Abstract
Background:
β-thalassemia, a monogenic autosomal recessive disorder, is prevalent in Middle East, particularly in Iran. In Iran, near to 20 mutations in the β-globin gene are introduced as common mutations with varying incidence frequencies in each city. Therefore, detection and screening for couples at high risk can help to solve the problems of this disease. In this study, optimized genotyping of two common mutations in Isfahan Province, IVSII-I (G-A) and FSC-8/9 insG, was performed using the T-ARMS method.
Methods:
In this case-control study, 10 healthy individuals and 30 patients affected by β-thalassemia major with a mean 24.76 ± 4.5 years were selected from Omid Hospital in Isfahan Province. After designing tetra primers for two prevalent mutations IVSII-I (G-A) and FSC-8/9 insG, samples were genotyped using tetra-primers ARMS PCR technique.
Results:
We have developed a sensitive single tube tetra-primers PCR assay to detect both IVSII-1 (G-A) and FS8-9 insG mutations. Moreover, we have distinguished homozygous and heterozygous forms of these mutations successfully. The frequency of IVSII-1 (G-A) mutation from 30 patients in Isfahan was 86.6% (33.3% heterozygote, and 53.3% mutant homozygote) and for FS8-9 insG mutation was 16.6% (13.3% heterozygote, and 3.3% mutant homozygote).
Conclusion:
Tetra-primers ARMS PCR could be a reliable, accurate and simple technique for genotyping SNP and different mutations. So far, no study was done on optimization methods for genotyping mutations in β-thalassemia by T-ARMS. Here, we successfully adjusted and enhanced this method for recognizing two common mutations (FSC-8/9 insG and IVSII-I (G-A)) of β-thalassemia in Isfahan population.
Keywords: β-thalassemia, IVSII-I mutation, FSC-8/9 mutation, Tetra primer amplification refractory mutation system (T-ARMS) method
Introduction
β-thalassemia syndromes are a group of hemoglobin disorders caused by mutations in the β-globin gene resulting in reduction (β + mutations) or loss (β 0 mutations) of β-globin chains synthesis (1, 2). β-thalassemia is a monogenic autosomal recessive disease fairly common worldwide but it is considered prevalent in the Middle East and especially Iran; close to 3 percent of the world population and an average of 5 percent of the Iranian population are carriers of β-thalassemia (2–6). Hence, detection and screening for couples at high risk can help to solve the problems of this disease. So far, close to 200 different mutations in the β-globin gene have been reported that associated with the onset of the disease (5). In Iran, near to 20 mutations are introduced as common mutations including varying incidence frequencies in each city (3, 7). There have been several studies in order to identify common mutations in different cities of Iran (3, 7–13).
In 2008, Derakhshandeh-Peykar et al. identified IVSII-I (G-A) with 41% frequency, FSC-8/9 (+G) with 10% frequency, IVS-I-5 (G-C) with frequency of 24%, FSC-36/37 (−T) with 29% frequency and IVS-I-110 (G-A) with 7% frequency as common mutations in Iran (7).
RFLP (Restriction Fragment Length Polymorphism), ASO-PCR (Allele Specific Oligo nucleotide-Polymerization Chain Reaction) and sequencing are used as a routine tests for genotyping and identifying β-thalassemia mutations in distinct populations, but each method still suffers from specific weaknesses and therefore, finding a simple, sensitive, accurate and inexpensive method for screening purposes is still one of the major problems in genetic diagnostic laboratories (14–16). In 2001, Shu Ye et al. introduced a simple and cost-effective method for genotyping single nucleotide polymorphisms called tetra-primers ARMS PCR (tetra primer amplification refractory mutation system) (17). This method includes a PCR reaction in a vial with two sets of primers followed by an electrophoresis analysis on agarose gel. One set of primers are specific for variety or polymorphism (inner primers) and the others one are outer primers that create control bond in PCR reaction. Inner primers are specific for mutant and wild type alleles and they are designed opposite of each other. PCR reaction using outer and inner primers were done in one vial, so two outer primers and inner-outer primers can react with each other and create different product with different length that they could be separated on gel electrophoresis. In Fig. 1, a schematic drawing for detecting single nucleotide polymorphisms by using the T-ARMS technique is presented. Unlike ASOPCR, in T-ARMS, for much specificity, two inner primers have another mismatch at second nucleotide near 3′ end.
Fig. 1:
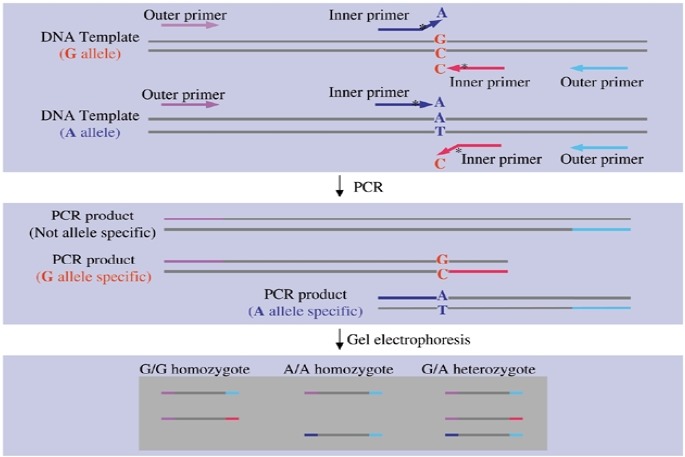
Schematic drawing of tetra-primers ARMS-PCR method. Genetic variation shown is a single nucleotide substitution. By using two pairs of primers, two different and specific segments are amplified for each nucleotide. Specificity of each of these two primers is because of a mispairing at the 3′ ends of the inner primers. However, to make the bindings more specific, a second mispairing is introduced at the second position at the 3′ end of each inner primer (17)
Rules for selecting a nucleotide for additional mismatch in T-ARMS are: a strong mismatch (G/A or C/T) at the 3′ end of an allele- specific primer will likely require a weak second mismatch (C/A or C/T) and vice versa, whereas a medium mismatch (A/A, C/C, G/G or T/T) at 3′ end will likely require a medium second mismatch. In some studies, T-ARMS PCR was used extensively for identification of mutations and polymorphisms and its accuracy and efficiency in genotyping of various mutations has been optimized and validated (18–20).
The aim of this study was designing and optimization of T-ARMS technique for genotyping IVSII-1 (G-A) and FSC-8-9 (+G) in β-thalassemia patients, which they are two of the most common mutations in the population of Isfahan (7).
Materials and Methods
Subjects and study design
The present study was performed in two-groups of case and control to evaluate accuracy and efficiency of tetra-primers ARMS PCR technique for detection and screening of patients.
Samples and DNA extraction:
After obtaining informed consent from healthy individuals and patients affected β-thalassemia major referred to the Isfahan’s Omid Hospital, between 2012 – 2014, 5ml of blood samples were taken from 10 healthy individuals and 30 patients (including 15 men and 15 women with a mean age of 24.76 ± 4.5 years). Then blood samples were collected in tubes containing EDTA. Leukocyte genomic DNA was extracted from blood samples using the standard method of salting out with slight modification (21). Genomics DNA were stored in −20°C after determining their relevant concentrations and analyses on gels electrophoresis.
Primer designing
In order to design the required primers, the sequence surrounding the β-globin gene was first taken referring to gene information bank located in the NCBI online databank (http://www.ncb-i.nlm.nih.gov). Subsequently, suitable primers were designed by using the Primer3 tool (http://frodo.wi.mit.edu/cgibin/primer3) and the final analyses were performed using the Oligo 7 software package. Table 1 shows the primers sequences, binding temperatures and length of PCR products for each allele. As described above, for each mutation genotyping, there are two pair primers sets, one set is the outer primers (outer forward (OF) and outer reverse (OR)) and the other one is inner primers (inner forward (IF) and inner reverse (IR)). In genotyping of IVSII-1 (GA) mutation, the IVSII-I OF and IVSII-I OR were used for amplification of the 391 bp fragment representing the control segment. IVSII-I OF and IVSII-I IR primers were used for amplification of the 268 bp fragment representing the healthy allele (G); and finally amplification of the 172 bp fragment, for identifying the mutant allele (A), was done by IVSII-I IF and IVSII-I OR primers. For the FSC-8-9 mutation, the FSC-8-9 OF and FSC-8-9 OR primers were used for amplification of the 268 bp control fragment; FSC-8-9 IF and FSC-8-9 OR were used for amplification of the healthy allele (−G) with the length of 108 bp; and FSC-8-9 OF and FSC-8-9 IR primers were used for identifying the mutant allele (+G) with the length of 216 bp.
Table 1:
List of primer sequences, binding temperature, and length of the amplified segment for genotyping of IVS-II-I and FSC 8/9 mutations
| Genetic mutation | Primer sequences | Tm | Annealing temperature | Amplified size |
|---|---|---|---|---|
| IVS II-I (G-A) | Forward inner primer (IVS II-I IF) (A allele): 5′-CGTGGATCCTGAGAACTTCATGA-3′ | 63 | 55 | 172 bp (A allele) |
| Reverse inner primer (IVS II-I IR) (G allele): 5′-AAACATCAAGCGTCCCATAGACTAAC-3′ | 62 | 55 | 268 bp (G allele) | |
| Forward outer primer (IVS II-I OF): 5′-TCTATTTTCCCACCCTTAGGCTG-3′ | 62 | 55 | 391 bp (form two outer primers) | |
| Reverse outer primer (IVS II-I OR): 5′-CTAAAACGATCCTGAGACTTCCACA-3′ | 62 | 55 | ||
| FSC 8/9 (+G) | Forward inner primer (FSC 8/9 IF) (−G allele): 5′-CCATGGTGCATCTGACTCCTGAGGAGACGT-3′ | 74 | 68 | 108 bp (−G allele) |
| Reverse inner primer (FSC 8/9 IR) (+G allele): 5’-CCCCACAGGGCAGTAACGGCAGTCC-3′ | 74 | 68 | 216 bp (+G allele) | |
| Forward outer primer (FSC 8/9 OF): 5′-TTAGACCTCACCCTGTGGAGCCACACCCT-3′ | 73.8 | 68 | 268 bp (form tow outer primers) | |
| Reverse outer primer (FSC 8/9 OR): 5′-CTTGATACCAACCTGCCCAGGGCCTCAC-3′ | 74 | 68 |
Polymerase chain reaction and electrophoresis
Each PCR reaction was performed in a final volume of 25 μl, including 100 ng of genomic DNA, 3.5 μl of 10x solution buffer, 1.5 μl of a 10 μM of four mixed dNTPs, 1.5 μl of 50 mM of Mgcl2, 0.25 μl of 5u/μl Taq DNA polymerase (Cinnagene, Co., Iran) and appropriate concentrations of each primer. In order to optimize for multiplex conditions, a temperature gradient along with a concentration gradient of MgCl2 was performed. The optimal time/temperature values for the experiments were as follows: the initial melting step of 95°C for 3 minutes; 30 cycles of thermal denaturation in 95°C for 30 seconds; annealing temperature of 55°C for IVSII-I mutation and 68°C for FSC-8-9 mutation for 35 seconds; initial replication in temperature of 72°C for 1 minute; and final elongation in 72°C for 10 minutes.
After completion of PCR reactions, the products were run on 2% agarose gels alongside a 100 bp marker. After electrophoresis and a dying step with ethidium bromide, bands were observed using a UV Gel Doc apparatus.
Results
Optimization of T-ARMS multiplex conditions for genotyping of IVSII-I mutation:
In order to optimize genotyping of IVSII-I mutation by using the T-ARMS technique, after designing the primers (Table 1), the temperature and concentration gradient of MgCl2 were performed for each pair of primers in separate vials using PCR technique. The temperature of 55°C with a concentration of 6mM of MgCl2 was chosen as optimum annealing temperature and concentration respectively. Control, wild type and mutant product of IVSII-I for patient and health control with optimal annealing temperature and Mg concentration are shown in Fig. 2. In the next step, polymerase chain reaction as a multiplex was performed in a vial with specific concentrations of each primer. Performing multiplex reaction with specific concentrations and different ratios of outer and inner primers showed the optimal concentration for IVSII-I should be 3 pM for outer primers (IVSII-I OF and IVSII-I OR) and 15 pM for inner primers (IVSII-I IR and IVSII-I IF; Fig. 3).
Fig. 2:
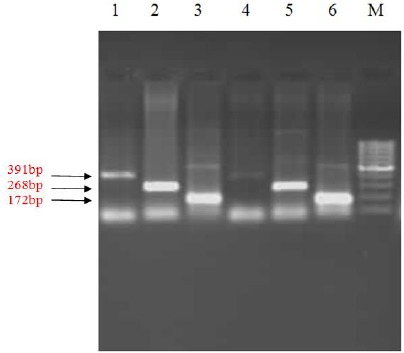
PCR amplification for detection of IVSII-I mutation in separate vials. In lanes 1 and 4, IVSII-I OF and IVSII-I OR primers are used, representing the control sequence. In lanes 2 and 5, IVSII-I OF and IVSII-I IR primers are used, representing the healthy sequence (allele G). Finally, in lanes 3 and 6, IVSII-I IF and IVSII-I OR) are used, representing the mutant sequence (allele A). M is a 100 bp ladder
Fig. 3:
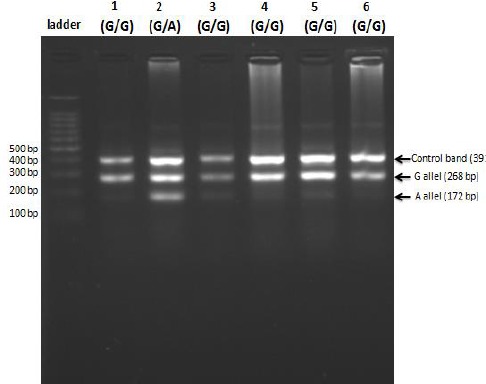
PCR amplification for detection of IVSII-I mutation in one vial using T-ARM technique (multiplex PCR). From left to right, the first lane contains a 100 bp ladder, followed by samples from patient individuals (lanes 1 to 6)
Optimization of T-ARMS multiplex conditions for genotyping of FSC-8/9 mutation
For FSC-8/9 mutation, optimum temperature and concentration gradients of MgCl2 50mM was 68°C and 1.5 μl which obtained by amplification of each fragment in separate vials (Fig. 4). In the next step, polymerase chain reaction as a multiplex was performed in a vial with specific concentration of each primers and it became evident that 5 pM of each primers is suitable for identifying the FSC-8/9 mutation. Results of optimal annealing temperature, primers and MgCl2 concentration for multiplex PCR have been shown in Fig. 5 and 6.
Fig. 4:
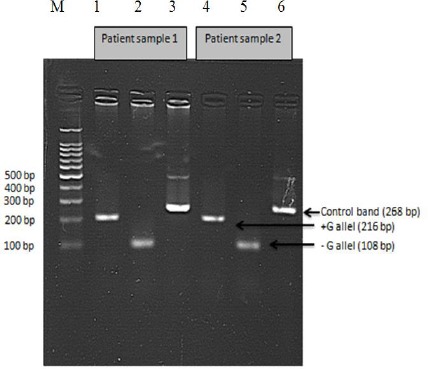
PCR amplification for detection of FSC-8/9 mutant in separate vials. From left to right, M is a 100 bp marker, then in the first three lanes are samples from patient 1 and the next three lanes are samples from patient 2. In lanes 1 and 4, FSC 8/9 IR and FSC 8/9 OF primers are used as indicators of mutant sequence (+G allele). In lanes 2 and 5, FSC 8/9 IF and FSC 8/9 OR primers are used as indicators of the healthy sequence (−G allele). In lanes 3 and 6, control primers (FSC 8/9 OR and FSC 8/9 OF) are used
Fig. 5:
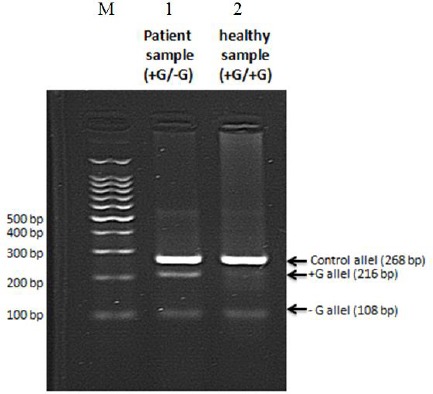
PCR amplification for detection of FSC 8/9 mutation in one vial using T-ARMS technique (multiplex PCR). From left to right, M is a 100 bp marker; lane 1 is a sample from patient; lane 2 is a sample from a healthy individual. The sample from patient is confirmed as heterozygous in multiplex conditions, as demonstrated previously in separate vials
Fig. 6:
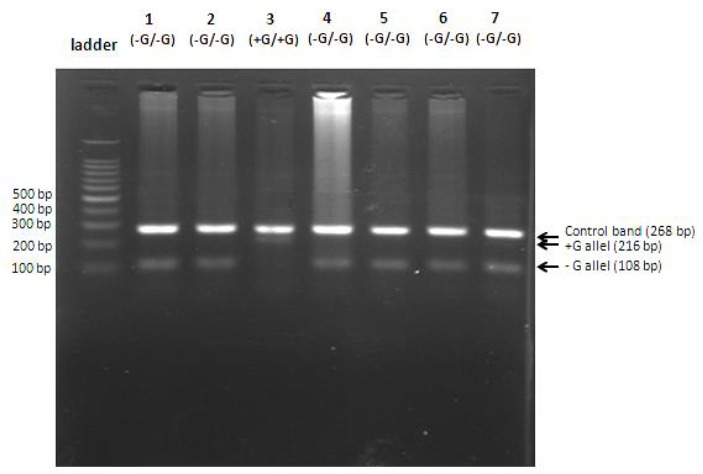
PCR amplification for detection of FSC 8/9 mutation in one vial using T-ARM technique (multiplex PCR). From left to right, in the first lane is a 100 bp marker, followed by samples from different patients (lanes 1 to 7). Except patient 3, having two alleles of FSC 8/9 mutant (homozygote for mutant), all other cases are healthy homozygote
Frequency of IVSII-I and FSC-8/9 mutations in population of Isfahan
Genotyping of 30 β-thalassemia major patients including 15 men and 15 women referred to Isfahan’s Omid Hospital, showed the presence of 18 mutant alleles for IVSII-I mutation including 10 heterozygous individuals (samples 1, 2, 4, 8, 13, 15, 16, 18, 22, and 25) and 4 homozygous individuals (samples 10, 21, 24, and 27) in the studied population.
So, it could be estimated that mutant allele frequency of this mutation in β-thalassemia major patients in Isfahan’s population to be 0.6. In addition, genotyping patients for FSC-8/9 mutation indicated the presence of 5 mutant alleles (frequency was 0.16) of this mutation including 1 homozygous individual (sample 3) and 4 heterozygous individuals (samples 2, 12, 13, and 23) in the studied population. Genotyping results from the cases studied here along with genotype distribution of each of the mutations in the related population are shown in table 2.
Table 2:
Frequency distribution of mutations IVS-II-I(G-A) and FSC 8/9 insG genotypes in both patients and healthy groups
| Genotype | IVS II-I | FSC 8/9 | ||||
|---|---|---|---|---|---|---|
| G/G (%) | A/G (%) | A/A (%) | −G/−G (%) | −G/+G (%) | +G/+G (%) | |
| Patients number | 16 (53.3%) | 10 (33.3%) | 4 (13.3%) | 25 (83.3%) | 4 (13.3%) | 1 (3.3%) |
Discussion
β-thalassemia is a monogenic disease prevalent in Iran that not only results in long term costs for the health care system but more importantly imposes significant difficulties for families and individuals involved. In Iran, approximately 20 common mutations are introduced for β-thalassemia, each having different frequencies in various areas (6). We have developed a tetra-primers ARMS PCR for detection and genotyping of two common mutations in β-thalassemia (IVSII-I (G-A) and FS8-9 insG) in Isfahan population. Current using methods for genotyping in genetics libraries still suffer from specific weaknesses. The use of RFLP, despite high accuracy has technical difficulties, such as unavailability of an enzyme with the desired recognition site or high cost of obtaining such enzymes. As the ASO-PCR method relies solely on a single mismatched base pair at the 3′ end, results in low accuracy and needs to be verified by a more precise method. Moreover, in this method, primers are poured into separate vials and a control band confirming the correct conditions for the PCR reaction is absent. Among these, the sequencing could be the gold standard but it is not 100% error prone and should be accompanied by a second method. Also, sequencing is expensive and time consuming (18, 22, 23). The T-ARMS is a simple and cost-effective method for genotyping of single nucleotide polymorphisms. This method includes a PCR reaction in a vial with two pairs of primers and is followed by electrophoresis on agaros gel. Unlike the ASO-PCR method, in this technique, during the designing of inner primers, the second mispairing is placed in a specific place near the 3′ end resulting in an increase in specificity and accuracy, so that other primers of the mutant allele are unable to pair with the normal sequence or vice versa.
So far, no study was done on optimization method for genotyping mutations in β-thalassemia by T-ARMS. Here, we successfully adjusted and enhanced this method for recognizing two common mutations (FSC-8/9 and IVSII-I) of β-thalassemia in population of Isfahan. For each mutation, the optimal temperature and concentration of MgCl2 were found using temperature and concentration gradients in separate vials obtaining control, healthy and mutant bands. Temperature and concentration of MgCl2 was then verified in a multiplex environment and by testing for different concentrations and ratios of outer and inner primers, optimal values for each were acquired. In this way, it was observed that decreasing concentrations of outer primers compared to inner primers, as shown by Shu Ye et al. (17), produced better results in multiplex conditions for IVSII-I but had no effect in FSC-8/9, in which, equal concentrations of primers appeared to be more suitable. This result suggests that the optimization procedure for the T-ARMS method can differs depending on the sequences of the target gene, single nucleotide variances, and the specific properties of the primers. Therefore, a common rule regarding primer concentration ratio does not exist. In fact, it seems appropriate that achieving a set of suitable values for concentrations of MgCl2 and proper annealing temperature both are effective in optimizing the T-ARMS method.
In this study, genotyping of 30 patients admitted to Isfahan’s Omid hospital with β-thalassemia major were performed for IVSII-I and FSC-8/9 using the T-ARMS method. Allele frequency of 60% for the IVSII-I mutation and allele frequency of 16.6% for FSC-8/9 in the studied population were obtained. Mutant allele frequencies for FSC-8/9 matched the previously reported valued by Derakhshandeh-Peykar et al. in population of Isfahan. However, observed allele frequency for the IVSIII is matched the reported value (7), and it is more than what had reported in other central cities of Iran (3). This difference, with respect to the number of patients admitted to Isfahan’s Omid Hospital from other towns of surrounding areas and considering the lower number of samples in this study is justifiable.
Conclusion
For the first time, genotyping of two common mutations in Isfahan’s population, IVSII-I and FSC-8/9 mutations, were performed using a simple, cost-effective, and accurate method of T-ARMS and allele frequency of these mutations in 30 β-thalassemia major patients were studied.
Ethical considerations
Ethical issues (Including plagiarism, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc.) have been completely observed by the authors.
Acknowledgment
This study was completed at University of Isfahan and supported by Departments of Research / Technology; and Graduate Studies. The authors are grateful to Dr. Hamid Hourfar for his clinical support and Omid hospital for Blood sampling and clinical data. The authors declare that there is no conflict of interests.
References
- 1. Galanello R, Origa R. ( 2010). Review Beta-thalassemia. Orphanet J Rare Dis, 21: 1172– 1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Weatherall DJ, Clegg JB. ( 2008). The β-thalassaemias. The Thalassaemia Syndromes, 5: 285– 356. [Google Scholar]
- 3. Najmabadi H, Karimi-Nejad R, Sahebjam S, Pour-farzad F, Teimourian S, Sahebjam F. ( 2001). The β-thalassemia mutation spectrum in the Iranian population. Hemoglobin, 25: 285– 96. [DOI] [PubMed] [Google Scholar]
- 4. Hashemi A, Abrishamkar M, Jenabzade AR, Eslami Z. ( 2009). Hydroxyurea Can Reduce or Eliminate Transfusion Requirements in Children with Major and Intermediate Thalassemia. Iran J Blood Cancer, 1: 147– 50. [Google Scholar]
- 5. Nozari G, Rahbar S, Golshaiyzan A, Rahmanzadeh S. ( 1995). Molecular Analyses of β-Thalassa in Iran. Hemoglobin, 19: 425– 31. [DOI] [PubMed] [Google Scholar]
- 6. Giardine B, van Baal S, Kaimakis P, Riemer C, Miller W, Samara M. ( 2007). HbVar database of human hemoglobin variants and thalassemia mutations. Hum Mut, 2: 206– 8. [DOI] [PubMed] [Google Scholar]
- 7. Derakhshandeh-Peykar P, Hourfar H, Heidari M, Kheirollahi M, Miryounesi M. ( 2008). The spectrum of β-thalassemia mutations in Isfahan Province of Iran. Iran J Public Health, 37: 2. [Google Scholar]
- 8. Najmabadi H, Pourfathollah AA, Neishabury M, Sahebjam F, Krugluger W, Oberkanins C. ( 2002). Rare and unexpected mutations among Iranian beta-thalassemia patients and prenatal samples discovered by reverse-hybridization and DNA sequencing. Haematologica, 87: 1113– 4. [PubMed] [Google Scholar]
- 9. Roudknar MH, Najmabadi H, Derakhshandeh P, Farhud DD. ( 2003). Detection of Rare and Unknown Mutations in β-tathalassemia Traits in Iran. Iran J Public Health, 32: 1. [Google Scholar]
- 10. Derakhshandeh-Peykar P, Akhavan-Niaki H, Tamaddoni A, Ghawidel-Parsa S, Holakouie Naieni K, Rahmani M. ( 2007). Distribution of β-thalassemia mutations in the northern provinces of Iran. Hemoglobin, 31: 351– 6. [DOI] [PubMed] [Google Scholar]
- 11. Yavarian M, Harteveld CL, Batelaan D, Bernini LF, Giordano PC. ( 2001). Molecular spectrum of β-thalassemia in the Iranian Province of Hormozgan. Hemoglobin, 25: 35– 43. [DOI] [PubMed] [Google Scholar]
- 12. Mahboudi F, Zeinali S, Merat A, Delmaghani S, Mostafavipour K, Moghaddam Z. ( 1996). The molecular basis of beta-thalassemia mutations in Fars province, Iran J Med Sci, 21: 104. [Google Scholar]
- 13. Merat A, Haghshenas M. ( 2000). the spectrum of beta thalassemia mutation in Iran. Med J Islamic Republic of Iran (MJIRI), 14: 103– 6. [Google Scholar]
- 14. Pirastu M, Ristaldi MS, Cao A. ( 1989). Prenatal diagnosis of beta thalassaemia based on restriction endonuclease analysis of amplified fetal DNA. J Med Genetics, 26: 363– 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Newton CR, Graham A, Heptinstall LE, Powell SJ, Summers C, Kalsheker N. ( 1989). Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res, 17: 2503– 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Old JM, Varawalla NY, Weatherall DJ. ( 1990). Rapid detection and prenatal diagnosis of β-thalassaemia: studies in Indian and Cypriot populations in the UK. The Lancet, 336: 834– 7. [DOI] [PubMed] [Google Scholar]
- 17. Ye S, Dhillon S, Ke X, Collins AR, Day INM. ( 2001). An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res, 29: e88– e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Piccioli P, Serra M, Gismondi V, Pedemonte S, Loiacono F, Lastraioli S. ( 2006). Multiplex tetra-primer amplification refractory mutation system PCR to detect 6 common germline mutations of the MUTYH gene associated with polyposis and colorectal cancer. Clin Che, 52: 739– 43. [DOI] [PubMed] [Google Scholar]
- 19. Shariati SAM, Behmanesh M, Galehdari H, Fathian A. ( 2008). Multiplex tetra-primer amplification refractory mutation system polymerase chain reaction to genotype SNP8NRG221533 of Neuregulin-1 gene. Iran J Biote (IJB), 6: 2. [Google Scholar]
- 20. Munoz C, Gomez Talquenca S, Volpe ML. ( 2009). Tetra primer ARMS-PCR for identification of SNP in β-tubulin of Botrytis cinerea, responsible of resistance to benzimidazole. J Micr Methods, 78: 245– 6. [DOI] [PubMed] [Google Scholar]
- 21. Miller SA, Dykes DD, Polesky HF. ( 1988). A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res, 16: 1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sanger F, Donelson JE, Coulson AR, Kssel H, Fischer D. ( 1973). Use of DNA polymerase I primed by a synthetic oligonucleotide to determine a nucleotide sequence in phage f1 DNA. Pro Nat Aca Sci, 70: 1209– 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sanger F, Nicklen S, Coulson AR. ( 1977). DNA sequencing with chain-terminating inhibitors. Pro Nat Aca Sci, 74: 5463– 7. [DOI] [PMC free article] [PubMed] [Google Scholar]


