Abstract
Summary: Parallel visualization of multiple individual human genomes is a complex endeavor that is rapidly gaining importance with the increasing number of personal, phased and cancer genomes that are being generated. It requires the display of variants such as SNPs, indels and structural variants that are unique to specific genomes and the introduction of multiple overlapping gaps in the reference sequence. Here, we describe GenPlay Multi-Genome, an application specifically written to visualize and analyze multiple human genomes in parallel. GenPlay Multi-Genome is ideally suited for the comparison of allele-specific expression and functional genomic data obtained from multiple phased genomes in a graphical interface with access to multiple-track operation. It also allows the analysis of data that have been aligned to custom genomes rather than to a standard reference and can be used as a variant calling format file browser and as a tool to compare different genome assembly, such as hg19 and hg38.
Availability and implementation: GenPlay is available under the GNU public license (GPL-3) from http://genplay.einstein.yu.edu. The source code is available at https://github.com/JulienLajugie/GenPlay
Contact: eric.bouhassira@einstein.yu.edu or julien.lajugie@gmail.com
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Allele-specific analyses, which are much more powerful when they are performed on phased genomes, can shed light onto myriad questions related to genotype/phenotype correlations, parental imprinting, X chromosome inactivation, DNA methylation, DNA replication and chromatin structure. Most genomes produced to date are unphased, but technology is now available to reliably generate phased genomes (Browning and Browning, 2009; Kitzman et al., 2011; Lajugie and Bouhassira, 2011; Peters et al., 2012; Roach et al., 2010; Yang et al., 2011). One current difficulty in performing allele-specific analysis is the lack of software to visualize and compare expression, epigenetic and other functional genomics data obtained from multiple phased genomes. Visualization of such data is difficult because of the presence of large number of indels and structural variants in humans. GenPlay Multi-Genome, a major update to GenPlay, was developed to help resolve some of these problems. GenPlay is a multi-platform active genome browser and analyzer written in Java. As other browsers (Blankenberg et al., 2010; Challis et al., 2012; Faith et al., 2007; Fernandez-Suarez and Schuster, 2010; Lukashin et al., 2011; Nicol et al., 2009; Sandve et al., 2010; Stein et al., 2002), GenPlay can display RNA-seq, ChIP-seq, Methyl-seq or TimEX-seq and a variety of DNA sequence and annotation tracks. In addition, GenPlay is unique because it can rapidly transform any loaded data using >100 pre-programmed operations (Lajugie and Bouhassira, 2011).
2 METHODS
To keep track and visualize all the deletions and insertions between genomes to be compared, GenPlay creates on the fly a global system of genomic coordinates that represents the sum of all the genomes to be analyzed in parallel in a multi-genome session (Supplementary Fig. S1). We termed this global system of coordinate a meta-reference genome. With this approach, two genomes (G1 and G2) mapped relative to hg19 are represented in memory by all the offsets between hg19 and the meta-reference-genome, G1 and the meta-reference-genome and G2 and the meta-reference genome. The meta-reference genome is created using user-provided variant calling format (VCF) files, which are text files containing the position, the sequence and genotype information for all the variants in one or more individuals (Danecek et al., 2011). At least 20 genomes can be loaded concurrently in GenPlay on a standard workstation with at least 4 GB of RAM. The meta-reference genome created by GenPlay is a simplified version of the threaded blockset concept that was developed by Blanchette et al. (2004) to generalize the classic notion of multiple alignments. Although software such as TBA/MultiZ is ideal for interspecies alignments, the meta-reference genome concept as implemented in GenPlay is most useful for comparison of phased or unphased human genomes and can also be used for interstrain or interspecies analysis provided that all genome studied are closely related.
3 RESULTS
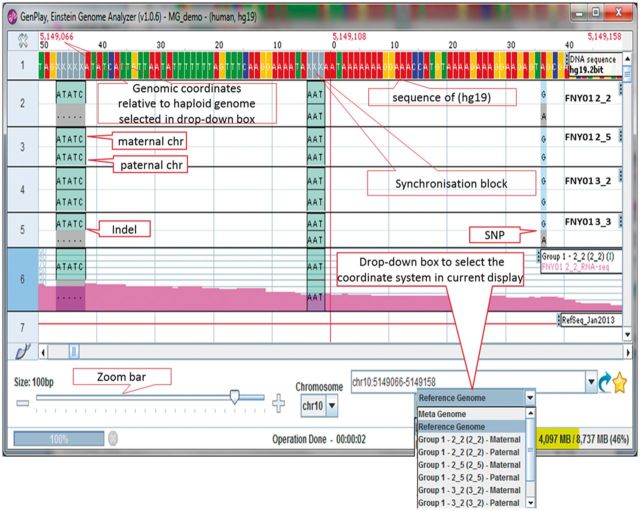
Once the meta-reference genome is computed, GenPlay can display the genetic variants present in all of the loaded genomes. Insertions that are present in only some of the genomes are replaced by synchronization blocks (Fig. 1 and Supplementary Fig. S2). Inversions are displayed as the sum of a deletion plus an insertion. Filters are available to select variants according to any of the fields present in the VCF file (genotype, phasing, allele depth, quality value, genotype probability, etc.). Filters can be combined to create complex filters. In multi-genome mode, GenPlay becomes a powerful VCF file browser that can be used to visualize the structure of multiple genomes in a graphical interface. GenPlay also allows users to visualize, analyze and transform expression and epigenetic data at the same time as the genetic data. In practice, the genetic variants are displayed as stripes, which can be superimposed to tracks containing expression, epigenetic data or annotations.
Fig. 1.
Screenshot of a multi-genome session illustrating the synchronization blocks
Data from VCFfiles obtained from RNA-seq, Chip-seq, Methyl-seq or any other experiments performed on phased genomes can be converted to allele-specific data tracks based on the phasing information present in the VCFfile. The converted tracks can be intersected with gene tracks or any other track to measure expression or any epigenetic mark in an allele-specific manner. An example of an allele-specific analysis performed in GenPlay can be found in a recent article exploring the regulation of DNA replication (Mukhopadhyay et al., 2014).
GenPlay Multi-Genome can be used to analyze data aligned on custom genomes. Currently, most expression and epigenetic analyses rely on alignment to a reference genome. This has two major drawbacks. First, blocks of polymorphic sequences are lost to the analysis because not all sequences that are present in humans exist in the reference genome. Second, genetic differences between the reference genomes and the sample being analyzed are a major cause of alignment errors that lead to inaccurate expression and epigenetic results. These problems can be eliminated by aligning RNA-seq, ChIP-seq or Methyl-seq experiments to the phased autologous genome from which the reads are derived, rather than to a reference genome. When a phased autologous genome is not available, an intermediate solution is to align the reads to the most closely related reference genome available. Recently, Dewey et al. have generated three race-specific reference sequences and have shown that alignment to a closely related genome decreases error rates (Dewey et al., 2011).
Alignments to autologous genomes can easily be performed using current aligners (Li and Durbin, 2009; Liu et al., 2012), but the results can be difficult to exploit because the resulting alignments are represented in genomic coordinate systems that are specific to each genome. This complicates interindividual comparisons and the exploitation of annotations and other reference data in public databases that are generally in the genomic coordinate system of the reference genome.
GenPlay Multi-Genome partly resolves these problems because data that are represented in any custom genomic coordinate system can be loaded concurrently with data represented in the reference genome coordinate system as long as a VCF file is available to create a meta-reference genome. In practice, users load a VCF file containing the genetic data specific to the individuals being studied and are then able to load expression and epigenetic data that have been aligned to these custom genomes. GenPlay can switch between genomic coordinates systems on the fly or search for variants or features represented in any of the coordinate systems summarized in the meta-genome. Users can also export their data into any of the coordinate systems that makeup the meta-reference genome, allowing for easy cross-referencing with other studies. A demo project that illustrates some of the capabilities of GenPlay Multi-Genome can be accessed at http://genplay.einstein.yu.edu/wiki/index.php/Projects#GenPlay_MultiGenome.2C_a_tool_to_compare_and_analyze_multiple_human_genomes_in_a_graphical_interface.
A currently useful application of GenPlay Multi-Genome is the visualization of all the sequence differences between the hg19 and hg38 assemblies. In an hg19/hg38 multi-genome session, annotation, expression or epigenetic data tracks in either the hg19 or the hg38 coordinate system can be loaded without any conversion. A tutorial describing this application can be accessed at http://genplay.einstein.yu.edu/wiki/index.php/GRCh37/hg19_GRCh38/hg38_Multi-Genome_Tutorial.
Generation of a VCF file comparing two or more genomes is not a trivial endeavor, but it can be accomplished using several approaches.
The VCF file describing the differences between the hg19 and hg38 assemblies was generated using the chain files used by the liftover software. Chain files describing the differences between assemblies are available for several species at the UCSC browser. We provide an application and a tutorial to convert chain files into VCF files. The tutorial, a link to the application and several links describing how to create chain files can be found here: http://genplay.einstein.yu.edu/wiki/index.php/How_to_Create_a_VCF_File_From_a_Chain_File
As discussed above, VCF files describing phased human genomes can be generated by quartet sequencing and other approaches. Once such a VCF file is created, it can be used to create a GenPlay Multi-Genome session that can be used to analyze functional genomic data aligned on any of the genome of the individuals described in the VCF file.
VCF files can also be generated from two FASTA files (created for instance with a de novo aligner) using genome aligners such as Last (Kielbasa et al., 2011) followed by Samtools (Li et al., 2009) to convert the *.maf fields into VCF files.
In conclusion, GenPlay Multi-Genome is an application that is useful for projects that require analysis and visualization of data expressed in multiple genomic coordinate systems.
Funding: J.L., N.F. and E.E.B. were supported by grants C024405 and C024172 from NYSTEM.
Conflict of interest: none declared.
Supplementary Material
REFERENCES
- Blanchette M, et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D, et al. Galaxy: a web-based genome analysis tool for experimentalists. Curr. Protoc. Mol. Biol. 2010 doi: 10.1002/0471142727.mb1910s89. Chapter 19, Unit 19 10 11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis D, et al. An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinformatics. 2012;13:8. doi: 10.1186/1471-2105-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey FE, et al. Phased whole-genome genetic risk in a family quartet using a major allele reference sequence. PLoS Genet. 2011;7:e1002280. doi: 10.1371/journal.pgen.1002280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, et al. Lightweight genome viewer: portable software for browsing genomics data in its chromosomal context. BMC Bioinformatics. 2007;8:344. doi: 10.1186/1471-2105-8-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Suarez XM, Schuster MK. Using the ensembl genome server to browse genomic sequence data. Curr. Protoc. Bioinform. 2010 doi: 10.1002/0471250953.bi0115s30. Chapter 1, Unit1 15. [DOI] [PubMed] [Google Scholar]
- Kielbasa SM, et al. Adaptive seeds tame genomic sequence comparison. Genome Res. 2011;21:487–493. doi: 10.1101/gr.113985.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzman JO, et al. Haplotype-resolved genome sequencing of a Gujarati Indian individual. Nat. Biotechnol. 2011;29:59–63. doi: 10.1038/nbt.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajugie J, Bouhassira EE. GenPlay, a multipurpose genome analyzer and browser. Bioinformatics. 2011;27:1889–1893. doi: 10.1093/bioinformatics/btr309. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CM, et al. SOAP3: ultra-fast GPU-based parallel alignment tool for short reads. Bioinformatics. 2012;28:878–879. doi: 10.1093/bioinformatics/bts061. [DOI] [PubMed] [Google Scholar]
- Lukashin I, et al. VISTA Region Viewer–a computational system for prioritizing genomic intervals for biomedical studies. Bioinformatics. 2011;27:2595–2597. doi: 10.1093/bioinformatics/btr440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R, et al. Allele-specific genome-wide profiling in primary erythroblasts reveal replication program organization. PLoS Genet. 2014;10:e1004319. doi: 10.1371/journal.pgen.1004319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol JW, et al. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics. 2009;25:2730–2731. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters BA, et al. Accurate whole-genome sequencing and haplotyping from 10 to 20 human cells. Nature. 2012;487:190–195. doi: 10.1038/nature11236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach JC, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–639. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandve GK, et al. The Genomic HyperBrowser: inferential genomics at the sequence level. Genome Biol. 2010;11:R121. doi: 10.1186/gb-2010-11-12-r121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, et al. Completely phased genome sequencing through chromosome sorting. Proc. Natl Acad. Sci. USA. 2011;108:12–17. doi: 10.1073/pnas.1016725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.