ABSTRACT
The accessory growth regulator (Agr)-like quorum sensing (QS) system of Clostridium perfringens controls the production of many toxins, including beta toxin (CPB). We previously showed (J. E. Vidal, M. Ma, J. Saputo, J. Garcia, F. A. Uzal, and B. A. McClane, Mol Microbiol 83:179–194, 2012, http://dx.doi.org/10.1111/j.1365-2958.2011.07925.x) that an 8-amino-acid, AgrD-derived peptide named 8-R upregulates CPB production by this QS system. The current study synthesized a series of small signaling peptides corresponding to sequences within the C. perfringens AgrD polypeptide to investigate the C. perfringens autoinducing peptide (AIP) structure-function relationship. When both linear and cyclic ring forms of these peptides were added to agrB null mutants of type B strain CN1795 or type C strain CN3685, the 5-amino-acid peptides, whether in a linear or ring (thiolactone or lactone) form, induced better signaling (more CPB production) than peptide 8-R for both C. perfringens strains. The 5-mer thiolactone ring peptide induced faster signaling than the 5-mer linear peptide. Strain-related variations in sensing these peptides were detected, with CN3685 sensing the synthetic peptides more strongly than CN1795. Consistent with those synthetic peptide results, Transwell coculture experiments showed that CN3685 exquisitely senses native AIP signals from other isolates (types A, B, C, and D), while CN1795 barely senses even its own AIP. Finally, a C. perfringens AgrD sequence-based peptide with a 6-amino-acid thiolactone ring interfered with CPB production by several C. perfringens strains, suggesting potential therapeutic applications. These results indicate that AIP signaling sensitivity and responsiveness vary among C. perfringens strains and suggest C. perfringens prefers a 5-mer AIP to initiate Agr signaling.
IMPORTANCE Clostridium perfringens possesses an Agr-like quorum sensing (QS) system that regulates virulence, sporulation, and toxin production. The current study used synthetic peptides to identify the structure-function relationship for the signaling peptide that activates this QS system. We found that a 5-mer peptide induces optimal signaling. Unlike other Agr systems, a linear version of this peptide (in addition to thiolactone and lactone versions) could induce signaling. Two C. perfringens strains were found to vary in sensitivity to these peptides. We also found that a 6-mer peptide can inhibit toxin production by some strains, suggesting therapeutic applications.
INTRODUCTION
In humans and livestock, Clostridium perfringens is a major cause of histotoxic infections, such as human gas gangrene, and intestinal infections, including enteritis and enterotoxemias (1, 2). The versatility of this Gram-positive anaerobic bacterium is largely attributable to its ability to produce ∼17 different toxins (3–5). However, individual strains produce only portions of this toxin repertoire; this toxin production variability is used in a classification system that assigns C. perfringens isolates to one of five types (A to E) based upon their production of alpha (CPA), beta (CPB), epsilon (ETX), and iota toxins (4, 5). Each C. perfringens type is associated with certain diseases (1, 4).
C. perfringens-induced diseases are not intoxications but rather true infections where the bacterium must grow and produce toxins in the host (1). There is only a relatively rudimentary understanding of toxin production regulation by C. perfringens. The production of many toxins that are expressed by C. perfringens during vegetative growth is controlled, at least in part, by a two-component regulatory system named VirS/VirR (6–8). In some cases, regulatory RNAs are involved in VirS/VirR-mediated regulation, although production of perfringolysin O (PFO) is directly regulated by the binding of phosphorylated VirR to DNA sequences, named VirR boxes, that are located directly upstream of the pfoA gene (6, 9). In contrast, the production of the enterotoxin (CPE) during sporulation is mediated by sporulation-specific alternative sigma factors (4, 10, 11).
The accessory growth regulator (Agr)-like quorum sensing (QS) system was first discovered in Staphylococcus aureus, where it controls gene expression via population density-induced autoinducer signaling molecules (8, 12, 13). Within the past 5 years, an Agr-like QS system has also been identified in C. perfringens, where this QS system was first demonstrated to regulate the production of CPA and PFO by type A strain 13 (12, 13). Since those initial studies, the C. perfringens Agr-like QS system was shown to regulate the production of ETX, PFO, and CPA in type D strain CN3718 (14), the production of CPB, PFO, and CPA (but not ETX) in type B strains CN1793 and CN1795 (15), the production of CPA, CPB, and PFO in type C strain CN3685 (16), and the production of CPE during sporulation by CPE-positive type A strain F5603 (17). In addition, the Agr-like system was found to be essential for type C strain CN3685 to cause necrotic enteritis in a rabbit small intestinal loop model or lethal enterotoxemia in a mouse oral challenge model (16). This requirement for a functional Agr-like QS system in CN3685 virulence was determined to involve the QS system controlling the intestinal production of CPB (16), which is essential for the virulence of type C strains (18).
Both similarities and differences exist between the Agr-like QS system of C. perfringens and the well-characterized, paradigm Agr system of Staphylococcus aureus (8, 12, 13, 19–21). For example, the C. perfringens Agr-like QS system includes an operon encoding AgrD and AgrB, where AgrD is the precursor peptide for the autoinducing peptide (AIP) that mediates Agr QS signaling and AgrB is the integral membrane endopeptidase that is involved in processing AgrD to the active AIP. However, C. perfringens lacks the AgrA/AgrC two-component system that responds to the AIP in S. aureus (6, 8, 19, 20). Some evidence suggests that the VirS/VirR two-component regulatory system may sometimes be the functional equivalent of AgrA/AgrC in C. perfringens, but this relationship has not yet been firmly proven and there appears to be exceptions since expression of the C. perfringens etx gene is regulated by the Agr-like QS system, yet etx expression is not affected by inactivation of the virS-virR operon (6, 8, 12, 14). Another difference is the absence of an identifiable RNAIII in C. perfringens, which is important because RNAIII is a major downstream effector that regulates the production of many virulence factors, including several toxins, in the S. aureus Agr QS system (6, 8, 19, 20).
There is considerable variation in the nature of the AIPs that activate the Agr systems of various Gram-positive bacteria (22). In S. aureus, at least 4 AIPs have been identified; these range from 7 to 10 amino acids, with all possessing a 5-mer thiolactone ring. In contrast, the AIP of Lactobacillus plantarum is only a 5-mer thiolactone ring with no tail amino acids (20, 21). The nature of the C. perfringens AIP has received only limited study to date. Our previous study (16) showed that the synthetic peptide 8-R, which contains an 8-amino-acid sequence based on the C. perfringens AgrD sequence and includes 5 of those amino acids in a thiolactone ring, can induce Agr signaling in an agrB null mutant of CN3685 (note that agrB mutants do not produce either AgrB or AgrD, because those proteins are both encoded within the same operon in C. perfringens [12, 13]). However, peptide 8-L, which is the linear version of 8-R without the thiolactone ring, failed to induce Agr signaling in this agrB mutant (16).
AgrD sequence-based synthetic peptides have provided important insights into the structure-function properties of AIPs from S. aureus and other species (20–22). Therefore, the current study prepared and characterized the signaling properties of several C. perfringens AgrD sequence-based synthetic peptides to gain insights into AIP signaling in C. perfringens.
MATERIALS AND METHODS
Bacteria, media, and reagents.
The C. perfringens strains used in this study are listed in Table 1. All isolates were stored as cooked meat medium (CMM) stock cultures at −20°C. Fluid thioglycolate medium (FTG) (Difco Laboratories) and TGY broth (3% tryptic soy broth [Becton-Dickinson], 2% glucose [Fisher Scientific], 1% yeast extract [Becton Dickinson], and 0.1% sodium thioglycolate [Sigma-Aldrich]) were used for broth cultures. Brain heart infusion (BHI) agar (Becton-Dickinson) plates containing 15 μg ml−1 chloramphenicol (Cm) were used for screening the CN1795 cpb-knockout mutant (CN1795cpbko) constructed in the current study. CN3685cpbko (an isogenic cpb null mutant of CN3685, also known as BMC100 [18]), CN3685agrBko (an agrB null mutant of CN3685, also known as BMJV10 [16]), and CN1795agrBko (an agrB null mutant of CN1795 [15]) had been previously constructed and characterized, both genotypically and phenotypically (Table 1).
TABLE 1.
C. perfringens isolates used in this study
| Isolate type and no. | Strain name | Description (disease, location of isolation, date of isolation) | Source or reference | Toxin gene(s)a |
|---|---|---|---|---|
| Type A | ||||
| 1 | SM101 | Transformable derivative of food poisoning strain, Europe, 1950s | 11 | cpe |
| 2 | Strain 13 | Soil | 12 | |
| Type B | ||||
| 1 | CN1795 | Vet lab, toxigenic, 1947 | 15 | cpb, etx |
| 2 | CN1795cpbko | CN1795 cpb null mutant | This study | etx |
| 3 | CN1795agrBko | CN1795 agrB null mutant | 15 | cpb, etx |
| 4 | PS49 | Unknown | 29 | cpb, etx |
| 5 | CN1793 | Vet lab, toxigenic, 1947 | 15 | cpb, etx |
| 6 | Bar2 | Sheep | 29 | cpb, etx |
| 7 | CN2003 | Vet lab, lamb ileum, Wales, 1947 | 29 | cpb, etx |
| Type C | ||||
| 1 | CN3685 | Sheep with struck | 18 | cpb |
| 2 | CN3685agrBko (BMJV10) | CN3685 agrB null mutant | 16 | cpb |
| 3 | CN3685cpbko (BMC100) | CN3685 cpb null mutant | 18 | |
| 4 | Bar3 | Pigbel, Argentina | 30 | cpb |
| 5 | CN5383 | Pigbel, Papua New Guinea, 1970s | 30 | cpb |
| 6 | CN3955 | Peritoneal fluid of ewe, Weybridge, United Kingdom, 1956 | 30 | cpb |
| 7 | CN3763 | Zeissler, Hamburg, Germany, 1955 | 30 | cpb |
| Type D | ||||
| 1 | CN2068 | Newcastle lamb | 31 | etx |
| 2 | CN3718 | Guinea pig heart blood | 14 | etx |
All strains carry the cpa gene, and all strains except SM101 and CN3763 carry the pfoA gene.
Synthetic peptides.
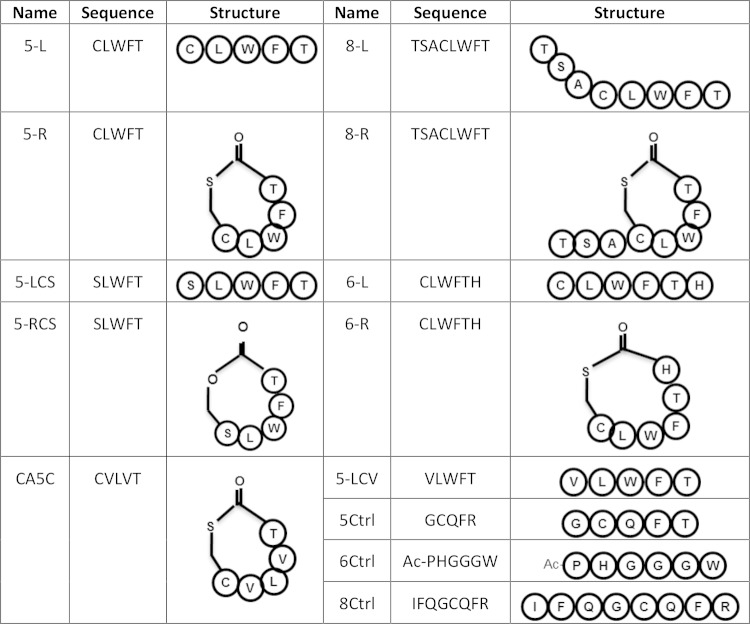
The synthetic peptides used in this study included 5-mer cyclic (5-R) and linear (5-L) peptides, as well as 8-mer cyclic (8-R) and linear (8-L) peptides, each designed based upon the previously demonstrated signaling ability of 8-R and containing sequences encoded within the C. perfringens CN3685 agrD open reading frame (ORF) (16). The sequences and schematic structures of each peptide are shown in Fig. 1. As a negative control, 5-mer (5Crtl), 6-mer (6Crtl), and 8-mer (8Crtl) peptides were also used (Fig. 1). Variant peptides and other peptides are also depicted in Fig. 1, and their use is described in Results.
FIG 1.
Peptides used in this study.
The synthesis of these signaling, control, and variant peptides were carried out by the Peptide and Peptoid Synthesis Core Facility Division of the Health Sciences Core Research Facilities (HSCRF) at the University of Pittsburgh. Synthesis was performed using standard 9-fluorenylmethoxy carbonyl (FMOC) chemistry cycles on a Liberty CEM microwave synthesizer and O-(benzotriazol-1-yl)-N,N,N′,N′-tetramethyluroniumtetrafluoroborate (TBTU)/1-hydroxybenzotriazole (Hobt) activation in dimethylformamide (DMF). Briefly, FMOC-Thr(tBu), FMOC-Ser(tBu), FMOC-Ala, FMOC-Cys(mmt), FMOC-Leu, FMOC-Trp(Boc), and FMOC-Phe were purchased from Peptides International and used for stepwise assembly of the linear sequences on H-Thr(tBu)-2-ClTrt resin (1% divinylbenzene [DVB], 200 to 400 mesh, 0.550 mmol g−1; EMD Chemicals, Inc.). In preparation for thiolactone ring formation following the completion of peptide chain assembly, the N-terminal amino group of each protected peptide resin was reversibly acylated using Boc anhydride–N,N-diisopropylethylamine (DIPEA)–DMF for 2 h at room temperature. Peptide resins were then divided into two equal portions in order to reserve material for the workup as the linear control sequences. In preparation for the thiolactone cyclization steps, cleavage of the fully protected peptide fragments from each 2-ClTrt resin was accomplished using 1% trifluoroacetic acid (TFA) in dichloromethane (DCM) and filtered into solutions of 10% pyridine in methanol. This process was performed a total of 5 times for each peptide sample followed by evaporation to dryness under high vacuum on a Buchi Rotavapor system at 37°C. Thiolactone formation was initiated by adding 10.2 mg of 1-hydroxy-7-azabenzotriazole (HoAt), 43.5 mg of 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (EDAC-HCl), 13.05 μl of DIPEA, and 4-(N,N-dimethylamino) pyridine (DMAP) to approximately 98 mg of each partially protected peptide fragment in 75 ml of DCM. The mixtures were then layered with nitrogen and stirred for 24 h at room temperature followed by evaporation to dryness under high vacuum. The fully side-chain-protected linear peptide resins along with the protected thiolactone-peptide fragments were then cleaved with TFA-H2O-Et3SiH-EtSMe (Sigma-Aldrich) (90:5:3:2) for 2 h at room temperature and then dried to a film under high vacuum. Films were resuspended in aqueous solvents and then purified on a Waters Prep 4000 series chromatography system using a Phenomenex Gemini (21.2- by 250-mm) 10-μm C18 column along with standard acetonitrile-0.1% TFA gradient conditions. The final analytical determination of peptide purity for all products was performed on a Waters Alliance chromatography system using a Phenomenex Gemini (4.6- by 250-mm) 5-μm C18 column along with standard acetonitrile-0.1% TFA gradient conditions. Matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) analysis on an Applied Biosystems Voyager workstation was used as a final confirmation of the expected target mass of each peptide.
Cell culture.
Human-derived enterocyte-like Caco-2 cells were routinely cultured in Eagle's minimum essential medium (MEM; Sigma) supplemented with heat-inactivated 10% fetal calf serum (FCS; Mediatech Incorporated), 1% glutamine (Sigma), 1% nonessential amino acids (Sigma), penicillin (100 U ml−1) (Fisher Scientific), and streptomycin (100 μg ml−1) (Fisher Scientific). Caco-2 cell cultures were incubated at 37°C in a 5% CO2 humidified atmosphere. MEM is Caco-2 cell culture medium without supplemental FCS and antibiotics (23).
CPB purification.
An isolated colony of CN3685 was cultured at 37°C for 8 h in 3,000 ml of TGY broth for CPB production. The toxin was then purified from the culture supernatant by anion-exchange chromatography using a DEAE-CL6B Sepharose column (Sigma) and an ÄKTA prime system (Amersham Bioscience), as previously described (18). The purity of the CPB preparation was evaluated by SDS-PAGE and Western blotting using a monoclonal anti-CPB antibody, followed by densitometric analysis. The final preparation was ∼95% homogeneous.
Construction of a CN1795 cpb null mutant (CN1795cpbko) using TargeTron technology.
The construction of this mutant was performed by targeted insertional mutagenesis using Clostridium-modified group II TargeTron technology (24). Inactivation of the cpb gene in strain CN1795 was achieved by specifically inserting, in the sense orientation, a group II intron sequence (∼900 bp) into the cpb ORF, as described previously (18). For this purpose, a previously constructed intron donor plasmid named pJIR750cpbi, which carries a cpb-targeted intron (18), was electroporated into the type B wild-type strain CN1795. Transformants were then plated onto BHI agar plates containing 15 μg ml−1 of Cm. Colony PCR was carried out for mutant screening using internal cpb primers cpbF and cpbR (18), which amplify a PCR product of ∼900 bp in the wild-type strain but amplify an ∼1.8-kb product from cpb null mutants due to the insertion of an ∼900-bp intron. One cpb gene null knockout mutant was subcultured on FTG culture for 10 days to cure the intron plasmid, creating the CN1795 cpb null mutant (CN1795cpbko). This mutant was then further characterized by PCR and intron Southern blotting analyses, as described below.
PCR analyses of the cpb gene.
Template DNA for PCR analysis was extracted from overnight TGY cultures and then purified using the MasterPure Gram-positive DNA purification kit (Epicentre). Each PCR mixture (50 μl) contained 5 μl of template DNA (100 ng), 25 μl of Taq complete 2× master mix (New England BioLabs), and 1 μl of each primer (1 μM final concentration). PCR amplification conditions applied for the PCR analysis were 1 cycle of 94°C for 2 min and 35 cycles of 94°C for 1 min, 55°C for 1 min, and 68°C for 1 min, followed by a final single extension of 5 min at 68°C. The PCR products were electrophoresed on a 1.5% agarose gel and stained with ethidium bromide for visualization.
Intron Southern blot analyses.
C. perfringens genomic DNA was isolated from wild-type strain CN1795 and the isogenic cpb gene null mutant strain CN1795cpbko using the MasterPure Gram-positive DNA purification kit. An aliquot (2.5 μg) of each isolated DNA sample was then digested overnight with EcoRI according to the manufacturer's (New England BioLabs) instructions. The digested DNA samples were electrophoresed on a conventional 1% agarose gel and then transferred onto a positively charged nylon membrane (Roche) for hybridization with an intron-specific probe (18). The probes were prepared using the PCR digoxigenin (DIG) probe synthesis kit (Roche) and intron primers (IBS and EBS2) (18). After hybridization, Southern blots were developed using reagents from the DIG DNA labeling and detection kit (Roche), according to the manufacturer's instructions.
Western blot analyses of CPB production by wild-type CN1795 and the CN1795cpbko mutant.
A 0.2-ml aliquot of each isolate from a CMM culture was inoculated into 10 ml of fresh FTG broth for overnight culture at 37°C. An aliquot (100 μl) of each FTG overnight culture was then inoculated into 10 ml of fresh TGY broth for a further overnight incubation at 37°C, followed by centrifugation at 13,000 × g for 3 min. Equal volumes of each culture supernatant were mixed with 5× SDS-PAGE loading buffer and boiled for 5 min. Twenty-five microliters of each sample was electrophoresed on a 12% SDS-PAGE gel, and the separated proteins were transferred onto a nitrocellulose membrane. The membrane was blocked with TBS-Tween 20 (0.05% [vol/vol]) and nonfat dry milk (5% [wt/vol]) for 1 h at room temperature, followed by probing with a mouse monoclonal anti-CPB antibody overnight at 4°C (18). Finally, bound antibody was detected with a horseradish peroxidase-conjugated secondary anti-mouse antibody (Sigma-Aldrich) and the addition of SuperSignal West Pico chemiluminescent substrate (Pierce).
Peptide signaling to induce CPB production by CN1795agrBko or CN3685agrBko.
Wild-type CN1795, the CN1795agrBko mutant, wild-type CN3685, and the CN3685agrBko mutant were each grown in 10 ml of FTG overnight at 37°C. An aliquot (200 μl) of each culture was inoculated into 5 ml of fresh TGY broth, and those cultures were incubated for 5 h at 37°C. A 15-μl aliquot of each agrB null mutant TGY culture was then inoculated into different wells of a 12-well plate, each containing 1 ml of TGY fresh medium and a final concentration of 100 μM one peptide (5-R, 5-L, 8-R, or 8-L). As a control, a 100 μM concentration of negative-control peptide (5Ctrl or 8Ctrl) was added to another well containing 1 ml of TGY freshly inoculated with the same amount of either CN1795agrBko or CN3685agrBko. As another control, wild-type CN1795 and CN3685, as well as their isogenic agrB gene-knockout mutants, were similarly cultured without peptide supplementation. The bacteria were anaerobically cultured overnight at 37°C. Supernatants of each sample were collected and boiled in 5× SDS-PAGE loading buffer; 25 μl of those samples was then used for Western blot detection of CPB production with a mouse monoclonal anti-CPB antibody, as described above.
To assess dose-response effects of signaling peptides, a 15-μl aliquot of a 5-h TGY culture of CN3685agrBko was inoculated into wells of a 12-well plate, with each well containing 1 ml of fresh TGY broth and various doses, ranging from a final concentration of 5 μM to 100 μM, of the 5-R, 5-L, or 8-R peptides. Wild-type CN3685 or CN3685agrBko cultures were similarly inoculated into a well containing 1 ml of fresh TGY broth without AIPs as a control. The cultures were then incubated anaerobically overnight at 37°C. After this incubation, 25 μl of the overnight cultures was collected and Western blotted for CPB production, as described above.
In another experiment, 15 μl of a 5-h TGY culture of CN3685agrBko or CN1795agrBko was inoculated into a well containing 1 ml of fresh TGY broth and a final 25 μM concentration of either the 5-R, 5-L, 5Ctrl, 8-R, 8-L, or 8Ctrl peptides. The culture was then incubated anaerobically overnight at 37°C. A 25-μl aliquot of supernatant from overnight cultures was collected and evaluated for CPB production with Western blotting, as described above.
Time course analysis of peptide signaling to induce CPB production by CN1795agrBko and CN3685agrBko.
Wild-type CN1795 and its isogenic CN1795agrBko mutant, or wild-type CN3685 and its isogenic CN3685agrBko mutant, were each grown in 10 ml of FTG overnight at 37°C. An aliquot (100 μl) of each of those cultures was then inoculated into 10 ml of TGY broth. After an overnight incubation at 37°C, 15 μl of each agrB null mutant culture was inoculated into one well of a 12-well plate, with each well containing 1 ml of fresh TGY and either the 5-R or the 5-L peptide at a final concentration of 100 μM. As controls, wild-type CN1795 and CN3685, or their agrB null mutants, were similarly cultured in both wells but without peptides. All bacteria were cultured for 3 h, for 6 h, or overnight (∼16 h) at 37°C under anaerobic conditions. Supernatants of each sample were then collected and boiled in 5× SDS-PAGE loading buffer. A 25-μl aliquot of each sample was used for Western blot detection of CPB production with a mouse monoclonal anti-CPB antibody, as described earlier.
Variant peptide signaling of the Agr-like QS system to produce CPB in vitro.
Several variant peptides were synthesized, including peptide 5-RCS that contains a C→S change relative to the 5-R peptide, the 5-LCS peptide that contains a C→S change relative to the 5-L peptide, and the 5-LCV peptide that contains a C→V change relative to 5-L (Fig. 1). In addition, a 5-mer peptide, i.e., Cac5c, was synthesized that contains the Clostridium acetobutylicum AgrD sequence-based sequence (25). To test the signaling abilities of these variant peptides, CN1795agrBko and CN3685agrBko were cultured in 10 ml of FTG broth overnight at 37°C. An aliquot (200 μl) of the overnight FTG culture was inoculated into 5 ml of fresh TGY medium, which was then cultured at 37°C for 5 h.
An aliquot (15 μl) from each of those 5-h CN3685agrBko or CN1795agrBko TGY cultures was inoculated into single wells of a 12-well plate, with each well containing 1 ml of fresh TGY medium and the 5-R, 5-L, 5-RCS, 5-LCS, 5-LCV, Ca5c, 6-R or 6-L, 6Ctrl, and 5Ctrl peptides at a final concentration of 100 μM. Supernatants were collected from the overnight cultures, and 25 μl of those samples was analyzed by Western blotting for the presence of CPB, as described above.
Comparison of signaling peptide sensing between different C. perfringens strains.
A Transwell plate was used to evaluate the responsiveness of different C. perfringens strains to natural Agr-like QS system-related signaling peptides. This Transwell system allows passage of small molecules between two chambers in a well containing a filter with pores (0.4-μm pore size; Corning) that allow free passage of polypeptides but are impermeable to bacteria, thus physically separating bacteria inoculated into the bottom chamber from those inoculated into the top chamber. For these experiments, 1 ml of fresh TGY was added to each of the two chambers in the 12-well Transwell plate. An aliquot (15 μl) of a 5-h TGY culture of the isogenic CN1795agrBko or CN3685agrBko mutant was then inoculated into the bottom chamber of each well. Another aliquot (15 μl) of a 5-h TGY culture of type A strain 13, SM101, type B cpb null mutant CN1795cpbko, type C cpb null mutant CN3685cpbko, and type D strains CN2068 and CN3718 was inoculated into the top chamber of each well (Table 1). As a control, wild-type CN3685, CN3685agrBko, wild-type CN1795, or CN1795agrBko was cultured separately in both chambers of a well, but without any peptides. These cultures were anaerobically incubated at 37°C overnight. Supernatants from the bottom chambers were then collected after centrifugation (13,000 × g for 3 min), and a CPB Western blot analysis was performed, as described above.
Evaluation of inactivation of the agrD sequence-based peptides in CN1795 and CN3685 cultures.
The CN1795cpbko, CN3685cpbko, CN1795agrBko, and CN3685agrBko mutants were each grown individually overnight at 37°C in 10 ml of TGY. An aliquot (100 μl) of each of those cultures was inoculated into 10 ml of TGY broth, and those cultures were then incubated overnight at 37°C. A 1-ml aliquot of the overnight CN1795cpbko and CN3685cpbko TGY cultures was collected and centrifuged for 3 min at 13,000 × g. A 100-μl aliquot of supernatant collected from each culture was then incubated at 37°C for 2 h with either the 5-R or 5-L peptides at a final concentration of 100 μM. An aliquot (900 μl) of fresh TGY broth was added to the 100-μl supernatant prior to inoculation with 15 μl from an overnight CN3685agrBko or CN1795agrBko TGY culture; those cultures were then incubated at 37°C for 5 h under anaerobic conditions. Wild-type CN3685, CN3685agrBko, wild-type CN1795, and CN1795agrBko were cultured similarly as controls, except for the absence of any added synthetic peptide. Supernatants of each culture were then collected and boiled in 5× SDS-PAGE loading buffer, and 25 μl of each sample was used for Western blot detection of CPB production using a mouse monoclonal anti-CPB antibody, as described above.
Evaluating the effects of synthetic signaling peptides on wild-type C. perfringens.
Wild-type CN1795 or CN3685 was serially cultured in FTG and then in fresh TGY broth, as described above. A final 100 μM concentration of the 5-LCV, Ca5c, 5Ctrl, 6-R, 6-L, 6Ctrl, 8-L, or 8Ctrl peptide was added into 1 ml of fresh TGY broth. A 15-μl aliquot of an overnight TGY culture of wild-type CN1795 or CN3685 was then inoculated into each peptide-containing TGY broth, followed by incubation of the cultures overnight at 37°C under anaerobic conditions. Supernatants were collected from those overnight cultures, or from control cultures (no peptide), and used for Western blot detection of CPB production, as described above.
The 6-R peptide was also tested for its ability to inhibit the Agr-like QS system of several other wild-type C. perfringens strains, including type B strains PS49, CN1793, Bar2, and CN2003 or type C strains Bar3, CN5383, CN3955, and CN3763 (Table 1). Bacterial cultures were prepared as described above for strain CN1795 or CN3685. Briefly, a 200-μl aliquot of an overnight FTG culture was subcultured in 5 ml of fresh TGY medium for 5 h at 37°C. The 6-R peptide, at a final concentration of 100 μM, was added to 1 ml of fresh TGY both in one well of a 12-well plate. A 15-μl aliquot of the 5-h TGY bacterial culture was then inoculated into that well, followed by overnight incubation at 37°C. The wild-type strains were also cultured similarly in the absence of 6-R. A 25-μl aliquot of each culture supernatant was then used for Western blot analysis of CPB production.
Effects of peptide 6-R on CPB production by C. perfringens incubated in the presence of host Caco-2 cells.
Wild-type CN1795 and CN3685 were each grown in 10 ml of FTG overnight, and 0.2-ml aliquots of those cultures were used to inoculate 10-ml tubes of TGY fresh broth, which were then grown overnight at 37°C as described above. These TGY culture cells were washed three times with prewarmed phosphate-buffered saline (PBS) buffer (PBS without calcium and magnesium; Corning Cellgro). A 20-μl aliquot of washed cells was added to 1 ml fresh MEM with a Caco-2 cell monolayer in a 12-well plate. MEM without Caco-2 cells was used as a negative control. At the same time, the 6-R peptide was also added to the same well at a final concentration of 100 μM. For comparison, the same treatments without 6-R peptide were used as a negative control. All treatment cultures were incubated for 2 h at 37°C under anaerobic conditions. Supernatants were collected from the 2-h cultures and prepared for Western blot detection of CPB, as described above.
Evaluating signaling competition between the 6-R and 5-R peptides.
The wild-type CN1795 and the CN1795agrBko mutant were each cultured at 37°C in 10 ml of FTG before those bacteria were subcultured at 37°C in 5 ml of TGY for 5 h, as described above. A 15-μl aliquot of those overnight TGY cultures was then inoculated into 1 ml of TGY present in one well of a 12-well plate. Similarly, a 15-μl aliquot of the CN1795agrBko TGY culture was also inoculated into a well containing 1 ml of TGY and a final 100 μM concentration of the 5-R or 6-R peptides alone or a mix of 100 μM the 5-R peptide plus 100 or 250 μM the 6-R peptide. The plate was then incubated overnight at 37°C under anaerobic conditions. Supernatants were collected from those overnight cultures, and Western blotting was performed to detect the presence of CPB, as described above.
Quantitative analysis of the presence of CPB in Western blotted samples.
NIH ImageJ software was used for all the quantitative comparisons in this study.
Statistical analysis.
The Friedman test was used for statistical analyses by GraphPad Prism software.
RESULTS
The 5-mer and 8-mer (with a thiolactone ring) synthetic peptides derived from the C. perfringens AgrD sequence can induce CPB production by Agr-like QS system null mutants of type B isolate CN1795 and type C isolate CN3685.
In our previous study (16), an 8-mer signaling peptide named 8-R, which possesses a 5-amino-acid thiolactone ring (Fig. 1), was synthesized based upon analogy to the proven AIPs of S. aureus and the deduced AgrD sequence of C. perfringens. This 8-R peptide was shown to induce CPB production by a CN3685 agrB null mutant (16). However, the 8-L synthetic peptide, corresponding to the linear version (no thiolactone ring) of 8-R, failed to induce CPB production by CN3685agrBko (16). This result could suggest that, as required for the well-characterized 7- to 10-amino-acid AIPs mediating signaling in the Agr system of S. aureus, a thiolactone ring is necessary for Agr-like QS system signaling (20, 21) in C. perfringens.
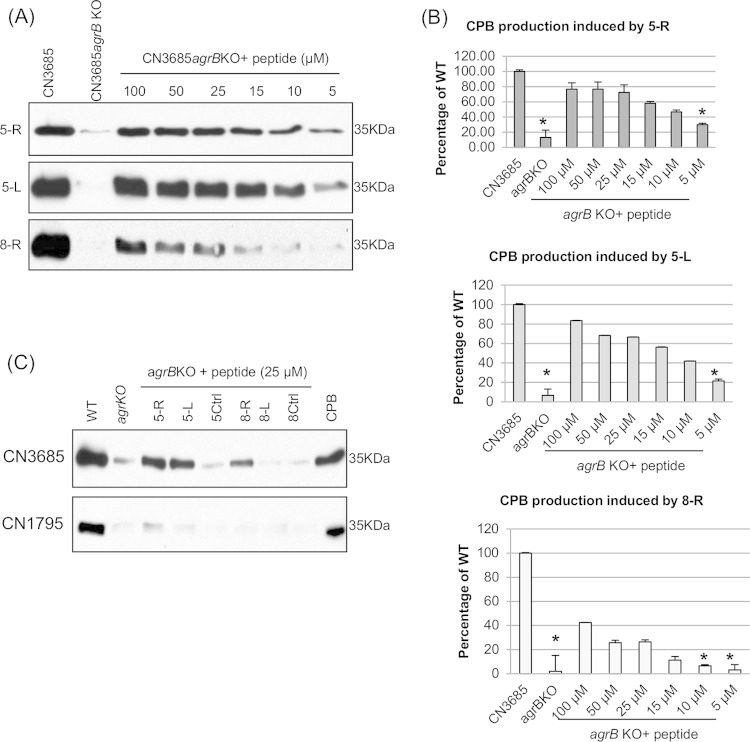
To test this hypothesis, the current study synthesized two peptides containing the central CLWFT sequence of 8-R without any amino acid tail (Fig. 1), i.e., peptide 5-R (which has a thiolactone ring formed by residues C and T) and peptide 5-L (the linear form of 5-R). When 100 μM concentrations of these 5-mer peptides were tested for their ability to signal the Agr-like QS system in C. perfringens (Fig. 2), both the cyclic and linear peptide forms of CLWFT induced strong CPB production by overnight TGY cultures of the agrB null mutants of CN1795 (CN1795agrBko) and CN3685 (CN3685agrBko). Furthermore, the linear 5-L peptide was nearly as efficient as the cyclic 5-R peptide in inducing overnight CPB production by these mutants (Fig. 2).
FIG 2.
CPB production by CN3685 or a CN3685 agrB null mutant (top of panel A) versus CN1795 or a CN1795 agrB null mutant (bottom of panel A) in the presence of 100 μM concentrations of different peptides. (A) Wild-type CN3685, the CN3685agrBko null mutant, wild-type CN1795, and the CN1795agrBko null mutant were used to compare natural CPB production levels by overnight TGY cultures. The last lanes of both panels show purified CPB, with a molecular mass of 35 kDa, as a positive control. In all other lanes, the indicated peptides were added overnight to TGY broth inoculated with CN3685agrBko (top) or CN1795agrBko (bottom). The image shows representative results that were reproducible over three repetitions. (B) Quantitative analysis of CPB production using ImageJ software, from panel A CPB Western blots. Points without error bars had errors too small to depict.
These studies also revealed that CN3685agrBko responds to the 5-L and 5-R signaling peptides better than CN1795agrBko (Fig. 2). Specifically, both the linear and ring 5-mer CLWFT peptides restored near-wild-type CPB production levels to overnight TGY cultures of CN3685agrBko. However, the same concentration of these 5-mer peptides restored only about half of wild-type CPB production to overnight TGY cultures of CN1795agrBko, indicating strain differences in sensing the signaling peptides.
Lastly, the 5-L and 5-R peptides were both better than the 8-R peptide at inducing CPB production by overnight TGY cultures of the type B and C Agr-like QS system null mutants (Fig. 2).
Signaling by 5-mer and 8-mer AgrD sequence-based peptides is dose dependent.
In S. aureus, 25 μM synthetic AIP has been used for Agr signaling to induce toxin production (26). Therefore, the current study tested different doses, ranging from 5 to 100 μM final culture concentrations, of the 5-R, 5-L, and 8-R signaling peptides for their ability to induce CPB production by overnight TGY cultures of CN3685agrBko and CN1795agrBko. The results demonstrated a clear dose-dependent relationship between the amount of signaling peptide present in the culture and the level of CPB production by CN3685agrBko (Fig. 3A). For the 5-mer peptides, a linear response was observed using between 5 and 25 μM peptide concentrations. As little as a 10 μM concentration of the 5-mer peptides significantly enhanced overnight CPB production by CN3685agrBko (Fig. 3B), with a calculated 50% effective concentration (EC50) of 11 μM for 5-R and 13 μM for 5-L. Signaling by the 8-R peptide was weaker than signaling by the 5-R or 5-L peptides (Fig. 3), with a relatively linear response noted by using concentrations between 5 and 100 μM 8-R and a calculated EC50 of 95 μM for this peptide. A 15 μM concentration of 8-R was sufficient to significantly enhance overnight CPB production by CN3685agrBko. The blot shown for Fig. 3A was intentionally exposed for a long period to demonstrate the weaker signaling effects observed using low peptide doses; however, the calculated EC50 for these peptides was similar using less exposed blots (data not shown).
FIG 3.
Restoration of CPB production to agrB null mutants using different doses of peptides (5-R, 5-L, and 8-R). (A) Western blot showing CPB production by CN3685agrBko inoculated into TGY-containing peptides at a final concentration of 5 to 100 μM, followed by overnight incubation at 37°C. (B) Quantitative Western blot of CPB production, using ImageJ software, by overnight cultures supplemented with different concentrations of signaling peptides (5-R, 5-L, and 8-R). Asterisks indicate a statistically significant difference (P < 0.05) by a treatment compared to wild-type CPB production. Results are the means plus standard deviation (SD) from three repetitions. Points without error bars had errors too small to depict. (C) Western blot detection of CPB production by the CN3685 agrB null mutant or CN1795 agrB null mutant after overnight TGY culture in the presence of a final concentration of 25 μM signaling peptides. Purified CPB was used as a positive control.
In contrast to the CN3685agrBko results, when 25 μM concentrations of 5-L or 5-R peptides were added to TGY cultures of CN1795agrBko, there was little, if any, overnight CPB production (Fig. 3C). These data indicated that higher concentrations of these three signaling peptides are needed for activation of the Agr-like QS system of CN1795 than for CN3685, again consistent with CN3685 sensing signaling peptides better than CN1795.
Time course of 5-R or 5-L peptide signaling in CN1795agrBko and CN3685agrBko.
A final 100 μM concentration of the 5-R or 5-L peptides was used to evaluate the time course of Agr-like QS system activation of CPB production in CN1795agrBko and CN3685agrBko. As shown in Fig. 4, a 3-h treatment with the 5-R peptide was sufficient to induce CPB production in CN3685agrBko but had a lesser effect on CPB production by CN1795agrBko, mirroring the naturally slower CPB production kinetics exhibited by wild-type CN1795 than by wild-type CN3685 in this experiment. However, in the presence of the 5-R peptide, CPB production by both agrB null mutants became strong by 6 h and then plateaued in overnight cultures. The 5-R peptide induced earlier CPB production than the 5-L peptide for both the CN3685agrBko and CN1795agrBko mutants (Fig. 4).
FIG 4.
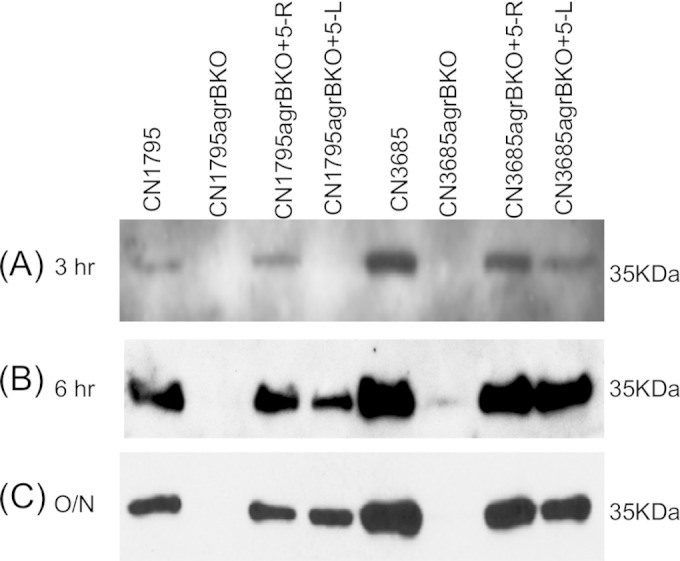
Time course study of CPB production by CN1795agrBko and CN3685agrBko in the presence of peptides 5-R and 5-L. Signaling peptide (100 μM) was added to the TGY broth followed by the inoculation of bacteria at 37°C. Samples were collected at 3 h, 6 h, and 16 h (overnight), and those supernatants were then used for Western blot detection for CPB production. The kinetics of natural CPB production by TGY cultures of wild-type CN3685 or CN1795, without added peptide, are shown for comparison. All experiments were repeated three times.
Effects of amino acid variations on CLWFT signaling.
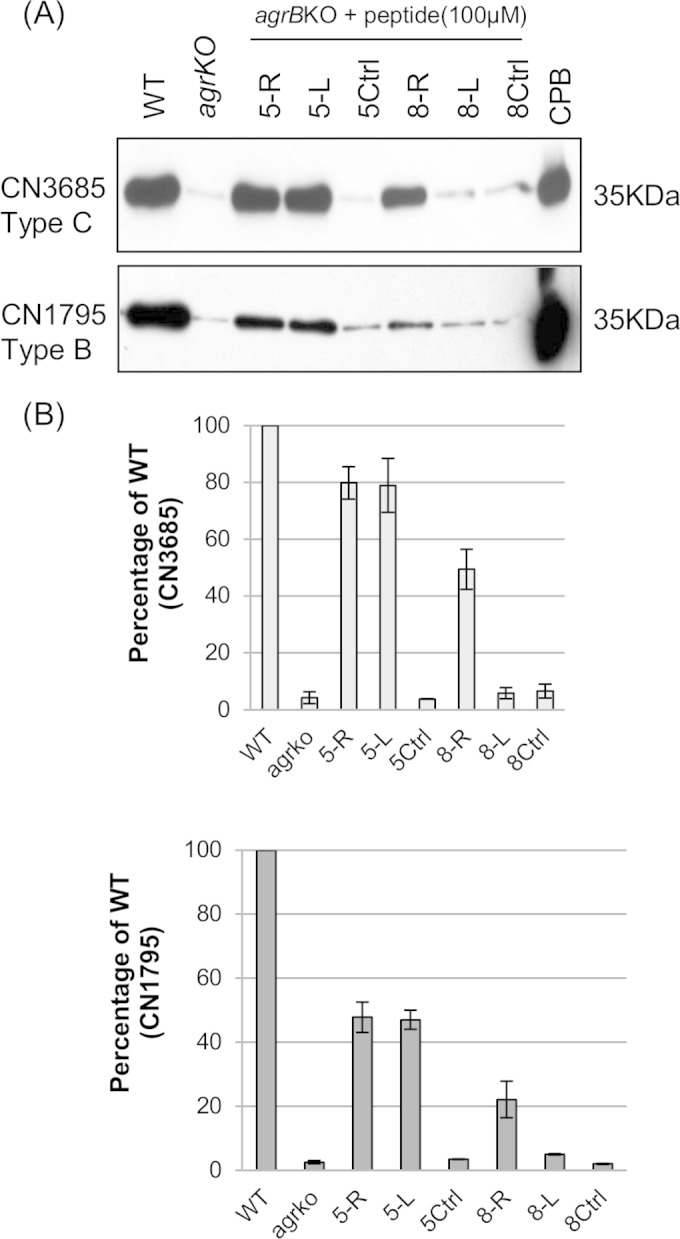
Since the cysteine residue in the CLWFT sequence is conserved among the AgrD sequences of both CN1795 and CN3685 (15, 16), this C residue could be essential for signaling activity. To test this hypothesis, synthetic peptides were prepared where the C residue was varied to create SLWFT or VLWFT peptides, both of which have a molecular weight similar to that of CLWFT (Fig. 1). To study if a thiolactone ring structure is essential for signaling, both cyclic and linear forms of SLWFT were synthesized (5-RCS and 5-LCS). Those variant peptides were then evaluated for their ability to restore CPB production by overnight TGY cultures of the CN3685agrBko and CN1795agrBko mutants.
The SLWFT peptides, in either cyclic (5-RCS) or linear (5-LCS) form, were found to induce CPB production by both CN3685agrBko and CN1795agrBko, similarly to peptides 5-R or 5-L (Fig. 5). However, the same concentration (100 μM) of 5-LCV (the variant VLWFT peptide) failed to induce CPB production by those two mutants (Fig. 5). In addition, a 5-mer peptide (CVLVT) containing AgrD sequences of C. acetobutylicum in a thiolactone ring (25) also failed to stimulate CPB production, indicating there is some peptide sequence specificity for stimulating the C. perfringens Agr-like QS system.
FIG 5.
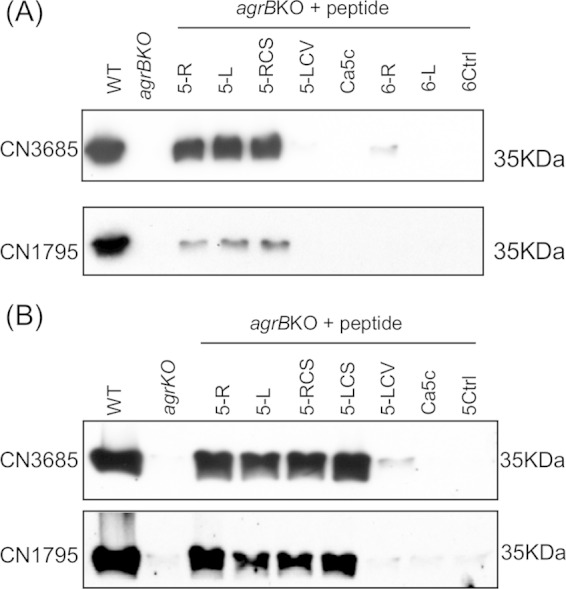
Structure-function analysis of the signaling peptides. (A) Western blot detection of overnight CPB production by TGY cultures of the CN3685 and CN1795 agrB gene null mutants after activation by 100 μM concentrations of AgrD sequence-based peptides (5-R and 5-L) or variant peptides (5-RCS, 5-LCV, Ca5c, 6-L, and 6-R). A 6-mer random sequence peptide (6Ctrl) was used as a negative control. (B) CPB production by overnight TGY cultures of the CN3685 agrB null mutant and the CN1795 agrB null mutant after treatment with 100 μM concentrations of 5-R, 5-L, 5-RCS, 5-LCS, 5-LCV, Ca5c. A similar concentration of a 5-mer random sequence peptide (5Ctrl) was used as a negative control. These experiments were repeated twice.
Comparison of signaling properties of 5-mer versus 6-mer C. perfringens AgrD sequence-based peptides.
While some 5-mer and 8-mer peptides can be sensed by CN3685agrBko to induce CPB production (Fig. 2), a recent study (25) demonstrated that a 6-mer cyclic peptide with the C. acetobutylicum AgrD-based sequence CVWVTH optimally signals the Agr-like QS system of that bacterium, although signaling was also observed when additional AgrD sequence-based amino acids were added to the ring as a tail. Therefore, the current study investigated whether a 6-mer cyclic peptide based on the C. perfringens AgrD sequence might also be optimally recognized by C. perfringens. For this purpose, both a C. perfringens AgrD sequence-based 6-mer cyclic peptide CLWFTH (6-R) and a linear form of this peptide (6-L) were synthesized (Fig. 1). Those variant peptides were then evaluated for their ability to restore CPB production by overnight TGY cultures of the CN3685agrBko and CN1795agrBko mutants.
As shown in Fig. 5, even 100 μM concentrations of the 6-mer cyclic form of the CLWFTH peptide (i.e., peptide 6-R) induced production of only small amounts of CPB by CN3685agrBko and no CPB production by CN1795agrBko, indicating that the 6-mer ring form of CLWFT is not well recognized by these C. perfringens strains. The linear CLWFTH (6-L) peptide, at 100 μM concentrations, also failed to induce any CPB production, in contrast to the strong CPB production that the linear 5-L peptide induced in CN3685agrBko or CN1795agrBko.
Comparison of peptide signaling among C. perfringens strains.
Our previous study indicated that when CN3685agrBko and a CN3685cpbko null mutant were cocultured, but physically separated in a Transwell, CN3685agrBko responds by producing CPB (16). Therefore, the current study examined whether CN3685agrBko and CN1795agrBko can also sense and produce CPB in response to the natural Agr-like QS system signaling peptide supplied by other C. perfringens strains, including type A wild-type strains 13 and SM101, a type B strain CN1795 cpb gene null mutant (CN1795cpbko), type C strain CN3685cpbko, and type D wild-type strains CN2068 and CN3718 (Table 1).
C. perfringens type A and type D strains do not produce CPB (4), so wild-type isolates of those types could be used as a source of Agr-like QS system signaling peptide in this experiment. However, both type B and C strains must produce CPB (4), which made it necessary to use isogenic cpb gene null mutants of these strains for this study. Therefore, the current study used an isogenic CN3685 cpb null mutant that had been constructed and characterized in previous research (18). However, the CN1795 cpb null mutant (named CN1795cpbko) had to be constructed in this study by using Clostridium-modified TargeTron technology (18), and this mutant was then characterized both genetically and phenotypically (Fig. 6). Compared to the ∼900-bp product that was amplified from the wild-type cpb gene in CN1795, PCR with internal cpb primers cpbF/R (18, 24) amplified an ∼1,800-bp band from the CN1795cpbko mutant, indicating successful insertion of the ∼900-bp intron (Fig. 6A). Southern hybridization with an intron-specific probe demonstrated the presence of a single intron insertion in CN1795cpbko (Fig. 6B). Moreover, Western blot hybridization using a CPB-specific antibody did not detect any CPB production by an overnight TGY culture of CN1795cpbko, although this Western blot anaylsis detected very strong CPB production by wild-type CN1795 cultured under the same conditions.
FIG 6.
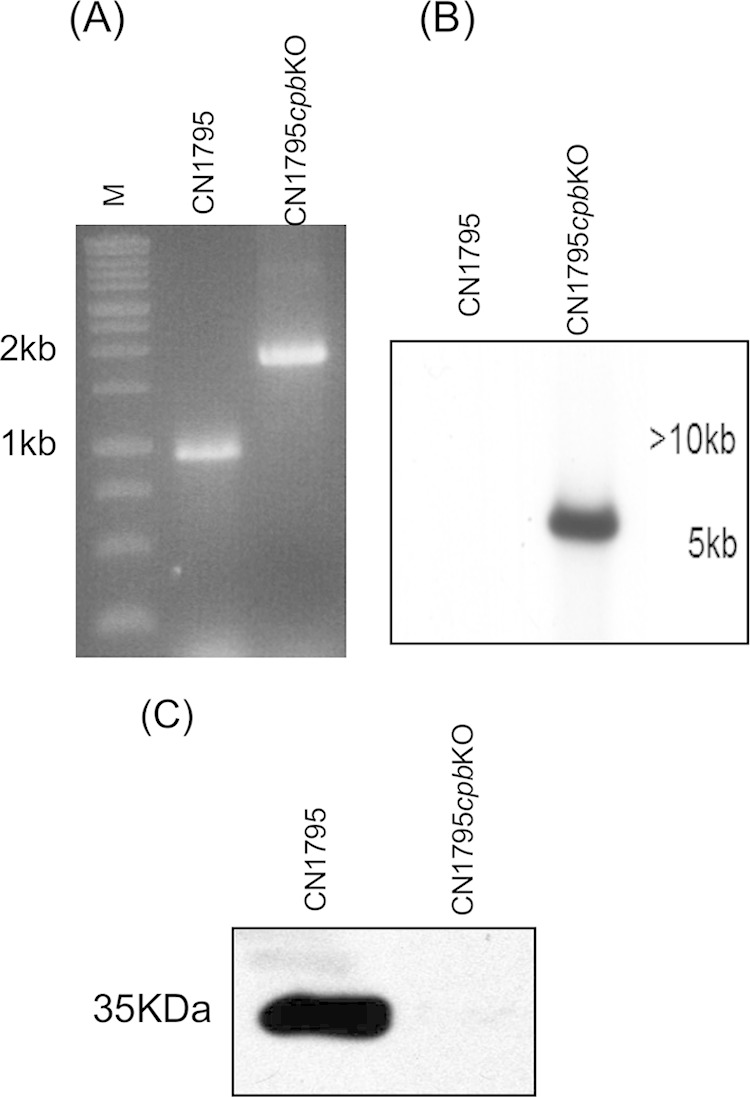
Inactivation of the cpb gene in C. perfringens type B strain CN1795 using the TargeTron gene-knockout system. (A) PCR confirmation of the isogenic cpb gene null mutant CN1795cpbko. Using primers (cpbF/cpbR) (18) that target the specific cpb gene in C. perfringens, a PCR assay using primers to an internal cpb sequence amplified an ∼900-bp product from the isolated DNA of wild-type CN1795. However, PCR using the same primers amplified an ∼1.8-kb product from the isolated DNA of CN1795cpbko, which indicated the insertion of the ∼900-bp group II intron Ltr. Lane M, 1-kb molecular mass marker. (B) Southern blot hybridization of a DIG-labeled intron-specific probe with DNA purified from wild-type CN1795 and cpb null mutant CN1795cpbko. DNA from each strain was digested with EcoRI and electrophoresed on a 1% agarose gel prior to blotting and hybridization with the intron probe. (C) Western blot for CPB production by overnight TGY cultures of wild-type CN1795 and cpb null mutant CN1795cpbko using an anti-CPB antibody.
A previously described (16) Transwell culture system was used to study whether the CN3685agrBko or CN1795agrBko mutants can recognize the natural Agr-like QS system signaling peptide made by the other C. perfringens strains described above. A membrane filter was present between the top and bottom chambers of each Transwell to prevent physical contact between the bacterial cells growing in the different chambers; therefore, only secreted small molecules could pass through the filter pores from one chamber to the other chamber. Using this system, CN3685agrBko could recognize the natural argD-encoded signaling peptides produced by all of the other tested donor strains, including CN1795cpbko (Fig. 7A). However, CN1795agrBko was not able to sense the signaling peptide produced by any of these donor isolates, including CN1795cpbko (Fig. 7B). These data demonstrated that CN3685 recognizes the natural Agr-like QS system signaling peptide, as well as AgrD sequence-based synthetic peptides, more sensitively than does CN1795.
FIG 7.
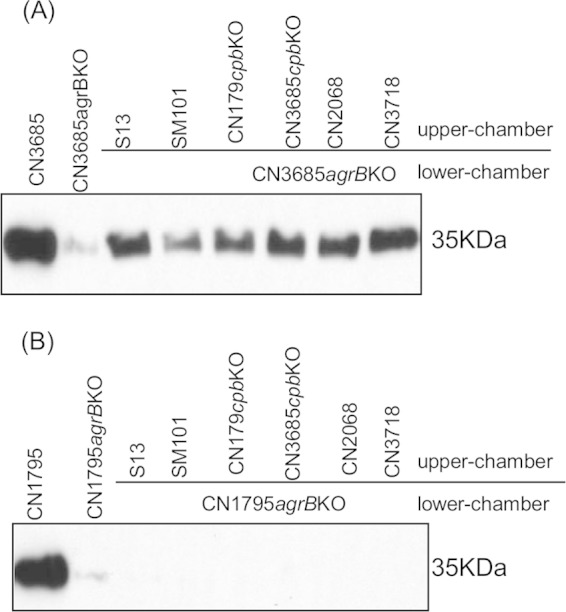
Transwell assay to determine if the natural signaling peptide produced by other C. perfringens strains can also activate the Agr-like system in CN3685agrBko and CN1795agrBko. The agrB null mutants (CN3685agrBko [A] and CN1795agrBko [B]) were inoculated into fresh TGY broth in the bottom chamber of Transwells as a recipient, while the donor strain was inoculated into fresh TGY broth in the top chamber. A membrane filter with 0.45-μm pores separated the two chambers, preventing bacterial passage between chambers but allowing the exchange of polypeptides. Donor strains included type A strain 13, SM101, type B strain CN1795cpbko, type C strain CN3685cpbko, and type D strains CN2068 and CN3718. The Transwell plate was anaerobically incubated overnight at 37°C, and culture supernatant was then collected for Western blot detection of CPB production. Wild-type strains CN3685 and CN1795 were used to compare CPB production in overnight TGY cultures. The image shows representative results that were reproducible over three repetitions.
Evaluation of the stability of signaling peptides in C. perfringens cultures.
Our data indicated that CN3685agrBko is better than CN1795agrBko at sensing Agr-like QS system signaling peptides, with higher concentrations of signaling peptides needed to induce equivalent CPB production by CN1795agrBko and CN3685agrBko. One possible explanation for this difference is that the signaling peptides are less stable, due to extracellular degradation or inactivation, in CN1795 cultures than in CN3685 cultures.
To test this hypothesis, the signaling peptides 5-R and 5-L were first preincubated for 2 h with sterile overnight supernatants of two cpb null mutants named CN1795cpbko and CN3685cpbko. CN3685agrBko produced similar levels of CPB when grown in supernatants removed from CN3685cpbko or CN1795cpbko cultures (Fig. 8A and B). Similarly, CN1795argBko produced equivalent amounts of CPB when grown in supernatants removed from CN3685cpbko and CN1795cpbko cultures. These data suggested that the different peptide sensing sensitivities for CN3685 and CN1795 were not due to stronger extracellular peptide degradation or inactivation by CN1795.
FIG 8.
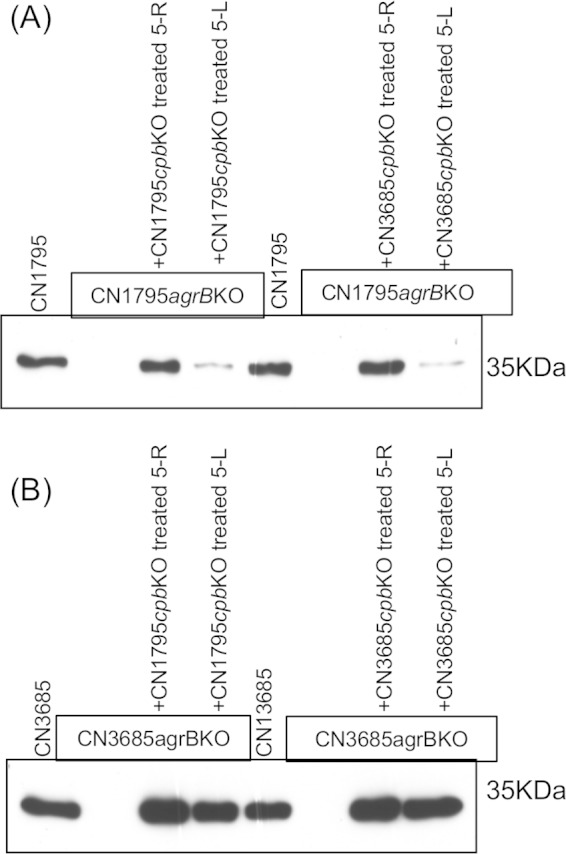
Investigation of the possible inactivation of signaling peptides by CN1795 or CN3685. A 100 μM concentration of peptides 5-R or 5-L was incubated for 2 h with 100 μl of supernatant from TGY broth cultures of CN1795cpbko or CN3685cpbko, followed by adding 900 μl of fresh TGY broth. CN1795agrBko or CN3685agrBko was then inoculated into this mixture. After a 5-h anaerobic culture at 37°C, the culture supernatants were collected for CPB detection using Western blot analysis. (A) CPB production by CN1795agrBko. Wild-type strain CN1795 was used as a positive control, while CN1795agrBko was used as a negative control. (B) CPB production by CN3685agrBko. Wild-type strain CN3685 was used as a positive control, while CN3685agrBko was used as a negative control.
The 6-R synthetic peptide inhibits signaling by the Agr-like QS system in some C. perfringens strains.
Some S. aureus AIPs can inhibit Agr signaling in other S. aureus strains (20, 21). Therefore, the 5-LCV, 5Ctrl, Ca5c, 6-R, 6-L, 6Ctrl, 8-L, and 8Ctrl synthetic peptides, which were unable to induce CPB production by CN3685agrBko or CN1795agrBko in Fig. 5, were tested for their potential ability to inhibit CPB production by wild-type CN3685 and CN1795.
Of those peptides, 6-R was found to decrease CPB production by both wild-type CN3685 and CN1795 (Fig. 9A). Given that result, several other type B and type C strains were tested to explore whether peptide 6-R can also inhibit their CPB production. Results from those experiments indicated that the reduction of overnight CPB production induced by this peptide varied among different C. perfringens strains. As noted for CN3685 and CN1795, 6-R blocked CPB production by PS49, CN2003, and CN3763 and greatly decreased CPB production by Bar2 and CN5383. However, this peptide did not affect CPB production by CN1793, Bar3, or CN3955 (Fig. 9B).
FIG 9.

Peptide 6-R shows an inhibitory effect on the native signaling peptide produced by some C. perfringens strains. (A) Competition between native AIP and the synthetic signaling peptide 6-R to inhibit overnight CPB production by agrB null mutants of CN1795 (top) or CN3685 (bottom). (B) Effects of 6-R on CPB toxin production by other strains, including type B PS49, CN1793, Bar2, and CN2003 and type C Bar3, CN5383, CN3955, and CN3763. A final concentration of 100 μM 6-R was added into fresh TGY broth, followed by inoculation of these wild-type strains and overnight anaerobic incubation at 37°C. Bacterial culture in the absence of 6-R was used as a control. The presence of CPB in the overnight culture was detected by CPB Western blotting. (C) CPB production was upregulated after 2 h in the presence of Caco-2 host cells, and the 6-R peptide could inhibit this toxin upregulation.
Our previous studies (16, 23) showed that contact with enterocyte-like Caco-2 cells triggers faster production of CPB by CN3685 and that this effect requires the Agr-like QS system. Therefore, the ability of the 6-R peptide to inhibit early (2-h) CPB production by CN3685 or CN1795 in the presence of Caco-2 cells was also explored. Results from these analyses revealed that the 6-R peptide could inhibit the earlier production of CPB by both CN1795 and CN3685 that is induced by the presence of Caco-2 cells (Fig. 9C).
The 6-R peptide can compete against the 5-R signaling peptide.
Data shown in Fig. 2, Fig. 9, and Fig. 10 indicated that the 5-R peptide induces strong overnight CPB production by the CN1795 agrB null mutant, while the 6-R peptide can inhibit overnight CPB production by wild-type CN1795. Therefore, we directly tested the possibility that the 6-R peptide can compete against the 5-R peptide and inhibit overnight CPB production by C. perfringens.
FIG 10.
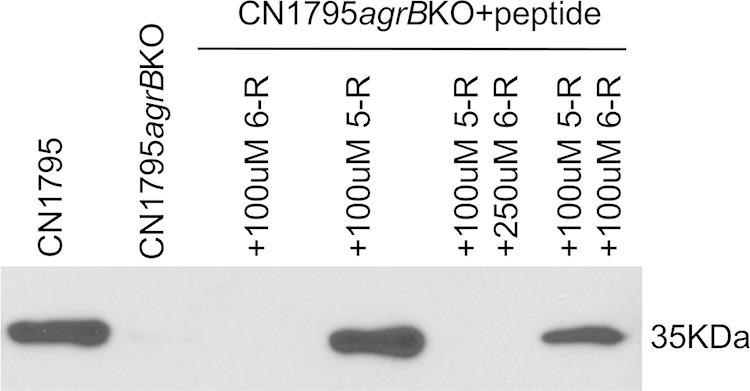
Peptide 6-R can compete with 5-R in the agrB-agrD system. Final 100 or 250 μM concentrations of peptide 6-R, along with a 100 μM concentration of peptide 5-R, were added into fresh TGY broth, followed by inoculation of that broth with CN1795agrBko. The culture was anaerobically incubated overnight at 37°C. Culture supernatant was collected for detection of CPB using a Western blot assay. Cultures of wild-type CN1795, CN1795agrBko, CN1795agrBko with the addition of 100 μM 5-R, and CN1795agrBko with the presence of 100 μM 6-R were tested similarly as controls.
Consistent with Fig. 9 results, the 6-R peptide did not induce CPB production by overnight cultures of CN1795agrBko, while the 5-R peptide induced strong overnight CPB production by this strain. However, when a 100 or 250 μM concentration of 6-R peptide and a 100 μM concentration of 5-R peptide were added together to CN1795agrBko, overnight CPB production was significantly decreased, indicating competition between these two peptides (Fig. 10). The extent of this competition was stronger using a 250 μM (95.8% ± 3.7% inhibition) than a 100 μM (60.5% ± 16.5% inhibition) concentration of the 6-R peptide.
DISCUSSION
Many Gram-positive pathogens use QS systems to control production of their virulence factors (22, 25, 27). These QS systems typically involve short signaling peptides, although the size, nature, and structure of those peptides vary widely. For example, Bacillus cereus and Streptococcus pyogenes regulate the expression of their virulence factors via QS systems that utilize linear signaling peptides ranging in length from 7 to 17 amino acids (22). Similarly, the AIP used by Gram-positive bacteria possessing an Agr-like QS system varies considerably in size, ranging from 5 to 12 amino acids (22, 27). However, it is generally accepted that Agr-like QS signaling in these bacteria can be triggered only by AIPs containing a thiolactone or, occasionally, lactone ring (20, 21).
Based upon analogy with natural S. aureus AIPs, but containing C. perfringens AgrD sequences, our previous studies (16) had synthesized an 8-mer peptide named 8-R which contains a 5-amino-acid thiolactone ring and a 3-amino-acid tail. This 8-R peptide was shown to induce CPB production by a CN3685 agrB null mutant (16). However, consistent with the consensus literature (20–22) for other Agr QS system-possessing bacteria, which indicated that AgrD sequence-based linear peptides do not signal, a peptide (named 8-L) corresponding to the linear form of the 8-R peptide was unable to induce CPB production. Those previous findings (16) were confirmed in the present study.
The 8-L and 8-R peptide signaling results might have indicated that the C. perfringens Agr-like QS system uses an AIP similar to the S. aureus AIPs, where the 3- to 4-amino-acid tails are thought to mediate the specificity of AIP binding to AgrC (20, 21). However, the current study also demonstrated that a 5-mer peptide (5-R) containing only a thiolactone ring could signal the CN3685 and CN1795 agrB null mutants. Furthermore, the current study determined that both linear and lactone ring-containing peptide versions of 5-R can also induce nearly as strong signaling as the 5-R thiolactone ring peptide, although for the 5-L peptide this signaling develops more slowly than for 5-R. Previous studies (20) showed that the S. aureus Agr system is activated only by thiolactone ring-containing peptides; however, both lactone ring-containing and thiolactone ring-containing peptides can induce Agr signaling in S. intermedius (20). To our knowledge, the clear ability of two linear peptides, i.e., 5-L and 5-LCS, to induce Agr signaling, as demonstrated in the current study, is a novel finding for any Agr-like QS system.
It has been suggested that the ability of S. intermedius to sense both thiolactone and lactone ring-containing peptides may reflect evolutionary memory (20). The versatile ability of C. perfringens to sense multiple 5-mer peptide variants might similarly involve evolutionary memory, where the AIP binding receptor of this bacterium has evolved flexibility to recognize 5-mer peptides in various linear or ring forms and 8-mer peptides in a thiolactone ring form; in this case, the 8-mer linear peptides might simply be too large and unfolded a structure to dock with the C. perfringens AIP receptor. In this regard, it should be mentioned again that C. perfringens does not encode the AgrC receptor used by S. aureus to bind its Agr QS system AIP. The functional equivalent(s) of AgrC in C. perfringens has yet to be conclusively determined. It has been suggested (12) that the C. perfringens AIP receptor is the VirS protein, but inactivation of the virS-virR operon does not affect production levels of all C. perfringens proteins that are regulated by the Agr-like QS system (8, 14).
Another observation from the current study suggests an intriguing alternative possibility to explain signaling by the 5-mer linear peptides in C. perfringens. In contrast to the linear signaling peptides 5-L and 5-LCS, the linear 5-mer LCV peptide was unable to induce CPB production by AgrB mutants. Since the 5-LCV peptide lacks the serine or cysteine residues required to form lactone or thiolactone rings, respectively, the inability of this peptide to induce signaling might indicate that C. perfringens can convert the linear 5-L and 5-LCS peptides to ring forms that then become active Agr signaling molecules. If so, this ring formation would have to be AgrB independent, since even the AgrB mutants responded to the linear 5-L and 5-LCS peptides. In this regard, it is notable that ring formation during AgrD processing is still a poorly understood process, even in the paradigm S. aureus system (20, 21). Determining why C. perfringens can respond to some linear 5-mer peptides is a goal of future investigations.
The 5-R, 5-L, 5-LCS, and 5-RCS peptides were considerably more efficient at inducing CPB production than peptide 8-R, suggesting that the natural AIP of C. perfringens may be a 5-mer peptide. A precedent already exists for tailless AIPs, since the AIP of the L. plantarum Agr system is a 5-mer thiolactone ring (20). Also, the AIP of C. acetobutylicum is proposed to be only a 6-mer thiolactone ring, without an amino acid tail (25).
In the current study, strong overnight CPB production could be induced in CN3685agrBko by even 10 μM concentrations of 5-R, which is less than the 25 μM amounts of a synthetic peptide corresponding to a proven natural S. aureus AIP that was used in previous studies to induce S. aureus toxin production (26), supporting possible physiologic relevance for our current findings. In addition, if C. perfringens and S. aureus are assumed to produce roughly similar amounts of their AIPs, the ability of 10 μM concentrations of 5-mer peptides to trigger Agr signaling in C. perfringens may further support physiologic relevance, since S. aureus naturally produces ∼10 μM concentrations of AIP (26).
Another novel finding of the current study is that C. perfringens strains CN3685 and CN1795 vary considerably in their sensitivity to signaling peptides, with CN3685 being much more sensitive. This does not appear to be attributable to CN1795 extracellularly inactivating AIPs. It is also notable that CN1795 was found to be even less sensitive than CN3685 to the natural CN1795 AIP. These results suggest either variability in AIP receptors or in AIP receptor density among C. perfringens strains. Precise discrimination of these two possibilities will require identification of the AIP receptor protein in C. perfringens.
The current study provides further insights into the possible role of the Agr-like QS system during C. perfringens pathogenesis. First, time course experiments performed in these studies indicate that some signaling peptides (e.g., 5-R) can induce CPB production within 3 h. This finding is in agreement with our previous work showing that, in the intestines, washed wild-type CN3685 cells can produce CPB and induce pathology within 5 to 6 h, while washed CN3685agrBko cells do not produce CPB or induce pathology within this time frame (16). Second, the demonstration that C. perfringens strains can respond to AIPs produced by other strains of this bacterium could indicate that AIPs supplied by normal gastrointestinal C. perfringens can signal disease strains to upregulate their production of toxins, like CPB, involved in intestinal infections. To this point, it is notable that previous studies (28) detected the presence of C. perfringens in the normal flora of approximately half of healthy American adults.
Finally, the ability of CN3685agrBko to respond to natural AIPs produced by a broad range of C. perfringens strains suggests limited diversity in the C. perfringens AIP. If so, this would contrast with the situation in S. aureus, where four different AIPs exist and some of these AIPs are capable of inhibiting Agr signaling in other S. aureus strains (20, 21). In that regard, it is notable that peptide 6-R was able to block or reduce CPB production by some C. perfringens strains. This finding supports diversity in C. perfringens AIP receptors or their density and also opens the possibility of using peptides to block C. perfringens toxin production during infections. The basis for 6-R peptide inhibition differences among C. perfringens strains is not yet clear and will require further study, but these differences do not obviously correlate with strain geographic origin, date of isolation, or disease involvement.
ACKNOWLEDGMENTS
This research was generously supported by grant R01 AI-056177 (B.A.M.) from the National Institute of Allergy and Infectious Diseases.
We thank P. Hauer for providing the CPB monoclonal antibodies used in the study and Kazi Islam and Raymond Yurko for peptide synthesis at the Peptide and Peptoid Synthesis Core Facility Division of the Health Sciences Core Research Facilities (HSCRF) at the University of Pittsburgh.
REFERENCES
- 1.McClane BA, Uzal FA, Miyakawa MF, Lyerly D, Wilkins TD. 2006. The enterotoxic clostridia, p 688–752. In Dworkin M, Falkow S, Rosenburg E, Schleifer H, Stackebrandt E (ed), The prokaryotes, 3rd ed Springer Press, New York, NY. [Google Scholar]
- 2.Rood JI. 2006. Clostridium perfringens and histotoxic disease, p 753–770. In Dworkin M, Falkow S, Rosenburg E, Schleifer H, Stackebrandt E (ed), The prokaryotes, 3rd ed Springer Press, New York, NY. [Google Scholar]
- 3.Hatheway C. 1990. Toxigenic clostridia. Clin Microb Rev 3:66–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li J, Adams V, Bannam TL, Miyamoto K, Garcia JP, Uzal FA, Rood JI, McClane BA. 2013. Toxin plasmids of Clostridium perfringens. Microbiol Mol Biol Rev 77:208–233. doi: 10.1128/MMBR.00062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McDonel JL. 1986. Toxins of Clostridium perfringens types A, B, C, D, and E, p 477–517. In Dorner F, Drews H (ed), Pharmacology of bacterial toxins. Pergamon Press, Oxford, United Kingdom. [Google Scholar]
- 6.Ohtani K, Shimizu T. 2014. Regulation of toxin gene expression in Clostridium perfringens. Res Microbiol doi: 10.1016/j.resmic.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 7.Ba-Thein W, Lyristis M, Ohtani K, Nisbet IT, Hayashi H, Rood JI, Shimizu T. 1996. The virR/virS locus regulates the transcription of genes encoding extracellular toxin production in Clostridium perfringens. J Bacteriol 178:2514–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J, Ma M, Uzal FA, McClane BA. 2014. Host cell-induced signaling causes Clostridium perfringens to upregulate production of toxins important for intestinal infections. Gut Microbes 5:96–107. doi: 10.4161/gmic.26419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohtani K, Hirakawa H, Tashiro K, Yoshizawa S, Kuhara S, Shimizu T. 2010. Identification of a two-component VirR/VirS regulon in Clostridium perfringens. Anaerobe 16:258–264. doi: 10.1016/j.anaerobe.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 10.Li J, McClane BA. 2010. Evaluating the involvement of alternative sigma factors SigF and SigG in Clostridium perfringens sporulation and enterotoxin synthesis. Infect Immun 78:4286–4293. doi: 10.1128/IAI.00528-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harry KH, Zhou R, Kroos L, Melville SB. 2009. Sporulation and enterotoxin (CPE) synthesis are controlled by the sporulation-specific factors SigE and SigK in Clostridium perfringens. J Bacteriol 191:2728–2742. doi: 10.1128/JB.01839-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohtani K, Yuan Y, Hassan S, Wang R, Wang Y, Shimizu M. 2009. Virulence gene regulation by the agr system in Clostridium perfringens. J Bacteriol 191:3919–3927. doi: 10.1128/JB.01455-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vidal JE, Chen J, Li J, McClane BA. 2009. Use of an EZ-Tn5-based random mutagenesis system to identify a novel toxin regulatory locus in Clostridium perfringens strain 13. PLoS One 4:e6232. doi: 10.1371/journal.pone.0006232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J, Rood JI, McClane BA. 2011. Epsilon toxin production by Clostridium perfringens type D strain CN3718 is dependent upon the agr operon but not the VirS/VirR. mBio 2(6):e00275-300275-11. doi: 10.1128/mBio.00275-11. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Chen J, McClane BA. 2012. Role of the Agr-like quorum-sensing system in regulating toxin production by Clostridium perfringens type B strains CN1793 and CN1795. Infect Immun 80:3008–3017. doi: 10.1128/IAI.00438-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vidal JE, Ma M, Saputo J, Garcia J, Uzal FA, McClane BA. 2012. Evidence that the Agr-like quorum sensing system regulates the toxin production, cytotoxicity and pathogenicity of Clostridium perfringens type C isolate CN3685. Mol Microbiol 83:179–194. doi: 10.1111/j.1365-2958.2011.07925.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Chen J, Vidal JE, McClane BA. 2011. The Agr-like quorum-sensing system regulates sporulation and production of enterotoxin and beta2 toxin by Clostridium perfringens type A non-food-borne human gastrointestinal disease strain F5603. Infect Immun 79:2451–2459. doi: 10.1128/IAI.00169-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sayeed S, Uzal FA, Fisher DJ, Saputo J, Vidal JE, Chen Y, Gupta P, Rood JI, McClane BA. 2008. Beta toxin is essential for the intestinal virulence of Clostridium perfringens type C disease isolate CN3685 in a rabbit ileal loop model. Mol Microbiol 67:15–30. doi: 10.1111/j.1365-2958.2007.06007.x. [DOI] [PubMed] [Google Scholar]
- 19.Painter KL, Krishna A, Wigneshweraraj S, Edwards AM. 2014. What role does the quorum-sensing accessory gene regulator system play during Staphylococcus aureus bacteremia? Trends in Microbiol 22:676–685. doi: 10.1016/j.tim.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 20.Novick RP, Geisinger E. 2008. Quorum sensing in staphylococci. Annu Rev Genet 42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 21.Thoendel M, Kavanaugh JS, Flack CE, Horswill AR. 2011. Peptide signaling in the staphylococci. Chem Rev 111:117–151. doi: 10.1021/cr100370n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gray B, Hall P, Gresham H. 2013. Targeting agr- and agr-like quorum sensing systems for development of common therapeutics to treat multiple Gram-positive bacterial infections. Sensors 13:5130–5166. doi: 10.3390/s130405130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vidal JE, Ohtani K, Shimizu T, McClane BA. 2009. Contact with enterocyte-like Caco-2 cells induces rapid upregulation of toxin production by Clostridium perfringens type C isolates. Cell Microbiol 11:363–369. doi: 10.1111/j.1462-5822.2008.01272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y, McClane BA, Fisher DJ, Rood JI, Gupta P. 2005. Construction of an alpha toxin gene knockout mutant of Clostridium perfringens type A by use of a mobile group II intron. Appl Environ Microbiol 71:7542–7547. doi: 10.1128/AEM.71.11.7542-7547.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steiner E, Scott J, Minton NP, Winzer K. 2012. An agr quorum sensing system that regulates granulose formation and sporulation in Clostridium acetobutylicum. Appl Environ Microbiol 78:1113–1122. doi: 10.1128/AEM.06376-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDowell P, Affas Z, Reynolds C, Holden MT, Wood SJ, Saint S, Cockayne A, Hill PJ, Dodd CE, Bycroft BW, Chan WC, Williams P. 2001. Structure, activity and evolution of the group I thiolactone peptide quorum-sensing system of Staphylococcus aureus. Mol Microbiol 41:503–512. doi: 10.1046/j.1365-2958.2001.02539.x. [DOI] [PubMed] [Google Scholar]
- 27.Olson ME, Todd DA, Schaeffer CR, Paharik AE, Van Dyke MJ, Buttner H, Dunman PM, Rohde H, Cech NB, Fey PD, Horswill AR. 2014. Staphylococcus epidermidis agr quorum-sensing system: signal identification, cross talk, and importance in colonization. J Bacteriol 196:3482–3493. doi: 10.1128/JB.01882-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carman RJ, Sayeed S, Li J, Genheimer CW, Hiltonsmith MF, Wilkins TD, McClane BA. 2008. Clostridium perfringens toxin genotypes in the feces of healthy North Americans. Anaerobe 14:102–108. doi: 10.1016/j.anaerobe.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sayeed S, Li J, McClane BA. 2010. Characterization of virulence plasmid diversity among Clostridium perfringens type B isolates. Infect Immun 78:495–504. doi: 10.1128/IAI.00838-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gurjar A, Li J, McClane BA. 2010. Characterization of toxin plasmids in Clostridium perfringens type C isolates. Infect Immun 78:4860–4869. doi: 10.1128/IAI.00715-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sayeed S, Li J, McClane BA. 2007. Virulence plasmid diversity in Clostridium perfringens type D isolates. Infect Immun 75:2391–2398. doi: 10.1128/IAI.02014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]