010-LC-L |
Store at −20°C
| Ligation mix | |||
|---|---|---|---|
| Component | Volume/reaction (µl) | Vendor | Catalog number |
| 2× Ligase buffer | 25 | Enzymatics | B1010L |
| Ligase storage buffer | 2 | - | - |
| T4 DNA ligase (Rapid) | 1 | Enzymatics | L6030-HC-L |
Store at −20°C
| PCR amplification mix | |||
|---|---|---|---|
| Component | Volume/reaction (µl) | Vendor | Catalog number |
| Kapa dNTP Mix | 1 | Kapa Biosystems | KK2101 |
| KAPA HiFi Fidelity Buffer (5×) | 10 | Kapa Biosystems | KK2101 |
| KAPA HiFi DNA Polymerase (1 U/µL) | 1 | Kapa Biosystems | KK2101 |
Store at −20°C
| Uracil DNA Glycosylase | |||
|---|---|---|---|
| Component | Volume/reaction (µl) | Vendor | Catalog number |
| Uracil DNA Glycosylase | 1 | Enzymatics | G0505L |
Store at −20°C
| Library dilution buffer | |||
|---|---|---|---|
| Component | Final concentration | Stock Solution | 100 ml |
| Tris-HCl, pH 8.0 | 10 mM | 1M | 1 ml |
| Tween-20 | 0.05% | 100% | 50 µL |
Store at room temperature
| Library normalization buffer | |||
|---|---|---|---|
| Component | Final concentration | Stock Solution | 100 ml |
| Tris-HCl, pH 8.5 | 10 mM | 1M | 1 ml |
| Tween-20 | 0.1% | 100% | 100 µL |
Store at room temperature
Adapters and Primers
-
3
Random hexamer (NNNNNN) was ordered as a ReadyMade™Primer from Integrated DNA Systems and reconstituted to 100 µM and then diluted to 50 µM to use for first strand synthesis.
-
4
The barcoded Y-shaped adapters are ordered from Bioo Scientific (Catalog # 512914). They are stored at −20°C.
-
5The PCR primers are ordered from Integrated DNA Systems and subsequently reconstituted at 100 µM and then diluted to 25 µM each and mixed in equal volume to make a 12.5µM PCR primer mix. Store at −20°C.
- PCR primer 1: 5'AATGATACGGCGACCACCGAGATCTACAC
- PCR primer 2: 5'CAAGCAGAAGACGGCATACGAGAT
Detailed Protocol
Step 1: mRNA isolation from total RNA
This step is performed to pull down the mRNA from the total RNA samples using magnetic beads that have oligo(dT) to select for poly(A) mRNA.
Bead washing
This procedure takes approximately 10 minutes and should be carried out before starting mRNA purification to remove sodium azide in which the beads are stored.
Resuspend the magnetic beads thoroughly in the vial to obtain a uniform suspension.
Transfer 20 µl of NEXTflex™ Poly(A) Beads to a fresh tube.
Place the tube on a DynaMag™-2 Magnet (Life Technologies Cat # 123-21D) / or / similar for 2 minutes.
Remove and discard the supernatant while the tube remains on the magnet.
Remove the tube from the magnet and add 100 µl of NEXTflex™ Poly(A) Binding Buffer to the tube, resuspending the beads thoroughly.
Place the tube on the magnet for 2 minutes.
Remove and discard the supernatant while the tube remains on the magnet.
Resuspend the beads in 100 µl of NEXTflex™ Poly(A) Binding Buffer.
mRNA pulldown
-
If your total RNA sample (1–10 µg) is below 100 µl in volume, adjust its volume to 98 µl using nuclease-free water. Add recommended dilution of ERCC Spike-in mix 1 and then add 100 µl of NEXTflex™ Poly(A) Binding Buffer.
E.g.: If using 5 µg of RNA, adjust its volume to 99 µl using nuclease-free water. Add 1 µl of 1:10 dilution of ERCC Spike-in mix 1 and then add 100 µl of NEXTflex™ Poly(A) Binding Buffer. For samples greater than 100 µl in starting volume, add an equal volume of NEXTflex™ Poly(A) Binding Buffer.
Heat the total RNA sample to 65°C for 2 minutes to disrupt secondary RNA structures. Immediately place on ice.
Add your total RNA to the 100 µl of washed beads (as previously described).
Mix thoroughly by rotating continuously on a Tube Rotator-unit for 5 minutes at room temperature.
Place the tube on the magnet for 2 minutes then carefully remove and discard the clear supernatant.
Separately aliquot 100 µl of NEXTflex™ Poly(A) Binding Buffer to a fresh 1.5 ml tube.
Remove the tube from the magnet, add 200 µl NEXTflex™ Poly(A) Washing Buffer and mix by pipetting. Place the tube on the magnet. Once the beads have pelleted, remove and discard the clear supernatant.
Repeat Step 7 for a total of two bead washes.
Resuspend the bead pellet with 50 µl of NEXTflex™ Poly(A) Elution Buffer.
Heat at 80°C for 2 min and place the tube immediately on the magnet. After the bead pellet forms, transfer the clear supernatant to the tube prepared in Step 6. Do not discard the used bead pellet.
Heat the supernatant sample to 65°C for 2 minutes to disrupt secondary structures. Immediately place on ice.
Add 200 µl of NEXTflex™ Poly(A) Washing Buffer to the bead pellet from Step 10. Mix by pipetting. Place the tube on the magnet. Once the beads have pelleted, remove and discard the clear supernatant.
Repeat Step 12 for a total of two bead washes.
Add the RNA sample from Step 11 to the washed beads from Step 13.
Mix thoroughly by rotating continuously on a tube rotator for 5 minutes at room temperature.
Place the tube on the magnet for 1–2 minutes then carefully remove and discard the clear supernatant.
Remove the tube from the magnet, add 200 µl NEXTflex™ Poly(A) Washing Buffer and mix by pipetting. Place the tube on the magnet. Once the beads have pelleted, remove and discard the clear supernatant.
Repeat Step 17 for a total of two bead washes.
Resuspend the bead pellet with 20 µl of NEXTflex™ Poly(A) Elution Buffer.
Heat the resuspended pellet to 80°C for 2 minutes, then place the tube immediately on the magnet. Transfer the mRNA to a fresh tube or plate. If needed, use 1 µl of eluted mRNA for quantification using a nanodrop or Qubit.
Step 2: RNA Fragmentation
This step is performed to fragment the mRNA to smaller fragments for cDNA synthesis and subsequent library preparation steps.
- For each reaction, mix in a PCR plate:
14 µl mRNA (10–100 µg) 5 µl RNA fragmentation buffer Mix well by pipetting and then incubate at 95°C for 10 minutes and then immediately place on ice.
STEP 3: Directional first strand synthesis
This step is performed to synthesize cDNA from the mRNA using random hexamer primers and reverse transcriptase enzyme.
For each reaction, add 1 µl random hexamer primer to the fragmented RNA (from Step 2)
Incubate at 65°C for 5 minutes, and then immediately place on ice.
Add 0.5 µl of EnzScript™ per reaction to 4 µl of First strand synthesis buffer (stranded). Add this mix to each reaction, mix gently and spin down.
Incubate at 25°C for 10 minutes, followed by 42°C for 50 minutes and then 70 °C for 15 minutes.
Step 4: Directional second strand synthesis
This step is performed to remove the RNA strand and synthesis the second DNA strand using the cDNA strand as a template while incorporating dUTP in the place of dTTP. The dUTP incorporation quenches second strand during amplification because the polymerase doesn’t incorporate past it.
- For each reaction, mix:
24.5 µl First strand synthesis product (from step 3) 5.5 µl Second strand synthesis mix (stranded) Mix well by pipetting and incubate at 16°C for 1 hour.
Step 5: Bead cleanup
Add 54 µL of well-mixed AMPure XP Beads to each sample and mix thoroughly by pipetting.
Incubate the plate for 5 minutes at room temperature.
Place the plate on the magnetic stand for 5 minutes at room temperature or until the liquid appears completely clear.
Remove and discard all of the supernatant from the plate taking care not to disturb the beads.
With plate on stand, add 200 µL of freshly prepared 80% ethanol to each well without disturbing the beads and incubate the plate for at least 30 seconds at room temperature. Carefully, remove and discard the supernatant.
Repeat step 5, for a total of two ethanol washes. Ensure the ethanol has been removed.
Remove the plate from the magnetic stand and let dry at room temperature for 2 minutes.
Resuspend dried beads in 18 µL Resuspension Buffer. Gently, pipette the entire volume up and down to mix thoroughly. Ensure that the beads are completely rehydrated and re-suspended.
Incubate resuspended beads at room temperature for 2 minutes.
Place the plate on the magnetic stand for 5 minutes at room temperature or until the supernatant appears completely clear.
Gently transfer 17 µL of the clear supernatant to a fresh well. The procedure can be safely stopped at this point and the samples stored at −80°C.
Step 3: A-tailing
A-tailing is performed by utilizing the polyermase activity of Klenow (3'–5' exo-) in presence of dATP to add a single ‘A’ to the 3' end of a blunt, double-stranded DNA. A-tailing prevents the blunt fragments from self ligating during the adapter ligation step.
- For each reaction, mix:
17 µl end repaired DNA 3.5 µl A-tailing mix Mix well by pipetting and then incubate at 37°C for 30 minutes followed by 70° for 5 minutes. Immediately proceed to adapter ligation
Step 4: Y-shaped adapter ligation
The ligation step ligates barcoded Y-shaped adapters to the ends of A-tailed DNA fragments. The adapters have a ‘T’ overhang, which is complementary to the adenylated DNA. The ligation step prepares the DNA fragments for subsequent hybridization onto the flow cells.
- For each reaction, mix:
20.5 µl Adenylated DNA 2 µl NEXTflex™ Barcoded Adapter (0.6 µM) 28 µl Ligation mix Mix well by pipetting and then incubate at 22°C for 15 minutes.
STEP 5: Double bead cleanup
Double-bead cleanup is performed at the end of ligation to remove any excess adapters that might have been self-ligated or be free floating and prevent them for getting amplified during PCR.
Add 50.5 µL of well-mixed AMPure XP Beads to each sample and mix thoroughly by pipetting.
Incubate the plate for 5 minutes at room temperature.
Place the plate on the magnetic stand for 5 minutes at room temperature or until the liquid appears completely clear.
Remove and discard all of the supernatant from the plate taking care not to disturb the beads.
With plate on stand, add 200 µL of freshly prepared 80% ethanol to each well without disturbing the beads and incubate the plate for at least 30 seconds at room temperature. Carefully, remove and discard the supernatant.
Repeat step 5, for a total of two ethanol washes. Ensure the ethanol has been removed.
Remove the plate from the magnetic stand and let dry at room temperature for 2 minutes.
Resuspend dried beads in 51 µL Resuspension Buffer. Gently, pipette the entire volume up and down to mix thoroughly. Ensure that the beads are completely rehydrated and re-suspended.
Incubate resuspended beads at room temperature for 2 minutes.
Place the plate on the magnetic stand for 5 minutes at room temperature or until the supernatant appears completely clear.
Gently transfer 50 µL of the clear supernatant to a fresh well.
Add 50 µL of well-mixed AMPure XP Beads to each well containing sample and mix thoroughly by pipetting.
Incubate the plate for 5 minutes at room temperature.
Place the plate on the magnetic stand for 5 minutes at room temperature or until the liquid appears completely clear.
Remove and discard all of the supernatant from the plate taking care not to disturb the beads.
With plate on stand, add 200 µL of freshly prepared 80% ethanol to each well without disturbing the beads and incubate the plate for at least 30 seconds at room temperature. Carefully, remove and discard the supernatant.
Repeat step 16, for a total of two ethanol washes. Ensure the ethanol has been removed.
Remove the plate from the magnetic stand and let dry at room temperature for 2 minutes.
Resuspend dried beads in 36 µL Resuspension Buffer. Gently, pipette the entire volume up and down to mix thoroughly. Ensure that the beads are completely rehydrated and re-suspended.
Incubate resuspended beads at room temperature for 2 minutes.
Place the plate on the magnetic stand for 5 minutes at room temperature or until the supernatant appears completely clear.
Gently transfer 35 µL of the clear supernatant to a fresh well.
The procedure may be stopped at this point and the reactions stored at −20°C.
STEP 6: Uracil-DNA Glycosylase (UDG) treatment and PCR amplification
In this step, the Uracil-DNA Glycosylase hydrolyzes the N-glycosylic bond between uracil and sugar in DNA, selectively degrading the dUTP marked strand and therefore the remaining strand is amplified to generate directional cDNA library. The PCR primers anneal in part to the adapter sequences.
For each reaction, mix:
| 35 µl | Adapter ligated DNA |
| 1 µl | Uracil DNA Glycosylase |
| 12 µl | PCR amplification mix |
| 2 µl | PCR primer mix (12.5 µM) |
Mix well by pipetting.
PCR cycling:
| 37°C | 2 minutes | |
| 98°C | 2 minutes | |
| 98°C | 30 seconds | |
| 65°C | 30 seconds | (repeat for 12–20 cycles) |
| 72°C | 60 seconds | |
| 72°C | 4 minutes | |
Tip
Always do the minimum number of PCR cycles possible.
STEP 7: Double bead cleanup
Post PCR amplification a double bead cleanup is performed to get rid of excess primer and primer dimers.
Add 50 µL of well-mixed AMPure XP Beads to each sample and mix thoroughly by pipetting.
Incubate the plate for 5 minutes at room temperature.
Place the plate on the magnetic stand for 5 minutes at room temperature or until the liquid appears completely clear.
Remove and discard all of the supernatant from the plate taking care not to disturb the beads.
With plate on stand, add 200 µL of freshly prepared 80% ethanol to each well without disturbing the beads and incubate the plate for at least 30 seconds at room temperature. Carefully, remove and discard the supernatant.
Repeat step 5, for a total of two ethanol washes. Ensure the ethanol has been removed.
Remove the plate from the magnetic stand and let dry at room temperature for 2 minutes.
Resuspend dried beads in 51 µL Resuspension Buffer. Gently, pipette the entire volume up and down to mix thoroughly. Ensure that the beads are completely rehydrated and re-suspended.
Incubate resuspended beads at room temperature for 2 minutes.
Place the plate on the magnetic stand for 5 minutes at room temperature or until the supernatant appears completely clear.
Gently transfer 50 µL of the clear supernatant to a fresh well.
Add 50 µL of well-mixed AMPure XP Beads to each well containing sample and mix thoroughly by pipetting.
Incubate the plate for 5 minutes at room temperature.
Place the plate on the magnetic stand for 5 minutes at room temperature or until the liquid appears completely clear.
Remove and discard all of the supernatant from the plate taking care not to disturb the beads.
With plate on stand, add 200 µL of freshly prepared 80% ethanol to each well without disturbing the beads and incubate the plate for at least 30 seconds at room temperature. Carefully, remove and discard the supernatant.
Repeat step 16, for a total of two ethanol washes. Ensure the ethanol has been removed.
Remove the plate from the magnetic stand and let dry at room temperature for 2 minutes.
Resuspend dried beads in 32 µL Resuspension Buffer. Gently, pipette the entire volume up and down to mix thoroughly. Ensure that the beads are completely rehydrated and re-suspended.
Incubate resuspended beads at room temperature for 2 minutes.
Place the plate on the magnetic stand for 5 minutes at room temperature or until the supernatant appears completely clear.
Gently transfer 30 µL of the clear supernatant to a fresh well.
The procedure may be stopped at this point and the libraries stored at −20°C until they are validated for quality and quantified for sequencing.
Quality control
Check the size and quality of the library by running it on a Bioanalyzer using the High Sensitivity DNA assay. If on the Bioanalyzer trace there are two bands, one of expected size and one of higher molecular weight, it’s indicative of a bubble product. This double product will not affect the outcome to the sequencing run as double stranded product is denatured prior to sequencing. As an extra verification step, a portion of this product (1–2 µL) can be denatured manually by heating the sample to 95°C for 5 minutes and then placing it on ice and subsequently be run on a Bioanalyzer RNA Pico 6000 Chip Kit. The denatured product should appear as a single band on a Pico 6000 chip.
Quantification of libraries
In order to get consistent number of reads across different samples it is important to accurately quantify the DNA library templates and then normalize all the samples before sequencing. To get the best sequencing results, it is important to get optimum cluster densities across every lane on every flow cell and this also makes quantification an important step. qPCR-based quantification is the most accurate method for quantifying library templates. Use the right Kapa library quantification kit by Kapa Biosystems for your qPCR instrument.
Setting up qPCR
Make a 1:1000 dilution of each of your library template for quantification in library dilution buffer.
Subsequently, use the 1:1000 dilution to make a 1:10,000 dilution.
Set up 10 µl qPCR reactions as described in the Technical Data Sheet for the Kapa kit
Analyze the qPCR data as described in the data sheet and then calculate the concentration of each of the library template.
Normalizing and pooling libraries for sequencing
If you have barcoded libraries, follow Bioo Scientific’s guidelines in the barcode manual for pooling normalized samples for sequencing
Normalize the concentration of each library to 20 nM using Library normalization buffer and then pool samples samples at equimolar concentration
Based on the coverage you want you can determine how many samples to pool per lane for 50 bp single end sequencing.
Data Analysis of RNA-seq data
Map the sequence reads to the mouse genome with Tophat (Kim et al., 2013). For strand-specific RNA-seq data, “--library-type fr-firststrand” is the parameter to use for Illumina.
Remove the reads mapped with low mapping quality (<5) using SAMtools (Li et al., 2009) to get rid of the reads mapped to multiple locations.
Use Homer (Heinz et al., 2010) to process the mapped reads. Homer includes tools to analyze RNA-seq, ChIP-seq, etc. First, tag directories for each sample will be created by a tool “makeTagDirectory”. Then, RNA expression is quantified using Perl scripts “analyzeRNA.pl” or “analyzeRepeats.pl”. The scripts have options to count reads mapped to intron, exon or gene body for each gene. For WT RNA-seq data, we interpret the intron signal as a representation of pre-mRNA expression or nascent transcription (Ameur et al., 2011) and the exon signal as representation of mRNA expression. The expression levels are normalized as reads per kilobase per million mapped reads (RPKM), because longer genes have chance to be mapped more reads. For gene annotation, we used the UCSC known canonical gene set to eliminate transcript variants. For gene annotation, Homer can use GTF files, which can be downloaded from UCSC Table browser.
Time Series Analysis for Circadian Cycling
Time series analysis of very short data sets is nontrivial. Ideally if one were to use Fourier Transform methods to assess the frequency and amplitude of time series data as in the case of locomotor activity data (Takahashi and Menaker, 1982), it would be necessary to analyze at least 10 cycles of the target periodicity at a sampling resolution that matches the Nyquist frequency (fs/2, where fs =sample rate). Because of the expense of ChIP-seq and RNA-seq samples, obtaining 10 cycles of molecular data is highly unlikely, and it is customary to assay only two cycles of circadian time series. Indeed analysis of only one cycle of data cannot reliably estimate period. A number of algorithms have been developed to estimate period and amplitude of short time series. Most use some type of fitting procedure to either sinusoidal or prespecified waveforms. Significance thresholds are usually then estimated using bootstrap methods. We have used three different programs for RNA cycling, COSOPT (Panda et al., 2002), JTK Cycle (Hughes et al., 2010) and ARSER (Yang and Su, 2010). COSOPT runs on Microsoft Windows, JTK requires R packages, and ARSER is implemented by a Python calling R program. For COSOPT and JTK Cycle analyses, data is detrended by linear regression. Previously (Koike et al., 2012) we considered a cycling gene if two out of three programs detected cycling with threshold of p<0.05. The period and phase from ARSER was used for further analysis. For ChIP-seq peak analysis, two cycles were concatenated and the cycling was analyzed with ARSER (p<0.05). Because COSOPT is slow and much less sensitive at detection of cycling transcripts compared to JTK Cycle and ARSER, we now routinely use the latter two programs for circadian cycling detection. While these programs are adequate, each has its propensity for false negative and false positive detection of cycling, and each is very sensitive to the details of sampling interval, number of replicates, and time series duration. We find that the sets of cycling genes detected by these programs is variable across experiments and can be discordant between programs because of the specific waveform features of the time series. Thus, the analysis of short circadian time series is clearly an area for future development.
References
- Ameur A, Zaghlool A, Halvardson J, Wetterbom A, Gyllensten U, Cavelier L, Feuk L. Total RNA sequencing reveals nascent transcription and widespread co-transcriptional splicing in the human brain. Nat Struct Mol Biol. 2011;18:1435–1440. doi: 10.1038/nsmb.2143. [DOI] [PubMed] [Google Scholar]
- Buck MJ, Lieb JD. ChIP-chip: considerations for the design, analysis, and application of genome-wide chromatin immunoprecipitation experiments. Genomics. 2004;83:349–360. doi: 10.1016/j.ygeno.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M, Kremmer E, Eick D. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science. 2007;318:1780–1782. doi: 10.1126/science.1145977. [DOI] [PubMed] [Google Scholar]
- Chaw YF, Crane LE, Lange P, Shapiro R. Isolation and identification of cross-links from formaldehyde-treated nucleic acids. Biochemistry. 1980;19:5525–5531. doi: 10.1021/bi00565a010. [DOI] [PubMed] [Google Scholar]
- Chen R, Schirmer A, Lee Y, Lee H, Kumar V, Yoo SH, Takahashi JS, Lee C. Rhythmic PER abundance defines a critical nodal point for negative feedback within the circadian clock mechanism. Mol Cell. 2009;36:417–430. doi: 10.1016/j.molcel.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckner R, Ludlow JW, Lill NL, Oldread E, Arany Z, Modjtahedi N, DeCaprio JA, Livingston DM, Morgan JA. Association of p300 and CBP with simian virus 40 large T antigen. Mol Cell Biol. 1996;16:3454–3464. doi: 10.1128/mcb.16.7.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- Gilmour DS, Lis JT. Detecting protein-DNA interactions in vivo: distribution of RNA polymerase on specific bacterial genes. Proc Natl Acad Sci U S A. 1984;81:4275–4279. doi: 10.1073/pnas.81.14.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatanaka F, Matsubara C, Myung J, Yoritaka T, Kamimura N, Tsutsumi S, Kanai A, Suzuki Y, Sassone-Corsi P, Aburatani H, et al. Genome-wide profiling of the core clock protein BMAL1 targets reveals a strict relationship with metabolism. Mol Cell Biol. 2010;30:5636–5648. doi: 10.1128/MCB.00781-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms. 2010;25:372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson V. Studies on histone organization in the nucleosome using formaldehyde as a reversible cross-linking agent. Cell. 1978;15:945–954. doi: 10.1016/0092-8674(78)90278-7. [DOI] [PubMed] [Google Scholar]
- Jackson V, Chalkley R. A new method for the isolation of replicative chromatin: selective deposition of histone on both new and old DNA. Cell. 1981;23:121–134. doi: 10.1016/0092-8674(81)90277-4. [DOI] [PubMed] [Google Scholar]
- Jones JC, Phatnani HP, Haystead TA, MacDonald JA, Alam SM, Greenleaf AL. C-terminal repeat domain kinase I phosphorylates Ser2 and Ser5 of RNA polymerase II C-terminal domain repeats. J Biol Chem. 2004;279:24957–24964. doi: 10.1074/jbc.M402218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpov VL, Preobrazhenskaya OV, Mirzabekov AD. Chromatin structure of hsp 70 genes, activated by heat shock: selective removal of histones from the coding region and their absence from the 5' region. Cell. 1984;36:423–431. doi: 10.1016/0092-8674(84)90235-6. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, Steeves TD, Vitaterna MH, Kornhauser JM, Lowrey PL, et al. Positional cloning of the mouse circadian clock gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- Kuo MH, Allis CD. In vivo cross-linking and immunoprecipitation for studying dynamic Protein:DNA associations in a chromatin environment. Methods. 1999;19:425–433. doi: 10.1006/meth.1999.0879. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Martelot G, Canella D, Symul L, Migliavacca E, Gilardi F, Liechti R, Martin O, Harshman K, Delorenzi M, Desvergne B, et al. Genome-wide RNA polymerase II profiles and RNA accumulation reveal kinetics of transcription and associated epigenetic changes during diurnal cycles. PLoS Biol. 2012;10 doi: 10.1371/journal.pbio.1001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- Lee C, Weaver DR, Reppert SM. Direct association between mouse PERIOD and CKIε is critical for a functioning circadian clock. Mol Cell Biol. 2004;24:584–594. doi: 10.1128/MCB.24.2.584-594.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey PL, Takahashi JS. Genetics of circadian rhythms in Mammalian model organisms. Adv Genet. 2011;74:175–230. doi: 10.1016/B978-0-12-387690-4.00006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee JD, von Hippel PH. Formaldehyde as a probe of DNA structure. I. Reaction with exocyclic amino groups of DNA bases. Biochemistry. 1975a;14:1281–1296. doi: 10.1021/bi00677a029. [DOI] [PubMed] [Google Scholar]
- McGhee JD, von Hippel PH. Formaldehyde as a probe of DNA structure. II. Reaction with endocyclic imino groups of DNA bases. Biochemistry. 1975b;14:1297–1303. doi: 10.1021/bi00677a030. [DOI] [PubMed] [Google Scholar]
- Menet JS, Rodriguez J, Abruzzi KC, Rosbash M. Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. Elife. 2012;1:e00011. doi: 10.7554/eLife.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak DE, Tian B, Brasier AR. Two-step cross-linking method for identification of NF-kappaB gene network by chromatin immunoprecipitation. Biotechniques. 2005;39:715–725. doi: 10.2144/000112014. [DOI] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reick M, Garcia JA, Dudley C, McKnight SL. NPAS2: an analog of clock operative in the mammalian forebrain. Science (New York, N Y) 2001;293:506–509. doi: 10.1126/science.1060699. [DOI] [PubMed] [Google Scholar]
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, Naef F. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011;9:e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon-Divon M, Dvinge H, Tammoja K, Bertone P. PeakAnalyzer: genome-wide annotation of chromatin binding and modification loci. BMC Bioinformatics. 2010;11:415. doi: 10.1186/1471-2105-11-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon MJ, Varshavsky A. Formaldehyde-mediated DNA-protein crosslinking: a probe for in vivo chromatin structures. Proc Natl Acad Sci U S A. 1985;82:6470–6474. doi: 10.1073/pnas.82.19.6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi JS, Menaker M. Role of the suprachiasmatic nuclei in the circadian system of the house sparrow, Passer domesticus. J Neurosci. 1982;2:815–828. doi: 10.1523/JNEUROSCI.02-06-00815.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmers C, Schmitz R, Nathanson J, Yeo G, Ecker J, Panda S. Circadian oscillations of protein-coding and regulatory RNAs in a highly dynamic mammalian liver epigenome. Cell metabolism. 2012;16:833–845. doi: 10.1016/j.cmet.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh JaC, CR Protein-DNA cross-linking. Trends in Biochemical Sciences. 1984;9:505–508. [Google Scholar]
- Yang R, Su Z. Analyzing circadian expression data by harmonic regression based on autoregressive spectral estimation. Bioinformatics. 2010;26:i168–i174. doi: 10.1093/bioinformatics/btq189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng PY, Vakoc CR, Chen ZC, Blobel GA, Berger SL. In vivo dual cross-linking for identification of indirect DNA-associated proteins by chromatin immunoprecipitation. Biotechniques. 2006;41 doi: 10.2144/000112297. 694, 696, 698. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]