Abstract
Lipoprotein biogenesis in Gram-negative bacteria occurs by a conserved pathway, each step of which is considered essential. In contrast to this model, LoVullo and colleagues demonstrate that the N-acyl transferase Lnt is not required in Francisella tularensis or Neisseria gonorrhoeae. This suggests the existence of a more flexible lipoprotein pathway, likely due to a modified Lol transporter complex, and raises the possibility that pathogens may regulate lipoprotein processing to modulate interactions with the host.
TEXT
Lipoproteins are a diverse class of multifunctional, membrane-associated molecules. Their contributions to the bacterial cell range from essential processes, such as maintaining envelope architecture and stability, to assisting with and mediating host-pathogen interactions (1–3). Lipoproteins constitute a significant fraction of the outer membrane (OM) of Gram-negative bacteria and are recognized as a pathogen-associated molecular pattern by host cells (3, 4). Due to the high cost associated with lipoprotein mislocalization, Gram-negative bacteria have evolved a conserved mechanism for the processing and sorting of these molecules, to ensure they correctly reach their final destination. In this issue of the Journal of Bacteriology, LoVullo et al. (5) challenge the current paradigm for lipoprotein processing and sorting in Gram-negative bacteria.
As with the majority of proteins destined for the periplasm or OM, the N terminus of a newly synthesized lipoprotein contains a cleavable signal peptide, which typically directs the preprolipoprotein to the Sec general secretory pathway for translocation across the cytoplasmic or inner membrane (IM) to the periplasm (Fig. 1) (2, 6). The C-terminal end of the signal peptide contains a 4-amino-acid lipobox motif, terminating with an invariant cysteine in the +1 position (the N terminus of the mature lipoprotein). This cysteine provides the acylation site and is required for lipoprotein processing. In addition, residues in the +2, +3, and +4 positions adjacent to the lipobox cysteine act as signals that determine whether the lipoprotein is sorted to the OM (the default pathway) or remains in the IM (7, 8). Finally, a flexible tethering sequence links the N-terminal processing and sorting determinants to the mature functional region of the protein (2).
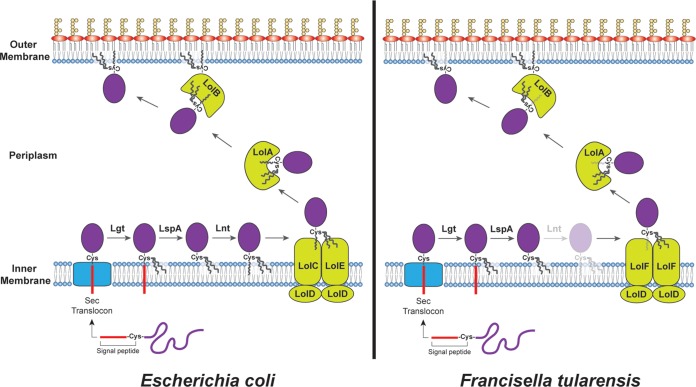
FIG 1.
Models for lipoprotein processing and sorting in Gram-negative bacteria. (Left) The current model for lipoprotein biogenesis in E. coli. The N-terminal signal peptide directs cytoplasmic preprolipoproteins to the Sec complex for translocation to the periplasm. Following passage through the Sec system, Lgt adds a diacylglyceride to the +1 cysteine, and LspA cleaves the signal peptide. Lnt then adds a third acyl chain to the newly available +1 cysteine amino group. The LolCDE transporter complex uses energy from ATP hydrolysis to extract the mature, triacylated lipoprotein from the IM. The LolA chaperone takes the lipoprotein from LolCDE and delivers it to the OM-anchored lipoprotein LolB, which then facilitates insertion of the lipoprotein into the OM. (Right) Lipoprotein biogenesis in F. tularensis, based on the results of LoVullo et al. (5). The Lol transporter complex in Francisella is composed of LolDF instead of LolCDE. The LolDF complex is able to recognize and extract diacylated as well as triacylated lipoproteins. Diacylated lipoproteins would result from loss of Lnt activity (indicated in the figure by increased transparency).
Following transport through the Sec translocon to the periplasm, the preprolipoprotein remains anchored to the IM by its N-terminal signal peptide (Fig. 1). Lgt, a preprolipoprotein diacylglyceryl transferase, catalyzes the addition of a diacylglyceride moiety to the sulfhydryl group of the +1 cysteine, forming a prolipoprotein (9). Next, the prolipoprotein signal peptidase Lsp cleaves the N-terminal amide bond of the +1 cysteine, releasing the signal peptide and leaving the lipoprotein anchored to the IM via its diacylated cysteine residue (10). With the +1 cysteine amino group now accessible, the final processing step requires the N-acyl transferase Lnt to catalyze the linkage of an additional acyl chain to the free amine, bringing the total number of acyl chains to three (Fig. 1) (11). The mature tricacylated lipoprotein can then be sorted to the OM via the Lol (Lipoprotein outer membrane localization) pathway or remain anchored in the IM, which is dictated by species-specific residues in the +2, +3, and +4 positions known as an Lol avoidance signal (12).
The Lol pathway is composed of three distinct components, an IM ATP-binding cassette (ABC) transporter-like complex (LolCDE in Escherichia coli), a periplasmic chaperone (LolA), and an OM lipoprotein (LolB) (Fig. 1). The LolCDE complex uses energy from ATP hydrolysis to extract mature lipoproteins (lacking an Lol avoidance signal) from the IM (13). The ATPase activity of the complex is provided by a homodimer of LolD, while LolC and LolE interact with LolD via their membrane-spanning domains. The periplasmic chaperone LolA captures lipoproteins from the LolCDE complex and then delivers its lipoprotein cargo to LolB in the OM (Fig. 1) (14, 15). Finally, LolB facilitates lipoprotein insertion into the inner leaflet of the OM. In some cases, the lipoprotein is transported across the OM bilayer to the cell surface.
The generally accepted model for lipoprotein processing and sorting in Gram-negative bacteria is based largely on experiments performed in E. coli. Given that lnt is essential in E. coli, addition of the third acyl chain is thought to be required for lipoprotein recognition by the Lol pathway. The study by LoVullo and colleagues (5) challenges this paradigm and suggests that the lipoprotein sorting pathway has greater flexibility than previously thought. LoVullo et al. began their study by analyzing a defined transposon mutant library of Francisella novicida (16), a close relative of the highly virulent human pathogen Francisella tularensis. F. tularensis is the causative agent of tularemia and a potential bioterrorism agent (17). They noted that the F. novicida transposon library did not contain insertions in the lgt, lsp, or lol genes, as expected for essential genes. However, two independent insertions were present in lnt, suggesting that, in contrast to E. coli, this gene is not essential in Francisella. LoVullo and colleagues confirmed this by constructing Δlnt deletion mutations in two different F. tularensis strains, including a human-pathogenic F. tularensis subsp. tularensis strain (5). The study further demonstrated that the known F. tularensis lipoprotein Tul4 (LpnA) shifts from a triacylated form in the wild-type strain to a diacylated form in the Δlnt background and that Tul4 and additional lipoproteins are still properly sorted to the OM in the Δlnt mutant. Moreover, the F. tularensis Δlnt mutants did not exhibit alterations in envelope integrity or other gross physiological defects. Thus, the lipoprotein sorting pathway in Francisella does not require Lnt activity and therefore presumably accepts diacylated substrates (Fig. 1).
LoVullo et al. (5) hypothesized that the Lol pathway of Francisella must contain some alternative functionality that allows it to recognize diacylated lipoproteins. Interestingly, the Lol system of F. tularensis lacks a gene for LolE, which in E. coli heterodimerizes with LolC to form the membrane component of the ABC transporter complex. Comparison of the lol genes present in various Gram-negative bacteria revealed that the absence of lolE is not unique to Francisella, but instead was found in more than half of the bacterial genomes they analyzed (5). Based on protein sequence analysis, LoVullo et al. concluded that the single LolC present in bacteria such as Francisella spp. contains features found in both LolC and LolE proteins. This suggests that the single LolC is a hybrid protein, which they renamed LolF. LoVullo et al. have proposed that a homodimer formed by LolF enables the Lol transporter complex of Francisella to recognize diacylated as well as triacylated lipoproteins and to transfer either type of substrate to LolA for sorting to the OM (Fig. 1) (5). To test the generality of their hypothesis, LoVullo et al. attempted to delete lnt from Neisseria gonorrhoeae, which has the same LolF arrangement as found in Francisella. Indeed, Δlnt mutants could be isolated in N. gonorrhoeae, and these mutants maintained proper lipoprotein-dependent functionality. Taken together, these results suggest that many bacteria may employ a more flexible lipoprotein sorting pathway than found in E. coli and that this increased flexibility may be due to an altered arrangement of the Lol ABC transporter complex. Confirmation of this intriguing idea awaits additional studies to directly compare the abilities of the E. coli LolCDE and F. tularensis LolFD transporters to bind and extract diacylated versus triacylated lipoproteins.
Lipoproteins are important contributors to Francisella-host interactions, and Toll-like receptor 2 (TLR2)-dependent sensing of lipoproteins is a key mediator of the host inflammatory response to F. tularensis infection (18, 19). Consistent with proper maintenance of lipoprotein function in the F. tularensis Δlnt mutant, LoVullo et al. found that the Δlnt mutant survived and replicated intracellularly in a macrophage-like cell line, similar to the wild-type strain. Although those authors did not assess virulence of the Δlnt mutant in the mouse model of tularemia, their results raise the interesting possibility that Francisella, and potentially other bacterial pathogens, may regulate lipoprotein acylation as a means to alter host responses during pathogenesis. Hints that this may be the case come from studies with Listeria monocytogenes and Staphylococcus aureus (20, 21). Future studies that examine pathogenesis of the F. tularensis Δlnt mutant in the host will be informative, as will examination of the acylation status of lipoproteins at different time points during infection.
The demonstration by LoVullo et al. (5) that lnt is not essential in Francisella spp. and N. gonorrhoeae, together with the finding that the Lol systems of many bacteria adopt the F. tularensis-like arrangement of a single lolF gene, represent an important shift in our current understanding of lipoprotein processing and sorting in Gram-negative bacteria. The work opens new questions about the mechanisms governing lipoprotein biogenesis and raises the possibility for unique functional roles held by diacylated versus triacylated lipoproteins. Such altered functionality might be particularly relevant for bacterial pathogens, which could regulate lipoprotein processing to modulate interactions with the host.
ACKNOWLEDGMENTS
Research in the Thanassi laboratory is supported by National Institutes of Health award R01GM062987. P.C. is supported by National Institutes of Health award T32AI007539.
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
REFERENCES
- 1.Okuda S, Tokuda H. 2011. Lipoprotein sorting in bacteria. Annu Rev Microbiol 65:239–259. doi: 10.1146/annurev-micro-090110-102859. [DOI] [PubMed] [Google Scholar]
- 2.Zuckert WR. 2014. Secretion of bacterial lipoproteins: through the cytoplasmic membrane, the periplasm and beyond. Biochim Biophys Acta 1843:1509–1516. doi: 10.1016/j.bbamcr.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakayama H, Kurokawa K, Lee BL. 2012. Lipoproteins in bacteria: structures and biosynthetic pathways. FEBS J 279:4247–4268. doi: 10.1111/febs.12041. [DOI] [PubMed] [Google Scholar]
- 4.Ray A, Cot M, Puzo G, Gilleron M, Nigou J. 2013. Bacterial cell wall macroamphiphiles: pathogen-/microbe-associated molecular patterns detected by mammalian innate immune system. Biochimie 95:33–42. doi: 10.1016/j.biochi.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 5.LoVullo ED, Wright LF, Isabella V, Huntley JF, Pavelka MS Jr. 2015. Revisiting the Gram-negative lipoprotein paradigm. J Bacteriol 197:1705–1715. doi: 10.1128/JB.02414-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lycklama a Nijeholt JA, Driessen AJ. 2012. The bacterial Sec-translocase: structure and mechanism. Philos Trans R Soc Lond B Biol Sci 367:1016–1028. doi: 10.1098/rstb.2011.0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Narita S, Tokuda H. 2007. Amino acids at positions 3 and 4 determine the membrane specificity of Pseudomonas aeruginosa lipoproteins. J Biol Chem 282:13372–13378. doi: 10.1074/jbc.M611839200. [DOI] [PubMed] [Google Scholar]
- 8.Seydel A, Gounon P, Pugsley AP. 1999. Testing the ‘+2 rule’ for lipoprotein sorting in the Escherichia coli cell envelope with a new genetic selection. Mol Microbiol 34:810–821. doi: 10.1046/j.1365-2958.1999.01647.x. [DOI] [PubMed] [Google Scholar]
- 9.Sankaran K, Wu HC. 1994. Lipid modification of bacterial prolipoprotein. Transfer of diacylglyceryl moiety from phosphatidylglycerol. J Biol Chem 269:19701–19706. [PubMed] [Google Scholar]
- 10.Tokunaga M, Tokunaga H, Wu HC. 1982. Post-translational modification and processing of Escherichia coli prolipoprotein in vitro. Proc Natl Acad Sci U S A 79:2255–2259. doi: 10.1073/pnas.79.7.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hillmann F, Argentini M, Buddelmeijer N. 2011. Kinetics and phospholipid specificity of apolipoprotein N-acyltransferase. J Biol Chem 286:27936–27946. doi: 10.1074/jbc.M111.243519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hara T, Matsuyama S, Tokuda H. 2003. Mechanism underlying the inner membrane retention of Escherichia coli lipoproteins caused by Lol avoidance signals. J Biol Chem 278:40408–40414. doi: 10.1074/jbc.M307836200. [DOI] [PubMed] [Google Scholar]
- 13.Narita S, Tokuda H. 2006. An ABC transporter mediating the membrane detachment of bacterial lipoproteins depending on their sorting signals. FEBS Lett 580:1164–1170. doi: 10.1016/j.febslet.2005.10.038. [DOI] [PubMed] [Google Scholar]
- 14.Tajima T, Yokota N, Matsuyama S, Tokuda H. 1998. Genetic analyses of the in vivo function of LolA, a periplasmic chaperone involved in the outer membrane localization of Escherichia coli lipoproteins. FEBS Lett 439:51–54. doi: 10.1016/S0014-5793(98)01334-9. [DOI] [PubMed] [Google Scholar]
- 15.Okuda S, Tokuda H. 2009. Model of mouth-to-mouth transfer of bacterial lipoproteins through inner membrane LolC, periplasmic LolA, and outer membrane LolB. Proc Natl Acad Sci U S A 106:5877–5882. doi: 10.1073/pnas.0900896106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallagher LA, Ramage E, Jacobs MA, Kaul R, Brittnacher M, Manoil C. 2007. A comprehensive transposon mutant library of Francisella novicida, a bioweapon surrogate. Proc Natl Acad Sci U S A 104:1009–1014. doi: 10.1073/pnas.0606713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oyston PC, Sjostedt A, Titball RW. 2004. Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat Rev Microbiol 2:967–978. doi: 10.1038/nrmicro1045. [DOI] [PubMed] [Google Scholar]
- 18.Thakran S, Li H, Lavine CL, Miller MA, Bina JE, Bina XR, Re F. 2008. Identification of Francisella tularensis lipoproteins that stimulate the toll-like receptor (TLR) 2/TLR1 heterodimer. J Biol Chem 283:3751–3760. doi: 10.1074/jbc.M706854200. [DOI] [PubMed] [Google Scholar]
- 19.Malik M, Bakshi CS, Sahay B, Shah A, Lotz SA, Sellati TJ. 2006. Toll-like receptor 2 is required for control of pulmonary infection with Francisella tularensis. Infect Immun 74:3657–3662. doi: 10.1128/IAI.02030-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurokawa K, Kim MS, Ichikawa R, Ryu KH, Dohmae N, Nakayama H, Lee BL. 2012. Environment-mediated accumulation of diacyl lipoproteins over their triacyl counterparts in Staphylococcus aureus. J Bacteriol 194:3299–3306. doi: 10.1128/JB.00314-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reglier-Poupet H, Frehel C, Dubail I, Beretti JL, Berche P, Charbit A, Raynaud C. 2003. Maturation of lipoproteins by type II signal peptidase is required for phagosomal escape of Listeria monocytogenes. J Biol Chem 278:49469–49477. doi: 10.1074/jbc.M307953200. [DOI] [PubMed] [Google Scholar]