Abstract
Successfully fighting infection requires a properly tuned immune system. Recent epidemiological studies link exposure to pollutants that bind the aryl hydrocarbon receptor (AHR) during development with poorer immune responses later in life. Yet, how developmental triggering of AHR durably alters immune cell function remains unknown. Using a mouse model, we show that developmental activation of AHR leads to long-lasting reduction in the response of CD8+ T cells during influenza virus infection, cells critical for resolving primary infection. Combining genome-wide approaches, we demonstrate that developmental activation alters DNA methylation and gene expression patterns in isolated CD8+ T cells prior to and during infection. Altered transcriptional profiles in CD8+ T cells from developmentally exposed mice reflect changes in pathways involved in proliferation and immunoregulation, with an overall pattern that bears hallmarks of T cell exhaustion. Developmental exposure also changed DNA methylation across the genome, but differences were most pronounced following infection, where we observed inverse correlation between promoter methylation and gene expression. This points to altered regulation of DNA methylation as one mechanism by which AHR causes durable changes in T cell function. Discovering that distinct gene sets and pathways were differentially changed in developmentally exposed mice prior to and after infection further reveals that the process of CD8+ T cell activation is rendered fundamentally different by early life AHR signaling. These findings reveal a novel role for AHR in the developing immune system: regulating DNA methylation and gene expression as T cells respond to infection later in life.
Introduction
A properly functioning immune system underlies multiple aspects of human health and well being, including elimination of pathogens without excessive damage to healthy tissues. Impaired immune responses leave individuals and populations vulnerable to disease. Many factors likely contribute to altered immune function. Several epidemiological studies reveal striking correlations between developmental exposures to anthropogenic chemicals and increasing incidence or severity of infections and poorer responses to routine immunizations (1–6). Although relatively few studies have examined this, they create a compelling case that developmental exposure to pollutants fundamentally alters the responsive capacity of the immune system, leading to long-lasting impairments that contribute to the burden of infectious disease. Maternal and early life exposures have enduring adverse effects on other systems, including nervous, cardiovascular, endocrine and reproductive, as well as cancer rates in offspring (7). Thus, it is not surprising that mounting evidence suggests developmental exposures also affect immune function; however, the factors that influence it are poorly understood.
One possible factor that links signals from the early life environment to the function of the immune system later in life is the aryl hydrocarbon receptor (AHR). AHR is a ligand-activated transcription factor that modulates function of the fully mature (adult) immune system (8). AHR ligands include numerous ubiquitous pollutants such as dioxins, polychlorinated biphenyls (PCBs) and polycyclic aromatic hydrocarbons (PAHs), as well as some naturally derived chemicals, such as tryptophan metabolites (9). Several studies indicate that early life exposure to commonly found AHR-binding pollutants alters immune function later in life (10). Recent studies using low, environmentally relevant maternal doses of AHR ligands demonstrate that persistent changes in host responses to influenza A virus (IAV) are observed in offspring, yet there are no differences in immune organ cellularity in naïve offspring (11, 12). These changes in immune function occur long after the window of developmental exposure (12, 13). Bone marrow cell transplantation further reveals that these diminished adaptive immune responses are intrinsic to hematopoietic cells (12). Yet, how triggering of AHR during development changes the function of the adult immune system remains undefined. Studies of developmental exposures in other organ systems suggest that alterations in epigenetic mechanisms may underlie persistent functional deregulation (14–17).
DNA methylation is one type of epigenetic regulation that influences gene expression and cellular function, is sensitive to environmental cues, and influences the normal development of the immune system (18, 19). Whether activation of AHR via developmental exposure to exogenous ligands alters DNA methylation in immune cells is unknown. Developmental exposure to the prototype AHR ligand, 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) alters DNA methylation patterns in testes, mammary tissue, muscle, and liver, and elevates DNA methyltransfersase (DNMT) activity in pre-implantation embryos (20–23), suggesting plausibility. Also, several studies demonstrate that developmental exposure to other agents, such as heavy metals, maternal smoking and air pollutants modulate DNA methylation in cord blood leukocytes (24–31). However, efforts to correlate changes in DNA methylation and gene expression profiles in mixed populations of blood cells with complex disease outcomes is confounded by the distinct DNA methylation pattern of each type of immune cell (32). Nevertheless, these prior studies collectively imply a connection between early life exposure to environmental agents and altered DNA methylation in mixed leukocyte populations; however, alterations in specific types of immune cells remain to be determined. Thus, triggering AHR during development could alter DNA methylation patterns in cells of the immune system, contributing to changes in gene expression patterns and cellular function later in life.
To address this issue, we selected CD8+ T cells because they are the principal means for successful host resistance in a primary IAV infection, and because their response to infection is profoundly and persistently changed by maternal exposure to the AHR agonist TCDD (12, 33). Although its role is not fully understood, alterations in global DNA methylation patterns occur as naïve CD8+ T cells differentiate and acquire effector function in response to infection, and we hypothesize that developmental activation of AHR alters this process (34). We first investigated whether the altered CD8+ T cell response to IAV is a result of developmental exposure diminishing the number of naïve, virus-specific T cells, and whether it requires AHR in the offspring. We then used unbiased, genome-wide approaches to investigate whether developmental AHR activation alters DNA methylation and gene expression profiles of purified CD8+ T cells prior to and during infection, and then assessed correlations between differentially methylated regions and differentially expressed genes. Our findings reveal new pathways through which environmentally derived AHR ligands influence the function of the immune system.
Materials and Methods
Developmental exposure and infection
Nulliparous C57BL/6 (B6) (National Cancer Institute, Frederick, MD) and B6.Ahrtm1Bra+/−(AHR+/−) females were paired with B6 or AHR+/− males, respectively. Pregnancy was determined by the presence of a vaginal plug (day 0 of gestation, GD0), and pregnant mice were individually housed for the remainder of the study. Offspring were weaned at 21 days of age. Offspring of AHR+/− breeders were genotyped as previously described (35). All mice were housed in a pathogen-free facility in microisolator cages, and food and water were provided ad libitum. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD, ≥99% pure, Cambridge Isotope Laboratories, Woburn, MA) was prepared and mice were dosed as previously described (12). Briefly, pregnant dams were given either TCDD (1 μg/kg body weight) or peanut oil vehicle (Veh) by gavage on GD0, GD7, and GD14 and on postnatal day 2 (PND2). This dose of TCDD is not overtly toxic to the dam or the pups, and does not lead to changes in the cellularity of the bone marrow, thymus, spleen or lymph nodes (11). Intranasal infection of adult offspring with influenza A virus (IAV) strain HKx31 (x31; H3N2) was performed as previously described (12). All procedures involving laboratory animals and infectious agents were reviewed and approved by the University of Rochester Institutional Animal Care and Use and Institutional Biosafety Committees. The University has accreditation through the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC). All guidelines from the U.S. Public Health Service Policy on Humane Care and Use of Laboratory Animals were followed in handling of vertebrate animals.
Isolation of immune cells and flow cytometry
Peripheral lymph nodes (inguinal, axillary, brachial, cervical, mediastinal and sacral), bone marrow, thymus, spleens and liver were isolated from naïve offspring either at PND7 or at maturity. Mediastinal lymph nodes (MLN) were collected from mature offspring nine days after IAV infection. Tissues were processed as previously described (11). Where indicated, total CD8+ T cells or naïve CD8+ T cells were enriched using MagCellect kits (R&D, Mineapolis, MN). For flow cytometry, cells were incubated with anti-mouse CD16/32, prior to incubation with the following fluorochrome-conjugated antibodies (BD Bioscience, San Jose, CA or eBioscience, San Diego, CA): CD8α (53-6.7), CD44 (IM7) and CD62L (MEL-14). MHC class I (MHCI) tetramers corresponding to two immunodominant IAV epitopes (Db/NP366–374 and Db/PA224–233) were used to identify virus specific CD8+ T cells (12). For the identification of virus-specific CD8+ T cells in naïve mice, CD3+ T cells were enriched from peripheral lymph nodes, and the frequency of NP+ and PA+ CD44loCD8+ T cells was determined by flow cytometry. LSR-II cytometers (BD Biosciences) were used for data acquisition (≥500,000 events were collected for cells from naïve animals, and 300,000 to 500,000 events were collected for analyses of cells from infected mice), and data analysis was performed using FlowJo software (Tree Star, Ashland, OR). Given that the dam, rather than the offspring, is directly exposed, the dam is defined as the statistical unit for all experiments, and each offspring was from a different treated dam. Data were analyzed using JMP software (SAS, Cary, NC). Differences between mean values from each exposure group were considered significant when p ≤ 0.05, as determined using a Student’s t-test.
RNA and DNA isolation, Immunoprecipiation (IP), and high throughput sequencing (seq)
RNA was isolated using the RNeasy Mini Kit (QIAGEN, Valencia, CA). RNA quality was verified on the 2100 Bioanalyzer (Agilent, Santa Clara, CA). TruSeq RNA Sample Preparation Kit V2 (Illumina, San Diego, CA) was used per manufacturer’s protocols. Methylated DNA IP (MeDIP) was performed as previously published (36). Briefly, DNA was extracted from enriched CD8+ T cells (≥90% purity) using the DNeasy Blood and Tissue Kit, (QIAGEN), digested with MseI, and further isolated with the QIAquick PCR Purification Kit (QIAGEN). Methyl-seq library construction was performed with NEXTflex Methyl-seq kit (Biooscientific, Austin, TA) per manufacturer’s recommendations. Briefly, 1200 ng of genomic DNA was sheared to an average size of 200–400bp with the Covaris S2 (Covaris, Woburn, MA). Sheared DNA sizing was confirmed with a Bioanlayzer 2100 (Agilent, Santa Clara, CA) and subsequent end repair, adenylation, and adaptor ligation was performed. DNA was heated to 95°C, and incubated with anti-5-methylcytosine antibody (Epigentek, Farmingdale, NY). Methylated DNA was immunoprecipitated with Dynabeads (Life Technologies, Grand Island, NY), and released from the beads using Proteinase K digestion. DNA was purified using the MinElute PCR Purification Kit (QIAGEN), followed by library PCR amplification and gel size selection. For both RNA-seq and MeDIP-seq, single end sequencing was performed using the Illumina GAIIx genome sequencer (Illumina, San Diego, CA) with an average of 20 million 72bp reads per sample. Chromatin IP (ChIP) was performed as previously published (37). Cells were harvested, cross-linked and lysed, and chromatin was sheared by sonication with a Bioruptor (Diagenode, Denville, NJ). Diluted chromatin was combined with 3 μg of the pertinent antibody for each ChIP assay and specific immune complexes were recovered by precipitation with 50:50 protein-A/G-agarose. After washes, chromatin complexes were released with NaHCO3-SDS by vigorous shaking. Cross-links were reversed in the presence of RNAse A at 65°C. ChIP-enriched DNA was purified and quantified using PicoGreen dsDNA dye (Invitrogen). DNA enrichment is represented as the percent of total input after subtracting the values of the relevant control antibodies. For some experiments, reverse transcription and real time PCR (RT-qPCR) was performed as previously described (38), and primers for Dnmt1, Dnmt3a, Dnmt3b and L13 were used (39, 40). For other experiments, gene expression on PND7 was examined. RNA was extracted from liver, reverse transcription was performed, and Cyp1a1 was measured by qPCR, with L13 used as a control (41). Data were analyzed using the ΔΔCT method (42).
MeDIP-seq and RNA-seq analysis
MeDIP-seq reads were aligned to mouse reference genome (mm9) using Bowtie (version 0.12.7 parameter set -n 2 -k 1 -m 1) allowing up to 2 nucleotide mismatches to the reference genome and considering only uniquely mappable reads for downstream analyses (43). MeDIP-seq data was analyzed with MEDIPS package in R (44). For each sample, the aligned reads were extended in the sequencing direction to a length of 400 nucleotides. The short read coverage of the extended reads was calculated at genome wide 50 bp bins. MeDIP-seq reads were counted in the gene bodies of Refseq genes and differential methylation in gene bodies was assessed with edgeR package in R (45). To identify the differentially methylated regions (DMRs) throughout the genome we used various functions of MEDIPS package in R. Mean reads per million (RPM) values were calculated for genome wide 500 bp non-overlapping windows using MEDIPS.methylProfiling function. For any given 500 bp non-overlapping window, mean RPM were calculated with statistically significant DMRs having p-value < 0.001 and a ratio of < 0.5 (hypomethylated) or >2 (hypermethylated) using MEDIPS.selectSignificants function of MEDIPS package. Continuous adjacent significant 500 bp DMRs were merged indicating an extended DMR. The DMRs identified for each comparison were annotated using CEAS. Wilcoxon test was used to determine if the differences between groups was statistically significant. For visualizing the methylation levels in the promoter (2-kb upstream to transcription start site (TSS)), gene body (TSS to transcription end site (TES)) and downstream (2-kb downstream to TES) we used metagene representation. Each region was divided into 20 equal non-overlapping windows and all genes were normalized for length and sequencing depth, and average methylation was plotted as a metagene. To visualize the coverage density of MeDIP-seq reads, we calculated coverage density across the chromosomes using Rsamtools library, density function in R, and custom-written scripts (46). Two-sample Kolmogorov-Smirnov test was used to determine if the differences between groups were statistically significant.
RNA-seq analysis was performed using Tuxedo programs with default parameters (47). Reads were aligned to the mouse reference genome (NCBI37, mm9) using TopHat (v1.4.1), then assembled into transcripts using Cufflinks (v.1.3.0). Differential analysis was performed using Cuffdiff (v.1.3.0). Data were analyzed through the use of IPA (Ingenuity® Systems, www.ingenuity.com). Differentially expressed genes (DEGs) between Veh and TCDD (p ≤ 0.05) were used. The p-values of canonical pathways were corrected with the Benjamini-Hochberg method for multiple testing, with FDR<5%.
Correlation between MeDIP-seq and RNA-seq data was calculated using Spearman’s rank correlation coefficient. Promoter regions (2-kb upstream, 500-bp downstream of TSS) of DEGs with overlapping DMRs (hyper- or hypo methylated) were binned into hypomethylated and/or hyper-methylated promoters. Isolated RNA and MeDIP-ed DNA samples from purified CD8+ T cells were used for gene-specific qPCR. RNA was amplified with WT-Ovation PicoSL Kit (Nugen, San Carlos, CA), converted to cDNA, and reverse-transcriptase (RT)-qPCR was performed using PrimeTime primers (IDT, Coralville, IA). L13 was used as a housekeeping gene, and analysis was performed by the ΔΔCt method (42) Immunoprecipitated DNA was prepared as above, followed by genome-wide amplification using GenomePlex Whole Genome Amplification Kit (Sigma-Aldrich, St. Louis, MO). qPCR on immunoprecipitated DNA was performed using EpiTect ChIP qPCR primers (QIAGEN, Valencia, CA). Primer location was chosen based on the location of the nearest differentially methylated region upstream of the TSS. Input values were used for normalization using the ΔΔCt method.
Results
Developmental exposure persistently changes CD8+ T cell responses to infection
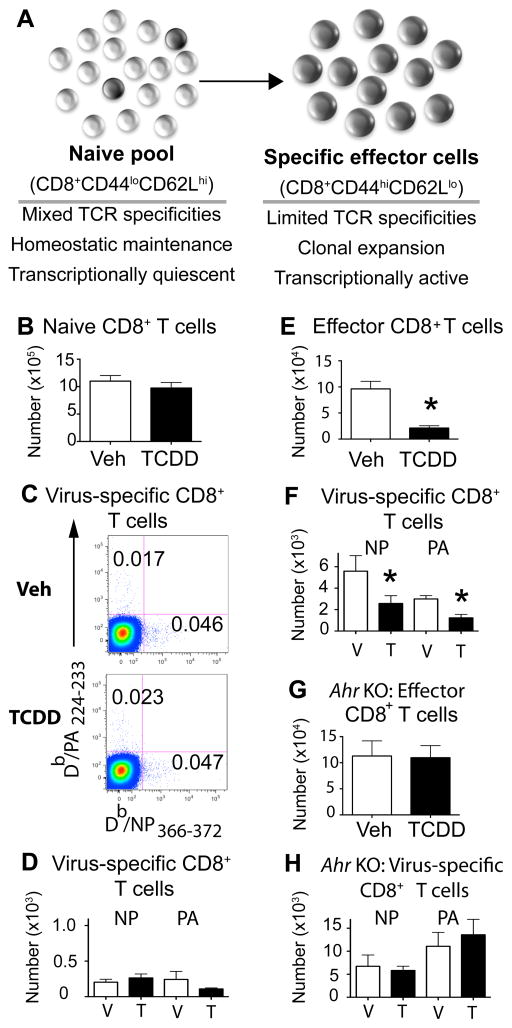
During viral infection, naive CD8+ T cells with T cell receptors (TCR) specific for IAV proliferate and differentiate, forming armed effector cytotoxic T lymphocytes (CTLe) (Fig. 1A). Prior to infection, adult offspring of vehicle control and TCDD-treated dams had a similar number of naïve (CD44lo) CD8+ T cells (Fig. 1B). MHCI restricted tetramers were utilized to identify and enumerate CD8+ T cells with TCR specific for two immunodominant peptide epitopes from IAV: nucleoprotein (NP366–372) and acid polymerase (PA224–233). We did not observe differences in the percent or number of virus-specific CD8+ T cells in uninfected developmentally exposed adult offspring, compared to offspring of control dams (Fig. 1C, 1D). This observation is consistent with prior reports that maternal exposure to this amount of TCDD did not alter thymic cellularity or the total number of T cells in lymphoid organs (11). However, following IAV infection there was a significantly blunted CD8+ T cell response in offspring of TCDD-treated dams, with 3-fold fewer CTLe and 50% fewer NP-specific and PA-specific CD8+ T cells in the developmentally exposed group (Fig. 1E, 1F). By dosing pregnant Ahr+/− dams and monitoring immune responses in Ahr+/+ (wild type) and Ahr−/− (KO) offspring, we found that the presence of the AHR in the fetus/offspring is required for maternal exposure to perturb CD8+ T cells later in life. Only infected Ahr+/+ offspring showed reduced CD8+ T cell responses to IAV, whereas Ahr−/− littermates did not (Fig. 1G, 1H). Thus, based on assessment of known immunodominant viral epitopes, developmental AHR activation did not discernably change the overall number of CD8+ T cells in naive animals; however, it altered their responsive capacity to infection. Further, this durable effect requires functional AHR protein in the developing offspring, demonstrating that differences in the reaction of these cells to infection are driven by early life signaling through AHR.
FIGURE 1.
Developmental exposure impairs the CTL response to IAV without altering frequency of naïve CD8+ T cells. (A) Key changes accompanying CD8+ T cells after infection. B–D: Data in the left column are from naïve offspring. Peripheral lymph nodes were harvested from uninfected adult mice developmentally exposed to vehicle control (Veh) or TCDD. Isolated cells were stained with MHC class I-restricted tetramers and antibodies, as described in the Materials and Methods. Doublet discrimination, exclusion of dead cells, and gating on CD3+ cells were performed, and then CD8+CD44lo T cells, Db/NP366–374+ CD8+ T cells, and Db/PA224–233+CD8+ T cells were defined based on FMO controls. (B) The average number of naïve (CD44lo) CD8+ T cells in naïve developmentally exposed mice is shown. (C) Representative dot plots depict Db/NP366–374+ and Db/PA224–233+CD8+ T cells in lymph nodes from each group; the number on the plots is the percentage in the gated region. (D) The average number of Db/NP366–374 and Db/PA224–233 specific CD8+ T cells per lymph node is shown. E–H: Data from infected offspring are presented the right column. (E,F) Developmentally exposed wild-type (Ahr+/+) mice were infected with IAV at maturity, and 9 days later the number of CTL effectors (CD44hiCD62Llo CD8+ T cells), and virus NP-specific and PA-specific CD8+ T cells in the mediastinal lymph nodes (MLN) were determined by flow cytometry. (G,H) Developmentally exposed Ahr−/− (KO) mice were infected with IAV at maturity, and 9 days later the number of CTL effectors, Db/NP366–374+ and Db/PA224–233+CD8+ T cells in the MLN were determined. Data are shown as mean ± SEM and depict findings from female offspring. Separate assessments using offspring of both sexes yielded similar results (data not shown). Evaluation of naïve mice (8–9 sex matched offspring from separate dams for each group) was performed once; data from infected mice (5–6 same sex offspring from separate dams) are from one experiment that is representative of at least two independent experiments. * indicates p ≤ 0.05 using a Student’s t test.
Developmental activation of AHR alters global DNA methylation profiles in CD8+ T cells
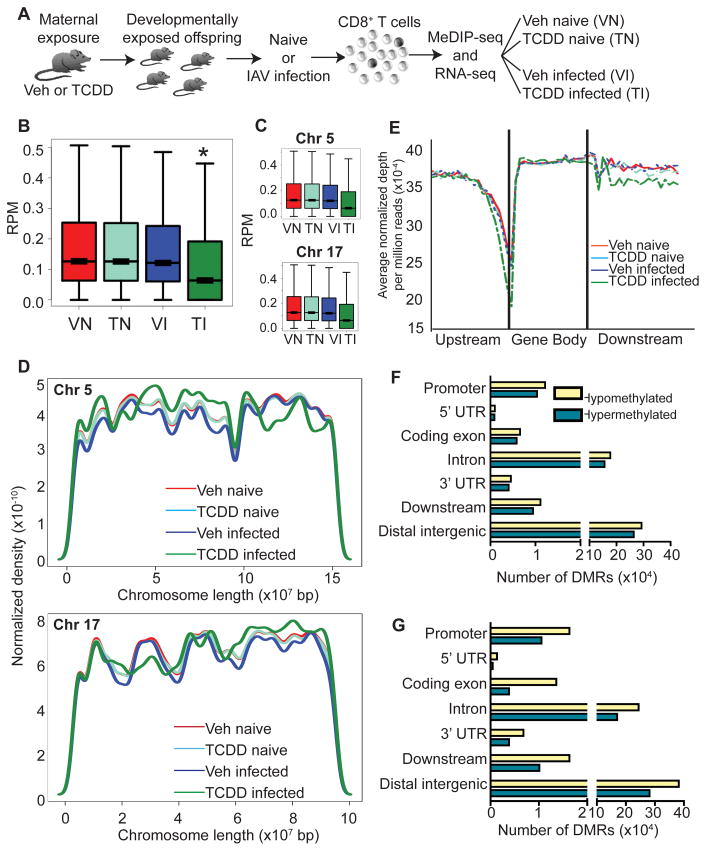
We next determined whether developmental exposure alters global DNA methylation profiles in CD8+ T cells isolated from the offspring, and how these profiles change following infection. We hypothesized that altered DNA methylation due to early life AHR activation contributes to the poorer ability of naïve CD8+ T cells to become activated. In order to test this idea fully, it is critical to determine the profile of DNA methylation from the entire pool of CD8+ T cells from developmentally exposed mice, rather than investigate changes in DNA methylation patterns only in cells that were already activated. DNA was isolated from CD8+ T cells purified from naïve and infected adult offspring of vehicle and TCDD-treated dams. We used methylated DNA immunoprecipitation followed by next generation DNA sequencing (MeDIP-seq) to enrich for and compare methylated DNA (Fig. 2A). MeDIP-seq generated 8–11 million reads per sample, with 70–82% alignment, high correlation between independent replicates (R > 0.996, p-value < 0.0001), and sufficient saturation (> 0.98) and coverage (Supplemental Fig. 1).
FIGURE 2.
DNA methylation is altered in CD8+ T cells following developmental AHR activation. (A) At maturity CD8+ T cells were isolated from naïve or IAV infected mice that were developmentally exposed to Veh or TCDD. To provide sufficient material, CD8+ T cells from up to 15 female littermates, derived from 2–3 dams, were pooled to create each sample. Methylated DNA immunoprecipitation and high-throughput sequencing (MeDIP-seq) was performed. (B,C) Box plots of (B) genome-wide and (C) representative chromosome-specific DNA methylation (chromosomes 5 and 17) for Veh naïve (VN), TCDD naïve (TN), Veh infected (VI) and TCDD infected (TI) are shown. Y-axis shows reads per million (RPM). (D) Coverage plots of selected chromosomes are shown. (E) Metagene analysis of upstream (2-kb upstream to transcription start site, TSS), gene body (TSS to transcription end site, TES) and downstream (TES to 2-kb downstream) regions, in which all genes were normalized for length and the average methylation is shown. (F,G) The genome was scanned with a 500-bp window, and DMRs were identified. Hypomethylated and hypermethylated DMRs located in various genomic features for the comparisons (F) Veh naïve vs. TCDD naïve and (G) Veh infected vs. TCDD infected are shown. * indicates p ≤ 0.00001 between treatment groups of similar infection status using the Wilcoxon rank sum test.
To identify differentially methylated regions (DMRs), we compared genome-wide DNA methylation in CD8+ T cells from each treatment group. The global distribution of DNA methylation was surprisingly similar between CD8+ lymphocytes from uninfected offspring of vehicle and TCDD treated dams (Veh naïve (VN) vs. TCDD naïve (TN), Fig. 2B). However, relative to IAV-infected offspring of control-treated dams, genome-wide DNA methylation was distinctly different in CD8+ T cells from infected offspring of TCDD treated dams (Fig. 2B, comparing TCDD infected (TI) vs. Veh infected (VI) groups), with overall skewing toward more hypomethylated DNA. Analysis of DNA methylation at individual chromosomes revealed a similar overall pattern, where TCDD infected offspring showed the most pronounced difference, with a general shift towards hypomethylation in CD8+ lymphocytes in this group compared to all other groups (representative chromosomes Chr 5 and Chr 17 in Fig. 2C; box plots for all chromosomes in Supplemental Fig. 2). However, analysis of CpG methylation along each chromosome revealed a more intricate pattern of altered methylation. Although the overall level of DNA methylation in CD8+ T cells from TCDD infected offspring was generally skewed toward hypomethylation, both hyper- and hypomethylated regions were observed along many chromosomes (Fig. 2D and Supplemental Fig. 3). Thus, developmental AHR activation followed by infection altered the DNA methylation landscape in CD8+ T cells. These changes covered large areas of chromosomes, were distributed across the whole genome, and resulted in a general shift towards hypomethylation.
While not fully understood, DNA methylation plays broad yet context specific roles in cellular function, which include maintenance of genome stability and regulation of gene expression. In addition to methylation at gene promoters, gene body and intergenic methylation may also play important regulatory roles (18). Metagene analyses, in which the length of upstream, gene body, and downstream regions for each gene were normalized and parsed into windows revealed that, compared to the other three groups, CD8+ T cells from TCDD infected offspring again stood out as distinct, with decreased methylation upstream and downstream of genes (Fig. 2E). Further refinement, in which the genome was scanned using 500 bp windows revealed that in CD8+ lymphocytes from developmentally exposed mice, DMRs occurred across all genomic features, with many found in promoter regions, as well as non-coding (intergenic) regions and gene bodies (introns and coding exons, Fig. 2F–G). While developmentally exposed naïve mice had very similar genome- and chromosome-wide DNA methylation profiles, this analysis revealed that many DMRs were present between Veh naïve and TCDD naïve, consisting of hypermethylated and hypomethylated regions (Fig. 2F). Infected mice also revealed significant differences based on developmental exposure, as developmental activation of AHR led to many DMRs in all genomic areas. While these DMRs represented both hyper- and hypomethylated regions, there were an increased number of hypomethylated DMRs in promoter regions, coding exons, introns, downstream and distal intergenic regions of CD8+ lymphocytes from the TCDD infected group, consistent with the genome-wide shift towards hypomethylation (Fig. 2G). Thus, developmental exposure to an exogenous AHR ligand caused some modest changes in DNA methylation patterns in naïve CD8+ T cells. Yet, when these cells responded to viral infection, substantive changes in DNA methylation were revealed, with a shift predominantly but not exclusively towards hypomethylation.
Gene expression profiles are altered by developmental activation of AHR
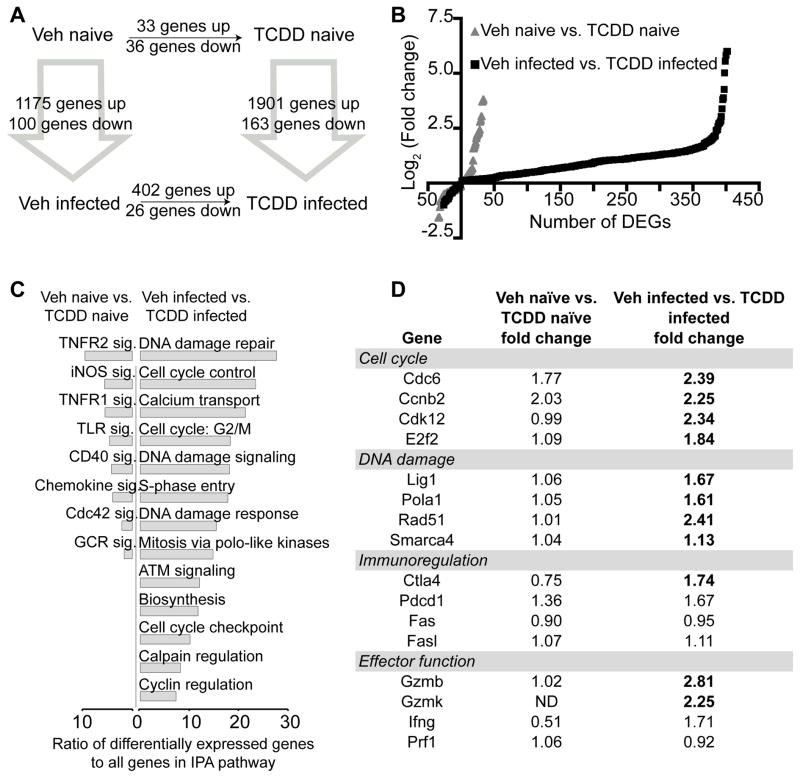
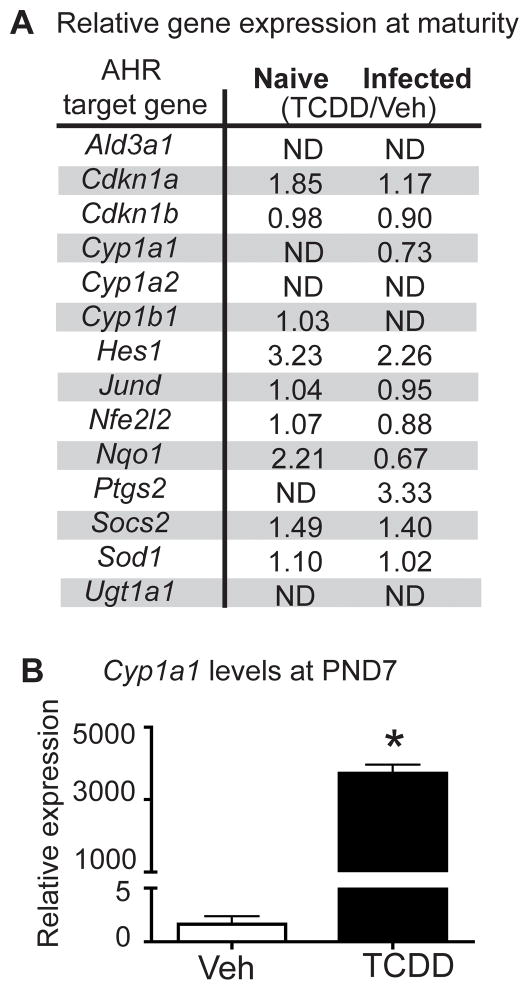
To examine whether developmental exposure also affects gene expression in CD8+ T cells, RNA-seq was performed on the same sample set created for MeDIP-seq (Fig. 2A). RNA-seq generated 15–20 million total reads per sample, with 87–89% of the reads aligning to the genome (Supplemental Fig. 1A). Consistent with the relatively quiescent state of naïve CD8+ T cells compared to CTL (48), IAV infection led to a marked upregulation of genes, regardless of maternal exposure (Fig. 3A). Comparing CD8+ T cells from Veh naïve to TCDD naïve groups, there were 69 differentially expressed genes (DEGs) (Fig. 3A–B, Supplemental Table I). Thirty-three genes had increased expression, whereas 36 had decreased expression. In contrast to uninfected mice, after infection, 428 DEGs were identified between CD8+ T cells from TCDD infected compared to Veh infected offspring, with almost all (402 genes) exhibiting increased expression in adult offspring of TCDD-treated dams (Fig. 3A–B, Supplemental Table I). Given that AHR is a transcription factor, increased transcription in CD8+ T cells, and ultimately impaired function, could reflect persistent up-regulation of defined AHR target genes. However, interrogation of the RNA-seq data set does not support this idea. Validated AHR target genes were not significantly differentially expressed in adult offspring of TCDD treated dams, compared to adult offspring of control dams (Fig. 4A). That is, statistically significantly enhanced expression of AHR target genes is observed in developmentally exposed mice neither prior to nor following infection. In contrast, developmental AHR activation caused transient and significant increases in AHR target genes in neonates. For example, enhanced expression of Cyp1a1 was observed in neonatal mice (i.e., shortly after maternal exposure to TCDD). Specifically, Cyp1a1 expression levels were 2292-times higher in the TCDD naïve group compared to the Veh naive group at PND7 (Fig. 4B). Thus, AHR activation induced expression of target genes during the exposure window, but the up-regulation of these genes was not sustained in CD8+ T cells in developmentally exposed mice. This indicates that the DEGs are likely not AHR target genes themselves, but rather reflect altered transcriptional regulation caused by early life AHR activation.
FIGURE 3.
Many genes are differentially expressed following developmental signaling of AHR. Gene expression was measured in CD8+ T cells by RNA-seq from the four groups depicted in Figure 2A. (A) The number of differentially expressed genes (DEGs) is shown for each comparison, at FDR<5%. (B) For the Veh naïve vs. TCDD naïve and Veh infected vs. TCDD infected comparisons, the log2 (fold change) is plotted for each DEG. (C) For these same comparisons, IPA analysis was performed. Only pathways significantly enriched (Benjamini-Hochberg method for multiple testing, FDR<5%) and relevant for the CD8+ T cell response to infection are shown. x-axis indicates the ratio of DEGs to all genes in the IPA gene set. (D) Fold change values for selected genes involved in the immune response to IAV are listed. Values in bold font are statistically significant (FDR<5%). Full list of DEGs is presented in Supplemental Table I.
FIGURE 4.
AHR target genes are modulated in neonates but not at maturity. (A) At maturity (6–8 weeks of age), CD8+ T cells were isolated from developmentally exposed mice and gene expression evaluated. Validated AHR target genes are listed, with the ratio of RNA-seq normalized reads for TCDD/Veh for naïve and infected shown. No statistically significant differences were observed at FDR <5%. Similar results were obtained using qPCR (not shown). ND = not detected. (B) Mean Cyp1a1 expression at PND7. Livers were removed from 7-day-old offspring of different dams that were treated Veh or TCDD. Nucleic acids were prepared, reverse transcribed, and used for qPCR. Data were analyzed using the ΔΔCT method, using L13 as an internal control. * indicates p ≤ 0.05 using a Student’s t test.
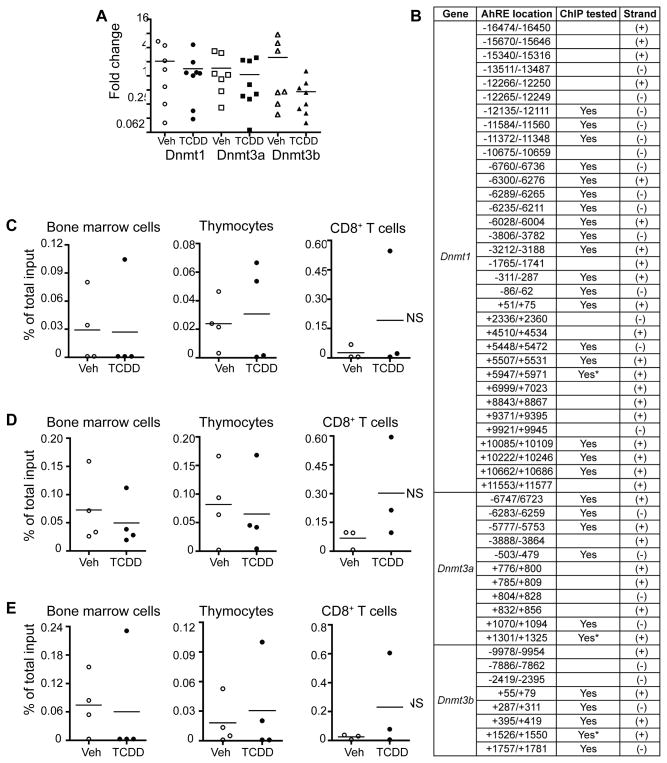
To identify pathways represented by these DEGs, Ingenuity Pathway Analysis (IPA) was performed comparing Veh naïve to TCDD naïve and Veh infected to TCDD infected. Of the few genes altered in the Veh naïve-TCDD naïve comparison, most were enriched in tumor necrosis factor receptor (TNFR) or other signaling pathways. Similar to DMRs, changes in gene expression in CD8+ lymphocytes from adult animals that were developmentally exposed became considerably more evident after infection. The major pathways enriched in CD8+ T cells from TCDD infected versus Veh infected included cell cycle regulation and DNA damage (Fig. 3C–D). Genes and pathways involved in immune responses to IAV were also differentially expressed (Fig. 3D). Deregulation of these pathways corresponds with altered CD8+ T cell clonal expansion and differentiation observed in infected developmentally exposed mice. Another key finding is that pathways involved in epigenetic regulation and DNA methylation were not altered in offspring of TCDD-treated dams (regardless of infection). Although Wu et al. (2004) reported increased DNMT activity in mouse embryos treated directly ex vivo with TCDD (20), we observed no differences in the relative level of expression of Dnmt1, Dnmt3a or Dnmt3b in purified CD8+ T cells from developmentally exposed mice (Fig. 5A). To further explore the idea that AHR may play a role in the regulation of DNMTs, we identified putative binding sites for AHR (AHR response elements, AHREs) in the upstream regulatory regions and gene bodies of all three Dnmt genes (Fig. 5B). Using chromatin immunoprecipitation, we next investigated whether maternal exposure leads to binding of AHR to these putative AHREs. We were unable to detect alterations in binding of AHR to these genes in bone marrow cells, thymocytes, or CD8+ T cells from developmentally exposed mice (Fig. 5C–E). Thus, while developmental exposure altered DNA methylation across the genome, and influenced expression of many other genes, we found no evidence that this was achieved through AHR persistently or directly altering Dnmt gene expression.
FIGURE 5.
Developmental activation of AHR does not lead to changes in Dnmt expression. (A) Dnmt expression levels were measured by RT-qPCR in CD8+ T cells isolated from developmentally exposed mice. Relative expression is presented as ΔΔCT normalized to L13. (B) Putative aryl hydrocarbon receptor elements (AHREs) were identified upstream and in the gene bodies of Dnmt1, Dnmt3a and Dnmt3b. (C–E) Binding of AHR to putative AHREs was measured by chromatin immunoprecipitation (ChIP) assay using isolated bone marrow cells, thymocytes, and CD8+ T cells. DNA enrichment is represented as the percent of total input after subtracting the values of the relevant control antibodies. Representative results are shown for the putative AHREs indicated by * in (B), and are consistent across the others tested.
Correlations between DMRs and gene expression were observed following infection
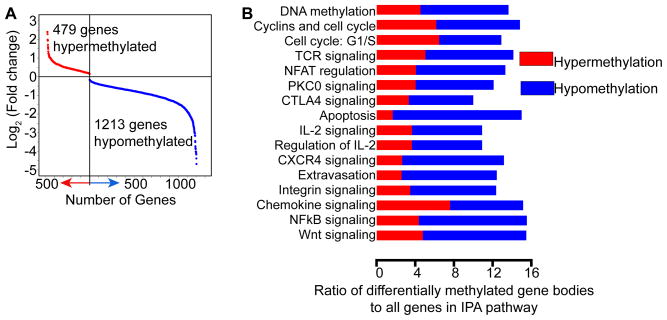
DNA methylation influences transcriptional regulation in a context-specific manner, and recent work indicates a role of DNA methylation in gene bodies as well as promoters (18). To directly examine correlations between DNA methylation and gene expression in these genomic locations, we integrated the MeDIP-seq and RNA-seq datasets. Consistent with other observations reported herein, differences in gene body methylation were more pronounced in CD8+ T cells from infected offspring. When comparing genes with differential gene body methylation in Veh infected and TCDD infected groups, we observed 1213 hypomethylated and 479 hypermethylated genes, with many involved in immune regulation, proliferation, transcription and epigenetic regulation differentially methylated (Fig. 6). However, there was little correlation between DMRs in gene bodies and gene expression.
FIGURE 6.
Differential gene body methylation is observed following developmental activation of AHR. (A) Differential gene body methylation is compared in CD8+ T cells from infected adult offspring of vehicle and TCDD treated dams. The graph depicts the log2 (fold change) of differentially methylated gene bodies for each gene, with hypermethylated genes shown in red and hypomethylated genes shown in blue. (B) IPA analysis was performed on these genes. Significantly different pathways relevant for the CD8+ T cell response to infection are shown, indicating the contribution from hyper- and hypomethylated genes. The x-axis indicates the ratio of genes with differential gene body methylation to all genes in the indicated IPA pathway.
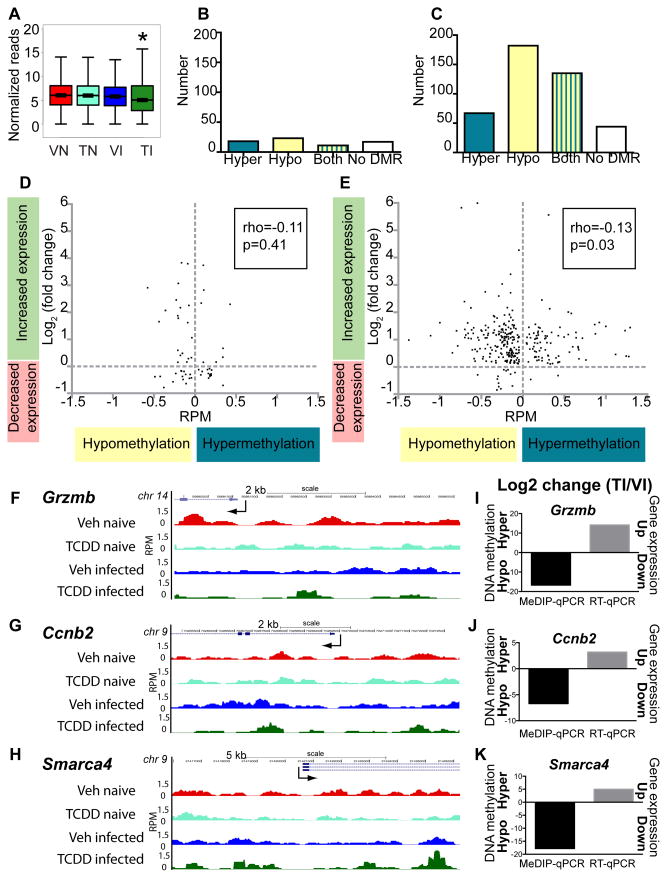
DNA methylation of promoter regions across the genome was significantly diminished in CD8+ T cells from TCDD infected offspring compared to all other groups (Fig. 7A). Considering DEGs in Veh naïve versus TCDD naïve and Veh infected versus TCDD infected, we determined whether the promoter region for each DEG was differentially methylated (Fig. 7B–C). The Veh naïve versus TCDD naïve comparison had few DEGs containing DMRs in their promoters (Fig. 7B). In contrast, the majority of DEGs in the Veh infected to TCDD infected comparison had either hypomethylation or a mixture of hyper- and hypomethylation at their promoters (Fig. 7C). We next examined whether differential promoter methylation (hypo- or hypermethylation) correlated with altered gene expression. For CD8+ T cells from Veh naïve versus TCDD naïve, changes in promoter methylation and gene expression had no correlation (Fig. 7D). However, following infection, changes in promoter methylation inversely correlated with gene expression in the Veh infected-TCDD infected comparison (Fig. 7E).
FIGURE 7.
Changes in gene expression correlate with promoter methylation in infected and developmentally exposed mice. (A) Genome-wide methylation at promoters (2 kb upstream to TSS) is shown for Veh naïve (VN), TCDD naïve (TN), Veh infected (VI) and TCDD infected (TI). (B,C) Differentially methylated regions (DMRs) at gene promoters were determined for the differentially expressed genes (DEGs) in the (B) Veh naïve vs. TCDD naïve and (C) Veh infected vs. TCDD infected comparisons. Promoters were classified as containing only hypermethylated DMRs (Hyper), only hypomethylated DMRs (Hypo), both hyper- and hypomethylated DMRs (Both), or lacking any DMRs (No DMR), and the number of DEG promoters in each category is shown. (D,E) For the DEGs in the (D) Veh naïve vs. TCDD naïve and (E) Veh infected vs. TCDD infected comparisons that had a unidirectional change in promoter methylation (Hyper only or Hypo only), the RPM difference in methylation is plotted on the x-axis, and the log2 (fold change) of gene expression is plotted on the y-axis. Correlation coefficient and p-value are indicated on each plot. * indicates p ≤ 0.00001 between treatment groups of similar infection status using the Wilcoxon rank sum test. (F–H) Visualization of promoter methylation using the UCSC browser was performed. Maximal height for visualization was set at rpm = 1.5 for all MeDIP-seq tracks. Traces show relative DNA methylation across the promoter regions for Grzmb, Ccnb2 and Smarca4; TSS indicated with an arrow. (I–J) CD8+ T cells were isolated from naïve (not shown) and infected adult mice that had been exposed during development to Vehicle or TCDD. RNA and DNA were prepared separately from the same pool of purified CD8+ T cells. Following MeDIP, qPCR was used to evaluate and compare promoter methylation (black bars). Input DNA values were used for normalization. Gene expression in CD8+ T cells was determined using RT-qPCR (grey bars). Data were analyzed using the ΔΔCT method, with L13 serving as the internal control. The qPCR data are presented as log2 fold change in the TCDD infected (TI) group relative to cells from the Vehicle infected (VI) exposure group.
To further affirm the observed DMRs and DEGs identified by high throughput sequencing, a subset of 25 genes from CD8+ T cells purified from developmentally exposed mice were examined. When visualized with a genome browser, the data show consistently that cells from the TCDD-exposed and infected group have a methylation pattern that is distinct from the other groups. For example, promoter methylation of three representative genes, Gzmb, Ccnb2, and Smarca4, which are involved in CD8+ T cell activation, cell cycle regulation and DNA damage repair, respectively, showed notable differences upstream of the TSS (Fig. 7F–H). Differential methylation and gene expression were independently examined by qPCR using CD8+ T cells isolated from all four groups. In agreement with sequencing data, genes such Gzmb, Ccnb2, and Smarca4 had diminished promoter methylation and elevated gene expression when isolated from infected offspring that were developmentally exposed to TCDD versus those from infected offspring of Veh treated dams (Fig. 7I–K). Thus, target gene approaches support the overall correlation between promoter methylation and gene expression that were observed using genome-wide approaches. Moreover, these data show that developmental activation of AHR modifies promoter methylation in CD8+ T cells that are responding to infection, and these changes correlate with altered gene transcription and with reduced clonal expansion and differentiation.
Discussion
The AHR has been extensively studied as an environmental sensor that promotes the metabolism and elimination of pollutants, yet it is now clear that it plays a much broader role, including regulation of the immune system (8). Although developmental exposure to environmental AHR ligands alters immune function in animal models and human populations, how AHR exerts these persistent changes has remained enigmatic (2, 6, 10). We demonstrated that early life activation of AHR fundamentally alters the responsive capacity of CD8+ T cells. Yet, this altered response was not due to fewer CD8+ T cells in developmentally exposed animals prior to infection, nor was it due to sustained expression of AHR target genes repressing CD8+ T cell function. Instead, developmental signaling through AHR reprogrammed DNA methylation and gene expression profiles of CD8+ T cells, which likely contributes to their diminished response following infection. Surprisingly, the mechanism is not simply AHR-mediated increases in DNA hypermethylation causing gene silencing in CD8+ T cells. Rather, this work reveals a more intricate mechanism. Changes in DNA methylation and gene expression in T cells from developmentally exposed mice were relatively modest prior to infection, but became more evident and complex following infection. This indicates that AHR modifies the responsive capacity of developing CD8+ T cells, and regulates their function by influencing multifaceted interactions between DNA methylation and gene expression during response to infection.
The underlying reason that developmental activation of AHR results in a poorer CD8+ T cell response to influenza virus infection later in life is not known. It could be due to changes within the effector cell subset, but could also be due to other changes, such as negative regulatory signals in the CD8+ T cells that are not CD44hiCD62Llo. Given that this is the first time a whole genome analysis of DNA methylation and gene expression of CD8+ T cells in mice following developmental activation of AHR has been performed, it was important to not exclude non-activated cells a priori, because doing so could have biased the information obtained and missed critical differences. Thus, in this study we interrogated DNA methylation and gene expression profiles of the total pool of CD8+ T cells from developmentally exposed mice, not just in the cells that had acquired an effector phenotype. Using this unbiased approach, we discovered that early life activation of AHR skews gene promoters toward hypomethylation following infection in CD8+ T cells, potentially causing the inappropriately enhanced transcription observed. Disrupted transcriptional potential may result from differential methylation at individual gene promoters, or from broader areas of differential methylation observed across chromosomes. For instance, it is possible that altered DNA methylation influences regulation of enhancer regions and other genomic elements, as our data reveal that triggering AHR during development changes DNA methylation in CD8+ T cells across many genomic features. Given emerging understanding of the complex ways that DNA methylation contributes to transcriptional regulation in other cell types (18), these changes outside of gene promoters may have broad and yet-unrealized implications as to how early life activation of AHR influences T cell function. Likewise, it will be interesting and important to determine which changes in DNA methylation reflect changes in distinct subsets of the total CD8+ T cell pool (e.g., naïve vs. effector), which may further reveal nuances in how developmental exposures leads to changes in DNA methylation patterns and how these are linked to an altered response later in life.
Combined with genome-wide changes in DNA methylation patterns, gene expression profiling demonstrates that early life AHR activation durably alters the responsive capacity of T cells. Based on the observed differences in gene expression in developmentally exposed infected mice, the defective CD8+ T cell response is not simply due to these cells remaining in a naïve state following infection, nor is it due to overt loss of expression of genes required for effector function. Instead, developmental AHR activation alters the transcriptional profile in CD8+ T cells, and the observed changes suggest several potential defects, which are not mutually exclusive. For instance, infected, developmentally exposed mice had fewer CTL and virus-specific CD8+ T cells, yet expression of effector genes remained high (for example, Gzmb), which is similar to exhausted T cells (49). Generally observed after prolonged exposure to antigen, such as in chronic infections, exhausted T cells paradoxically lose effector function but retain expression of genes for key effector molecules. Consistent with this overall idea, AHR regulates, either directly or indirectly, proliferation capacity and exhaustion in hematopoietic stem cells, as well as cellular senescence in other cell types (50–55). Inappropriate early life AHR signaling could also lead to an immune system that is improperly poised toward tolerance, an essential regulatory mechanism that prevents self-reactive lymphocytes from responding. Indeed, deregulated tolerance underlies imbalanced immunity and contributes to disease (56). Interestingly, CD8+ T cells from developmentally exposed and infected mice and tolerized CD8+ T cells share a similar pattern of differentially expressed genes, including those involved in cell cycle regulation and DNA repair (56). Related to tolerance and culling of inappropriately activated T cells are apoptosis and deletion of improperly activated T cells, such as via activation induced cell death (AICD). At higher doses of TCDD, enhanced thymocyte apoptosis has been observed (57, 58). However, we did not detect an increase in apoptotic CD8+ T cells from developmentally exposed mice (data not shown), nor did we find differences in DNA methylation or expression of genes associated with apoptosis or AICD. However, the overall shift toward DNA hypomethylation could signify that developmental exposure leads to greater genomic instability in T cells. Genomic instability can enhance DNA damage (59, 60), potentially contributing to the reduced clonal expansion of CD8+ T cells after infection. Pathways involved in DNA damage repair and cell cycle regulation were enriched among the DEGs. Moreover, while not examined in the context of developmental exposure, AHR agonists influence DNA stability (61). These observations suggest early life AHR activation influences CD8+ T cell function later in life without causing overt immunotoxicity. Consistent with these potential mechanisms discussed here, recent human transcriptomic data show correlations between fetal exposure to AHR-binding pollutants, including TCDD, and altered expression of genes involved in immune regulation, cell cycle and antigen processing and presentation in cord blood cells (62).
We showed that developmental exposure to an AHR ligand disrupts DNA methylation and gene expression profiles in CD8+ T cells. Yet, in order to fully understand how the early life environment shapes the function of the immune system later in life, it will be important to investigate how AHR affects other epigenetic regulatory mechanisms, and to integrate these changes in the context of cell-cell interactions that are the foundation of antiviral T cell responses. For instance, in addition to DNA methylation, DNA hydroxymethylation, histone modifications, and microRNAs (miRNAs) shape the epigenetic landscape to influence transcription and cellular function. Although not examined in the context of developmental exposures, in non-immune cells, AHR ligands alter regulation of histone modifications (63–66). AHR activation in fetal thymocytes alters levels of many miRNAs that might be involved in T cell activation (67, 68). Thus, in addition to DNA methylation, developmental AHR activation may alter other epigenetic mechanisms. Furthermore, developmental exposure could influence different epigenetic mechanisms in different immune cells at the same time, such that altered response reflects the net effect of multiple epigenetic changes in more than one cell type. For example, as naïve CD8+ T cells differentiate into CTL, accessory cells, such as dendritic cells (DCs) and CD4+ T cells deliver key co-stimulatory and regulatory signals. Our data demonstrate that changes in DNA methylation and gene expression in CD8+ T cells from developmentally exposed mice are most pronounced following infection, implicating altered signaling from accessory cells as a potential mechanism by which AHR alters CD8+ T cell function. Developmental exposure could reduce CD8+ T cell activation not only by altering DNA methylation patterns in CD8+ lymphocytes, but by influencing epigenetic mechanisms in DCs and/or CD4+ T cells, thus impairing their function. In support of this idea, developmental AHR activation was recently shown to impair CD4+ T cell activation and subset specification following IAV infection, including skewing towards more T regulatory cells (35). Other types of early life exposures also impinge on CD4+ T cell-dependent function in offspring (69, 70). The consequences of developmental exposure to AHR ligands on DC function have yet to be explored, but early life exposure to other environmental agents affects DCs (71). Moreover, in the fully mature immune system, AHR modulates DC frequency and function (38, 72, 73). Thus, it will be important to interrogate AHR-mediated alterations in other immune cell types, and integrate this information to establish the contribution of intrinsic changes, such as alterations in DNA methylation and gene expression, with changes in key accessory cells to fully understand all the pathways that lead to impaired CD8+ T cell responses to infection.
DNA methylation patterns are sensitive to early life environmental insults, although the functional consequences of such changes are not fully understood, particularly in the context of immune responses to infection. Early life exposure to AHR binding pollutants causes enduring changes in the responsive capacity of the immune system, including reduced CD8+ T cell clonal expansion and differentiation in response to IAV. As CD8+ T cells respond to acute primary infection, their DNA methylation profiles change, and this is thought to influence acquisition of effector function (34). However, mechanisms that lead to long-lasting functional deficits following early life exposures are unknown. Here we demonstrate that activation of AHR during development alters DNA methylation and gene expression profiles in CD8+ T cells of adult offspring. Changes in DNA methylation are relatively subtle in the absence of infection. However, as CD8+ T cells respond to viral infection, pronounced differences in DNA methylation are revealed across the genome. These findings reveal that early life AHR activation alters the interaction between DNA methylation and gene expression, which could contribute to the reduced responsive capacity of CD8+ T cells. The implications of this include a better understanding of how early life exposures to chemicals that bind AHR durably shape immunity later in life, and provide a new line of thinking with regard to AHR as a key regulator of the development and function of the immune system.
Supplementary Material
Acknowledgments
The authors would like to thank Shauna Marr, Dr. Steven Welle and Michelle Zanche for technical advice and assistance, and Lisbeth Boule, Dr. Mike Bulger, and Dr. Steve Georas for critical evaluation of the manuscript.
This work was supported by grants from the National Institutes of Health (R01-ES017250, R01-ES023260, R01-HL097141, T32-ES07026, and P30-ES01247). W.L.K. was supported by R01-DK069710. C-I.K. and A.P. were supported by R01-ES006273 and P30-ES06096.
Abbreviations
- AHR
aryl hydrocarbon receptor
- AICD
activation induced cell death
- ChIP
chromatin immunoprecipitation
- DC
dendritic cell
- DEG
differentially expressed gene
- DNMT
DNA methyltransferase
- DMR
differentially methylated region
- FMO
fluorescence minus one
- GD
gestation day
- IAV
influenza A virus
- IPA
Ingenuity Pathway Analysis
- KO
Ahr−/− knockout
- MeDIP-seq
methylated DNA immunoprecipitation-high throughput sequencing
- miRNA
microRNA
- MLN
mediatstinal lymph node
- NP
nucleoprotein
- PA
acid polymerase
- PAH
polycyclic aromatic hydrocarbon
- PCB
polychlorinated biphenyl
- PND
post natal day
- qPCR
real time PCR
- RNA-seq
RNA-high throughput sequencing
- RPM
reads per million
- TCDD
2,3,7,8-tetrachlorodibenzo-p-dioxin
- TES
transcription end site
- TI
developmentally exposed to TCDD and infected
- TN
developmentally exposed to TCDD and naive
- TSS
transcription start site
- Veh
vehicle control
- VI
developmentally exposed to Veh and infected
- VN
developmentally exposed to Veh and naive
- WT
Ahr+/+ wild type
Footnotes
Conflict of interest: The authors declare they have no actual or potential competing financial interests.
Methylated DNA immunoprecipitation sequencing and RNA sequencing data presented in this article have been submitted to the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/); accession number GSE60115.
References
- 1.Dallaire F, Dewailly E, Vezina C, Muckle G, Weber JP, Bruneau S, Ayotte P. Effect of prenatal exposure to polychlorinated biphenyls on incidence of acute respiratory infections in preschool Inuit children. Environ Health Perspect. 2006;114:1301–1305. doi: 10.1289/ehp.8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heilmann C, Budtz-Jorgensen E, Nielsen F, Heinzow B, Weihe P, Grandjean P. Serum concentrations of antibodies against vaccine toxoids in children exposed perinatally to immunotoxicants. Environ Health Perspect. 2010;118:1434–1438. doi: 10.1289/ehp.1001975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heilmann C, Grandjean P, Weihe P, Nielsen F, Budtz-Jorgensen E. Reduced antibody responses to vaccinations in children exposed to polychlorinated biphenyls. PLoS Med. 2006;3:e311. doi: 10.1371/journal.pmed.0030311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glynn A, Thuvander A, Aune M, Johannisson A, Darnerud PO, Ronquist G, Cnattingius S. Immune cell counts and risks of respiratory infections among infants exposed pre- and postnatally to organochlorine compounds: a prospective study. Environmental Health. 2008;7:62. doi: 10.1186/1476-069X-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stolevik SB, Nygaard UC, Namork E, Haugen M, Kvalem HE, Meltzer HM, Alexander J, van Delft JH, Loveren H, Lovik M, Granum B. Prenatal exposure to polychlorinated biphenyls and dioxins is associated with increased risk of wheeze and infections in infants. Food Chem Toxicol. 2011;49:1843–1848. doi: 10.1016/j.fct.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 6.Stolevik SB, Nygaard UC, Namork E, Haugen M, Meltzer HM, Alexander J, Knutsen HK, Aaberge I, Vainio K, van Loveren H, Lovik M, Granum B. Prenatal exposure to polychlorinated biphenyls and dioxins from the maternal diet may be associated with immunosuppressive effects that persist into early childhood. Food Chem Toxicol. 2013;51:165–172. doi: 10.1016/j.fct.2012.09.027. [DOI] [PubMed] [Google Scholar]
- 7.Barouki R, Gluckman PD, Grandjean P, Hanson M, Heindel JJ. Developmental origins of non-communicable disease: implications for research and public health. Environ Health. 2012;11:42. doi: 10.1186/1476-069X-11-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quintana FJ, Sherr DH. Aryl hydrocarbon receptor control of adaptive immunity. Pharmacol Rev. 2013;65:1148–1161. doi: 10.1124/pr.113.007823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen LP, Bradfield CA. The search for endogenous activators of the aryl hydrocarbon receptor. Chem Res Toxicol. 2008;21:102–116. doi: 10.1021/tx7001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winans B, Humble MC, Lawrence BP. Environmental toxicants and the developing immune system: a missing link in the global battle against infectious disease? Reprod Toxicol. 2011;31:327–336. doi: 10.1016/j.reprotox.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vorderstrasse BA, Cundiff JA, Lawrence BP. Developmental exposure to the potent aryl hydrocarbon receptor agonist 2,3,7,8-tetrachlorodibenzo-p-dioxin Impairs the cell-mediated immune response to infection with influenza a virus, but enhances elements of innate immunity. J Immunotoxicol. 2004;1:103–112. doi: 10.1080/15476910490509244. [DOI] [PubMed] [Google Scholar]
- 12.Hogaboam JP, Moore AJ, Lawrence BP. The aryl hydrocarbon receptor affects distinct tissue compartments during ontogeny of the immune system. Toxicol Sci. 2008;102:160–170. doi: 10.1093/toxsci/kfm283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luebke RW, Chen DH, Dietert R, Yang Y, King M, Luster MI. The comparative immunotoxicity of five selected compounds following developmental or adult exposure. J Toxicol Environ Health B Crit Rev. 2006;9:1–26. doi: 10.1080/15287390500194326. [DOI] [PubMed] [Google Scholar]
- 14.Szyf M, Bick J. DNA methylation: a mechanism for embedding early life experiences in the genome. Child Dev. 2013;84:49–57. doi: 10.1111/j.1467-8624.2012.01793.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dolinoy DC, Weidman JR, Jirtle RL. Epigenetic gene regulation: linking early developmental environment to adult disease. Reprod Toxicol. 2007;23:297–307. doi: 10.1016/j.reprotox.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 16.Daxinger L, Whitelaw E. Transgenerational epigenetic inheritance: more questions than answers. Genome Res. 2010;20:1623–1628. doi: 10.1101/gr.106138.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 19.Suarez-Alvarez B, Baragano Raneros A, Ortega F, Lopez-Larrea C. Epigenetic modulation of the immune function: a potential target for tolerance. Epigenetics. 2013;8:694–702. doi: 10.4161/epi.25201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Q, Ohsako S, Ishimura R, Suzuki JS, Tohyama C. Exposure of mouse preimplantation embryos to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) alters the methylation status of imprinted genes H19 and Igf2. Biol Reprod. 2004;70:1790–1797. doi: 10.1095/biolreprod.103.025387. [DOI] [PubMed] [Google Scholar]
- 21.Manikkam M, Tracey R, Guerrero-Bosagna C, Skinner MK. Dioxin (TCDD) induces epigenetic transgenerational inheritance of adult onset disease and sperm epimutations. PLoS One. 2012;7:e46249. doi: 10.1371/journal.pone.0046249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papoutsis AJ, Selmin OI, Borg JL, Romagnolo DF. Gestational exposure to the AhR agonist 2,3,7,8-tetrachlorodibenzo-p-dioxin induces BRCA-1 promoter hypermethylation and reduces BRCA-1 expression in mammary tissue of rat offspring: Preventive effects of resveratrol. Mol Carcinog. 2013 doi: 10.1002/mc.22095. [DOI] [PubMed] [Google Scholar]
- 23.Somm E, Stouder C, Paoloni-Giacobino A. Effect of developmental dioxin exposure on methylation and expression of specific imprinted genes in mice. Reprod Toxicol. 2013;35:150–155. doi: 10.1016/j.reprotox.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 24.Intarasunanont P, Navasumrit P, Waraprasit S, Chaisatra K, Suk WA, Mahidol C, Ruchirawat M. Effects of arsenic exposure on DNA methylation in cord blood samples from newborn babies and in a human lymphoblast cell line. Environ Health. 2012;11:31. doi: 10.1186/1476-069X-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kile ML, Houseman EA, Baccarelli AA, Quamruzzaman Q, Rahman M, Mostofa G, Cardenas A, Wright RO, Christiani DC. Effect of prenatal arsenic exposure on DNA methylation and leukocyte subpopulations in cord blood. Epigenetics. 2014;9:774–782. doi: 10.4161/epi.28153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koestler DC, Avissar-Whiting M, Houseman EA, Karagas MR, Marsit CJ. Differential DNA methylation in umbilical cord blood of infants exposed to low levels of arsenic in utero. Environ Health Perspect. 2013;121:971–977. doi: 10.1289/ehp.1205925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pilsner JR, Hu H, Ettinger A, Sanchez BN, Wright RO, Cantonwine D, Lazarus A, Lamadrid-Figueroa H, Mercado-Garcia A, Tellez-Rojo MM, Hernandez-Avila M. Influence of prenatal lead exposure on genomic methylation of cord blood DNA. Environ Health Perspect. 2009;117:1466–1471. doi: 10.1289/ehp.0800497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanders AP, Smeester L, Rojas D, Debussycher T, Wu MC, Wright FA, Zhou YH, Laine JE, Rager JE, Swamy GK, Ashley-Koch A, Lynn Miranda M, Fry RC. Cadmium exposure and the epigenome: Exposure-associated patterns of DNA methylation in leukocytes from mother-baby pairs. Epigenetics. 2014;9:212–221. doi: 10.4161/epi.26798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joubert BR, Haberg SE, Nilsen RM, Wang X, Vollset SE, Murphy SK, Huang Z, Hoyo C, Midttun O, Cupul-Uicab LA, Ueland PM, Wu MC, Nystad W, Bell DA, Peddada SD, London SJ. 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ Health Perspect. 2012;120:1425–1431. doi: 10.1289/ehp.1205412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang IJ, Chen SL, Lu TP, Chuang EY, Chen PC. Prenatal smoke exposure, DNA methylation, and childhood atopic dermatitis. Clin Exp Allergy. 2013;43:535–543. doi: 10.1111/cea.12108. [DOI] [PubMed] [Google Scholar]
- 31.Tang WY, Levin L, Talaska G, Cheung YY, Herbstman J, Tang D, Miller RL, Perera F, Ho SM. Maternal exposure to polycyclic aromatic hydrocarbons and 5′-CpG methylation of interferon-gamma in cord white blood cells. Environ Health Perspect. 2012;120:1195–1200. doi: 10.1289/ehp.1103744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reinius LE, Acevedo N, Joerink M, Pershagen G, Dahlen SE, Greco D, Soderhall C, Scheynius A, Kere J. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS One. 2012;7:e41361. doi: 10.1371/journal.pone.0041361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kohlmeier JE, Woodland DL. Immunity to Respiratory Viruses. Annual Review of Immunology. 2009;27:61–82. doi: 10.1146/annurev.immunol.021908.132625. [DOI] [PubMed] [Google Scholar]
- 34.Scharer CD, Barwick BG, Youngblood BA, Ahmed R, Boss JM. Global DNA methylation remodeling accompanies CD8 T cell effector function. J Immunol. 2013;191:3419–3429. doi: 10.4049/jimmunol.1301395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boule LA, Winans B, Lawrence BP. Effects of Developmental Activation of the AhR on CD4 T-Cell Responses to Influenza Virus Infection in Adult Mice. Environ Health Perspect. 2014 doi: 10.1289/ehp.1408110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Down TA, Rakyan VK, Turner DJ, Flicek P, Li H, Kulesha E, Graf S, Johnson N, Herrero J, Tomazou EM, Thorne NP, Backdahl L, Herberth M, Howe KL, Jackson DK, Miretti MM, Marioni JC, Birney E, Hubbard TJ, Durbin R, Tavare S, Beck S. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol. 2008;26:779–785. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ko CI, Wang Q, Fan Y, Xia Y, Puga A. Pluripotency factors and Polycomb Group proteins repress aryl hydrocarbon receptor expression in murine embryonic stem cells. Stem Cell Res. 2014;12:296–308. doi: 10.1016/j.scr.2013.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin GB, Winans B, Martin KC, Lawrence BP. New insights into the role of the aryl hydrocarbon receptor in the function of CD11c cells during respiratory viral infection. Eur J Immunol. 2014;44:1685–1698. doi: 10.1002/eji.201343980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Attwood J, Richardson B. Relative quantitation of DNA methyltransferase mRNA by real-time RT-PCR assay. Methods Mol Biol. 2004;287:273–283. doi: 10.1385/1-59259-828-5:273. [DOI] [PubMed] [Google Scholar]
- 40.Phillips JM, Burgoon LD, Goodman JI. The constitutive active/androstane receptor facilitates unique phenobarbital-induced expression changes of genes involved in key pathways in precancerous liver and liver tumors. Toxicol Sci. 2009;110:319–333. doi: 10.1093/toxsci/kfp108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wheeler JL, Martin KC, Resseguie E, Lawrence BP. Differential consequences of two distinct AhR ligands on innate and adaptive immune responses to influenza A virus. Toxicol Sci. 2014;137:324–334. doi: 10.1093/toxsci/kft255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 43.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chavez L, Jozefczuk J, Grimm C, Dietrich J, Timmermann B, Lehrach H, Herwig R, Adjaye J. Computational analysis of genome-wide DNA methylation during the differentiation of human embryonic stem cells along the endodermal lineage. Genome Res. 2010;20:1441–1450. doi: 10.1101/gr.110114.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morgan M. An Introduction to Rsamtools 2010 [Google Scholar]
- 47.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaech SM, Wherry EJ, Ahmed R. Effector and memory T-cell differentiation: implications for vaccine development. Nat Rev Immunol. 2002;2:251–262. doi: 10.1038/nri778. [DOI] [PubMed] [Google Scholar]
- 49.Wherry EJ. T cell exhaustion. Nat Immunol. 2011;12:492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- 50.Singh KP, Bennett JA, Casado FL, Walrath JL, Welle SL, Gasiewicz TA. Loss of aryl hydrocarbon receptor promotes gene changes associated with premature hematopoietic stem cell exhaustion and development of a myeloproliferative disorder in aging mice. Stem Cells Dev. 2014;23:95–106. doi: 10.1089/scd.2013.0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boitano AE, Wang J, Romeo R, Bouchez LC, Parker AE, Sutton SE, Walker JR, Flaveny CA, Perdew GH, Denison MS, Schultz PG, Cooke MP. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science. 2010;329:1345–1348. doi: 10.1126/science.1191536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu P, Herrmann R, Bednar A, Saloupis P, Dwyer MA, Yang P, Qi X, Thomas RS, Jaffe GJ, Boulton ME, McDonnell DP, Malek G. Aryl hydrocarbon receptor deficiency causes dysregulated cellular matrix metabolism and age-related macular degeneration-like pathology. Proc Natl Acad Sci U S A. 2013;110:E4069–4078. doi: 10.1073/pnas.1307574110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jablonska O, Shi Z, Valdez KE, Ting AY, Petroff BK. Temporal and anatomical sensitivities to the aryl hydrocarbon receptor agonist 2,3,7,8-tetrachlorodibenzo-p-dioxin leading to premature acyclicity with age in rats. Int J Androl. 2010;33:405–412. doi: 10.1111/j.1365-2605.2009.01031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marlowe JL, Fan Y, Chang X, Peng L, Knudsen ES, Xia Y, Puga A. The aryl hydrocarbon receptor binds to E2F1 and inhibits E2F1-induced apoptosis. Mol Biol Cell. 2008;19:3263–3271. doi: 10.1091/mbc.E08-04-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sartor MA, Schnekenburger M, Marlowe JL, Reichard JF, Wang Y, Fan Y, Ma C, Karyala S, Halbleib D, Liu X, Medvedovic M, Puga A. Genomewide analysis of aryl hydrocarbon receptor binding targets reveals an extensive array of gene clusters that control morphogenetic and developmental programs. Environ Health Perspect. 2009;117:1139–1146. doi: 10.1289/ehp.0800485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schietinger A, Delrow JJ, Basom RS, Blattman JN, Greenberg PD. Rescued tolerant CD8 T cells are preprogrammed to reestablish the tolerant state. Science. 2012;335:723–727. doi: 10.1126/science.1214277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Camacho IA, Nagarkatti M, Nagarkatti PS. Evidence for induction of apoptosis in T cells from murine fetal thymus following perinatal exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) Toxicol Sci. 2004;78:96–106. doi: 10.1093/toxsci/kfh048. [DOI] [PubMed] [Google Scholar]
- 58.Weinstein DA, Gogal RM, Jr, Mustafa A, Prater MR, Holladay SD. Mid-gestation exposure of C57BL/6 mice to 2,3,7,8-tetrachlorodibenzo-p-dioxin causes postnatal morphologic changes in the spleen and liver. Toxicol Pathol. 2008;36:705–713. doi: 10.1177/0192623308320276. [DOI] [PubMed] [Google Scholar]
- 59.Bull CF, Mayrhofer G, O’Callaghan NJ, Au AY, Pickett HA, Low GK, Zeegers D, Hande MP, Fenech MF. Folate Deficiency Induces Dysfunctional Long and Short Telomeres; Both States Are Associated with Hypomethylation and DNA Damage in Human WIL2-NS Cells. Cancer Prev Res (Phila) 2014;7:128–138. doi: 10.1158/1940-6207.CAPR-13-0264. [DOI] [PubMed] [Google Scholar]
- 60.Li J, Harris RA, Cheung SW, Coarfa C, Jeong M, Goodell MA, White LD, Patel A, Kang SH, Shaw C, Chinault AC, Gambin T, Gambin A, Lupski JR, Milosavljevic A. Genomic hypomethylation in the human germline associates with selective structural mutability in the human genome. PLoS Genet. 2012;8:e1002692. doi: 10.1371/journal.pgen.1002692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Korzeniewski N, Wheeler S, Chatterjee P, Duensing A, Duensing S. A novel role of the aryl hydrocarbon receptor (AhR) in centrosome amplification - implications for chemoprevention. Mol Cancer. 2010;9:153. doi: 10.1186/1476-4598-9-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hochstenbach K, van Leeuwen DM, Gmuender H, Gottschalk RW, Stolevik SB, Nygaard UC, Lovik M, Granum B, Namork E, Meltzer HM, Kleinjans JC, van Delft JH, van Loveren H. Toxicogenomic profiles in relation to maternal immunotoxic exposure and immune functionality in newborns. Toxicol Sci. 2012;129:315–324. doi: 10.1093/toxsci/kfs214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gomez-Duran A, Ballestar E, Carvajal-Gonzalez JM, Marlowe JL, Puga A, Esteller M, Fernandez-Salguero PM. Recruitment of CREB1 and Histone Deacetylase 2 (HDAC2) to the Mouse Ltbp-1 Promoter Regulates its Constitutive Expression in a Dioxin Receptor-dependent Manner. Journal of Molecular Biology. 2008;380:1–16. doi: 10.1016/j.jmb.2008.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ovesen JL, Schnekenburger M, Puga A. Aryl Hydrocarbon Receptor Ligands of Widely Different Toxic Equivalency Factors Induce Similar HistoneMarksin Target Gene Chromatin. Toxicol Sci. 2011 doi: 10.1093/toxsci/kfr032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kurita H, Schnekenburger M, Ovesen JL, Xia Y, Puga A. The Ah Receptor Recruits IKKalpha to Its Target Binding Motifs to Phosphorylate Serine-10 in Histone H3 Required for Transcriptional Activation. Toxicol Sci. 2014 doi: 10.1093/toxsci/kfu027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schnekenburger M, Peng L, Puga A. HDAC1 bound to the Cyp1a1 promoter blocks histone acetylation associated with Ah receptor-mediated trans-activation. Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression. 2007;1769:569–578. doi: 10.1016/j.bbaexp.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Singh NP, Singh UP, Guan H, Nagarkatti P, Nagarkatti M. Prenatal exposure to TCDD triggers significant modulation of microRNA expression profile in the thymus that affects consequent gene expression. PLoS One. 2012;7:e45054. doi: 10.1371/journal.pone.0045054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Podshivalova K, Salomon DR. MicroRNA regulation of T-lymphocyte immunity: modulation of molecular networks responsible for T-cell activation, differentiation, and development. Crit Rev Immunol. 2013;33:435–476. doi: 10.1615/critrevimmunol.2013006858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Niedzwiecki M, Zhu H, Corson L, Grunig G, Factor PH, Chu S, Jiang H, Miller RL. Prenatal exposure to allergen, DNA methylation, and allergy in grandoffspring mice. Allergy. 2012;67:904–910. doi: 10.1111/j.1398-9995.2012.02841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schaub B, Liu J, Hoppler S, Schleich I, Huehn J, Olek S, Wieczorek G, Illi S, von Mutius E. Maternal farm exposure modulates neonatal immune mechanisms through regulatory T cells. J Allergy Clin Immunol. 2009;123:774–782 e775. doi: 10.1016/j.jaci.2009.01.056. [DOI] [PubMed] [Google Scholar]
- 71.Fedulov AV, Kobzik L. Allergy risk is mediated by dendritic cells with congenital epigenetic changes. Am J Respir Cell Mol Biol. 2011;44:285–292. doi: 10.1165/rcmb.2009-0400OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nguyen NT, Kimura A, Nakahama T, Chinen I, Masuda K, Nohara K, Fujii-Kuriyama Y, Kishimoto T. Aryl hydrocarbon receptor negatively regulates dendritic cell immunogenicity via a kynurenine-dependent mechanism. Proc Natl Acad Sci U S A. 2010;107:19961–19966. doi: 10.1073/pnas.1014465107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Simones T, Shepherd DM. Consequences of AhR activation in steady-state dendritic cells. Toxicol Sci. 2011;119:293–307. doi: 10.1093/toxsci/kfq354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.