Abstract
Introduction
Commensal gut microbiota play an important role in regulating metabolic and inflammatory conditions. Reshaping intestinal microbiota through pharmacologic means may be a viable treatment option. We sought to delineate the functional characteristics of glucocorticoid-mediated alterations on gut microbiota and their subsequent repercussions on host mucin regulation and colonic inflammation.
Methods
Adult male C57Bl/6 mice, germ-free (GF), Muc2-heterozygote (+/−), or Muc2-knockout (−/−) were injected with dexamethasone, a synthetic glucocorticoid, for four weeks. Fecal samples were collected for gut microbiota analysis via 16S rRNA T-RFLP and amplicon sequencing. Intestinal mucosa was collected for mucin gene expression studies. GF mice were conventionalized with gut microbes from treated- and non-treated groups to determine their functional capacities in recipient hosts.
Results
Exposure to DEX in WT mice led to substantial shifts in gut microbiota over a four-week period. Furthermore, a significant down-regulation of colonic Muc2 gene expression was observed after treatment. Muc2-knockout mice harbored a pro-inflammatory environment of gut microbes, characterized by the increase or decrease in prevalence of specific microbiota populations such as Clostridiales and Lactobacillaceae, respectively. This colitogenic phenotype was transmissible to IL10-knockout (IL10-KO) mice, a genetically susceptible model of colonic inflammatory disorders. Microbiota from donors pre-treated with DEX, however, ameliorated symptoms of inflammation.
Conclusions
Commensal gut bacteria may be a key mediator of the anti-inflammatory effects observed in the large intestine after GC exposure. These findings underscore the notion that intestinal microbes comprise a “microbial organ” essential for host physiology that can be targeted by therapeutic approaches to restore intestinal homeostasis.
Keywords: Animal models of IBD, Inflammation in IBD, Microbiology of IBD, Steroids in IBD
INTRODUCTION
Commensal gut bacteria provide a multitude of beneficial effects to the host which, include producing essential nutrients, metabolizing otherwise indigestible food components, educating the host immune system, strengthening gut barrier integrity, and other functions.1 On the other hand, gut microbiota can also cause or contribute to a variety of metabolic, inflammatory, and immunologic diseases.2–4 Many studies have reported that alterations in the gut microbiota induce both pro- and anti-inflammatory effects in the host.5–7 Thus, the delicate balance between host and gut microbes determines commensalism or pathogenicity, the latter triggered by even the slightest perturbations caused by internal (host) or external (environmental) factors. For instance, inflammatory bowel diseases (IBD) are believed to arise from an unfortunate combination of dysbiosis and underlying genetic susceptibility. This concept, which broadly applies to many “Western” disorders, is an example often used to support the “hygiene hypothesis”, where rapid societal changes in environment, diet, and lifestyle over the past century have perturbed evolutionarily-determined host-microbe relationships critical for normal immune and physiological development.8,9 In support of this hypothesis, antibiotics provide some IBD patients with therapeutic benefit,10 underscoring the notion that gut bacteria play a key role in the etiopathology of this disease. In addition, the consumption of a high-fat “Western” diet promotes the growth of otherwise rare pathobionts that can upset normal host-microbe relationships to trigger the development of experimental colitis in genetically-susceptible hosts.11
The mucus layer that overlies the gut epithelium plays a key role in establishing this mutualistic relationship between host and microbe. It helps protect the mucosa from chemical, enzymatic, and microbial damage and, at the same time, can be a major determinant of microbial assemblage and food supply. Mucins constitute the major component of this mucus layer12 and in the colon, Muc2 is the most prominent mucin expressed and secreted by goblet cells.13 The importance of Muc2 in protecting the integrity of the gut epithelium is exemplified by the development of spontaneous and experimentally-induced colitis in mice lacking the Muc2 gene (Muc2-KO).14 Additionally, decreased Muc2 expression has been implicated in the etiopathogenesis of sporadic and IBD-associated colon cancers.14,15
Many factors can influence intestinal mucin gene expression and function. Physiological and pharmacological levels of glucocorticoids (GCs) can profoundly affect mucus production and secretion. GCs are currently one of the most widely used first-line agents in the treatment of IBD.16 Dexamethasone (DEX), a synthetic, more potent GC, has been routinely used as an anti-inflammatory, immunosuppressive agent.17 Previous work has shown that one mechanism by which DEX resolves its anti-inflammatory effects is through a decrease in NF-κB activity.18 However, prolonged exposure to GCs can result in various side effects, including central adiposity, insulin resistance and gastrointestinal complications.19,20 Thus, due to its pleiotropic nature, many GC-mediated effects have not been well-characterized. Recent studies have suggested that GC signaling derives a portion of its effects through gut microbes.21,22 Since many mechanisms of GC action are not well described, it was of interest to explore the cross-talk between GCs, the intestinal microbiota, and colonic physiology. Herein, the effects of chronic GC exposure (using DEX) on shaping the gut microbiota and their roles in regulating both mucin expression and inflammation in the colon were investigated.
MATERIALS AND METHODS
Animals and experimental treatments
All wild-type (WT) and Muc2-KO specific pathogen free (SPF) male mice used in this study were of the C57Bl/6 strain, bred and housed at the University of Chicago. GF mice were bred and maintained in sterile flexible film isolators within the University of Chicago Gnotobiotic Research Animal Facility (GRAF). All mice were entrained on a 12-h light/dark cycle with lights on beginning at 6AM. Mice were given ad libitum access to irradiated standard rodent purified diet (for SPF mice: #2016S, Harlan-Teklad, Madison, WI, USA; for GF mice: autoclaved #5K67, LabDiet, St. Louis, MO) as well as sterile autoclaved water. 7- to 10-week old male wild-type C57Bl/6 mice were injected intra-peritoneally (i.p.) with pharmaceutical-grade DEX: 1 mg/kg, once per day (acute GC exposure) or 5 mg/kg, once per 3 days (chronic GC exposure) (Fig. 1A). Control mice were injected with an equivalent dose of sterile saline. Fecal samples were periodically collected throughout the study for bacterial DNA analysis. All animal protocols and experiments were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Chicago.
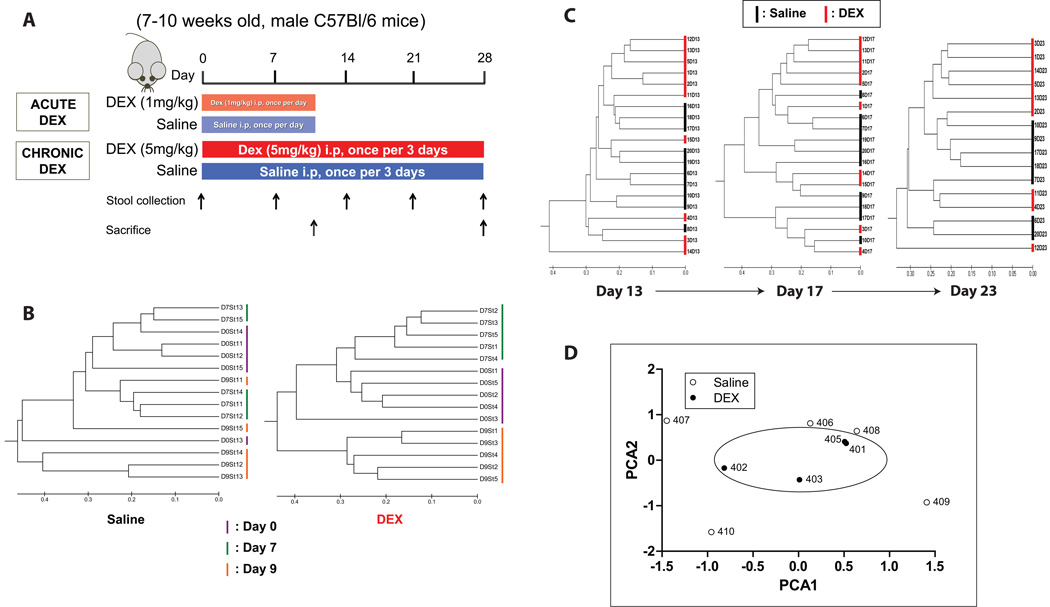
FIGURE 1. Schematic of DEX experiment and effects on gut microbiota.
(A) Adult male C57Bl/6 mice were injected with either dexamethasone (DEX, 1 mg/kg or 5 mg/kg) or an equivalent amount of sterile saline (controls). Stool samples were collected periodically for T-RFLP analysis. Bray-Curtis dendrograms of T-RFLP sequences from DEX- and saline-treated mice after (B) 10 days, and (C) 4 weeks of treatment. (D) Principle Component Analysis (PCA) plot of microbiota from DEX and saline-treated mice. i.p.: intraperitoneally. DEX: dexamethasone. SAL: saline.
Bacterial DNA isolation and Polymerase Chain Reaction (PCR)
Isolation of bacterial DNA was performed as previously described.23 In brief, mouse stool and cecal contents were collected and placed in 1mL of T.N.E.S. extraction buffer. After addition of 0.1-mm-diameter zirconia/silica beads (BioSpec Products, Bartlesville, OK, USA), samples were disrupted using a Mini-Beadbeater-8k Cell Disrupter (BioSpec Products) and incubated overnight at 55°C. Supernatants were then extracted with an equal volume of Phenol:Chloroform:Isoamylalcohol (25:24:1; Ambion, Austin, TX, USA) and DNA precipitated using an equal volume of 100% ethanol. The resulting pellet was centrifuged (13,000 rpm for 5 min), washed with 70% ethanol, dried, and reconstituted in nuclease-free water.
Polymerase chain reaction was performed as follows: 5µL of 10× Ex Taq buffer containing 20mM MgCl2 (Takara, Tokyo, Japan), 4µL of 2.5mM dNTP Mixture (Takara), 1µL each of FAM-labeled forward (27F, 5'-AGA GTT TGA TCC TGG CTC AG-3') and reverse (1492R, GGT TAC CTT GTT ACG ACT-3') primer (10mM each), 0.25µL of Taq polymerase (Takara), 36.75µL nuclease-free water, and 2µL of DNA template. The PCR conditions were: 94°C for 5 min followed by 30 cycles of amplification consisting of denaturation at 94°C for 30 sec, annealing at 58°C for 1 min, and extension at 72°C for 1.5 min.
DNA Repurification and Terminal Restriction Fragment Length Polymorphism (T-RFLP) sequence analysis
The resulting PCR product was then repurified using 3M sodium acetate (pH 5) and restriction digested with the MspI enzyme. Samples were then dialyzed on a HAWP membrane filter (Millipore, Billerica, MA, USA) to remove excess salts and submitted to the Cancer Research Center DNA Sequencing Facility at the University of Chicago for sequence analysis. Using these fluorescently labeled 5’-terminal restriction fragments, Bray-Curtis dendrograms and Principal Component Analysis (PCA) plots were then generated with the Molecular Evolutionary Genetics Analysis (MEGA) software platform (www.megasoftware.net) or through the T-RFLP analysis (T-REX) website.24
16S rRNA-based Illumina library preparation and data analysis
PCR primers used were specific for the 515–806 bp region of the 16S rRNA encoding gene (338F: 5’-GTGCCAGCMGCCGCGGTAA-3’ and 806R: 5’-GGACTACHVGGGTWTCTAAT-3’) and contained Illumina 3' adapter sequences as well as a 12-bp barcode. This barcode-based primer approach allowed sequencing of multiple samples in a single sequencing run without the need for physical partitioning. Sequencing was performed by an Illumina MiSeq DNA sequencer at Argonne National Laboratory. Sequences were then trimmed and classified with the QIIME toolkit. Using the QIIME wrappers, OTUs were picked at 97% sequence identity using cdhit and a representative sequence was then chosen for each OTU by selecting the most abundant sequence in that OTU. These representative sequences were aligned using PyNAST and taxonomy was assigned to them using the RDP Classifier. The PyNAST-aligned sequences were also used to build a phylogenetic tree with FastTree and unweighted UniFrac distances then computed between all samples for additional ecological analyses.
Mucosal scrapings, RNA extraction, cDNA synthesis, and quantitative real-time PCR
After termination of the experiment, mice were sacrificed via CO2 asphyxiation and the abdominal cavity immediately opened. The large intestine was dissected and splayed on a chilled glass surface. Using sterile surgical scissors, the intestinal lumen was exposed and gently scraped to collect mucosal contents for protein and/or RNA analysis. Mesenteric lymph nodes (MLNs) were excised and homogenized for protein analysis via ELISA.
Colonic mucosal scrapings were homogenized in TRIzol reagent (Ambion) and mixed with chloroform. After centrifugation (10,000 rpm for 15 min), the top aqueous phase was mixed with 100% isopropanol to precipitate RNA. Samples were centrifuged (10,000 rpm for 10 min) and pellets were washed (75% ethanol), dried, and reconstituted in nuclease-free water. RNA purity and concentration were assessed through UV-Vis spectrophotometry using the Nanodrop Lite (Thermo Scientific, Wilmington, DE, USA). 1µg of total RNA was reverse-transcribed to complementary DNA (cDNA) using the Transcriptor First Strand cDNA Synthesis Kit (Roche, Indianapolis, IN, USA) according to manufacturer’s instructions. The relative quantitation of gene expression was performed using the LightCycler 480 Real-Time PCR System (Roche). Primers used were: Muc1 (F: gcagtcctcagtggcacctc, R: caccgtgggctactggagag), Muc2 (F: gctgacgagtggttggtgaatg, R: gatgaggtggcagacaggagac), Muc3 (F: cgtggtcaactgcgagaatgg, R: cggctctatctctacgctctcc), GAPDH (F: ggcaaattcaacggcacagt, R: agatggtgatgggcttccc). Gene expression data are presented as 2−ΔCt (target gene – housekeeping gene).
Conventionalization of germ-free mice
To reintroduce gut microbiota into GF mice, termed “conventionalization” (CONV), donor mice were sacrificed as described above. Cecal contents were quickly excised and placed in a tube containing 1 mL sterile PBS. The contents were mixed vigorously for 30 seconds and centrifuged briefly. The supernatant was then transferred to 1-cc syringes and orally gavaged (150 µL) to GF WT or IL10-KO mice.
Protein isolation and BCA analysis
Freshly excised protein samples were placed in cell lysis buffer (20mM Tris-HCl, 150mM NaCl, 1mM Na2 EDTA, 1mM EGTA, 1% Triton, 2.5mM sodium pyrophosphate, 1mM β-glycerophosphate, 1mM Na3VO4, 1µg/mL leupeptin, 1mM PMSF) (Cell Signaling Technology, Danvers, MA, USA). Samples were homogenized, centrifuged (13,000 rpm for 5 min) and supernatant collected. Protein concentrations were quantified using the BCA protein assay.25
Enzyme-linked immuno sorbent assay (ELISA)
To characterize gut inflammation between treatment groups and controls, a colorimetric sandwich ELISA was performed on mucosal scrapings according to manufacturer’s instructions (eBioscience, San Diego, CA, USA). In brief, target-specific capture antibodies were coated on 96-well plates (Corning Inc, Corning, NY, USA) and incubated overnight at 4°C. The following day, plates were washed, blocked for one hour at room temperature and washed again. Diluted standards and unknown samples were then added and incubated overnight at 4°C. On the final day, plates were washed and incubated in secondary antibody for one hour at room temperature. Plates were then washed, avidin-HRP added, washed again, and incubated in TMB substrate solution for 15 min at room temperature. Stop solution (2N H2SO4) was then added to discontinue HRP activity. Absorbances were measured on a plate reader (VersaMax, Sunnyvale, CA, USA) at a wavelength of 450 nm to determine cytokine concentration.
Statistical analyses
All bar graphs are expressed as mean ± standard error of the mean (SEM). Student’s t-test was used for two-group comparisons and one-way analysis of variance (ANOVA) used for multiple group (>2) comparative analysis. Tukey post-hoc analyses were applied for multiple comparisons, with statistical significance established at P-values < 0.05.
RESULTS
DEX treatment alters gut microbiota communities
Adult male C57Bl/6 mice were injected i.p. with DEX or saline, once per day for 10 days (acute treatment), or once per 3 days over a four-week period (chronic treatment). T-RFLP analysis of fecal samples revealed a substantial shift in gut microbiota after DEX treatment. This shift was evident in both acute (10 days) and chronic (28 days) models of DEX treatment and persisted throughout the duration of the study (Fig. 1B,C). Principal Component Analysis (PCA) revealed a microbial-based clustering in DEX-treated animals, but not in saline-treated controls (Fig. 1D).
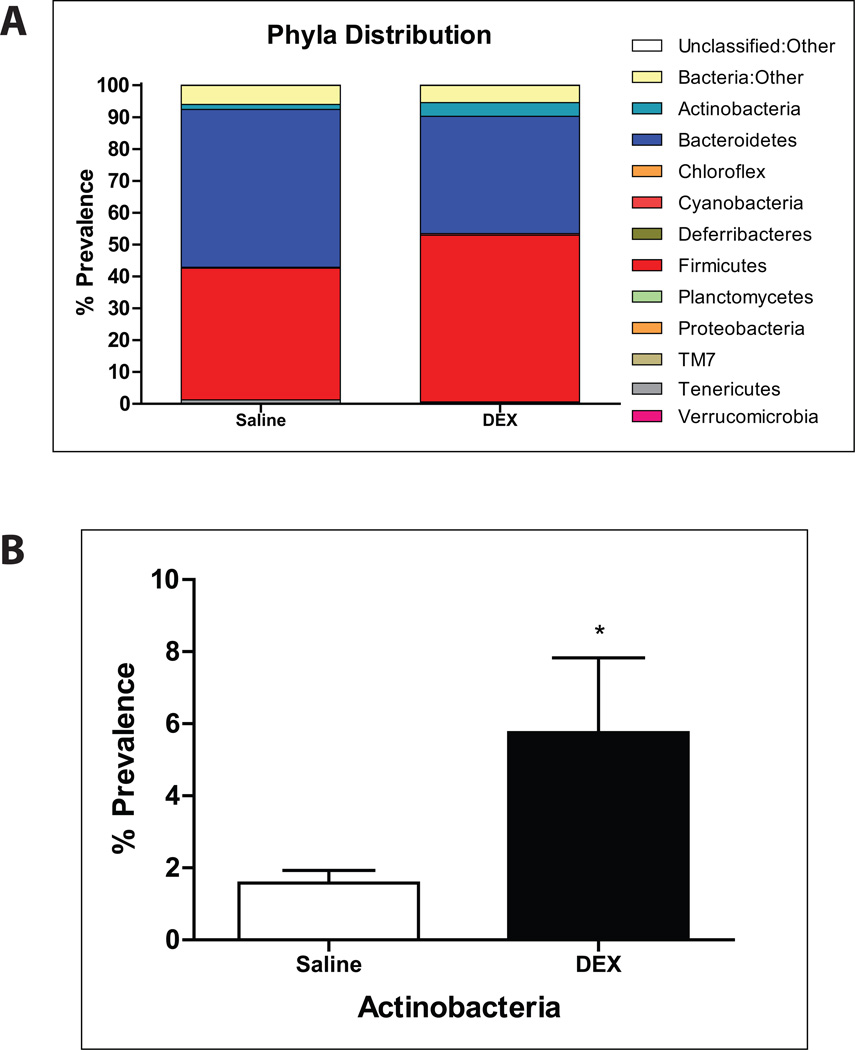
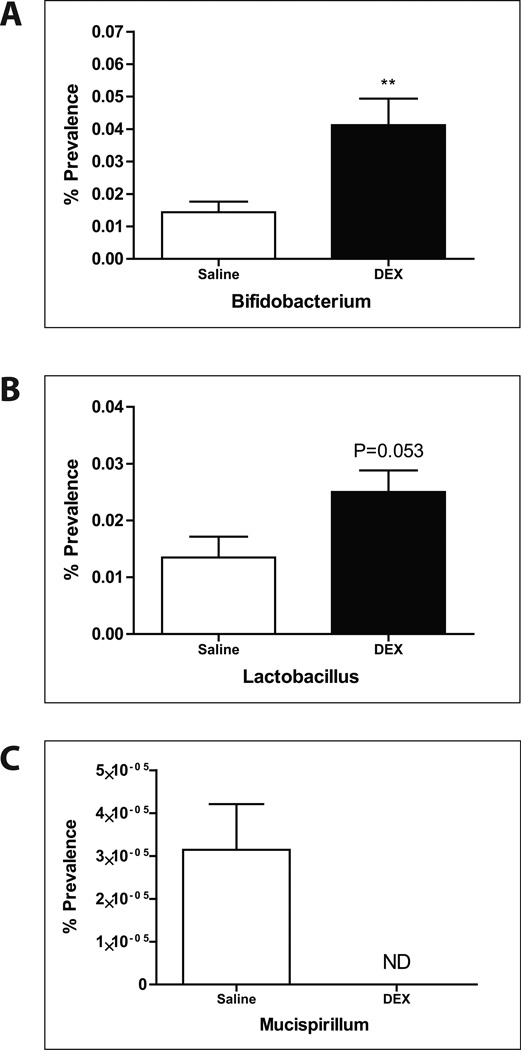
Amplicon sequencing of the 16S rRNA V4–5 region from fecal samples revealed phylogenetic shifts in gut microbiota after DEX exposure, notably in Actinobacteria (Fig. 2A,B). The Actinobacteria genus, Bifidobacterium, was significantly elevated after DEX treatment (Fig. 3A). Furthermore, Lactobacillus levels were also increased when compared to controls (Fig. 3B). On the other hand, a specific genus of gut bacteria known to be a colonic mucin degrader, Mucispirillum, was noticeably absent after treatment with DEX (Fig. 3C), suggesting a possible relationship between GC, altered gut microbiota, and mucin regulation in the intestine.
FIGURE 2. 16S Illumina sequencing analysis of gut microbiota.
(A) Phyla distribution of fecal microbiota after chronic DEX treatment. (B) Bar graph showing the percent prevalence of Actinobacteria. Values are expressed as mean ± SEM. *p<0.05 vs. saline-treated controls. DEX: dexamethasone.
FIGURE 3. Deep sequencing analysis (genus level) of fecal microbiota in DEX- and control-treated mice.
Percent prevalence of (A) Bifidobacterium, (B) Lactobacillus, and (C) Mucispirillum. Values are expressed as mean ± SEM. **p<0.01 vs. saline controls. DEX: dexamethasone. ND: non-detectable.
Mucin gene expression decreases after exposure to DEX
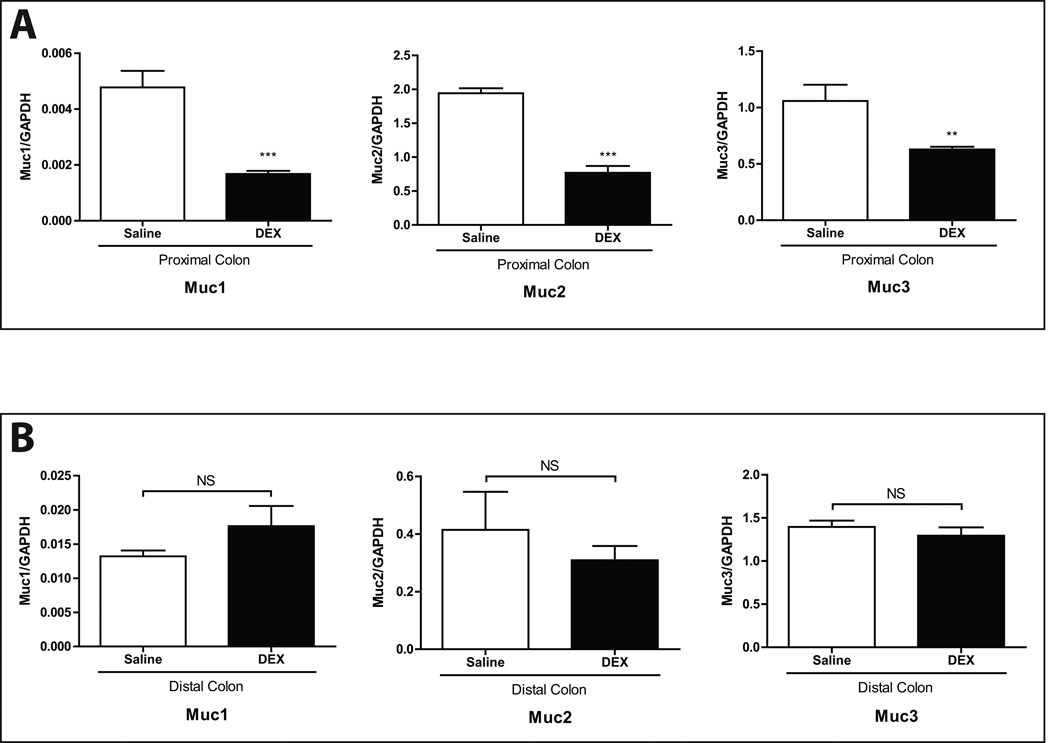
Muc2 is the predominant mucin gene expressed in colonic goblet cells26,27 and is a key factor in the maintenance of colonic health.28 Quantitative real-time PCR results showed that Muc2 gene expression was significantly lower in DEX-treated mice. Gene expression levels of Muc1 and Muc3, which together constitute a minority of gut mucins, were also significantly lower after DEX. However, these differences were only evident in the proximal colon (Fig. 4A), as mucin gene expression in the distal colon was not significantly different between DEX- and saline-treated mice (Fig. 4B).
FIGURE 4. Colonic mucin gene expression levels after chronic exposure to DEX.
Gene expression levels of Muc1, 2, and 3 in (A) proximal and (B) distal large intestine. GAPDH expression was used as an internal control. Values are expressed as mean ± SEM. **p<0.01 and ***p<0.001 vs. saline (controls). NS: not statistically significant. DEX: dexamethasone.
Conventionalization with DEX-treated donor microbiota upregulates mucin gene expression
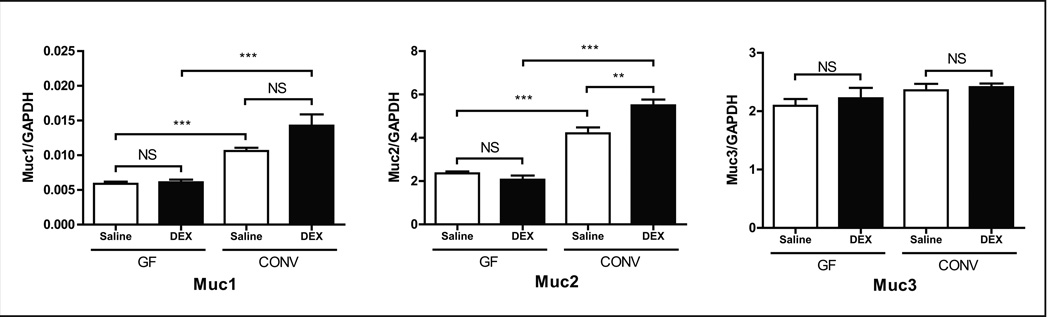
To determine the direct effects of DEX on host-mediated mucin synthesis in the proximal colon, GF mice were chronically treated with DEX. Interestingly, no significant changes in mucin gene expression were observed between DEX and control groups. However, conventionalizing GF mice with microbiota from DEX-treated donors led to a significant increase in both Muc1 and Muc2 expression in the proximal large intestine (Fig. 5) suggesting a key role for gut microbiota in the regulation of colonic mucin synthesis.
FIGURE 5. Colonic mucin gene expression levels in germ-free and conventionalized wild-type mice.
Gene expression levels of Muc1, 2, and 3 in the proximal colon of GF and CONV wild-type mice. GAPDH expression was used as an internal control. Values are expressed as mean ± SEM. **p<0.01 and ***p<0.001 between bracketed groups. GF: germ-free. CONV: conventionalized. NS: not statistically significant. DEX: dexamethasone.
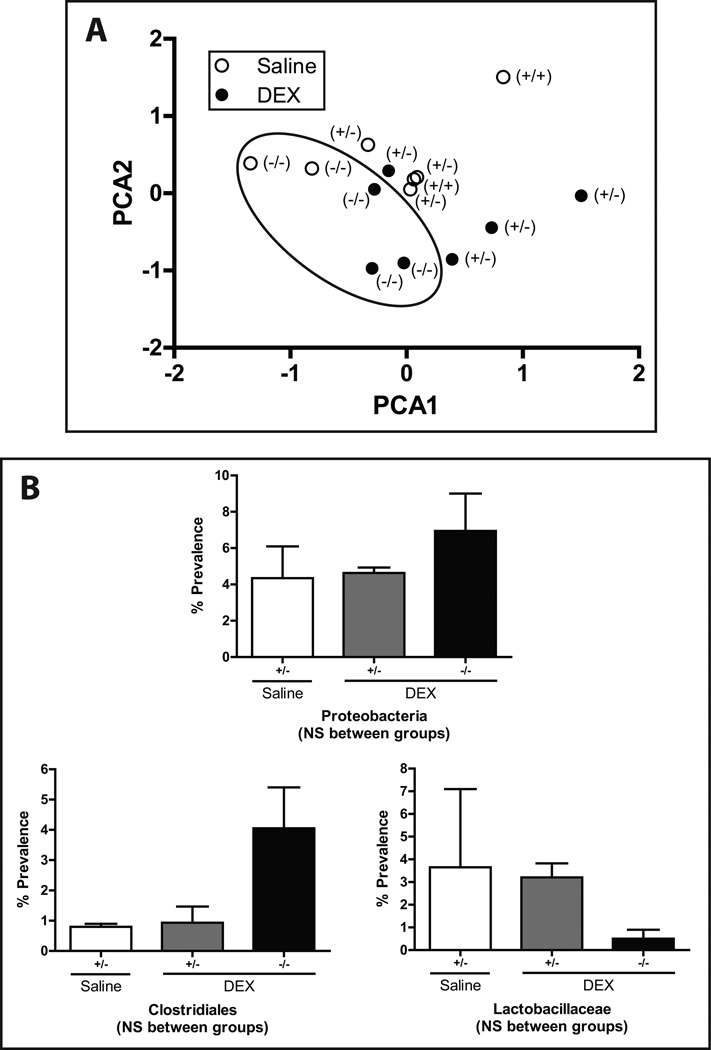
Microbiota of Muc2-KO mice cluster distinctly from heterozygote and wild-type mice
To further explore the relationship between DEX and Muc2, mice possessing the wild-type (+/+), heterozygote (+/−), or knockout (−/−) allele for Muc2 were chronically treated with DEX. Similar to earlier findings, microbiota from mice exposed to DEX clustered distinctly from controls. However, Muc2-KO mice retained a microbiota distinct from those of their heterozygote and wild-type counterparts (circled, Fig. 6A). A trend toward a greater relative proportion of Proteobacteria, a phyla commonly associated with inflammation, was observed in Muc2-KO mice despite DEX treatment. Deeper phylogenetic analysis showed increasing Clostridiales in DEX-treated Muc2-KO mice compared to DEX-treated Muc2-heterozygotes and a concomitant decrease in Lactobacillaceae (Fig. 6B), though these patterns were not statistically significant. Our observations suggest that Muc2-KO microbiota may convey a potentially proinflammatory milieu to the host through the expansion and/or reduction of certain gut-associated bacteria.
FIGURE 6. Muc-KO mice possess microbiota distinct from those of their wild-type and heterozygote counterparts.
(A) PCA graph of relative microbiota distribution between DEX- and saline-treated mice with the Muc2 (+/+, +/−, −/−) genotype. Muc2-KO (−/−) are denoted with a circle. (B) Percent prevalence of Proteobacteria, Clostridiales, and Lactobacillaceae in Muc2-heterozygote and knockout mice. Values are expressed as mean ± SEM. NS: not statistically significant. +/+: wild-type. +/−: Muc2 heterozygote. −/−: Muc2 knockout. DEX: dexamethasone.
Microbiota from Muc2-KO mice are colitogenic while microbiota from DEX-treated mice are anti-inflammatory
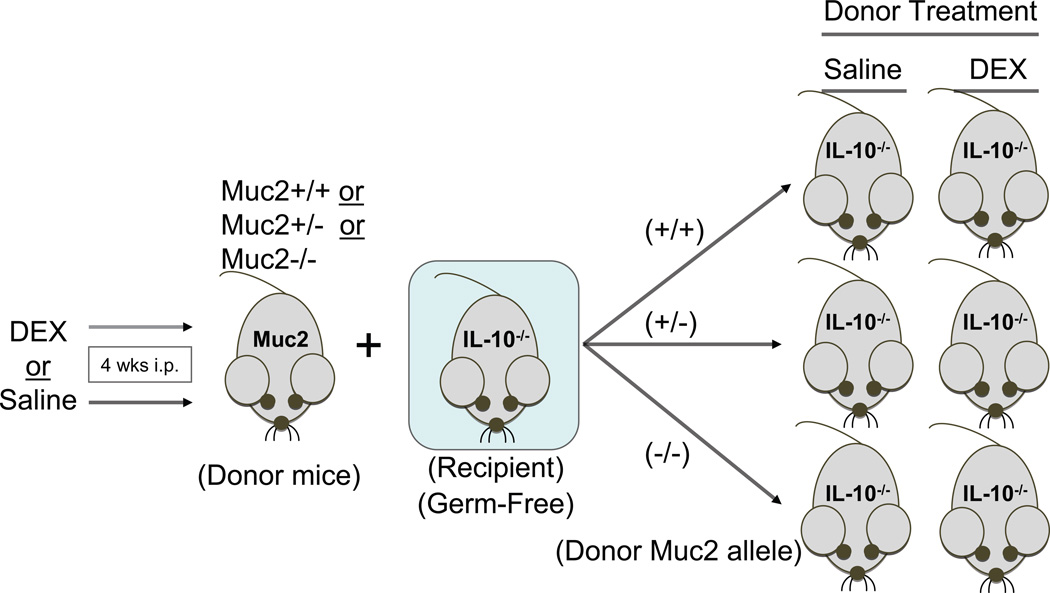
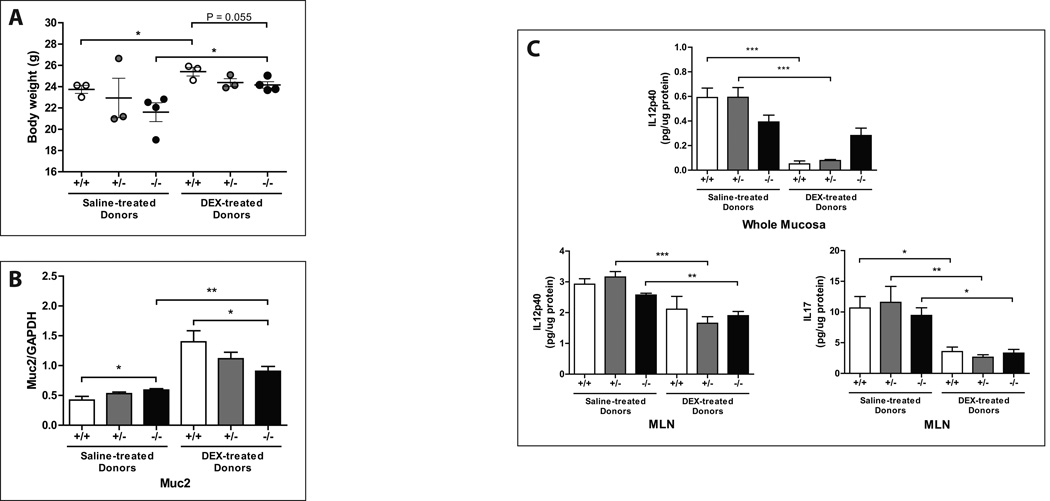
To characterize the functional repercussions of glucocorticoid-altered gut bacteria on colonic inflammation, cecal contents from DEX- or saline-treated Muc2-KO mice were conventionalized into GF IL10-KO mice (Fig. 7). These conventionalized recipient mice are used as a model of genetic susceptibility to colitis, as they spontaneously develop colitis at a rate of 20–30% in our SPF animal facility. However, germ-free IL10-KO mice do not develop colitis unless they are exposed to bacteria, suggesting a microbe-driven component mediating this effect. IL10-KO mice conventionalized with saline-treated cecal contents exhibited a marked decrease in body weight when compared to their respective DEX-conventionalized counterparts (Fig. 8A). Mice conventionalized with Muc2-KO microbiota, irrespective of the donor’s treatment (DEX or saline), had lower body weights relative to those conventionalized with wild-type or heterozygote microbiota (Fig. 8A). Interestingly, conventionalization with saline-treated microbiota led to a pronounced decrease in Muc2 gene expression relative to DEX-treated donor microbiota (Fig. 8B). When inflammatory cytokine levels in MLNs and mucosal scrapings were examined, significant increases in IL12p40 and IL17 were observed in all recipients that were conventionalized with saline-treated microbiota. In contrast, DEX-treated donors did not confer an inflammatory milieu to the same degree (Fig. 8C). Altogether, these results suggest that in the absence of Muc2, gut microbiota confer a pro-inflammatory, colitogenic phenotype. In contrast, DEX treatment in mice expressing the Muc2 gene alters gut microbiota to reduce inflammation in a genetically susceptible model of colitis.
FIGURE 7. Experimental design for the conventionalization of germ-free IL10-knockout mice.
Muc2-KO (−/−), heterozygote (+/−), and wild-type (+/+) donor mice were injected with DEX as outlined previously. Cecal contents were then extracted and used to conventionalize germ-free IL10-KO recipient mice. These recipients were then housed for 3 weeks prior to sacrifice.
FIGURE 8. DEX-treated donor microbiota allays symptoms of colonic inflammation after conventionalization.
Graphs are arranged according to the Muc2 genotype and treatment (DEX or saline) of their respective donors. (A) Body weights of recipient mice at the time of sacrifice and (B) Muc2 gene expression in the proximal colon of conventionalized IL10-KO recipient mice. (C) Mucosal IL12p40 and IL17 content measured by ELISA. Values are expressed as mean ± SEM. *p<0.05, **p<0.01, and ***p<0.001 between bracketed groups. +/+: wild-type. +/−: Muc2 heterozygote. −/−: Muc2 knockout. NS: not statistically significant. DEX: dexamethasone. MLN: mesenteric lymph nodes.
DISCUSSION
The Center for Disease Control and Prevention classifies IBD as one of the five most prevalent disease burdens in the United States, costing roughly $1.7 billion dollars in healthcare overall.29 While the etiology of IBD remains to be fully characterized, aberrations in homeostatic balance between host biology, genetics, and environmental factors such as the gut microbiota are keys to the pathogenesis of these diseases. Previous studies have reported a multitude of genetic polymorphisms that either increase or lower the risk of developing IBD,30 while others have demonstrated a considerable role for intestinal microbes in their pathogenesis.1,31,32
Our studies show that the balance between host and bacteria can be mitigated through certain circumstances. Both endogenous and therapeutic GCs can affect this relationship. For instance, a link between steroid hormones and gut microbiota has been implicated by the observation that altered gut microbiota can lead to substantial shifts in the hypothalamic-pituitary-adrenal (HPA) axis response to stress.33 A study by Ünsal and colleagues showed that rodents treated with a single high-dose injection of DEX increased the number of ileal anaerobic bacteria while low-dose GC produced an increase in coliform bacteria,34 demonstrating a significant, acute effect of GC exposure on gut microbes. However, these studies have not identified specific gut microbes affected by GC exposure nor addressed the functional repercussions of GCs and altered gut microbiota. Our results demonstrate that exposing healthy SPF mice to the GC DEX, both acutely (10 days) or chronically (over 4 weeks), leads to a dramatic shift in gut microbiota that persist throughout the length of the study. Furthermore, these alterations in gut bacteria regulate the colonic inflammatory state in a genetically susceptible mouse model of colitis, demonstrating a gut microbe-mediated mechanism by which GCs apply their anti-inflammatory effects in the large intestine. GCs have potent anti-inflammatory benefits,35,36 so the presence of Bifidobacteria and Lactobacillus, both previously associated with anti-inflammatory effects,37–39 may point toward a microbe-centric mechanism through which these effects occur in our experimental model. Interestingly, we observed a noticeable absence of Mucispirillum, a mucin-reliant gut microbe, in DEX-treated mice. Since mucins play a considerable role in the gut, namely in colonic protection,28,40 it was of significant interest to determine the effects of GC exposure on mucin gene expression in the colon.
Glucocorticoid exposure, gut microbiota, and Muc2 gene expression
Our results showed a significant decrease in mucin gene expression after DEX treatment. Given the protective nature of mucins, this observed decrease in mucin gene expression initially seemed counter-intuitive, as GCs tend to confer protective, anti-inflammatory effects.36,41–43 However, previous studies have demonstrated an up-regulation of mucins under pro-inflammatory settings,44,45 while others report a down-regulation of their expression.14 In the context of inflammatory conditions where mucins are over-expressed, such as in asthma, GCs decrease mucin synthesis as part of their anti-inflammatory mechanism.46 Based on these and other reports, we surmise that the impact of GC-mediated effects on host and microbiota is highly contextual. In healthy mice without pre-existing inflammatory or genetic conditions, mucin expression even if decreased by DEX may be amply sufficient to maintain intestinal homeostasis. Additionally, counter-regulatory pathways for mucosal homeostasis are more likely to be intact and able to compensate for reduced mucin expression.
Interestingly, GF mice treated with DEX alone exhibited no significant difference in mucin expression relative to controls. This lack of change in GF mice suggests to us that DEX alone is not sufficient to regulate mucin synthesis in the absence of gut microbes. One possible explanation for this may be due to incomplete development or the hypothalamic-pituitary-adrenal (HPA) axis activity in GF mice.47–49 The HPA axis is critical to proper glucocorticoid signaling,50 so it is possible that the GF response to DEX exposure is quite different from that of the SPF mouse. When GF mice were conventionalized with microbiota from DEX-treated donors, we observed a significant increase in Muc2 expression relative to controls. Previous studies have reported that the reintroduction of microbes via conventionalization elevates mucin levels in the intestine.51,52 In concert with our previous 16S rRNA amplicon data showing a lack of mucin-utilizing microbes in DEX-treated mice, these results are consistent with our hypothesis that the up-regulation of mucin expression due to conventionalization is now uninhibited due to the lack of mucophilic bacteria in these mice. Our results suggest that exposure to DEX leads to divergent effects on mucin regulation in the proximal large intestine of SPF and GF mice. These results also suggest that while exposure to DEX under germ-free conditions is insufficient in altering colonic mucins, the presence of gut microbes in the absence of DEX can substantially affect their expression. Further studies delineating the complex regulatory network of mucin synthesis and expression are clearly warranted.
Gut microbiota in Muc2-KO mice
The Muc2-KO mouse possesses a distinct gut-associated bacterial community that is influenced by its altered genetic and immune states, as has been shown in other genetic knockout mouse models.4,53–55 To explore the effects of GC signaling on gut microbiota in the absence of Muc2, we examined the effects of DEX treatment on colonic microbiota in WT and in Muc2 heterozygote and KO mice. While DEX-treated mice clearly clustered from saline-treated controls, Muc2-KO mice also clustered distinctly from their heterozygote and wild-type counterparts. One possible explanation is that the variability in microbial community structure due to DEX is considerably less than the genetic and immune-mediated alterations of the Muc2-KO mouse. Host biology and environmental cues are both substantial contributors toward shaping gut-associated bacterial communities.3,56,57 However, our results point toward a greater effect of genetics in coordinating the host-microbe dynamic in this mouse model. Since mucin expression is critical for the growth and survival of gut bacteria,58,59 it is of little surprise that the complete ablation of the predominant mucin expressed in the colon (Muc2) imparts considerable pressures on the dynamics of bacterial colonization.
Bacterial 16S rRNA amplicon sequencing revealed a trend toward greater prevalence of Proteobacteria in Muc2-KO mice. This phyla has been shown to be intimately tied to a pro-inflammatory state.11,60,61 Furthermore, these mice also exhibited increased prevalence of Clostridiales alongside a concomitant decrease in Lactobacillaceae, though these patterns were not statistically significant. Previous studies have shown a close association between inflammation and the presence or absence of these microbes, respectively,62–64 altogether supporting the presence of a pro-inflammatory milieu within the microbiota of Muc2-KO mice.
IL10-KO mice conventionalized with altered microbiota from DEX- or saline-treated donors
To characterize the inflammatory potential of these altered microbes, the microbiota of DEX- and saline-treated donor mice were conventionalized into GF IL10-KO mice, a genetically susceptible mouse model of colitis. Notably, the IL10-KO mouse model only develops colitis in the presence of microbes,65 underscoring the importance of the enteric bacteria in the pathogenesis of gut inflammation. The onset and severity of colitis upon conventionalization thus provides a functionally relevant readout to assess the pathogenic potential of each microbiota community. Declining trends in both body weight as well as colonic Muc2 expression levels, among other traits, are hallmark characteristics of colitis.28 IL10-KO mice conventionalized with saline-treated donor microbiota exhibited both a considerable decrease in body weight as well as significantly lower Muc2 gene expression levels relative to their DEX-treated donor counterparts. We reported earlier that DEX treatment in SPF mice significantly lowered Muc2 gene expression but show here that this microbiota retains the greatest level of Muc2 expression after conventionalization (Fig. 8B). This finding corroborates our previous observations that conventionalizing GF mice with DEX-treated donor microbiota resulted in a significant up-regulation of Muc2 gene expression. Taken together, we hypothesize that glucocorticoids may serve to “balance” Muc2 expression as part of its anti-inflammatory mechanisms, decreasing its expression under healthy conditions in SPF mice, while preventing further reduction to critically damaging levels under diseased conditions to prevent the exacerbation of colitogenic symptoms in IL10-KO mice.
It is possible that the decrease in Muc2 expression in the IL10-KO mice is a consequence of the colonic inflammation brought about by the conventionalization process,66 in which case the anti-inflammatory effects of DEX-treated donor microbiota actually prevent further dampening of Muc2 gene expression. In support of the anti-inflammatory effects of DEX-altered microbiota, we found pro-inflammatory cytokine levels (IL12p40, IL17) in both mucosal and mesenteric lymph nodes to be significantly lower in donors exposed to DEX relative to controls. Notably, DEX-treated Muc2-KO donor microbiota were more colitogenic compared to both wild-type and Muc2-heterozygote cohorts, reinforcing our earlier findings that Muc2-KO mice are comparatively refractory to DEX-induced changes in gut microbiota.
In summary, we have shown that chronic exposure to DEX shifts gut microbiota in varying degrees to regulate colonic mucin gene expression in mice under both healthy and diseased conditions. Our results highlight the notion that gut microbiota are necessary and sufficient to regulate mucin gene expression. In contrast, DEX exposure in the absence of microbes is not presently sufficient to produce such an effect. We also report that the inherent microbiota from Muc2-KO mice is acutely colitogenic, pointing toward a possible mechanism through which these mice spontaneously develop colitis. However, DEX-mediated shifts in gut bacteria confer a protective effect on the development of colonic inflammation when these microbes are conventionalized into the IL10-KO mouse model. These findings add to the ever-expanding belief that gut microbiota wield considerable influence in the body and underscores their abundant potential as complex mechanistic regulators of gastrointestinal biology.
ACKNOWLEDGMENTS
The authors would like to thank Dr. Leonard Augenlicht from Albert Einstein College of Medicine for providing the Muc2-knockout mouse model. This work was supported by the grants: NIDDK Digestive Disease Research Center DK-42086 (EBC), NIH grants DK097268, T32 DK07074, and DK47722 (EBC), the Gastrointestinal Research Foundation, NIDDK UH3DK083993 (EBC), and the Harry and Leone Helmsley Charitable Trust.
REFERENCES
- 1.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Devaraj S, Hemarajata P, Versalovic J. The human gut microbiome and body metabolism: implications for obesity and diabetes. Clinical Chemistry. 2013;59:617–628. doi: 10.1373/clinchem.2012.187617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cerf-Bensussan N, Gaboriau-Routhiau Vr. The immune system and the gut microbiota: friends or foes? Nat Rev Immunol. 2010;10:735–744. doi: 10.1038/nri2850. [DOI] [PubMed] [Google Scholar]
- 4.Vijay-Kumar M, Aitken JD, Carvalho FA, et al. Metabolic syndrome and altered gut microbiota in mice lacking toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heimesaat MM, Fischer A, Siegmund B, et al. Shift towards pro-inflammatory intestinal bacteria aggravates acute murine colitis via Toll-like receptors 2 and 4. PLoS One. 2007;2:e662. doi: 10.1371/journal.pone.0000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Packey CD, Sartor RB. Commensal bacteria, traditional and opportunistic pathogens, dysbiosis and bacterial killing in inflammatory bowel diseases. Current opinion in infectious diseases. 2009;22:292. doi: 10.1097/QCO.0b013e32832a8a5d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cerf-Bensussan N, Gaboriau-Routhiau V. The immune system and the gut microbiota: friends or foes? Nat Rev Immunol. 2010;10:735–744. doi: 10.1038/nri2850. [DOI] [PubMed] [Google Scholar]
- 8.Bloom Seth M, Bijanki Vinieth N, Nava Gerardo M, et al. Commensal bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host & Microbe. 2011;9:390–403. doi: 10.1016/j.chom.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eckburg PB, Relman DA. The role of microbes in Crohn's disease. Clinical Infectious Diseases. 2007;44:256–262. doi: 10.1086/510385. [DOI] [PubMed] [Google Scholar]
- 10.Ohkusa T, Kato K, Terao S, et al. Newly developed antibiotic combination therapy for Ulcerative Colitis: a double-blind placebo-controlled multicenter trial. Am J Gastroenterol. 2010;105:1820–1829. doi: 10.1038/ajg.2010.84. [DOI] [PubMed] [Google Scholar]
- 11.Devkota S, Wang Y, Musch MW, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/−mice. Nature. 2012;487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forstner JF, Forstner GG. Gastrointestinal mucus. Physiology of the gastrointestinal tract. 1994:1255–1283. [Google Scholar]
- 13.Tytgat KMAJ, Buller HA, Opdam FJM, et al. Biosynthesis of human colonic mucin: Muc2 is the prominent secretory mucin. Gastroenterology. 1994;107:1352–1363. doi: 10.1016/0016-5085(94)90537-1. [DOI] [PubMed] [Google Scholar]
- 14.Velcich A, Yang W, Heyer J, et al. Colorectal cancer in mice genetically deficient in the mucin Muc2. Science. 2002;295:1726–1729. doi: 10.1126/science.1069094. [DOI] [PubMed] [Google Scholar]
- 15.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 16.Faubion WA, Jr, Loftus EV, Jr, Harmsen WS, et al. The natural history of corticosteroid therapy for inflammatory bowel disease: a population-based study. Gastroenterology. 2001;121:255–260. doi: 10.1053/gast.2001.26279. [DOI] [PubMed] [Google Scholar]
- 17.Friend. Review article: issues in oral administration of locally acting glucocorticosteroids for treatment of inflammatory bowel disease. Alimentary Pharmacology & Therapeutics. 1998;12:591–603. doi: 10.1046/j.1365-2036.1998.00348.x. [DOI] [PubMed] [Google Scholar]
- 18.Scheinman RI, Gualberto A, Jewell CM, et al. Characterization of mechanisms involved in transrepression of NF-kappa B by activated glucocorticoid receptors. Mol Cell Biol. 1995;15:943–953. doi: 10.1128/mcb.15.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallant C, Kenny P. Oral glucocorticoids and their complications. A review. J Am Acad Dermatol. 1986;14:161–177. doi: 10.1016/s0190-9622(86)70018-2. [DOI] [PubMed] [Google Scholar]
- 20.Heimdal K, Hirschberg H, Slettebà H, et al. High incidence of serious side effects of high-dose dexamethasone treatment in patients with epidural spinal cord compression. Journal of Neuro-Oncology. 1992;12:141–144. doi: 10.1007/BF00172664. [DOI] [PubMed] [Google Scholar]
- 21.Lutgendorff F, Akkermans LMA, Soderholm JD. The role of microbiota and probiotics in stress-induced gastrointestinal damage. Current Molecular Medicine. 2008;8:282–298. doi: 10.2174/156652408784533779. [DOI] [PubMed] [Google Scholar]
- 22.O'Mahony SM, Marchesi JR, Scully P, et al. Early life stress alters behavior, immunity, and microbiota in rats: implications for Irritable Bowel Syndrome and psychiatric illnesses. Biological Psychiatry. 2009;65:263–267. doi: 10.1016/j.biopsych.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 23.Huang EY, Leone VA, Devkota S, et al. Composition of Dietary Fat Source Shapes Gut Microbiota Architecture and Alters Host Inflammatory Mediators in Mouse Adipose Tissue. Journal of Parenteral and Enteral Nutrition. 2013 doi: 10.1177/0148607113486931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Culman S, Bukowski R, Gauch H, et al. T-REX: software for the processing and analysis of T-RFLP data. BMC Bioinformatics. 2009;10:171. doi: 10.1186/1471-2105-10-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Redinbaugh MG, Turley RB. Adaptation of the bicinchoninic acid protein assay for use with microtiter plates and sucrose gradient fractions. Analytical Biochemistry. 1986;153:267–271. doi: 10.1016/0003-2697(86)90091-6. [DOI] [PubMed] [Google Scholar]
- 26.Tytgat KM, Bovelander F-J, Opdam FJ, et al. Biosynthesis of rat MUC2 in colon and its analogy with human MUC2. Biochemical Journal. 1995;309:221. doi: 10.1042/bj3090221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johansson MEV, Phillipson M, Petersson J, et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proceedings of the National Academy of Sciences. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van der Sluis M, De Koning BAE, De Bruijn ACJM, et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131:117–129. doi: 10.1053/j.gastro.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 29.CDC. Inflammatory Bowel Disease (IBD) [Accessed April 14, 2013];2011 Available at: [Google Scholar]
- 30.Lees CW, Barrett JC, Parkes M, et al. New IBD genetics: common pathways with other diseases. Gut. 2011;60:1739–1753. doi: 10.1136/gut.2009.199679. [DOI] [PubMed] [Google Scholar]
- 31.Loh G, Blaut M. Role of commensal gut bacteria in inflammatory bowel diseases. Gut microbes. 2012;3:503–514. doi: 10.4161/gmic.22156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 33.Forsythe P, Kunze WA, Bienenstock J. On communication between gut microbes and the brain. Current Opinion in Gastroenterology. 2012;28:557–562. doi: 10.1097/MOG.0b013e3283572ffa. [DOI] [PubMed] [Google Scholar]
- 34.Ünsal H, Balkaya M, Ünsal C, et al. The short-term effects of different doses of dexamethasone on the numbers of some bacteria in the ileum. Digestive Diseases and Sciences. 2008;53:1842–1845. doi: 10.1007/s10620-007-0089-6. [DOI] [PubMed] [Google Scholar]
- 35.Barnes PJ. Anti-inflammatory actions of glucocorticoids: molecular mechanisms. Clin Sci. 1998;94:557–572. doi: 10.1042/cs0940557. [DOI] [PubMed] [Google Scholar]
- 36.Smoak KA, Cidlowski JA. Mechanisms of glucocorticoid receptor signaling during inflammation. Mechanisms of Ageing and Development. 2004;125:697–706. doi: 10.1016/j.mad.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 37.Furrie E, Macfarlane S, Kennedy A, et al. Synbiotic therapy (Bifidobacterium longum/Synergy 1) initiates resolution of inflammation in patients with active ulcerative colitis: a randomised controlled pilot trial. Gut. 2005;54:242–249. doi: 10.1136/gut.2004.044834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riedel CU, Foata F, Philippe D, et al. Anti-inflammatory effects of bifidobacteria by inhibition of LPS-induced NF-kappaB activation. World J Gastroenterol. 2006;12:3729–3735. doi: 10.3748/wjg.v12.i23.3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Madsen KL, Doyle JS, Jewell LD, et al. Lactobacillus species prevents colitis in interleukin 10 gene-deficient mice. Gastroenterology. 1999;116:1107–1114. doi: 10.1016/s0016-5085(99)70013-2. [DOI] [PubMed] [Google Scholar]
- 40.Kim YS, Byrd JC. Ulcerative colitis: a specific mucin defect? Gastroenterology. 1984;87:1193. [PubMed] [Google Scholar]
- 41.Nanthakumar NN, Young C, Ko JS, et al. Glucocorticoid responsiveness in developing human intestine: possible role in prevention of necrotizing enterocolitis. Am J Physiol Gastrointest Liver Physiol. 2005;288:G85–G92. doi: 10.1152/ajpgi.00169.2004. [DOI] [PubMed] [Google Scholar]
- 42.Auphan N, DiDonato JA, Rosette C, et al. Immunosuppression by glucocorticoids: inhibition of NF-kB activity through induction of IkB synthesis. Science. 1995;270:286–290. doi: 10.1126/science.270.5234.286. [DOI] [PubMed] [Google Scholar]
- 43.Crinelli R, Antonelli A, Bianchi M, et al. Selective inhibition of NF-kB activation and TNF-a production in macrophages by red blood cell-mediated delivery of dexamethasone. Blood Cells, Molecules, and Diseases. 2000;26:211–222. doi: 10.1006/bcmd.2000.0298. [DOI] [PubMed] [Google Scholar]
- 44.Blank M, Klussmann E, Krüger-Krasagakes S, et al. Expression of MUC2-mucin in colorectal adenomas and carcinomas of different histological types. International Journal of Cancer. 1994;59:301–306. doi: 10.1002/ijc.2910590302. [DOI] [PubMed] [Google Scholar]
- 45.Hanski C, Hofmeier M, Schmitt-Gräff A, et al. Overexpression or ectopic expression of MUC2 is the common property of mucinous carcinomas of the colon, pancreas, breast, and ovary. The Journal of Pathology. 1997;182:385–391. doi: 10.1002/(SICI)1096-9896(199708)182:4<385::AID-PATH861>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 46.Liu JB, Zhang ZX, Xu YJ, et al. Effects of glucocorticoid on airway mucus secretion in asthma: experiment with asthmatic mouse model. Zhonghua yi xue za zhi. 2006;86:2491. [PubMed] [Google Scholar]
- 47.Sudo N. Stress and gut microbiota: Does postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response? International Congress Series. 2006;1287:350–354. [Google Scholar]
- 48.Sudo N, Chida Y, Aiba Y, et al. Postnatal microbial colonization programs the hypothalamic–pituitary–adrenal system for stress response in mice. The Journal of Physiology. 2004;558:263–275. doi: 10.1113/jphysiol.2004.063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neufeld KM, Kang N, Bienenstock J, et al. Reduced anxiety-like behavior and central neurochemical change in germ-free mice. Neurogastroenterology & Motility. 2011;23:255-e119. doi: 10.1111/j.1365-2982.2010.01620.x. [DOI] [PubMed] [Google Scholar]
- 50.Pariante CM, Lightman SL. The HPA axis in major depression: classical theories and new developments. Trends in Neurosciences. 2008;31:464–468. doi: 10.1016/j.tins.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 51.Meslin J-C, Fontaine N, Andrieux C. Variation of mucin distribution in the rat intestine, caecum and colon: effect of the bacterial flora. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology. 1999;123:235–239. doi: 10.1016/s1095-6433(99)00056-2. [DOI] [PubMed] [Google Scholar]
- 52.Deplancke B, Gaskins HR. Microbial modulation of innate defense: goblet cells and the intestinal mucus layer. The American Journal of Clinical Nutrition. 2001;73:1131S–1141S. doi: 10.1093/ajcn/73.6.1131S. [DOI] [PubMed] [Google Scholar]
- 53.Elinav E, Strowig T, Kau Andrew L, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Henao-Mejia J, Elinav E, Jin C, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wen L, Ley RE, Volchkov PY, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lozupone CA, Stombaugh JI, Gordon JI, et al. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ley RE, Backhed F, Turnbaugh P, et al. Obesity alters gut microbial ecology. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hoskins LC. Mucin degradation in the human gastrointestinal tract and its significance to enteric microbial ecology. European Journal of Gastroenterology & Hepatology. 1993;5:205–213. [Google Scholar]
- 59.Cummings JH, Macfarlane GT. The control and consequences of bacterial fermentation in the human colon. Journal of Applied Bacteriology. 1991;70:443–459. doi: 10.1111/j.1365-2672.1991.tb02739.x. [DOI] [PubMed] [Google Scholar]
- 60.Mukhopadhya I, Hansen R, El-Omar EM, et al. IBD-what role do Proteobacteria play? Nat Rev Gastroenterol Hepatol. 2012;9:219–230. doi: 10.1038/nrgastro.2012.14. [DOI] [PubMed] [Google Scholar]
- 61.Ramer-Tait A, Overstreet A-M, Hostetter J, et al. Colonization with distinct Proteobacteria provocateurs induces multifactorial gastrointestinal inflammation that is regulated by CD4+ T cells: P-194. Inflammatory Bowel Diseases. 2012;18:S92. [Google Scholar]
- 62.de La Serre CB, Ellis CL, Lee J, et al. Propensity to high-fat diet-induced obesity in rats is associated with changes in the gut microbiota and gut inflammation. Am J Physiol Gastrointest Liver Physiol. 2010;299:G440–G448. doi: 10.1152/ajpgi.00098.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hoffmann C, Hill DA, Minkah N, et al. Community-wide response of the gut microbiota to enteropathogenic Citrobacter rodentium infection revealed by deep sequencing. Infection and Immunity. 2009;77:4668–4678. doi: 10.1128/IAI.00493-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ventura M, O'Flaherty S, Claesson MJ, et al. Genome-scale analyses of health-promoting bacteria: probiogenomics. Nat Rev Micro. 2009;7:61–71. doi: 10.1038/nrmicro2047. [DOI] [PubMed] [Google Scholar]
- 65.Sellon RK, Tonkonogy S, Schultz M, et al. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infection and Immunity. 1998;66:5224–5231. doi: 10.1128/iai.66.11.5224-5231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van der Sluis M, Bouma J, Vincent A, et al. Combined defects in epithelial and immunoregulatory factors exacerbate the pathogenesis of inflammation: mucin 2-interleukin 10-deficient mice. Laboratory Investigation. 2008;88:634–642. doi: 10.1038/labinvest.2008.28. [DOI] [PubMed] [Google Scholar]