Abstract
Background
The 20–25% of the inflammatory bowel disease (IBD) patients presents the disease before the age of 18–20, with worse extent and severity, compared to adult-onset IBD. We sought to identify the differential expression of microRNAs in pediatric ulcerative colitis (UC) and their association with different clinical phenotypes.
Methods
MicroRNA expression analysis was performed in colonic tissues derived from pediatric UC patients and no-IBD controls. MiR-4284 levels were verified by real-time quantitative PCR in two additional cohorts of pediatric UC patients. Bioinformatics analysis was performed to predict the targets of miR-4284. In vitro experiments using luciferase reporter assays and real-time PCR evaluated the direct effect of miR-4284 on CXCL5 mRNA. In vivo experiments were performed in two mouse models of experimental colitis.
Results
A 24-microRNA signature was identified in colonic tissues derived from pediatric-UC patients. The most down-regulated microRNA in the tissue of pediatric UC patients, relative to non-IBD controls, was miR-4284. In situ hybridization revealed that miR-4284 is present in colonic epithelial cells and its levels correlate with the disease activity. Furthermore, we found that miR-4284 regulates CXCL5 mRNA expression through binding to its 3’UTR. CXCL5 had increased mRNA levels in colonic tissue from pediatric-UC patients and correlated with disease activity. Furthermore, we found an inverse correlation between miR-4284 and CXCL5 levels in the colonic pediatric UC tissues and in two mouse models of experimental colitis.
Conclusions
Our data reveal a novel microRNA pediatric-UC signature, provide evidence that miR-4284 directly regulates CXCL5 and correlates with the disease activity.
Keywords: non-coding RNA, colonic mucosa, inflammatory bowel disease
Introduction
The pathogenesis of inflammatory bowel diseases (IBD) involves the response of the immune system to intestinal microbiota in genetically predisposed individuals. The genetic component of this predisposition has been intensely investigated but only a small proportion of the disease can be attributed to the identified genetic factors.1, 2 Moreover, 25% of IBD patients present during childhood, and have significantly greater disease extent, severity and worse progression compared to adult-onset IBD.3 This population also has fewer co-morbid conditions providing an advantage in investigating the genetic component of the disease.
MicroRNAs are small non-coding RNA oligonucleotides that add an additional mode of regulation of gene expression and have been involved in the pathogenesis of a variety of human inflammatory diseases.4–5 We recently identified microRNAs (let-7 and miR-155) that are essential regulators of Toll-like receptor signaling.5 Additionally, we reported that microRNAs miR-21 and miR-181b target the signal transducer and activator of transcription 3 (STAT3), a known major factor activated during the inflammatory response.6
Previous studies have identified specific microRNAs to be deregulated in the context of IBD.7 MiR-192 levels, which can inhibit chemokine production, are decreased in tissue samples from ulcerative colitis (UC) patients.8 In addition, certain microRNAs are deregulated in adult ileal and colonic Crohn’s disease (CD).9 Very recently, a study identified a number of deregulated microRNAs in the rectal tissue of pediatric patients.10 Our recent studies revealed that miR-124 is epigenetically regulated specifically in pediatric patients with active UC and this leads to increased STAT3 activity in the colonic tissue of these patients.11 However, further investigation is needed to define the role and function of the microRNA-signature specific to pediatric UC patients.
In the present study, microRNA profiling analysis was performed and identified miR-4284 as the most down-regulated microRNA in colonic biopsies from pediatric UC patients. This deregulation was validated in additional cohorts of patients and was associated with the disease activity. Furthermore, we present evidence that miR-4284 directly targets CXCL5, binding to the 3’ untranslated region (3’UTR) of CXCL5 mRNA. CXCL5 is known to participate in the inflammatory response of colonic epithelial cells by facilitating the recruitment of neutrophils12, 13 and has been previously implicated in the pathogenesis of UC.14, 15 This miR-4284/CXCL5 deregulation identified in pediatric UC patients was also replicated in two mouse models of experimental colitis.
Materials and Methods
Study Population/Patient Characteristics
Pediatric tissue samples were obtained from subjects enrolled in the Massachusetts General Hospital for Children Pediatric IBD bio-repository with informed consent/assent from subjects or their legal guardians and approval by the Partners Health Care Institutional Review Board (Protocol #2009P001287). Sigmoid colon mucosal biopsies were collected at the time of colonoscopy, snap frozen in liquid nitrogen and stored at −80 °C. Matched sigmoid biopsies were also obtained and sent to pathology for routine histologic evaluation. Sigmoid biopsies from a total of 26 subjects diagnosed with IBD before the age of 17, and 19 non-IBD controls, were analyzed. The IBD group included 18 pediatric subjects with active-UC and 8 with inactive disease. Patient clinical characteristics are listed in Tables 1–3.
TABLE 1.
Clinical characteristics of patients for microRNA profiling.
| Clinical characteristics | 1* | 2* | 3* | 4 | 5* | 6 | 7* | 8* | 9* | 10* |
|---|---|---|---|---|---|---|---|---|---|---|
| Diagnosis | UC | UC | UC | UC | UC | No IBD | No IBD | No IBD | No IBD | No IBD |
| IBD Treatment Naïve | No | No | No | Yes | Yes | N/A | N/A | N/A | N/A | N/A |
| Age at collection | 16 | 17 | 16 | 18 | 14 | 18 | 7 | 10 | 18 | 16 |
| Gender | Female | Male | Female | Male | Female | Male | Male | Female | Male | Female |
| Gross sigmoid disease | Yes | Yes | Yes | N/A | Yes | N/A | N/A | N/A | N/A | No |
| Microscopic Sigmoid inflammation |
Yes | Yes | Yes | N/A | Yes | N/A | N/A | N/A | N/A | Yes, focal active, no chronicity |
| IBD Flare (clinical) | Yes | No | Yes | N/A | Yes | N/A | N/A | N/A | N/A | N/A |
| Paris Disease Location (UC)a | E1 | E4 | E2 | N/A | E3 | N/A | N/A | N/A | N/A | N/A |
| UC Paris Disease severityb | S0 | S1 | S0 | N/A | S1 | N/A | N/A | N/A | N/A | N/A |
| Paris Age at IBD Diagnosisc | A1a | A2 | A1b | N/A | A1b | N/A | N/A | N/A | N/A | N/A |
| IBD Medications | Mesalamine, prednisone |
Prednisone, mesalamine, metronidazole |
Mesalamine, balsalazide |
None | None | N/A | N/A | N/A | N/A | N/A |
| PUCAId (on day of collection) | 30, PUCAI | 0, PUCAI | 35, PUCAI | N/A | 70, PUCAI | N/A | N/A | N/A | N/A | N/A |
E1 - Ulcerative proctitis, E2 - Left sided (distal to splenic flexure), E3 - Extensive (hepatic flexure distally), E4 - Pancolitis (proximal to hepatic fixture), N/A: Not available or not applicable (if no IBD)
S0: never severe, S1: ever severe (PUCAI > or = 65 ever), N/A: Not available or Not applicable (if no IBD)
A1a: 0–9 years old (yo), A1b: 10–16 yo, A2: >16 yo, N/A = Not available or Not applicable (if no IBD)
Pediatric Ulcerative Colitis Activity Index
Samples used for subsequent validation by rt-PCR
TABLE 3.
Clinical characteristics of the cohort of pediatric patients with Inactive or Active ulcerative colitis, provided by the Pediatric IBD Center, Mass General Hospital for Children.
| Sample # | IBD Treatment Naïve |
Age at collection |
Gender | Hispanic/non | Race | Gross sigmoid disease |
Paris Disease Location |
PUCAI (on day of sample collection) |
|---|---|---|---|---|---|---|---|---|
| Active UC | ||||||||
| 1 | No | 23 | Female | Non-Hispanic | White | Yes (mild) | E4 | 0 |
| 2 | No | 12 | Female | Non-Hispanic | White | yes | E4 | 30 |
| 3 | No | 21 | Male | non-Hispanic | White | yes | E4 | 60 |
| 4 | No | 10 | Female | Non-Hispanic | White/Asian Indian |
yes | E4 | 25 |
| 5 | No | 11 | Female | Hispanic | White | Yes-mild chronic | E4 | Unable to ascertain |
| 6 | Yes | 17 | Male | Non-Hispanic | White | Yes | E3 | 35 |
| 7 | Yes | 13 | Female | Non-Hispanic | White | Yes | E4 | 45 |
| 8 | No | 16 | Male | Non-Hispanic | White/Asian | yes | E4 | 40 |
| Inactive UC | ||||||||
| 1 | No | 19 | Male | Non-Hispanic | White | No | E3 | 0 |
| 2 | No | 17 | Male | Non-Hispanic | White | Yes (mild) | E4 | 0 |
| 3 | No | 22 | Female | Non-Hispanic | White | No | E4 | 15 |
| 4 | No | 21 | Male | Non-Hispanic | White | No | E3 | 0 |
| 5 | No | 22 | Female | Non-Hispanic | White | No | E4 | 0 |
| 6 | No | 25 | Male | Non-Hispanic | White | No | E4 | 0 |
| 7 | Yes | 14 | Female | Non-Hispanic | White | No | E4 | 0 |
| 8 | No | 17 | Male | Non-Hispanic | White | No | E4 | 5 |
Colonic biopsies from adult patients with active (n = 16) or inactive UC (n = 16) were obtained from the bio-repository of the Department of Gastroenterology and Hepatology, Leiden University Medical Center (see patient clinical phenotypes, Table 4).
TABLE 4.
Clinical characteristics of the cohort comprised of adult patients with Inactive (N1-N16) or Active (I1-I16) disease.
| Sample | Gender | Age | Remarks |
|---|---|---|---|
| N1 | Female | 73 | CECUM |
| N2 | Male | 40 | COL. ASCENDING |
| N3 | Female | 42 | CECUM |
| N4 | Female | 41 | CECUM |
| N5 | Male | 26 | RECTUM |
| N6 | Male | 50 | COL. ASCENDING |
| N7 | Female | 40 | COLON |
| N8 | Female | 40 | COLON |
| N9 | Male | 45 | COLON |
| N10 | Male | 66 | COLON |
| N11 | Male | 29 | COLON |
| N12 | Female | 73 | COLON ASCENDING. |
| N13 | Male | 60 | COLON |
| N14 | Male | 65 | COLON |
| N15 | Male | 58 | COLON |
| N16 | Male | 48 | COLON |
| I1 | Female | 73 | COL.TRANSVERSE |
| I2 | Male | 40 | COL. DESCENDING |
| I3 | Female | 42 | COL.ASCENDING |
| I4 | Female | 41 | COL. DESCENDING |
| I5 | Male | 26 | COL. DESCENDING |
| I6 | Male | 50 | COL. DESCENDING |
| I7 | Female | 40 | COLON |
| I8 | Female | 40 | COLON |
| I9 | Male | 45 | COLON |
| I10 | Male | 66 | COLON |
| I11 | Male | 29 | COLON |
| I12 | Female | 73 | COLON ASCENDING. |
| I13 | Male | 60 | COLON |
| I14 | Male | 65 | COLON |
| I15 | Male | 58 | COLON |
| I16 | Male | 48 | COLON |
Matched biopsies from inflamed and non-inflamed areas of the colon of pediatric patients (n = 5) with UC were obtained from the IBD Biobank of the UCLA, Center for Inflammatory Bowel Diseases (see patient clinical phenotypes, Table 5).
TABLE 5.
Clinical characteristics of ulcerative colitis pediatric patients with matched biopsies from non-inflamed and inflamed areas.
| Patient | Non-Inflamed | Inflamed | Age | Gender |
|---|---|---|---|---|
| 1 | Terminal ileum | Terminal ileum | 13 | Female |
| 2 | Terminal ileum | Cecum | 13 | Female |
| 3 | Terminal ileum | Rectum | 17 | Male |
| 4 | Terminal ileum | Cecum | 17 | Male |
| 5 | Terminal ileum | Rectum | 16 | Female |
RNA isolation
RNA was extracted from colonic biopsies and mouse colonic tissues, after homogenization using Trizol® (Invitrogen), according to the manufacturer’s instructions, with some modifications. Specifically, two extra washing steps of the RNA pellet with 70% ethanol were added, in order to ensure minimal phenol contamination of the sample. The purity of the samples was evaluated by estimating the ratios of A260nm/A230nm and A260nm/A280nm for phenol and protein contamination, respectively, using a microplate spectrophotometer (Synergy HT, BioTek); only samples with ratios ≥2.0 were used in our analyses. Furthermore, the integrity of the RNA was evaluated by the Bioanalyzer 2100 (Agilent) and samples with RNA integrity number (RIN) higher than 7.0 were included in the study.
In situ hybridization
For the localization of the microRNA miR-4284, paraffin embedded sections of biopsies provided by MGH biorepository were used for in situ hybridization. Custom miRCURY LNA™ microRNA detection probes for miR-4284 were designed and labeled with DIG at both 3’- and 5’-ends, and used according to the manufacturer’s instructions (Exiqon).
Cell transfections
Cells were seeded in 6 well plates (250,000 cells/well) and transfected with the mirVana miR-4284 mimic (Ambion) or its negative control, at a final concentration of 50 nM, using the transfection reagent FuGENE6 (Promega), according to the manufacturer’s guidelines. Cells were harvested 42 hours post-transfection and RNA was isolated.
Luciferase reporter plasmid psiCheck2-CXCL5-3’UTR carrying the 3’UTR of CXCL5 was used. Sequential transfection was performed; first, the reporter plasmid (4 µg plasmid per 1×106 cells) was transfected using FuGENE6 (Promega) and 24 hours later, cells were transfected with a final concentration of 50 nM miRNA mimics/inhibitors or the respective negative controls, provided by the manufacturer, as described above.
MicroRNA expression analysis
MicroRNA expression analysis was performed in sigmoid biopsies from 5 pediatric UC patients and 5 non-IBD pediatric patients (controls) using the miRCURY microRNA Array Profiling (Exiqon Inc.). These samples were provided from the bio-repository at MassGeneral Hospital for Children Pediatric inflammatory bowel disease. Primer sets for the microRNAs, as well as primers for the reference genes U6 snRNA and 5S rRNA, were provided by Exiqon and used according to manufacturer’s instructions (Exiqon Inc.).
Real-time quantitative PCR analysis
mRNA levels were assessed by real-time PCR on a CFX384 detection system (Bio-Rad). Real-time PCR for the mRNA levels of CXCL5, Cxcl5 and GAPDH was performed using iQ SYBR Green supermix (BioRad). Primer sequences are provided in Supplementary Data.
Bioinformatics analysis
Bioinformatics analyses were performed using microRNA databases Targetscan16 (www.targetscan.org, release 6.2) and microRNA.org17 (http://www.microrna.org, August 2010 release), based on the sequence of hsa-miR-4284.
Mouse models of experimental colitis
Eight to ten weeks old, C57BL/6 male mice (The Jackson Laboratory, ME) were treated with an established protocol for experimental chronic colitis. Specifically, for the dextran sulfate sodium (DSS)-colitis mouse model, mice were maintained on 3% DSS-water for 5 days with an interval of 14 days on regular water, provided ad libitum, for a total of three cycles. Mice were then sacrificed and colonic tissues were collected.
IL-10 knockout (IL10-KO) mice on the C57BL/6 background were purchased from Jackson Laboratory. For the experiments, 8 week-old male IL-10 KO mice and their wild type littermates were single housed and treated with piroxicam (Sigma) (80 mg per 250 gr of food) for two weeks. Piroxicam, is a non-steroidal anti-inflammatory drug used to accelerate the development of colitis in the genetically susceptible IL10-KO mice, as previously described.18 Tissues were harvested two weeks following piroxicam cessation.
Results
Identification of a 24-microRNA signature in colonic tissues from pediatric UC patients
We first sought to identify the microRNA profile of colonic tissues derived from pediatric UC patients as compared to non-IBD pediatric controls (Controls). A cohort of 10 sigmoid biopsies, five for each group, was used for this analysis (Table 1). Our analysis revealed 24 microRNAs to be significantly deregulated between the two groups and the average levels of differential expression are shown here on a heatmap format (Figure 1A). Interestingly, a novel microRNA, miR-4284, was identified as the most deregulated microRNA (~12-fold reduction) in the colonic biopsies of pediatric patients, compared to non-IBD controls.
Figure 1.
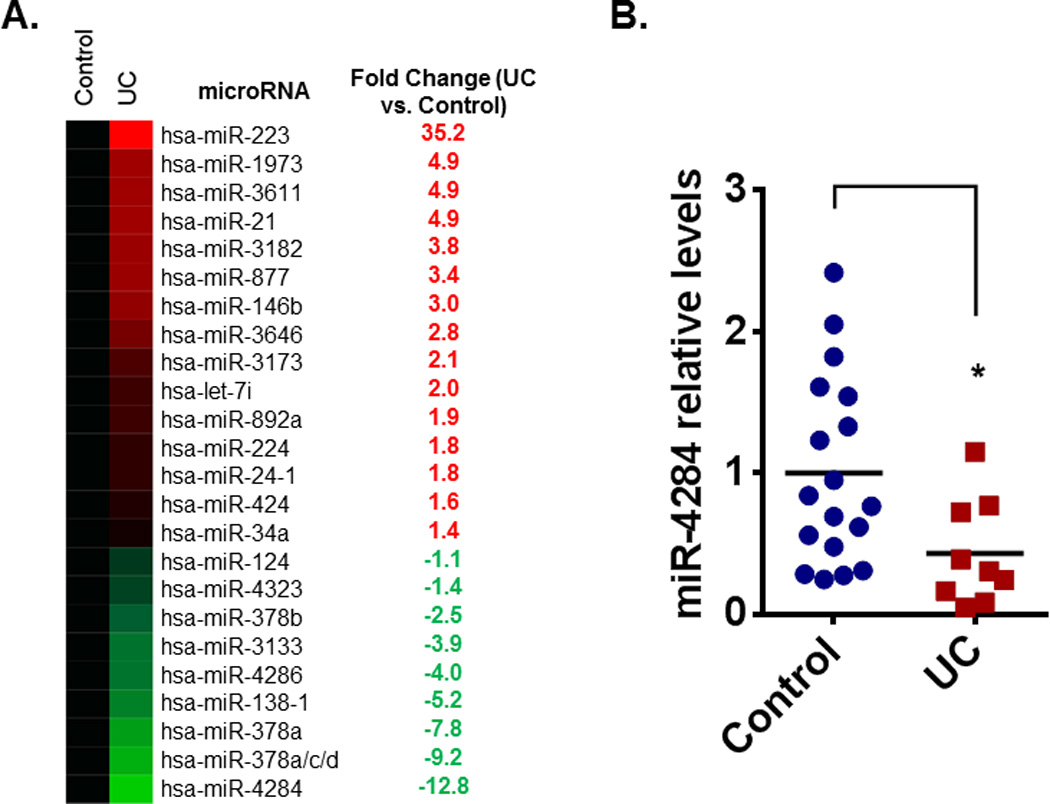
(A) MicroRNA profiling in biopsies from pediatric UC patients (UC, n = 5) revealed a signature of deferentially regulated microRNAs, as compared to non-IBD pediatric patients (Control, n = 5). Heatmap representation and the fold-change in the relative levels of each deregulated microRNA (red indicates up-regulation, green down-regulation). (B) Real-time PCR for miR-4284 levels was performed in biopsies from pediatric patients with no-IBD (Control, n = 18) and with UC (n = 9). Data are presented as scatter plots, lines represent the mean. *p < 0.05, t-test, Prism6 (GraphPad Software Inc.).
To verify the profiling results, we performed real-time qPCR analysis for miR-4284 in a second cohort of patient samples. Specifically, we used 18 colonic biopsies from non-IBD pediatric patients (control group) and 9 biopsies from pediatric patients with active UC. In agreement with the microRNA profiling data results, miR-4284 levels were significantly decreased in the pediatric UC samples, compared to controls (Figure 1B).
miR-4284 is specifically deregulated in pediatric UC and its levels correlate with disease activity
To further investigate the association of miR-4284 levels with disease activity we used a cohort comprised of biopsies from patients that were in remission (Inactive UC, n = 8) and patients with active UC (n = 8). RNA extracted from the biopsies was analyzed by real-time PCR and the levels of miR-4284 were estimated. We found that miR-4284 levels were significantly decreased in the active UC samples, with an observed 7-fold reduction, compared to the inactive UC controls (Inactive UC 0.15 vs. Active UC, set to 1.00) (Figure 2A).
Figure 2.
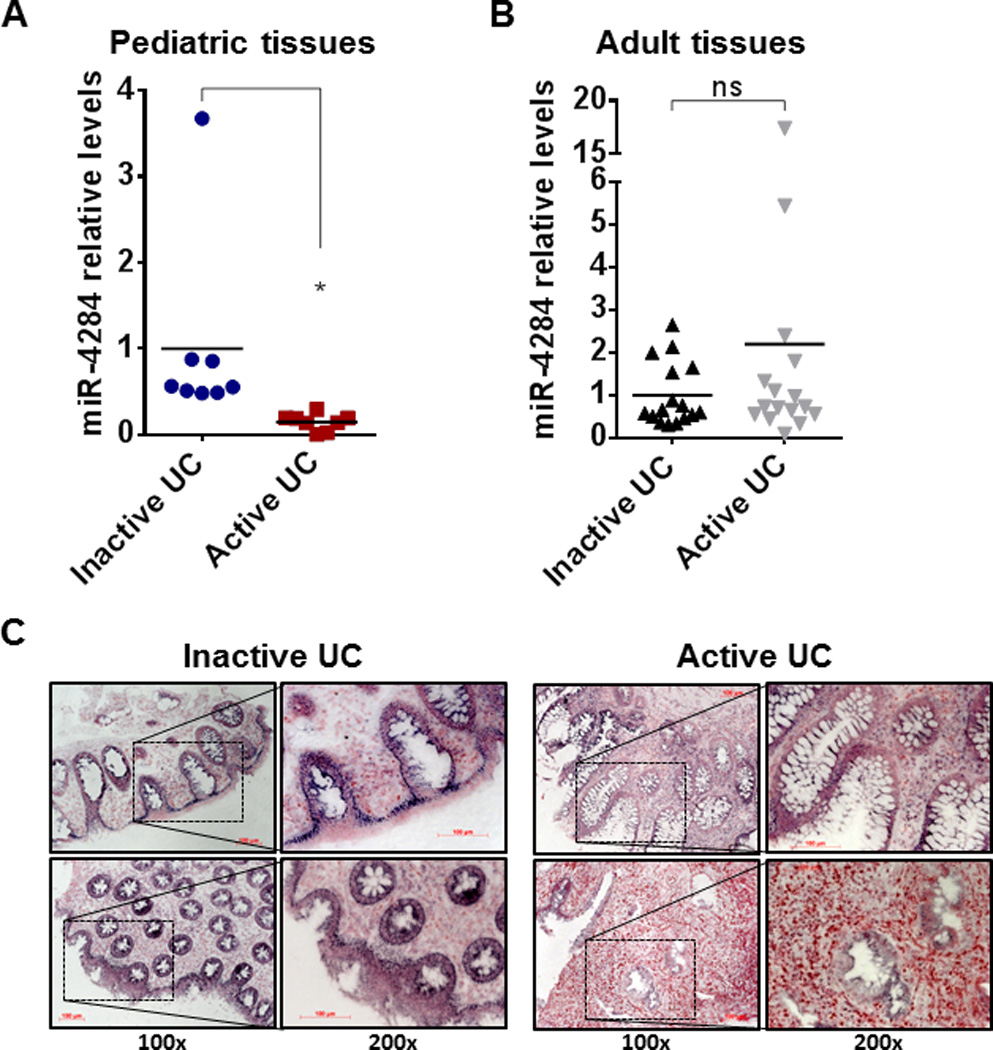
Real-time PCR for miR-4284 in biopsies from pediatric and adult patients with inactive or active UC. (A) Pediatric tissues: Inactive UC: n = 8, Active UC: n = 8 and (B) Adult tissues: Inactive UC: n = 16, Active UC: n = 16. Data are presented as scatter plots, line represents the mean. *p < 0.05, ns: not statistically significant, t-test, Prism6 (GraphPad Software Inc.). (C) In situ hybridization for miR-4284 (blue stain) in sections of colonic biopsies from pediatric patients with inactive or active UC, as indicated. Representative microscopy pictures are shown in magnification 100x and 200x (insert), as indicated. Scale bars represent 100 µm. Nuclear Fast Red was used for nuclear counter staining (red stain).
To address the specificity of this deregulation, we estimated the levels of miR-4284 in a group of adult patients in remission (Inactive adult, n = 16) and another group of adult patients with active UC (Active adult, n = 16). As shown in Figure 2B, there is no significant difference between the two groups in the adult tissues, indicating that the deregulation of miR-4284 is specific to pediatric UC patients.
The clinical characteristics of both of the cohorts described are available in Tables 3 and 4, respectively.
miR-4284 is localized in colonic epithelial cells derived from pediatric colonic tissues
To gain insight into the cell population in which miR-4284 deregulation takes place, we applied in situ hybridization on sections of biopsies from pediatric UC patients with inactive and active disease, using DIG-labeled probes specific for miR-4284. As shown in representative pictures (Figure 2C), miR-4284 (blue stain) is abundant specifically in the epithelial cells of the biopsies derived from patients with inactive UC, while it is not detected in the sections from patients with active UC. This provides strong evidence that the deregulation of miR-4284 that we identified in the previous analyses is mainly due to deregulation of this microRNA in the epithelial population of the colon of pediatric patients.
miR-4284 regulates directly CXCL5 expression through binding in its 3’UTR
Bioinformatics analysis using databases Targetscan (www.targetscan.org, release 6.2) and microRNA.org, based on the sequence of miR-4284, predicted possible targets of this microRNA. We selected candidate targets known to be involved in pathways relevant to IBD pathogenesis. Interestingly, CXCL5, a chemokine reported to be involved in the pathogenesis of adult UC14, was one of the predicted targets (Figure 3A). Furthermore, it has been previously shown that CXCL5 is mainly expressed in the colonic epithelial cells of UC patients.15 These observations make CXCL5 an attractive target for miR-4284 and can explain the role of this microRNA in the pathogenesis of the disease.
Figure 3.
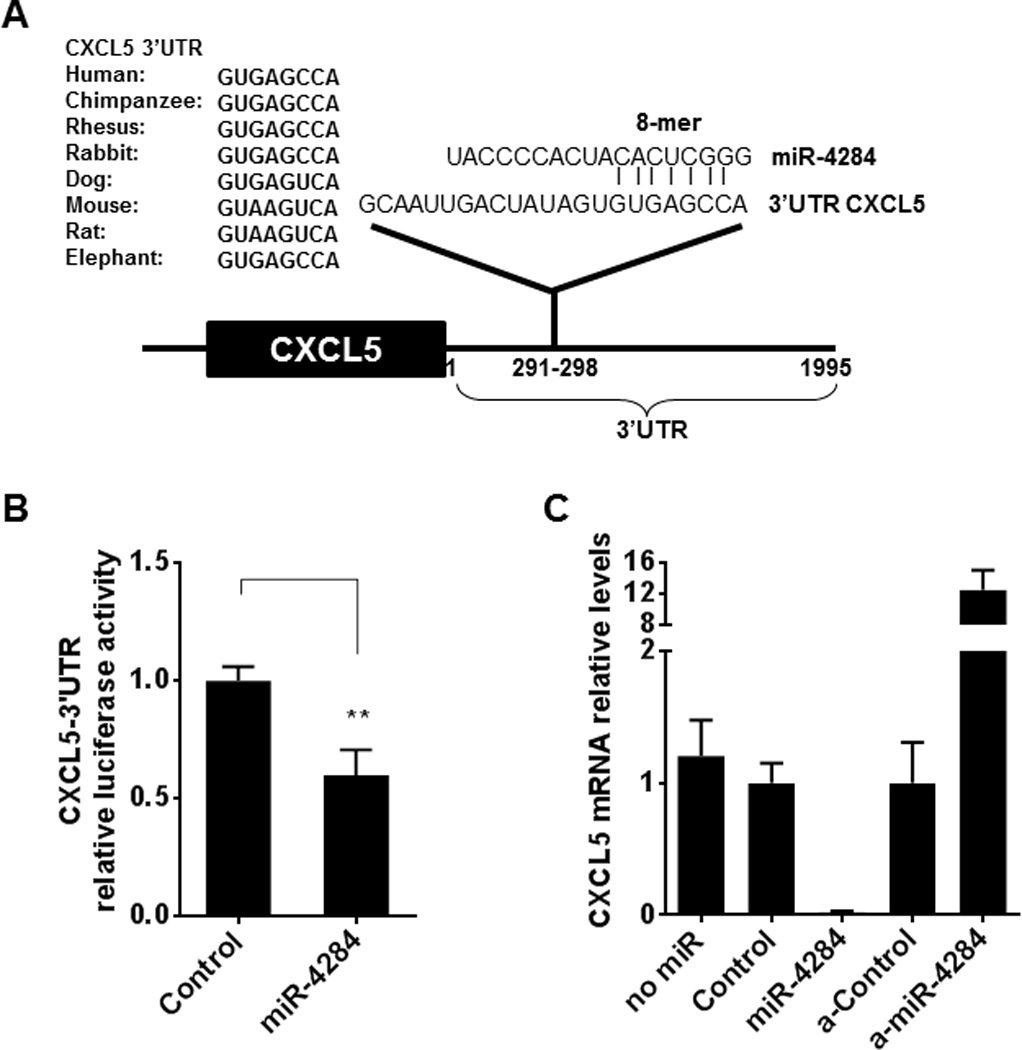
(A) Predicted miR-4284 binding sites in the 3’UTR of CXCL5 with sequence complementarity and phylogenetic conservation of the 8 nt seed-sequence indicated, as predicted by miRNA databases Targetscan and microRNA.org. (B) Luciferase assays using a reporter vector (psiCHECK2) carrying the 3’UTR of CXCL5 inHEK293T cells transfected with negative control microRNA mimics (Control, 50 nM) or miR-4284 mimics (50 nM), for 36 hours. Experiments were performed in triplicates; values represent mean ±SD, **p < 0.01, t-test analysis, Prism6 (GraphPad Software Inc.). (C) Real-time PCR for endogenous CXCL5 mRNA levels in NCM460 colonic epithelial cells transfected with 50 nM mimics (miR-4284) or inhibitors of miR-4284 (a-miR- 4284), as compared to their respective negative controls. Values represent mean ±SD.
To verify the direct interaction of miR-4284 with the 3’UTR of CXCL5, luciferase activity reporter assays were performed using a plasmid containing the luciferase gene, followed by the 3’UTR of CXCL5. Co-transfection experiments of the reporter plasmid and miR-4284 mimics - or the corresponding negative control - in HEK293T cells, resulted in a significant decrease in luciferase activity, compared to the negative control mimics (Control), indicating the direct interaction of miR-4284 with CXCL5 (Figure 3B).
Additionally, we determined the endogenous levels of CXCL5 mRNA after introduction of mimics or inhibitors of miR-4284 in non-transformed human colonic epithelial NCM460 cells. Real-time PCR for the levels of CXCL5 mRNA revealed a dramatic decrease (~50-fold) of CXCL5 in the presence of miR-4284 mimics and conversely a ~12.5-fold increase of CXCL5 when inhibitors of miR-4284 were introduced into the cells, compared to their corresponding negative controls (Figure 3C). Thus, these findings strongly support the direct regulation of CXCL5 by miR-4284 in colonic epithelial cells.
CXCL5 mRNA is significantly increased in the colonic biopsies from pediatric UC patients and correlates with disease activity and inflammatory status
To verify the clinical relevance of the miR-4284/CXCL5 interaction, the mRNA levels of CXCL5 in the biopsies from the different cohorts of pediatric patients were estimated. CXCL5 mRNA levels were significantly increased when comparing colonic biopsies of pediatric UC to non-IBD patients (Control) (Figure 4A). The same holds true for the colonic biopsies from pediatric patients with active UC when compared to pediatric patients in remission (Inactive UC) (Figure 4B).
Figure 4.
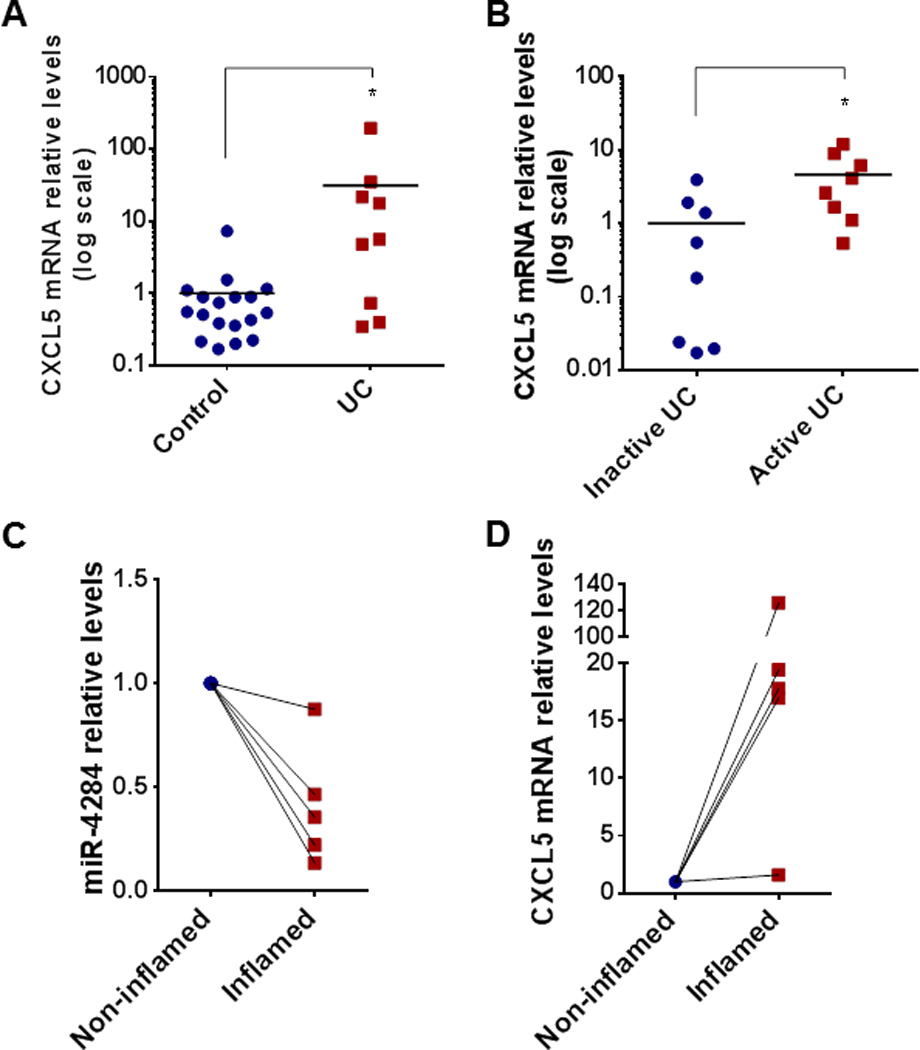
(A) Real-time PCR for CXCL5 mRNA levels in biopsies from pediatric no-IBD (Controls, n = 18) patients vs. UC patients (n = 9). (B) Real-time PCR for CXCL5 mRNA levels in a cohort of biopsies from UC patients with inactive (n = 8) or active disease (n = 8). *p < 0.05, t-test, Prism6 (GraphPad Software Inc.). Real-time PCR for the levels of (C) miR-4284 and (D) CXCL5 mRNA in matched samples of biopsies acquired from non-inflamed and inflamed colonic areas of five pediatric patients with ulcerative colitis, collected at the same visit for each patient. Data represent the levels of each target, relative to the levels of the matched non-inflamed biopsy.
To further establish the correlation of the deregulation of miR-4284 with the levels of CXCL5 we analyzed a set of five pairs of biopsies, acquired at the same visit, from inflamed and non-inflamed areas of the large intestine of each pediatric UC patient. There was an significant decrease in miR-4284 levels (more than 2-fold, in 4 out of 5 pairs) (Figure 4C), combined with an increase in CXCL5 mRNA levels (more than 15-fold in 4 out of 5 pairs) in biopsies obtained from the inflamed area, as compared to their matched biopsies from non-inflamed areas (Figure 4D).
The above findings, when combined with the decreased levels of miR-4284 we observed for all the different cohorts analyzed, support a novel functional role of miR-4284 in pediatric UC patients by regulating the levels of CXCL5 in colonic epithelial cells.
The miR-4284/CXCL5 axis in two mouse models of experimental colitis
To validate the relationship of miR-4284/CXCL5 deregulation with the development of colitis, two different mouse models of experimental colitis were used; the chemically induced DSS-chronic colitis mouse model and the genetically modified IL10-deficient (IL10-KO) mouse model. The levels of miR-4284 and CXCL5 mRNA were estimated in the colonic tissues of the mice, after establishing colitis. In the DSS model of chronic colitis, miR-4284 levels in the colonic tissues of the DSS-treated mice were decreased 2-fold (Figure 5A), while CXCL5 mRNA levels were increased 19-fold (Figure 5B), compared to untreated mice (Control). Similarly, in the IL10-KO mouse model, miR-4284 levels were decreased 2.3-fold (Figure 5C), while CXCL5 mRNA levels were increased 3.9-fold (Figure 5D), compared to wild-type controls.
Figure 5.
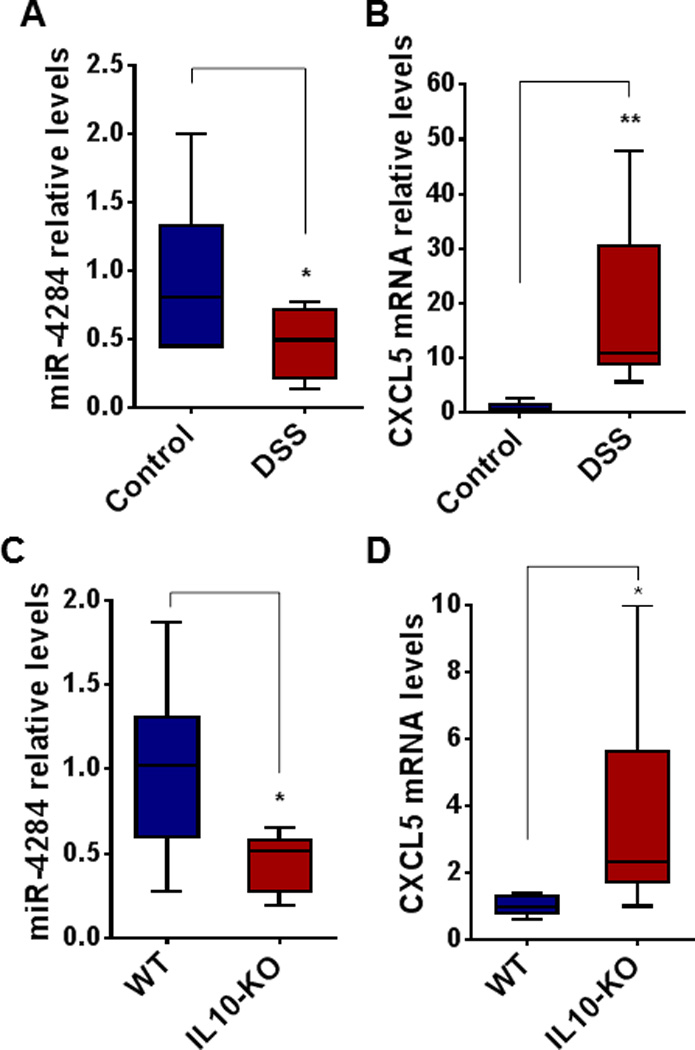
Real-time PCR for miR-4284 and CXCL5 mRNA levels in colonic tissues from two mouse models of experimental colitis. (A–B) Chronic colitis DSS-induced mouse model. Wild type, C57BL/6J, mice were kept on 3% DSS-containing water ad libitum (DSS, n = 7), for 3 cycles of 5 days, interrupted by 16 days of regular water, as described in the methods section. Mice kept on regular water served as controls (n = 7). (C–D) IL10-deficient mice (IL10-KO, n = 7) and wild-type controls (WT, n = 7) were fed piroxicam-containing diet for 2 weeks, followed by 2 weeks of regular chow. Colonic tissues were collected and RNA was extracted. Data are presented as boxes with whiskers, from minimum to maximum. *p < 0.05, **p < 0.01, t-test, Prism6 (GraphPad Software Inc.).
Discussion
A lot of effort has been invested to identify susceptibility loci in patients with IBD.19, 20 It is evident that a complex disease such as IBD cannot be understood unless multiple layers of immunoregulation and all of the genetic contributors to its pathogenesis are identified and viewed in context.21 In addition to standard treatments with 5-aminosalicylates, corticosteroids and immunomodulators, the use of anti-TNF monoclonal antibodies for the treatment of IBD has been effective and benefited exponentially more patients.22 However, the substantial costs and a number of treatment failures associated with these approaches, manifested as primary nonresponse or loss of response, indicate the need for new therapeutic modalities and better methods for monitoring disease activity in IBD patients.23 MicroRNAs, providing a previously unappreciated level of transcriptional modulation, can offer the potential missing link between known immunologic factors, may reveal new targets that contribute to pathogenesis and can provide new biomarkers for monitoring disease activity.24
In the present study we took the approach of molecular profiling analysis in biopsies of pediatric patients with UC, followed by validation using real-time PCR in another cohort of patients. Our analysis revealed a 24-microRNA signature in pediatric UC patients, compared to pediatric patients that underwent colonoscopy but were not diagnosed with IBD. Among the deregulated microRNAs identified was miR-124, which we recently reported to be epigenetically down-regulated in pediatric UC samples.11 Furthermore, several microRNAs (miR-21, miR-146b, miR-24 and let-7) known to regulate the inflammatory response were found to be deregulated in pediatric UC tissues.25–28
On the other hand, the microRNA with the largest down-regulation was miR-4284. Mir-4284 has never before implicated in IBD or any other inflammatory disease, and has only very recently reported to be deregulated in clear cell papillary renal cell carcinoma (CCPRCC).29 The functional role and the target/s of miR-4284 in the above reports were not investigated. Interestingly, in the present study, we found miR-4284 to be down-regulated in a UC-patient group with active disease, compared to subjects with inactive UC. The same analysis was performed in colonic tissues from adult patients with active and inactive UC, where no significant difference in the levels of miR-4284 was detected. Collectively, our findings provide evidence that a novel microRNA, miR-4284, in the colonic tissue is a potential marker of UC specifically in pediatric patients and that the down-regulation of this microRNA can also indicate the activity of the disease in these patients.
It is known that microRNAs exert their function(s) through direct regulation of gene expression binding to the 3’UTR.4 We explored for possible direct miR-4284 gene targets that have been implicated in IBD pathogenesis. Among others, CXCL5 was predicted by sequence complementarity algorithms as a possible target of miR-4284. Interestingly, CXCL5 has been reported to be up-regulated in UC patients and shown to be specifically expressed by colonic epithelial cells of these patients.14, 15 CXCL5 has been established as a facilitator of neutrophil recruitment to sites of epithelial injury along with IL-8.30 Of note, previous studies have shown that inhibition of Cxcl5 or its receptor CXCR2,31, 32 can affect the development of experimental colitis in mouse models, supporting the therapeutic potential of targeting CXCL5 through miR-4284.
The human relevance of miR-4284/CXCL5 interaction was established by analyses of the same pediatric biopsies in which the decreased miR-4284 levels were identified. We were able to verify this inverse correlation of miR-4284/CXCL5 experimentally in two different models of experimental colitis. The present study supports even further the notion that these mouse models may more closely mimic the pathogenesis of pediatric rather than adult UC, at least at the microRNA-level. If this is verified, it could allow for considering these models a better approximation of pediatric UC and allow investigation of the function and the potential therapeutic role of microRNAs through the modulation of their levels in the gut mucosa of these mouse models.
Overall, this study presents evidence for the role of a novel microRNA, miR-4284, in the pathogenesis of pediatric UC, modulating the levels of CXCL5 expression in the colonic epithelial cells, associated with pediatric UC disease activity (Figure 6). The specificity of miR-4284 down-regulation in pediatric UC colonic tissues makes it a potential biomarker as a monitoring tool for disease progression.
Figure 6.
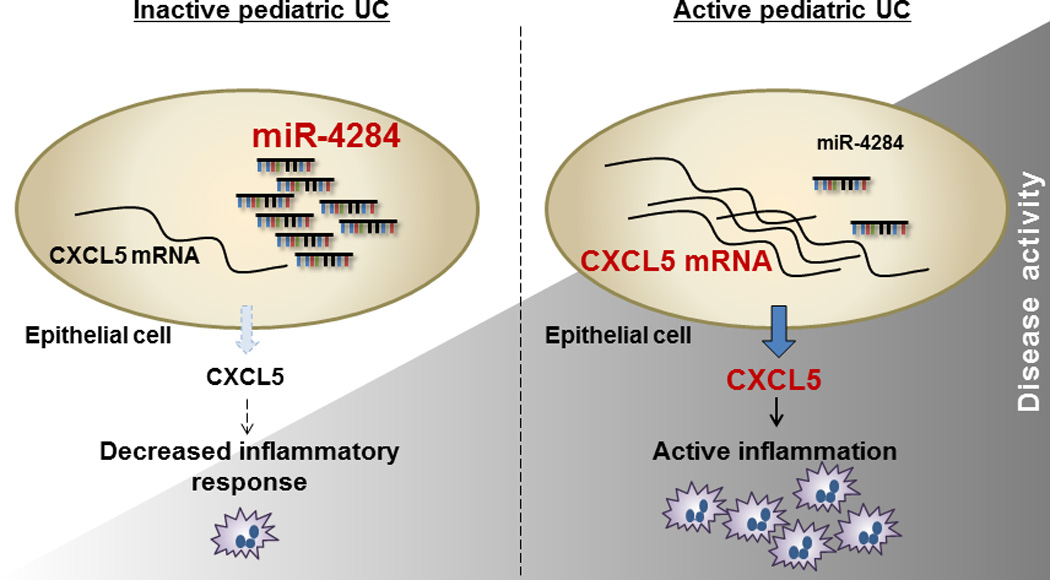
Schematic representation of the proposed role of miR-4284 in the pathogenesis of ulcerative colitis in pediatric patients. Our data suggest that miR-4284 controls the expression of CXCL5 in the colonic epithelial cells of pediatric patients following the disease activity. In remission (inactive disease) miR-4284 is abundant, leading to low levels of CXCL5 mRNA and subsequently lower expression of CXCL5. During the active state of the disease, decreased levels of miR-4284 allow for higher expression of CXCL5 resulting in neutrophil recruitment and the subsequent inflammation in that area of the gut.
Supplementary Material
TABLE 2.
Clinical characteristics of the second cohort from the Pediatric IBD Center, Mass General Hospital for Children
| Sample # | Age at collection |
Gender | Paris Disease Location |
UC Paris Disease severity |
IBD Medications | Symptoms | PUCAI (on day of sample collection) |
|---|---|---|---|---|---|---|---|
| No-IBD (Controls) | |||||||
| 1 | 18 | Male | N/A | N/A | N/A | N/A | N/A |
| 2 | 9 | Female | N/A | N/A | N/A | Poor weight gain | N/A |
| 3* | 7 | Male | N/A | N/A | N/A | N/A | N/A |
| 4* | 10 | Female | N/A | N/A | N/A | N/A | N/A |
| 5* | 18 | Male | N/A | N/A | N/A | N/A | N/A |
| 6 | 16 | Male | N/A | N/A | N/A | N/A | N/A |
| 7* | 16 | Female | N/A | N/A | N/A | Abdominal pain, diarrhea, oral ulcers, |
N/A |
| 8 | 14 | Female | N/A | N/A | N/A | Abdominal pain | N/A |
| 9 | 19 | Male | N/A | N/A | N/A | Weight loss | N/A |
| 10 | 14 | Female | N/A | N/A | N/A | Abdominal pain, weight loss | N/A |
| 11 | 11 | Male | N/A | N/A | N/A | Chronic diarrhea, short stature | N/A |
| 12 | 14 | Male | N/A | N/A | N/A | Nausea, abd pain | N/A |
| 13 | 7 | Female | N/A | N/A | N/A | Increased rectal bleeding, abd pain, gas, hard stools |
N/A |
| 14 | 18 | Male | N/A | N/A | N/A | N/A | N/A |
| 15 | 8 | Male | N/A | N/A | N/A | Bloody diarrhea | N/A |
| 16 | 10 | Male | N/A | N/A | N/A | Diarrhea, bloody stool, abd pain | N/A |
| 17 | 16 | Female | N/A | N/A | N/A | 10–15 stools/day; blood in stool | N/A |
| 18 | 14 | Male | N/A | N/A | N/A | N/A | N/A |
| Ulcerative colitis (Pediatric UC) | |||||||
| 1 | 17 | Male | E4 | S0 | Balsalazide | None | 0, PUCAI |
| 2 | 17 | Male | E4 | S0 | Balsalazide | Rectal bleeding | 35, PUCAI |
| 3 | 12 | Male | E3 | S0 | Infliximab | Joint pain | 0, PUCAI |
| 4* | 16 | Female | E2 | S0 | Mesalamine, balsalazide | Bloody diarrhea | 35, PUCAI |
| 5* | 16 | Female | E1 | S0 | Mesalamine, Prednisone | Rectal bleeding, abdominal pain | 30, PUCAI |
| 6* | 17 | Male | E4 | S1 | Prednisone, mesalamine, metronidazole |
None | 0, PUCAI |
| 7 | 17 | Male | |||||
| 8 | 7 | Male | E2 | S0 | Mesalamine enemas | None | 15, PUCAI |
| 9* | 14 | Female | E3 | S1 | None | Abdominal pain, bloody diarrhea, weight loss |
70, PUCAI |
Indicates subjects common with those of TABLE 1 used in the array analysis.
Acknowledgment
Conflicts of Interest and Source of Funding:
This work was supported in part by NIH grants DK60729 (C.P. & D.I.) and DK080058 (E.K.).
We would like to thank Dr. Alessio Morley-Fletcher for helping with the colonic tissue processing.
Footnotes
For the remaining authors none were declared.
References
- 1.Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruemmele FM, Turner D. Differences in the management of pediatric and adult onset ulcerative colitis--lessons from the joint ECCO and ESPGHAN consensus guidelines for the management of pediatric ulcerative colitis. J Crohns Colitis. 2014;8:1–4. doi: 10.1016/j.crohns.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Androulidaki A, Iliopoulos D, Arranz A, et al. The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity. 2009;31:220–231. doi: 10.1016/j.immuni.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iliopoulos D, Jaeger SA, Hirsch HA, et al. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell. 2010;39:493–506. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalal SR, Kwon JH. The Role of MicroRNA in Inflammatory Bowel Disease. Gastroenterol Hepatol (N Y) 2010;6:714–722. [PMC free article] [PubMed] [Google Scholar]
- 8.Wu F, Zikusoka M, Trindade A, et al. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology. 2008;135:1624–1635. doi: 10.1053/j.gastro.2008.07.068. e24. [DOI] [PubMed] [Google Scholar]
- 9.Wu F, Zhang S, Dassopoulos T, et al. Identification of microRNAs associated with ileal and colonic Crohn's disease. Inflamm Bowel Dis. 2010;16:1729–1738. doi: 10.1002/ibd.21267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zahm AM, Hand NJ, Tsoucas DM, et al. Rectal microRNAs are perturbed in pediatric inflammatory bowel disease of the colon. J Crohns Colitis. 2014 doi: 10.1016/j.crohns.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koukos G, Polytarchou C, Kaplan JL, et al. MicroRNA-124 regulates STAT3 expression and is down-regulated in colon tissues of pediatric patients with ulcerative colitis. Gastroenterology. 2013;145:842–852. doi: 10.1053/j.gastro.2013.07.001. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walz A, Burgener R, Car B, et al. Structure and neutrophil-activating properties of a novel inflammatory peptide (ENA-78) with homology to interleukin 8. J Exp Med. 1991;174:1355–1362. doi: 10.1084/jem.174.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koch AE, Kunkel SL, Harlow LA, et al. Epithelial neutrophil activating peptide-78: a novel chemotactic cytokine for neutrophils in arthritis. J Clin Invest. 1994;94:1012–1018. doi: 10.1172/JCI117414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keates S, Keates AC, Mizoguchi E, et al. Enterocytes are the primary source of the chemokine ENA-78 in normal colon and ulcerative colitis. American Journal of Physiology - Gastrointestinal and Liver Physiology. 1997;273:G75–G82. doi: 10.1152/ajpgi.1997.273.1.G75. [DOI] [PubMed] [Google Scholar]
- 15.Z'Graggen K, Walz A, Mazzucchelli L, et al. The C-X-C chemokine ENA-78 is preferentially expressed in intestinal epithelium in inflammatory bowel disease. Gastroenterology. 1997;113:808–816. doi: 10.1016/s0016-5085(97)70175-6. [DOI] [PubMed] [Google Scholar]
- 16.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 17.Betel D, Wilson M, Gabow A, et al. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berg DJ, Zhang J, Weinstock JV, et al. Rapid development of colitis in NSAID-treated IL-10-deficient mice. Gastroenterology. 2002;123:1527–1542. doi: 10.1053/gast.2002.1231527. [DOI] [PubMed] [Google Scholar]
- 19.Christodoulou K, Wiskin AE, Gibson J, et al. Next generation exome sequencing of paediatric inflammatory bowel disease patients identifies rare and novel variants in candidate genes. Gut. 2013;62:977–984. doi: 10.1136/gutjnl-2011-301833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beaudoin M, Goyette P, Boucher G, et al. Deep resequencing of GWAS loci identifies rare variants in CARD9, IL23R and RNF186 that are associated with ulcerative colitis. PLoS Genet. 2013;9:e1003723. doi: 10.1371/journal.pgen.1003723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polytarchou C, Koukos G, Iliopoulos D. Systems biology in inflammatory bowel diseases: ready for prime time. Curr Opin Gastroenterol. 2014;30:339–346. doi: 10.1097/MOG.0000000000000081. [DOI] [PubMed] [Google Scholar]
- 22.Arijs I, Li K, Toedter G, et al. Mucosal gene signatures to predict response to infliximab in patients with ulcerative colitis. Gut. 2009;58:1612–1619. doi: 10.1136/gut.2009.178665. [DOI] [PubMed] [Google Scholar]
- 23.Ben-Horin S, Chowers Y. Tailoring anti-TNF therapy in IBD: drug levels and disease activity. Nat Rev Gastroenterol Hepatol. 2014;11:243–255. doi: 10.1038/nrgastro.2013.253. [DOI] [PubMed] [Google Scholar]
- 24.Barnard JA. Recent advances in pediatric gastroenterology, hepatology and nutrition. F1000Prime Rep. 2013;5:25. doi: 10.12703/P5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi C, Liang Y, Yang J, et al. MicroRNA-21 knockout improve the survival rate in DSS induced fatal colitis through protecting against inflammation and tissue injury. PLoS One. 2013;8:e66814. doi: 10.1371/journal.pone.0066814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taganov KD, Boldin MP, Chang KJ, et al. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hatziapostolou M, Polytarchou C, Aggelidou E, et al. An HNF4alpha-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell. 2011;147:1233–1247. doi: 10.1016/j.cell.2011.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munari E, Marchionni L, Chitre A, et al. Clear cell papillary renal cell carcinoma: micro-RNA expression profiling and comparison with clear cell renal cell carcinoma and papillary renal cell carcinoma. Hum Pathol. 2014;45:1130–1138. doi: 10.1016/j.humpath.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacDermott RP. Chemokines in the inflammatory bowel diseases. J Clin Immunol. 1999;19:266–272. doi: 10.1023/a:1020583306627. [DOI] [PubMed] [Google Scholar]
- 31.Buanne P, Di Carlo E, Caputi L, et al. Crucial pathophysiological role of CXCR2 in experimental ulcerative colitis in mice. J Leukoc Biol. 2007;82:1239–1246. doi: 10.1189/jlb.0207118. [DOI] [PubMed] [Google Scholar]
- 32.Kwon JH, Keates AC, Anton PM, et al. Topical antisense oligonucleotide therapy against LIX, an enterocyte-expressed CXC chemokine, reduces murine colitis. Am J Physiol Gastrointest Liver Physiol. 2005;289:G1075–G1083. doi: 10.1152/ajpgi.00073.2005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


