Abstract
Background:
HNF4A-p.I463Vvariant, reported previously in two distinct families suspected of MODY-1, is assessed in this report to determine whether it is a mutation or a polymorphism (frequency >1%).
Methods:
200 Tunisian healthy people were screened for the presence of HNF4A-p.I463V variant, using RFLP-PCR technique and sequencing. Then, the frequency of this variant was estimated in the Tunisian population and compared to other populations registered in genetic databases. We also performed in-silico analysis using PolyPhen2 and Mutation T@sting softwares to assess the probable effect of HNF4A-p.I463V variant.
Results:
HNF4A-p.I463V had a rare frequency in different populations and was found in 3 control subjects (1.5%) of the studied population. PolyPhen2 predicted that it is a polymorphism, whereas mutation T@sting suggested a probably affected mutant protein.
Conclusion:
HNF4A-p.I463V has a relatively high frequency (>1%) in our control cohort. It is also present in different ethnicities and in- silico analysis showed conflicting results. For these reasons, HNF4A-p.I463V should not be considered as a mutation responsible for MODY-1.
Keywords: MODY, HNF4A-p.I463V, Polymorphism, Mutation, Tunisia, in-silico, Type 2 diabetes, rs147638455
Introduction
Hepatocyte nuclear factor 4a (HNF4A) is an orphan member of the nuclear receptor family of transcription factors (1). It is expressed primarily in the liver, gut, kidney and pancreas and has important role during embryonic life in pancreatic development and maintaining of β cell function (2). Indeed, knockout studies found that Hnf4a-/- mice revealed impaired gastrulation and died around day E9 (3). The HNF4A protein consists of five functional domains: an N-terminal activation function (AF-1, also referred to as A/B domain), two zinc fingers responsible for DNA binding (C domain), a potential ligand binding domain (E domain) and the F domain (1).
HNF4A gene (ENSG00000101076) has many transcripts. This gene could produce nine potential isoforms (HNF4A1–HNF4A9) (4). The ENST00000316099 transcript is considered the canonical transcript that codes the isoform HNF4-Alpha-1 with 474 amino-acids (P41235-1). Mutations in HNF4A gene are responsible of MODY-1 (Maturity-onset diabetes of the young type 1) (OMIM#125850) and neonatal hyperinsulinism hypoglycemia (5, 6). To date, about 103 HNF4A inactivating mutations were reported in 173 MODY families (7). Some HNF4A variants were found to be associated with type 2 diabetes (T2D) (8).
T2D is a chronic disorder of glucose metabolism, characterized by a state of hyperglycemia due to defective insulin secretion, peripheral insulin resistance and increased hepatic glucose production (9). Genetic and environmental factors, contribute to the appearance of T2D (10). Whereas, MODY is a dominantly inherited form of early-onset type 2 diabetes (T2D), generally diagnosed in childhood, adolescence or young adulthood (11). It is a rare monogenic disease caused by primary defects of insulin secretion with a lack of auto-antibodies against the pancreatic cells and is rarely associated with obesity (11).
The NM_000457.4: c.1387A>G or HNF4A - p.I463V mutation (rs147638455 or CM993900), also known as p.I431V and p.I453V depending on the corresponding gene’s isoform, was previously found in a Caucasian and a Tunisian families suspected of MODY (12, 13).
But, it remains unclear whether this variant is a mutation responsible for MODY-1 or a polymorphism. In the case of a molecular diagnosis of MODY-1, finding HNF4A-p.I463V would be confusing: should we consider it as a mutation?
In this report, we present a simple and easy protocol to screen HNF4A-p.I463V variant. We also assessed its frequency in Tunisian North-African population and discussed its probable effect.
Materials and Methods
DNA samples
Two hundred anonymous unrelated healthy controls were recruited in 2013 from the Blood Transfusion Center of Sousse (Tunisia). They were examined by the physician of the center and were found not diabetic and did not present any other disease. The mean age of the control subjects was 27.23 ± 10.28 years old. All these individuals were screened for the presence or absence of HNF4A -p.I463V variant.
The study was carried out after taking informed written consent from all individuals and after approval of the Ethics Committee of Farhat Hached University Hospital.
HNF4A -p.I463V mutation analysis
Genomic DNA was extracted from peripheral blood using Flexigene DNA kit (QiagenInc). The HNF4A -p.I463V mutation analysis was carried out by RFLP-PCR method. First, HNF4A exon 10 including exon-intron boundaries was amplified by PCR using the following primers F: 5′ TTT ACT CCC ACA AAG GCT GG 3′ and R: 5′ ATC ACC AGG TGC TCT CTT AG 3′. PCR was performed in a 50 μl volume containing 50 ng of genomic DNA, 20 pmol of each primer and 1 U of Recombinant DNA polymerase (Invitrogen, Carlsbad, CA, USA). PCR conditions and thermocycler program are available on request.
Fifteen μl of PCR product were mixed with 2μl BSA, 2μl buffer, 5.75 μl sterilized water and digested by 2.5 U of FOKI restriction enzyme (TaKaRa Bio Inc, Otsu, Shiga, Japan).The digested PCR products were then separated on 2.5% agarose gel.
The HNF4A-p.I463V substitution abolishes the restriction site of FOKI. Thus, the wild fragment (266 bp) is cut into 164 bp and102 bp fragments. Finally, in case of mutation detection, direct sequencing was used to confirm it.
Statistical analysis
The studied population was checked for Hardy-Weinberg equilibrium.
Prediction of p.I463V-HNF4A mutation effect in-silico
The p.I463V substitution was studied in-silico to predict the putative effect on protein function. We performed in-silico analysis using two different types of software: PolyPhen2 (http://genetics.bwh.harvard.edu/pph2/index.shtml) and Mutation T@sting (http://www.mutationtaster.org/). These bioinformatics tools enable high-throughput prediction of the potential impact of residue changes and large-scale polymorphism analyses. PolyPhen and Mutation T@sting predict possible impact of an amino acid substitution on the structure and function of a human protein using straight forward physical and comparative considerations (14, 15).
Results
Carrier Frequency of p.I463V
The assessment of HNF4A-p.I463V variant in 200 healthy Tunisian people by RLFP-PCR method found that 3 controls (1.5%) were carriers of this mutation. The results were also confirmed by direct sequencing of HNF4A exon 10 (Fig. 1).The healthy carriers were aged 20, 26 and 37 years old.
Fig. 1:
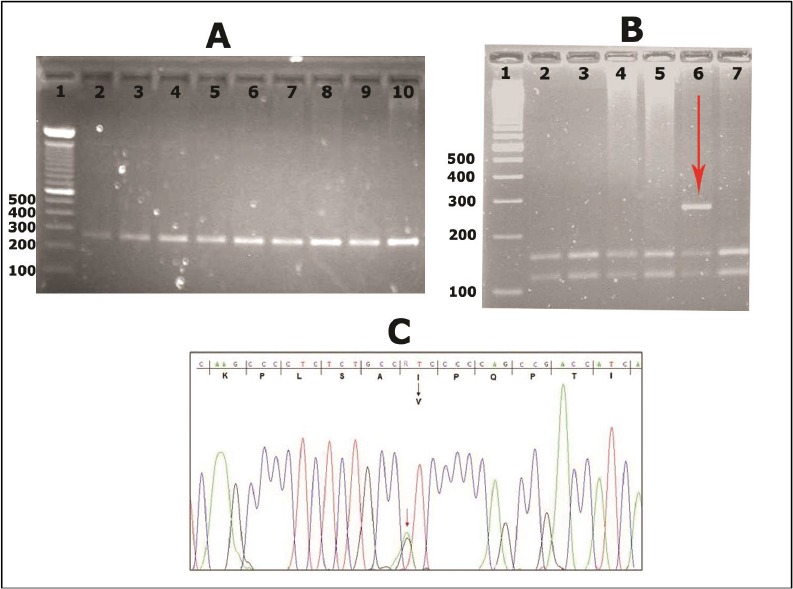
HNF4A-p.I463V variant detected after sequencing and enzymatic digestion by FOKI restriction enzyme A: Gel electrophoresis of HNF4A exon 10 (266 bp) before digestion by FOKI restriction enzyme in 9 controls. B: Gel electrophoresis of HNF4A exon 10 after digestion by FOKI restriction enzyme into 2 fragments (164 and 102 bp) in 7 controls. Well 6 shows a heterozygote person for HNF4A p.I463V mutation. C: Chromatograph of a part of HNF4A exon 10 sequence showing the c.1387A>G/p.I463V mutation
Comparison with other populations
The Tunisian population of this study is in Hardy-Weinberg equilibrium (Table 1). The frequency of HNF4A-p.I463V is rare in the other populations. Indeed, this variant is more frequent in the Tunisian population than the European and African American populations.
Table 1:
rs147638455 frequency in different populations
| Population | Source | Sample size | Chrom sample | Mean age (years old) | Genotypes | ||
|---|---|---|---|---|---|---|---|
| G/G | G/A | A/A | |||||
| European American | ESP project* | 4300 | 8600 | NA*** | 0 | 9.3 10−4 | 0.99 |
| African American | ESP project | 2203 | 4406 | NA | 0 | 0 | 1 |
| European American | Clinseq project** | 662 | 1324 | NA | 0 | 0.002 | 0.998 |
| North African-Tunisiana | This study | 200 | 400 | 27.23 ± 10.28 | 0 | 0.015 | 0.985 |
Exome sequencing project,
CSAgilent (dbSNP 138),
Not applicable,
: the Tunisian population is in Hardy-Weinberg equilibrium (P=0.91 > 0.05)
Results of in Silico Analysis
PolyPhen2 predicted that this nucleotide change is a “polymorphism” with a score of 0.144 [0–1]. While, the Mutation T@sting software indicated that this variant could be responsible of a probably affected protein.
Discussion
Performing a molecular diagnosis of monogenic diabetes is not an easy task because of its clinical and genetic heterogeneity (16). Some genetic variants could also be problematic such as HNF4A-p.I463V. In an earlier study, we found that HNF4A-p.I463V cosegregated with diabetes phenotype in a Tunisian family (Fig. 2 and Table 2). It was present in three patients from this family and was absent in two other non-affected relatives (13). Contrariwise, this variant didn’t show a perfect cosegregation in a Caucasian family (12). Hitherto, there is not enough information in the literature about HNF4A-p.I463V variant (rs147638455). It is not clear whether it is a mutation directly causing MODY-1 or just a non-synonymous polymorphism with low effect.
Fig. 2:
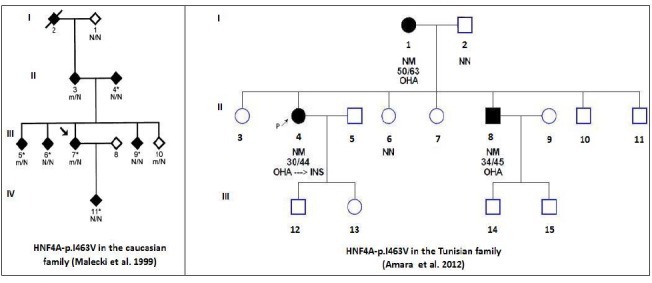
The diabetic Caucasian and Tunisian families having HNF4A-p.I463V variant Filled and open symbols represent diabetic subjects and normal glucose tolerance individuals, respectively. The numbers under the symbols are the identification numbers. Below the numbers, it is the genotype at codon I463V: N, normal allele (Isoleucine); m, mutant allele (valine). Below the genotype is the diagnostic age of diabetes for affected members and age at examination, followed by the treatment for diabetes (OHA: Oral hypoglycemic agents and INS: Insulin). An arrow with P letter indicates the proband.
Table 2:
The clinical characteristics of the reported HNF4A-p.I463V carriers
| Group | Individual identifier | Diabetes | Diagnosis Age | Examination Age | Fasting glycaemia | Hb1Ac | BMI | Treatment | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Tunisian family | II-1 | Yes | 50 | 63 | NF | NF | NF | OHA | (12) |
| II-4 | Yes | 30 | 44 | 17.2 | 13.4 | 35.6 | OHA→INS | ||
| II-8 | Yes | 34 | 45 | NF | NF | 30.3 | OHA | ||
| Mean | NA | 38 | 50.6 | NA | NA | 32.9 | NA | ||
| Caucasian family | 3 | Yes | 64 | 64 | 5.43 | 6.2 | NF | Diet | (13) |
| 10 | No | NA | 31 | NA | 5.7 | NF | NA | ||
| Mean | NA | 64 | 47.5 | NA | NA | NF | NA | ||
| Tunisian Healthy cohort | H1 | No | NA | 20 | NA | NA | NA | NA | This study |
| H2 | No | NA | 26 | ||||||
| H3 | No | NA | 37 | ||||||
| Mean | NA | NA | 27.6 |
NA: not applicable, NF: not found, OHA = Oral hypoglycemic agents, INS: Insulin
HNF4A-p.I463V a variant with interesting characteristics
This variant occurs in a conserved region across species (PhyloP score = 4.56 [−14.1; 6.4]) and is localized in the F domain of HNF4A (Fig. 3). This domain plays a role in the protein transactivation activity (17).
Fig. 3:
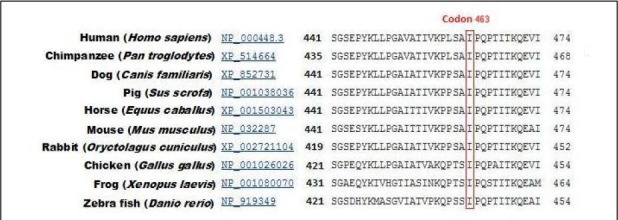
Isoleucine amino-acid in position 463 is conserved across species A portion of HNF4A exon 10 amino-acid sequence is compared between various species. The human Isoleucine at codon 463 and Isoleucine of other species are framed in a red box. The species are annotated in bold and the NCBI accession numbers in blue
HNF4A-p.I463V variant could not be responsible for MODY-1
Using a simple protocol of screening for HNF4A-p.I463V variant (rs147638455), we detected it in three healthy controls out of 200 Tunisian unrelated healthy individuals (400 chromosomes). Indeed, it is an unexpected high carrier frequency (1.5%) in this sample compared with the other populations (Table 1).
Commonly, MODY develops before the age of 25 years old (11). The healthy controls carriers of rs147638455 in our population are aged 20, 26 and 37 years (Table 2). Even if we omit the 20 year-old control individual, the rs147638455 frequency would remain high for a mutation (1%). In other ethnic populations, rs14763845 was rarely found in healthy individuals (Table 1). If we suppose that this variant is responsible for MODY-1, which is a rare monogenic disease, Tunisian frequency, argues against this hypothesis.
On the other hand, the in silico analysis using mutation T@ster and polyphen2 showed conflicting results. In parallel, physicochemical difference between Isoleucine and Valine amino-acids is small (Grantham Distance = 29 [0–215]). In addition to the absence of a cosegregation of HNF4A-p.I463V variant in the Caucasian family, the Tunisian family suspected of MODY had a mean age of diabetes diagnosis of 38 ± 10.58 years old (13) (Table 2 and Fig. 2). Even familial, the diabetes observed in this family, could be T2D. Otherwise, a previous study showed that patients with mutations in HNF4A exons 9 and 10 developed diabetes later than those with mutations in exons 2–8 did(4). So, mutations in exons 9 and 10 could not be responsible for MODY-1 (1).
Speculations about the effect of this variant and its implication in type 2 diabetes
HNF4A-p.I463V could act in an interactive polymorphisms network leading to T2D. At that time, the relatively young age of our healthy carriers (20, 26 and 37 years old) does not definitely mean that they will not present T2D later.
A similar missense mutation in the F domain (HNF4A-p.V393I localized in exon 9) was found in a French diabetic family diagnosed at the age of 45 years. HNF4A-p.V393I was associated with a marked reduction of transactivation activity responsible for impaired insulin secretion and co-segregated with diabetes (18).
It is of note that some non diabetic individuals aged between 30 and 40 year-old were carriers of HNF4A-p.V393I (18). Thus, HNF4A-p.I463V could have a similar effect to HNF4A-p.V393I. Indeed, most of the patients that carry HNF4A-p.I463V variant are over 30 years (Table 2). This could be a supplementary argument to associate HNF4A-p.I463V with T2D.
Association and functional studies should be carried out, to assess the exact effect of HNF4A-p.I463V variant. Meantime, we recommend to do not consider HNF4A-p.I463V as a mutation in case of molecular diagnostic of MODY.
This recommendation is based on the high frequency of this variant in the Tunisian population, its presence in healthy persons and the conflicting results of the in silico analysis.
Conclusion
We can speculate that HNF4A-p.I463V is a polymorphism with possible role in T2D and not a causal mutation implicated directly in MODY-1.
Ethical considerations
Ethical issues (Including plagiarism, informed consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc.) have been completely observed by the authors.
Acknowledgments
The authors thank all the volunteers and the staff of the Regional Center of Blood Transfusion of Sousse for their contribution to this work. Special thanks to Mrs Sihem SASSI, Mrs Ahlem M’SAKNI, Miss Safa BOUKER and Mr Hedi LAATIRI for their excellent technical assistance. The authors declare that there is no conflict of interests.
Abbreviations
- MODY
Maturity-Onset diabetes of the young
- T2D
Type 2 diabetes
- HNF4A
Hepatocyte nuclear factor 4a
References
- 1. Ryffel GU. (2001). Mutations in the human genes encoding the transcription factors of the hepatocyte nuclear factor (HNF)1 and HNF4 families: functional and pathological consequences. J Mol Endocrinol, 27: 11–29. [DOI] [PubMed] [Google Scholar]
- 2. Stoffel M, Duncan SA. (1997). The maturity-onset diabetes of the young (MODY1) transcription factor HNF4alpha regulates expression of genes required for glucose transport and metabolism. Proc Natl Acad Sci U S A, 94: 13209–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen WS, Manova K, Weinstein DC, Duncan SA, Plump AS, Prezioso VR, Bachvarova RF, Darnell JE., Jr. (1994). Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes Dev, 8: 2466–77. [DOI] [PubMed] [Google Scholar]
- 4. Harries LW, Locke JM, Shields B, Hanley NA, Hanley KP, Steele A, Njolstad PR, Ellard S, Hattersley AT. (2008). The diabetic phenotype in HNF4A mutation carriers is moderated by the expression of HNF4A isoforms from the P1 promoter during fetal development. Diabetes, 57: 1745–52. [DOI] [PubMed] [Google Scholar]
- 5. Yamagata K, Furuta H, Oda N, Kaisaki PJ, Menzel S, Cox NJ, Fajans SS, Signorini S, Stoffel M, Bell GI. (1996). Mutations in the hepatocyte nuclear factor-4alpha gene in maturity-onset diabetes of the young (MODY1). Nature, 384: 458–60. [DOI] [PubMed] [Google Scholar]
- 6. Kapoor RR, Locke J, Colclough K, Wales J, Conn JJ, Hattersley AT, Ellard S, Hussain K. (2008). Persistent hyperinsulinemic hypoglycemia and maturity-onset diabetes of the young due to heterozygous HNF4A mutations. Diabetes, 57: 1659–63. [DOI] [PubMed] [Google Scholar]
- 7. Colclough K, Bellanne-Chantelot C, Saint-Martin C, Flanagan SE, Ellard S. (2013). Mutations in the Genes Encoding the Transcription Factors Hepatocyte Nuclear Factor 1 Alpha (HNF1A) and 4 Alpha (HNF4A) in Maturity-Onset Diabetes of the Young (MODY) and Hyperinsulinaemic Hypoglycaemia. Hum Mutat. [DOI] [PubMed] [Google Scholar]
- 8. Love-Gregory L, Permutt MA. (2007). HNF4A genetic variants: role in diabetes. Curr Opin Clin Nutr Metab Care, 10: 397–402. [DOI] [PubMed] [Google Scholar]
- 9. Narayan KM, Boyle JP, Geiss LS, Saaddine JB, Thompson TJ. (2006). Impact of recent increase in incidence on future diabetes burden: U.S., 2005–2050. Diabetes Care, 29: 2114–6. [DOI] [PubMed] [Google Scholar]
- 10. Stumvoll M, Goldstein BJ, van Haeften TW. (2005). Type 2 diabetes: principles of pathogenesis and therapy. Lancet, 365: 1333–46. [DOI] [PubMed] [Google Scholar]
- 11. Vaxillaire M, Froguel P. (2008). Monogenic diabetes in the young, pharmacogenetics and relevance to multifactorial forms of type 2 diabetes. Endocr Rev, 29: 254–64. [DOI] [PubMed] [Google Scholar]
- 12. Malecki MT, Yang Y, Antonellis A, Curtis S, Warram JH, Krolewski AS. (1999). Identification of new mutations in the hepatocyte nuclear factor 4alpha gene among families with early onset Type 2 diabetes mellitus. Diabet Med, 16: 193–200. [DOI] [PubMed] [Google Scholar]
- 13. Amara A, Chadli-Chaieb M, Ghezaiel H, Philippe J, Brahem R, Dechaume A, Saad A, Chaieb L, Froguel P, Gribaa M, Vaxillaire M. (2012). Familial early-onset diabetes is not a typical MODY in several Tunisian patients. Tunis Med, 90: 882–7. [PubMed] [Google Scholar]
- 14. Roumen L, Sanders MP, Pieterse K, Hilbers PA, Plate R, Custers E, de Gooyer M, Smits JF, Beugels I, Emmen J, Ottenheijm HC, Leysen D, Hermans JJ. (2007). Construction of 3D models of the CYP11B family as a tool to predict ligand binding characteristics. J Comput Aided Mol Des, 21: 455–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang LL, Li Y, Zhou SF. (2009). A bioinformatics approach for the phenotype prediction of nonsynonymous single nucleotide polymorphisms in human cytochromes P450. Drug Metab Dispos, 37: 977–91. [DOI] [PubMed] [Google Scholar]
- 16. Amara A, Chadli-Chaieb M, Chaieb L, Saad A, Gribaa M. (2014). Challenges for Molecular Diagnosis of Familial Early-Onset Diabetes in Unexplored Populations. Iran J Public Health, 43: 1011–1013. [PMC free article] [PubMed] [Google Scholar]
- 17. Eeckhoute J, Briche I, Kurowska M, Formstecher P, Laine B. (2006). Hepatocyte nuclear factor 4 alpha ligand binding and F domains mediate interaction and transcriptional synergy with the pancreatic islet LIM HD transcription factor Isl1. J Mol Biol, 364: 567–81. [DOI] [PubMed] [Google Scholar]
- 18. Hani EH, Suaud L, Boutin P, Chevre JC, Durand E, Philippi A, Demenais F, Vionnet N, Furuta H, Velho G, Bell GI, Laine B, Froguel P. (1998). A missense mutation in hepatocyte nuclear factor-4 alpha, resulting in a reduced transactivation activity, in human late-onset non-insulin-dependent diabetes mellitus. J Clin Invest, 101: 521–6. [DOI] [PMC free article] [PubMed] [Google Scholar]


