Abstract
Aedes aegypti mosquitoes vector several arboviruses of global health significance, including dengue viruses and chikungunya virus. RNA interference (RNAi) plays an important role in antiviral immunity, gene regulation and protection from transposable elements. Double-stranded RNA binding proteins (dsRBPs) are important for efficient RNAi; in Drosophila functional specialization of the miRNA, endo-siRNA and exo-siRNA pathway is aided by the dsRBPs Loquacious (Loqs-PB, Loqs-PD) and R2D2, respectively. However, this functional specialization has not been investigated in other dipterans. We were unable to detect Loqs-PD in Ae. aegypti; analysis of other dipteran genomes demonstrated that this isoform is not conserved outside of Drosophila. Overexpression experiments and small RNA sequencing following depletion of each dsRBP revealed that R2D2 and Loqs-PA cooperate non-redundantly in siRNA production, and that these proteins exhibit an inhibitory effect on miRNA levels. Conversely, Loqs-PB alone interacted with mosquito dicer-1 and was essential for full miRNA production. Mosquito Loqs interacted with both argonaute 1 and 2 in a manner independent of its interactions with dicer. We conclude that the functional specialization of Loqs-PD in Drosophila is a recently derived trait, and that in other dipterans, including the medically important mosquitoes, Loqs-PA participates in both the miRNA and endo-siRNA based pathways.
INTRODUCTION
Aedes aegypti mosquitoes are vectors of many significant arboviruses, including the dengue viruses, chikungunya virus, and yellow fever virus. Approximately 50–100 million cases of dengue occur every year and an estimated 2.5 billion people are at risk (1). Recent outbreaks of chikungunya virus have raised concern over its re-emergence and spread to previously non-endemic areas in both Europe (2) and the Americas (3). In addition, an estimated 200 000 cases of yellow fever are thought to occur worldwide (4). Despite the existence of an effective vaccine, the prevalence of yellow fever has been increasing over the last two decades (4).
RNA interference mechanisms are used by eukaryotic organisms for gene regulation, protection from transposable elements, and defense from viral infection [reviewed in (5)]. In general, RNA interference involves the processing of double stranded RNA precursors into small RNA duplexes, which are then loaded into an effector complex, unwound, and used to detect homologous mRNAs for targeted degradation [reviewed in (6)]. While the importance of mosquito RNAi for innate immunity and vector competence has been heavily studied over the last decade (7–9), considerably less is known about the mechanisms involved in mosquito RNAi and the degree of similarity between the mosquito and the drosophilid silencing pathways.
The short interfering (si)RNA pathway is important for regulating gene expression, silencing transposable elements, and inhibiting viral replication (10). The siRNAs derived from genomic origin, such as from convergent or hairpin transcripts, or from transposable elements are known as endo-siRNAs, while those of viral origin or experimentally introduced long dsRNAs are known as exo-siRNAs. This distinction is important because biogenesis and processing of miRNAs, endo-siRNAs, and exo-siRNAs depend on different dsRBPs functioning as Dicer binding partners. In Drosophila, alternative splicing of loquacious (loqs) mRNA results in four distinct dsRBP isoforms known as Loqs-PA, -PB, -PC and -PD. Both Loqs-PA and -PB partner with Dicer-1 (11), though binding with Loqs-PB appears to be preferred (12). Transgenic expression of Loqs-PB is sufficient to rescue defects in both viability and fertility in a loqs null background, while Loqs-PA is only able to rescue viability (13). Loqs-PD partners with Dicer-2 and is important to endo-siRNA biogenesis and RISC (RNA-induced silencing complex) loading (13–16).
Another dsRBP, known as R2D2, also partners with Dcr2 and facilitates dsRNA recognition and siRNA RISC loading (17). However, it is unclear if R2D2 is important for loading both endo- and exo-siRNAs (18,19), or only exo-siRNAs (16). Furthermore, the specifics of interactions between R2D2, Dcr2 and Loqs-PD remain uncertain. Marques et al. (18) utilized loqs and r2d2 knockout Drosophila mutants to develop a model in which R2D2 and Loqs-PD act sequentially in both siRNA pathways (18). Their results suggested Loqs-PD functions alongside Dcr2 to process long exogenous dsRNAs and endogenous hairpin RNAs into siRNA duplexes, after which R2D2 facilitates loading these siRNAs into RISC. In an alternative model, R2D2 and Loqs-PD may compete for Dcr2 binding and act independently in exo- and endo-siRNA pathways, respectively (16).
Little is known about mosquito dsRBPs and their roles in the various RNAi pathways. Studies involving knockdown of Aedes aegypti R2D2 have indicated that this dsRBP plays a role in limiting dengue virus replication, presumably due to its involvement in the exo-siRNA pathway (8). However, R2D2's association with mosquito exo-siRNA components, such as Dcr2 and Ago2, remains to be studied. Likewise, while the distinct drosophilid Loqs isoforms are known to associate with different Dicer and Argonaute proteins (11,14–16,18), nothing is known about the mosquito Loqs orthologs.
The objective of this study was to determine the role of dsRBPs R2D2 and Loqs in the endo-siRNA, exo-siRNA and miRNA pathways of the mosquito Ae. aegypti, a critical vector of human pathogens and a model for other important mosquito vectors. We present evidence that drosophilids are unique in encoding a functionally specialized Loqs-PD isoform, and that in mosquitoes and likely other dipterans, the Loqs-PA isoform serves this role through interactions with both miRNA and siRNA components. RNAi-based depletion of R2D2 or Loqs reduced small RNA levels from exo-siRNA and endo-siRNA sources, indicating these gene products act non-redundantly in siRNA production; depletion of Loqs-PB did not substantially affect siRNA production but did result in a strong loss of miRNAs. Overexpression of mosquito Loqs-PA was found to increase the efficiency of silencing triggered by an inverted repeat construct, but not exogenous dsRNA. Taken together, these data suggest that in mosquitoes the miRNA and siRNA pathways converge on the hub protein loquacious, a situation reminiscent of the human orthologs TRBP and PACT.
MATERIALS AND METHODS
RACE and cDNA sequencing
Transcript sequencing was performed using RNA templates isolated from the Liverpool and khw strains of Ae. aegypti. Transcript initiation and termination sites for each gene were determined via 3′ and 5′ RACE using the Smart RACE cDNA kit (Clontech, Mountain View, CA, USA) and primers listed in Supplementary Table S1. For both r2d2 and loqs, full-length cDNA clones were generated from Liverpool strain adults using the High Capacity Reverse Transcriptase cDNA synthesis kit (Applied Biosystems, Grand Island, NY, USA); these cDNAs were used as templates to amplify the individual dsRBP sequences using the Platinum Pfx PCR Kit (Life Technologies, Grand Island, NY, USA). The primers used for each PCR reaction are listed in Supplementary Table S1. Products from both the RACE and cDNA amplification reactions were cloned into TOPO vector (Life Technologies, Grand Island, NY, USA) and sequenced with M13F (5′-GTAAAACGACGGCCAGT-3′) and M13R (5′-AACAGCTATGACCATG-3′) primers. Full-length cDNA sequences were deposited in GenBank (KJ598053-5).
RNA isolation and reverse transcriptase PCR
Total RNA was isolated using TRIzol Reagent (Life Technologies, Grand Island, NY, USA), per the manufacturer's instructions. For RNA extraction from cell culture, TRIzol Reagent was added directly to the cell culture plates for lysis and processing. For analysis of whole mosquitoes or tissues, samples were frozen in liquid nitrogen and stored at −80°C until RNA extraction. Reverse-transcriptase PCR (RT-PCR) was used for analysis of tissue-specific gene expression using 1 μg of each RNA template and the One-Step RT-PCR kit (Qiagen, Germantown, MD, USA), followed by gel electrophoresis. Quantitative PCR was performed as previously described (20) with Power SYBR Green PCR Mastermix on the StepOne or 7300 Real-time PCR System (Life Technologies, Grand Island, NY, USA); samples were compared with the levels of actin mRNA. All oligonucleotide primers used for RT-PCR and qPCR reactions are listed in Supplementary Table S2.
Plasmid construction
Oligonucleotides encoding FLAG or HA epitopes flanked by NdeI and SacI restriction enzyme sites (Supplementary Table S3) were annealed and ligated into NdeI and SacI sites of a pSLfa plasmid, immediately downstream of the Ae. aegypti polyubiquitin (PUb) promoter sequence (21) and upstream of a SV40 3′UTR polyadenylation sequence. The resulting plasmids were named PUb-HA-MCS and PUb-FLAG-MCS. The open reading frames (ORFs) for r2d2, loqs-ra and loqs-rb were amplified using the One-Step Reverse Transcriptase PCR Kit (Qiagen, Germantown, MD, USA) and primers designed to add NdeI and SalI sites to the 5′ and 3′ ends, respectively (Supplementary Table S3). The PCR products were digested, purified by low melt agarose gel extraction, and ligated into the NdeI and SalI sites in the MCS of PUb-HA-MCS and/or PUb-FLAG-MCS vector plasmids. The resulting plasmids were: PUb-HA-R2D2, PUb-HA-Loqs-PA, PUb-HA-Loqs-PB, PUb-HA-LoqsΔ258, PUb-HA-LoqsΔ226, PUb-FLAG-Loqs-PA, and PUb-FLAG-Loqs-PB.
For expressing the tagged dsRBPs via recombinant Sindbis viruses, each of the ORFs were amplified from the above plasmids using Platinum Pfx (Life Technologies, Grand Island, NY, USA) and primers designed to add AscI and PacI restriction enzyme recognition sites to the 5′ end of the tag and 3′ end of the ORF, respectively (Supplementary Table S3). After restriction digestion and gel extraction, each tagged dsRBP was ligated into the TE/3′2J double subgenomic Sindbis virus vector (22) using AscI and PacI restriction enzyme recognition sites. The resulting plasmids were named: pTE/3′2J-HA-R2D2, pTE/3′2J-HA-Loqs-PA, pTE/3′2J-HA-Loqs-PB, pTE/3′2J-FLAG-Loqs-PA and pTE/3′2J-FLAG-Loqs-PB.
For endo-siRNA and exo-siRNA sensor experiments, pSLfa PUb-MCS was generated by digesting pSLfa-PUb-GFP-SV40 (21) with NcoI/NotI and ligated with annealed oligos (Supplementary Table S3) forming a multiple cloning site (MCS). The resultant plasmid was digested with BamHI/SalI and the BamHI/SalI Renilla hairpin fragment from pRmHa-MCS-Renilla-IR (14) was ligated, to form pSLfa-PUb-Renilla-IR. Similarly the Renilla ORF was digested from pKhsp82-Renilla (23) with BamHI/SalI and ligated into the same pSLfa-PUb-MCS vector to generate pSLfa-PUb-Renilla. pGL3-PUb-FF is as previously described (21).
Cell culture, transfection, luciferase assays and infection
BHK-21 and Vero cells were maintained at 37°C, 5% CO2 in Dulbecco's modified Eagle's medium (Cellgro, Tewksbury, MA, USA), supplemented with 10% fetal bovine serum (FBS), 1% penicillin–streptomycin and 1% l-glutamine. Aag2 cells were maintained at 28°C in Schneider's Drosophila medium (Lonza BioWhittaker, Basel, Switzerland) supplemented with 10% FBS, 1% penicillin–streptomycin and 1% l-glutamine. pTE/3′2J plasmids were linearized using XhoI prior to in-vitro transcription. Viral RNAs were transcribed in-vitro using SP6 RNA polymerase, and electroporated into BHK-21 cells, as previously described (24). Infectious viruses were harvested, aliquoted and stored at −80°C until use. Viruses were titered by plaque assay in Vero cells. For SINV infections, Aag2 cells were seeded into 25 cm2 flasks and allowed to grow to ∼80% confluency. After removing the growth medium from the cells, virus was added to the flask at an MOI of 5 or higher and the volume was brought up to 1 ml using Schneider's medium. Cells were incubated with the virus on a rocker platform for 1 h, after which an additional 10 ml of growth medium was added. Cells were incubated at 28°C until ready to harvest.
DsRNAs were prepared following the Replicator RNAi Kit (Thermo Scientific, Waltham, MA, USA) and primers indicated in Supplementary Table S4. For EGFP and Firefly Luciferase, dsRNAs were produced directly from gene specific amplicons. For ago2, r2d2, loqs and loqs-rb, gene specific amplicons were first cloned into plasmid pGEM T-easy (Promega, Madison WI, USA). The resultant clones were sequence confirmed and re-amplified using a common set of primers (T7 UPR and Phi6 UPF) for dsRNA generation. All transcription reactions were incubated overnight at 37°C, DNaseI/RNaseA treated and then purified with the MEGAClear kit (Life Technologies, Grand Island, NY, USA).
For IP and fractionation experiments, cells were transfected in 25 cm2 flasks (2.5 μg of plasmid DNA, 20 μl enhancer, 220 μl buffer EC, 62.5 μl effectene transfection reagent) according to the manufacturer's protocol (Qiagen, Germantown, MC, USA). For reporter assays, cells were transfected in 96-well plates using 20 ng of reporter construct DNA (pGL3-PUb-FL for exo-siRNA experiments, pSLfa-PUb-RL for endo-siRNA experiments), 6 ng control DNA (pSLfa-PUb-RL or pGL3-PUb-FL) and either 20 ng of double-stranded RNA targeting firefly luciferase or 20 ng of pSLfa-PUb-RL-IR. At 24 h post-transfection, cells were washed once with 100 μl phosphate buffered saline (PBS), then lysed in 45 μl 1× Passive Lysis Buffer (Promega, Madison, WI, USA). Lysates were incubated 30 min at room temperature with rocking and then frozen at −80°C until luciferase assays were performed. Luciferase assays were performed on 20 μl of lysate using the Dual-Luciferase Reporter Assay System according to the manufacturer's protocol using a GloMax Multi-Detection System (Promega, Madison WI, USA).
Co-immunoprecipitation, cell fractionation and immunoblotting
For experiments involving only HA/FLAG overexpression construct, transfected Aag2 cells were harvested by scraping into PBS and pelleted at 500 x g for 10 min at 4°C. Cell pellets were lysed in native lysis buffer (20 mM HEPES, pH 7.0, 150 mM NaCl, 2.5 mM MgCl2, 0.3% Triton X-100, 30% glycerol) treated with ethylenediaminetetraacetic acid (EDTA)-free protease inhibitor (Roche, Indianapolis, IN, USA) and rotated for 30 min at 4°C. Three micrograms of anti-HA or anti-FLAG mouse monoclonal antibody (GenScript) was incubated with 50 μl Protein G Dynabeads (Life Technologies, Grand Island, NY, USA) and rotated for 30 min at 4°C. Excess antibody was removed by washing the beads once with 200 μl PBS-T (0.02% Tween-20). The Aag2 cell lysates were incubated with the antibody–Dynabead complex for 1 h on a rotator at 4°C. Once the lysate was removed from the beads, the Dynabead complex was washed three times with 200 μl IP Wash Buffer (20 mM HEPES, pH 7.0, 150 mM NaCl, 2.5 mM MgCl2, 0.3% Triton X-100) and once with 100 μl IP Wash Buffer. The entire complex was transferred to a clean tube during this last wash. After removing the remaining wash buffer, the bound complex was denatured in Laemmli sample buffer (Bio-Rad, Hercules, CA, USA) and stored at −20°C.
For experiments involving detection of endogenous Dcr and Ago proteins, co-IP assays were performed using the Pierce Magnetic HA-Tag IP/Co-IP Kit (Thermo Scientific, Waltham, MA, USA) per the manufacturer's instructions. Briefly, cells were harvested as above and lysed in 500 μl of lysis/wash Buffer supplemented with Halt Protease Inhibitor Cocktail per 50 mg of wet cell pellet, rotated for 30 min at 4°C, and cell debris removed by centrifugation at 13 000 x g for 10 min. Magnetic beads were washed with lysis/wash buffer and incubated with lysates on a rotator for 1 h at room temperature. Beads were then washed three times with 300 μl lysis/wash buffer and once with 300 μl water. Bound fractions were eluted from beads with 1× non-reducing sample buffer, heated to 95°C for 5 min, then supplemented with Dithiothreitol (DTT) to a final concentration of 50mM.
Co-IP samples were resolved by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. For anti-HA or anti-FLAG blots, we used 1:10 000 dilutions of horseradish peroxidase (HRP)- conjugated mouse monoclonal primary antibodies (GenScript, Piscataway, NJ, USA). This facilitated detection of dsRBPs, which migrate near the heavy antibody chain present in the immunoprecipitates. Rabbit polyclonal antibodies to detect Ae. aegypti Dcr1, Dcr2, Ago1 and Ago2 were obtained from GenScript. A complete list of the primary antibody epitopes and dilutions used for immunoblotting is provided in Supplementary Table S5. All primary antibodies were diluted in 3% non-fat dry milk/TBS-T. For the secondary antibody incubations, we diluted 1:50 000 HRP-conjugated goat anti-rabbit antibody (GenScript, Piscataway, NJ, USA) in TBS-T. Chemiluminescent detection was performed using ECL Prime Reagent (Amersham, GE Healthcare, Pittsburgh, PA, USA) and radiographic film.
For cell compartment localization assays, transfected and 2 dpi SINV-infected Aag2 cells were fractionated into cytoplasmic, membrane, nuclear and cytoskeletal fractions using the QProteome Cell Compartment Fractionation Kit (Qiagen, Germantown, MD, USA), per the manufacturer's instructions. Successful sub-cellular fractionation was verified by immunoblotting with antibodies detecting either β-actin or heterochromatin protein 1 (HP1), which are cytoplasmic and nuclear proteins, respectively. The anti-β-actin antibody was an HRP-conjugated mouse polyclonal antibody obtained from GenScript (diluted 1:5000), while the anti-Drosophila HP1 antibody was a mouse monoclonal antibody (diluted 1:500) obtained from the Developmental Studies Hybridoma Bank, developed under the auspices of the NICHD and maintained by The University of Iowa. Following detection of anti-HP1 antibody binding, incubation with HRP-conjugated goat anti-mouse secondary antibody was performed, using a 1:50 000 dilution. Anti-Dcr, Ago, FLAG and HA immunoblots were performed as described above.
Small RNA sequencing and analysis
Aag2 cells were seeded in six-well plates; triplicate wells were transfected after 24 h with 2.5 μg dsRNA against EGFP, r2d2, loqs or loqs-rb. At 3 days post-transfection cells from each well were re-seeded into 25 cm2 flasks. At 24 h all flasks were transfected with 1.25 μg firefly luciferase dsRNA and 1.25 μg pSLfa-PUb-RL-IR. RNA was harvested 24 h after this second transfection (5 days post-dsRNA treatment) using Trizol. Libraries were prepared with the TruSeq small RNA sample preparation kit (Illumina, San Diego, CA, USA), according to the manufacturer's instructions with minor modifications. Briefly, small RNAs were first isolated by PAGE, selecting ∼18–35 bp RNAs and PCR amplification was increased from 11 cycles to 15. All 12 libraries were indexed separately, pooled and sequenced on a single lane of an Illumina HiSeq2500; sequencing was performed by Beckman Coulter Genomics (Danvers, MA, USA). Following bioinformatic separation of reads based on barcodes, small RNAs were analyzed essentially as described by (25). Adapter sequences were removed bioinformatically (FASTX toolkit); reads containing ambiguous bases or where the adapter could not be identified were discarded. Trimmed reads were mapped using bowtie (26) to a non-redundant set of sequences including Ae. aegypti transcripts (AaegL3.2), transposons (TEfam; http://tefam.biochem.vt.edu/tefam/index.php), persistently infecting viruses (CFAV; NC_001564) and synthetic constructs (Firefly, Renilla luciferase). Only perfectly matching (−v 0), unique (−m 1) reads were accepted. Reads that did not map to the initial set were re-mapped to the Ae. aegypti genome assembly (AaegL3). Only mapped reads with lengths consistent with miRNAs or siRNAs (21–24nt) were selected for statistical analysis, the number of such reads was summed for each sequence prior to analysis with EdgeR (27). Target sequences were classified as either siRNA-like loci or miRNA-like loci based on the following criteria. siRNA-like loci were required to (1) possess a significant peak length of 21nt [# of 21nt > (20nt + 22nt)]; (2) derive relatively equally from both strands (ratio of 21nt from sense and antisense strands no greater than 3:1 in either direction); and (3) derive relatively randomly from the target sequence (<25% of reads mapping to the same start position). For miRNA-like loci, >90% of reads were required to map to a single start position at the peak length, a criteria sufficient to identify 99% of known miRNAs. Small RNA data is available for download from the GEO (GSE65070); raw and normalized read count data is presented in Supplementary Table S6.
RESULTS
Gene structure and tissue distribution of Ae. aegypti dsRNAbps
We first characterized the gene structure and major splicing variants for both Ae. aegypti r2d2 and loqs (Figure 1A). For r2d2 (VectorBase gene AAEL011753), 5′ RACE (n = 6) revealed a start of transcription 32 bp upstream from the computer-predicted start (AAEL011753-RA), while 3′ RACE suggested that transcription termination occurs much sooner that the computational predicted gene model suggests. Poly-A tails for five of the six clones sequenced occurred between 255 and 290 bp upstream from the predicted stop, and 413 bp upstream for one of the clones. No additional exons or splice variants were detected.
Figure 1.
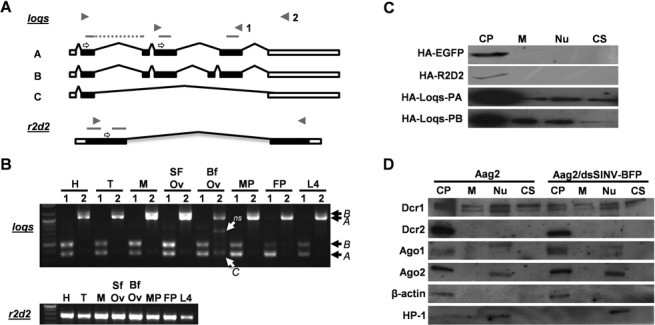
Characterization of dsRBP gene structure, expression and localization. (A) Structures of loqs-ra, loqs-rb, and loqs-rc splice variants and r2d2 mRNA. Solid boxes represent ORFs, unfilled boxes represent UTRs, and gray bars represent predicted DRMs. Primer locations used for RT-PCR and cDNA sequencing are marked by block arrows; 3′ RACE primers indicated by open arrows. (B) One-step RT-PCR using head (H), thorax (T), midgut (M), sugar-fed ovaries (SFO), blood-fed ovaries (BFO), male pupae (MP), female pupae (FP) and L4 larvae (L4) total RNA as templates to detect dsRBP transcripts. (C) Localization of overexpressed HA or FLAG-tagged dsRBPs in Aag2 cells. HA-EGFP and HA-R2D2 were expressed via dsSINV; HA-Loqs-PA and HA-Loqs-PB were expressed via plasmid transfection. (D) Localization of mosquito Dcr and Ago proteins in uninfected and infected Aag2 cell fractions: cytoplasm (CP), membrane (M), nucleus (N), and cytoskeleton (CS). Antibodies recognizing β-actin (cytoplasmic) and heterochromatin protein 1 (HP1, nuclear) were used to verify the success of each fractionation experiment.
For loqs (AAEL008687), 3′ and 5′ RACE sequencing revealed variability in both transcription start and stop locations. Of the fifteen 5′ RACE clones sequenced, thirteen began transcription 69 bp upstream of the predicted start, one 111 bp upstream and one 34 bp upstream. Of the initial twelve 3′ RACE loqs clones sequenced, one clone ended at exactly the predicted stop, three clones ended 332 bp upstream, three clones between 1052 and 1123 bp upstream, four clones between 1304 and 1356 bp and one clone ended 1832 bp upstream. Primers were designed at the 5′ and 3′ ends and through cDNA sequencing we confirmed the presence of three predominant mRNA splice variants, which we refer to as loqs-ra, -rb and -rc (Figure 1A). The loqs-ra isoform matched the exon structure of AAEL008687-RA. The loqs-rb isoform is similar, but includes an additional exon (exon 5) that increases the distance between the last two of the three predicted dsRNA binding motifs (DRMs 2 and 3). These isoforms correspond to those encoding Drosophila melanogaster Loqs-PA and Loqs-PB, both of which partner with Dicer-1 (11). The drosophilid isoform Loqs-PD includes only the first two DRMs and is important to endo-siRNA biogenesis (11). However, the third Ae. aegypti isoform we detected, loqs-rc, does not resemble either of the two remaining drosophilid isoforms, Loqs-PC or -PD, as Ae. aegypti loqs-rc includes only the first DRM. Since we did not recover an loqs-rd isoform, we repeated 3′ RACE experiments using a primer located further upstream in exon 2, rather than exon 4. Again, we were unable to recover a loqs-rd form, suggesting that Ae. aegypti may not make Loqs-PD. Data mining from several recent RNA-seq studies (28–31) confirmed that expressed transcripts from a wide array of tissues/developmental stages do not map past the splice donor site at the end of exon 4, further suggesting that Ae. aegypti does not make Loqs-PD (Supplementary Figure S1), though loqs-ra and loqs-rb forms were readily recovered (28).
To determine the timing and pattern of r2d2 and loqs mRNA expression in the mosquito body, we performed One-Step RT-PCR reactions using total RNA isolated from Ae. aegypti heads, thorax, midguts, sugar-fed ovaries, blood-fed ovaries, male pupae, female pupae and L4 larvae (Figure 1B). The approximate primer locations are indicated in Figure 1A. Our results indicate that r2d2 is uniformly expressed in all of the tissues analyzed. Likewise, both loqs-ra and loqs-rb isoforms are detectable in all tissues analyzed at approximately the same abundance as each other, with overall expression higher in ovaries, while loqs-rc expression is much weaker and appears slightly stronger in blood-fed ovaries. Similar results were recently reported via mRNA-seq experiments by Akbari et al. (28).
Sub-cellular localization of Ae. aegypti RNAi components
Cellular compartment fractionation assays were used to determine the intracellular localization of R2D2, Loqs-PA and Loqs-PB (Figure 1C) in relation to Dcr1, Dcr2, Ago1 and Ago2 proteins (Figure 1D) in cultured mosquito cells. For the dsRBPs, HA or FLAG-tagged EGFP, R2D2 and Loqs were individually expressed through infection of Aag2 cells with recombinant double-subgenomic Sindbis virus (dsSINV, TE/3′2J) designed to express the tagged protein. Both EGFP and R2D2 were detected only in cytoplasmic fractions; Loqs-PA and Loqs-PB were primarily cytoplasmic proteins, but were detectable in all sub-cellular fractions as well (Figure 1C). Similarly, the siRNA component Dcr2 was only detected in the cytoplasmic fraction, while Ago2 appeared to localize in both cytoplasmic and nuclear fractions (Figure 1D). In contrast, the miRNA component Dcr1 was detectable in all sub-cellular fractions, similar to Loqs-PA and Loqs-PB. Ae. aegypti Ago1 appears as doublet band around 112 kilodaltons (kD); these doublets were detectable in the cytoplasmic fractions, while a slightly larger band was also consistently detected in the nuclear fraction (three independent replicates). As we expressed the dsRBPs with a viral expression system, we determined whether virus infection altered the localization pattern of miRNA or siRNA gene products; no difference in the localization of any Dcr or Ago proteins was detected following infection with Sindbis virus (SINV, Figure 1D).
Loqs-PD isoforms are conserved amongst drosophilids, but not in other dipterans
Our failure to identify a loqs-rd transcript in Ae. aegypti suggests that this isoform may not be conserved amongst all dipterans. To determine the potential for dipterans outside of Drosophila to encode Loqs-PD, we performed a two-step blast-based search of various dipteran genome assemblies. In the first step, the D. melanogaster Loqs protein sequence was used to identify loqs orthologs in the relevant genomes via blastp or tblastn. In the second step, 15–20 residues corresponding to the end of exon 4 for each species were used to query its own genome assembly (tblastn). After manual inspection of the aligned regions, the coding potential of read-through into the intronic region was determined. As shown in Figure 2A, the unique Loqs-PD tail region is conserved in almost all drosophilids, with only D. willistoni containing a premature stop codon limiting potential translation to just six additional amino acids. In contrast, the ability of other dipterans to generate Loqs-PD tail regions appeared to be limited, with little apparent conservation (Figure 2B). While the malaria mosquito, Anopheles gambiae, has the potential to encode an additional 68 amino acids following the exon 4 splice donor site, this region is not conserved amongst other Anophelines (Supplementary Figure S2). As stated earlier, while Ae. aegypti has the potential to encode an additional 41 amino acids, no evidence of such a transcript could be found. We conclude that Ae. aegypti, and likely most non-drosophilid dipterans, do not encode a Loqs-PD isoform and thus must use an alternative strategy to coordinate processing and/or loading of endo-siRNAs.
Figure 2.
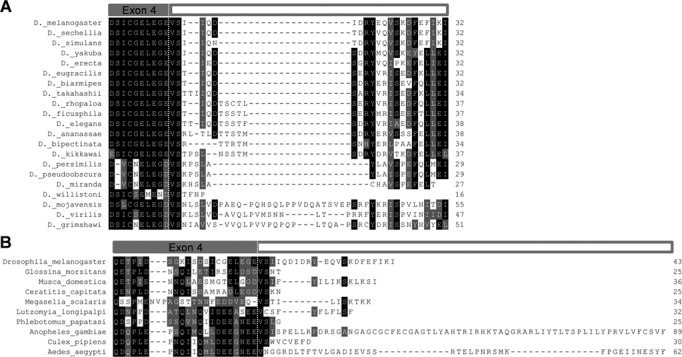
Loqs-PD is not conserved throughout Diptera. (A) Loqs-PD tail regions from 21 Drosophila species. (B) Loqs-PD tail region from D. melanogaster compared to hypothetical Loqs-PD tails from nine other non-drosophilid dipterans. Dotted line indicates the boundary between Loqs exon 4 and the unique -PD tail generated from read-through into the intronic region. In all cases, the last amino acid listed is followed by a stop codon. Identical (black) and similar (gray) amino acids are indicated by highlighting.
Ae. aegypti Loqs interacts with both siRNA and miRNA components
To further explore the relationships of Ae. aegypti R2D2, Loqs-PA and Loqs-PB to siRNA and miRNA factors, we employed co-immunoprecipitation (co-IP) assays to test for protein–protein interactions between each dsRBP and Dcr1, Dcr2, Ago1 and Ago2. As both the Drosophila (Loqs) and human (TRBP) orthologs of Ae. aegypti Loqs have been shown to bind Dcr1 through an interaction mediated by the 3rd DRM at the C-terminus of the protein (19,32), two deletion constructs were included with a truncation immediately preceding the third DRM (Δ258) or immediately following the second DRM (Δ226) (Figure 3A). Co-IP experiments confirmed a strong association between R2D2 and Dcr2; this interaction was not affected by pre-treatment with RNaseA and is consistent with the role of this protein in the siRNA pathway (Figure 3B). Likewise, only Loqs-PB interacted with DCR1, consistent with its role in miRNA biogenesis. Interestingly, both Loqs-PA and Loqs-PB interacted with Dcr2, Ago1 and Ago2. Deleting the third DRM had no effect on the interaction between Loqs and Ago proteins. However, this interaction was lost when the 32 a.a. spacer separating the 2nd and 3rd DRMs was deleted (Figure 3B). Neither deletion had any effect on the ability of Loqs to interact with Dcr2.
Figure 3.
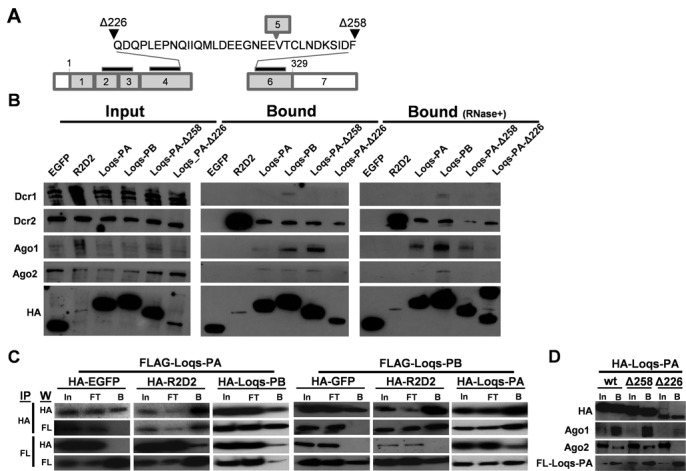
Interactions between mosquito dsRBPs and RNAi/miRNA components. (A) Schematic representation of the Loqs-PA exon structure showing the location of the two Loqs-PA truncations used (Δ258 and Δ226). Start (1) and Stop (329) positions in the ORF are indicated; gray bars above indicate the locations of the three DRMs. The 32 amino acid spacer between DRMs 2 and 3 is highlighted, along with the location of exon 5 when spliced into Loqs-PB. (B) Co-immunoprecipitation of Dcr and Ago proteins with HA-tagged dsRBPs in Aag2 cells. HA-dsRBPs were expressed in Aag2 cells by transfection of plasmid DNA. (C) Co-Immunoprecipitation of FLAG-Loqs-PA or FLAG-Loqs-PB with HA-tagged proteins. Column headings indicate the overexpressed HA-tagged protein, row headings indicate the antibody used in the corresponding western blot. Input (In), flow-through (FT) and bound (B) fractions are indicated. To increase the amount of detectable R2D2, HA-R2D2 proteins were expressed via infection with dsSINV; all others were expressed by plasmid transfection. Anti-HA and anti-FLAG co-IP assays were run 24 h post-infection (48 h post-transfection). (D) Dimerization of Loqs-PA is unaffected by both the Δ258 and Δ226 deletions as shown by co-IP.
Both the Drosophila (Loqs) and human (TRBP) orthologs of Ae. aegypti Loqs are capable of forming homo- and heterodimers with themselves and related dsRBPs. To test whether R2D2, Loqs-PA, and Loqs-PB are capable of interacting, additional co-IP assays were run on lysates from Aag2 cells overexpressing both HA and FLAG-tagged proteins. All three dsRBPs were found to interact with each other when overexpressed, though an association between HA-R2D2 and FLAG-Loqs-PB could only be detected from the anti-HA IP, but not the anti-FLAG IP (Figure 3C). Deletion of the third DSRM and/or the spacer sequence did not affect dimerization of Loqs (Figure 3D), similar to human TRBP (32,33).
An in silico prediction using Pepfold (34) of the 32 a.a. spacer region suggests this region should adopt a coil–helix–coil motif (Supplementary Figure S3). Interestingly, the 21 a.a. tail region of Drosophila Loqs-PD, critical for binding DmDcr2 (34), is predicted to form a similar coil–helix–coil structure (Supplementary Figure S3). While we could not detect read-through of the 4th exon that might correspond to an Ae. aegypti Loqs-PD isoform, structural modeling of the predicted peptide sequence that would result from such a hypothetical translation revealed an unrelated structure. This suggests that the spacer region between the second and third dsRBMs of Ae. aegypti Loqs may perform a similar function to the unique tail of Drosophila Loqs-PD.
Functional role of Loqs in the mosquito siRNA and miRNA pathways
To examine the functional role of Ae. aegypti R2D2, Loqs-PA and Loqs-PB in mosquito RNA interference, we first overexpressed each protein in mosquito cells and measured the effect on either the endo-siRNA-based silencing of an inverted-repeat (Figure 4A), or exo-siRNA-based silencing of double-stranded RNA using luciferase-based reporter assays (Figure 4B). Of the three proteins, only overexpression of Loqs-PA increased the effectiveness of endo-siRNA silencing (Figure 4C). This effect could be nullified by co-overexpression of R2D2, and could be exasperated by co-overexpression of both R2D2 and Loqs-PB, most likely due to dominant negative effects of heterodimer formation (16). Conversely, only overexpression of R2D2 increased the ability to silence exogenous dsRNA, an effect that was also eliminated by co-overexpression of either Loqs-PA or Loqs-PB (Figure 4D). Though knockdown of each gene was successful (Figure 4E), double-stranded RNA treatments targeting each dsRBP failed to reveal a significant effect on our silencing reporters (Supplementary Figure S4A). Follow-up experiments suggested that substantial overexpression of Dcr2 in these cells (Supplementary Figure S4B) might mask an effect of loss of Loqs, which is dispensable for dicing activity but has been shown to increase the ability Dcr2 to process dsRNA (13,35). Alternatively, redundancy between silencing factors may also mask an effect. Thus, to determine the effect of loss of Ae. aegypti R2D2, Loqs-PA and Loqs-PB on small RNAs directly, we sequenced the small RNA fraction from mosquito cells treated with dsRNA RNA targeting egfp (control), r2d2, the unique exon 5 only present in loqs-rb, or loqs exon 2 (present in both isoforms); both the endo-siRNA (IR-construct) and exo-siRNA (dsRNA) reporters were transfected into all cells. The experiment was performed with three biological replicates per treatment, yielding 12 small RNA libraries.
Figure 4.
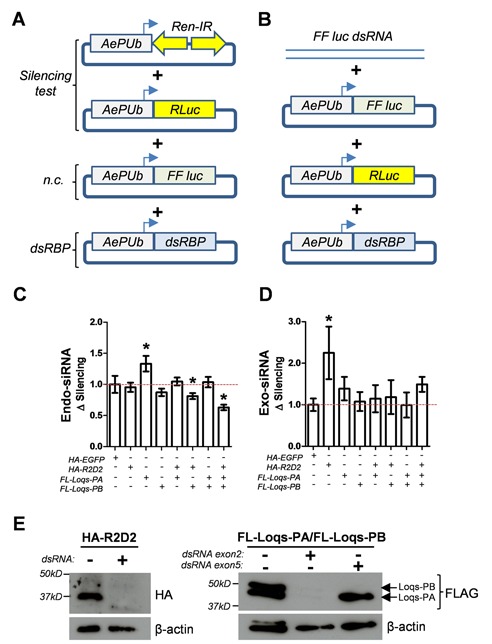
Functional role of AaLoqs and AaR2D2 in siRNA-based silencing in mosquito cells. Schematic representations of endo-siRNA (A) and exo-siRNA (B) based reporters transfected into Aag2 mosquito cells and evaluated for their ability to silence a hairpin construct (C) or exogenous dsRNA (D) to silence a corresponding reporter gene (Firefly luciferase). Silencing activity was measured by comparing normalized firefly luciferase values (using an internal Renilla reporter) in the absence or presence of the silencing construct/dsRNA. Each experiment was repeated at least three times, with each experiment consisting of eight biological replicates. For all treatments, the highest/lowest values were removed prior to statistical analysis (ANOVA, Bonferroni's Multiple Comparison Test). In each case the ANOVA was significant (P < 0.05), with individual samples significantly different from HA-EGFP transfected cells indicated (*). (E) Western blot confirming overexpression of each dsRBP in the presence (+) or absence (-) of the indicated dsRNA.
We first confirmed the specificity of each dsRNA treatment, as siRNAs mapping to EGFP, r2d2, loqs (exon 2) and loqs-rb (exon 5) were only identified in the respective treatment groups (Figure 5A). Depletion of either R2D2 or both Loqs-PA/PB resulted in a significant decrease in small RNAs derived from the reporter dsRNA molecule (Figure 5B). This effect was not seen upon depletion of Loqs-PB, indicating that of the two, Loqs-PA is likely responsible. Interestingly, loss of R2D2 also decreased small RNA levels derived from the IR-repeat reporter, indicating a role for this protein in endo-siRNA production as well. Depletion of both Loqs isoforms decreased production of IR-derived small RNAs from the antisense, but not sense strands in a manner dependent on Loqs-PB (Figure 5B). Globally, depletion of R2D2 resulted in a reduction of siRNAs from a subset of transposable elements, with the two reporters being amongst the most significant to lose siRNAs (Figure 5C). Surprisingly, siRNAs derived from a persistently infecting RNA flavivirus (Cell Fusing Agent virus, CFAV) did not change in the absence of R2D2. Depletion of all Loqs isoforms, but not Loqs-PB alone, resulted in a substantial increase in siRNAs derived from both protein-coding genes and transposable elements, but a loss of siRNAs derived from CFAV, indicating a complex role for Ae. aegypti Loqs-PA in siRNA production (Figure 5C). Depletion of Loqs-PB alone resulted in only minor changes in siRNA production, including a significant increase in siRNAs derived from the plasmid backbone of the inverted repeat construct (but not from the inverted repeat itself). Consistent with its known role in miRNA processing, depletion of Loqs-PB resulted in a significant reduction in 39/82 (48%) miRNAs, compared with only 15 (18%) miRNAs that increased in expression (Figure 5C). Conversely, depletion of R2D2 resulted in an increase in abundance of 31/82 (38%) miRNAs, compared to just 3 (4%) that decreased, suggesting that R2D2 antagonizes miRNA production. Most interestingly, depletion of both Loqs isoforms essentially restored the abundance of miRNAs, suggesting that the ratio of Loqs-PA to Loqs-PB may be the more significant factor in miRNA biogenesis than the absolute amount of each dsRBP.
Figure 5.
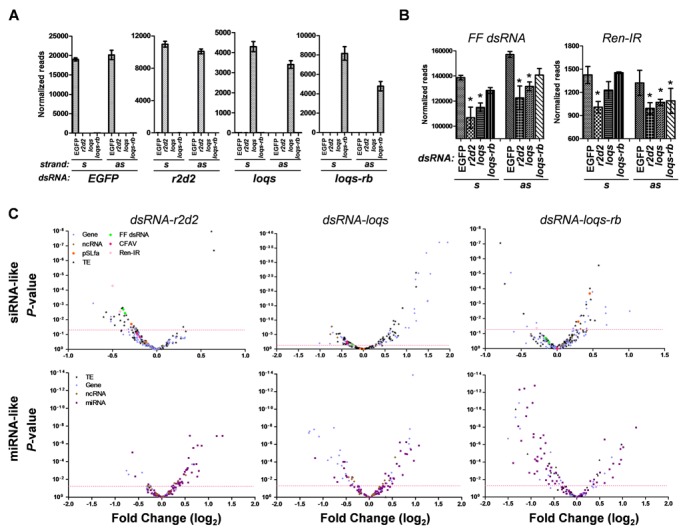
Depletion of mosquito dsRBPs perturbs small RNA levels as revealed through small RNA sequencing. (A) Normalized read counts for small RNAs derived from each treatment dsRNA. (B) Normalized read counts for small RNAs mapping to Firefly luciferase (FF dsRNA) or the Renilla luciferase inverted repeat (Ren-IR) in each treatment group. Statistical analysis is from EdgeR considering the entire dataset. (C) Volcano plots showing the fold change of small RNA abundance for each parent sequence (Gene, protein-coding gene; ncRNA, non-coding RNA; TE, transposable element; CFAV, cell-fusing agent virus; pSLfa, plasmid backbone of the Ren-IR construct; FF dsRNA, Firefly luciferase dsRNA; Ren-IR, Renilla inverted repeat) as compared to EGFP dsRNA-treated cells. Red dotted line indicates P-value = 0.05.
If R2D2 and Loqs act in independent pathways, we would expect little correlation between the small RNAs that change in abundance when each of these proteins is depleted. Conversely, if these proteins act non-redundantly in the same pathway, we would expect a strong correlation between changes in small RNA levels. We compared the change in abundance of small RNAs for all 104 targets whose small RNA levels were significantly altered in R2D2-depleted cells upon knockdown of all Loqs isoforms or just Loqs-PB. Strikingly, we observed a highly significant correlation in the fold change of small RNAs derived from protein-coding genes, transposons and miRNAs between R2D2-depleted and Loqs-depleted cells (Figure 6, Table 1). This correlation was not observed between R2D2-depleted and Loqs-PB-depleted cells, indicating that indeed Loqs-PA and R2D2 act non-redundantly in both the production of siRNAs and the suppression of miRNAs. A significant correlation was also found when comparing Loqs-depleted to Loqs-PB-depleted cells [as expected since Loqs-PB is knocked down in both cases (Figure 6, Table 1)].
Figure 6.
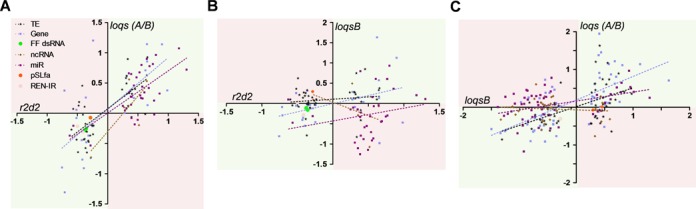
Small RNA changes are highly correlated upon R2D2 or Loqs depletion in mosquito cells. Fold change of small RNA abundance for each parent sequence (Gene, protein-coding gene; ncRNA, non-coding RNA; TE, transposable element; CFAV, cell-fusing agent virus; pSLfa, plasmid backbone of the Ren-IR construct; FF dsRNA, Firefly luciferase dsRNA; Ren-IR, Renilla inverted repeat) for pairwise comparisons between r2d2:loqs (A), r2d2:loqs-rb (B) and loqs-rb:loqs (C). Quadrants are shaded based on a cooperative (green) or antagonistic (pink) relationship.
Table 1. DsRBPs R2D2 and Loqs-PA are non-redundant and cooperate in small RNA production/stability.
| Category (n) | Slope | R2 | P -value | |
|---|---|---|---|---|
| r2d2 vs loqs | Gene | 0.85+0.19 | 0.46 | 0.0002 |
| miR | 0.70+0.13 | 0.47 | <0.0001 | |
| TE | 0.77+0.12 | 0.52 | <0.0001 | |
| ncRNA | 1.18+0.56 | 0.59 | 0.1272 | |
| r2d2 vs loqs-rb | Gene | 0.40+0.19 | 0.16 | 0.0467 |
| miR | 0.32+0.25 | 0.04 | 0.2116 | |
| TE | 0.11+0.13 | 0.02 | 0.3887 | |
| ncRNA | −0.67+0.86 | 0.17 | 0.4952 | |
| loqs-rb vs loqs | Gene | 0.62+0.09 | 0.42 | <0.0001 |
| miR | 0.24+0.07 | 0.19 | 0.0011 | |
| TE | 0.50+0.13 | 0.25 | 0.0005 | |
| ncRNA | −0.05+0.10 | 0.01 | 0.6306 |
Taken together, we conclude that in mosquitoes, and potentially most other dipterans, in the absence of an ortholog of the Drosophila Loqs-PD isoform it is Loqs-PA that plays a complex role in siRNA based silencing, important for the production of some siRNAs in coordination with R2D2, while also antagonizing the generation of other siRNAs and some miRNAs. The role of Loqs-PB appears to be conserved in miRNA biogenesis and this isoform also appears to largely antagonize siRNA production.
DISCUSSION
Over the past few years, substantial evidence has accumulated that the model dipteran, D. melanogaster, uses alternative splicing to functionally segregate miRNA- and endo-siRNA-based responsibilities of the hub protein, Loqs. DmLoqs-PA and -PB isoforms both partner with DmDcr1 in miRNA biogenesis, while DmLoqs-PD partners with DmDcr2 to process various classes of endo-siRNAs. Loqs null flies are not viable; while Loqs-PA can rescue viability, only Loqs-PB can rescue fly fertility (13), and this isoform is thought to be more essential for miRNA processing (10,14), as it interacts much more readily with Dcr1 (12). In contrast, DmLoqs-PD protein partners with DmDcr2 and is important to endo-siRNA biogenesis (13–15,19). We examined the conservation of these segregated functions in other diptera. Surprisingly, genomic comparisons revealed that the ability to generate Loqs-PD isoforms was not conserved in other dipterans, and despite repeated attempts, a Loqs-PD form could not be detected in the mosquito Ae. aegypti. Given its absence in both mosquitoes and sandflies, two of the oldest dipteran groups, along with the fact that drosophilids are known to be a much more recent radiation (36), our results suggest that the functional segregation of loqs’ responsibilities is a derived trait, restricted to drosophilids.
Instead, we observed that in mosquito cells, Loqs-PA plays a complex role in regulating endo-siRNA, exo-siRNA and miRNA levels but largely cooperates non-redundantly with R2D2. Overexpression of Loqs-PA, but not Loqs-PB, increased the ability of mosquito cells to silence an inverted repeat construct, while loss of all Loqs isoforms disrupted small RNA levels from exogenous and endogenous sources in a manner similar to R2D2 depletion. Biochemical evidence has shown that in Drosophila, both R2D2 and Loqs-PD can decrease the substrate concentration at which Dicer effectively processes dsRNA (13,35). Interestingly, overexpression of R2D2 increased the ability of mosquito cells to silence a target triggered by exogenous dsRNA. This is somewhat surprising, given that R2D2 is not stable unless bound by Dcr2 (37). However, we found that Dcr2 was substantially overexpressed in Aag2 cells, while levels of R2D2 mRNA were much lower than found in adult mosquitoes. Thus, our overexpression experiment more closely resembled that of a rescue of R2D2, and likely resulted in additional R2D2/Dcr2 heterodimers in place of unpaired Dcr2. Combined with our observations that R2D2 and Loqs-PA are both required for proper small RNA levels, our overexpression data may suggest that silencing of our endo-siRNA and exo-siRNA reporters are limited by independent bottlenecks. It has been long known that single-stranded ends on double-stranded RNA molecules inhibit processing by Dcr (38). Thus, the bottleneck in processing endo-siRNAs may be more effectively relieved by Loqs, consistent with the mechanism proposed by Marques (18). Processing perfect dsRNA may not require this extra help, resulting in a bottleneck in loading- a role primarily assigned to R2D2 (37).
In Drosophila, R2D2 partners with Dcr2 and enables loading of small interfering viral RNAs (viRNAs) in the Ago2 effector complex. Co-immunoprecipitation of AaDcr2 with AaR2D2 supports conservation of this function in mosquitoes and agrees with previous observations that depletion of AaR2D2 can increase viral replication in infected mosquitoes (8) and is critical for the anti-viral exo-siRNA response in flies (39). However, depletion of R2D2 did not alter the abundance of small RNAs derived from a persistently infecting flavivirus (CFAV), whereas CFAV small RNAs did decrease upon knockdown of Loqs, suggesting a potential role for mosquito Loqs in antiviral immunity.
Loss of R2D2 has been reported to increase the abundance of miRNAs, suggesting a potential antagonistic relationship between these two dsRBPs (18). However, the authors of that study were guarded in their interpretation due to a lack of biological replicates and appropriate normalization methods. Using replicate libraries and modern normalization methods, we also observed a strong antagonistic relationship between R2D2 and Loqs-PB concerning the generation of miRNAs. Thus, we conclude that R2D2 restricts miRNA production, potentially though increasing the selectivity of Dcr2 for long dsRNA (35). Interestingly, depletion of all Loqs isoforms was not nearly as disruptive to miRNA levels as depletion of Loqs-PB alone. This suggests an additional layer of competition between Loqs-PA and Loqs-PB, with Loqs-PA able to inhibit miRNA production similar to R2D2. Hartig and Forstemann (16) reported competition between Loqs-PA/PB and Loqs-PD as overexpression of the former decreased the production of endo-siRNA CG4068B, while overexpression of Loqs-PD increased endo-siRNA expression. An additional complication is that depletion of Loqs, but not Loqs-PB or R2D2 resulted in a substantial increase in siRNA production from both protein-coding genes and transposons. This suggests that Loqs-PA may also antagonize the endo-siRNA pathway in some cases, essentially serving as gatekeeper to prevent the dicing of unintended substrates. Both AaLoqs-PA and -PB likely participate in other protein-protein interactions that further regulate their role in RNAi, as both the Drosophila Loqs protein and the human orthologs PACT/TRBP are known hub proteins with large interaction networks (32,40).
Both AaLoqs isoforms were able to interact with native Dcr2, Ago1 and Ago2, whereas only AaLoqs-PB was observed to interact with Dcr1, again suggesting a role for AaLoqs in both the miRNA and the siRNA pathways of mosquitoes. The interactions we observed between Loqs and AaAgo1/AaAgo2 suggest a role beyond simply processing dsRNA. As in Drosophila, we did not recover AaR2D2 in complex with AaAgo2, suggesting that its well-defined role in siRNA loading does not require a stable interaction with Ago (this is mediated through Dcr2). Drosophila Loqs also interacts with Ago1 (41) and human TRBP is a well-established component of the RISC (32). We were able to map the interaction domain of Ae. aegypti Loqs for Ago1/Ago2 to the short linker sequence between the second and third dsRNA-binding domains. This appears to be independent from the interaction domain between DmLoqs and Dcr1 (third DSRM) (14) and the interaction domain we observed for binding Dcr2, the latter of which was still able to bind Loqs with just the first and second DSRMs present.
The third loqs isoform we identified, loqs-rc, consisting of exons 1, 2, 6 and 7, does not resemble any of the previously described drosophilid isoforms. While we were able to recover several cDNA clones of the loqs-rc splice variant, we were unable to express an HA-tagged version of this protein in Aag2 cells, either through recombinant virus or through plasmid transfection. This is somewhat reminiscent of the drosophilid loqs-rc splice variant, which was only detectable in S2 cells and had no detectable protein product (11,15). As this product was also barely detectable by PCR of various tissues, its biological significance is questionable.
In addition to their roles in post-transcriptional gene silencing, several RNAi proteins perform functions in the cell nucleus [reviewed by Castel and Martienssen (42)]. In plants and fungi, RNAi-based silencing inhibits not only translation, but also occurs at the transcriptional level by regulating heterochromatin formation. Transcriptional gene silencing (TGS) occurs when epigenetic modifications, such as histone methylation, occur at target genomic loci in response to nuclear RNAi. The specific mechanisms by which RNAi can regulate TGS are not completely known and likely vary by species. Cernilogar et al. (43) found that both Ago2 and Dcr2 associate with RNA polymerase II and transcriptionally active loci in euchromatin to negatively regulate transcription by inhibiting RNA polymerase II activity. In particular, their research revealed a role for these RNAi components in the heat shock response (43). Additionally, a recent study by Taliaferro et al., suggested that depletion of Ago2 affected pre-mRNA splicing patterns, based on genome-wide screens (44). Our subcellular fractionation assays support a potential role for the RNAi component Ago2 in the mosquito cell nucleus, as well as the miRNA components Ago1/Dcr1, and both isoforms of Loqs. In contrast, the RNAi components Dcr2 and R2D2 were restricted to the cell cytoplasm, suggesting their roles might be more limited in mosquitoes. Further studies are needed to clarify the roles of both siRNA and miRNA factors in the mosquito nucleus.
In summary, our experiments have revealed an unexpected twist in the story of how double-stranded RNA binding proteins interact with RNAi factors across various invertebrate taxa. Our results suggest that the well-characterized Drosophila Loqs-PD isoform is a derived trait, essentially a specialized form of the ancestral Loqs-PA isoform. In other diptera, Loqs-PA has maintained a generally cooperative role with R2D2 and Dcr2 in siRNA production (exogenous and endogenous) while largely antagonizing Loqs-PB in miRNA production. Loqs-PA also appears to serve as a gatekeeper, keeping protein-coding mRNAs from entering the siRNA pathway. How Loqs-PA balances synergistic and antagonistic functions related to RNAi remains unknown. The ability to perform site-specific gene editing (23) should allow us to address the functional role of these proteins in RNAi directly in other dipterans such as mosquitoes and resolve these interesting questions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank members of the Adelman lab for technical assistance, and Dr Rui Zhou for generously providing the Renilla inverted repeat construct (pRmHa-MCS-Renilla-IR)
FUNDING
National Institutes of Health [AI085091 to Z.A., GM072767 to E.S.]. Funding for open access charge: NIH [AI085091] and the Fralin Life Science Institute at Virginia Tech.
Conflict of interest statement. None declared.
REFERENCES
- 1.WHO. Dengue and dengue haemorrhagic fiver – Fact Sheet. 2009 [Google Scholar]
- 2.Enserink M. Infectious diseases. Chikungunya: no longer a third world disease. Science. 2007;318:1860–1861. doi: 10.1126/science.318.5858.1860. [DOI] [PubMed] [Google Scholar]
- 3.Enserink M. Infectious diseases. Crippling virus set to conquer Western Hemisphere. Science. 2014;344:678–679. doi: 10.1126/science.344.6185.678. [DOI] [PubMed] [Google Scholar]
- 4.Gubler D.J. The changing epidemiology of yellow fever and dengue, 1900 to 2003: full circle. Comp. Immunol. Microbiol. Infect. Dis. 2004;27:319–330. doi: 10.1016/j.cimid.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 5.Li L., Liu Y. Diverse small non-coding RNAs in RNA interference pathways. Methods Mol. Biol. 2011;764:169–182. doi: 10.1007/978-1-61779-188-8_11. [DOI] [PubMed] [Google Scholar]
- 6.Jinek M., Doudna J.A. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457:405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- 7.Myles K.M., Wiley M.R., Morazzani E.M., Adelman Z.N. Alphavirus-derived small RNAs modulate pathogenesis in disease vector mosquitoes. Proc. Natl. Acad. Sci. U.S.A. 2008;105:19938–19943. doi: 10.1073/pnas.0803408105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sánchez-Vargas I., Scott J.C., Poole-Smith B.K., Franz A.W., Barbosa-Solomieu V., Wilusz J., Olson K.E., Blair C.D. Dengue virus type 2 infections of Aedes aegypti are modulated by the mosquito's RNA interference pathway. PLoS Pathog. 2009;5:e1000299. doi: 10.1371/journal.ppat.1000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanchez-Vargas I., Travanty E.A., Keene K.M., Franz A.W., Beaty B.J., Blair C.D., Olson K.E. RNA interference, arthropod-borne viruses, and mosquitoes. Virus Res. 2004;102:65–74. doi: 10.1016/j.virusres.2004.01.017. [DOI] [PubMed] [Google Scholar]
- 10.Czech B., Malone C.D., Zhou R., Stark A., Schlingeheyde C., Dus M., Perrimon N., Kellis M., Wohlschlegel J.A., Sachidanandam R., et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forstemann K., Tomari Y., Du T., Vagin V.V., Denli A.M., Bratu D.P., Klattenhoff C., Theurkauf W.E., Zamore P.D. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye X., Paroo Z., Liu Q. Functional anatomy of the Drosophila microRNA-generating enzyme. J. Biol. Chem. 2007;282:28373–28378. doi: 10.1074/jbc.M705208200. [DOI] [PubMed] [Google Scholar]
- 13.Fukunaga R., Han B.W., Hung J.H., Xu J., Weng Z., Zamore P.D. Dicer partner proteins tune the length of mature miRNAs in flies and mammals. Cell. 2012;151:533–546. doi: 10.1016/j.cell.2012.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou R., Czech B., Brennecke J., Sachidanandam R., Wohlschlegel J.A., Perrimon N., Hannon G.J. Processing of Drosophila endo-siRNAs depends on a specific Loquacious isoform. RNA. 2009;15:1886–1895. doi: 10.1261/rna.1611309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartig J.V., Esslinger S., Bottcher R., Saito K., Forstemann K. Endo-siRNAs depend on a new isoform of loquacious and target artificially introduced, high-copy sequences. EMBO J. 2009;28:2932–2944. doi: 10.1038/emboj.2009.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartig J.V., Forstemann K. Loqs-PD and R2D2 define independent pathways for RISC generation in Drosophila. Nucleic Acids Res. 2011;39:3836–3851. doi: 10.1093/nar/gkq1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X., Jiang F., Kalidas S., Smith D., Liu Q. Dicer-2 and R2D2 coordinately bind siRNA to promote assembly of the siRISC complexes. RNA. 2006;12:1514–1520. doi: 10.1261/rna.101606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marques J.T., Kim K., Wu P.H., Alleyne T.M., Jafari N, Carthew R.W. Loqs and R2D2 act sequentially in the siRNA pathway in Drosophila. Nat. Struct. Mol. Biol. 2010;17:24–30. doi: 10.1038/nsmb.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyoshi K., Miyoshi T., Hartig J.V., Siomi H., Siomi M.C. Molecular mechanisms that funnel RNA precursors into endogenous small-interfering RNA and microRNA biogenesis pathways in Drosophila. RNA. 2010;16:506–515. doi: 10.1261/rna.1952110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adelman Z.N., Anderson M.A., Morazzani E.M., Myles K.M. A transgenic sensor strain for monitoring the RNAi pathway in the yellow fever mosquito, Aedes aegypti. Insect. Biochem. Mol. Biol. 2008;38:705–713. doi: 10.1016/j.ibmb.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson M.A., Gross T.L., Myles K.M., Adelman Z.N. Validation of novel promoter sequences derived from two endogenous ubiquitin genes in transgenic Aedes aegypti. Insect. Mol. Biol. 2010;19:441–449. doi: 10.1111/j.1365-2583.2010.01005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hahn C.S., Hahn Y.S., Braciale T.J., Rice C.M. Infectious Sindbis virus transient expression vectors for studying antigen processing and presentation. Proc. Natl. Acad. Sci. U.S.A. 1992;89:2679–2683. doi: 10.1073/pnas.89.7.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aryan A., Anderson M.A., Myles K.M., Adelman Z.N. TALEN-Based Gene Disruption in the Dengue Vector Aedes aegypti. PLoS One. 2013;8:e60082. doi: 10.1371/journal.pone.0060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Myles K.M., Kelly C.L., Ledermann J.P., Powers A.M. Effects of an opal termination codon preceding the nsP4 gene sequence in the O'Nyong-Nyong virus genome on Anopheles gambiae infectivity. J. Virol. 2006;80:4992–4997. doi: 10.1128/JVI.80.10.4992-4997.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adelman Z.N., Anderson M.A., Wiley M.R., Murreddu M.G., Samuel G.H., Morazzani E.M., Myles K.M. Cooler temperatures destabilize RNA interference and increase susceptibility of disease vector mosquitoes to viral infection. PLoS Neglect. Trop. Dis. 2013;7:e2239. doi: 10.1371/journal.pntd.0002239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akbari O.S., Antoshechkin I., Amrhein H., Williams B., Diloreto R., Sandler J., Hay B.A. The developmental transcriptome of the mosquito Aedes aegypti, an invasive species and major arbovirus vector. G3. 2013;3:1493–1509. doi: 10.1534/g3.113.006742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gibbons J.G., Janson E.M., Hittinger C.T., Johnston M., Abbot P., Rokas A. Benchmarking next-generation transcriptome sequencing for functional and evolutionary genomics. Mol. Biol. Evol. 2009;26:2731–2744. doi: 10.1093/molbev/msp188. [DOI] [PubMed] [Google Scholar]
- 30.Bonizzoni M., Dunn W.A., Campbell C.L., Olson K.E., Dimon M.T., Marinotti O., James A.A. RNA-seq analyses of blood-induced changes in gene expression in the mosquito vector species, Aedes aegypti. BMC Genomics. 2011;12:82. doi: 10.1186/1471-2164-12-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biedler J.K., Hu W., Tae H., Tu Z. Identification of early zygotic genes in the yellow fever mosquito Aedes aegypti and discovery of a motif involved in early zygotic genome activation. PLoS One. 2012;7:e33933. doi: 10.1371/journal.pone.0033933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daniels S.M., Gatignol A. The multiple functions of TRBP, at the hub of cell responses to viruses, stress, and cancer. Microbiol. Mol. Biol. Rev.: MMBR. 2012;76:652–666. doi: 10.1128/MMBR.00012-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laraki G., Clerzius G., Daher A., Melendez-Pena C., Daniels S., Gatignol A. Interactions between the double-stranded RNA-binding proteins TRBP and PACT define the Medipal domain that mediates protein-protein interactions. RNA Biol. 2008;5:92–103. doi: 10.4161/rna.5.2.6069. [DOI] [PubMed] [Google Scholar]
- 34.Thevenet P., Shen Y., Maupetit J., Guyon F., Derreumaux P., Tuffery P. PEP-FOLD: an updated de novo structure prediction server for both linear and disulfide bonded cyclic peptides. Nucleic Acids Res. 2012;40:W288–W293. doi: 10.1093/nar/gks419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cenik E.S., Fukunaga R., Lu G., Dutcher R., Wang Y., Tanaka Hall T.M., Zamore P.D. Phosphate and R2D2 restrict the substrate specificity of Dicer-2, an ATP-driven ribonuclease. Mol. Cell. 2011;42:172–184. doi: 10.1016/j.molcel.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wiegmann B.M., Trautwein M.D., Winkler I.S., Barr N.B., Kim J.W., Lambkin C., Bertone M.A., Cassel B.K., Bayless K.M., Heimberg A.M., et al. Episodic radiations in the fly tree of life. Proc. Natl. Acad. Sci. U.S.A. 2011;108:5690–5695. doi: 10.1073/pnas.1012675108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomari Y., Matranga C., Haley B., Martinez N., Zamore P.D. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- 38.Elbashir S.M., Lendeckel W., Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marques J.T., Wang J.P., Wang X., de Oliveira K.P., Gao C., Aguiar E.R., Jafari N., Carthew R.W. Functional specialization of the small interfering RNA pathway in response to virus infection. PLoS Pathog. 2013;9:e1003579. doi: 10.1371/journal.ppat.1003579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murali T., Pacifico S., Yu J., Guest S., Roberts G.G. 3rd, Finley R.L. Jr. DroID 2011: a comprehensive, integrated resource for protein, transcription factor, RNA and gene interactions for Drosophila. Nucleic Acids Res. 2011;39:D736–D743. doi: 10.1093/nar/gkq1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang F., Ye X., Liu X., Fincher L., McKearin D., Liu Q. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev. 2005;19:1674–1679. doi: 10.1101/gad.1334005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castel S.E., Martienssen R.A. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 2013;14:100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cernilogar F.M., Onorati M.C., Kothe G.O., Burroughs A.M., Parsi K.M., Breiling A., Lo Sardo F., Saxena A., Miyoshi K., Siomi H., et al. Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature. 2011;480:391–395. doi: 10.1038/nature10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taliaferro J.M., Aspden J.L., Bradley T., Marwha D., Blanchette M., Rio D.C. Two new and distinct roles for Drosophila Argonaute-2 in the nucleus: alternative pre-mRNA splicing and transcriptional repression. Genes Dev. 2013;27:378–389. doi: 10.1101/gad.210708.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


