Abstract
Reliably marking larvae and reidentifying them after metamorphosis is a challenge that has hampered studies on recruitment, dispersal, migration and survivorship of amphibians for a long time, as conventional tags are not reliably retained through metamorphosis. Molecular methods allow unique genetic fingerprints to be established for individuals. Although microsatellite markers have successfully been applied in mark–recapture studies on several animal species, they have never been previously used in amphibians to follow individuals across different life cycle stages. Here, we evaluate microsatellites for genetic across-stages mark–recapture studies in amphibians and test the suitability of available software packages for genotype matching. We sampled tadpoles of the dendrobatid frog Allobates femoralis, which we introduced on a river island in the Nature Reserve ‘Les Nouragues’ in French Guiana. In two subsequent recapture sessions, we searched for surviving juveniles and adults, respectively. All individuals were genotyped at 14 highly variable microsatellite loci, which yielded unique genetic fingerprints for all individuals. We found large differences in the identification success of the programs tested. The pairwise-relatedness-based approach, conducted with the programs kingroup or ML-Relate, performed best with our data set. Matching ventral patterns of juveniles and adult individuals acted as a control for the reliability of the genetic identification. Our results demonstrate that microsatellite markers are a highly powerful tool for studying amphibian populations on an individual basis. The ability to individually track amphibian tadpoles throughout metamorphosis until adulthood will be of substantial value for future studies on amphibian population ecology and evolution.
Keywords: amphibians, genetic identification, life cycle, mark–recapture, metamorphosis, microsatellites
Introduction
Precise identification and recognition of individuals is important in several research fields. Repeated captures of single individuals allow researchers to estimate species abundance (Nichols 1992), gain information on life history parameters such as growth rate, age at sexual maturity, reproduction and survivorship (Lebreton et al. 1992; Pradel 1996; Schmidt et al. 2002) and to investigate aspects of behavioural ecology, such as dispersal and migration (Schaub et al. 2001).
Many animals do not feature individually distinct colour patterns that allow for unambiguous identification when individuals are recaptured, at least not throughout their entire life. To enable individual identification, several techniques have been developed: attached tags, injectable microchips and elastomers, external colour marks, tattoos and brandings, as well as the removal of parts of tissue from the ears, tail or digits to create individual ‘clipping codes’ (Clark 1971; Donnelly et al. 1994; Frederick 1997; Olsen & Vøllestad 2001; Gibbons & Andrews 2004). Ideally, marks should be long-lasting, remain attached reliably, be easy to apply and read and inexpensive (Nietfeld et al. 1994). Besides these logistic requirements, marks should have only minimal impact on the animals in terms of affecting their survival, physiology, growth, predation risk and behaviour (Wood & Slade 1990; Dussault & Rodríguez 1997; Wilson et al. 2011; Carlson et al. 2013). This is of major importance, as any adverse effect of marking could compromise the actual studies and lead to incorrect conclusions and management decisions. Not all markers are suitable for all animal species. Particularly for small animals, it is often challenging to find the optimal marking technique; for example, the weight of available markers could add too much load, and/or the required handling procedure might cause severe difficulties or injuries. Hardly any marking technique meets all requirements ideally, and thus, researchers often have to trade-off costs and benefits when deciding on a technique.
Besides conventional marking, individuals can also be identified from genetic data, for example by their compound genotype at multiple polymorphic loci (Jeffreys et al. 1985; Beckmann & Soller 1990; Palsbøll 1999). Such ‘genetic markers’ are conceptually similar to conventional marking: individuals are identified by their unique genetic fingerprint (Palsbøll 1999). Hypervariable DNA sequences are ubiquitously present in eukaryote genomes (Tautz & Renz 1984), and thus, it is generally possible to (re-)identify individuals from any eukaryote species. The required DNA sample can be obtained from tissue such as blood and skin or via cells obtained from hair or faeces. Genetic markers have been implemented in capture–recapture studies of various animal populations, for example humpback whales (Palsboll et al. 1997), bears (Taberlet et al. 1997; Woods et al. 1999; Boulanger et al. 2004), wombats (Sloane et al. 2000), holothurians (Uthicke & Benzie 2002), giant salamanders (Unger et al. 2012) and fish (Andreou et al. 2012).
Several marking techniques are used for amphibians, including toe clipping, tattooing, branding, passive transponders and other types of body implants (Ferner 1979; Donnelly et al. 1994; Jehle & Hödl 1998; Courtois et al. 2013). For species with highly variable colour patterns, photographs may be sufficient for mark–recapture studies of adult individuals (e.g. Caorsi et al. 2012). While coded toe clipping was commonly applied to individually mark adult amphibians for many decades, the method is currently being questioned on ethical grounds because several studies found adverse effects on recapture rates and survival for some species (Davis & Ovaska 2001; May 2004; McCarthy & Parris 2004; Funk et al. 2005).
Moreover, amphibians are characterized by a complex life cycle. The drastic morphological changes from tadpole to the adult stage, including the development of limbs, resorption of the tail, rapid growth and the development of adult skin, as well as the generally high regenerative abilities of amphibians (Brockes 1997; Brockes & Kumar 2002) make it almost impossible to permanently mark individuals across life cycle stages. This circumstance has hampered studies on recruitment, dispersal, migration and survivorship in amphibian populations and limited mark–recapture studies to either the larval (Jung et al. 2002; Martin 2011; Ribeiro & Rebelo 2011) or the postmetamorphic stage (Smith 1987; but see Sinsch 1997; Altwegg & Reyer 2003). So far, none of the conventional markers are reliably maintained through metamorphosis (Grant 2008; Martin 2011; Courtois et al. 2013).
Survival rates of aquatic larvae play a crucial role in amphibian population dynamics, as the highest loss of individuals occurs at the larval stage (Vonesh & De la Cruz 2002). Particularly when considering the current amphibian extinction crisis, comprehensive data on demography and ecological characteristics of amphibian larvae are urgently needed. In amphibians, polymorphic genetic markers are frequently used for the estimation of genealogical relationships or to calculate the relatedness between individuals of unknown ancestry (Jehle & Arntzen 2002; Blouin 2003). Although genetic tracking has been suggested as a suitable method to study animals that are difficult to observe or identify (Hoffman et al. 2006), to date, the method has not been previously applied successfully in amphibians to track larvae until adulthood.
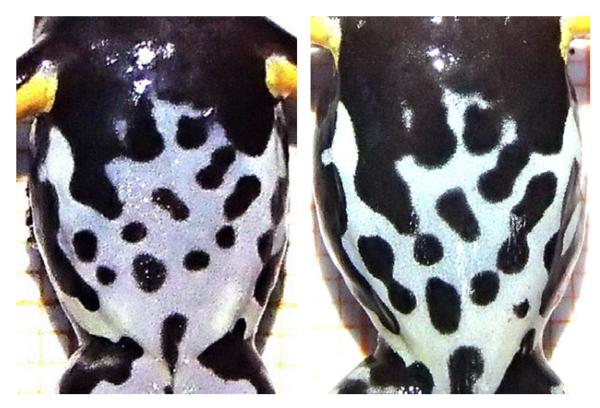
For this study, we used samples of Allobates femoralis, a small diurnal poison frog (Dendrobatidae) that occurs over entire Amazonia, which were collected in the course of a long-term study on juvenile dispersal patterns and general population ecology in this species. Allobates femoralis is characterized by male territoriality, female site fidelity, a prolonged breeding period, a polygynandrous mating system and male tadpole transport to water bodies (Roithmair 1992; Ringler et al. 2009, 2011, 2012, 2013a; Montanarin et al. 2011). Adult individuals can be distinguished via unique ventral patterns that are already present in juvenile individuals from approximately 6 months of age on (see Fig. 1). A large number of highly polymorphic microsatellite markers are already available for this species (Jehle et al. 2008; Ursprung et al. 2011a; Ringler et al. 2013b).
Fig. 1.
Example of corresponding juvenile (left) and adult (right) ventral patterns in one Allobates femoralis individual.
The aim of this study was to assess the suitability of microsatellite markers for genetic mark–recapture studies in amphibians across life cycle stages and to evaluate available software packages for matching the microsatellite genotypes.
Materials and methods
In March 2012, we released 1800 tadpoles of the dendrobatid frog Allobates femoralis on a 5-ha river island that is located in the immediate vicinity of the CNRS research station ‘Saut Parar e’ in the Nature Reserve ‘Les Nouragues’ in French Guiana (3°59′N, 52°35′W; Ringler et al., 2014) and was previously uninhabited by this species. The tadpoles were sampled at random from artificial pools which had been used previously in an experiment on resource supplementation in a nearby autochthonous population on the mainland (Ringler M, Hödl W, Ringler E, in preparation). We photographed the tadpoles digitally on scale paper for later size measurements, clipped a piece of the tail for genotyping and finally distributed them in semi-random order in 20 artificial pools (volume ~25 L, interpool distance ~10 m, 90 tadpoles per pool) on the island.
During September 2012, we surveyed the island for juvenile A. femoralis and found 42 individuals. From January to March 2013, we searched for individuals that had reached sexual maturity. To this end, we conducted extensive surveys on the island during periods of calling activity (08.00–12.00 h, 14.00–19.00 h), where we encountered 36 males and 31 females. All spatial locations of frogs were recorded on pocketPCs (MobileMapper 10; Ashtech/Spectra Precision) in ArcPAD 10 (ESRI) using a highly detailed background map (Ringler et al., 2014).
Standardized digital photographs were taken of all juvenile and adult A. femoralis for identification by their ventral coloration patterns. We used the pattern-matching software wild-id (Bolger et al. 2012) to speed up subsequent visual matching of juvenile and adult frogs. Adult individuals were sexed by the presence (male) or absence (female) of vocal sacs.
Tissue samples were obtained by removing the third toe of both hind limbs of all newly encountered adults and juveniles (Ursprung et al. 2011b) and were immediately preserved in 96% ethanol. All samples were genotyped at 14 highly variable microsatellite loci. Ambiguous loci were genotyped up to three times. For detailed protocols and characteristics of the microsatellite loci, see the studies by Jehle et al. (2008), Ursprung et al. (2011a) and Ringler et al. (2013b). We used cervus 3.0.3 (Kalinowski et al. 2007) to determine the number of alleles, observed and expected heterozygosities, and PIC (mean polymorphic information content). The probabilities of identity for random samples (PID) and for full-siblings (PSIB) were calculated with genecap (Wilberg & Dreher 2004).
We tested the suitability of available Freeware software packages for matching the microsatellite genotypes. On the one hand, we used programs that are specifically aimed for genotype matching, such as identity (Wagner & Sefc 1999), genecap (Wilberg & Dreher 2004), genalex 6.5 (Peakall & Smouse 2006) and allelematch (Galpern et al. 2012). On the other hand, we also tried an indirect approach using pairwise relatedness values to assess genotype identity, as provided by the programs kingroup (Konovalov et al. 2004) and ml-relate (Kalinowski et al. 2006). All programs were tested twice, first with a reduced data set containing only the genotype data from all juvenile and adult frogs and then with the full data set of all tadpoles, juveniles and adults. We evaluated the performance of the programs based on the following points:
Number of correct matches between corresponding juveniles and adults
Number of false matches between noncorresponding juveniles and adults (α-error)
Number of undetected matches between corresponding juveniles and adults (β-error)
Consistency of trios (i.e. whether matching juveniles and adult individuals were assigned to the same tadpole genotype)
Number of unambiguous, singular adult–tadpole matches
Effect of missing loci and genotyping errors on matching success
Ability to handle large data sets (>1000 genotypes)
Genetic assignments were evaluated based on the known corresponding juvenile–adult pairs that were inferred from the unique ventral patterns (Fig. 1). The relatedness-based programs provide pairwise relatedness values across all given genotypes. We tested whether pairwise relatedness values of known juvenile–adult matches were significantly different from pairwise relatedness values to the next most closely related genotype. We then defined a minimum threshold to accept genotype matches for all unknown assignments (juveniles–tadpoles, adults–tadpoles).
Results and discussion
The samples yielded on average 24 alleles per locus (Table 1) across the 14 microsatellite loci in the genetic analysis. For details on the characteristics of the used microsatellite loci, see the studies by Jehle et al. (2008), Ursprung et al. (2011a) and Ringler et al. (2013b). The data set contained unique genetic fingerprints for all 1800 tadpole samples, with an average probability of identity of 1.15 × 10−22 for random pairs and of 1.73 × 10−7 for full-siblings (Table 1).
Table 1.
Variability of the 14 microsatellite markers used to determine individual identity
| Locus | A | H O | H E | PIC | P ID | P SIB |
|---|---|---|---|---|---|---|
| Afem05 | 24 | 0.557 | 0.639 | 0.620 | 0.149 | 0.468 |
| Afem12 | 14 | 0.825 | 0.876 | 0.864 | 0.027 | 0.319 |
| Afem09 | 24 | 0.515 | 0.861 | 0.846 | 0.034 | 0.328 |
| Afem03 | 12 | 0.839 | 0.834 | 0.816 | 0.046 | 0.345 |
| Afem15 | 22 | 0.585 | 0.857 | 0.842 | 0.035 | 0.330 |
| Afem13 | 17 | 0.616 | 0.851 | 0.837 | 0.036 | 0.334 |
| Afem16 | 20 | 0.841 | 0.871 | 0.858 | 0.029 | 0.322 |
| Afem20 | 10 | 0.731 | 0.715 | 0.667 | 0.129 | 0.425 |
| Afem27 | 37 | 0.891 | 0.939 | 0.935 | 0.007 | 0.282 |
| Afem24 | 30 | 0.856 | 0.937 | 0.934 | 0.008 | 0.283 |
| Afem04 | 27 | 0.739 | 0.895 | 0.887 | 0.019 | 0.308 |
| Afem17 | 24 | 0.570 | 0.866 | 0.854 | 0.030 | 0.325 |
| Afem25 | 46 | 0.796 | 0.923 | 0.918 | 0.011 | 0.291 |
| Afem22 | 27 | 0.903 | 0.921 | 0.916 | 0.011 | 0.292 |
| Mean | 23.857 | 0.733 | 0.856 | 0.842 | ||
| Overall probability of identity | 1.15 × 10−22 | 1.15 × 10−22 | ||||
A, number of alleles; HO, observed heterozygosity; HE, expected heterozygosity; PIC, polymorphic information content; PID, probability of identity; PSIB, probability of sibling identity; values are based on the 1800 tadpole genotypes.
Based on their unique ventral pattern, we identified 20 adults in spring 2013 that corresponded to one of the 42 juvenile individuals from fall 2012 (Figs 1 and S1, Supporting information). The survival rate of the introduced tadpoles on the island thus was 3.72% (67 adult individuals from 1800 tadpoles). Our sample of juveniles was smaller than the number of identified adults because sexually immature juveniles are much harder to detect as they do not yet call and engage in courtship. The unique ventral patterns of Allobates femoralis postmetamorphs served as independent controls to test the reliability of the assignments in the different software packages. Patterns were already present at this early life stage and could unambiguously be reidentified in adult individuals a few months later (see Figs 1 and S1, Supporting information).
When comparing the genotypes of the 20 known matches locus by locus, we detected the presence of missing loci in at least one of the two matching samples in 19 cases (one missing locus: 12 cases, two loci: four cases, three loci: three cases). Genotyping errors occurred less often but nevertheless happened with five individuals (one allele differing: three cases, two alleles: one case, three alleles: one case). For an overview of the missing loci and genotyping errors, see Table S1 (Supporting information). Based on the observed cases of mismatching alleles, we estimated a genotyping error rate of 0.007 (i.e. 8 errors of 1120 total alleles in 40 samples with 14 loci) for our entire study.
The tested programs showed a wide variation in their performance with our data set. While none of the programs produced false assignments (α-error), the rate of β-errors when matching adults or juveniles and tadpoles was quite high in most programs.
IDENTITY
identity is a command line program for the analysis of microsatellite data. We used the program to screen the data set for identical genotypes. The program detected the fewest matches as even a single missing locus prevented the program from identifying corresponding genotypes. As a consequence, the program identified least of the known juvenile–adult matches. Only the four pairs with exact identical genotypes (same nonamplifying loci in both samples) were found. The program failed to run the analysis on the full data set (1909 genotypes), as this data set contained more genotypes than the program was able to handle (the computation crashed when processing more than 1002 individuals). Consequently, no information on the effect of ‘genotypic noise’ and the completeness of trios could be obtained. Therefore, we cannot recommend the program for genetic mark–recapture studies.
GENECAP
genecap is a Microsoft EXCEL macro for analysing multilocus genotype data. The program can account for missing loci and genotyping errors; however, the output only lists those pairs with no more than ‘two alleles different’. The program correctly identified 15 of the 20 known pairs as matches (i.e. all pairs without genotype errors) and correctly ‘suggested’ four more (three pairs differing only in one allele, one pair differing in two alleles). The correct juvenile–adult pair that differed in more than two alleles remained undetected. The results remained the same for the reduced and the full data set. For the 20 juvenile–adult pairs, 12 unambiguous trios (tadpole–juvenile–adult) were found. Corresponding juveniles and adults were never assigned to different tadpole genotypes. However, for seven adults and six juveniles, no matching tadpole genotype was found. Of the 67 adults in total, only 46 could be assigned to a tadpole. The main disadvantage of the program is that it does not provide information on lower rank matches beyond the category ‘two alleles different’, unlike genalex (for example see next section). Based only on the genecap analysis, it is therefore not possible to improve the matching success by repeated genotyping of ambiguous samples.
GENALEX
This program is also a Microsoft EXCEL macro and is designed for analysing a wide range of population genetic data. In contrast to gencap, genalex provides the user with a comprehensive output that lists pairs of individuals in categories based on the degree of genotype concordance (i.e. in ascending order of locus mismatches). Thereby, we obtained an output where all dyads that match in at least four loci are given. However, the program does not indicate which of the listed pairs should actually be considered as identical genotypes. Thus, the researcher has to define a threshold (i.e. the number of locus mismatches) up to which all given dyads should be considered as identical genotypes. In our case, when analysing the reduced data set, all correct pairs were listed in the first 5 categories. The four pairs with exactly identical genotypes were listed in the category ‘Matching at All Loci’, and the remaining 16 correct juvenile–adult pairs were listed in the following lower-congruence categories: seven pairs were matching ‘All But 1 Locus’, six pairs ‘All But 2 Loci’, two pairs ‘All But 3 Loci’ and one pair ‘All But 4 Loci’. Locus mismatches identified by the program likewise resulted from genotyping errors and missing alleles. Only correct juvenile–adult matches appeared until the level ‘All But 4 Loci’. In the following categories ‘All But 5 Loci’ and ‘All But 6 Loci’, only genotype dyads were listed that belonged to the same sampling cohort (i.e. two adult individuals). The next juvenile–adult assignment did not occur until the category ‘All But 8 Loci’. Hence, we could have used this ‘gap’ for separating ‘matches’ from all other dyads. However, in cases where sampling cohorts are not discrete (e.g. not of different life stages), researchers will have to set an arbitrary threshold limit for defining genotype matches.
The program failed to run the analysis on the full data set, as the computation crashed when processing more than 1440 individuals. Therefore, no information on the effect of ‘genotypic noise’ and the completeness of trios could be obtained. Another difference between genecap and genalex is that the former considers genotype differences at the level of alleles, while genalex compares genotypes at the level of locus mismatches (i.e. the difference in even one of the two alleles within a given locus leads to a whole locus mismatch). Thus, the resolution of analysis should be slightly higher in genecap. As genalex could not finish the computation of the full data set, we cannot directly compare the performance of both programs.
ALLELEMATCH
allelematch is a package of functions for the statistical software environment r (http://www.r-project.org). It conducts matching and clustering of multilocus genotype data to find unique individuals and identify potential genotyping errors. In a first step, the program estimates the number of likely genotyping errors in the sample, which can then be incorporated in the subsequent analysis. For both data sets, allelematch estimated a mismatch rate of 3 alleles per sample pair that should be allowed. The program subsequently identified all but one juvenile–adult match in both data sets. Surprisingly, the undetected juvenile–adult pair was not the same for the reduced and full data set, respectively. Although the matching of juveniles and adults was good, the success rate when matching juveniles and adults to tadpoles was quite low: consistent trios were only found for 11 juvenile–adult pairs, while for nine juveniles and eight adults, no matching tadpole was found. In total, the program assigned tadpoles to only 36 of the 67 adults. Unfortunately, the format of the output was rather inconvenient for our purpose: results are given as an html document, which contains a list of all individual genotypes in the order of the input file, without any markup or grouping of identified matches. With a data set containing many individuals, the resulting output file is highly cluttered, and identifying all genotype matches found by the program is time-consuming. Nonetheless, allelematch proved to be quite powerful for the matching of juveniles and adults.
KINGROUP
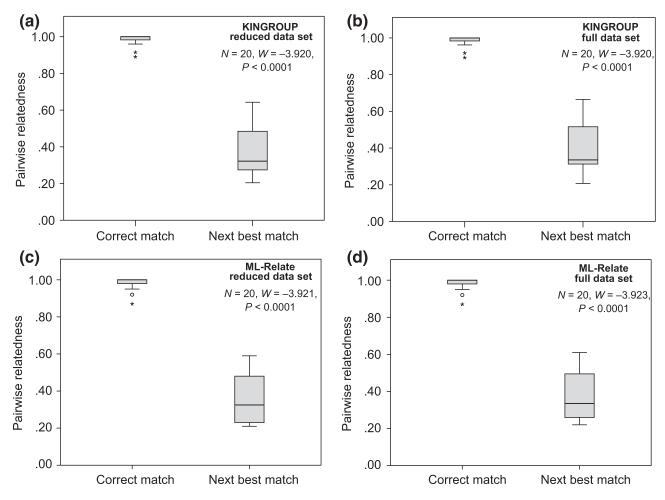
kingroup is an open source java program using a maximum-likelihood approach for pedigree reconstruction and kin group assignment. It calculates relatedness coefficients that can be interpreted as a continuous measure of the overall genetic similarity between two individuals within a population. Values range from −1 to +1, with negative and positive values indicating that two individuals have a lower and higher probability, respectively, of recent coalescence than random dyads within the population (Queller & Goodnight 1989; Konovalov & Heg 2008). Matching genotypes were unambiguously identified: there was a significant difference between the relatedness of correct (known) juvenile–adult pairs and the relatedness measures of the next best matches (Wilcoxon signed-rank test, reduced data set: W = −3.920, P < 0.0001, Fig. 2a; full data set: W = −3.920, P < 0.0001, Fig. 2b). Consequently, no false matches were found (i.e. no β-error). Because the lowest juvenile–adult pair had a relatedness value of r = 0.89, we thus set the cut-off value for accepting genotype matches at r = 0.8. The program also managed to unambiguously identify all trios (i.e. juveniles and adults were assigned to the same tad-pole). Only one juvenile was assigned to two tadpoles, according to our cut-off criterion of r = 0.8. However, here, the best matching tadpole had a much higher r score than the other one (r = 0.9571 vs. 0.8176) and was also exclusively assigned to the known corresponding adult. Furthermore, the remaining adult individuals that did not have corresponding juvenile genotypes were almost all assigned unambiguously to one single tadpole; only for three adults multiple (two or three) tadpole genotypes had r-values above 0.8. However, one of the tadpoles always had a much higher r-score than the other one, and we considered the one with the higher score to be the correct tadpole match.
Fig. 2.
Box plots showing the distribution of pairwise relatedness values of correctly identified corresponding juvenile–adult pairs (left bars) and values of the respective next best matches (right bars); (a) kingroup reduced data set, (b) kingroup full data set, (c) ML-Relate reduced data set, (d) ML-Relate full data set. Outlier values are indicated by * and °.
ML-RELATE
Like kingroup, ml-relate also calculates maximum-likelihood estimates of relatedness (Wagner et al. 2006), but here, the coefficient of relatedness ranges from 0 (unrelated) to 1 (identical genotype). Matching genotypes were unambiguously identified regardless of the data set used: there was a significant difference between the relatedness of known matches and that of the next best match (Wilcoxon signed-rank test, reduced data set: W = −3.921, P < 0.0001, Fig. 2c; full data set: W = −3.923, P < 0.0001, Fig. 2d). The lowest r-value for known juvenile–adult pairs was r = 0.87; thus, the threshold for accepting genotype matches was again set at r = 0.8. Under this criterion, the program managed to unambiguously identify all trios (i.e. juveniles and adults were assigned to the same tadpole). Also, all remaining adult individuals that did not have corresponding juvenile genotypes were almost all assigned unambiguously to one single tadpole; for three adults, more than one tadpole genotype had an r-value above 0.8 (same adults and tadpoles as in the kingroup analysis). For three adults, the best matching tadpoles were slightly below the threshold of r = 0.8.
Generally, the relatedness-based programs performed best for our data set and at the same time provided the highest flexibility to identify matching genotypes. Both kingroup and ml-relate were able to unambiguously identify all juvenile–adult matches. Furthermore, it was possible to assess the resolving power of the microsatellite markers because the full set of relatedness values for all pairs of individuals are computed and listed in the output. We therefore compared the relatedness values of best matches (i.e. corresponding juvenile–adult pairs) against their next best matches to test if matches were significantly different to the next best match, which was the case in our study (see Fig. 2). Even if some adult individuals could not be unambiguously assigned to a single tadpole immediately, in general, this approach at least reduced the number of candidate matches for a given individual. Repeated PCR runs of ambiguous loci might then potentially help to reduce the number of missing loci or to detect previous genotyping errors, to enhance the resolution of the relatedness estimation. In contrast to all other tested programs that only provide a list of ‘good’ matches in the output, the information gained from the kingroup and ml-relate computations actually provides sufficient information for such a post hoc refinement.
Problems and constraints
The main drawback of genetic tracking is the need for a tissue sample from each capture. While the removal of multiple toes is required when toe clipping is applied as marking method, for DNA sampling the removal of one or two toes or even only toe pads is sufficient. Negative effects on individual recapture rates and survival significantly increase with the number of toes clipped (McCarthy & Parris 2004). A previous study on toe regeneration in A. femoralis found no difference in survival rates between clipped (two toes) and unclipped A. femoralis populations (Ursprung et al. 2011b). Thus, we assume only marginal effects of clipping only one or two toes or toe pads for DNA sampling. However, if DNA can only be obtained in an invasive way (e.g. blood sampling, toe clipping), this might become problematic in studies where multiple identifications are required within a short time frame. But noninvasive techniques can also cause problems: noninvasively obtained samples often yield only small quantities of DNA that will result in low amplification success (Taberlet & Luikart 1999; Lukacs & Burnham 2005), and the resulting genotype mismatches can then lead to high rates of misidentification (Goossens et al. 1998; Creel et al. 2003).
Another problem associated with genetic tracking is the required polymorphism of the markers used to unambiguously identify also closely related genotypes. The probability that two unrelated individuals have identical genotypes correlates inversely with the number of loci analysed and the number of alleles per locus (Waits et al. 2001). In our study, the discriminatory power of the markers was well beyond the variability needed for individual identification (cf. Waits et al. 2001). However, for amphibian populations where variability is considerably reduced due to recent bottlenecks or high rates of inbreeding, more microsatellite markers might be needed to reach a satisfying level of resolution (Selkoe & Toonen 2006). For such populations, we recommend the stepwise selective increase of markers as proposed by Rew et al. (2011) to minimize laboratory work and expenses. To maximize the resolution of the genetic analysis, we decided to use a relatively large number of microsatellite markers in this study. Several authors have strongly advocated the use of small numbers of microsatellite loci, as genotyping errors increase together with the numbers of loci (Creel et al. 2003; Paetkau 2004; Kolodziej et al. 2012, 2013). Nonetheless, we decided to use all available markers available for our study population because we expected a high number of full-siblings in our tadpole sample (cf. Waits et al. 2001). Therefore, we aimed for a maximum of genotypic resolution in our data set. The fact that none of the programs produced wrong assignments (α-error) and that adult individuals were assigned almost exclusively to only one single tadpole genotype actually indicates that our resolution was good enough to overcome this problem. However, β-errors did occur in some of the tested programs (see Table 2), which potentially could have resulted from a high genotyping error rate. But when comparing genotypes of known corresponding juvenile–adult pairs, we found that most of the ‘mismatches’ were actually due to amplification failure (i.e. missing loci) rather than genotyping errors (genotyping error rate of 0.007; Table S1, Supporting information). Thus, we conclude that the β-error rate actually reflects the large impact of missing loci on the assignment success of the various programs. While the programs that use a full genotype approach evaluate the similarity between two sample genotypes locus by locus (or allele by allele), the relatedness-based programs also incorporate the entire allelic frequencies in their computation and are thus apparently less susceptible to missing data. Most programs succeeded in correctly identifying corresponding juvenile and adult genotypes, while the matching between adults and tadpoles was less satisfying. We hypothesize that the lower DNA concentrations in the fin clips of tadpole compared to toes of juveniles and adults may have resulted in lower amplification success in tadpoles, which ultimately might have led to the lower matching success rates for adults/juveniles and tadpoles.
Table 2.
Results of the genetic matching of juveniles and adult Allobates femoralis when using the reduced and the full data set, respectively
| Reduced dataset |
Full dataset |
|||||||
|---|---|---|---|---|---|---|---|---|
| Program | Correct | α-error | β-error | Correct | α-error | β-error | Trios | Adult-Tp |
| identity | 4/20 | 0 | 16 | n/a | n/a | n/a | n/a | n/a |
| genecap | 19/20 | 0 | 1 | 19/20 | 0 | 1 | 12/20 | 46/67 |
| genalex | 20/20 | 0 | 0 | n/a | n/a | n/a | n/a | n/a |
| allelematch | 19/20 | 0 | 1 | 19/20 | 0 | 1 | 11/20 | 36/67 |
| kingroup | 20/20 | 0 | 0 | 20/20 | 0 | 0 | 19/20 | 64/67 |
| ml-relate | 20/20 | 0 | 0 | 20/20 | 0 | 0 | 20/20 | 61/67 |
Correct, number of correctly identified juvenile–adult matches; α-error, false matches; β-error, undetected matches; Trios, number of correctly identified corresponding adult, juvenile and tadpole genotypes; Adult-Tp, Number of unambiguous singular adult–tadpole matches; n/a, not available.
Conclusions
In this study, we evaluated the use of microsatellite markers for genetic mark–recapture studies in amphibians across life cycle stages and tested the suitability of software packages available for this task. Repeated sampling and genotyping allowed us to match the microsatellite genotypes of tadpoles, juveniles and adult frogs in an introduced experimental island population of the Neotropical poison frog Allobates femoralis. We thereby overcame the long-standing problem of long-term monitoring of individual amphibians throughout their entire life cycle.
Our study demonstrates that microsatellite markers can be used to reliably track tadpoles through metamorphosis until maturity. The combination of genetic tagging and visual pattern matching proved to be extremely powerful in our study species, as the comparison of unique belly patterns was sufficient for individual identification of juvenile and adult individuals. The pairwise-relatedness-based approach, conducted with the programs kingroup and ml-relate, performed best with our data set, as they accounted best for genotyping errors and missing values. To the best of our knowledge, this is the first study that successfully and completely tracked individual tadpoles until maturity. This data set of individually unique microsatellite genotypes will be used in further analyses to assess factors that determine fitness, in terms of survival and reproductive success in this semi-natural closed study population of A. femoralis. Furthermore, this approach will enable us to reconstruct dispersal trajectories from the natal pool of tadpole development over juvenile encounter locations to the final territory and perching sites of males and females.
Supplementary Material
Fig. S1 Ventral patterns of all corresponding juveniles and adults.
Table S1 Genotype comparisons of all corresponding juveniles and adults.
Acknowledgements
This study was financed by the Austrian Science Fund (FWF) through the project P24788-B 22 (PI Eva Ringler). The authors are grateful to Walter Hödl, Andrius Pašukonis, Kristin Gyimesi, Gerhard Rainer and students of the field course 2013 who assisted with fieldwork in French Guiana. Thanks to Gesche Westphal-Fitch for language editing.
Footnotes
Data Accessibility The full data set with all microsatellite genotypes of tadpole, juvenile and adult A. femoralis is available from Dryad: doi:10.5061/dryad.db800.
Supporting Information Additional Supporting Information may be found in the online version of this article:
References
- Altwegg R, Reyer H. Patterns of natural selection on size at metamorphosis in water frogs. Evolution. 2003;57:872–882. doi: 10.1111/j.0014-3820.2003.tb00298.x. [DOI] [PubMed] [Google Scholar]
- Andreou D, Vacquie-Garcia J, Cucherousset J, Blanchet S, Gozlan RE, Loot G. Individual genetic tagging for teleosts: an empirical validation and a guideline for ecologists. Journal of Fish Biology. 2012;80:181–194. doi: 10.1111/j.1095-8649.2011.03165.x. [DOI] [PubMed] [Google Scholar]
- Beckmann JS, Soller M. Toward a unified approach to genetic mapping of eukaryotes based on sequence tagged microsatellite sites. Nature Biotechnology. 1990;8:930–932. doi: 10.1038/nbt1090-930. [DOI] [PubMed] [Google Scholar]
- Blouin MS. DNA-based methods for pedigree reconstruction and kinship analysis in natural populations. Trends in Ecology & Evolution. 2003;18:503–511. [Google Scholar]
- Bolger DT, Morrison TA, Vance B, Lee D, Farid H. A computer-assisted system for photographic mark–recapture analysis. Methods in Ecology and Evolution. 2012;3:813–822. [Google Scholar]
- Boulanger J, McLellan BN, Woods JG, Proctor MF, Strobeck C. Sampling design and bias in DNA-based capture-mark-recapture population and density estimates of Grizzly Bears. Journal of Wildlife Management. 2004;68:457–469. [Google Scholar]
- Brockes JP. Amphibian limb regeneration: rebuilding a complex structure. Science. 1997;276:81–87. doi: 10.1126/science.276.5309.81. [DOI] [PubMed] [Google Scholar]
- Brockes JP, Kumar A. Plasticity and reprogramming of differentiated cells in amphibian regeneration. Nature Reviews Molecular Cell Biology. 2002;3:566–574. doi: 10.1038/nrm881. [DOI] [PubMed] [Google Scholar]
- Caorsi VZ, Santos RR, Grant T. Clip or snap? An evaluation of toe-clipping and photo-identification methods for identifying individual Southern Red-Bellied Toads, Melanophryniscus cambaraensis. South American Journal of Herpetology. 2012;7:79–84. [Google Scholar]
- Carlson BE, Langkilde T, Foster S. A common marking technique affects tadpole behavior and risk of predation. Ethology. 2013;119:167–177. [Google Scholar]
- Clark DR. Branding as a marking technique for amphibians and reptiles. Copeia. 1971;1971:148. [Google Scholar]
- Courtois EA, Lelong C, Calvez O, Loyau A, Schmeller DS. The use of visible implant alpha tags for anuran tadpoles. Herpetological Review. 2013;44:230–233. [Google Scholar]
- Creel S, Spong G, Sands JL, et al. Population size estimation in Yellowstone wolves with error-prone noninvasive microsatellite genotypes. Molecular Ecology. 2003;12:2003–2009. doi: 10.1046/j.1365-294x.2003.01868.x. [DOI] [PubMed] [Google Scholar]
- Davis TM, Ovaska K. Individual recognition of amphibians: effects of toe clipping and fluorescent tagging on the salamander Plethodon vehiculum. Journal of Herpetology. 2001;35:217. [Google Scholar]
- Donnelly MA, Guyer C, Juterbock JE, Alford RA. Techniques for marking amphibians. In: Heyer WR, Donnelly MA, McDiarmid RW, Hayek LC, Foster MS, editors. Measuring and Monitoring Biological Diversity. Standard Methods for Amphibians. Smithsonian Institution Press; Washington, District of Columbia: 1994. pp. 277–284. [Google Scholar]
- Dussault C, Rodríguez MA. Field trials of marking stream salmonids by dye injection and coded-wire-tagging. North American Journal of Fisheries Management. 1997;17:451–456. [Google Scholar]
- Ferner JW. A review of marking techniques for amphibians and reptiles. Herpetological Circular. 1979;9:1–41. [Google Scholar]
- Frederick JL. Evaluation of fluorescent elastomer injection as a method for marking small fish. Bulletin of Marine Science. 1997;61:399–408. [Google Scholar]
- Funk WC, Donnelly MA, Lips KR. Alternative views of amphibian toe-clipping. Nature. 2005;433:193. doi: 10.1038/433193c. [DOI] [PubMed] [Google Scholar]
- Galpern P, Manseau M, Hettinga P, Smith K, Wilson P. Allele-match: an R package for identifying unique multilocus genotypes where genotyping error and missing data may be present. Molecular Ecology Resources. 2012;12:771–778. doi: 10.1111/j.1755-0998.2012.03137.x. [DOI] [PubMed] [Google Scholar]
- Gibbons JW, Andrews KM. PIT tagging: simple technology at its best. BioScience. 2004;54:447. [Google Scholar]
- Goossens B, Waits LP, Taberlet P. Plucked hair samples as a source of DNA: reliability of dinucleotide microsatellite genotyping. Molecular Ecology. 1998;7:1237–1241. doi: 10.1046/j.1365-294x.1998.00407.x. [DOI] [PubMed] [Google Scholar]
- Grant EHC. Visual implant elastomer mark retention through metamorphosis in amphibian larvae. Journal of Wildlife Management. 2008;72:1247–1252. [Google Scholar]
- Hoffman JI, Trathan PN, Amos W. Genetic tagging reveals extreme site fidelity in territorial male Antarctic fur seals Arctocephalus gazella. Molecular Ecology. 2006;15:3841–3847. doi: 10.1111/j.1365-294X.2006.03053.x. [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Wilson V, Thein SL. Individual-specific ‘fingerprints’ of human DNA. Nature. 1985;316:76–79. doi: 10.1038/316076a0. [DOI] [PubMed] [Google Scholar]
- Jehle R, Arntzen JW. Review: microsatellite markers in amphibian conservation genetics. Herpetological Journal. 2002;12:1–9. [Google Scholar]
- Jehle R, Hödl W. PITs versus patterns: effects of transponders on recapture rate and body condition of Danube crested newts (Triturus dobrogicus) and common spadefoot toads (Pelobates fuscus) Herpetological Journal. 1998;8:181–186. [Google Scholar]
- Jehle R, Gasser H, Pfunders M, Amézquita A, Lima Pimentel A, Hödl W. Ten polymorphic microsatellite loci for Allobates femoralis, an Amazonian dendrobatoid frog. Molecular Ecology Resources. 2008;8:1326–1328. doi: 10.1111/j.1755-0998.2008.02304.x. [DOI] [PubMed] [Google Scholar]
- Jung RE, Dayton GH, Williamson SJ, Sauer JR, Droege S. An evaluation of population index and estimation techniques for tadpoles in desert pools. Journal of Herpetology. 2002;36:465. [Google Scholar]
- Kalinowski ST, Wagner AP, Taper ML. ML-relate: a computer program for maximum likelihood estimation of relatedness and relationship. Molecular Ecology Notes. 2006;6:576–579. [Google Scholar]
- Kalinowski ST, Taper ML, Marshall TC. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Molecular Ecology. 2007;16:1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x. [DOI] [PubMed] [Google Scholar]
- Kolodziej K, Theissinger K, Brün J, Schulz HK, Schulz R. Determination of the minimum number of microsatellite markers for individual genotyping in wild boar (Sus scrofa) using a test with close relatives. European Journal of Wildlife Research. 2012;58:621–628. [Google Scholar]
- Kolodziej K, Schulz HK, Theissinger K, Ebert C, Hohmann U, Schulz R. Comparison of established methods for quantifying genotyping error rates in wildlife forensics. Conservation Genetics Resources. 2013;5:287–292. [Google Scholar]
- Konovalov DA, Heg D. A maximum-likelihood relatedness estimator allowing for negative relatedness values. Molecular Ecology Resources. 2008;8:256–263. doi: 10.1111/j.1471-8286.2007.01940.x. [DOI] [PubMed] [Google Scholar]
- Konovalov DA, Manning C, Henshaw MT. kingroup: a program for pedigree relationship reconstruction and kin group assignments using genetic markers. Molecular Ecology Notes. 2004;4:779–782. [Google Scholar]
- Lebreton J, Burnham KP, Clobert J, Anderson DR. Modeling survival and testing biological hypotheses using marked animals: a unified approach with case studies. Ecological Monographs. 1992;62:67. [Google Scholar]
- Lukacs PM, Burnham KP. Review of capture-recapture methods applicable to noninvasive genetic sampling. Molecular Ecology. 2005;14:3909–3919. doi: 10.1111/j.1365-294X.2005.02717.x. [DOI] [PubMed] [Google Scholar]
- Martin RA. Evaluating a novel technique for individual identification of anuran tadpoles using coded wire tags. Herpetological Conservation and Biology. 2011;6:155–160. [Google Scholar]
- May RM. Ethics and amphibians. Nature. 2004;431:403. doi: 10.1038/431403a. [DOI] [PubMed] [Google Scholar]
- McCarthy MA, Parris KM. Clarifying the effect of toe clipping on frogs with Bayesian statistics. Journal of Applied Ecology. 2004;41:780–786. [Google Scholar]
- Montanarin A, Kaefer IL, Lima Pimentel A. Courtship and mating behaviour of the Brilliant-thighed Frog Allobates femoralis from Central Amazonia: implications for the study of a species complex. Ethology Ecology & Evolution. 2011;23:141–150. [Google Scholar]
- Nichols JD. Capture-recapture models. BioScience. 1992;42:94–102. [Google Scholar]
- Nietfeld MT, Barrett MW, Silvy N. Wildlife marking techniques. In: Bookhout TA, editor. Research and Management Techniques for Wildlife and Habitats. The Wildlife Society; Bethesda, Maryland: 1994. pp. 140–168. [Google Scholar]
- Olsen EM, Vøllestad LA. An evaluation of visible implant elastomer for marking age-0 Brown Trout. North American Journal of Fisheries Management. 2001;21:967–970. [Google Scholar]
- Paetkau D. The optimal number of markers in genetic capture-mark-recapture studies. Journal of Wildlife Management. 2004;68:449–452. [Google Scholar]
- Palsbøll PJ. Genetic tagging: contemporary molecular ecology. Biological Journal of the Linnean Society. 1999;68:3–22. [Google Scholar]
- Palsboll PJ, Allen J, Berube M, et al. Genetic tagging of humpback whales. Nature. 1997;388:767–769. doi: 10.1038/42005. [DOI] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradel R. Utilization of capture-mark-recapture for the study of recruitment and population growth rate. Biometrics. 1996;52:703. [Google Scholar]
- Queller DC, Goodnight KF. Estimating relatedness using genetic markers. Evolution. 1989;43:258–275. doi: 10.1111/j.1558-5646.1989.tb04226.x. [DOI] [PubMed] [Google Scholar]
- Rew MB, Robbins J, Mattila D, Palsbøll PJ, Bérubé M. How many genetic markers to tag an individual? An empirical assessment of false matching rates among close relatives. Ecological Applications. 2011;21:877–887. doi: 10.1890/10-0348.1. [DOI] [PubMed] [Google Scholar]
- Ribeiro J, Rebelo R. Survival of Alytes cisternasii tadpoles in stream pools: a capture-recapture study using photo-identification. Amphibia-Reptilia. 2011;32:365–374. [Google Scholar]
- Ringler M, Ursprung E, Hödl W. Site fidelity and patterns of short- and long-term movement in the Brilliant-thighed Poison Frog Allobates femoralis (Aromobatidae) Behavioral Ecology and Sociobiology. 2009;63:1281–1293. [Google Scholar]
- Ringler M, Ringler E, Magaña Mendoza D, Hödl W. Intrusion experiments to measure territory size: development of the method, tests through simulations, and application in the frog Allobates femoralis. PLoS One. 2011;6:e25844. doi: 10.1371/journal.pone.0025844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringler E, Ringler M, Jehle R, Hödl W. The female perspective of mating in A. femoralis, a territorial frog with paternal care—a spatial and genetic analysis. PLoS One. 2012;7:e40237. doi: 10.1371/journal.pone.0040237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringler E, Pašukonis A, Hödl W, Ringler M. Tadpole transport logistics in a Neotropical poison frog: indications for strategic planning and adaptive plasticity in anuran parental care. Frontiers in Zoology. 2013a;10:1–10. doi: 10.1186/1742-9994-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringler E, Pašukonis A, Hödl W, Ringler M. Characterization of seven new polymorphic microsatellite loci in the Brilliant-thighed Poison Frog Allobates femoralis (Dendrobatidae), and their cross-species utility in three other dendrobatid species. Herpetological Journal. 2013b;23:175–178. [PMC free article] [PubMed] [Google Scholar]
- Ringler M, Mangione R, Pašukonis A. High-resolution forest mapping for behavioural studies in the Nature Reserve ‘Les Nouragues’, French Guiana. Journal of Maps. 2014 doi: 10.1080/17445647.2014.972995. doi:10.1080/17445647.2014.972995 (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roithmair ME. Territoriality and male mating success in the dart-poison frog, Epipedobates femoralis (Dendrobatidae, Anura) Ethology. 1992;92:331–343. [Google Scholar]
- Schaub M, Pradel R, Jenni L, Lebreton J. Migrating birds stop over longer than usually thought: an improved capture-recapture analysis. Ecology. 2001;82:852. [Google Scholar]
- Schmidt BR, Schaub M, Anholt BR. Why you should use capture-recapture methods when estimating survival and breeding probabilities: on bias, temporary emigration, overdispersion, and Common Toads. Amphibia-Reptilia. 2002;23:375–388. [Google Scholar]
- Selkoe KA, Toonen RJ. Microsatellites for ecologists: a practical guide to using and evaluating microsatellite markers. Ecology Letters. 2006;9:615–629. doi: 10.1111/j.1461-0248.2006.00889.x. [DOI] [PubMed] [Google Scholar]
- Sinsch U. Postmetamorphic dispersal and recruitment of first breeders in a Bufo calamita metapopulation. Oecologia. 1997;112:42–47. doi: 10.1007/s004420050281. [DOI] [PubMed] [Google Scholar]
- Sloane MA, Sunnucks P, Alpers D, Beheregaray LB, Taylor AC. Highly reliable genetic identification of individual Northern Hairynosed Wombats from single remotely collected hairs: a feasible censusing method. Molecular Ecology. 2000;9:1233–1240. doi: 10.1046/j.1365-294x.2000.00993.x. [DOI] [PubMed] [Google Scholar]
- Smith DC. Adult recruitment in chorus frogs: effects of size and date at metamorphosis. Ecology. 1987;68:344. [Google Scholar]
- Taberlet P, Luikart G. Non-invasive genetic sampling and individual identification. Biological Journal of the Linnean Society. 1999;68:41–55. [Google Scholar]
- Taberlet P, Camarra J, Griffin S, et al. Noninvasive genetic tracking of the endangered Pyrenean Brown Bear population. Molecular Ecology. 1997;6:869–876. [PubMed] [Google Scholar]
- Tautz D, Renz M. Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucleic Acids Research. 1984;12:4127–4138. doi: 10.1093/nar/12.10.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger SD, Burgmeier NG, Williams RN. Genetic markers reveal high PIT tag retention rates in Giant Salamanders (Cryptobranchus alleganiensis) Amphibia-Reptilia. 2012;33:313–317. [Google Scholar]
- Ursprung E, Ringler M, Jehle R, Hödl W. Strong male/male competition allows for nonchoosy females: high levels of polygynandry in a territorial frog with paternal care. Molecular Ecology. 2011a;20:1759–1771. doi: 10.1111/j.1365-294X.2011.05056.x. [DOI] [PubMed] [Google Scholar]
- Ursprung E, Ringler M, Jehle R, Hödl W. Toe regeneration in the neotropical frog Allobates femoralis. Herpetological Journal. 2011b;21:83–86. [Google Scholar]
- Uthicke S, Benzie JAH. A genetic fingerprint recapture technique for measuring growth in ‘unmarkable’ invertebrates: negative growth in commercially fished holothurians (Holothuria nobilis) Marine Ecology Progress Series. 2002;241:221–226. [Google Scholar]
- Vonesh JR, De la Cruz O. Complex life cycles and density dependence: assessing the contribution of egg mortality to amphibian declines. Oecologia. 2002;133:325–333. doi: 10.1007/s00442-002-1039-9. [DOI] [PubMed] [Google Scholar]
- Wagner HW, Sefc KM. IDENTITY. Centre for Applied Genetics. University of Agricultural Sciences; Vienna, Austria: 1999. http://www.uni-graz.at/~sefck/ [Google Scholar]
- Wagner AP, Creel S, Kalinowski ST. Estimating relatedness and relationships using microsatellite loci with null alleles. Heredity. 2006;97:336–345. doi: 10.1038/sj.hdy.6800865. [DOI] [PubMed] [Google Scholar]
- Waits LP, Luikart G, Taberlet P. Estimating the probability of identity among genotypes in natural populations: cautions and guidelines. Molecular Ecology. 2001;10:249–256. doi: 10.1046/j.1365-294x.2001.01185.x. [DOI] [PubMed] [Google Scholar]
- Wilberg MJ, Dreher BP. genecap: a program for analysis of multilocus genotype data for non-invasive sampling and capture-recapture population estimation. Molecular Ecology Notes. 2004;4:783–785. [Google Scholar]
- Wilson CD, Arnott G, Reid N, Roberts D. The pitfall with PIT tags: marking freshwater bivalves for translocation induces short-term behavioural costs. Animal Behaviour. 2011;81:341–346. [Google Scholar]
- Wood MD, Slade NA. Comparison of ear-tagging and toe-clipping in Prairie Voles, Microtus ochrogaster. Journal of Mammalogy. 1990;71:252. [Google Scholar]
- Woods JG, Paetkau D, Lewis D, McLellan BN, Proctor M, Strobeck C. Genetic tagging of free-ranging Black and Brown Bears. Wildlife Society Bulletin. 1999;27:616–627. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Ventral patterns of all corresponding juveniles and adults.
Table S1 Genotype comparisons of all corresponding juveniles and adults.