Abstract
Alzheimer’s disease (AD) is a neurodegenerative disease characterized by severe cognitive deterioration. While causes of AD pathology are debated, a large body of evidence suggests that increased cleavage of Amyloid Precursor Protein (APP) producing the neurotoxic Amyloid-β (Aβ) peptide plays a fundamental role in AD pathogenesis. One of the detrimental behavioral symptoms commonly associated with AD is the fragmentation of sleep-activity cycles with increased nighttime activity and daytime naps in humans. Sleep-activity cycles, as well as physiological and cellular rhythms, which may be important for neuronal homeostasis, are generated by a molecular system known as the circadian clock. Links between AD and the circadian system are increasingly evident but not well understood. Here we examined whether genetic manipulations of APP-like (APPL) protein cleavage in Drosophila melanogaster affect rest-activity rhythms and core circadian clock function in this model organism. We show that the increased β-cleavage of endogenous APPL by the β-secretase (dBACE) severely disrupts circadian behavior and leads to reduced expression of clock protein PER in central clock neurons of aging flies. Our data suggest that behavioral rhythm disruption is not a product of APPL-derived Aβ production but rather may be caused by a mechanism common to both α and β-cleavage pathways. Specifically, we show that increased production of the endogenous Drosophila Amyloid Intracellular Domain (dAICD) caused disruption of circadian rest-activity rhythms, while flies overexpressing endogenous APPL maintained stronger circadian rhythms during aging. In summary, our study offers a novel entry point toward understanding the mechanism of circadian rhythm disruption in Alzheimer’s disease.
Keywords: Circadian rhythms, period gene, Amyloid precursor protein, Amyloid intracellular domain, BACE, Alzheimer’s disease, Drosophila
Introduction
Alzheimer’s disease (AD) is characterized by progressive neurodegeneration resulting in the loss of cognitive ability. The exact pathology of AD is not well understood and is widely debated. The amyloid cascade hypothesis suggests that abnormal production of neurotoxic Amyloid-beta (Aβ) in combination with tangles of phosphorylated Tau-microtubule associated protein lead to neuronal dysfunction, cell loss and thus cognitive decline (Hardy and Selkoe, 2002; Mandelkow and Mandelkow, 1998; Selkoe, 2000). The crucial enzyme in the production of Aβ is the rate-limiting Beta-site Amyloid Precursor Protein Cleaving Enzyme (BACE), which shows elevated expression in AD patients (Fukumoto et al., 2002; Vassar et al., 2009). Recently, knock-in of human BACE was shown to recapitulate many behavioral and physiological phenotypes of AD in a murine model (Plucinska et al., 2014). In addition to BACE, Amyloid Precursor Protein (APP) is cleaved by an α-secretase. Both cleavage pathways yield a secreted N-terminal fragment known as sAPP, sAPPβ for β-cleavage and sAPPα for α-cleavage. Subsequent ϒ-cleavage following β or α-cleavage results in either the Aβ or P3 fragment, respectively (De Strooper and Annaert, 2000; Selkoe, 2000; Turner et al., 2003). In addition to those two fragments, ϒ-cleavage also produces the APP intracellular domain (AICD), which has been shown to affect transcriptional regulation (Belyaev et al., 2010; Kimberly et al., 2001; Pardossi-Piquard and Checler, 2012; Turner et al., 2003). Both detrimental and positive effects of the AICD have been reported in cultured cells (Lu et al., 2000; Zhou et al., 2012); however, the physiological function of the AICD remains poorly understood.
One of the detrimental behavioral symptoms commonly associated with AD is the fragmentation of sleep-activity cycles with increased nighttime activity and daytime naps (Harper et al., 2005; Volicer et al., 2001; Wu and Swaab, 2007). Impaired rest-activity rhythms have also been reported in experimental AD model mice (Roh et al., 2012; Sterniczuk et al., 2010). Rest-activity cycles are generated by a molecular system known as the circadian clock. The circadian clock mechanism is based on negative feedback loops involving transcriptional activators and repressors, which are largely conserved from Drosophila to humans (Hardin and Panda, 2013). Circadian clocks generate daily rhythms in expression of many output genes leading to cellular, physiological, and behavioral rhythms. Loss of circadian rhythms is detrimental to health and adversely affects neuronal homeostasis in both, murine (Hastings and Goedert, 2013; Kondratova and Kondratov, 2012; Reddy and O'Neill, 2010) and Drosophila models (Krishnan et al., 2012). Therefore, it is important to understand the connections between the circadian system and AD pathology.
The links between AD and circadian rhythms were inferred from transgenic model animals expressing pathogenic versions of human genes (Rezaval et al., 2008; Roh et al., 2012; Sterniczuk et al., 2010). Recent reports showed that pathogenic human amyloid peptides disrupt behavioral rest-activity rhythms in Drosophila but are not sufficient to disrupt molecular oscillations of PERIOD (PER) in central pacemaker neurons (Chen et al., 2014; Long et al., 2014). Here, we manipulated AD-related genes in the fruit fly, Drosophila melanogaster in order to understand endogenous pathways that may interfere with the circadian system. APP has a functional ortholog in Drosophila called Amyloid Precursor Protein-Like (APPL) (Luo et al., 1992; Luo et al., 1990; Rosen et al., 1989). In addition, orthologs of BACE, the α-secretase, and the ϒ-secretase complexes have been identified in Drosophila. They are known as dBACE, Kuzbanian (KUZ) and Presenillin (PSN), respectively, and were shown to process APPL resulting in peptide fragments comparable to APP fragments (Bolkan et al., 2012; Carmine-Simmen et al., 2009; Greeve et al., 2004). This conservation made Drosophila a suitable model for our study. Because BACE is the rate-limiting enzyme in the production of Aβ, we first investigated if over-expression of dBACE is involved in the rest-activity disturbances characteristic of AD.
We found that increasing dBACE expression disrupted endogenous rest-activity rhythms. This effect was most severe in aged flies suggesting an age-dependent mechanism. Furthermore, dBACE expression resulted in the dampened oscillation of the core clock protein PER in central pacemaker neurons, which are master regulators of rest-activity rhythms. Surprisingly, over-expression of Kuzbanian (KUZ) also disrupted rest-activity rhythms suggesting a mechanism independent of endogenous Drosophila Aβ peptide (dAβ) production, which like its vertebrate ortholog has been shown to be neurotoxic (Carmine-Simmen et al., 2009). This suggested that rhythm deficits were due to a cleavage product of APPL conserved in both α and β cleavage pathways. Therefore, we expressed the APPL intracellular domain (dAICD) and this caused severe disruption of behavioral rest-activity rhythms. In contrast, expression of full-length APPL in central clock neurons protected against an age-dependent decline in rest-activity rhythms. Taken together, these findings suggest that APPL and specifically the dAICD can affect circadian behavior by interfering with molecular clock oscillations.
Materials and methods
Fly stocks
D. melanogaster were reared on diet containing 1% agar, 6.25% cornmeal, 6.25% molasses, and 3.5% Red Star yeast at 25 °C. Flies were entrained to 12-hour light:dark (LD, 12:12) cycles (with an average light intensity of ~1500 lx). All experiments were performed on mated male flies of different ages, as specified in the results. We used the binary UAS-GAL4 system to express specific genes using transgenic flies carrying the following constructs: UAS-dBACE, UAS-APPL, UAS-dAICD (Carmine-Simmen et al., 2009), and UAS-KUZ (Bloomington Stock Center). The drivers used to express these responders in a tissue specific manner were: tim-GAL4 active in all clock cells (Kaneko and Hall, 2000), elav-GAL4 pan-neuronal driver from the Bloomington Stock Center, and the pdf-GAL4 active in clock pacemaker neurons (Renn et al., 1999). Full genotypes of flies used are given in supplemental data Table S4. To express the dAICD in salivary glands, we used P{GawB}AB1 provided by the Bloomington stock center. Control flies were generated by crossing tissue specific drivers to Green Fluorescent Protein (UAS-GFP) for all behavioral experiments. For Immunocytochemistry (ICC), we used UAS-dBACE flies crossed to white (wild-type) flies as a control to avoid GFP fluorescence.
Locomotor activity analysis
Flies were entrained in LD 12:12 at 25 °C. Locomotor activity for 5d, 35d, and 50d old flies was recorded for 3d in LD 12:12 followed by 7d in constant darkness (DD) using the Trikinetics locomotor activity monitor (Waltham, MA) following a protocol described previously (Rakshit et al., 2012). Rhythmic locomotor activity was determined by fast Fourier transform (FFT) values and periodograms created using ClockLab software (Actimetrics; Wilmette, IL). Flies with FFT values below 0.04 and a periodogram peak that did not break the line of significance were considered arrhythmic. Flies with FFT values between 0.04 and 0.08 were considered weakly rhythmic, while FFT values of 0.08 and above were designated as strongly rhythmic. Period length calculations were based on rhythmic flies only. For statistical analysis, an unpaired t-test with Welch’s correction was used to compare experimental genotypes to age-matched controls (GraphPad Prism v5.0; GraphPad Software Inc. San Diego, CA).
Rapid iterative negative geotaxis (RING) assay
The RING assay was used to test the climbing ability in age 5d, 15d, 35d, and 50d flies as previously described (Gargano et al., 2005). Briefly, 25 flies of a given genotype were placed in empty vials and tapped in rapid succession to the base of the vial. This action initiated a rapid negative geotaxis response. Still frame images were captured four seconds following the final tap. These images were converted to gray scale and analyzed using NIH Image J software to calculate the average climbing distance in each vial following a negative geotaxis response. Statistical significance of average distance climbed based on at least 3 vials per age and genotype was determined by two-way ANOVA with Bonferroni’s post-test using GraphPad Prism5 software, as previously described (Rakshit et al., 2012).
Longevity assay
Flies were held in 240ml round bottom polypropylene ventilated bottles (Genesee Scientific, San Diego, CA). Each bottle contained 50 mated male flies, 4 bottles for each genotype. These bottles were inverted on 35 mm Falcon Prinaria tissue culture dishes (Becton, Dickinson and Company) each filled with 15ml of diet. Fresh diet was provided and mortality recorded 3 times a week. Lifespan curves were plotted using Kaplan-Meier survival curves and statistical analysis of median lifespan was done by the log-rank test (Mantel-Cox) using GraphPad Prism5 software, as described (Rakshit et al., 2012).
Immunocytochemistry
Pigment Dispersing Factor (PDF) and PERIOD (PER) immunostaining was used to examine the pacemaker neurons of flies held in constant darkness for 72 hours. Identification of specific central clock cells expressing PER was accomplished by co-staining for the PDF neuropeptide. Samples were collected at circadian time (Ct) 22 and Ct10 which correspond to the relative peak (Ct22) and trough (Ct10) of PER protein levels in wild-type flies in DD. Flies were fixed in freshly prepared 4% paraformaldehyde in Phosphate Buffer Saline (PBS) with 0.1% Triton-X100 (PBS-T 0.1%) for 30 minutes. Fly brains were then dissected in PBS-T 0.1% and post-fixed in 4% paraformaldehyde in PBS-T 0.1%, then rinsed with PBS with 0.5% Triton-X100 (PBS-T 0.5%). Brains were blocked overnight in 5% normal goat serum (NGS) in PBS-T 0.5%, and incubated for 48 hours in primary 1:500 mouse nb33 monoclonal anti-PDF (Developmental Studies Hybridoma Bank) and 1:10,000 rabbit anti-PER (kindly provided by R. Stanewsky). Following incubation, brains were rinsed 6 times in PBS-T 0.5%, and incubated overnight in secondary Alexa Fluor 555 anti-mouse and 488 anti-rabbit, both 1:500 (Life Technologies). Following incubation, brains were rinsed 6 times in PBS-T 0.5% and mounted on microscope slides in Vectashield™ mounting media with DAPI (Vector Laboratories, Burlingame, California). Images were acquired via an Olympus Fluoview 5 confocal microscope on an OlympusBX51 platform housed at Oregon Health and Science University. Images were acquired using the 40× oil immersion objective.
Relative immunofluorescence intensity was calculated using Fiji (available at Fiji.sc/Fiji) with a 30-pixel circle region of interest (ROI) corresponding to the size of PER-positive nuclei. The mean intensity of these 30 pixels was quantified for individual cells in each neuronal group: small lateral ventral (sLNv), large lateral ventral (lLNv), and lateral dorsal (LNd) neurons. Nuclear PER signal from individual cells was then averaged for each neuronal group to yield one value for each group of neurons in each hemisphere (n=1); 5–22 brain hemispheres were quantified for a given neuronal group. To obtain the mean staining intensity for each neuronal group in each hemisphere we utilized a formula described previously I= 100 × (S-B)/B where mean relative intensity per cell group (I) is equal to, 100 multiplied by mean relative signal per cell group (S) subtracted from average background signal (B) divided by average background signal (B) (Yoshii et al., 2009). Statistical comparison of average PER signal intensity was calculated by two-way ANOVA with Bonferroni’s post-test using GraphPad Prism5 software. For the immunocytochemistry on salivary glands, glands were dissected from 3rd instar larvae, fixed in 4% paraformaldehyde, blocked in 5% NGS overnight and incubated in anti-C-terminal APPL (kindly provided by P. Copenhaver) at 1:4000 for 4 hours. Staining was detected with FITC conjugated anti-chicken 1:1000 (Jackson ImmunoResearch). Nuclear stain ToPro3 was obtained from Molecular Probes and diluted 1 µM in PBS. Staining was done for 30min.
Results
Over-expression of dBACE in clock cells accelerates aging phenotypes and disrupts rest-activity rhythms
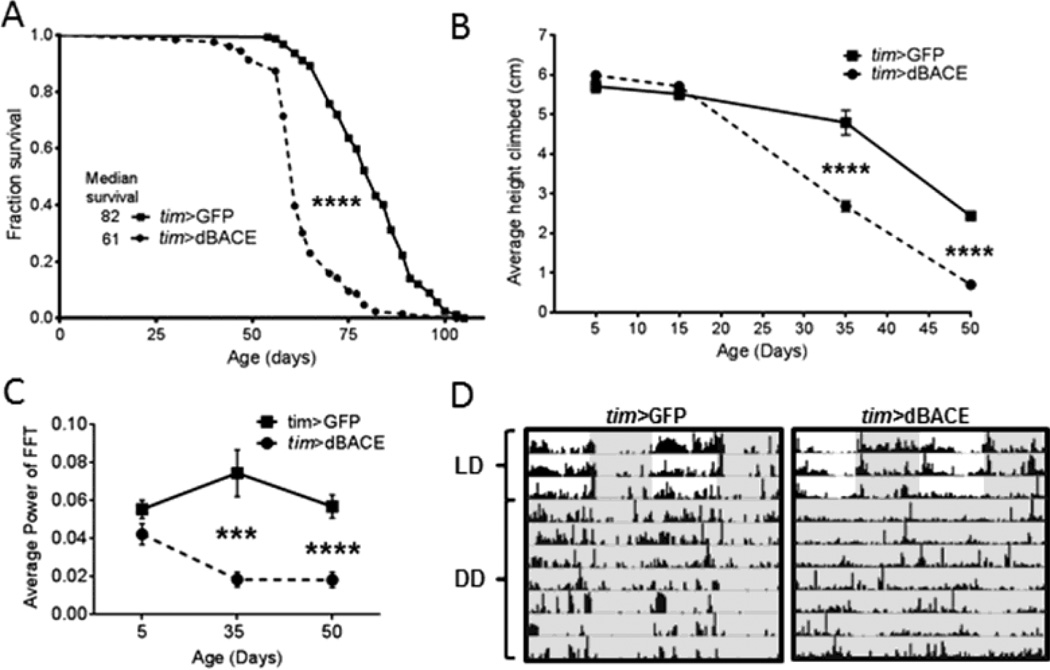
To examine effects of elevated dBACE in flies, we expressed dBACE in all clock cells using the timeless (tim-GAL4) driver. We first tested whether this affected lifespan, and found a significant reduction (p<0.0001) in tim>dBACE flies with a median lifespan of 61d compared to 82d for tim>GFP controls (Fig. 1A). Aging also was accelerated in tim>dBACE flies as they had significantly reduced climbing ability (p<0.0001) at 35d and 50d compared to age-matched controls (Fig. 1B). We next investigated the status of behavioral rest-activity rhythms in tim>dBACE flies. In LD, age 5d tim>BACE flies had the characteristic bimodal activity pattern with morning and evening peaks preceded by anticipatory activity, but these peaks were attenuated in age 35d and 50d flies relative to controls (Fig. S1 A). To test endogenous rest-activity rhythms, we next monitored flies in DD and found that free-running rhythms were disrupted such that flies had short bouts of activity and rest around the clock. The average rhythm strength (calculated by FFT) was significantly reduced at age 35d (p<0.001), and at age 50d (p<0.0001) compared to age-matched controls (Fig. 1C and 1D). At age 50d, only 11% of tim>dBACE flies remained rhythmic compared to 64% of controls (Table 1).
Figure 1.
Overexpression of dBACE in all clock cells with tim-GAL4 accelerates aging phenotypes. (A) Median lifespans show a significant reduction in tim>dBACE flies (p<0.0001). Statistical analysis by log-rank test. (B) Climbing ability is significantly reduced in age 35d and 50d tim>dBACE flies (p<0.0001). Statistical analysis by two-way ANOVA. (C) The strength of rest-activity rhythms is significantly reduced in tim>dBACE flies compared to controls, at age 35d (p<0.001) and 50d (p<0.0001). Statistical analysis by Welch’s unpaired t-test. (D) Representative actograms demonstrate deterioration of rest-activity rhythms in age 50d tim>dBACE flies compared to age-matched tim>GFP controls. Darkness indicated by gray shading.
Table 1.
Effects of dBACE over-expression on locomotor activity profiles#
| Age | Genotype | n | % Rhythmicity (Strong + Weak) |
Rhythm Strength (Average FFT±SEM) |
Period (DD) |
|---|---|---|---|---|---|
| Day 5 | tim-GAL4 > UAS-GFP | 42 | 64% (24% + 40%) | 0.056 ± 0.005 | 23.69 |
| tim-GAL4 > UAS-dBACE | 39 | 44% (15% + 29%) | 0.042 ± 0.005 | 23.59 | |
| elav-GAL4 > UAS-GFP | 41 | 88% (42% + 46%) | 0.077 ± 0.006 | 23.42 | |
| elav-GAL4 > UAS-dBACE | 59 | 76% (25% + 51%) | 0.067 ± 0.005 | 23.26 | |
| pdf-GAL4 > UAS-GFP | 57 | 93% (67% + 26%) | 0.114 ± 0.008 | 23.67 | |
| pdf-GAL4 > UAS-dBACE | 51 | 84% (65% + 19%) | 0.101 ± 0.010 | 23.33 | |
| Day 35 | tim-GAL4 > UAS-GFP | 45 | 78% (27% + 51%) | 0.075 ± 0.012 | 23.89 |
| tim-GAL4 > UAS-dBACE | 36 | 8% ( 3% + 5% ) | 0.019 ± 0.004*** | 23.46 | |
| elav-GAL4 > UAS-GFP | 42 | 86% (43% + 43%) | 0.074 ± 0.005 | 23.35 | |
| elav-GAL4 > UAS-dBACE | 44 | 39% ( 7% + 32%) | 0.041 ± 0.006**** | 23.38 | |
| pdf-GAL4 > UAS-GFP | 55 | 64% (20% + 44%) | 0.054 ± 0.005 | 23.61 | |
| pdf-GAL4 > UAS-dBACE | 57 | 48% (14% + 34%) | 0.048 ± 0.004 | 23.53 | |
| Day 50 | tim-GAL4 > UAS-GFP | 25 | 64% (16% + 48%) | 0.056 ± 0.007 | 23.87 |
| tim-GAL4 > UAS-dBACE | 19 | 11% ( 0% + 11%) | 0.018 ± 0.004**** | 23.61 | |
| elav-GAL4 > UAS-GFP | 29 | 86% (52% + 34%) | 0.081 ± 0.006 | 23.41 | |
| elav-GAL4 > UAS-dBACE | 33 | 18% ( 3% + 15%) | 0.030 ± 0.004**** | 23.33 | |
| pdf-GAL4 > UAS-GFP | 30 | 90% (37% + 53%) | 0.076 ± 0.007 | 24.05 | |
| pdf-GAL4 > UAS-dBACE | 33 | 27% ( 3% + 24%) | 0.036 ± 0.004*** | 23.83 |
Percent rhythmicity was determined by FFT values. FFT values from 0.04 to 0.08 were defined as weakly rhythmic and FFT values>0.08 defined as strongly rhythmic. Additionally to be considered rhythmic flies had to break the line of significance on their respective periodograms. Average period was calculated using only rhythmic flies. Significance was determined by Welch’s unpaired t-test with comparison to age–matched GFP-expressing controls.
denote p<0.001,
denotes p<0.0001.
Neuronal over-expression of dBACE disrupts behavioral rest-activity rhythms
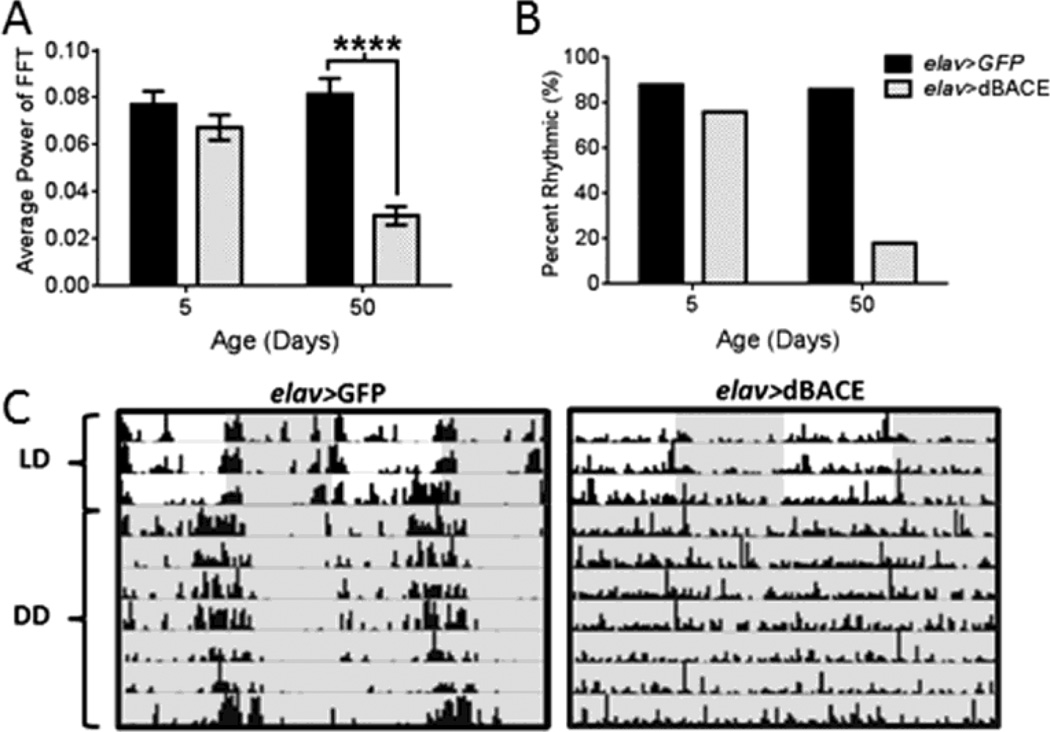
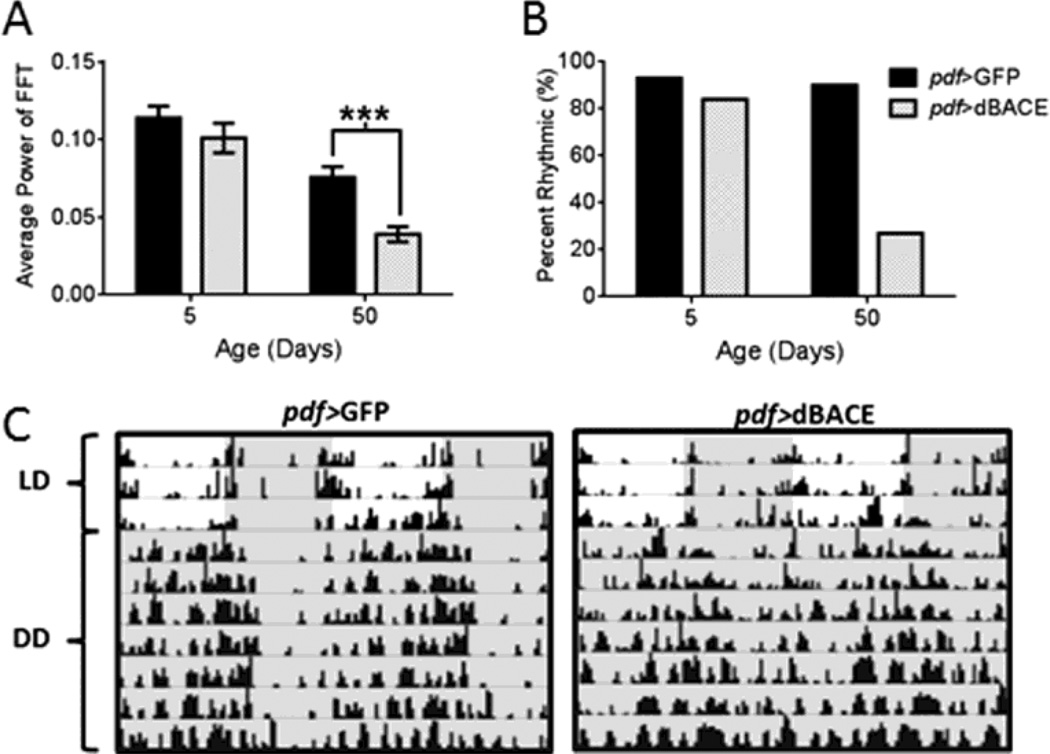
Circadian clocks are found throughout the nervous system and peripheral organs, and the tim-GAL4 driver is active in all these clock cells. To determine whether rhythm disruption in tim>dBACE flies was due to dBACE elevation in the nervous system, we expressed dBACE with the pan-neuronal elav-GAL4 driver. This also disrupted behavioral rest-activity rhythms in aging flies. In LD, morning and evening activity peaks were attenuated in age 35d and 50d elav>dBACE flies relative to controls (Fig. S1 B). FFT analysis of locomotor activity in DD showed significant weakening of endogenous rest-activity rhythms at age 35d and 50d (p<0.0001) (Table 1, Fig. 2A). At age 50d only 18% of elav>dBACE flies remained rhythmic, compared to controls that maintained 86% rhythmicity (Table 1 and Fig. 2B). The arrhythmic flies were active around the clock without consolidated rest periods (Fig. 2C). To determine if over-expression of dBACE in central pacemaker neurons is sufficient to induce this phenotype, we over-expressed dBACE in the PDF-positive central pacemaker neurons (pdf-GAL4). Resulting pdf>dBACE flies showed distinct but attenuated activity peaks in LD (Fig. S1 C), and severe rhythm disruption in DD. Average rhythm power was significantly reduced in age 50d pdf>dBACE flies (p<0.001) when compared to pdf>GFP control flies (Table 1 and Fig. 3A). Only 27% of pdf>dBACE flies remained rhythmic at age 50d compared to 90% in controls (Table 1 and Fig. 3B). As shown in Figure 3C, pdf>dBACE flies were fairly active in old age with activity bouts distributed around the clock without consolidated rest, in contrast to pdf>GFP control flies, which maintained consolidated rhythms. Thus, dBACE over-expression in central pacemaker neurons is sufficient to induce circadian behavioral deficits.
Figure 2.
Pan-neuronal over-expression of dBACE disrupts rhythmic locomotor activity in aging flies. (A) Average FFT values are significantly reduced in age 50d elav>dBACE flies in DD compared to elav>GFP controls (p<0.0001). Statistical analysis by Welch’s unpaired t-test. (B) Most elav>dBACE flies become arrhythmic at age 50d. (C) Representative actograms illustrate deterioration of rest-activity rhythms in age 50d elav>dBACE flies compared to age-matched elav>GFP controls. Darkness indicated by gray shading.
Figure 3.
Over-expression of dBACE in central clock neurons disrupts rhythmic locomotor activity. (A) Average FFT values are significantly reduced in age 50d pdf>dBACE flies compared to pdf>GFP controls (p<0.001). Statistical analysis by Welch’s unpaired t-test (B) Most pdf>dBACE flies become arrhythmic at age 50d. (C) Representative actograms illustrate the deterioration of rest-activity rhythms in age 50d pdf>dBACE flies compared to age-matched pdf>GFP controls. Darkness indicated by gray shading.
Over-expression of dBACE dampens the cycling of PER in central pacemaker neurons
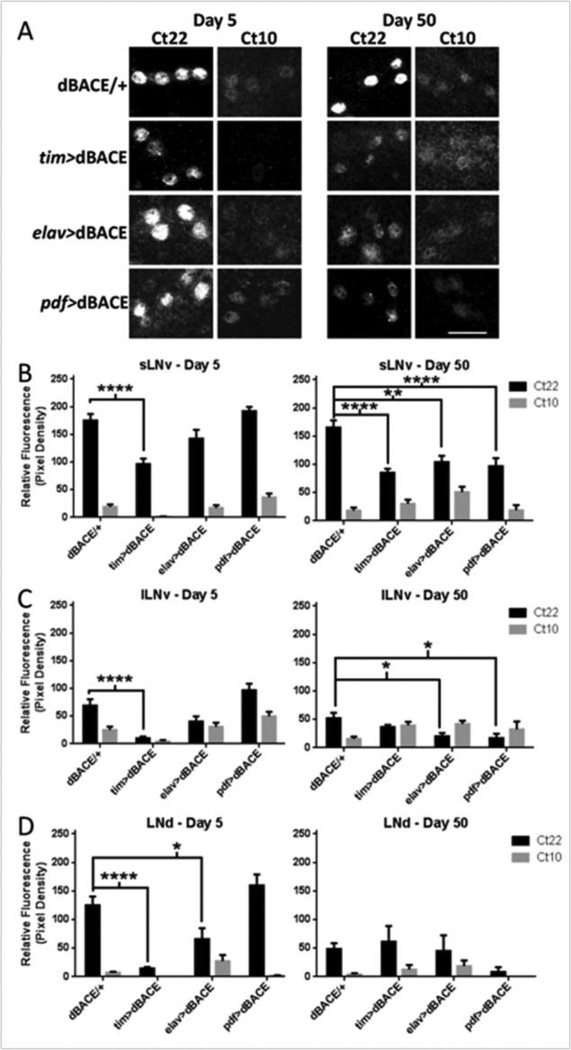
Given that increased β-cleavage is sufficient to disrupt behavioral rhythmicity, we next asked whether this phenotype is a product of alterations in central clock neurons. Expression of the core clock gene period (per) in these neurons is essential to generate oscillations in gene expression that result in behavioral rhythmicity (Hall, 2005; Helfrich-Forster, 2005). To determine if the disruption of behavioral rest-activity rhythms was associated with disruption of per in these neurons, we examined PER protein levels using an anti-PER antibody in three distinct groups of lateral pacemaker neurons: the small ventral Lateral Neurons (sLNv), the large ventral Lateral Neurons (lLNv), and the dorsal Lateral Neurons (LNd) (Helfrich-Forster et al., 2007). The sLNv and lLNV are easily identified by co-staining with PDF, a neuropeptide present in high concentrations in the cytoplasm of each cell, with the exception of the 5th sLNv and LNd which are PDF-negative. These cells can only be identified by the expression of PER. PER oscillations were examined following 3 consecutive days in DD because dBACE over-expression most severely disrupts behavioral rest-activity rhythms in DD. Because the levels of PER protein peaks around Ct22 and are lowest at Ct10 in wild type flies, we examined these two time points. As a control, we used transgenic UAS-dBACE flies crossed to white flies in order to avoid background fluorescent signal from a GFP-expressing control.
In analyzing the pattern of PDF-expressing cells, we did not observe changes in the morphology of dorsal or optic lobe arborizations, or the number of these cells, suggesting that they are intact (data not shown). Although the LNd do not express PDF, these cells were easily identifiable by PER expression and their proximity to LNv and they also appeared intact. Despite this, expression of dBACE via tim-GAL4 significantly reduced PER protein expression in the central pacemaker neurons. In age 5d tim>dBACE flies, PER cycling was significantly dampened (p<0.0001) in sLNv, lLNv, and LNd (Fig. 4A–D). These data are in line with behavioral rest-activity disruption in age 5d in tim>dBACE flies, which was observable as a trend but did not reach statistical significance when compared to control flies (Table 1.)This reduction in PER levels persisted in age 50d flies in the sLNv (p<0.0001) but we could not detect a significant difference in lLNv and LNd due to the dampened PER-expression in controls (Fig. 4A–D). elav>dBACE flies also revealed a modest but significant (p<0.05) dampening in PER expression in the LNd at age 5d (Fig. 4D) but not in the other examined cell types. At age 50d, elav>dBACE flies showed significant attenuation of PER levels in sLNv (p<0.01) and lLNv (p<0.05) (Fig. 4A–C). PER oscillations in pdf>dBACE flies were not significantly altered at age 5d (Fig. 4A–D). However, by age 50d, PER oscillations are significantly attenuated in both sLNv (p<0.0001) and lLNv (p<0.05). As expected, PER levels were not affected in LNd of pdf>dBACE flies (Fig. 4A–D) because pdf-GAL4 does not result in dBACE over-expression in these cells (Fig. 4D) (Helfrich-Forster et al., 2007).
Figure 4.
Over-expression of dBACE affects the levels of PER in the small ventral Lateral Neurons (sLNv), large ventral Lateral Neurons (lLNv), and dorsal Lateral neurons (LNd). Measurements of PER expression were taken at Circadian time (Ct) 22 and Ct 10. (A) Representative pictures showing PER levels at age 5d and 50d in sLNv relative to dBACE/+ controls. Scale bar = 25 µm. (B) Quantification of PER expression in sLNv. At age 5d tim>dBACE flies show significantly dampened PER oscillation (p<0.0001). By age 50d all genotypes show significant dampening in PER oscillation, tim>dBACE and pdf>dBACE (p<0.0001), and elav>dBACE (p<0.01) compared to age-matched controls. (C) Quantification of PER expression in lLNv. At age 5d tim>dBACE flies show significantly dampened PER oscillation (p<0.0001) compared to age-matched controls. By age 50d significant dampening in PER oscillation occurs in elav>dBACE and pdf>dBACE (p<0.05) compared to age-matched controls. (D) Quantification of PER expression in the LNd. At age 5d tim>dBACE and elav>dBACE flies show significantly dampened PER oscillation, p<0.0001 and p<0.05 respectively. At age 50d no significant difference in PER oscillation is observed compared to age-matched controls.
Rest-activity rhythms are disrupted by KUZ over-expression
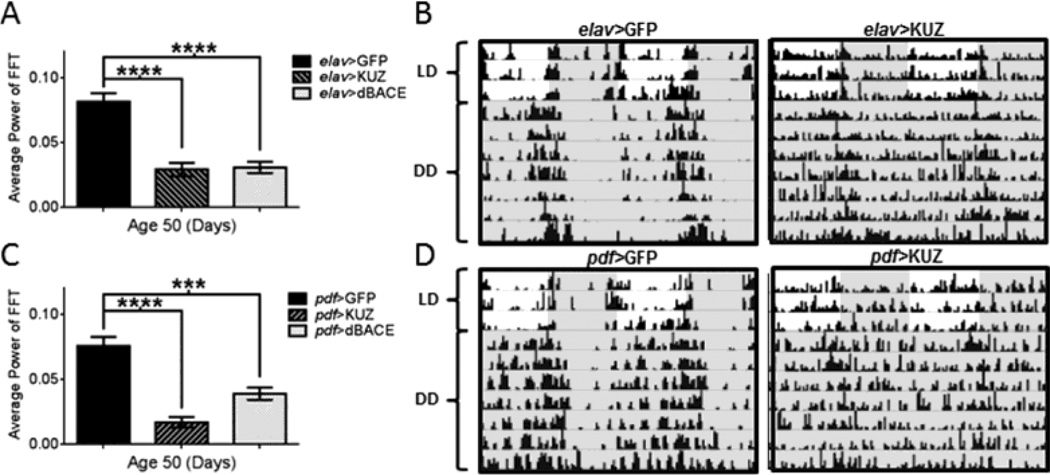
To determine whether effects of dBACE expression are related to a cleavage product specific to the β-cleavage pathway, we monitored rest-activity rhythms in flies with over-expressed KUZ. KUZ is an ortholog of human ADAM10 and has been shown to have α-secretase activity against APPL in flies (Carmine-Simmen et al., 2009). To our surprise, expression of KUZ had similar effects as expression of dBACE. At age 50d, only 25% of elav>KUZ flies and only 8% of pdf>KUZ flies remained rhythmic in DD compared to 86% and 90% in respective age-matched controls (Table S1, Table S2). Overall, over-expression of KUZ led to continuous activity without consolidated rest, in contrast to GFP-expressing control flies, which maintained consolidated rhythms (Fig. 5B and 5D). The phenotype of KUZ over-expression was comparable to dBACE expression at age 50d; in both pan-neuronal (elav-GAL4) and central clock (pdf-GAL4) neurons, KUZ expression resulted in a significant disruption of rest-activity rhythms (p<0.0001) (Fig. 5A and 5C). In fact, the phenotype was even more severe with KUZ expression than dBACE expression. Already at age 5d elav>KUZ were only 54% rhythmic while elav>dBACE were 76% rhythmic and not different from controls at this age (Table S1). Similarly, over-expression of KUZ in the central clock neurons via pdf-GAL4 resulted in severely dampened rest-activity rhythms already at age 5d. At age 5d pdf>KUZ flies were only 25% rhythmic compared to pdf>dBACE, which at 84% were not different from pdf>GFP controls (Table S2). Together, these data suggested that the rhythm disruption observed is likely caused by a cleavage product that can be generated by both α and β-cleavage.
Figure 5.
Rhythmic locomotor activity is disrupted by over-expression of KUZ. (A) Rest-activity rhythms in age 50d elav>KUZ flies are significantly reduced (similarly to elav>dBACE flies), compared to elav>GFP (p<0.0001). Statistical analysis by Welch’s unpaired t-test. (B) Representative actograms illustrate rhythmic locomotor activity disruption by age 50d in elav>KUZ. (C) Expression of KUZ in central clock neurons also significantly reduces power of activity rhythms in pdf>KUZ flies at age 50d (p<0.0001), similarly to expression of pdf>dBACE (p<0.001), compared to age-matched pdf>GFP controls. Statistical analysis by Welch’s unpaired t-test. (D) Representative actograms illustrate rhythmic locomotor activity disruption by age 50d in pdf>KUZ flies compared to age-matched pdf>GFP controls. Darkness indicated by gray shading. Percent rhythmic flies are indicated in Table S1 and S2.
Expression of AICD disrupts rest-activity rhythms
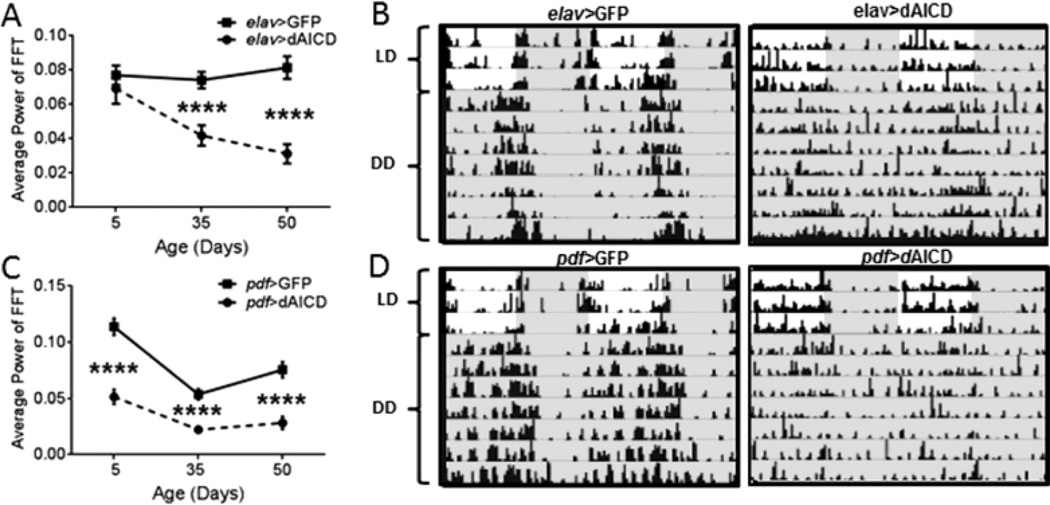
Because over-expression of dBACE and KUZ similarly affected rest-activity rhythms, we hypothesized that an excess of APPL-intracellular domain (dAICD) could interfere with behavioral rhythmicity. Like the APP cleavage pathway in mammals, in Drosophila cleavage of APPL by KUZ or dBACE results in a C-terminal fragment (CTF) that is subsequently cleaved by the ϒ-secretase complex resulting in the production of dAICD (Carmine-Simmen et al., 2009). Indeed, pan-neuronal expression of dAICD resulted in an age-dependent decline in rhythmic locomotor activity (Table S1 and Fig. 6A). Beginning at age 35d, elav>dAICD flies had significantly reduced rhythmic locomotor activity (p<0.0001) and the phenotype was more severe at age 50d (p<0.0001) (Table S1, Fig. 6A–B). This phenotype nearly mimics the dampening of rhythmic locomotor activity seen in both elav>dBACE flies and elav>KUZ flies at age 50d (Table S1). Expression of dAICD in central clock neurons also resulted in dampened rest-activity rhythms beginning at age 5d (p<0.0001), continuing through 35d (p<0.0001) and 50d (p<0.0001) (Fig. 6C–D). This phenotype is more severe than that observed in age 5d pdf>dBACE. At age 50d expression of dAICD, KUZ and dBACE in the central clock neurons yields a similar phenotype with all groups showing a significant dampening of rest-activity rhythms compared to controls (p<0.0001 and p<0.001) (Table S2). This strongly suggests that dAICD is disrupting rhythmicity and that the effect of dBACE and KUZ is mediated by increased production of dAICD.
Figure 6.
Rhythmic locomotor activity is disrupted by the expression of dAICD. (A) elav>dAICD flies show a significant reduction in rhythm power at age 35d and 50d (p<0.0001) compared to elav>GFP controls. Statistical analysis by Welch’s unpaired t-test. (B) Representative actograms illustrate the disruption of rhythmic locomotor activity in elav>dAICD flies at age 50d compared to elav>GFP controls. (C) pdf>dAICD flies show significant reduction in rhythmicity at all ages examined (p<0.0001), compared to pdf>GFP controls. Statistical analysis by Welch’s unpaired t-test. (D) Representative actograms illustrate the disruption of rhythmic locomotor activity in pdf>dAICD flies at age 50d compared to pdf>GFP controls. Darkness indicated by gray shading. Percent rhythmic flies are indicated in Table S1 and S2.
dAICD is capable of entering the nucleus, but is not toxic to central pacemaker neurons
In mammals, the AICD is transported to the nucleus where it exerts transcriptional activity (Ghosal et al., 2010; Goodger et al., 2009; Maulik et al., 2012; Robinson et al., 2014). To determine whether dAICD also undergoes nuclear translocation, we expressed it in salivary glands, which due to their large cells and nuclei provide better resolution than neuronal cells. Indeed, we could detect dAICD in the nuclei of these cells (Fig. S2 A) which were co-stained with the nuclear marker To-Pro3 (Fig. S2 B). This suggests that the dAICD could be involved in the transcriptional regulation of the circadian clock.
Because the AICD was reported to have toxic effects on cells, we tested the status of central pacemaker neurons in age 50d pdf>dAICD flies using the anti-PDF antibody. Both sLNv and lLNv neurons were present in these flies (Fig. S3), suggesting that the PDF-neurons remain viable in the presence of elevated dAICD.
Rest-activity rhythms are strengthened by APPL expression in central pacemaker neurons
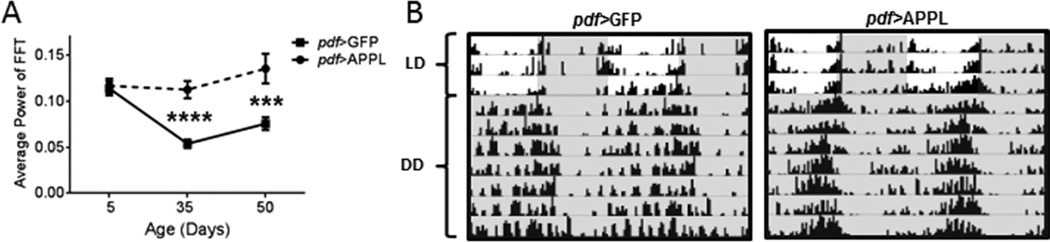
Increased α or β cleavage by KUZ or dBACE not only results in an increase of dAICD but also a decrease of full-length APPL, which could affect behavioral rhythms. We therefore tested whether over-expression of full-length APPL had an effect on rest-activity rhythms. Pan-neuronal expression of full-length APPL resulted in rest-activity rhythms similar to elav>GFP controls at age 5d and 35d but became slightly but significantly deteriorated by age 50d, p<0.05 (Table S3). In contrast, we detected robust rhythm strengthening when APPL was expressed only in central pacemaker neurons. Induction of full-length APPL in PDF-positive central pacemaker neurons resulted in strengthened rest-activity rhythms compared to pdf>GFP controls beginning at age 35d (p<0.0001) (Table S3 and Fig. 7A). This effect continued to age 50d with average FFT values significantly higher than controls (p<0.001) and more highly consolidated rest-activity rhythms in pdf>APPL flies (Table S3 and Fig. 7A and 7B). These data suggest that the presence of full-length APPL may be beneficial to maintain strong behavioral rhythmicity specifically when expressed in PDF-positive central pacemaker neurons.
Figure 7.
Over-expression of full-length APPL in central clock neurons prevents age-related decline in rhythmic locomotor activity. (A) The power of locomotor activity rhythms is significantly improved in pdf>APPL flies at age 35d (p<0.001) and age 50d (p<0.0001) compared to age-matched pdf>GFP controls. Statistical analysis by Welch’s unpaired t-test. (B) Representative actograms illustrate the strengthening of activity rhythms in pdf>APPL flies at age 50d. Darkness indicated by gray shading. Percentage of rhythmic flies is shown in Table S3.
Discussion
Loss of rest-activity rhythms is a well-established early symptom of AD in humans. Because disruption of circadian rhythms is detrimental to neuronal homeostasis (Kondratova and Kondratov, 2012; Krishnan et al., 2012), it is important to understand relationships between AD and circadian rhythms at the cellular and molecular levels. To address this question, we examined how manipulations of the fly ortholog of APP and its cleaving enzymes affect endogenous rest-activity rhythms and clock mechanism in Drosophila. We report here that the over-expression of dBACE disrupts behavioral rest-activity rhythms, and that this effect was most severe in aged flies suggesting an age-dependent mechanism. Furthermore, dBACE expression resulted in the dampened oscillation of core clock protein PER in central pacemaker neurons, which are master regulators of rest activity rhythms. Significantly reduced PER levels were observed in the sLNv and lLNv neurons of age 50d flies expressing dBACE in all clock cells (including glia), all neurons, or only in PDF-positive sLNv and lLNv neurons. These data suggest that manipulation of APP-cleavage by dBACE over-expression directly affects the oscillation of PER protein in central pacemaker neurons in a cell-autonomous manner. Since a functional clock mechanism in sLNv is necessary and sufficient to maintain free running activity rhythms, reduced oscillations of PER in these neurons could be responsible for the loss of activity rhythms in age 50d flies. Importantly, the decline in PER levels occurred only in flies with manipulated dBACE, not in old control flies. This is in agreement with recent findings that aging does not dampen PER oscillations in pacemaker neurons of wild type flies (Luo et al., 2012) (but see also (Umezaki et al., 2012)), while it reduces clock oscillations in peripheral clocks (Luo et al., 2012; Rakshit et al., 2012).
While we report here that the loss of behavioral rhythms after manipulation of dBACE is associated with reduced expression of clock genes in the central pacemaker, recent work showed that expression of human Aβ peptides leads to disruption of rest activity rhythms without interfering with PER oscillations in the central pacemaker (Chen et al., 2014; Long et al., 2014). Even strongly neurotoxic Aβ peptides, such as Aβ42 arctic, did not cause rhythm disruption when expressed in central pacemaker neurons; rather, pan-neuronal expression was required (Chen et al., 2014; Long et al., 2014). The fact that even the most neurotoxic Aβ peptides are not capable of dampening PER oscillation in pacemaker neurons suggests that Aβ production does not affect clock oscillations and that it is not Aβ production that causes the phenotype we observe upon over-expression of dBACE. We confirmed this by expression of KUZ, whose activity does not increase dAβ production; however, it also led to disruption of rest-activity rhythms. Similar rhythm disruption by dBACE and KUZ suggested that an excess cleavage product of both pathways might be responsible for the disruption. Like in the mammalian APP cleavage pathway, in Drosophila cleavage of APPL by KUZ or dBACE results in a C-terminal fragment (CTF) that is subsequently cleaved by the ϒ-secretase resulting in the production of dAICD (Carmine-Simmen et al., 2009). Indeed, we show that expression of dAICD resulted in an age-dependent decline in rhythmic locomotor activity. As with dBACE and KUZ expression, dAICD expression caused weakening or complete loss of behavioral rhythms while age-matched control flies remained highly rhythmic. In this context, it is worth noting that α-secretase activators are considered for clinical trials to reduce Aβ production in AD patients (Epis et al., 2012; Panza et al., 2009). However, according to our results this could lead to disruptions of circadian rhythms and sleep patterns thus negatively impacting the live of patients and their caretakers.
Our data suggest that increased dAICD may be the proximal cause of decay in rest-activity rhythms. The role of AICD in AD is increasingly evident but poorly understood (Vassar et al., 2009). AICD is able to enter the nucleus and has been implicated in transcriptional regulation that may affect cell death, neurite outgrowth and neuronal excitability (Ghosal et al., 2010; Goodger et al., 2009; Maulik et al., 2012; Robinson et al., 2014). Interestingly, transgenic mice expressing AICD have increased activity of GSK-3 (Ryan and Pimplikar, 2005), which in flies affects the circadian clock. Over-expression of GSK-3 in Drosophila leads to altered circadian behavior by hyper-phosphorylation of TIMELESS (TIM), a key circadian protein which forms dimers with PER that enter the nucleus and regulate the clock mechanism (Martinek et al., 2001). Of further interest, increased GSK-3 activity has been implicated in AD, and it was shown in Drosophila that increased GSK-3 activity mediates the toxicity of Aβ peptides (Sofola et al., 2010).
Cleavage of APPL likely results in a significant decline in intact APPL, and this could be detrimental as APPL has neuroprotective effects (Wentzell et al., 2012). It was also recently shown that loss of full-length APPL induces cognitive deficits in memory (Goguel et al., 2011). We report here that flies over-expressing full-length APPL in central pacemaker neurons maintained stronger behavioral rest-activity rhythms during aging than control flies; however this effect was not observed when APPL was expressed pan-neuronally. This could be caused by negative effects of APPL when expressed in other unspecified neurons, or could be related to driver strength. Overall, our data suggest that the loss of full-length APPL might negatively affect circadian behavior by way of the central pacemaker neurons.
Over-expression of dAICD induced a severe phenotype, disrupting rest-activity rhythms as early as age 5d when expressed in central pacemaker neurons and by age 35d with pan-neuronal expression. Taken together these results suggest that while loss of full-length APPL by over-expression of its secretases might negatively impact circadian behavior, the cleavage product dAICD induces the most severe behavioral rest-activity disruption. Interestingly, the observed effect is not likely a product of neurodegeneration as it was previously shown that dAICD has no effect on neurodegeneration (Wentzell et al., 2012), and we show in this study that the pacemaker cells appear intact in pdf>dAICD flies. In addition, we show that dAICD, like the vertebrate AICD, can be found in the nucleus. Therefore, our study suggests that dAICD may directly or indirectly affect the expression of clock genes. This offers a novel entry point toward understanding the mechanism of circadian rhythm disruption in Alzheimer’s disease.
Supplementary Material
Highlights.
dBACE overexpression accelerates decline of rest-activity rhythm in aging Drosophila.
PER oscillations in pacemaker neurons are dampened by elevated dBACE.
Kuzbanian overexpression induces age-related rest-activity rhythm decline in flies.
Neuronal dAICD expression induces age-related rest-activity rhythm decline.
APPL expression in pacemaker neurons strengthens rest-activity rhythms in aging flies.
Acknowledgments
We thank Dr. Taishi Yoshii for advice on immunocytochemistry, Dr. R Stanewsky for anti-PER antibody, Dr. Philip Copenhaver for anti-AICD antibody, and Dani Long for reading the manuscript. Research reported in this publication was supported by the National Institute of Aging of the National Institutes of Health under award number R01 AG045830 to JMG and by a pilot project grant from the Oregon Institute of Occupational Health Sciences to DK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Belyaev ND, et al. The transcriptionally active amyloid precursor protein (APP) intracellular domain is preferentially produced from the 695 isoform of APP in a {beta}-secretase-dependent pathway. J Biol Chem. 2010;285:41443–41454. doi: 10.1074/jbc.M110.141390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolkan BJ, et al. beta-secretase cleavage of the fly amyloid precursor protein is required for glial survival. J Neurosci. 2012;32:16181–16192. doi: 10.1523/JNEUROSCI.0228-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmine-Simmen K, et al. Neurotoxic effects induced by the Drosophila amyloid-beta peptide suggest a conserved toxic function. Neurobiol Dis. 2009;33:274–281. doi: 10.1016/j.nbd.2008.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KF, et al. The central molecular clock is robust in the face of behavioural arrhythmia in a Drosophila model of Alzheimer's disease. Dis Model Mech. 2014;7:445–458. doi: 10.1242/dmm.014134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Strooper B, Annaert W. Proteolytic processing and cell biological functions of the amyloid precursor protein. J Cell Sci. 2000;113(Pt 11):1857–1870. doi: 10.1242/jcs.113.11.1857. [DOI] [PubMed] [Google Scholar]
- Epis R, et al. Alpha, beta-and gamma-secretases in Alzheimer's disease. Front Biosci (Schol Ed) 2012;4:1126–1150. doi: 10.2741/s322. [DOI] [PubMed] [Google Scholar]
- Fukumoto H, et al. Beta-secretase protein and activity are increased in the neocortex in Alzheimer disease. Arch Neurol. 2002;59:1381–1389. doi: 10.1001/archneur.59.9.1381. [DOI] [PubMed] [Google Scholar]
- Gargano JW, et al. Rapid iterative negative geotaxis (RING): a new method for assessing agerelated locomotor decline in Drosophila. Exp Gerontol. 2005;40:386–395. doi: 10.1016/j.exger.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Ghosal K, et al. APP intracellular domain impairs adult neurogenesis in transgenic mice by inducing neuroinflammation. PLoS One. 2010;5:e11866. doi: 10.1371/journal.pone.0011866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goguel V, et al. Drosophila amyloid precursor protein-like is required for long-term memory. J Neurosci. 2011;31:1032–1037. doi: 10.1523/JNEUROSCI.2896-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodger ZV, et al. Nuclear signaling by the APP intracellular domain occurs predominantly through the amyloidogenic processing pathway. J Cell Sci. 2009;122:3703–3714. doi: 10.1242/jcs.048090. [DOI] [PubMed] [Google Scholar]
- Greeve I, et al. Age-dependent neurodegeneration and Alzheimer-amyloid plaque formation in transgenic Drosophila. J Neurosci. 2004;24:3899–3906. doi: 10.1523/JNEUROSCI.0283-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JC. Systems approaches to biological rhythms in Drosophila. Methods Enzymol. 2005;393:61–185. doi: 10.1016/S0076-6879(05)93004-8. [DOI] [PubMed] [Google Scholar]
- Hardin PE, Panda S. Circadian timekeeping and output mechanisms in animals. Curr Opin Neurobiol. 2013;23:724–731. doi: 10.1016/j.conb.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- Harper DG, et al. Disturbance of endogenous circadian rhythm in aging and Alzheimer disease. Am J Geriatr Psychiatry. 2005;13:359–368. doi: 10.1176/appi.ajgp.13.5.359. [DOI] [PubMed] [Google Scholar]
- Hastings MH, Goedert M. Circadian clocks and neurodegenerative diseases: time to aggregate? Current Opinion in Neurobiology. 2013 doi: 10.1016/j.conb.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Forster C. Neurobiology of the fruit fly's circadian clock. Genes Brain Behav. 2005;4:65–76. doi: 10.1111/j.1601-183X.2004.00092.x. [DOI] [PubMed] [Google Scholar]
- Helfrich-Forster C, et al. The lateral and dorsal neurons of Drosophila melanogaster: new insights about their morphology and function. Cold Spring Harb Symp Quant Biol. 2007;72:517–525. doi: 10.1101/sqb.2007.72.063. [DOI] [PubMed] [Google Scholar]
- Kaneko M, Hall JC. Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J Comp Neurol. 2000;422:66–94. doi: 10.1002/(sici)1096-9861(20000619)422:1<66::aid-cne5>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Kimberly WT, et al. The intracellular domain of the beta-amyloid precursor protein is stabilized by Fe65 and translocates to the nucleus in a notch-like manner. J Biol Chem. 2001;276:40288–40292. doi: 10.1074/jbc.C100447200. [DOI] [PubMed] [Google Scholar]
- Kondratova AA, Kondratov RV. The circadian clock and pathology of the ageing brain. Nat Rev Neurosci. 2012;13:325–335. doi: 10.1038/nrn3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan N, et al. Loss of circadian clock accelerates aging in neurodegeneration-prone mutants. Neurobiol Dis. 2012;45:1129–1135. doi: 10.1016/j.nbd.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long DM, et al. Relationships between the circadian system and Alzheimer's disease-like symptoms in Drosophila. PLoS One. 2014;9:e106068. doi: 10.1371/journal.pone.0106068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu DC, et al. A second cytotoxic proteolytic peptide derived from amyloid beta-protein precursor. Nat Med. 2000;6:397–404. doi: 10.1038/74656. [DOI] [PubMed] [Google Scholar]
- Luo L, et al. Human amyloid precursor protein ameliorates behavioral deficit of flies deleted for Appl gene. Neuron. 1992;9:595–605. doi: 10.1016/0896-6273(92)90024-8. [DOI] [PubMed] [Google Scholar]
- Luo LQ, et al. Identification, secretion, and neural expression of APPL, a Drosophila protein similar to human amyloid protein precursor. J Neurosci. 1990;10:3849–3861. doi: 10.1523/JNEUROSCI.10-12-03849.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, et al. Old flies have a robust central oscillator but weaker behavioral rhythms that can be improved by genetic and environmental manipulations. Aging Cell. 2012;11:428–438. doi: 10.1111/j.1474-9726.2012.00800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelkow EM, Mandelkow E. Tau in Alzheimer's disease. Trends Cell Biol. 1998;8:425–427. doi: 10.1016/s0962-8924(98)01368-3. [DOI] [PubMed] [Google Scholar]
- Martinek S, et al. A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell. 2001;105:769–779. doi: 10.1016/s0092-8674(01)00383-x. [DOI] [PubMed] [Google Scholar]
- Maulik M, et al. Mutant human APP exacerbates pathology in a mouse model of NPC and its reversal by a beta-cyclodextrin. Hum Mol Genet. 2012;21:4857–4875. doi: 10.1093/hmg/dds322. [DOI] [PubMed] [Google Scholar]
- Panza F, et al. Disease-modifying approach to the treatment of Alzheimer's disease: from alpha-secretase activators to gamma-secretase inhibitors and modulators. Drugs Aging. 2009;26:537–555. doi: 10.2165/11315770-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Pardossi-Piquard R, Checler F. The physiology of the beta-amyloid precursor protein intracellular domain AICD. J Neurochem. 2012;120(Suppl 1):109–124. doi: 10.1111/j.1471-4159.2011.07475.x. [DOI] [PubMed] [Google Scholar]
- Plucinska K, et al. Knock-in of human BACE1 cleaves murine APP and reiterates Alzheimer-like phenotypes. J Neurosci. 2014;34:10710–10728. doi: 10.1523/JNEUROSCI.0433-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakshit K, et al. Effects of aging on the molecular circadian oscillations in Drosophila . Chronobiol Int. 2012;29:1–10. doi: 10.3109/07420528.2011.635237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AB, O'Neill JS. Healthy clocks, healthy body, healthy mind. Trends Cell Biol. 2010;20:36–44. doi: 10.1016/j.tcb.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renn SC, et al. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila [published erratum appears in Cell 2000 Mar 31;101(1):following 113] Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- Rezaval C, et al. A functional misexpression screen uncovers a role for enabled in progressive neurodegeneration. PLoS One. 2008;3:e3332. doi: 10.1371/journal.pone.0003332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson A, et al. Upregulation of PGC-1alpha expression by Alzheimer's disease-associated pathway: presenilin 1/amyloid precursor protein (APP)/intracellular domain of APP. Aging Cell. 2014;13:263–272. doi: 10.1111/acel.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh JH, et al. Disruption of the sleep-wake cycle and diurnal fluctuation of beta-amyloid in mice with Alzheimer's disease pathology. Sci Transl Med. 2012;4:150ra122. doi: 10.1126/scitranslmed.3004291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen DR, et al. A Drosophila gene encoding a protein resembling the human beta-amyloid protein precursor. Proc Natl Acad Sci U S A. 1989;86:2478–2482. doi: 10.1073/pnas.86.7.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan KA, Pimplikar SW. Activation of GSK-3 and phosphorylation of CRMP2 in transgenic mice expressing APP intracellular domain. J Cell Biol. 2005;171:327–335. doi: 10.1083/jcb.200505078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe DJ. Toward a comprehensive theory for Alzheimer's disease. Hypothesis: Alzheimer's disease is caused by the cerebral accumulation and cytotoxicity of amyloid beta-protein. Ann N Y Acad Sci. 2000;924:17–25. doi: 10.1111/j.1749-6632.2000.tb05554.x. [DOI] [PubMed] [Google Scholar]
- Sofola O, et al. Inhibition of GSK-3 ameliorates Abeta pathology in an adult-onset Drosophila model of Alzheimer's disease. PLoS Genet. 2010;6:e1001087. doi: 10.1371/journal.pgen.1001087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterniczuk R, et al. Characterization of the 3xTg-AD mouse model of Alzheimer's disease: Part 1. Circadian changes. Brain Res. 2010;1348:139–148. doi: 10.1016/j.brainres.2010.05.013. [DOI] [PubMed] [Google Scholar]
- Turner PR, et al. Roles of amyloid precursor protein and its fragments in regulating neural activity, plasticity and memory. Prog Neurobiol. 2003;70:1–32. doi: 10.1016/s0301-0082(03)00089-3. [DOI] [PubMed] [Google Scholar]
- Umezaki Y, et al. Pigment-Dispersing Factor Is Involved in Age-Dependent Rhythm Changes in Drosophila melanogaster. J Biol Rhythms. 2012;27:423–432. doi: 10.1177/0748730412462206. [DOI] [PubMed] [Google Scholar]
- Vassar R, et al. The beta-secretase enzyme BACE in health and Alzheimer's disease: regulation, cell biology, function, and therapeutic potential. J Neurosci. 2009;29:12787–12794. doi: 10.1523/JNEUROSCI.3657-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volicer L, et al. Sundowning and circadian rhythms in Alzheimer's disease. Am J Psychiatry. 2001;158:704–711. doi: 10.1176/appi.ajp.158.5.704. [DOI] [PubMed] [Google Scholar]
- Wentzell JS, et al. Amyloid precursor proteins are protective in Drosophila models of progressive neurodegeneration. Neurobiology of Disease. 2012;46:78–87. doi: 10.1016/j.nbd.2011.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YH, Swaab DF. Disturbance and strategies for reactivation of the circadian rhythm system in aging and Alzheimer's disease. Sleep Med. 2007;8:623–636. doi: 10.1016/j.sleep.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Yoshii T, et al. The neuropeptide pigment-dispersing factor adjusts period and phase of Drosophila's clock. J Neurosci. 2009;29:2597–2610. doi: 10.1523/JNEUROSCI.5439-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, et al. The APP intracellular domain (AICD) inhibits Wnt signalling and promotes neurite outgrowth. Biochim Biophys Acta. 2012;1823:1233–1241. doi: 10.1016/j.bbamcr.2012.05.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.