Abstract
AIM: To investigate the expressions of microRNA-20a (miR-20a) and let-7a in esophageal squamous cell carcinoma (ESCC) and their diagnostic value.
METHODS: Seventy patients with ESCC and 40 healthy subjects were enrolled to investigate the expression of miR-20a and let-7a using quantitative real-time PCR. The expression of miR-20a and let-7a was compared between ESCC patients and healthy subjects. The plasma levels of miR-20a and let-7a in relation to patient clinicopathologic parameters, the receiver operating characteristic (ROC) curve, and the sensitivity and specificity of miR-20a and let-7a in ESCC diagnosis were analyzed.
RESULTS: Plasma levels of miR-20a were significantly higher in ESCC patients than in healthy controls, and plasma levels of let-7 were lower in ESCC patients than in healthy controls (both P < 0.05). The area under the ROC curve of miR-20a was 0.767 (95%CI: 0.677-0.857; P < 0.001), when the cut-off value was set at 4.77, the sensitivity and specificity were 64.3% and 75.0%, respectively. The area under the ROC curve of let-7a was 0.829 (95%CI: 0.754-0.904; P < 0.001), when the cut-off value was set at 6.22, the sensitivity and specificity were 74.3% and 85.0%, respectively. Thus, the sensitivity and specificity of let-7a were higher than those of miR-20a. The median relative plasma expression of let-7a in clinical stage III/IV (0.24) was lower than that in stage I/II (0.42), while the expression of miR-20a according to stage was not statistically different. The expressions of miR-20a and let-7a were not related to gender, age, tumor diameter, tumor grade, or pathologic stage.
CONCLUSION: Plasma miR-20a and let-7a levels are significantly altered in patients with ESCC and can be used as potential biomarkers in the diagnosis of ESCC.
Keywords: Esophageal squamous cell carcinoma, MicroRNA-20a, Let-7a, Plasma
Core tip: The levels of microRNA-20a (miR-20a) and let-7a are reportedly changed in tumors. However, plasma levels of miR-20a and let-7a in esophageal squamous cell carcinoma (ESCC) have not yet been determined. In this study, we found that both miR-20a and let-7a levels changed in ESCC patients compared with healthy controls, and thus may serve as biomarkers in the diagnosis of ESCC.
INTRODUCTION
Esophageal squamous cell carcinoma (ESCC) is among the ten most frequent cancers worldwide with a high incidence and poor prognosis[1,2]. Although patients with ESCC undergo appropriate surgery, relapse and metastasis can still occur in 90% of patients. Therefore, early diagnosis of ESCC is extremely important.
MicroRNAs (miRNAs) are small, noncoding RNAs which can influence target gene expression through mRNA degradation and translation inhibition[3]. MiRNAs can also act as oncogenes or tumor suppressors and have different biologic functions for the hundreds of downstream genes[4]. miR-20a belongs to the miR-17-92 cluster, which is a widely overexpressed oncogene in diverse cancer subtypes[5]. Let-7a belongs to the let-7 family, is a well-known miRNA, and is an important tumor suppressor in various cancers. let-7 and its family members are highly conserved across species in sequence and function, and misregulation of let-7 leads to a less-differentiated cellular state and the development of cell-based diseases[6]. It has been reported that miR-20a is upregulated in ESCC tissues[7]. As one of the most extensive miRNAs, the expression level of let-7 is decreased in many types of cancers, such as of the lung, stomach, and colon[8-10]. However, the circulating levels of miR-20a and let-7a in ESCC are not known.
The aim of this study was to determine whether the expression of miR-20a and let-7a in plasma is altered in ESCC patients compared with healthy controls, and to evaluate whether miR-20a and let-7a are associated with the clinicopathologic features of ESCC.
MATERIALS AND METHODS
Study population
Seventy patients hospitalized from May 2013 to May 2014 at The First Affiliated Hospital of Zhengzhou University (Zhengzhou, China) were enrolled in this study. In addition, 40 age- and sex-matched healthy volunteers participated in this study. All patients were untreated prior to surgery. The diagnosis of ESCC was based on the examination of tumor specimens using the 7th Edition of the AJCC Cancer Staging Manual[11]. A single-blind research design was used in this study.
Plasma samples and miRNA isolation
Fasting venous blood samples from ESCC patients and healthy controls were collected and placed in tubes containing EDTA-K2. The samples were centrifuged at 3000 × g for 10 min at 4 °C, and the supernatant was then isolated and centrifuged at 12000 × g for 10 min at 4 °C. Plasma was collected and stored in aliquots at -80 °C until analysis. Clinicopathologic information was available for all patients. Two hundred microliters of plasma were spiked with mi-Script miRNA mimic SV40 (Qiagen, Venlo, Limburg, Netherlands) (2 μmol/L, 1 μL/100 μL plasma). Total RNA was extracted from these plasma samples using TRI reagent BD TB-126 (Molecular Research Center Inc., Cincinnati, OH, United States) according to the manufacturer’s protocol, and dissolved in 10 μL of RNase-free water. The concentration and quality of the RNA samples were determined using a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States).
Quantitative real-time polymerase chain reaction
Total RNA (0.5 μg) from each sample was reverse transcribed using miRNA-specific stem-loop RT primer. The PrimeScript RT reagent kit with gDNA Eraser (DRR047S; TaKaRa Bio Inc., Shiga, Japan) was used according to the kit instructions. In brief, the 20 μL reactions were incubated for 15 min at 42 °C, followed by 5 s at 85 °C, and the resulting cDNA was stored at -20 °C for subsequent quantitative real-time PCR (qRT-PCR) on an ABI 7500 fast RT-PCR system (Applied Biosystems of Thermo Fisher Scientific) using SYBR Premix Ex TaqTMII (DRR820S; TaKaRa Bio Inc.) according to the manufacturer’s instructions. Briefly, the thermal cycling consisted of an initial denaturation at 95 °C for 30 s, followed by 40 cycles of 95 °C for 3 s and 60 °C for 30 s. A melt curve was constructed after each reaction. All PCRs yielded a single peak on the melt curve, indicating acceptable specificity of the primers. Assays were performed in triplicate. Data were analyzed with 7500 Fast System SDS Software version 1.4.0.25 (Applied Biosystems) with the automatic Ct setting for assigning baseline and threshold for Ct determination. Ct values > 36 were considered to represent no expression. miRNA relative expression levels were calculated using the following equation: 2-∆Ct (∆Ct = Ct target - Ct spiked-in SV40). Fold changes in miRNA levels were calculated using the 2-∆∆Ct method[12].
Ethics statement
The study protocol was carried out according to the principles of the Declaration of Helsinki and approved by the Medical Ethics Committee of The First Affiliated Hospital of Zhengzhou University. Written informed consent was obtained from all the participants before enrollment.
Statistical analysis
All experiments were repeated independently at least three times, and the data are presented as mean ± SD or median. The results were calculated using the Student’s t test, Mann-Whitney U test or Kruskal-Wallis test. In addition, a Spearman’s correlation test was performed. Receiver operating characteristic (ROC) curve analysis and the derived area under the curve (AUC) were used to estimate the ability of the biomarkers to distinguish ESCC patients from healthy controls. All statistical analyses were performed using SPSS 17.0 (SPSS Inc., Chicago, IL, United States) and results were considered significant when P < 0.05. We set two-fold as the threshold to measure miR-20a and let-7a as up- or downregulated.
RESULTS
Levels of plasma miR-20a and let-7a in ESCC patients and healthy controls
To assess the value of plasma miR-20a and let-7a for predicting ESCC, plasma miRNA levels were measured using qRT-PCR in ESCC patients and healthy controls. The results are summarized in Table 1. Independent-samples t tests showed statistically significant differences in plasma miR-20a and let-7a levels between ESCC patients and healthy controls: higher miR-20a plasma levels (P = 0.000) and lower plasma levels of let-7a (P = 0.000) were observed in ESCC patients (Table 1). Data on 2-∆∆Ct analysis showed that plasma concentrations of miR-20a and let-7a were markedly aberrant in ESCC patients relative to those in healthy controls, fold-change 2.09 ± 1.39 and 0.53 ± 0.43, respectively (Figure 1).
Table 1.
Relative miR-20a and let-7a levels
| Group | Age (yr) | miR-20a | let-7a |
| ESCC patients | 60.48 ± 8.40 | 2.09 ± 1.39 | 0.53 ± 0.43 |
| Healthy controls | 61.72 ± 6.93 | 1.12 ± 0.68 | 1.13 ± 0.58 |
| P-value | 0.431 | 0.000 | 0.000 |
ESCC: Esophageal squamous cell carcinoma.
Figure 1.
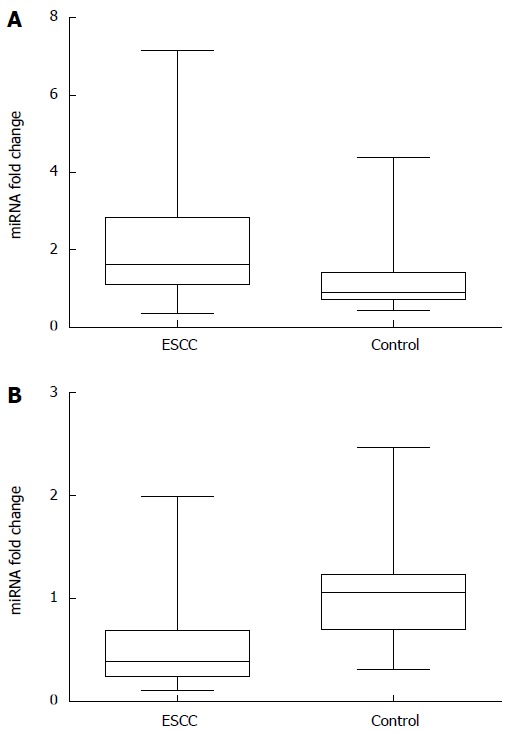
Relative plasma miRNA levels. A: miR-20a; B: let-7a.
Correlation between plasma miRNAs and clinicopathologic features of ESCC
The plasma levels of miR-20a and let-7a were then compared with patient clinicopathologic features. miR-20a had no significant associations with any of the clinicopathologic features (Table 2). The level of let-7a had no significant associations with gender, age, tumor diameter, or pathologic stage, but increased with tumor stage (P < 0.05).
Table 2.
Correlation between miR-20a and let-7a levels and clinicopathologic features in patients with esophageal squamous cell carcinoma
| Characteristics | n |
Fold change1 |
|
| miR-20a | let-7a | ||
| Gender | |||
| Male | 46 | 1.61 (0.95-2.82) | 0.33 (0.22-0.53) |
| Female | 24 | 1.63 (1.33-2.93) | 0.45 (0.29-0.89) |
| P-value | 0.481 | 0.107 | |
| Age (yr) | |||
| ≤ 60 | 37 | 1.62 (1.24-2.89) | 0.39 (0.23-0.79) |
| > 60 | 33 | 1.62 (1.06-2.82) | 0.35 (0.24-0.64) |
| P-value | 0.463 | 0.900 | |
| Diameter of tumor | |||
| < 4 cm | 36 | 1.51 (0.99-2.47) | 0.40 (0.23-0.79) |
| ≥ 4 cm | 34 | 2.10 (1.33-3.08) | 0.36 (0.24-0.62) |
| P-value | 0.077 | 0.961 | |
| TNM stage | |||
| I/II | 49 | 1.52 (0.95-2.80) | 0.42 (0.28-0.88) |
| III/IV | 21 | 2.04 (1.40-3.40) | 0.24 (0.19-0.39) |
| P-value | 0.118 | 0.009 | |
| Distant metastasis | |||
| M0 | 61 | 1.62 (1.10-2.82) | 0.39 (0.24-0.79) |
| M1 | 9 | 1.64 (1.21-4.45) | 0.35 (0.12-0.44) |
| P-value | 0.300 | 0.174 | |
| Histologic grade | |||
| Well differentiated | 25 | 1.50 (1.10-2.41) | 0.45 (0.25-0.59) |
| Moderately differentiated | 28 | 1.76 (1.04-2.82) | 0.33 (0.26-0.97) |
| Poorly differentiated | 17 | 1.75 (1.21-3.29) | 0.30 (0.22-0.62) |
| P-value | 0.632 | 0.849 | |
Median of relative expression with 25th-75th percentile; Mann-Whitney U test waws performed for comparisons between two groups and Kruskall-Wallis test for comparisons among three groups.
Diagnostic accuracy of circulating miR-20a and let-7a
To evaluate plasma miR-20a and let-7a as potential biomarkers of ESCC, ROC curve analyses were carried out and the values for the AUC were calculated. As shown in Figure 2, the ROC curves of plasma miR-20a and let-7a significantly discriminated between ESCC patients and healthy controls with an AUC of 0.767 (95%CI: 0.677-0.857; P < 0.001) and 0.829 (95%CI: 0.754-0.904; P < 0.001), respectively. These results showed that plasma miR-20a and let-7a had accurate diagnostic value for ESCC. When the cutoff value of miR-20a was set at 4.77, the sensitivity and specificity were 64.3% and 75.0%, respectively; when the cutoff value of let-7a was set at 6.22, the sensitivity and specificity were 74.3% and 85.0%, respectively. Thus, the sensitivity and specificity of let-7a were higher than those of miR-20a.
Figure 2.
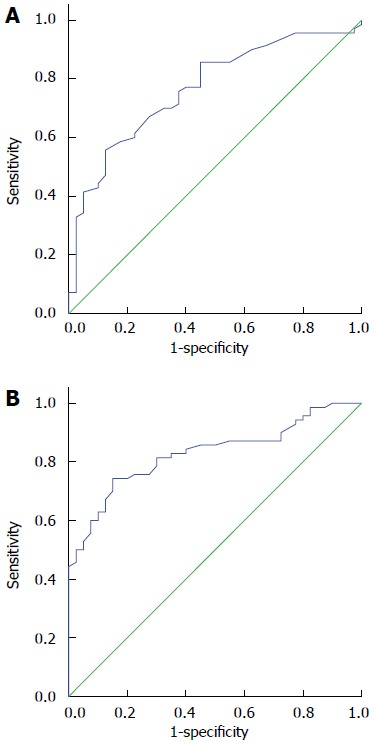
Plasma miRNA levels associated with diagnosis of esophageal squamous cell carcinoma. Receiver operating characteristic curves for plasma A: miR-20a; and B: let-7a.
DISCUSSION
ESCC is diverse in regions with different environmental risks and genetic factors[13-15]. Despite recent advances in diagnosis and treatment strategies, the prognosis of ESCC across all stages remains poor. Therefore, the diagnosis of ESCC is important. To date, hundreds of human miRNAs have been identified. miRNAs, which are implicated in key cellular processes, play important roles in cancer[16]. It is known that one single miRNA can influence the expression of several thousands of genes, thus miRNAs control one-third of the human genome[17]. Many studies have found that miRNAs can act as biomarkers for the diagnosis, prognosis, and treatment evaluation of cancers. However, it can sometimes be difficult to obtain tissue for diagnosis, particularly in patients with ESCC. Recent studies have demonstrated that specific expression profiles of circulating miRNAs could be promising blood-based noninvasive biomarkers for the detection and prognosis of different types of cancer, and human serum or plasma contains a large number of intact and stable miRNAs, which can be detected using a simple assay such as qRT-PCR[18,19].
MiR-20a has been found to be upregulated in colon adenocarcinoma and gliomas and downregulated in breast cancer and pancreatic carcinoma[20,21]. On the other hand, miR-20a has been shown to inhibit proliferation and metastasis of pancreatic carcinoma cells by directly downregulating Stat3, the cyclin-dependent kinase inhibitor that is activated in primary pancreatic cancer and is involved in various physiologic functions, including apoptosis, cell cycle regulation, angiogenesis, and metastasis. Findings have clearly shown that while miR-20a is overexpressed in human ovarian cancer tissues and enhances long-term cellular proliferation and invasion capabilities, it also mediates immune evasion in vivo and downregulates MICA/B expression, which can decrease natural killer cell cytotoxicity[22]. Gene-expression data have shown that several tumors overexpress miR-20a, and E2F and Myc are inhibited at the post-transcriptional step by miR-17-92 and can induce the transcription of miR-17-92, which forms a negative feedback loop in the interaction network[23,24]. It has been confirmed that plasma miR-20a is highly expressed in lung cancer and can predict survival in patients with squamous cell carcinoma[25]. miR-20a can also regulate CDKN1a/p21, and transforming growth factor-β receptor 2[23]. However, miR-20a expression in primary hepatocellular carcinoma is lower than in normal liver and predicts poor survival in patients, and directly regulates Mcl-1 expression, which is an antiapoptotic member of the Bcl-2 family[26]. Therefore, miR-20a can be an oncogene or a tumor-suppressor gene. Compared with miR-20a, let-7 expression is low in various tumor types. let-7 controls cell differentiation and proliferation, and many oncogenes are regulated by let-7, such as RAS, HMGA2, and MYC[27].
In this study, we investigated the expression levels of miR-20a and let-7a in plasma. The levels of miR-20a and let-7a showed a clear distinction between ESCC patients and healthy individuals. miR-20a was highly expressed in ESCC patients, whereas let-7a showed low expression. Furthermore, plasma miR-20a and let-7a had significant diagnostic value for ESCC as shown by ROC curve analyses. Thus, circulating miR-20a and let-7a may be useful diagnostic biomarkers in patients with ESCC. Although the expression of let-7a in plasma had no significant association with gender, age, tumor diameter, or pathologic stage, let-7a expression increased with tumor stage. This finding was also observed in other studies in which let-7 targeted HmgA2 and RAS, and hence inhibited cancer growth[28,29]. However, no significant association was found between the level of miR-20a and gender, age, tumor diameter, tumor grade, or pathologic stage. There were occasional errors in the limited samples included in this study. Therefore, a larger population is needed to validate the specificity and sensitivity of miR-20a and let-7a. Furthermore, additional investigations are required in the near future to determine the mechanism of miR-20a and let-7a in ESCC.
In conclusion, our results suggest that miR-20a and let-7a expression in plasma could serve as biomarkers in the diagnosis of ESCC. The level of let-7a was associated with tumor stage. However, further study is needed to elucidate the mechanism of miR-20a and let-7a, which may become potential therapeutic targets of ESCC.
COMMENTS
Background
Esophageal squamous cell carcinoma (ESCC) has a marked regional variation in terms of incidence and mortality with a high incidence and poor prognosis. Therefore, early diagnosis of ESCC is important.
Research frontiers
MicroRNAs (miRNAs) play an important role in cancer and can often act as biomarkers in diagnosis. Circulating miRNAs are stable and can be detected by quantitative real-time polymerase chain reaction, and thus could be promising blood-based noninvasive biomarkers for cancer.
Innovations and breakthroughs
This study showed that circulating miR-20a and let-7a levels can be detected easily and noninvasively. Plasma levels of miR-20a and let-7a were significantly altered and may be used in diagnosis.
Applications
miR-20a was upregulated and let-7a was downregulated in ESCC and may be used as potential biomarkers in the diagnosis of ESCC.
Terminology
Receiver operating characteristic curve is a composite index reflecting the sensitivity and specificity. Sensitivity: the proportion of individuals with ESCC who had a positive test result; Specificity: the proportion of subjects without ESCC who had a negative test result.
Peer-review
This is a well-designed and well-written study in which the authors show that circulating miRNA-20a and let-7 can be detected in plasma and change significantly in patients with ESCC in relation to healthy individuals.
Footnotes
Supported by Medical Scientific Research Foundation of Health Department of Henan Province of China, No. 201403077.
Ethics approval: The study was reviewed and approved by The First Affiliated Hospital of Zhengzhou University Institutional Review Board.
Informed consent: All study participants, or their legal guardian, provided informed written consent prior to study enrollment.
Conflict-of-interest: The authors report no conflict of interest.
Data sharing: No additional data are available.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: November 3, 2014
First decision: December 26, 2014
Article in press: February 11, 2015
P- Reviewer: Liu X, Velayos B, Wang LS S- Editor: Ma YJ L- Editor: AmEdior E- Editor: Zhang DN
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Ke L. Mortality and incidence trends from esophagus cancer in selected geographic areas of China circa 1970-90. Int J Cancer. 2002;102:271–274. doi: 10.1002/ijc.10706. [DOI] [PubMed] [Google Scholar]
- 3.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 5.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 7.Ogawa R, Ishiguro H, Kuwabara Y, Kimura M, Mitsui A, Katada T, Harata K, Tanaka T, Fujii Y. Expression profiling of micro-RNAs in human esophageal squamous cell carcinoma using RT-PCR. Med Mol Morphol. 2009;42:102–109. doi: 10.1007/s00795-009-0443-1. [DOI] [PubMed] [Google Scholar]
- 8.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 9.Zhang HH, Wang XJ, Li GX, Yang E, Yang NM. Detection of let-7a microRNA by real-time PCR in gastric carcinoma. World J Gastroenterol. 2007;13:2883–2888. doi: 10.3748/wjg.v13.i20.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29:903–906. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- 11.Rice TW, Blackstone EH, Rusch VW. 7th edition of the AJCC Cancer Staging Manual: esophagus and esophagogastric junction. Ann Surg Oncol. 2010;17:1721–1724. doi: 10.1245/s10434-010-1024-1. [DOI] [PubMed] [Google Scholar]
- 12.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 13.Morita M, Kumashiro R, Kubo N, Nakashima Y, Yoshida R, Yoshinaga K, Saeki H, Emi Y, Kakeji Y, Sakaguchi Y, et al. Alcohol drinking, cigarette smoking, and the development of squamous cell carcinoma of the esophagus: epidemiology, clinical findings, and prevention. Int J Clin Oncol. 2010;15:126–134. doi: 10.1007/s10147-010-0056-7. [DOI] [PubMed] [Google Scholar]
- 14.Yang CX, Wang HY, Wang ZM, Du HZ, Tao DM, Mu XY, Chen HG, Lei Y, Matsuo K, Tajima K. Risk factors for esophageal cancer: a case-control study in South-western China. Asian Pac J Cancer Prev. 2005;6:48–53. [PubMed] [Google Scholar]
- 15.Sun G, Wang S, Hu X, Su J, Huang T, Yu J, Tang L, Gao W, Wang JS. Fumonisin B1 contamination of home-grown corn in high-risk areas for esophageal and liver cancer in China. Food Addit Contam. 2007;24:181–185. doi: 10.1080/02652030601013471. [DOI] [PubMed] [Google Scholar]
- 16.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 18.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 19.Boeri M, Verri C, Conte D, Roz L, Modena P, Facchinetti F, Calabrò E, Croce CM, Pastorino U, Sozzi G. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci USA. 2011;108:3713–3718. doi: 10.1073/pnas.1100048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan H, Wu J, Liu W, Zuo Y, Chen S, Zhang S, Zeng M, Huang W. MicroRNA-20a overexpression inhibited proliferation and metastasis of pancreatic carcinoma cells. Hum Gene Ther. 2010;21:1723–1734. doi: 10.1089/hum.2010.061. [DOI] [PubMed] [Google Scholar]
- 21.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie J, Liu M, Li Y, Nie Y, Mi Q, Zhao S. Ovarian tumor-associated microRNA-20a decreases natural killer cell cytotoxicity by downregulating MICA/B expression. Cell Mol Immunol. 2014;11:495–502. doi: 10.1038/cmi.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aguda BD, Kim Y, Piper-Hunter MG, Friedman A, Marsh CB. MicroRNA regulation of a cancer network: consequences of the feedback loops involving miR-17-92, E2F, and Myc. Proc Natl Acad Sci USA. 2008;105:19678–19683. doi: 10.1073/pnas.0811166106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanfiorenzo C, Ilie MI, Belaid A, Barlési F, Mouroux J, Marquette CH, Brest P, Hofman P. Two panels of plasma microRNAs as non-invasive biomarkers for prediction of recurrence in resectable NSCLC. PLoS One. 2013;8:e54596. doi: 10.1371/journal.pone.0054596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan MQ, Huang CB, Gu Y, Xiao Y, Sheng JX, Zhong L. Decrease expression of microRNA-20a promotes cancer cell proliferation and predicts poor survival of hepatocellular carcinoma. J Exp Clin Cancer Res. 2013;32:21. doi: 10.1186/1756-9966-32-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Büssing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008;14:400–409. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 28.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 29.Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci USA. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]


