Abstract
Background:
The functions of long noncoding RNAs (lncRNAs) have been identified in several cancers, but the roles of lncRNAs in colorectal cancer (CRC) are less well understood. The transcription factor MYC is known to regulate lncRNAs and has been implicated in cancer cell proliferation and tumorigenesis.
Methods:
CRC cells and tissues were profiled to identify lncRNAs differentially expressed in CRC, from which we further selected MYC-regulated lncRNAs. We used luciferase promoter assay, ChIP, RNA pull-down assay, deletion mapping assay, LC-MS/MS and RNA immunoprecipitation to determine the mechanisms of MYC regulation of lncRNAs. Moreover, soft agar assay and in vivo xenograft experiments (four athymic nude mice per group) provided evidence of MYC-regulated lncRNAs in cancer cell transformation and tumorigenesis. The Kaplan-Meier method was used for survival analyses. All statistical tests were two-sided.
Results:
We identified lncRNAs differentially expressed in CRC (P < .05, greater than two-fold) and verified four lncRNAs upregulated and two downregulated in CRC cells and tissues. We further identified MYC-regulated lncRNAs, named MYCLos. The MYC-regulated MYCLos may function in cell proliferation and cell cycle by regulating MYC target genes such as CDKN1A (p21) and CDKN2B (p15), suggesting new regulatory mechanisms of MYC-repressed target genes through lncRNAs. RNA binding proteins including HuR and hnRNPK are involved in the function of MYCLos by interacting with MYCLo-1 and MYCLo-2, respectively. Knockdown experiments also showed that MYCLo-2, differentially expressed not only in CRC but also in prostate cancer, has a role in cancer transformation and tumorigenesis.
Conclusions:
Our results provide novel regulatory mechanisms in MYC function through lncRNAs and new potential lncRNA targets of CRC.
Mammalian genomes encode a large number of noncoding RNAs from small regulatory RNAs such as microRNAs and Piwi-associated RNAs (piRNAs) to long noncoding RNAs (lncRNAs; longer than 200bp) (1,2). To date, more than 30 000 long noncoding RNAs (lncRNAs) have been annotated or identified (3–6). A few lncRNAs such as CCAT1 (CARLo-5) and CCAT2 have been also reported in colorectal cancer (CRC) (7–9). However, an enormous number of lncRNAs still remain to be elucidated and characterized.
The proto-oncogene MYC is frequently amplified in many types of cancer, including CRC (10,11). The gene product of MYC is a transcription factor that regulates transcription of many protein-coding genes and noncoding RNAs such as microRNAs, and the downstream genes are involved in various cellular processes including cell cycle, differentiation, cell growth, metabolism, cell adhesion, angiogenesis, chromosome instability, and cell transformation (10). To date, a large number of MYC-regulated genes including CDKN2B (p15) and CDKN1A (p21) have been identified (10,12–14). Those target genes such as CDKN1A and CDKN2B are critical intermediators of MYC-mediated tumorigenesis (13,15).
Methods
LncRNA Microarray
Total RNA extracted from four normal colon-derived fibroblast cell lines (CCD-18Co, CCD-33Co, CCD-112CoN, CCD-841CoN), four colorectal cancer cell lines (HT29, SW620, HCT116, RKO), three primary CRC tissue samples and their matched normal adjacent tissues (NATs) (Origene), HCT116 cells treated with siCTRL or siMYC and RKO cells treated with siCTRL or siMYC that was subjected to lncRNA microarray. Further details are available in Supplementary Methods (available online).
Patients and Primary Colorectal and Prostate Tissue Samples
Primary colorectal and prostate tissue samples were provided by the Department of Pathology at The Ohio State University (OSU). All human tissues were obtained according to a protocol approved by the Ohio State Institutional Review Board. Tissue samples were fresh-frozen in liquid nitrogen after surgery and kept at -80℃. Frozen tissue samples were homogenized using the Tissue Ruptor (QIAGEN) before RNA extraction. Total RNA was extracted using Trizol (Invitrogen) in accordance with manufacturer’s instructions.
Xenograft and Tumor-Free Survival Analysis
Animal experiments were approved by The Ohio State University animal care and use committee and conducted following The Ohio State University animal policy in accordance with National Institutes of Health guidelines. 0.5 million cells transfected with indicated siRNAs 24 hours before injection were subcutaneously injected into the right flanks of five-week-old female athymic nude mice (Jackson laboratory, four mice per group). Further details are available in Supplementary Methods (available online).
NanoString Gene Expression Assay and Data Analysis
For NanoString Gene expression assay analyses, the nCounter Virtual Cell Cycle Gene Set was used, following manufacturer’s instructions (NanoString Technologies). Briefly, total RNA (100ng) was used as input for nCounter mRNA sample preparation reactions. All sample preparation was performed according to manufacturer’s instructions (NanoString Technologies). Further details are available in Supplementary Methods (available online).
Rapid Amplification of cDNA Ends (RACE)
5’ and 3’ RACE were performed using the SMARTer RACE cDNA Amplification Kit (Clontech). All procedures were done in accordance with manufacturer’s instruction. Total RNA from HT29 or SW620 was used. Polymerase chain reaction (PCR) of the internal region was performed when starting points of 5’ and 3’ RACE had an unamplified gap. All primers used for RACE are presented in Supplementary Table 5 (available online).
Cells, Oligonucleotides, and Transfection
All cell lines were purchased from American Type Culture Collection (ATCC) and cultured as recommended by the ATCC. All experiments with cells were done between passage 4–9. All custom siRNAs were designed by the Dharmacon custom siRNA design tool based on sequence information identified by RACE. For each MYCLo, two kinds of custom siRNAs were designed and used. Further details are available in the Supplementary Methods (available online).
Quantitative Real-Time PCR
Total RNA was prepared from cells using TRIZOL (Invitrogen) in accordance with manufacturer’s instructions. Total RNA was subjected to quantitative real-time PCR (qRT-PCR). RNA levels were analyzed using TaqMan Gene Expression Assays, in accordance with manufacturer’s instructions (Applied Biosystems). All RT reactions, including no-template controls and RT minus controls, were run in a GeneAmp PCR 9700 Thermocycler (Applied Biosystems). Further details are available in the Supplementary Methods (available online).
Cell Proliferation Assay
For cell proliferation assay, the MTS assay from Promega (CellTiter 96 AQueous One Solution Cell Proliferation Assay) was used following manufacturer’s instruction. Briefly, cells in a 96-wells plate were incubated for 72 hours or 96 hours in a humidified 5% CO2 atmosphere after transfection with indicated siRNAs, followed by addition of 20 μL CellTiter 96 AQueous One Solution and one- to four-hour incubation in humidified 5% CO2 atmosphere. The absorbance at 490nm was recorded.
Flow Cytometry Analysis
For DNA content analysis, cells were fixed in methanol at -20° C, washed again, rehydrated, resuspended in phosphate-buffered saline containing 2 μg/mL propidium iodide (PI), and 5 μg/mL RNase A and analyzed by BD FACS Calibur Flow Cytometer.
Luciferase Reporter Assay
Cells were plated in 24-well plates 24 hours before transfection. All plasmids constructed with pGL4-luc2P vectors were cotransfected with control Renilla luciferase plasmid (pGL4-hRluc/TK) and siRNAs, as indicated by using Lipofectamine 2000 (Invitrogen). Further details are available in the Supplementary Methods (available online).
Soft Agar Colony Formation Assay
Soft agar colony formation assay was performed with the CytoSelect cell transformation assay (Cell Biolab, Inc.) in accordance with the manufacturer’s instructions. The cells transfected with indicated siRNA were incubated for 24 hours, followed by a soft agar colony formation assay.
RNA Pull-Down Assay, Deletion Mapping, RIP, ChIP, and Antibodies
RNA pull-down and deletion mapping were performed using a Magnetic RNA-Protein Pull-Down kit (Pierce) in accordance with the manufacturer’s instructions. The full-length or deleted RNA samples were prepared using the LITMUS28i system and T7 in vitro transcription kit (NEB) followed by biotin-labeling at the 3’ end of in vitro transcribed RNA using the RNA 3’ End Desthiobiotinylation kit (Pierce). Further details are available in the Supplementary Methods (available online).
Data Access
The raw data of microarray analysis are deposited in the Gene Expression Omnibus (GEO) under accession number GSE39846. Primary data of Nanostring gene expression analysis are deposited in the GEO under accession number GSE39077. Sequence information of MYCLos is available in GenBank (Accession number: MYCLo-1, JX046909; MYCLo-2, JX046910; MYCLo-3 v1, JX046911; MYCLo-3 v2, JX628584).
Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS)
LC-MS/MS was performed by the Campus Chemical Instrument Center (CCIC) Mass Spectrometry and Proteomics Facility at The Ohio State University (OSU). Sample preparation and mass spectrometry are fully described in the Supplementary Methods (available online). Briefly, capillary-liquid chromatography tandem mass spectrometry (Cap-LC/MS/MS) was performed on a Thermo Finnigan LTQ mass spectrometer equipped with a CaptiveSpray source (Bruker Michrom Billerica, MA) operated in positive ion mode. The LC system was an UltiMate 3000 system from Dionex (Sunnyvale, CA). Capillary-liquid chromatography-nanospray tandem mass spectrometry (Capillary-LC/MS/MS) of global protein identification was performed on a Thermo Finnigan LTQ orbitrap mass spectrometer equipped with a microspray source (Michrom Bioresources Inc., Auburn, CA) operated in positive ion mode. Samples were separated on a capillary column (0.2X150mm Magic C18AQ 3µ 200A, Michrom Bioresources Inc, Auburn, CA) using an UltiMate 3000 HPLC system from LC-Packings A Dionex Co (Sunnyvale, CA).
Statistical Analysis
All graph values represent means ± SD from three independent experiments with each measured in triplicate. The differences between two groups were analyzed with the unpaired two-sided Student’s t test. A P value of less than .05 was considered statistically significant and indicated with asterisks, as described in figure legends. The Kaplan-Meier method was used to estimate survival curves, and the log-rank test was used to test for differences between curves using SPSS Statistical Software (SPSS Inc., Chicago, IL). All statistical tests were two-sided.
Results
Identification of lncRNAs Dysregulated in CRC
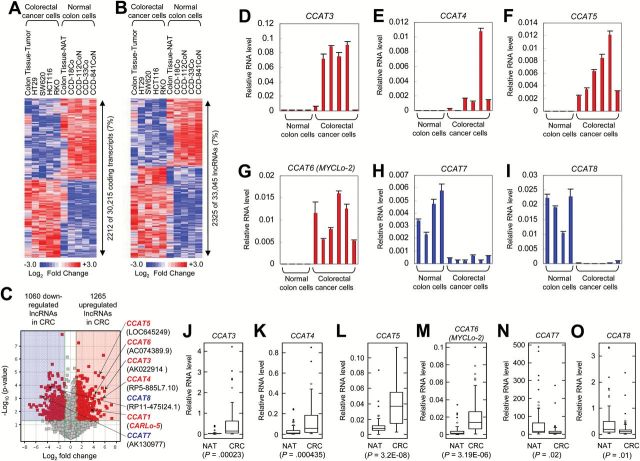
To identify lncRNAs that are dysregulated in colorectal cancer, we employed a lncRNA microarray to profile lncRNAs in normal colon-derived and CRC-derived cells and tissues. From the microarray analysis covering 30 215 protein-coding transcripts and 33 045 annotated and/or known lncRNAs, we observed that a similar percentage (7%) of protein-coding transcripts (2212 of 30215) and lncRNAs (2325 of 33045) are differentially expressed (P < .05, greater than two-fold) in CRC (Figure 1, A and B; Supplementary Table 1, available online). Of the identified 2325 lncRNAs (1265 upregulated and 1060 downregulated lncRNAs in CRC) (Figure 1C), we selected top candidates with our criteria (greater than 15-fold change and P < .01) and then performed 5’ and 3’ rapid amplification of cDNA ends (RACE) (Supplementary Figure 1, A and B, available online). Based on the sequence information from RACE, we tested the expression levels of the lncRNAs by using quantitative real-time PCR analysis in CRC cells and tissues. Of the selected lncRNAs, four lncRNAs named CCAT3, CCAT4, CCAT5, and CCAT6 (also named MYCLo-2) are upregulated in CRC cell lines (Figure 1, D-G; Supplementary Figure 1C, available online), and two lncRNAs named CCAT7 and CCAT8 are downregulated in CRC cell lines (Figure 1, H and I; Supplementary Figure 1C, available online). The results were also confirmed by using 52 CRC tissues and their matched NATs (Figure 1, J-O).
Figure 1.
Identification of long noncoding RNAs (lncRNAs) differentially expressed in colorectal cancer (CRC). A and B) Heatmaps of protein-coding transcripts (A) and lncRNAs (B) dysregulated in CRC (HCT116, RKO, SW620, HT29, and CRC tissues), compared with normal colon (CCD-18Co, CCD-33Co, CCD112CoN, CCD-841-CoN and colon normal adjacent tissues [NATs]). C) Volcano map indicating up- or downregulation of CRC-associated lncRNAs (CCAT1, CCAT3~8). D-G) Verification of induced expression of CCAT3 (D), CCAT4 (E), CCAT5 (F), and CCAT6 (G) in CRC cell lines by using quantitative real-time polymerase chain reaction (qRT-PCR). Data are means ± SD of three independent experiments and each measured in triplicate. H and I) Verification of repressed expression of CCAT7 (H) and CCAT8 (I) in CRC cell lines by using qRT-PCR. Data are means ± SD of three independent experiments and each measured in triplicate. J-O) Differential expression of CCAT3 (J), CCAT4 (K), CCAT5 (L), CCAT6 (M), CCAT7 (N), and CCAT8 (O) in 52 matched CRC and NAT tissues. Shown are box plots with lower and upper bounds of the boxes representing the 25th and 75th quartiles, respectively; whiskers demarcate values within the 10th to 90th percentiles, and solid circles indicate values less than the 10th percentile and greater than the 90th percentile.
Further Identification and Characterization of a Subset of CRC-Associated lncRNAs That Are Regulated by Proto-Oncogene MYC
We tested MYC mRNA levels in multiple normal colorectal cell lines (CCD-18Co, CCD-33Co, CCD-112CoN, CCD-841CoN), CRC-derived cell lines (HT29, SW620, HCT116, RKO), CRC tissues, and their matched NATs that were used in lncRNA microarray (Figure 1, A and B). As previously reported (16), MYC mRNA levels were statistically significantly higher in the CRC samples than in the normal colon samples (CCD18-Co, mean = 1, SD = .00925; HCT116, mean = 17.21, SD = .40947; P = .0025) (Figure 2A). This result suggests that MYC could be involved in the regulation of a cohort of lncRNAs that are dysregulated in CRC (Figure 1B).
Figure 2.
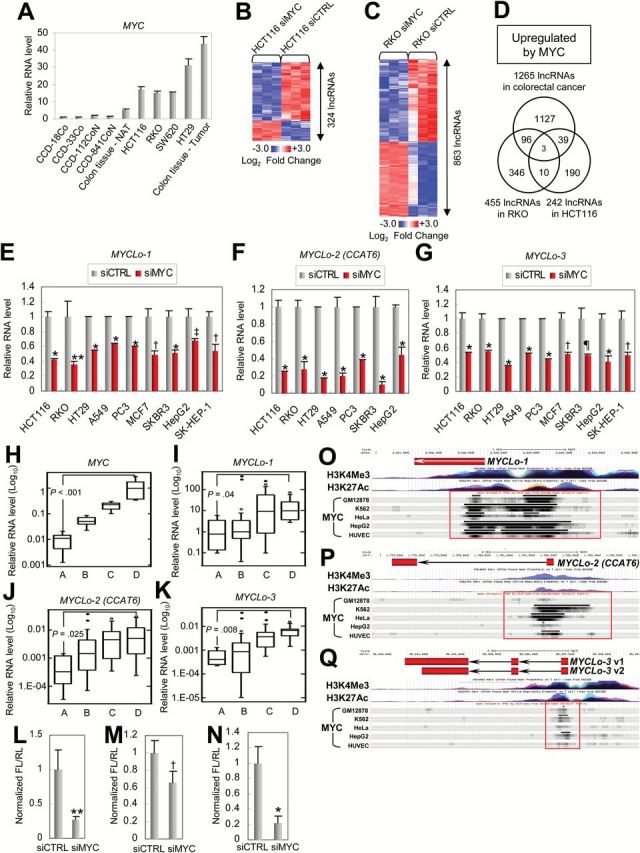
Identification of colorectal cancer [CRC]–associated lncRNAs directly regulated by MYC. A) Comparison of mRNA levels of MYC in normal colon–derived (CCD-18Co, CCD-33Co, CCD112CoN, CCD-841-CoN, and colon normal adjacent tissue [NAT] samples) and CRC cells/tissues (HCT116, RKO, SW620, HT29, and CRC tissue samples). Data are means ± SD of three independent experiments and each measured in triplicate. B and C) Heatmaps of lncRNAs dysregulated by MYC knockdown in HCT116 (B) and RKO (C). D) Venn diagrams showing the number of upregulated long noncoding RNAs (lncRNAs) in a MYC-dependent manner. Hypergeometric probability of three lncRNAs commonly found in all criteria is P(X=3) = 0.01. E-G) Quantitative real-time polymerase chain reaction (qRT-PCR) results showing MYC-mediated positive regulation of MYCLo-1 (E), -2 (F), and -3 (G) in various types of cancer cells. Data are means ± SD of three independent experiments and each measured in triplicate (**P = .0062; †P = .001; ‡P = .0079; ¶P = .004; *P < .001). Statistical significance determined by unpaired two-sided Student’s t test. H-K) Expressional associations between MYC (H) and MYCLos (MYCLo-1 [I], -2 [J], and -3 [K]) in 50 colorectal tissue samples. The number of samples in each group is eight (Group A), 17 (Group B), 15 (Group C), and 10 (Group D). The P values between Group A and Group D are indicated. Shown are box plots with lower and upper bounds of the boxes represent the 25th and 75th quartiles, respectively; whiskers demarcate values within the 10th to 90th percentiles, and solid circles indicate values less than the 10th percentile and greater than the 90th percentile. Statistical significance was determined by the unpaired two-sided Student’s t test. L-N) Luciferase assays showing MYC-dependent regulation of 5’ promoter activities of MYCLo-1 (L), -2 (M), and -3 (N). The promoter region of each MYCLo for luciferase assay is indicated with a red box in (O-Q), respectively. Data are means ± SD of three independent experiments and each measured in triplicate (**P = .01; †P = .03; *P = .004). Statistical significance was determined by the unpaired two-sided Student’s t test. O-Q) The database of open chromatin TFBS by ChIP-seq from ENCODE/Open Chrom (UT Austin) shows that MYC binds to 5’ promoter regions of MYCLo-1 (O), -2 (P), and -3 (Q) in various types of cells such as GM12878, K562, HeLa, HepG2, and HUVEC. MYC-binding regions are indicated with red boxes.
To further identify MYC-regulated lncRNAs among the lncRNAs dysregulated in CRC, we screened lncRNAs in CRC-derived cells treated with MYC-targeting siRNAs (siMYC) by using the lncRNA microarray (Supplementary Figure 2A, available online). As a result (P < .05, greater than two-fold), we identified 324 MYC-regulated lncRNAs in HCT116 cells (Figure 2B; Supplementary Table 2, available online) and 863 MYC-regulated lncRNAs in RKO cells (Figure 2C; Supplementary Table 3, available online). To find out the most interesting candidates that are regulated by MYC and dysregulated in CRC, we compared the lists of lncRNAs differentially expressed in CRC-derived cells (Figure 1B), siMYC-treated HCT116 cells (Figure 2B), and siMYC-treated RKO cells (Figure 2C). By selecting lncRNAs commonly found in the three groups, we finally isolated three lncRNAs (AK021907, AC074389.9, and KTN1-AS1) upregulated by MYC (Figure 2D). These MYC-regulated lncRNAs are named MYCLos (AK021907, MYCLo-1; AC074389.9 [CCAT6], MYCLo-2; KTN1-AS1, MYCLo-3). Of the three lncRNAs, MYCLo-2 is identical with CCAT6 that we identified and verified as the lncRNA upregulated in CRC (Figure 1, G and M; Supplementary Figure 1B, available online).
By using 5’ and 3’ rapid amplification of cDNA ends (RACE), we determined the presence and full length of the MYC-regulated lncRNAs (Supplementary Figure 2B, available online; MYCLo-2 is shown in Supplementary Figure 1B, available online). The expression of the lncRNAs was also confirmed by Northern blot (Supplementary Figure 2C, available online). The results show that actual sequences of the lncRNAs are a little different from the predicted sequences (Supplementary Figure 2D, available online) and MYCLos are localized in various genomic loci such as chromosome 20, 7 and 14 (Supplementary Figure 2, E-G, available online).
We next determined if MYC-mediated regulation of the MYCLos is common in various types of cancer cells. MYC-mediated regulation of MYCLos was observed in the cells from various cancer types, indicating that MYC-mediated regulation of MYCLos is conserved in many types of cancer and suggesting a universal role of these lncRNAs in various MYC-driven cancer cells (Figure 2, E-G). By using an additional siRNA-targeting MYC, we confirmed the MYC-mediated regulation of the MYCLos (Supplementary Figure 2, H and I, available online).
To further verify the relationships between MYC and MYCLos, we also examined their expression levels in 50 human primary colorectal tissue samples (25 normal colon and 25 CRC tissue samples) classified into four groups by MYC expression level (Figure 2H). By comparing the expression levels of MYCLo-1, -2, and -3 in the four groups, we found that expression levels of MYCLo-1, -2, and -3 are statistically significantly associated with MYC expression level (Figure 2, I-K). In addition, MYCLos are highly expressed in CRC cell lines (Figure 1G; Supplementary Figure 2, J and K, available online) that harbor MYC overexpression (Figure 2A).
To test whether MYC regulates MYCLo-1, -2, and -3 at the transcription level, we exploited a luciferase reporter assay. The promoter activities of the MYCLos were reduced by MYC knockdown, indicating that MYC induces the MYCLos expression at the transcription level (Figure 2, L-N). Using the database of open chromatin TFBS by ChIP-seq from ENCODE/Open Chrom (UT Austin) (17), we also observed strong and common binding of MYC to the promoters of the MYCLos in various cells (Figure 2, O-Q). These results suggest that the MYC-induced MYCLos are directly regulated by the transcription enhancer MYC at the transcription level.
Effect of MYC-Induced MYCLos on Cell Proliferation and Expression of MYC Target Genes Such as CDKN1A and CDKN2B
The proto-oncogene MYC accelerates cell proliferation by regulating many cell cycle regulator genes (10). To determine whether MYC-induced MYCLos are involved in the MYC function, the effect of MYCLos in cell proliferation was examined using siRNAs targeting MYCLos (siMYCLos). In HCT116 (colorectal cancer) and PC3 (prostate cancer) cells, knockdown of MYCLo-1, -2, or -3 as well as MYC resulted in decreased cell proliferation (siCTRL, 3.63056±0.20821, siMYCLo-2, 2.61959±0.0482, P < .001) (Figure 3A) (siCTRL, 3.94523±0.2959, siMYCLo-2, 3.07757±0.26933, P = .0048) (Figure 3B). In addition, we exogenously introduced the MYC-induced MYCLos to confirm their function in cell proliferation (Supplementary Figure 3A, available online). Consistent with the knockdown experiments, overexpression of the MYCLos in normal colon-derived fibroblasts resulted in enhanced cell proliferation (siCTRL, 2.77902±0.13299, siMYCLo-2, 3.56119±0.18969, P < .001) (Figure 3C). These results indicate that the MYC-induced MYCLos such as MYCLo-1, -2, and -3 are supportive to MYC function by enhancing cell proliferation.
Figure 3.
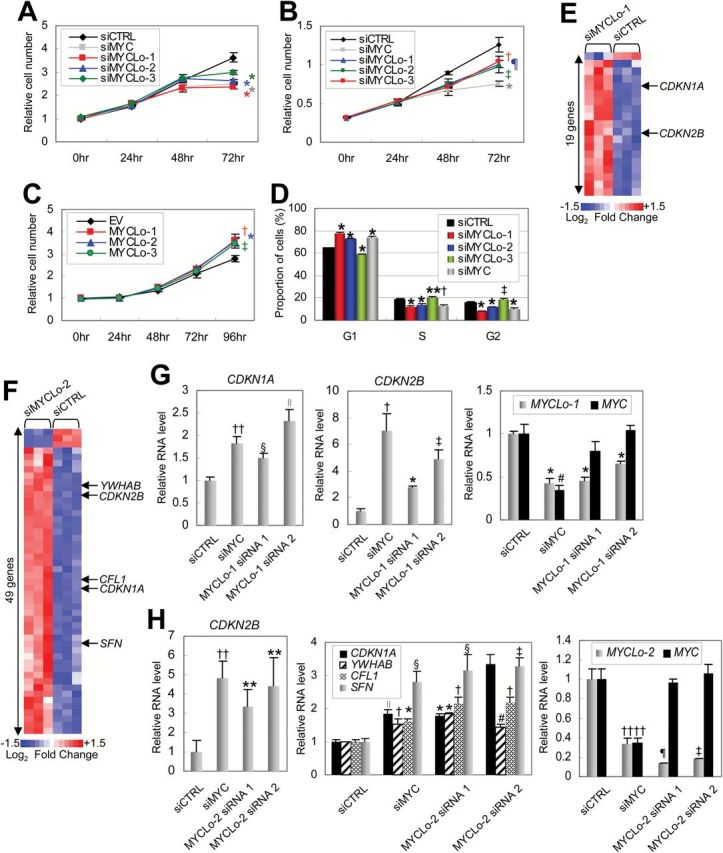
Effect of MYCLos knockdown on cell proliferation and cell cycle progression. A and B) Cell proliferation assays. HCT116 (n = 5, A) or PC3 (n = 4, B) cells were treated with siRNAs (final concentration: 50nM), as indicated and subjected to proliferation assay every 24 hours (†P = .013; ‡P = .0048; ¶P = .009; *P < .001). Error bars represent standard deviation. C) Cell proliferation assay. CCD-18Co cells were transfected with pcDNA3.3 plasmids (EV) expressing MYCLo-1, -2, or -3 and subjected to a cell proliferation assay every 24 hours (n = 3) (*P = .001; †P = .0045; ‡P = .0025). Error bars represent standard deviation. D) Proportion of cells in each cell cycle phase determined by flow cytometry analysis. HCT116 cells were treated with siRNAs (50nM) as indicated and analyzed 48 hours after transfection. Data are mean ± SD of three independent experiments and each measured in triplicate (**P = .047; †P = .0085; ‡P = .00662; *P < .001). E) Heatmap representing cell cycle regulator genes dysregulated by MYCLo-1. The genes that are commonly regulated by MYC are indicated. Expression values displayed in gradient of red and blue are Log2-transformed fold change. The list of the dysregulated genes is available in Supplementary Table 4 (available online). F) Heatmap representing cell cycle regulator genes dysregulated by MYCLo-2. The genes that are commonly regulated by MYC are indicated. Expression values displayed in gradient of red and blue are Log2-transformed fold change. The list of the dysregulated genes is available in Supplementary Table 4 (available online). G) Quantitative real-time polymerase chain reaction (qRT-PCR) results showing MYC-independent regulation of CDKN1A (left) and CDKN2B (middle) expression by MYCLo-1 knockdown (right). HCT116 cells were treated with 50nM siRNAs targeting MYC or MYCLo-1 for 72 hours. Data are mean ± SD of three independent experiments, measured in triplicate (††P = .003; §P = .0017; ∥P = .013; †P = .013; ‡P = .001; #P = .002; *P < .001). H) qRT-PCR results showing MYC-independent regulation of CDKN2B (left), CDKN1A, YWHAB, CFL1, and SFN (middle) expression by MYCLo-2 knockdown (right). HCT116 cells were treated with 50nM siRNAs targeting MYC or MYCLo-2 for 72 hours. Data are mean ± SD of three independent experiments and each measured in triplicate (**P = .03; ††P = .002; ∥P = .003; §P = .01; †P = .001; ‡P = .005; #P = .016; ¶P = .004; *P < .001). Statistical significance was determined by the unpaired two-sided Student’s t test.
MYC enhances cell cycle progression by regulating cell cycle arrests, leading to an increase of cell proliferation rate (10). To investigate if MYCLos also have a function in cell cycle regulation, we analyzed the proportion of cell populations in each cell cycle phase (G1, S, and G2). Knockdown of MYCLo-1 or -2 resulted in accumulation of cells in the G1 phase, suggesting that MYCLo-1 and -2 are involved in the G1/S transition (Figure 3D). Knockdown of MYCLo-3 led to accumulation of cells in the S and G2 phases, suggesting the function of MYCLo-3 in G2 phase regulation (G1 phase, siCTRL, 64.58±0.34771, siMYCLo-1, 77.97333±0.70501, P < .001, siMYCLo-2, 73.00667±0.4517, P < .001, siMYCLo-3, 59.25±0.42884, P < .001, siMYC, 74.32333±0.97552, P < .001; S phase, siCTRL, 18.82±0.4613, siMYCLo-1, 12.11333±0.50501, P < .001, siMYCLo-2, 13.95±0.5656, P < .001, siMYCLo-3, 20.55333±0.82779, P = .047, siMYC, 12.93667±1.26508, P < .0085; G2 phase, siCTRL, 15.85667±0.35233, siMYCLo-1, 8.59333±0.02887, P < .001, siMYCLo-2, 11.82±0.31512, P < .001, siMYCLo-3, 10.35667±0.37634, P = .00662, siMYC, 18.80667±0.67419, P < .001) (Figure 3D).
To identify cell cycle regulator genes modulated by the MYC-induced MYCLos (MYCLo-1, -2, and -3), we used nCounter Virtual Cell Cycle Gene Sets of the NanoString Gene Expression Assay. Of 183 cell cycle regulator genes, a cohort of genes were detected as downstream genes of MYCLos (P < .05, greater than 1.5-fold change): 19 genes regulated by MYCLo-1 (Figure 3E; Supplementary Table 4-2, available online), 49 genes regulated by MYCLo-2 (Figure 3F; Supplementary Table 4-3, available online), and 13 genes regulated by MYCLo-3 (Supplementary Figure 3B and Supplementary Table 4-4, available online). We also found a subset of genes that are commonly regulated by MYC and the MYCLos, by profiling cell cycle regulator genes dysregulated by MYC knockdown (Supplementary Figure 3C and Supplementary Table 4-1, available online).
Using qRT-PCR, we confirmed that elevated expression levels of CDKN1A and CDKN2B by knockdown of MYCLo-1 or -2 as well as MYC knockdown (Figure 3, G and H, left and middle panels; Supplementary Figure 3D, available online). We further verified a few more cell cycle regulator genes such as YWHAB (14-3-3 alpha), SFN (14-3-3 sigma), and CFN (cofilin) that are also regulated by MYC and MYCLo-2 (Figure 3H, middle panel). These elevations are consistent with the effect of MYC knockdown, albeit independent of MYC, Miz-1, and p53 expression levels (Figure 3, G and H, right panels; Supplementary Figure 3D, available online).
MYCLos-Mediated Transcriptional Regulation of CDKN1A and CDKN2B in the Distal Promoter Regions
To demonstrate the mechanism by which the MYC-regulated lncRNAs repress CDKN1A and CDKN2B expression, we amplified two different regions in the promoters of CDKN1A and CDKN2B genes, respectively, one including the proximal MYC-binding (MB) region and the other including the upstream region without the MB region (Supplementary Figure 4, A and B, available online), and exploited the amplified regions for testing promoter activity using a luciferase reporter assay (Figure 4, A and B; Supplementary Figure 4, C and D, available online).
Figure 4.
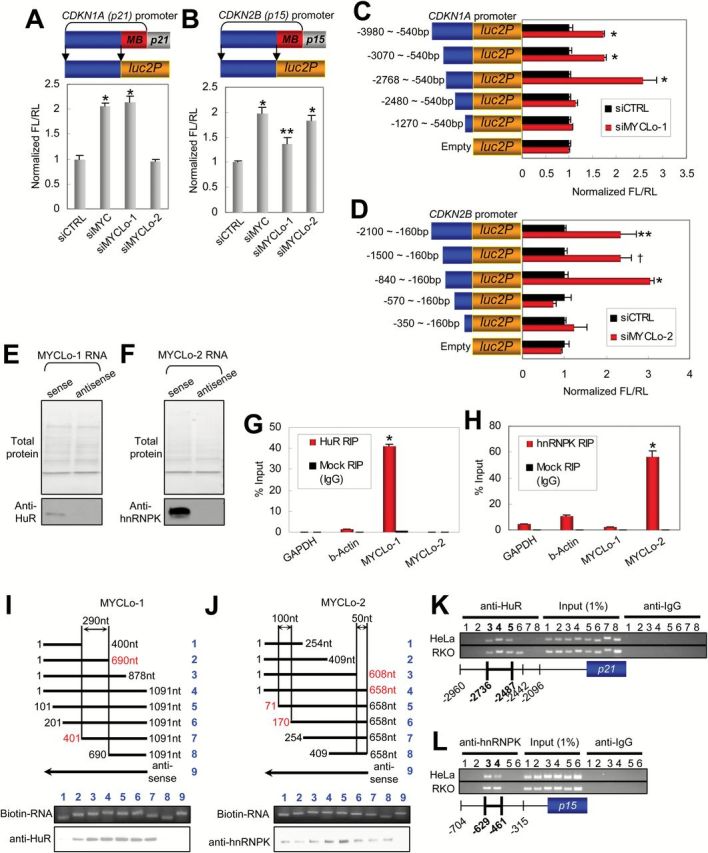
Interactions between MYCLo-1/2 and RNP binding proteins (HuR and hnRNPK) and their involvement in the regulation of 5’ promoter activities of CDKN1A and CDKN2B. A and B) Schematics (top) of pGL4 luciferase vectors inserted with distal 5’ promoter regions (blue bars) of CDKN1A (A) and CDKN2B (B). Red bars depict proximal promoter regions where MYC binds (MB). Luciferase reporter assays (bottom) representing transcription activity of the distal 5’ promoter regions of CDKN1A (A) and CDKN2B (B) in HCT116 cells transfected with the pGL4 luciferase vectors and siRNAs as indicated. Data are mean ± SD of three independent experiments and each measured in triplicate (**P = .024; *P < .001). C and D) Luciferase reporter assays to identify specific 5’ promoter regions of CDKN1A (C) and CDKN2B (D), where MYCLo-1 and -2 regulate their promoter activities, respectively. Data are mean ± SD of three independent experiments and each measured in triplicate (**P = .027; †P = .014; *P < .001). E and F) RNA pull-down assays showing the interactions between MYCLo-1 and HuR (E), and MYCLo-2 and hnRNPK (F). Total protein in SDS-PAGE gel was analyzed by liquid chromatography–tandem mass spectrometry. Western blot data are representative of two independent experiments. G and H) RIP assays and quantitative real-time polymerase chain reaction detection demonstrating the specific interactions between HuR and MYCLo-1 (G), and hnRNPK and MYCLo-2 (H). Data are mean ± SD of three independent experiments and each measured in triplicate (*P < .001). I and J) Deletion mapping analyses showing the importance of central region of MYCLo-1 (I, 401~690bp) and of marginal regions of MYCLo-2 (J, 71~170bp and 608~658bp) in the interactions with HuR and hnRNPK, respectively. Data are representative of three independent experiments. K and L) ChIP assays showing that HuR binds to the specific 5’ promoter region of CDKN1A, where MYCLo-1 regulates its transcription activity (K) and that hnRNPK binds to the specific 5’ promoter region of CDKN2B, where MYCLo-2 regulates its transcription activity (L). Data are representative of three independent experiments.
We first sought to test if MYCLo-1 and -2 are involved in the transcriptional regulation of CDKN1A and CDKN2B in the proximal MB regions on which MYC binds with the other MYC-associated factors. Knockdown of MYC induced promoter activities of CDKN1A and CDKN2B genes; however, their promoter activities were not statistically significantly affected by knockdown of MYCLo-1 or -2 (Supplementary Figure 4, C and D, available online). Interestingly, the knockdown of MYC also induced their promoter activities in the distal promoter regions without the MB region, and knockdown of MYCLo-1 or -2 also induced the promoter activities of CDKN1A and/or CDKN2B (Figure 4, A and B, available online). These results indicate that MYC can control transcription activity of CDKN1A and CDKN2B not only in the proximal region but also in the distal regions of their promoters. In addition, the results also demonstrate that MYCLo-1 and -2 are implicated in the MYC-regulated transcription activity of CDKN1A and CDKN2B genes in their distal promoter regions.
To further identify the specific promoter regions of CDKN1A and CDKN2B regulated by MYCLo-1 and -2, we serially deleted the promoter region from the distal side and tested their promoter activities (Figure 4, C and D, left panels). The luciferase reporter assay using the deleted genomic regions of CDKN1A promoter indicated that the CDKN1A promoter region approximately between -2768bp and -2480bp is critical for CDKN1A regulation by MYCLo-1 (Figure 4 C). The luciferase reporter assay using the deleted genomic regions of CDKN2B promoter also demonstrated that the CDKN2B promoter region approximately between -840bp and -570bp is critical for CDKN2B regulation by MYCLo-2 (Figure 4 D). These results reveal the specific promoter regions that involve MYCLos in the MYC-mediated transcriptional regulation of CDKN1A and CDKN2B.
Role of Physical Interactions Between MYCLos and RNA Binding Proteins Such as HuR and hnRNPK
To investigate the mechanism by which MYCLos function, we sought to identify the RNA-binding proteins for MYCLo-1 and -2 using RNA pull-down assays. By incubating biotinylated RNA or antisense RNA of the MYCLos with streptavidin beads and nuclear cell extracts, we isolated proteins specifically associated with MYCLo-1 or -2 and resolved them on an SDS-PAGE gel followed by mass spectrometry of the RNA-associated total proteins (Figure 4, E and F, upper panels). As a result, we identified 10 proteins associated with MYCLo-1 and 8 proteins associated with MYCLo-2 (Supplementary Figure 4, E and F, available online). Of the identified proteins, we found RNA-binding protein ELAVL1 (HuR) in the MYCLo-1 pull-down assay (Supplementary Figure 4E, available online). In addition, we found hnRNPK and hnRNPR as RNA-binding proteins interacting with MYCLo-2 (Supplementary Figure 4F, available online). The counted number of peptides indicates that MYCLo-2 more preferentially interacts with hnRNPK (46 peptides) than hnRNPR (6 peptides) (Supplementary Figure 4F, available online). By using an RNA-Protein Interaction Prediction program (RPISeq) (18), we also observed the high probability of HuR-MYCLo-1 and hnRNPK-MYCLo-2 interactions (Figure 4, G and H). The interactions were also verified by using Western blot analysis (Figure 4, E and F, lower).
To further confirm the interactions (HuR-MYCLo-1 and hnRNPK-MYCLo-2), we conducted RNA immunoprecipitation (RIP) using antibodies against HuR or hnRNPK. Consistent with the results of RNA pull-down assay, MYCLo-1 is abundant in RIP samples using HuR antibodies as compared with the samples using nonspecific (negative control using IgG antibody) antibodies (Figure 4G), and MYCLo-2 is enriched by RIP using hnRNPK antibodies (Figure 4H). To determine which specific regions of MYCLo-1 and -2 interact with HuR or hnRNPK, we performed deletion-mapping experiments using truncated MYCLo-1 or MYCLo-2 (Figure 4, I and J, upper panels). The analyses suggest that HuR interacts with the central region (401~690bp) of MYCLo-1 (Figure 4I, lower panel). On the other hand, the various 5’ or 3’ deletions of MYCLo-2 do not affect the interaction between MYCLo-2 and hnRNPK (Figure 4J, lower panel). However, the enrichment of hnRNPK in full-length (#4) and partially deleted (#5, deleted in 1-70nt) RNAs of MYCLo-2 suggests that hnRNPK might interact with at least two regions of MYCLo-2 (71-170nt and 608-658nt). The prediction analysis (RPISeq) using the sequence of the central regions of MYCLo-1 and -2 also shows that the regions are strongly predicted to interact with the RNA-binding proteins, supporting the conclusion (Supplementary Figure 4, G and H, available online).
We identified the specific regions in the promoters of CDKN1A (-2768bp~-2480bp) and CDKN2B (-840bp~-570bp) genes that implicate MYCLo-1 and -2 in transcriptional regulation of the genes (Figure 4, C and D). To determine whether MYCLo-1/HuR and MYCLo-2/hnRNPK complexes are involved in the regulation at the specific promoter regions by direct interaction, we performed a chromosome immunoprecipitation (ChIP) assay. By using antibodies against HuR, we observed that HuR interacts with the specific region (-2736bp~-2487bp) in the promoter of CDKN1A (Figure 4K). The result of the ChIP assay using antibodies against hnRNPK also shows that hnRNPK interacts with the specific region (-629bp~-461bp) of the CDKN2B promoter (Figure 4L). The coincidence of MYCLos function and HuR/hnRNPK binding strongly suggests that HuR is implicated in the transcriptional repression of CDKN1A through interaction with MYCLo-1 and that MYCLo-2 represses CDKN2B transcription by interacting with hnRNPK.
Oncogenic Function of MYCLo-2
By using northern blot, we confirmed the overexpression of MYCLo-2 (CCAT6) in CRC cell lines (Supplementary Figure 5A, available online). In addition, the comparison of individual pairs of the tissue samples clearly shows that most CRC tissues harbor very high expression levels of MYCLo-2 and that MYCLo-2 is more highly expressed in 46 CRC tissues of the 52 pairs of the matched tissue samples (Figure 5A). In addition, MYCLo-2 is also overexpressed in PC cells and tissues, compared with normal prostate-derived cells (Figure 5, B and C).
Figure 5.
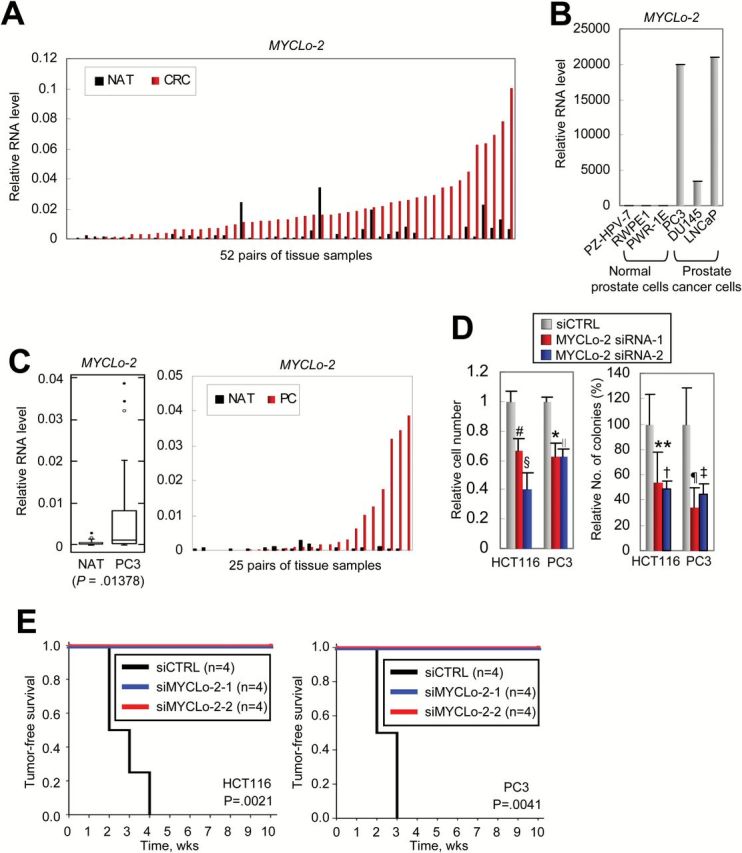
Function of MYCLo-2 in cancer cell transformation and tumorigenesis. A) Comparison of expression level of MYCLo-2 in each pair of CRC tissues (red) and their matched normal adjacent tissues (NATs) (black). B) Quantitative real-time polymerase chain reaction (qRT-PCR) results showing significant overexpression of MYCLo-2 in PC-derived cell lines, compared with normal prostate-derived cell lines. Data are mean ± SD of three independent experiments and each measured in triplicate. C) Box and whisker plot (left) showing the significant overexpression of MYCLo-2 in PC, compared with their matched NAT (25 pairs of primary tissues). Comparison of expression level (right) of MYCLo-2 in each pair of PC tissues (red) and their matched NATs (black). D) Soft agar colony formation assays indicating that knockdown of MYCLo-2 causes a decreased number of cells (left) and colonies (right) in CRC-derived (HCT116) and PC-derived (PC3) cells. Data are mean ± SD of three independent experiments, each measured in triplicate (#P = .0066; §P = .00315; *P = .01185; ∥P = .00151; **P = .003322; †P = .001821; ¶P = .0107; ‡P = .02474). E) Kaplan-Meier plot of tumor-free survival analysis in athymic nude mice xenografted with siRNA-treated HCT116 (left) or PC3 (right) cells as indicated. The Kaplan-Meier method was used to estimate survival curves, and the log-rank test was used to test for differences between curves using SPSS Statistical Software (SPSS Inc., Chicago, IL).
The knockdown of MYCLo-2 statistically significantly reduced the number of cells and colonies in colony formation assays in vitro, indicating that MYCLo-2 has a role in tumor cell transformation (HCT116 cell number, siCTRL, 1±0.06391, siMYCLo-2-1, 0.66259±0.08495, P = .0066, siMYCLo-2-2, 0.66259±0.08495, P = .00315; PC3 cell number, siCTRL, 1±0.02433, siMYCLo-2-1, 0.62727±0.08494, P = .01185, siMYCLo-2-2, 0.6278±0.04929, P = .00151; HCT116 colony number, siCTRL, 100±23.52415, siMYCLo-2-1, 53.90625±23.85053, siMYCLo-2-2, 48.4375±6.50521; PC3 colony number, siCTRL, 100±28.11947, siMYCLo-2-1, 34.04255±15.53823, siMYCLo-2-2, 44.68085±8.14832) (Figure 5D; Supplementary Figure 5B, available online). We also examined the knockdown effect of MYCLo-2 in tumorigenesis by using in vivo xenografts of tumor cells transfected with MYCLo-2 siRNAs. Consistent with our data supporting the oncogenic function of MYCLo-2, tumor development was statistically significantly inhibited by MYCLo-2 depletion in both CRC-derived (HCT116) and PC-derived (PC3) cells (Figure 5E; Supplementary Figure 5C, available online). These results demonstrate that MYCLo-2 is not only overexpressed in tumors but also involved in tumorigenesis and tumor growth.
Discussion
In this work, we identified novel lncRNAs differentially regulated in CRC by profiling CRC cells and tissues. In addition, MYC-regulated lncRNAs named MYCLos were further identified. MYCLos participate in cell proliferation and cell cycle regulation by modulation of expression of known MYC target genes such as CDKN1A and CDKN2B. We further found that MYCLo-1 physically interacts with HuR and MYCLo-2 with hnRNPK. Further data support that these interactions might be involved in the regulation of MYC targets such as CDKN1A and CDKN2B. Lastly, we show that knockdown of MYCLo-2 inhibits cancer cell transformation and tumorigenesis.
In addition to the recent studies that identified a couple of CRC-associated lncRNAs such as CCAT1 and CCAT2, we discovered and verified more novel CRC-associated lncRNAs (7–9). In addition, we identified the lncRNAs that are downregulated in CRC, suggesting their putative tumor suppressor function. The up- or downregulation of a newly identified lncRNA named CCAT3-8 suggests the function of lncRNAs in CRC. Furthermore, the dysregulation of CCAT3-8 in CRC tissue samples supports our suggestion for putative function of CCAT3-8 in cancer pathogenesis. Indeed, our results show that CCAT6 (MYCLo-2) have a role in oncogenesis. To elucidate the oncogenic or tumor-suppressive function of other lncRNAs, further studies are required.
By further identification of MYC-regulated lncRNAs in CRC-associated lncRNAs, we also revealed an unknown mechanism of MYC function through lncRNAs. MYC-regulated lncRNAs, such as MYCLo-1 and MYCLo-2, regulate known MYC target genes, such as CDKN1A and CDKN2B, through the interactions with HuR and hnRNPK. To date, many studies have reported a potential role of the RNA-binding proteins such as hnRNPK and HuR in cancer (19–21). However, no notable mechanism of their function in cancer has been reported yet. Here, our results raise the possibility that lncRNAs associated with hnRNPK or HuR might be related to their function in cancer.
Recent reports show that CDKN1A (p21) is a critical target gene of MYC. MYC-repressed CDKN1A expression is involved in various MYC functions such as cell cycle, cell death, cellular senescence, tumorigenesis, epithelial-mesenchymal transition, and stemness (15,22–25). Our data show that MYC-regulated lncRNAs are involved in transcriptional repression of CDKN1A, suggesting lncRNA involvement in the versatile functions of MYC. Although we tried to establish stable knockdown cells of MYCLo-2, the cells harboring MYCLo-2 knockdown disappeared from the mixed cell population in a couple of weeks and single clonal cells with MYCLo-2 knockdown were extinct, suggesting that knockdown of MYCLo-2 inhibits cell cycle progression at the early stage; however, it may have a role in other processes such as cellular senescence and cell death.
Although we show that lncRNAs such as MYCLo-2 are regulated by MYC in a wide range of cancer types, we verified their overexpression and oncogenic function in colorectal and prostate cancer. To expand the knowledge of MYCLo-2 in other types of cancer, further investigation is required. We identified the lncRNAs in human cancers. Therefore, their homologous lncRNAs such as Myclos in rodents are not yet known. For better understanding of the lncRNAs in vivo, the identification of their counterparts in rodents is highly needed. Finally, other lncRNAs identified in this study also remain to be characterized in future studies.
The transcription factor MYC is a well-known regulator of cell stemness (26). It was recently reported that dicer deficiency may cause deregulation of lncRNA through MYC dysregulation in embryonic stem cells (27). The results also support that MYC-regulated lncRNAs have a critical role not only in oncogenesis but also in other MYC functions. Although we identified and characterized several lncRNAs associated with CRC and MYC, more lncRNAs remain to be explored. Our findings suggest new targets of cancer and MYC-associated pathogenesis.
Funding
This work was supported by grants from the National Cancer Institute (EDRN U01 CA152758).
Supplementary Material
TK and CMC contributed to project coordination. TK conceived the idea. TK and YJJ designed the project. TK identified and registered CCATs and MYCLos. TK, YJJ and RC performed experiments. TK, YJJ, JHL, YP, SHK, and ET analyzed data. HA provided quantitative real-time polymerase chain reaction and NanoString Gene Expression assays data. TK and CMC prepared the manuscript.
We especially thank Dr. Robert J. Coffey (Vanderbilt University Medical Center) for the support and advice that contributed to this manuscript. We also thank Dr. Thomas D. Schmittgen (The Ohio State University) for the revision of this manuscript.
References
- 1. Mattick JS, Makunin IV. Small regulatory RNAs in mammals. Hum Mol Genet. 2005;14 Spec No 1:R121–R132. [DOI] [PubMed] [Google Scholar]
- 2. Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Khalil AM, Guttman M, Huarte M, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106(28):11667–11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Orom UA, Derrien T, Beringer M, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143(1):46–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Calin GA, Liu CG, Ferracin M, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12(3):215–229. [DOI] [PubMed] [Google Scholar]
- 7. Ling H, Spizzo R, Atlasi Y, et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013;23(9):1446–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nissan A, Stojadinovic A, Mitrani-Rosenbaum S, et al. Colon cancer associated transcript-1: a novel RNA expressed in malignant and pre-malignant human tissues. Int J Cancer. 2012;130(7):1598–1606. [DOI] [PubMed] [Google Scholar]
- 9. Kim T, Cui R, Jeon YJ, et al. Long-range interaction and correlation between MYC enhancer and oncogenic long noncoding RNA CARLo-5. Proc Natl Acad Sci U S A. 2014;111(11):4173–4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dang CV. MYC on the Path to Cancer. Cell. 2012;149(1):22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Muzny DM, et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Seoane J, Pouponnot C, Staller P, Schader M, Eilers M, Massague J. TGFbeta influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nat Cell Biol. 2001;3(4):400–408. [DOI] [PubMed] [Google Scholar]
- 13. Inoue S, Hao Z, Elia AJ, et al. Mule/Huwe1/Arf-BP1 suppresses Ras-driven tumorigenesis by preventing c-Myc/Miz1-mediated down-regulation of p21 and p15. Genes Dev. 2013;27(10):1101–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Staller P, Peukert K, Kiermaier A, et al. Repression of p15INK4b expression by Myc through association with Miz-1. Nat Cell Biol. 2001;3(4):392–399. [DOI] [PubMed] [Google Scholar]
- 15. Oskarsson T, Essers MA, Dubois N, et al. Skin epidermis lacking the c-Myc gene is resistant to Ras-driven tumorigenesis but can reacquire sensitivity upon additional loss of the p21Cip1 gene. Genes Dev. 2006;20(15):2024–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rochlitz CF, Herrmann R, de Kant E. Overexpression and amplification of c-myc during progression of human colorectal cancer. Oncology. 1996;53(6):448–454. [DOI] [PubMed] [Google Scholar]
- 17. Lee BK, Bhinge AA, Battenhouse A, et al. Cell-type specific and combinatorial usage of diverse transcription factors revealed by genome-wide binding studies in multiple human cells. Genome Res. 2011;22(1):9–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Muppirala UK, Honavar VG, Dobbs D. Predicting RNA-protein interactions using only sequence information. BMC Bioinformatics. 2011;12:489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gao R, Yu Y, Inoue A, Widodo N, Kaul SC, Wadhwa R. Heterogeneous nuclear ribonucleoprotein K (hnRNP-K) promotes tumor metastasis by induction of genes involved in extracellular matrix, cell movement, and angiogenesis. J Biol Chem. 2006;288(21):15046–15056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carpenter B, McKay M, Dundas SR, Lawrie LC, Telfer C, Murray GI. Heterogeneous nuclear ribonucleoprotein K is over expressed, aberrantly localised and is associated with poor prognosis in colorectal cancer. Br J Cancer. 2006;95(7):921–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang J, Guo Y, Chu H, Guan Y, Bi J, Wang B. Multiple Functions of the RNA-Binding Protein HuR in Cancer Progression, Treatment Responses and Prognosis. Int J Mol Sci. 2013;14(5):10015–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Newbold A, Salmon JM, Martin BP, Stanley K, Johnstone RW. The role of p21 and p27 in HDACi-mediated tumor cell death and cell cycle arrest in the Emu-myc model of B-cell lymphoma. Oncogene. 2013;33:5415–5423. [DOI] [PubMed] [Google Scholar]
- 23. Zhang J, Lou X, Yang S, et al. BAG2 is a target of the c-Myc gene and is involved in cellular senescence via the p21(CIP1) pathway. Cancer Lett. 2011;318(1):34–41. [DOI] [PubMed] [Google Scholar]
- 24. Liu M, Casimiro MC, Wang C, et al. p21CIP1 attenuates Ras- and c-Myc-dependent breast tumor epithelial mesenchymal transition and cancer stem cell-like gene expression in vivo. Proc Natl Acad Sci U S A. 2009;106(45):19035–19039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Carnero A, Beach DH. Absence of p21WAF1 cooperates with c-myc in bypassing Ras-induced senescence and enhances oncogenic cooperation. Oncogene. 2004;23(35):6006–6011. [DOI] [PubMed] [Google Scholar]
- 26. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. [DOI] [PubMed] [Google Scholar]
- 27. Zheng GX, Do BT, Webster DE, Khavari PA, Chang HY. Dicer-microRNA-Myc circuit promotes transcription of hundreds of long noncoding RNAs. Nat Struct Mol Biol. 2014;21(7):585–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.