Abstract
Signaling lymphocyte activation molecules (SLAMs) play an integral role in immune regulation. Polymorphisms in the SLAM family receptors are implicated in human and mouse model of lupus disease. The lupus-associated, somatically mutated and class-switched pathogenic autoantibodies are generated in spontaneously developed germinal centers (Spt-GCs) in secondary lymphoid organs. The role and mechanism of B cell-intrinsic expression of polymorphic SLAM receptors that affect B cell tolerance at the GC checkpoint is not clear. Here, we generated several bacterial artificial chromosome (BAC) transgenic mice that overexpress B6 alleles of different SLAM family genes in autoimmune-prone B6.Sle1b mice. B6.Sle1b mice overexpressing B6-derived Ly108 and CD84 exhibit a significant reduction in the Spt-GC response and autoantibody production compared to B6.Sle1b mice. These data suggest a prominent role of Sle1b-derived Ly108 and CD84 in altering the GC checkpoint. We further confirm that expression of lupus-associated CD84 and Ly108 specifically on GC B cells in B6.Sle1b mice is sufficient to break B cell tolerance leading to an increase in autoantibody production. In addition, we observe that B6.Sle1b B cells have reduced BCR signaling, and a lower frequency of B cell-T cell conjugates, which are reversed in B6.Sle1b mice overexpressing B6 alleles of CD84 and Ly108. Finally, we find a significant decrease in apoptotic GC B cells in B6.Sle1b mice compared to B6 controls. Our study establishes the central role of GC B cell-specific CD84 and Ly108 expression in maintaining B cell tolerance in GCs and in preventing autoimmunity.
INTRODUCTION
The SLAM (signaling lymphocyte activation molecule) family receptors play critical roles in immune regulation and are required for an effective humoral response (1, 2). The Sle1b sublocus derived from the lupus-prone NZM2410/NZW strain harbors the SLAM family (Slamf) genes (3). Previous studies have suggested the Slamf genes to be major players in mediating loss of tolerance to nuclear antigens and in the development of autoimmunity in B6.Sle1b mice (3). Polymorphisms in the Ly108/Slamf6 gene are implicated in the loss of early B cell (4) as well as peripheral T cell-mediated tolerance (5). Several independent studies have suggested the contribution of three other SLAMF genes, Slamf1 (SLAM/CD150), Slamf2 (CD48) and Slamf3 (Ly9) in autoimmunity (6–8). However, the role of B cell-intrinsic expression of different isoforms of Ly108 or other SLAM receptors in the regulation of B cell tolerance at the germinal center (GC) checkpoint remains unclear. Elucidation of the mechanism by which SLAM receptors regulate GCs is important as GCs are spontaneously developed in autoimmune mice and humans and generate somatically mutated and class-switched pathogenic autoantibodies (9–11).
Because Slamf genes are genetically linked, it has been difficult to determine the potential role of polymorphisms in a specific family member in autoimmunity using knockout mice generated in an autoimmune 129 background that are subsequently backcrossed to B6 (6–8, 12, 13). It is also unclear whether non-Slamf genes located in Sle1b contribute to a break in B cell tolerance given the presence of non-synonymous mutations in these genes in autoimmune B6.Sle1b mice (3). To definitively determine the role of particular Slamf and/or non-Slamf genes within the Sle1b sublocus in the development of autoimmunity and to study the mechanisms by which these genes affect B cell tolerance at the GC checkpoint, we used a bacterial artificial chromosome (BAC) transgenic rescue approach. We generated 6 BAC transgenic mouse lines expressing B6 alleles of the different genes spanning the entire Sle1b sublocus and then bred them onto B6.Sle1b mice. The B6.Sle1b BAC transgenic mouse line containing B6 alleles of Ly108 and CD84 (designated Sle1b-BAC90) showed a significant reduction in anti-nuclear Ab (ANA) titers and positivity (penetrance) and a decreased spontaneously developed GC (Spt-GC) response, indicating a critical role of Ly108 and CD84 in maintaining GC B cell tolerance.
Both CD84 and Ly108 are essential in maintaining tolerance because B6.Sle1b BAC transgenic mice expressing the B6 allele of Ly108 alone showed only a partial restoration of tolerance to ANA. Using a conditional deletion (Cre-LoxP) system, we showed that GC B cell-specific expression of autoimmune-prone CD84 and Ly108 genes is sufficient for the loss of B cell tolerance. B cell function assays revealed that polymorphisms in the CD84 and Ly108 proteins in B6.Sle1b B cells helped B cells escape tolerance by lowering BCR signaling, decreasing apoptosis and attenuating B cell-T cell interactions. Normalization of B6.Sle1b-derived CD84 and Ly108 by overexpression of wild type CD84 and Ly108 restored BCR signaling and B cell-T cell interactions. Our results highlight the vital role of GC B cell specific Ly108 and CD84 expression and function in the maintenance of B cell tolerance at the GC checkpoint.
MATERIALS AND METHODS
Mice
Breeding pairs for C57BL/6 (B6), GC B cell Cre (C.129P2(Cg)-Ighg1tm1(IRES-cre)Cgn/J) and OT-II transgenic (B6.Cg-Tg (TcraTcrb)425Cbn/J) mice were originally purchased from the Jackson Laboratory and bred in house. Derivation of B6 mice congenic for the Sle1b sub-locus (named B6.Sle1b) was previously described (14). All animals were housed in specific pathogen-free animal facility at Penn State Hershey Medical Center and all procedures were performed in accordance with the guidelines approved by our Institutional Animal Care and Use Committee.
Generation of BAC transgenic mice
Bacterial Artificial Chromosomes (BACs) were identified from the B6-derived BAC library (BACPAC Resources, Oakland, CA). The following BACs were used: RPCI-23 194D6 (BAC41), RPCI-23 48O11 (BAC47) RPCI-23 171K8 (BAC25), RPCI-23 145F9 (BAC40), RPCI-23-388C4 (BAC90), RPCI-23 462J8 (BAC95), and RPCI-23 438K9 (BAC-Ly108). Bacterial cultures from these BAC clones were grown in LB with chloramphenicol (12.5 μg/mL) overnight at 37°C. Qiagen Midi Kit solutions, P1, P2, and P3, were used to isolate BAC DNA (Qiagen, Inc., Valencia, CA). The DNA was then further purified by phenol-chloroform extraction followed by cesium chloride gradient ultracentrifugation. BAC DNA was then diluted to 50 ng/μL in TE buffer (10mM TrisHCl pH 7.5, 0.1mM EDTA) and injected into fertilized-oocytes derived from B6. Founder lines were identified by PCR specific for the vector ends and then bred on to respective genetic backgrounds.
Generation of the floxed BAC90 mice
The BAC (RPCI-23-388C4) used to generate the floxed BAC90 mice was purchased from the BACPAC resources (Oakland, CA). The BAC vector backbone already contained LoxP sites that flanked the insert (B6-derived CD84 and Ly108). No additional manipulation was made to the backbone. BAC vector DNA amplification, purification and injection into B6-derived fertilized-oocytes were performed as described above in the “Generation of BAC transgenic mice”.
Flow cytometry
The following antibodies were utilized for flow cytometric analysis of mouse splenocytes or bone marrow cells: PacBlue-anti-B220 (RA3-6B2); Alexa Fluor 700-anti-CD4 (RM4-5); PE-anti-PD-1 (29F.1A12); PerCP-Cy5.5-anti-CD69 (H1.2F3); APC-Cy7- anti-CD25 (PC61); Cy5-anti-CD86 (GL1); PeCy7-anti-CD95 (FAS, Jo2); PeCy7-anti-MHC-II (M5/114.15.2); APC-anti-CD24 (HSA) (M1/69); Biotin-anti-Ly5.1 (BP-1) (6C3); FITC-anti-CD23 (B3B4); PE-Cy5-streptavidin (SA) were from purchased from BioLegend, San Diego, CA. Biotin-anti-CXCR5 (2G8) from BD Pharmingen, San Diego, CA. FITC-peanutagglutinin (PNA) from Vector Labs, Burlingame, CA. PE-anti-IgM (eB121-15F9); APC anti-CD93 (AA4.1); FITC-anti-F4/80 (BM8) from eBiosciences, San Diego, CA. The following antibodies were utilized for phosphoflow analysis of purified mouse B cells: PE-anti-Btk (pY223)/ItK (pY180) (N35-86); Alexa Fluor 647-anti-ERK1/2 (pT202/pY204) (20A); Pacific Blue-anti-p38 MAPK (pT180/pY182); Alexa Fluor 488-anti-Syk (pY348) (I120-722). Stained cells were analyzed using the BD LSR II flow cytometer (BD Biosciences, Franklin lakes, NJ). Data were acquired using FACSDiva software (BD Biosciences, San Jose, CA) and analyzed using FlowJo software (Tree Star, San Carlos, CA).
Immunohistology
The following antibodies and reagents were utilized for immunohistochemical analysis of mouse spleen sections: PE-anti-CD4 (GK1.5); FITC-GL7 (RA3-6B2); APC-anti-IgD (all from BD Biosciences). Spleen cryostat sections (5–6 um) were prepared as previously described (15). Immunohistochemistry was performed as described (15) and stained sections were analyzed with a Leica DM4000 fluorescence microscope and Leica software (Leica Microsystems, Buffalo Grove, IL). The color intensity of the images was slightly enhanced using Adobe Photoshop CS4 (Adobe Systems, San Jose, CA) for better visualization and was carried out consistently between all sections while maintaining the integrity of the data.
ELISpot assays
ELISpot assays were performed as previously described (16). Briefly, splenocytes in 10% RPMI were plated at a concentration of 1 × 105 cells/well onto anti-IgM or anti-IgG (Invitrogen, Grand Island, NY) coated, or at 1 × 106 cells/well on dsDNA-, histone-, or nucleosome-coated multiscreen 96-well filtration plates (Millipore, Bedford, MA). Serially diluted (1:2) cells were incubated for 6 h at 37°C. IgM-producing AFCs were detected using biotinylated anti-mouse IgM (Jackson Immunoresearch, West Grove, PA) and streptavidin (SA)-alkaline phosphatase (Vector Laboratories, Burlingame, CA). IgG-producing AFCs were detected using alkaline phosphatase-conjugated anti-mouse IgG (Molecular Probes, Grand Island, NY). dsDNA-, histone-, and nucleosome-specific AFCs were detected by biotinylated anti-kappa Ab (Invitrogen, Grand Island, NY) and streptavidin (SA)-alkaline phosphatase (Vector Laboratories, Burlingame, CA) or alkaline phosphatase-conjugated anti-mouse IgG (Molecular Probes, Grand Island, NY). Plates were developed using the Vector Blue Alkaline phosphatase Substrate Kit III (Vector Laboratories, Burlingame, CA). ELISpots were enumerated and analyzed using a computerized imaging/analysis system (Cellular Technology, Shaker Heights, OH).
Serology: Ig and autoAb titers
ELISA plates were coated with anti-IgM or anti-IgG capture antibodies (from Invitrogen, Grand Island, NY) and detected using biotinylated anti-mouse IgM (Jackson Immunoresearch, West Grove, PA) or alkaline-phosphatase conjugated anti-mouse IgG (Molecular Probes, Grand Island, NY). Total IgG autoAb titers were measured in ELISA plates coated with dsDNA, histone, nucleosome, Sm/RNP or cardiolipin and detected with biotinylated anti-kappa Ab (Invitrogen, Grand Island, NY). IgG subtype-specific autoAb titers were detected by biotinylated-IgG1, biotinylated IgG2b, and AP-IgG2c Abs (Southern Biotech, Birmingham, AL). Biotinylated antibodies were detected by streptavidin (SA)-alkaline phosphatase (Vector Laboratories, Burlingame, CA). The plates were developed by the PNPP (p-Nitrophenyl Phosphate, Disodium Salt) (Thermo Fisher Scientific, Rockford, IL) substrates for alkaline phosphatase.
In vitro B cell proliferation assay
Naïve B cells were purified from indicated mice with mouse anti-CD43 (Ly-48) microbeads. Purified B cells were stained with 3 μM CFSE (Sigma Aldrich, St. Louis, MO) in PBS with 5% FBS for 15 min at RT. Stained B cells were cultured with 25 μg/ml soluble anti-IgM (Jackson ImmunoResearch Laboratories) and 20 μg/ml soluble anti-CD40 Ab (BioLegend, San Diego, CA). After 72 hours of stimulation, cells were acquired through the BD LSR II flow cytometer.
Ex-vivo apoptosis detection assay
Total splenocytes from 3–4 mo old female mice of the indicated strains were stained with SR-FLICA in-vitro poly-caspase detection reagent (AbD Serotec, Kidlington, U.K.) for 30 min at 37°C in a water bath, followed by staining with indicated GC B cell markers. Cells were acquired immediately after staining on the BD LSRII cytometer. DAPI positive dead cells and doublets were gated out during the analysis.
Cell cycle analysis
B cells were cultured with anti-IgM (25 μg/ml) and anti-CD40 (20 μg/ml) for indicated time periods, harvested and washed with chilled PBS, and fixed with chilled 70% ethanol overnight at −20 °C. Subsequently, cells were centrifuged at 1000 X g for 10 min at 4 °C, washed with PBS and stained with PI staining solution containing 50 μg/ml Propidium Iodide (PI), 50 μg/ml RNase A (Roche Applied Sciences, Indianapolis, IN) and 100 μM EDTA in PBS, for 1–2 h at 42 °C. Cells were analyzed by flow cytometry.
Calcium flux assay
Naïve B cells were purified from each indicated group of mice with mouse anti-CD43 (Ly-48) microbeads. Purified B cells were stained with Fura Red (10 uM) and Fluo-3 (5 uM) (Invitrogen, Carlsbad, CA) in dyeless RPMI (Mediatech) for 45 minutes at 37 °C. A baseline calcium reading was acquired for 1 minute before addition of 5 or 10 μg/ml of anti-IgM (Jackson ImmunoResearch Laboratories), followed immediately by a three-minute acquisition through the BD LSR II flow cytometer.
B cell-T cell in vitro conjugate assay
Naïve B cells from each indicated group of mice were purified with mouse anti-CD43 (Ly-48) microbeads, stained with 5 uM Cell Trace Violet (Invitrogen) and activated with 25 μg/ml soluble anti-IgM and 1 μg/ml LPS for 24 hours at 37 °C. Naïve T cells from OT-II transgenic mice were purified with a pan-T isolation kit (Miltenyi Biotech), stained with 3 uM CFSE, and activated with 10 μg/ml anti-CD3 and 2 μg/ml anti-CD28 for 24 hours at 37°C. 10 μg/ml OVA peptide (Invivogen, San Diego, CA) was added to the B cells three hours prior to the start of the assay. 1×106 B cells and 1×106 T cells were placed together for the stipulated time intervals, fixed with 4% paraformaldehyde after the appropriate time concluded and acquired through the BD LSR II.
Phosphoflow analysis
Splenocytes isolated from each indicated mouse group were activated with 25 ug/ml of anti-IgM for the indicated time intervals at 37 °C. Following activation, cells were fixed with Lyse/Fix (BD) for 10 minutes at 37 °C. Following washes, cells were permeabilized with Perm/Wash Buffer (BD) for 30 minutes at RT. Following 2 washes with Perm/Wash Buffer, cells were stained for anti-B220 and phosphoflow antibodies (BD) described above for 1 hour. Samples were acquired through the BD LSRII flow cytometer.
Assessment of germinal center sizes
Immunohistochemistry was performed using the Abs listed in the Immunohistology section and stained sections were analyzed with a Leica DM4000 fluorescence microscope and Leica software (Leica Microsystems, Buffalo Grove, IL). A total of 10 randomly selected germinal centers per mouse from the indicated mouse groups (5 mice from each group) were assessed for total area (uM2) using the Leica Microsystems Software.
In vitro B cell functional assays
B cells were purified from naïve B6, B6.Sle1b or Sle1b-BAC90 mice with mouse anti-CD43 (Ly-48) microbeads, stimulated with 25 ug/ml anti-IgM, 10 ug/ml anti-CD40, and/or 5 ug/ml of anti-Ly108 (13G3-19D) (ebioscience) or 2 ug/ml of recombinant CD84 agonist (R&D Systems) for 24 hours, and acquired with BD LSRII flow cytometer.
DNA/RNA-preparation and Real time RT-PCR
Total DNA was isolated with DNeasy Blood & Tissue kit (Qiagen, Hilden, Germany) following the manufacturer’s guidelines from the tails of the indicated mice. Gene expression was quantified using Taqman master mix by the Applied Biosystems StepOne Plus real time PCR system. Primers for CD84 and Ly108 copy number assay were purchased from Life Technologies. Amplification conditions for all primer sets were; one cycle at 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 sec and 60 °C for 1 min. Mouse Tert was used as the reference gene.
Total RNA was isolated from purified B cells and T cells, or GC B cells and Tfh cells sorted with FACS Aria Sorter by using the RNeasy mini kit (Qiagen), following the manufacturer’s guidelines. RNA was reverse transcribed using the High-Capacity Reverse Transcription kit (Applied Biosystems, Grand Island, NY). Gene expression was quantified using Taqman PCR Master Mix kit or Power SYBR Green PCR Master Mix kit (Applied Biosystems, Grand Island, NY) by the Applied Biosystems StepOne Plus real-time PCR system. Primers for the SLAM genes were purchased from Life Technologies and Ly108 isoform primers were designed using Primer3 software and synthesized by IDT technologies (Coralville, Iowa). Primer sequences – 1.) Ly108.1: forward 5′ –ctcgtccaatgcaggaaatg -3′, reverse 5′ – agatgtgttctccctggatttc -3′ 2.) Ly108.2: forward 5′ – ctcgtccaatgcaggaaatg -3′, reverse 5′-aggagttatagttgattaag -3′ 3.) Ly108-H1: forward 5′-agcacagatggcccagg -3′, reverse 5′-aggagttatagttgattaaag -3′. Amplification conditions for all primer sets were: one cycle at 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 sec and 60 °C for 1 min. GAPDH was used as the reference gene for sample normalization.
Extracellular flux analysis
Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were measured in a XF96 extracellular flux analyzer by using a kit as per manufacturer’s instructions (Seahorse Bioscience, Boston, MA). Briefly, B cells from B6, B6.Sle1b and Sle1b-BAC90 mice were purified by negative selection using CD43 microbeads by MACS purification (Miltenyi Biotech). Purified B cells were stimulated with 25 μg/ml anti-IgM and 20 μg/ml anti-CD40 for 18h in RPMI+ 10% FCS. Thereafter, 6 × 105 activated B cells were plated in XF seahorse media with 25mM glucose onto a Cell-TAK (BD Biosciences) coated XF96 plate. Oligomycin, 1 μM; FCCP, 2 μM; rotenone and antimycin, 1 μM each were added sequentially and measurements of extracellular flux were recorded. Glycolysis was calculated from ECAR values at the basal respiration phase and glycolytic capacity was calculated from ECAR values after the inhibition of ATP synthase by the addition of oligomycin, when cells resort to meet their energy demands by using their maximum glycolytic capacity. OCR and ECAR data were analyzed using the XF wave software and plotted using GraphPad prism v6.0.
2D Live cell microscopy, image acquisition and cell tracking analysis
For the live cell tracking measurements, the confocal technique (Leica AOBS SP8 laser scanning confocal microscope, Heidelberg, Germany) was utilized along with a heated live cell stage equipped with a humidified 5% CO2 perfusion system (Tokai Hit, Japan). The live cell data mode (xyt) in the image acquisition software (Leica Confocal Software LAF) was used for the 2D imaging of live cells. 2D live cell images of activated B cells loaded with OVA peptide and labeled with Cell Trace (Invitrogen) and activated OT-II T cells stained with CFSE (Invitrogen) were mixed in 0.5mg/ml collagen solution, pH 7.0 (TelCol, type1 acid soluble collagen solution from Advanced BioMatrix, San Diego, CA) and acquired sequentially using the 63X/1.2 NA high numerical aperture apochromatic water immersion objective at every 30 sec for 1 hour. The laser lines for excitation included UV diode, 80 MHz white light laser (Leica AOBS SP8 module) and the emission signals were collected sequentially using AOBS tunable filters to eliminate cross-talk and cross-excitation. All of the fluorescent images as a function of time were collected using highly sensitive HyD detectors (with time gated option). The images (8 bit) were line-averaged to minimize the random noise, and a pixel format of 512X512 was selected. For each sample, 2D time series images were compiled and cell tracking was performed using Bitplane Imaris (Switzerland) and Bitplane XT functions based on the Matlab program. The data were compiled in excel sheets and final plots, slope measurements and statistical analysis were performed either in Graphpad Prism Ver. 6 or Origin Lab Ver. 9.1.
Statistical analysis
Unpaired, non-parametric, Mann-Whitney, student’s t-test -GraphPad Prism 6 analytical tool (La Jolla, CA) or Microsoft Excel (Redmond, WA) were used. Wherever indicated, NS = non-significant, * p < 0.05, ** p < 0.01, and ***p < 0.001. Error bars reflect mean values unless otherwise mentioned.
RESULTS
A major role of SLAM family receptors CD84 and Ly108 in maintaining tolerance
The SLAM family genes, located in the telomeric end of chromosome 1 in autoimmune mice (i.e., NZM2410/NZW-derived Sle1b interval) and human SLE patients, are implicated in altering B cell tolerance (3, 16–18). To definitively determine the contribution of particular Slamf genes within the Sle1b sublocus in autoimmunity, a panel of B6-derived BACs spanning the entire Sle1b interval was used to generate a series of transgenic mouse lines (Figure 1A).
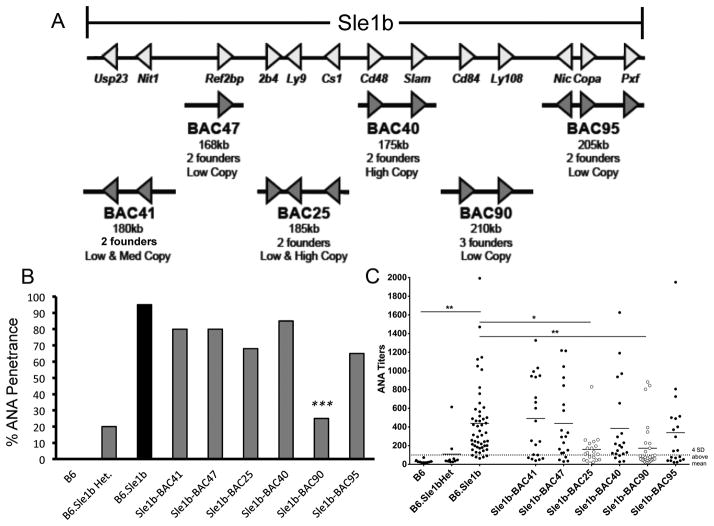
Figure 1. Suppression of ANA production in B6.Sle1b mice overexpressing B6-derived CD84 and Ly108.
(A) Schematic of the six B6-derived BACs (Bacterial Artificial Chromosomes) spanning the Sle1b interval. Each BAC harbors B6 alleles that genetically complement the indicated region in the Sle1b interval. Founders had variable copy numbers (low copy (1–2), med copy (3–4), and high copy (>10)). (B) Six BACs carrying the B6 alleles of the Sle1b genes were crossed to B6.Sle1b mice and two-three founder lines for each BAC were analyzed for IgG-specific ANA positivity at 7 months of age. Statistical analysis was performed using logistic regression (SAS version 9.1). (C) IgG ANA titers were assessed in 7 month old BAC transgenic mice. Dotted line represents the value that is four standard deviations above the mean titer from B6 mice. ANA titers in Sle1b-BAC25 and Sle1b-BAC90 mice were significantly higher than B6.Sle1b (p<0.01 and p<0.001, respectively). *p <0.05, **p <0.01, ***p <0.001.
Multiple founder lines carrying different copy numbers were generated for each BAC (Figure 1A). Of the 6 BAC transgenic mouse lines generated, only the BAC transgene carrying B6 alleles of CD84 and Ly108 (referred to as BAC90) was able to significantly suppress both anti-nuclear antibody (ANA) positivity/penetrance (Figure 1B) and titers (Figure 1C) when bred to B6.Sle1b mice (referred to as Sle1b-BAC90). The BAC transgene carrying 2B4 (Slamf4), Ly9 (Slamf3) and Cs1 (Slamf7) (referred to as BAC25) also had lower ANA titers (Figure 1C) but 70% of B6.Sle1b mice expressing BAC25 transgene (designated Sle1b-BAC25) still remained positive for ANA (Figure 1B). When we analyzed ANA titers in B6.Sle1b mice expressing each founder line for a given BAC transgenic strain, we did not find any significant difference among the individual founder lines except for BAC25 and BAC90 (Supplementary Figure 1). Of the three BAC90 transgenic founder lines generated, two (BAC90-4389 and BAC90-4390) had significantly lower ANA titers than the third one (BAC90-4270). A significant difference in ANA titers was observed between BAC90-4270 and BAC90-4389 lines (Supplementary Figure 1). We also observed a difference between the two BAC25 lines (Supplementary Figure 1).
To determine whether Ly108 alone was able to mediate the suppression of ANA production in Sle1b-BAC90 mice, a BAC transgenic mouse carrying only the B6 allele of Ly108 was generated and bred onto the B6.Sle1b background (referred to as Sle1b-BAC-Ly108). Sle1b-BAC-Ly108 mice displayed a reduction in serum ANA penetrance and titers (Supplementary Figure 2A–D), but not to the extent that was observed in aged Sle1b-BAC90 mice that carry both CD84 and Ly108 (Figure 1B–C). These data indicate that allelic variations in both CD84 and Ly108 were required to break tolerance to nuclear self-antigens in B6.Sle1b mice.
Restoration of B cell tolerance at the GC checkpoint by overexpression of B6-derived CD84 and Ly108
Sle1b alters peripheral B cell tolerance at the GC checkpoint, leading to elevated spontaneously developed GC (Spt-GC) and follicular helper T cell (Tfh) responses (16). Here, we asked whether introduction of the B6 wild type alleles of CD84 and Ly108 into B6.Sle1b mice could restore tolerance by regulating the GC checkpoint in B6.Sle1b mice. We first evaluated the surface expression and found that CD84 and Ly108 levels were higher in B cells from B6.Sle1b and Sle1b-BAC90 mice compared to B6 mice at 2 months of age (Supplementary Figure 3A). However, B6.Sle1b B cells exhibited a significant reduction in both CD84 and Ly108 surface expression by 8 months compared to B6 and Sle1b-BAC90 B cells (Supplementary Figure 3B). A similar temporally biphasic expression profile for CD84 and Ly108 was observed when GC B cells were analyzed at 2 and 8 mo of age (data not shown). CD84 expression on CD4+ T cells remained similar among the three strains, but Ly9 and Ly108 levels on CD4+ T cells were higher in B6.Sle1b and Sle1b-BAC90 mice than B6 mice at both 2 and 8 mo of age (data not shown). We observed no difference in SLAM adaptor (SAP) molecule transcript levels in B cells and T cells from these mice (data not shown).
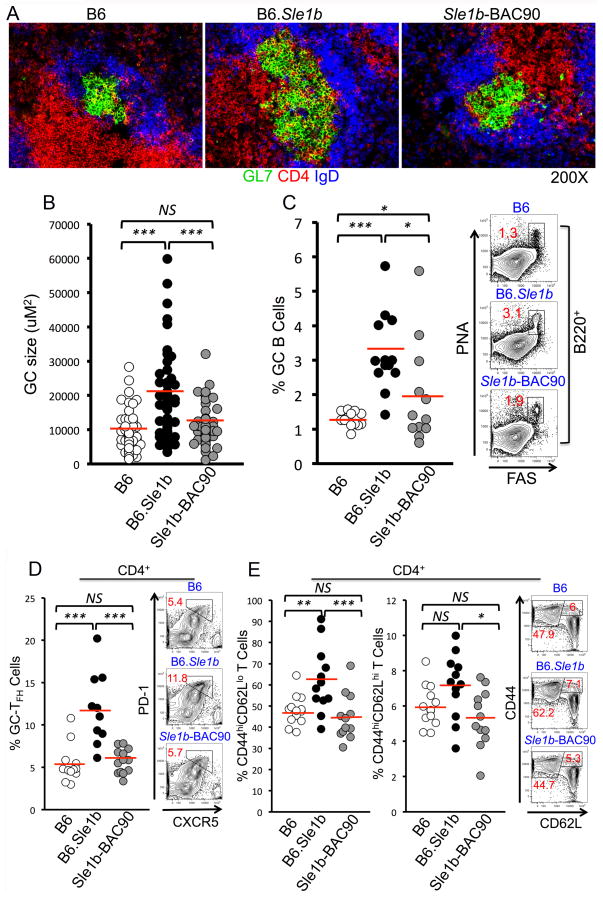
Immunohistological analysis of spleens revealed smaller and less frequent Spt-GCs in B6 and Sle1b-BAC90 mice compared to B6.Sle1b mice (Figure 2A–B). This decrease in Spt-GCs was confirmed by flow cytometric analysis of splenocytes, which showed a significant reduction in the percentage of B220+FashiPNAhi Spt-GC B cells in 6–8 mo old Sle1b-BAC90 female mice compared to age-matched B6.Sle1b females (Figure 2C). There was a strong inverse correlation between the percentage of GC B cells and the levels of Ly108 and CD84 expression on GC B cells (data not shown). Next, we asked whether the decrease in Spt-GCs in Sle1b-BAC90 mice resulted in a decline in the Tfh response. B6 and Sle1b-BAC90 mice had similar percentages of CD4+CXCR5hiPD-1hi Tfh cells, CD4+CD44hiCD62Llo short-lived effector/effector memory T cells and CD4+CD44hiCD62Lhi central memory T cells, all of which were significantly lower than B6.Sle1b mice (Figure 2D–E). These data suggest the cooperative function of CD84 and Ly108 in maintaining GC B cell tolerance.
Figure 2. Restoration of B cell tolerance in GCs in Sle1b-BAC90 mice expressing wild type CD84 and Ly108.
(A) Representative immunohistological images of spleen sections from 3–5 mice per indicated group, stained with GC B cell marker GL7 (green), CD4 T cell marker (red) and follicular B cell marker IgD (blue). (B) Scatterplot displays the range of GC sizes from each mouse strain. (C) Flow cytometric analysis of percentage of B220+FashiPNAhi GC B cells. Each circle represents an individual mouse. (D–E) Percentage of Tfh cells (CD4+CXCR5hiPD-1hi), short-lived effector/effector memory T cells (CD4+CD44hiCD62Llo) and central memory T cells (CD4+CD44hiCD62Lhi). Each circle represents an individual mouse. *p <0.05, **p <0.01, ***p <0.001.
Wild type CD84 and Ly108 prevent development of ANA-specific antibody-forming cells (AFCs) and serum autoantibody titers
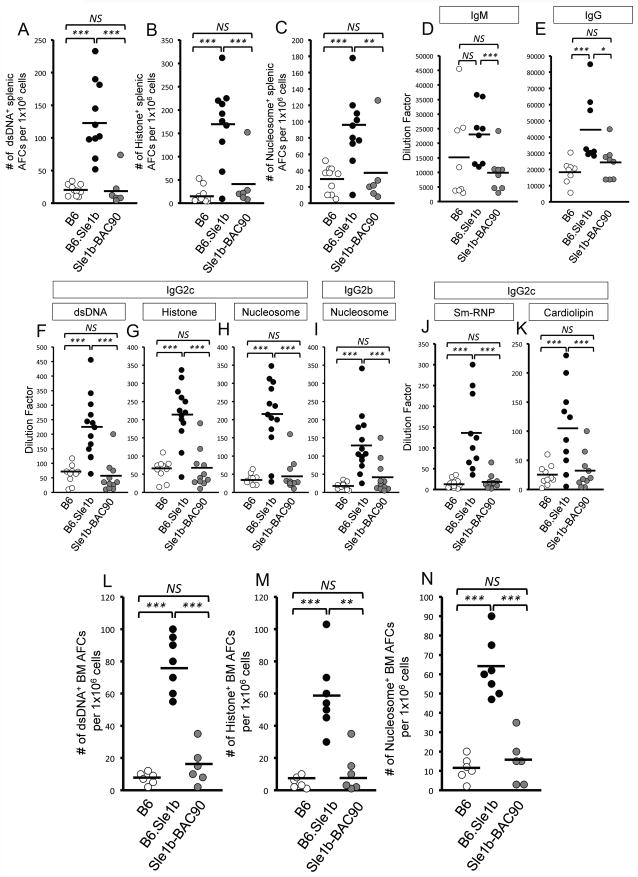
Next, we asked whether the reduction in Spt-GC responses in Sle1b-BAC90 mice resulted in decreased ANA-specific AFCs and Ab titers. B6 and Sle1b-BAC90 mice had significantly lower numbers of dsDNA-, histone-, nucleosome-specific AFCs in the spleen (Figure 3A–C), as well as total serum IgM and IgG titers (Figure 3D–E) than B6.Sle1b mice. Sle1b-BAC90 mice also exhibited significantly reduced levels of IgG2c-specific antibodies against dsDNA, histone, nucleosome (Figure 3F–H), Sm/RNP, and cardiolipin (Figure 3J–K) as well as IgG2b-specific anti-nucleosome antibodies (Figure 3I) compared to B6.Sle1b mice. The reduced autoantibody levels in Sle1b-BAC90 mice strongly correlated with the lower percentage of Spt-GC B cells (data not shown).
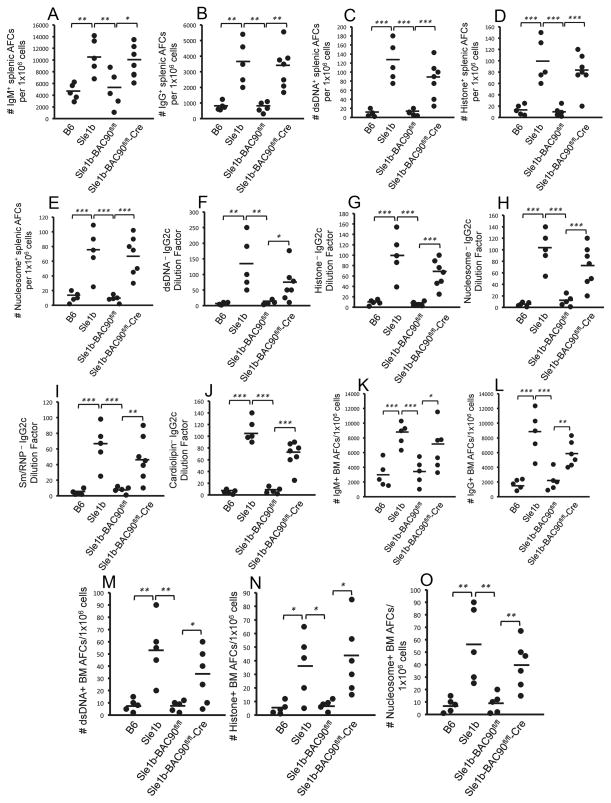
Figure 3. Reduction of ANA-specific AFCs and Ab titers in Sle1b-BAC90 mice.
ELISPOT analysis of (A) dsDNA-, (B) histone-, and (C) nucleosome-specific splenic AFCs in 6–8 mo old B6, B6.Sle1b and Sle1b-BAC90 mice. Total IgM (D) and IgG (E) serum titers at 6–8 mo of age in each indicated group of mice. (F–K) ELISA analysis of IgG2c specific antibodies against dsDNA, histone, nucleosome, Sm/RNP, cardiolipin as well as IgG2b specific antibodies against nucleosome at 6–8 months of age. (L–N) ELISPOT analysis of dsDNA-, histone-, and nucleosome-specific long-lived bone marrow AFCs in 6–8 mo old mice of indicated genotypes. Each circle represents an individual mouse. *p <0.05, **p <0.01, ***p <0.001.
The cardinal function of GCs is the generation of long-lived AFCs, which mainly reside in the bone marrow (BM) (19). We asked whether elevated Spt-GC and Tfh responses in B6.Sle1b mice resulted in an increased number of ANA-specific, long-lived BM AFCs, and if the elevated ANA-specific BM AFC response in B6.Sle1b mice could be reversed by the introduction of B6 alleles of CD84 and Ly108 in Sle1b-BAC90 mice. While there were significantly higher numbers of ANA-specific AFCs in the BM of B6.Sle1b mice, their numbers were markedly reduced in Sle1b-BAC90 mice (Figure 3L–N). The reduction in the Spt-GC, Tfh and ANA responses in Sle1b-BAC90 mice was not the result of global suppression of the ability of B cells to mount an immune response, because no differences were observed in the foreign Ag-specific GC, Tfh and Ab responses among B6, B6.Sle1b and Sle1b-BAC90 mice immunized with SRBCs (data not shown).
Overexpression of B6-derived CD84 and Ly108 on GC B cells is sufficient to restore tolerance in B6.Sle1b mice
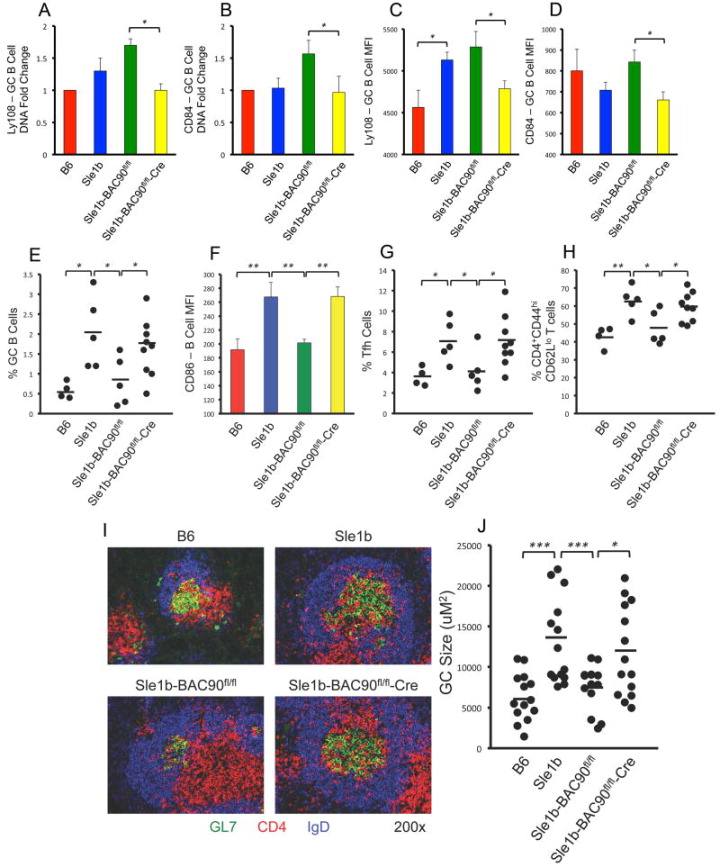
To determine if the overexpression of B6 alleles of CD84 and Ly108 within GC B cells could restore tolerance, we generated Sle1b-BAC90fl/fl mice and crossed them with GC B cell specific Cg-Ighg1-Cre mice (20, 21). Q-PCR analysis of gene copy number and flow cytometric analysis of surface expression confirmed significantly reduced expression levels of the Ly108 and CD84 genes specifically in GC B cells (Figure 4A–D) and not in non-GC naïve B cells (data not shown), confirming GC B cell specific deletion of BAC90 transgene containing B6-CD84 and Ly108 genes in Sle1b-BAC90fl/fl-Cre mice.
Figure 4. GC B cell-intrinsic effect of overexpression of B6 Ly108 and CD84 on restoration of tolerance.
(A–B) GC B cells were sorted (FACS Aria) and analyzed for genomic expression of Ly108 and CD84, which were normalized to B6 control. (C–D) Flow cytometric analysis of Ly108 and CD84 expression levels on GC B cells shown in MFI. Scatter plots showing the percentages of GC B cells (E). (F) MFI of CD86 levels on B cells. Scatter plots showing the percentages of Tfh cells (G), and short-lived effector/effector memory T cells (H) in 4–5 mo old mice. (I) Representative images of spleen sections from 4–5 mo old mice stained with GL-7 (green), anti-CD4 (red), and anti-IgD (blue). These data represent randomly picked 3–5 mice per group. (J) Scatter plot showing GC sizes for each indicated group of mice at 4–5 months of age. *p <0.05, **p <0.01, ***p <0.001.
Flow cytometric analysis of 4–5 month old Sle1b-BAC90fl/fl-Cre mice showed increased percentages of GC B cells, Tfh cells, and short-lived effector T cells, as well as increased CD86 expression on B cells compared to B6 and Sle1b-BAC90fl/fl (Cre−/−) controls (Figure 4E–H). These elevated responses in Sle1b-BAC90fl/fl-Cre mice were similar to increased responses seen in B6.Sle1b mice (Figure 4E–H). Immunofluorescent staining of spleen sections with GL7, anti-IgD and anti-CD4 confirmed the flow cytometry data, showing increased number and size of IgDnegGL7+ Spt-GCs in B6.Sle1b and Sle1b-BAC90fl/fl-Cre mice compared to B6 and Sle1b-BAC90fl/fl controls (Figure 4I–J).
This increase in Spt-GC response in Sle1b-BAC90fl/fl-Cre mice resulted in elevated numbers of IgM-, IgG-, dsDNA-, histone-, and nucleosome-specific AFCs in the spleen (Figure 5A–E) as well as serum IgG2c titers specific for dsDNA, histone, nucleosome, Sm/RNP and cardiolipin (Figure 5F–J). Cre-mediated excision of transgenically expressed wild type Ly108 and CD84 from GC B cells in Sle1b-BAC90fl/fl-Cre mice resulted in elevated numbers of IgM-, IgG-, dsDNA-, histone-, and nucleosome-specific long lived AFCs in the BM (Figure 5K–O), confirming that GC B cell specific B6 wild type CD84 and Ly108 expression was sufficient for the restoration of tolerance in B6.Sle1b mice.
Figure 5. GC B cell specific excision of B6 Ly108 and CD84 in Sle1b-BAC90 mice reinstates loss of tolerance to ANA.
(A–E) ELISPOT analysis of IgM-, IgG-, dsDNA-, histone-, and nucleosome-specific splenic AFCs in 4–5 mo old mice. (F–J) IgG2c specific antibodies against dsDNA, histone, nucleosome, Sm/RNP, cardiolipin, and IgG2b-specific antibodies against nucleosome were measured through ELISA. (K–O) ELISPOT analysis of IgM-, IgG-, dsDNA-, histone-, and nucleosome-specific long-lived bone marrow AFCs at 4–5 months. Each circle represents an individual mouse. *p <0.05, **p <0.01, ***p <0.001.
Wild type CD84 and Ly108 expression normalizes BCR signaling, proliferation and apoptotic capacity of B cells in B6.Sle1b mice
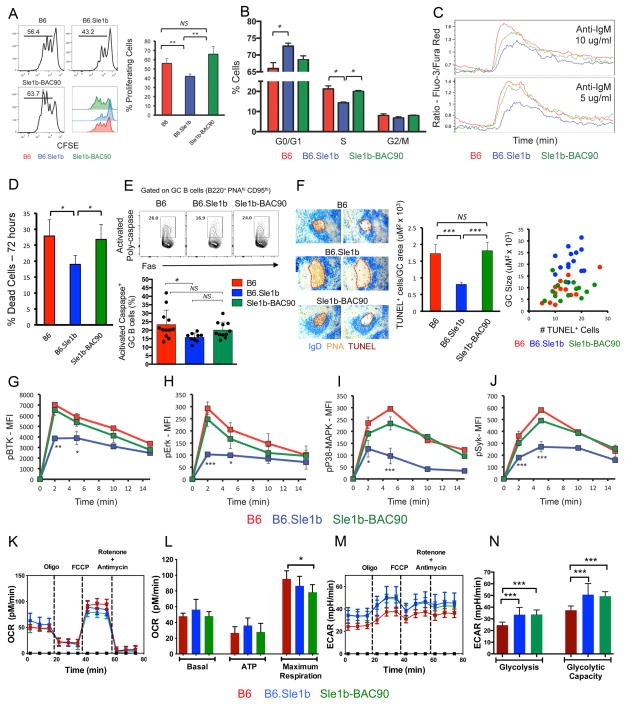
Next, we determined whether elevated Spt-GC responses in B6.Sle1b mice compared to B6 controls correlated with increased proliferation of B cells that can potentially drive the autoimmune response. In vitro stimulation of B cells with anti-IgM and anti-CD40 unexpectedly resulted in a reduced number of proliferating B cells at 72 hours in B6.Sle1b mice than B6 controls (Figure 6A). Moreover, a significantly higher percentage of B cells remained in the G0/G1 phase in B6.Sle1b mice resulting in a lower fraction of cells in S phase than B6 controls (Figure 6B). This reduced proliferation in B6.Sle1b mice was corrected in Sle1b-BAC90 mice overexpressing B6-CD84 and Ly108 (Figure 6A–B). In line with the lower proliferation rate, stimulation with anti-IgM caused a lower Ca2+ flux in B6.Sle1b B cells than in B6 B cells (Figure 6C). Notably, Sle1b-BAC90 mice exhibited Ca2+ flux similar to B6 mice (Figure 6C), highlighting a potential crosstalk between BCR and SLAM signaling.
Figure 6. Analysis of B cell proliferation, apoptosis, signaling, and cellular metabolism.
(A-left panel) In vitro proliferation of CFSE-labeled B cells stimulated with anti-IgM and anti-CD40. (A-right panel) Bar graphs display the percentages of proliferating B cells. (B) Cell cycle analysis of purified B cells after anti-IgM and anti-CD40 stimulation by PI staining. (C) Calcium flux of B cells after anti-IgM stimulation. (D) Bar graphs show the percentage of dead cells after anti-IgM and anti-CD40 stimulation. (E) Top panel, Contour plots show representative gating strategy for GC B cells stained ex-vivo with SR-VAD-FMK, a polycaspase probe. Lower panel, Bar graph shows the percentage of GC B cells positive for activated poly-caspases in the indicated strains. Each circle represents an individual mouse. Error bars show mean with SD. (F) Left panel, consecutive spleen sections were stained for IgD (blue), PNA (red), and TUNEL (brown). Data is representative of 5 mice per group. Middle panel shows bar graphs depicting the ratio of TUNEL+ cells and GC size (uM2 × 103). Right panel shows correlation between the number of TUNEL+ cells and the GC size. (G–J) Flow cytometric analysis to evaluate the kinetics of phosphorylation of BtK, Erk, p38/MAPK, and Syk after anti-IgM stimulation of B cells. (K and M) Representative plots show the measurement of OCR and ECAR, respectively, in B6, B6.Sle1b and Sle1b-BAC90 B cells stimulated with anti-IgM and anti-CD40 for 18h. (L) Histograms show the average basal oxygen consumption; ATP production through ATP synthase and the maximal electron transport capacity of activated B cells from 3–4 mice per indicated group. (N) Histograms depict average ECAR values for glycolysis and the glycolytic capacity of activated B cells from 3–4 mice per strain. * p =≤0.05, ** p =≤0.01, *** p =≤0.001.
Next, to determine if the decrease in proliferative capacity of B6.Sle1b B cells resulted in an increase in apoptosis of B6.Sle1b B cells, we evaluated B cell survival upon stimulation. In vitro stimulation of B cells with anti-IgM and anti-CD40 showed a lower percentage of apoptotic B6.Sle1b B cells compared to B6 and Sle1b-BAC90 B cells (Figure 6D). We also determined the percentage of B cells undergoing apoptosis in GCs by measuring the caspase activity in DAPInegB220+FashighPNAhigh GC B cells. We found a significant decrease in B220+PNAhighFashigh Caspase+ apoptotic GC B cells in B6.Sle1b mice compared to B6 controls (Figure 6E). The percentage of apoptotic GC B cells in Sle1b-BAC90 mice overall was higher than B6.Sle1b mice even though the difference was not statistically significant (Figure 6E). We observed no significant difference in the percentage of apoptotic GC B cells between B6 and Sle1b-BAC90 mice. Consistent with this finding, staining for apoptotic TUNEL+ cells on spleen sections showed fewer apoptotic cells per μμ2 of GC area in B6.Sle1b mice than B6 and Sle1b-BAC90 mice (Figure 6F). Supporting the above findings our gene expression analysis revealed significant upregulation of several anti-apoptotic genes in B6.Sle1b B cells compared to B6 B cells (Table I).
Table I.
List of anti-apoptotic genes upregulated in B cells from B6.Sle1b mice
| Gene name | Fold change | P value |
|---|---|---|
| BCL2L1 | 2.2 | 0.03 |
| SPHK1 | 2.2 | 0.007 |
| HMGB2 | 2.5 | 0.048 |
| BIRC5 | 2.6 | 0.011 |
| RRM2 | 1.9 | 0.018 |
| METAP2 | 1.6 | 0.031 |
Tabulated anti-apoptotic genes were significantly upregulated in sorted B cells derived from 3 mo old B6.Sle1b mice as determined by Illumina Bead Chip arrays (Mouse WG-6 V2). Gene expression is shown in terms of increased fold change in B6.Sle1b mice compared to B6 controls. Statistical analysis was performed by pairwise homogeneous t-test.
To determine if there were defects in the BCR signaling pathway in B6.Sle1b B cells, we evaluated the phosphorylation status of BCR proximal signaling molecules (BTK and Syk) and downstream molecules (p-38-MAPK and Erk) after BCR activation with anti-IgM using phospho flow analysis. Consistent with the Ca2+ flux results, B6.Sle1b B cells exhibited decreased phosphorylation of all of these molecules (Figure 6G–J). Introduction of the B6 alleles of CD84 and Ly108 in Sle1b-BAC90 mice rescued this signaling defect, further supporting the possibility for a crosstalk between BCR and SLAM signaling, and indicating a role of Ly108 and CD84 in the restoration of peripheral B cell tolerance. Our results also show that B cells from B6.Sle1b mice were not anergic and that the reduced signaling activity exhibited by these B cells was not due to primary B cell developmental defects (Supplementary Figure 4).
Several genes encoding enzymes required for various metabolic pathways, including those involved in cellular energy metabolism are modulated by the activation of B cells through BCR, CD40L and TLR ligands (22). Additionally, it has been demonstrated that signaling events, which lead to variations in cellular energy metabolism during GC reactions are necessary for the maintenance and differentiation of GC B cells and plasma cells (21). Given the aforementioned differences in BCR signaling (Figure 6G–J), we then asked if B cells from B6, B6.Sle1b and Sle1b-BAC90 mice were metabolically different in responding to energy demands upon activation. No significant differences were observed in the baseline oxygen consumption among B cells from B6, B6.Sle1b and Sle1b-BAC90 mice following activation with anti-IgM and anti-CD40 (Figure 6K and L). ATP production after exposure to oligomycin was also comparable among these three stains (Figure 6K and L). In addition, B6 and B6.Sle1b B cells showed similar maximum respiration while Sle1b-BAC90 B cells showed marginally reduced maximal respiration compared to B6 controls (Figure 6K and L). Notably, B6.Sle1b and Sle1b-BAC90 B cells had significantly higher glycolysis and glycolytic capacity than B6 B cells (Figure 6M and N). These results indicate that reduced calcium mobilization and lower BCR signaling in B6.Sle1b B cells did not dampen the energy metabolism in these cells.
Sle1b expressing B cells have a lower frequency of conjugate formation with wild type T cells, which is reversed by the overexpression of wild type CD84 and Ly108
Previous studies showed that SAP expression in T cells is required for effective B and T cell conjugation leading to optimal GC responses during T cell-dependent antigenic stimulation or viral infection (23, 24). The effect of B cell-intrinsic expression of lupus-associated SLAM family receptors on conjugate formation in the context of autoimmunity is not clear. We asked whether polymorphic SLAM expression in B cells from B6.Sle1b mice led to differential interaction with T cells, affecting their ability to form B cell: T cell conjugates. We further asked whether overexpression of B6-derived CD84 and Ly108 in B6.Sle1b B cells could normalize such potential differences in the ability of B6.Sle1b B cells to form conjugates with T cells.
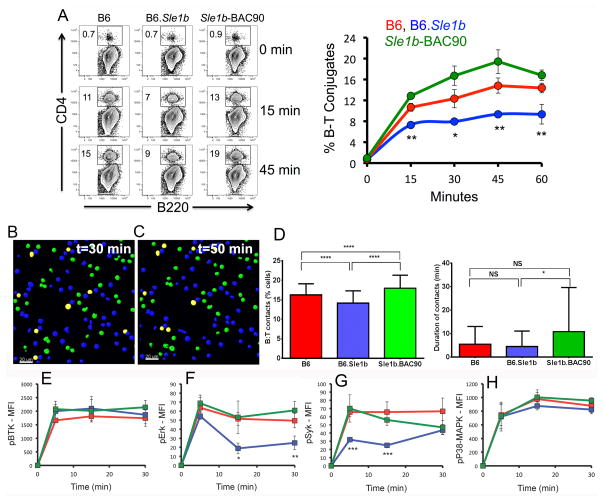
We utilized an in vitro flow cytometry-based B and T cell conjugation assay where purified B cells from B6, B6.Sle1b and Sle1b-BAC90 mice were activated with anti-IgM and LPS and loaded with OVA peptide to present to B6 OT-II transgenic T cells specific for OVA323–339. B6.Sle1b B cells formed a significantly reduced number of conjugates over the course of 60 minutes compared to B cells from B6 mice, which was corrected when B cells were from Sle1b-BAC90 mice (Figure 7A).
Figure 7. Reduced B cell-T cell conjugate formation in B6.Sle1b mice.
(A) Representative flow cytometry plots of B220+ B cell and CD4+ OT-II T cell conjugate formation. Conjugate formation was calculated as B220+CD4+ double positive for each indicated mouse group over the course of 60 minutes. Kinetics of B–T conjugate formation is depicted in the graph on the right (A). (B–C) Representative 2D confocal images of B6 B cells (blue) and OT-II T cells (green) in co-culture at 30 min and 50 min, respectively. The B and T cells in contact with each other are shown in yellow. (D) Averaged percentage (left panel) and duration (right panel) of contacts between OT-II T cells and B cells from indicated strains over a period of 60 min. The analysis was performed twice, with B cells pooled from 2–3 mice per group. (E–H) Kinetics of phosphorylation of BtK, Erk, p38/MAPK, and Syk in B cells during conjugate formation with T cells over the course of 30 minutes. *p <0.05, **p <0.01, ***p <0.001.
To track B cell-T cell interactions in real time, 2D confocal images of conjugates between activated B cells and OT-II T cells were acquired sequentially at every 30 seconds for 60 min (Figure 7B–C). Consistent with the flow cytometry data, the percentage of B cell-OT-II T cell conjugates was significantly lower over time when B cells were from B6.Sle1b mice compared to B cells from B6 or Sle1b-BAC90 mice (Figure 7D, left panel). We also measured the duration of contacts between OT-II T cells and B cells from each strain over a period of 60 min. We found similar B:T contact duration time between B cells from B6 and B6.Sle1b mice. The contact duration time was higher with B cells from Sle1b-BAC90 mice compared to B6 and B6.Sle1b (Figure 7D, right panel).
Phosphoflow analysis of B cells after conjugate formation with OT-II T cells showed similar phosphorylation levels of BTK and p38-MAPK among all 3 strains (Figure 7E and H), but reduced phosphorylation of Erk and Syk in B cells from B6.Sle1b mice than B6 and Sle1b-BAC90 mice (Figure 7F and G). These data suggest that differential expression of CD84 and Ly108 on B cells could alter B and T cell interactions resulting in reduced BCR signaling. It is plausible that such variations in the frequency of B and T cell interactions and lower BCR signaling could potentially facilitate autoimmune B cells to escape the GC tolerance checkpoint.
DISCUSSION
Polymorphisms in the SLAM family genes are implicated in both murine and human lupus (1, 2, 17, 25–27). Using B6.Sle1b mice that express the NZM2410/NZW lupus strain-derived SLAM family genes, we previously showed that this sublocus alters B cell tolerance at the GC checkpoint, leading to increased ANA-specific AFCs and ANA titers (3, 16). However, the mechanisms by which B cell-intrinsic expression of polymorphic SLAM family genes may alter B cell tolerance at the GC checkpoint and the SLAM family members required for mediating this process were not previously addressed. Here, by analyzing seven different B6.Sle1b mouse lines carrying various BAC transgenes of B6 alleles of SLAM and non-SLAM family genes spanning the entire Sle1b locus we identified both Ly108 and CD84 as playing essential roles in the loss of B cell tolerance in B6.Sle1b mice. Overexpression of B6 wild type CD84 and Ly108 can restore B cell tolerance at the GC checkpoint, leading to reduced numbers of ANA-specific long-lived AFCs in the BM. Subsequently, taking advantage of GC B cell-specific Cre mice in which Cre conditionally removed transgenically expressed B6 alleles of CD84 and Ly108 on GC B cells, we further demonstrate that the expression of polymorphic CD84 and Ly108 on GC B cells is critical for the loss of tolerance to nuclear Ags in B6.Sle1b mice. These results highlight the importance of CD84 and Ly108 expression on GC B cells in regulating Spt-GC and Tfh responses and in preventing autoimmunity.
Our in vitro functional assays revealed lower Ca2+ flux, reduced proliferation and apoptosis of B cells expressing Sle1b compared to B6, all of which were restored in Sle1b-BAC90 mice overexpressing B6 alleles of CD84 and Ly108. Consistent with these data we found reduced levels of phosphorylation of several molecules in the BCR signaling pathway in B cells from B6.Sle1b mice whereas B6 and Sle1b-BAC90 mice showed comparable levels of phosphorylation of these molecules. Our results are consistent with a previous report (4) showing lower Ca2+ flux and less cell death of HEL transgenic B cells expressing Sle1b (Sle1b-HEL Tg) upon BCR stimulation. However, Kumar et al. reported increased proliferation of Sle1b-HEL Tg B cells whereas we found a modest reduction in proliferation of B cells expressing Sle1b (4). This discrepancy in proliferation of Sle1b B cells may have resulted from the difference in antigen specificity (i.e., HEL Tg vs. polyreactive).
The mechanism by which autoreactive B cells in B6.Sle1b mice are selected into autoantibody producing long-lived AFCs or memory B cells is not clear. Based on our data showing reduced proliferation and apoptosis of B cells expressing Sle1b and the reversal of these phenotypes with overexpression of B6-derived CD84 and Ly108, we suggest that increased spontaneous GC, Tfh and long-lived AFC responses in B6.Sle1b mice may not be the result of an enhanced positive selection of autoreactive B cells in GCs. Rather, we believe that autoreactive B cell development in GCs of B6.Sle1b mice is due to altered negative selection of GC B cells. We propose two plausible mechanisms by which polymorphic SLAM expression in B cells from B6.Sle1b mice may help autoreactive B cells escape negative selection in GCs. One mechanism is that polymorphic SLAM expression in B6.Sle1b B cells helps reduce BCR signaling upon engagement of self-Ags, which allows these cells to survive in the germinal center and not undergo negative selection.
We propose a second scenario where GC B cells interact with T cells. Given an increased frequency of Tfh cells in B6.Sle1b mice, the chances of B cell-T cell interaction in GCs is very high. Therefore, given our data from the conjugate assay, we suggest that reduced B:T conjugates due to B cell-intrinsic expression of polymorphic SLAMs helps maintain reduced BCR signaling in B cells expressing Sle1b. This differential frequency of interaction of B6.Sle1b B cells with T cells on one hand may help autoreactive B cells to escape negative selection in GCs by reducing BCR signaling and on the other hand provide sufficient survival signals to these B cells to proliferate (albeit at a slower rate than B6 controls) and differentiate into autoantibody-producing AFCs. Consistent with this idea, we previously observed augmented GC responses of DNA and p-azophenylarsonate (Ars) dual-reactive B cells from Ig VH knock-in mice (named HKIR) expressing Sle1b (16). Under non-autoimmune conditions, due to their DNA-reactivity HKIR Ars-DNA-dual reactive B cells are negatively regulated in GCs presumably by the GC tolerance checkpoint (28–30). However, autoimmune susceptibility, as in B6.Sle1b mice alters their negative selection in GCs (16). Recently, Sinai et al. also have reported altered B and T cell interactions to be responsible for increased GC B cell phenotype in vitro in autoimmune prone B6.Sle1 mice (31).
Anergic B cells have arrested development, are excluded from B cell follicles, and are unable to mobilize calcium (32). Anergic B cells are also unable to proliferate, upregulate activation markers, mediate BCR signaling or mount an immune response after BCR engagement (32). Our results show that B6.Sle1b B cells exhibit normal B cell development in the bone marrow and spleen, are present in B cell follicles and seed GCs, and thus do not exhibit an anergic state. Analysis of the cellular energy metabolism also indicates that reduced calcium mobilization and lower BCR signaling upon stimulation of B6.Sle1b B cells do not dampen the energy metabolism in these cells. B6.Sle1b B cells appear to be comparable, if not better, in responding to the high-energy demands upon activation. In a recent comparative analysis, Rathmell and co-workers found anergic B cells to be metabolically suppressed while B cells from autoimmune BAFF Tg mice to be metabolically more active with increased glycolytic efficiency, which was necessary for autoantibody production (33). This study is in line with our data showing that B6.Sle1b B cells are not anergic, and rather can undergo optimal metabolic reprogramming upon activation to produce high titers of autoantibodies.
The functional and mechanistic roles of Ly108 and CD84 in the context of GC are unclear. Ly108 has been proposed to act as a molecular rheostat that determines the stringency through which autoreactive B cells are purged during primary development (4) while other studies have implicated similar functions for Ly108 in T cells (5, 34). Although our data indicate that polymorphisms in CD84 and Ly108 in B cells determine efficiency of tolerance, we cannot exclude a possible contribution of Ly108 expression by T cells particularly with respect to production of IFN-γ (35). IFN-γ has been demonstrated to drive pathogenic accumulation of Tfh cells in Roquinsan mice and promote antibody class switching to pathogenic IgG2c, both of which are observed in B6.Sle1b mice (36–38).
Although CD84 and Ly108 were previously demonstrated to be integral for optimal GC response against protein immunization (23), no such defect was observed in mice deficient in CD84 in the context of an acute LCMV and VACV infection (24). Our analysis also revealed no defect in spontaneous or SRBC-induced GC and Tfh responses in mice deficient in either CD84 or Ly108 (data not shown). These data indicate redundancy in functions of SLAM family receptors. However, these previous studies did not investigate B cell-intrinsic expression of polymorphic CD84 and Ly108 in the Spt-GC response. Our data suggest that CD84 and Ly108 function in a regulatory capacity in B cells as Spt-GC response inversely correlated with CD84 and Ly108 expression specifically on GC B cells (data not shown). In addition, in vitro stimulation with anti-Ly108 resulted in increased CD84 levels on activated B6 B cells, which was defective in Sle1b B cells but mostly restored in Sle1b-BAC90 B cells (data not shown), indicating that functions of these two SLAM family genes may be tightly linked. These data suggest that altered signaling in B cells mediated by Sle1b-derived CD84 and Ly108 may lead to a modulation in B and T cell interactions and subsequent selection in GCs. Consistent with this idea, we observed impaired B and T cell conjugation when B cells expressed Sle1b, which was restored by B cell expression of wild type CD84 and Ly108.
Earlier studies have focused on SLAM signaling in T cells and NKT cells owing to the discovery of SLAM adaptor molecule SAP that is highly expressed in these cell types (2). SAP regulates T cell-mediated help (39), Tfh cell development (40), cytokine production (41) and GC response (42). SAP expression in T cells, not in B cells, is required for humoral immune response (43). Ly108 was also shown to regulate T cell signaling via the SAP-Fyn pathway (34). Crotty and colleagues have further suggested that Ly108 regulates T cell and B cell interactions by modulating the ratio of positive and negative signals mediated by SAP and SHP-1 (24). The requirement of SAP and T cells in autoimmune responses has also been described in lupus-prone MRL-FASlpr (44), Sanroque (45) and B6.Sle1b mice (46).
While previous studies highlighted the importance of SLAM-SAP signaling in T cells for shaping B cell responses, whether similar signaling mechanism exits in B cells has not been previously defined. Detre et al. recently reported the role of SAP in modulating B cell functions (47). Because B cells do not express SAP, the mechanism by which SAP deficiency alters B cell response is not clear. We observed no differences in SAP transcript levels in T and B cells (including Tfh and GC B cells) among B6, B6.Sle1b and Sle1b-BAC90 mice. A recent study has implicated the role of SAP-related molecule EAT-2 in mediating its effects in NK cells by linking SLAM family receptors to phospholipase Cγ, calcium flux and Erk kinase (48). Whether signaling through this or other unknown adaptor molecules is compromised in B6.Sle1b B cells, resulting in lower calcium flux and reduced Erk phosphorylation, remains to be determined.
In summary, we show that CD84 and Ly108 expression on GC B cells are critical for maintaining peripheral B cell tolerance at the GC checkpoint. Our data further highlight the importance of the GC pathway in autoimmune disease pathogenesis, and suggest that GC B cell specific CD84 and Ly108 may be therapeutic targets for SLE.
Supplementary Material
Acknowledgments
These studies were supported by a grant from the NIH (AI091670) to Z.S.M.R.
We thank Drs. Aron Lukacher, Tim Manser and Milena Bogunovic for the critical reading of the manuscript and helpful discussions. We thank Ms. Stephanie Schell for proofreading the manuscript. Finally, we thank Dr. Jose Casco (the University of Texas Southwestern Medical Center) for maintenance and transfer of BAC transgenic mice, and the Penn State Hershey flow cytometry core facility for their service.
Non standard abbreviations
- GC
germinal center
- Spt-GC
spontaneous germinal center
- AFC
antibody forming cell
- PNA
peanut agglutinin
- ANA
anti-nuclear antibody
- SRBC
sheep red blood cells
- Tfh
follicular helper T cells
- SLE
systemic lupus erythematosus
- Anti-Sm/RNP
anti-smith antigen/ribonucleoprotein
- BM
bone marrow
- CFSE
carboxyfluorescein succinimidyl ester
References
- 1.Wang A, Batteux F, Wakeland EK. The role of SLAM/CD2 polymorphisms in systemic autoimmunity. Curr Opin Immunol. 2010;22:706–714. doi: 10.1016/j.coi.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 2.Cannons JL, Tangye SG, Schwartzberg PL. SLAM Family Receptors and SAP Adaptors in Immunity. Immunology. 2011;29:665–705. doi: 10.1146/annurev-immunol-030409-101302. [DOI] [PubMed] [Google Scholar]
- 3.Wandstrat AE, Nguyen C, Limaye N, Chan AY, Subramanian S, Tian X-H, Yim Y-S, Pertsemlidis A, Garner HR, Morel L, Wakeland EK. Association of Extensive Polymorphisms in the SLAM/CD2 Gene Cluster with Murine Lupus. Immunity. 2004;21:769–780. doi: 10.1016/j.immuni.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 4.Kumar KR, Li L, Yan M, Bhaskarabhatla M, Mobley AB, Nguyen C, Mooney JM, Schatzle JD, Wakeland EK, Mohan C. Regulation of B cell tolerance by the lupus susceptibility gene Ly108. Science. 2006;312:1665–1669. doi: 10.1126/science.1125893. [DOI] [PubMed] [Google Scholar]
- 5.Keszei M, Detre C, Rietdijk ST, Muñoz P, Romero X, Berger SB, Calpe S, Liao G, Castro W, Julien A, Wu Y-Y, Shin D-M, Sancho J, Zubiaur M, Morse HC, Morel L, Engel P, Wang N, Terhorst C. A novel isoform of the Ly108 gene ameliorates murine lupus. J Exp Med. 2011;208:811–822. doi: 10.1084/jem.20101653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koh AE, Njoroge SW, Feliu M, Cook A, Selig MK, Latchman YE, Sharpe AH, Colvin RB, Paul E. The SLAM family member CD48 (Slamf2) protects lupus-prone mice from autoimmune nephritis. J Autoimmun. 2011;37:48–57. doi: 10.1016/j.jaut.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keszei M, Latchman YE, Vanguri VK, Brown DR, Detre C, Morra M, Arancibia-Carcamo CV, Arancibia CV, Paul E, Calpe S, Castro W, Wang N, Terhorst C, Sharpe AH. Auto-antibody production and glomerulonephritis in congenic Slamf1−/− and Slamf2−/− [B6.129] but not in Slamf1−/− and Slamf2−/− [BALB/c.129] mice. Int Immunol. 2011;23:149–158. doi: 10.1093/intimm/dxq465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Salort J, Cuenca M, Terhorst C, Engel P, Romero X. Ly9 (CD229) Cell-Surface Receptor is Crucial for the Development of Spontaneous Autoantibody Production to Nuclear Antigens. Front Immunol. 2013;4:225–225. doi: 10.3389/fimmu.2013.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cappione A, Anolik JH, Pugh-Bernard A, Barnard J, Dutcher P, Silverman G, Sanz I. Germinal center exclusion of autoreactive B cells is defective in human systemic lupus erythematosus. J Clin Invest. 2005;115:3205–3216. doi: 10.1172/JCI24179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vinuesa CG, Sanz I, Cook MC. Dysregulation of germinal centres in autoimmune disease. Nat Rev Immunol. 2009;9:845–857. doi: 10.1038/nri2637. [DOI] [PubMed] [Google Scholar]
- 11.Luzina IG, Atamas SP, Storrer CE, daSilva LC, Kelsoe G, Papadimitriou JC, Handwerger BS. Spontaneous formation of germinal centers in autoimmune mice. J Leukoc Biol. 2001;70:578–584. [PubMed] [Google Scholar]
- 12.Bygrave AE, Rose KL, Cortes-Hernandez J, Warren J, Rigby RJ, Cook HT, Walport MJ, Vyse TJ, Botto M. Spontaneous autoimmunity in 129 and C57BL/6 mice-implications for autoimmunity described in gene-targeted mice. PLoS Biol. 2004;2:E243. doi: 10.1371/journal.pbio.0020243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carlucci F, Cortes-Hernandez J, Fossati-Jimack L, Bygrave AE, Walport MJ, Vyse TJ, Cook HT, Botto M. Genetic dissection of spontaneous autoimmunity driven by 129-derived chromosome 1 Loci when expressed on C57BL/6 mice. J Immunol. 2007;178:2352–2360. doi: 10.4049/jimmunol.178.4.2352. [DOI] [PubMed] [Google Scholar]
- 14.Morel L, Blenman KR, Croker BP, Wakeland EK. The major murine systemic lupus erythematosus susceptibility locus, Sle1, is a cluster of functionally related genes. Proc Natl Acad Sci USA. 2001;98:1787–1792. doi: 10.1073/pnas.031336098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rahman ZSM, Rao SP, Kalled SL, Manser T. Normal induction but attenuated progression of germinal center responses in BAFF and BAFF-R signaling-deficient mice. J Exp Med. 2003;198:1157–1169. doi: 10.1084/jem.20030495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong EB, Khan TN, Mohan C, Rahman ZSM. The lupus-prone NZM2410/NZW strain-derived Sle1b sublocus alters the germinal center checkpoint in female mice in a B cell-intrinsic manner. J Immunol. 2012;189:5667–5681. doi: 10.4049/jimmunol.1201661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cunninghame Graham DS, Vyse TJ, Fortin PR, Montpetit A, Cai Y-C, Lim S, McKenzie T, Farwell L, Rhodes B, Chad L, Hudson TJ, Sharpe A, Terhorst C, Greenwood CMT, Wither J, Rioux JD CaNIOS GenES Investigators. Association of LY9 in UK and Canadian SLE families. Genes Immun. 2008;9:93–102. doi: 10.1038/sj.gene.6364453. [DOI] [PubMed] [Google Scholar]
- 18.Jørgensen TN, Alfaro J, Enriquez HL, Jiang C, Loo WM, Atencio S, Bupp MRG, Mailloux CM, Metzger T, Flannery S, Rozzo SJ, Kotzin BL, Rosemblatt M, Bono MR, Erickson LD. Development of murine lupus involves the combined genetic contribution of the SLAM and FcgammaR intervals within the Nba2 autoimmune susceptibility locus. J Immunol. 2010;184:775–786. doi: 10.4049/jimmunol.0901322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chu VT, Berek C. The establishment of the plasma cell survival niche in the bone marrow. Immunol Rev. 2013;251:177–188. doi: 10.1111/imr.12011. [DOI] [PubMed] [Google Scholar]
- 20.Casola S, Cattoretti G, Uyttersprot N, Koralov SB, Seagal J, Segal J, Hao Z, Waisman A, Egert A, Ghitza D, Rajewsky K. Tracking germinal center B cells expressing germ-line immunoglobulin gamma1 transcripts by conditional gene targeting. Proc Natl Acad Sci USA. 2006;103:7396–7401. doi: 10.1073/pnas.0602353103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heise N, De Silva NS, Silva K, Carette A, Simonetti G, Pasparakis M, Klein U. Germinal center B cell maintenance and differentiation are controlled by distinct NF-κB transcription factor subunits. J Exp Med. 2014;211:2103–2118. doi: 10.1084/jem.20132613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu X, Hart R, Chang MS, Kim J-W, Lee SY, Cao YA, Mock D, Ke E, Saunders B, Alexander A, Grossoehme J, Lin K-M, Yan Z, Hsueh R, Lee J, Scheuermann RH, Fruman DA, Seaman W, Subramaniam S, Sternweis P, Simon MI, Choi S. Analysis of the major patterns of B cell gene expression changes in response to short-term stimulation with 33 single ligands. J Immunol. 2004;173:7141–7149. doi: 10.4049/jimmunol.173.12.7141. [DOI] [PubMed] [Google Scholar]
- 23.Cannons JL, Qi H, Lu KT, Dutta M, Gomez-Rodriguez J, Cheng J, Wakeland EK, Germain RN, Schwartzberg PL. Optimal Germinal Center Responses Require a Multistage T Cell:B Cell Adhesion Process Involving Integrins, SLAM-Associated Protein, and CD84. Immunity. 2010;32:253–265. doi: 10.1016/j.immuni.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kageyama R, Cannons JL, Zhao F, Yusuf I, Lao C, Locci M, Schwartzberg PL, Crotty S. The receptor Ly108 functions as a SAP adaptor-dependent on-off switch for T cell help to B cells and NKT cell development. Immunity. 2012;36:986–1002. doi: 10.1016/j.immuni.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chatterjee M, Hedrich CM, Rauen T, Ioannidis C, Terhorst C, Tsokos GC. CD3-T cell receptor co-stimulation through SLAMF3 and SLAMF6 receptors enhances RORγt recruitment to the IL17A promoter in human T lymphocytes. J Biol Chem. 2012;287:38168–38177. doi: 10.1074/jbc.M112.415067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chatterjee M, Rauen T, Kis-Toth K, Kyttaris VC, Hedrich CM, Terhorst C, Tsokos GC. Increased expression of SLAM receptors SLAMF3 and SLAMF6 in systemic lupus erythematosus T lymphocytes promotes Th17 differentiation. J Immunol. 2012;188:1206–1212. doi: 10.4049/jimmunol.1102773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chatterjee M, Kis-Toth K, Thai T-H, Terhorst C, Tsokos GC. SLAMF6-driven co-stimulation of human peripheral T cells is defective in SLE T cells. Autoimmunity. 2011;44:211–218. doi: 10.3109/08916934.2010.530627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Notidis E, Heltemes L, Manser T. Dominant, hierarchical induction of peripheral tolerance during foreign antigen-driven B cell development. Immunity. 2002;17:317–327. doi: 10.1016/s1074-7613(02)00392-8. [DOI] [PubMed] [Google Scholar]
- 29.Heltemes-Harris L, Liu X, Manser T. Progressive surface B cell antigen receptor down-regulation accompanies efficient development of antinuclear antigen B cells to mature, follicular phenotype. J Immunol. 2004;172:823–833. doi: 10.4049/jimmunol.172.2.823. [DOI] [PubMed] [Google Scholar]
- 30.Rahman ZSM, Alabyev B, Manser T. FcgammaRIIB regulates autoreactive primary antibody-forming cell, but not germinal center B cell, activity. J Immunol. 2007;178:897–907. doi: 10.4049/jimmunol.178.2.897. [DOI] [PubMed] [Google Scholar]
- 31.Sinai P, Dozmorov IM, Song R, Schwartzberg PL, Wakeland EK, Wülfing C. T/B cell interactions are more transient in response to weak stimuli in SLE-prone mice. Eur J Immunol. 2014;44:3522–3531. doi: 10.1002/eji.201444602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cambier JC, Gauld SB, Merrell KT, Vilen BJ. B-cell anergy: from transgenic models to naturally occurring anergic B cells? Nat Rev Immunol. 2007;7:633–643. doi: 10.1038/nri2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caro-Maldonado A, Wang R, Nichols AG, Kuraoka M, Milasta S, Sun LD, Gavin AL, Abel ED, Kelsoe G, Green DR, Rathmell JC. Metabolic reprogramming is required for antibody production that is suppressed in anergic but exaggerated in chronically BAFF-exposed B cells. J Immunol. 2014;192:3626–3636. doi: 10.4049/jimmunol.1302062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhong M-C, Veillette A. Control of T lymphocyte signaling by Ly108, a signaling lymphocytic activation molecule family receptor implicated in autoimmunity. J Biol Chem. 2008;283:19255–19264. doi: 10.1074/jbc.M800209200. [DOI] [PubMed] [Google Scholar]
- 35.Howie D, Okamoto S, Rietdijk S, Clarke K, Wang N, Gullo C, Bruggeman JP, Manning S, Coyle AJ, Greenfield E, Kuchroo V, Terhorst C. The role of SAP in murine CD150 (SLAM)-mediated T-cell proliferation and interferon gamma production. Blood. 2002;100:2899–2907. doi: 10.1182/blood-2002-02-0445. [DOI] [PubMed] [Google Scholar]
- 36.Peng SL, Szabo SJ, Glimcher LH. T-bet regulates IgG class switching and pathogenic autoantibody production. Proc Natl Acad Sci USA. 2002;99:5545–5550. doi: 10.1073/pnas.082114899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baudino L, Azeredo da Silveira S, Nakata M, Izui S. Molecular and cellular basis for pathogenicity of autoantibodies: lessons from murine monoclonal autoantibodies. Springer Semin Immunopathol. 2006;28:175–184. doi: 10.1007/s00281-006-0037-0. [DOI] [PubMed] [Google Scholar]
- 38.Lee SK, Silva DG, Martin JL, Pratama A, Hu X, Chang P-P, Walters G, Vinuesa CG. Interferon-γ excess leads to pathogenic accumulation of follicular helper T cells and germinal centers. Immunity. 2012;37:880–892. doi: 10.1016/j.immuni.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 39.Cannons JL, Yu LJ, Jankovic D, Crotty S, Horai R, Kirby M, Anderson S, Cheever AW, Sher A, Schwartzberg PL. SAP regulates T cell-mediated help for humoral immunity by a mechanism distinct from cytokine regulation. J Exp Med. 2006;203:1551–1565. doi: 10.1084/jem.20052097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCausland MM, Yusuf I, Tran H, Ono N, Yanagi Y, Crotty S. SAP regulation of follicular helper CD4 T cell development and humoral immunity is independent of SLAM and Fyn kinase. J Immunol. 2007;178:817–828. doi: 10.4049/jimmunol.178.2.817. [DOI] [PubMed] [Google Scholar]
- 41.Czar MJ, Kersh EN, Mijares LA, Lanier G, Lewis J, Yap G, Chen A, Sher A, Duckett CS, Ahmed R, Schwartzberg PL. Altered lymphocyte responses and cytokine production in mice deficient in the X-linked lymphoproliferative disease gene SH2D1A/DSHP/SAP. Proc Natl Acad Sci USA. 2001;98:7449–7454. doi: 10.1073/pnas.131193098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qi H, Cannons JL, Klauschen F, Schwartzberg PL, Germain RN. SAP-controlled T-B cell interactions underlie germinal centre formation. Nature. 2008;455:764–769. doi: 10.1038/nature07345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Veillette A, Zhang S, Shi X, Dong Z, Davidson D, Zhong M-C. SAP expression in T cells, not in B cells, is required for humoral immunity. Proc Natl Acad Sci USA. 2008;105:1273–1278. doi: 10.1073/pnas.0710698105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Komori H, Furukawa H, Mori S, Ito MR, Terada M, Zhang M-C, Ishii N, Sakuma N, Nose M, Ono M. A signal adaptor SLAM-associated protein regulates spontaneous autoimmunity and Fas-dependent lymphoproliferation in MRL-Faslpr lupus mice. J Immunol. 2006;176:395–400. doi: 10.4049/jimmunol.176.1.395. [DOI] [PubMed] [Google Scholar]
- 45.Linterman MA, Rigby RJ, Wong RK, Yu D, Brink R, Cannons JL, Schwartzberg PL, Cook MC, Walters GD, Vinuesa CG. Follicular helper T cells are required for systemic autoimmunity. J Exp Med. 2009;206:561–576. doi: 10.1084/jem.20081886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keszei M, Detre C, Castro W, Magelky E, O’Keeffe M, Kis-Toth K, Tsokos GC, Wang N, Terhorst C. Expansion of an osteopontin-expressing T follicular helper cell subset correlates with autoimmunity in B6.Sle1b mice and is suppressed by the H1-isoform of the Slamf6 receptor. FASEB J. 2013;27:3123–3131. doi: 10.1096/fj.12-226951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Detre C, Yigit B, Keszei M, Castro W, Magelky EM, Terhorst C. SAP modulates B cell functions in a genetic background-dependent manner. Immunol Lett. 2013;153:15–21. doi: 10.1016/j.imlet.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pérez-Quintero LA, Roncagalli R, Guo H, Latour S, Davidson D, Veillette A. EAT-2, a SAP-like adaptor, controls NK cell activation through phospholipase Cγ, Ca++, and Erk, leading to granule polarization. J Exp Med. 2014;211:727–742. doi: 10.1084/jem.20132038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.