Abstract
SFPQ, (a.k.a. PSF), is a human tumor suppressor protein that regulates many important functions in the cell nucleus including coordination of long non-coding RNA molecules into nuclear bodies. Here we describe the first crystal structures of Splicing Factor Proline and Glutamine Rich (SFPQ), revealing structural similarity to the related PSPC1/NONO heterodimer and a strikingly extended structure (over 265 Å long) formed by an unusual anti-parallel coiled-coil that results in an infinite linear polymer of SFPQ dimers within the crystals. Small-angle X-ray scattering and transmission electron microscopy experiments show that polymerization is reversible in solution and can be templated by DNA. We demonstrate that the ability to polymerize is essential for the cellular functions of SFPQ: disruptive mutation of the coiled-coil interaction motif results in SFPQ mislocalization, reduced formation of nuclear bodies, abrogated molecular interactions and deficient transcriptional regulation. The coiled-coil interaction motif thus provides a molecular explanation for the functional aggregation of SFPQ that directs its role in regulating many aspects of cellular nucleic acid metabolism.
INTRODUCTION
Splicing Factor Proline and Glutamine Rich (SFPQ) is an abundant ubiquitous nuclear protein that is essential for life in vertebrates (1,2). In mammals, SFPQ regulates key pathways such as circadian rhythms (3) and pluripotency in hESCs (4). First identified as a splicing factor associated with polypyrimidine tract-binding protein (5), SFPQ has been implicated in myriad nuclear functions, including DNA repair, transcriptional regulation, splicing and RNA transport (6). A common feature in these varied molecular functions is the ability to interact with nucleic acids. SFPQ interacts with dsDNA at the promoters of a number of genes, where it exerts control over transcriptional regulation (7–9). SFPQ is also a key player in double-strand break repair pathways, interacting directly with DNA and DNA repair proteins (10,11) and rapidly localizing to sites of DNA damage in cells (12). SFPQ also interacts with long non-coding RNAs (lncRNAs) in many scenarios: with NEAT1 lncRNA as an essential factor for subnuclear ‘paraspeckle’ formation (knockdown of SFPQ results in the loss of paraspeckles (13)), with CTBP1-AS in advanced prostate cancer (14), with MALAT1 in metastasis (15) and VL30-1 retroelement lncRNA in oncogenesis (7).
SFPQ is one of the three homologous mammalian proteins that belong to the ‘Drosophila-Behavior, Human-Splicing (DBHS)’ family, the other two being NONO and PSPC1. The three DBHS proteins share a common core structural ‘DBHS’ region of ∼300 amino acids, which encompasses two RNA recognition motif domains (RRM1 and RRM2), a conserved region termed NONO/paraspeckle (NOPS) domain and an extensive region of coiled-coil. Outside this conserved region, the three proteins contain varying quantities of low complexity sequence (SFPQ is 707 amino acids long and contains proline/glutamine- and glycine-rich regions; NONO contains histidine/glutamine-rich and proline-rich regions; PSPC1 contains proline/alanine- and glycine-rich regions) and a C-terminal nuclear localization signal.
The DBHS proteins are obligate dimers, capable of homo- and heterodimerization in vitro (16–18). Dimerization is mediated by RRM2, NOPS and part of the coiled-coil, regions which are also required for stable protein structure. We have previously identified and characterized a heterodimer of PSPC1 with NONO, that included the dimerization domain and RRM1 for both molecules (18). We observed that the NOPS domain forms an extensive peptide-like interaction with RRM2 of the other subunit in the dimer and the final 50 amino acids of these truncated proteins form an unusual antiparallel right-handed heterodimeric coiled-coil. Studies inspired by this unusual structure provided a hint that predicted coiled-coil beyond the dimerization domain could be important for the ability of DBHS proteins to be targeted to paraspeckles (18).
It is rapidly emerging that the ability of RNA-binding proteins to form and be targeted to RNA–protein granules, such as paraspeckles in the nucleus and stress-granules in the cytoplasm, is directly linked to their propensity for ‘functional aggregation’ (19). Indeed, a number of proteins that are critical components of paraspeckles have been identified to have low complexity domains with a propensity for aggregation in addition to their nucleic acid-binding domains, and mutation of these proteins and concomitant misregulation of gene expression in motor neurons, has been linked to the cause of amyotrophic lateral sclerosis (ALS) (19,20).
In order to investigate the precise role of coiled-coil mediated aggregation of DBHS proteins in nuclear organization and gene regulation, we embarked on a study in which we have solved the first crystal structures of SFPQ including a remarkable extended coiled-coil polymerization domain. We complement this work with studies of its solution structure and in vitro DNA binding activity and a suite of cellular assays, all of which combine to provide a molecular basis for SFPQ's multifunctional role in the nucleus, with a common theme of functional aggregation via reversible and dynamic polymerization.
MATERIALS AND METHODS
Construct design and plasmid construction for expression
Human SFPQ constructs were cloned into pCDF-11 (EMBL) using standard molecular cloning techniques. The full-length cDNA of human SFPQ was used as a template in polymerase chain reaction (PCR). Five constructs were generated, coding for the following truncated proteins: SFPQ-276–535, SFPQ-276–598, SFPQ-369–598, SFPQ-214–535 and SFPQ-214–598. Each construct was cloned into pCDF-11 using NcoI and XhoI sites. pCDF11-SFPQ-214–598 quadruple mutant (L535A, L539A, L546A, M549A) was generated by QuikChange site-directed mutagenesis (Agilent) using complementary oligonucleotides containing the base-pair substitutions. All constructs were verified by DNA sequencing. The construction of pETDuet-1-NONO-53–312 was described elsewhere (17).
Protein expression and purification
Each protein was expressed in Rosetta™ 2 (DE3) (Merck Millipore) and purified using nickel-affinity chromatography followed by size exclusion chromatography. The cells were grown in LB broth with 50 μg/ml spectinomycin and 50 μg/ml chloramphenicol at 37°C and induced with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at an Abs600 of 0.6–0.8. After further incubation at 25°C for 16 h, the cells were harvested by centrifugation at 3000 × g. The cell pellet was resuspended in buffer A (20 mM Tris–HCl (pH 7.5), 250 mM NaCl, 10% glycerol), disrupted using an Avestin C5 homogenizer and clarified by centrifugation at 30 000 × g. The soluble fraction was then applied to a nickel-chelating column (Hi-Trap HP, GE healthcare). SFPQ was eluted using a gradient of 25–500 mM imidazole in buffer A. Pooled fractions were subjected to tobacco etch virus (TEV) protease digestion with 1:100 ratio (w/w) at RT overnight. His-tagged TEV protease and uncleaved SFPQ were removed by nickel-affinity chromatography. Concentrated flow-through fractions were then applied to a 16/60 Superdex 200 column and developed with 20 mM Tris–HCl (pH 7.5), 250 mM NaCl. For co-expression of SFPQ with NONO-53–312, pETDuet-1-NONO-53–312 was co-transformed with the appropriate pCDF-11-SFPQ constructs into Rosetta™ 2 (DE3) (Merck Millipore) and the SFPQ/NONO heterodimer purified as described above. Typically, the His-SFPQ was expressed at higher levels than the untagged NONO, resulting in two distinct peaks (SFPQ/NONO heterodimer and SFPQ homodimer) eluting the Ni-affinity purification. The fractions of the first peak that contained the heterodimer were pooled and further purified as above.
Crystallization and data collection
Crystals for SFPQ-276–535 were obtained from hanging-drop vapor diffusion experiments at 20°C using a reservoir solution of 1.9 M Ammonium sulfate, 0.2 M Tris–HCl (pH 8.5) and 20% (v/v) ethylene glycol. Typically 2 μl protein (4 mg/ml) was mixed with 2 μl reservoir solution and equilibrated against 1 ml reservoir solution. Crystals for SFPQ-276–598 were obtained from sitting-drop vapor method at 20°C using a reservoir solution of 26% 1,4-dioxane, 50 mM MOPS (pH 7.0), 5 mM MgCl2 and 1 mM spermine. Six microliters protein (8.7 mg/ml) was mixed with 3 μl reservoir solution and equilibrated against 0.5 ml reservoir solution. Crystals for SFPQ-369–598 were also optimized using sitting-drop vapor method at 20°C. Six microliters protein (6.7 mg/ml) was mixed with 3 μl reservoir solution (22% PEG3350 and 0.2 M Na2HPO4, pH 9.1) and equilibrated against 0.5 ml reservoir solution. Diffraction data were recorded at the Australian Synchrotron on crystals cooled to 100 K. Datasets for SFPQ-276–598 and SFPQ-369–598 (Crystal 1 and Crystal 2) were collected on beamline MX2 and SFPQ-276–535 (Crystal 3) was collected on MX1. The data were processed using XDS (21); the space group evaluated with POINTLESS (22) and the data merged and scaled using SCALA (22). SFPQ-276–598 (Crystal 1) was crystallized in space group P43212 with unit cell dimensions a = b = 66.58 Å, c = 398.02 Å. SFPQ-369–598 (Crystal 2) belonged to space group C2221 with unit cell dimensions a = 127.40 Å, b = 180.42 Å, c = 57.03 Å. SFPQ-276–535 (Crystal 3) belonged to space group P212121 with unit cell dimensions a = 63.45 Å, b = 67.61 Å, c = 119.53 Å. Data collection and processing statistics are summarized in Table 1.
Table 1. Crystallographic data collection and refinement statistics.
| SFPQ-276–598 (Crystal 1) | SFPQ-369–598 (Crystal 2) | SFPQ-276–535 (Crystal 3) | |
|---|---|---|---|
| Data collection | |||
| Wavelength (Å) | 0.9537 | 0.9537 | 0.9686 |
| Space group | P43212 | C2221 | P212121 |
| Cell dimensions | |||
| a, b, c (Å) | 66.6, 66.6, 398.0 | 127.4, 180.4, 57.0 | 63.5, 67.6, 119.5 |
| Resolution (Å) | 47.08–3.49 | 48.20–3.00 | 19.52–2.05 |
| (3.82–3.49) | (3.24–3.00) | (2.12–2.05) | |
| Rmerge | 0.100 (0.307) | 0.083 (0.694) | 0.102 (1.051) |
| <I/σI> | 10.8 (4.3) | 8.7 (1.9) | 13.1 (2.1) |
| Completeness (%) | 99.4 (98.1) | 98.6 (99.8) | 99.6 (100.0) |
| Redundancy | 6.8 (6.6) | 4.1 (4.1) | 7.1 (6.9) |
| Refinement | |||
| Unique reflections | 12 261 (2757) | 13 384 (2716) | 32 776 (2924) |
| R | 0.272 (0.309) | 0.242 (0.261) | 0.199 (0.231) |
| Rfree | 0.335 (0.337) | 0.291 (0.298) | 0.235 (0.284) |
| B-factors (Å2) | |||
| Protein | 127.4 | 96.4 | 43.6 |
| Ligand | 61.5 | ||
| Water | 44.1 | ||
| Ramachandran Plot† | |||
| Favored region (%) | 96.8 | 97.8 | 99.0 |
| Disallowed region (%) | 0.0 | 0.0 | 0.0 |
| R.m.s. deviations | |||
| Bond length (Å) | 0.007 | 0.008 | 0.010 |
| Bond angles (°) | 0.88 | 0.98 | 1.04 |
| PDB code | 4WIJ | 4WIK | 4WII |
¶Values in parentheses are for the highest-resolution shell.
†Calculated using MolProbity (26).
Crystallographic structure solution, refinement and validation
The crystal structures were solved by molecular replacement using PHASER (23). For SFPQ-276–535 (Crystal 3), the structure of PSPC1/NONO (PDB code: 3SDE (18)) was used as search model. Two monomers (one dimer) were found in the rotational search (Z-score 25) and the translation search confirmed the space group to be P212121. At this stage, the residues which differ in identity from the search model were mutated to alanine and the resulting model was subjected to iterative model building with COOT (24) and refinement with autoBUSTER (25). The final model includes two chains with residues 276–528 in Chain A and 285–529 in Chain B, one sulfate, eight ethylene glycol and 176 water molecules. Structure solution for SFPQ-276–598 (Crystal 1) and SFPQ-369–598 (Crystal 2) utilized the structure of SFPQ-276–535 (Crystal 3) as search model in molecular replacement. In the case of SFPQ-369–598 (Crystal 2), the search model was further modified by deleting the first RRM. The extended coiled-coil helices were built in manually with help of COOT. Iterative model building with COOT and refinement with autoBUSTER was performed. The final model for SFPQ-276–598 includes two chains with residues 292–589 in Chain A and 292–592 in Chain B. SFPQ-369–598 also has two chains in the asymmetric unit modeled with residues 370–597 in Chain A and residues 370–594 in Chain B. All final models were validated using MOLPROBITY (26). The refinement statistics are included in Table 1. Coordinates were manipulated with PDB-MODE (27). Sequence alignments were edited with ALINE (28).
Small-angle X-ray scattering data collection
SAXS experiments were carried out at the SAXSWAXS beamline of the Australian Synchrotron on a dilution series of protein sample. Scattering from a dilution series [6.0, 3.0, 1.5, 0.75, 0.38, 0.19 mg/ml of SFPQ-276–598/NONO-53–312 in 20 mM Tris–HCl (pH 7.5) and 250 mM NaCl] was carried out. Sample flowed through a glass capillary at a constant rate of 5 μl/s with data from 10 consecutive 1 s exposures collected on a Pilatus 1M detector and processed using ScatterBrain software (written and provided by the Australian Synchrotron; available at http://www.synchrotron.org.au/). All data were placed on an absolute scale using an empirical calibration factor derived from scattering by water in the identical experimental setup and normalized to the beamstop intensity. Solvent-subtracted and averaged data were exported for analysis.
SAXS data analysis
Data were analyzed with the ATSAS suite (29) (Supplementary Figure S1 and Supplementary Table S1). Guinier and P(r) analyses were performed using PRIMUS and GNOM, model fits to the scattering profiles used CRYSOL and ab initio structure calculations used DAMMIF. The lowest concentration shows flattening in the lowest q-regime of a log–log plot characteristic of a uniform sized scattering particle and the I(q) vs q profile shown (χ value of 0.970) demonstrates excellent fit to a model of the heterodimer derived directly from the SFPQ crystal structure. Further, the Guinier and P(r) distribution determined Rg and I(0) values are in excellent agreement (Supplementary Table S1) and the P(r) distribution has the characteristic shape of a mostly globular molecule with an extended tail; a single peak at 30–40 Å with a long tail extending to a dmax of 140 Å. For ab initio shape reconstruction, the 0.38 mg/ml data was used, as its Porod volume (1093310 Å3) is commensurate with the molecular mass of the heterodimer (Porod 65.6 kDa versus calculated 68.5 kDa). The filtered average model from all 10 independent calculations (normalized spatial discrepancy (NSD) values of 0.96 +/− 0.5) shows the expected globular body with an elongated tail. With increasing protein concentration, the flattening of the lowest q-regime is lost, Rg values steadily increase (Supplementary Table S1), the heterodimer model fit is poor with χ-values increasing sharply and I(0)/c also steadily increasing. These results indicate steadily increasing concentration-dependent protein associations. Somewhere between the 1.5 and 3.0 mg/ml samples, the value of I(0)/c doubles, consistent with the formation of tetramers. Increasing protein concentration also gives rise to a secondary peak in P(r) around 100 Å that increases in intensity along with the steadily increasing maximum r-value. These changes in P(r) are consistent with the formation of an increasing population of extended tetramers in which the globular domains of each dimer are separated in space. The secondary peak in P(r) near 100 Å would correspond to the approximate separation of the centers of scattering density (mass) for these globular domains, as is observed in the crystal structures. At the highest concentration measured, higher order associations are indicated by the continuing increases structural parameters, including I(0)/c. The presence of multiple species in the higher concentration solutions prohibits more detailed modeling of the tetramer species.
Electrophoretic mobility shift assay
A 6-carboxyfluorecein (6-FAM™)-labeled 61-bp DNA probe (the promoter region of the human proto-oncogene GAGE6 (7) was chemically synthesized (Integrated DNA Technologies). A total of 50 fmol of the FAM-labeled DNA probe was incubated with SFPQ/NONO in a volume of 10 μl in electrophoretic mobility shift assay (EMSA) buffer (20 mM HEPES (pH 7.5), 50 mM KCl, 5 mM MgCl2, 0.5 mM ethylenediaminetetraacetic acid (EDTA), 1 mM dithiothreitol (DTT), and 5% glycerol) at RT for 30 min. The reactions were analyzed by 4–20% gradient polyacrylamide gel electrophoresis (PAGE) (BioRad) in Tris-glycine buffer (25 mM Tris, 0.192 M glycine, pH 8.3). Fluorescence was detected using Typhoon Trio (GE healthcare) with excitation and emission at 488 and 520 nm, respectively.
Transmission electron microscopy
Samples were prepared in 20 mM Tris–HCl (pH 7.5), 250 mM NaCl, 0.5 mM EDTA at the following concentrations: double-stranded DNA; pCDF11 plasmid linearized with XhoI (New England Biolabs); 10 nM. Protein alone (SFPQ-214–598 or SFPQ-214–598mut or SFPQ-276–598); 3 μM. Protein/DNA complex: 600 nM protein and 16 nM DNA. Freshly prepared samples were incubated at 4°C for 30 min before application to glow-discharged carbon-covered 150-mesh copper grids. Grids were negatively stained with 1% (w/v) uranyl acetate. Images were obtained with a JEOL 2100 transmission electron microscope operated at 120 keV.
Plasmids for fluorescent protein fusions
Plasmids for fluorescent protein fusion experiments were generated with the Gibson Assembly cloning method (New England Biolabs) using the pEYFP-NLS plasmid in the case of truncated proteins and the pEGFP-C1 (Clontech) vector in the case of full-length proteins. pEYFP-NLS vector was generated by inserting the SV40 large T-antigen nuclear localization signal into pEYFP-C1 (Clontech). pEYFP-NLS-SFPQ-276–555, 276–565, 276–585 and 276–598 were made by assembling the PCR products of SFPQ encoding the corresponding residues into the linearized pEYFP-NLS by EcoRI and BamHI. The same strategy was applied to generate pEYFP-NLS-SFPQ-276–598mut (L535A, L539A, L546A, M549A) with the exception of using pCDF11-SFPQ-214–598mut as the PCR template in place of the wild-type construct. To make the siRNA-resistant SFPQ plasmids, silent mutations in residues 446–451 were introduced (this region is the target for the SFPQ siRNA used in this study, see below). Two sets of PCR primers were designed to amplify two halves of the full-length SFPQ. The reverse primer for the first half (1–451) and the forward primer for the second half (446–707) had 18 overlapping nucleotides with the designated silent mutations in residues 446–451. The two PCR fragments were then cloned into the linearized pEGFP-C1 vector by EcoRI and BamHI using the Gibson Assembly cloning method (New England Biolabs). The quadruple mutant version of siRNA-resistant SFPQ was also generated by the same strategy, using the quadruple mutant SFPQ plasmid as the PCR template. pEYFP-NLS-PSPC1-61-340 was made previously (18). peYFP-NLS-PSPC1-61-340 mutant (M320A, L324A, L331A, M334A) was generated by QuikChange site-directed mutagenesis (Agilent) using complementary oligonucleotides containing the base-pair substitutions. All constructs were verified by DNA sequencing.
SFPQ and PSPC1 localization assay
HeLa cells (cultured in DMEM supplemented with 10% fetal calf serum (FCS)) were seeded at 1.5 × 105/well on coverslips in a 12-well plate. Twenty-four hours later, transfection with plasmids encoding the various peYFP-C1-NLS-SFPQ truncations was carried out using Lipofectamine 2000 (Life Technologies) according to the manufacturer's instructions, using 300 ng of plasmid DNA per well. Sixteen hours post-transfection, cells were washed with phosphate buffered saline (PBS), fixed with cold 4% paraformaldehyde for 10 min, washed with PBS and permeabilized with cold 70% ethanol for 16–24 h at 4°C. Cells fixed on coverslips were then subjected to NEAT1 FISH (Fluorescence in situ hybridization) using Stellaris FISH probe—human Neat1 5′ Segment with Quasar 570 Dye (Biosearch Technologies) according to the manufacturer's instructions. Cell imaging and capture (z-stacks) were carried out using a 60× objective and the DAPI, FITC and TRITC filters on the NIKON Ti-E wide-field fluorescent microscope, followed by analysis with NIS elements software. Only cells with very low levels of expression were selected for imaging to avoid overexpression artifacts.
siRNA treatment, qPCR and western blotting analysis
For knockdown experiments, all transfections were done in triplicate with at least three biological replicates. 2.5 × 105 HeLa cells (cultured in DMEM supplemented with 10% FCS) were seeded in each well of a 6-well plate and siRNA transfection was carried out with the ‘reverse’ technique using 4 μl Lipofectamine 2000 (Life Technologies) and either 24 pmol scrambled siRNA (Stealth siRNA Negative control Med GC, Life Technologies) or SFPQ Stealth siRNA (UGUUCAAGUGGUUCCACAAUGACUG, Life Technologies). In some transfections, 500 ng of plasmid encoding green fluorescent protein (GFP), siRNA-resistant GFP-SFPQ or siRNA-resistant GFP-SFPQmut were also added to the transfection mix. Four hours post-transfection, medium was replaced with fresh DMEM/10% FCS. At 48 h post-transfection either RNA or protein purification of the samples was carried out. RNA was purified using TRISURE reagent (BIOLINE, according the manufacturer's instructions). A total of 500 ng of RNA was then reverse transcribed (QuantiTect Reverse Transcription Kit; Qiagen) according to the manufacturer's instructions. The relative expression levels of SFPQ (qPCR primers: forward GAGGAGAAGATCTCGGACTCG, reverse: CGACATCGCTGTGTGTAAGTTT), ADARB2 (qPCR primers: forward ATATTCGTGCGGTTAAAAGAAGGTG, reverse ATCTCGTAGGGAGAGTGGAGTCTTG) and GAPDH (qPCR primers: forward ATGGGGAAGGTGAAGGTCG, reverse GGGGTCATTGATGGCAACAATA) were measured by qPCR using SYBRgreen dye (BIOLINE) on the Rotor-Gene cycler. GAPDH expression levels were used to normalize the SFPQ and ADARB2 expression levels in all samples. In experiments in which plasmids had been co-transfected, cells were visualized under a fluorescent microscope with a FITC filter to assess the transfection efficiency. To assess relative protein expression levels, lysates were prepared from transfected HeLa cells by first rinsing with PBS, followed by cell disruption in 2× sodium dodecyl sulphate (SDS) loading dye (100 mM Tris pH 6.8, 4% w/v sodium, dodecyl sulfate, 0.2% w/v bromophenol blue, 20% v/v glycerol, 200 mM β-mercaptoethanol), followed by removal of the DNA in a QIAshredder column (Qiagen). Samples were heated at 100°C for 3 min and used for SDS-PAGE protein gel electrophoresis (NUPAGE 4–12% gradient, MOPS gels, Life Technologies) and subject to western blotting with anti-GFP (Roche) and anti-Beta Actin (AC-15, Abcam) followed by anti-mouse 800 Fluorescent Secondary Antibody (Licor). Western blots were visualized using the Odyssey System (Licor) and Analysis Software.
For SFPQ siRNA knockdown and paraspeckle rescue experiments, cells were grown on coverslips in 6-well plates, then transfected with siRNA against SFPQ (as above) using the RNAimax reagent according to the manufacturer's instructions (Life Technologies). Forty-eight hours following siRNA transfection, cells were then transfected with plasmids encoding GFP, GFP-SFPQ or GFP-SFPQ mutant, followed by fixation and processing for NEAT1 FISH 14 h later. Following imaging with identical exposure times, the number of NEAT1 foci (as defined by Softworx ‘define polygon’ function with constant thresholds) per nucleus was counted for each condition (n = 30).
RESULTS
The crystal structure of SFPQ reveals a novel coiled-coil interaction motif
Three truncated versions of SFPQ were generated, encompassing various regions of the key domains: the DNA-binding domain (DBD), RRMs, a protein interaction domain (NOPS) and an extended coiled-coil domain (Figure 1a). Crystal structures of three different length SFPQ proteins, termed Crystal 1, 2 and 3, were solved by molecular replacement to 3.5, 3.0 and 2.1 Å resolution, respectively, using the structure of a PSPC1/NONO heterodimer (PDB 3SDE) as a search model (Table 1). These structures demonstrate that SFPQ forms symmetrical dimers (Figure 1b, d and e), where the overall conformation and principal dimer interface is similar to that observed in the structure of a PSPC1/NONO heterodimer in our previous study (18) (Figure 1c). A root-mean-square-deviation of 1.8 Å for 442/494 Cα atoms reflects the 70% sequence identity in this conserved region of the family. The highly unusual spatial arrangement of RRM domains is therefore confirmed as being a common feature of DBHS protein homo- and heterodimers.
Figure 1.
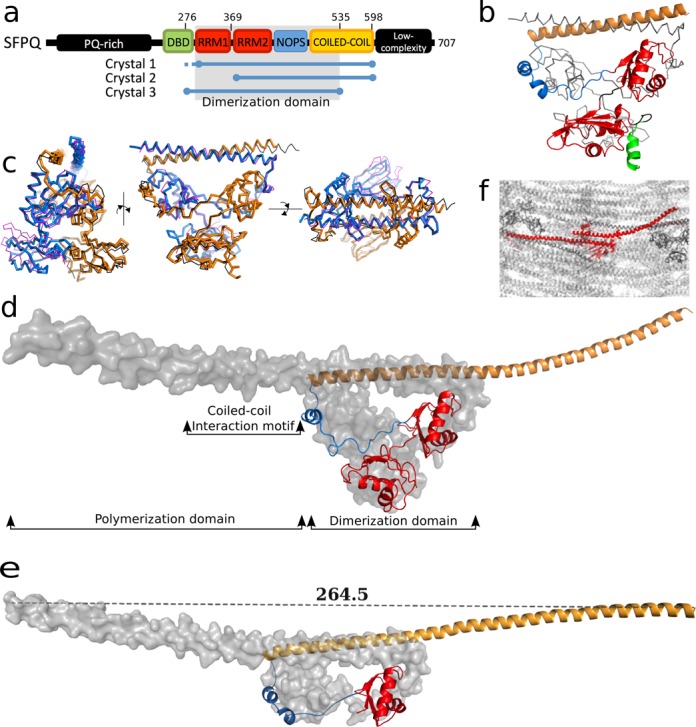
Crystal structure of human SFPQ. (a) Schematic presentation of human SFPQ domains discussed in this study. (b) Crystal structure of SFPQ-276–535 (Crystal 3, PDB code 4WII; chain A, gray trace; chain B, cartoon colored according to Figure 1a). (c) Superposition of SFPQ-276–535 homodimer (chain A blue; chain B gold) with PSPC1/NONO heterodimer (PDB 3SDE (18); magenta and black, respectively). (d) Crystal structure of SFPQ-276–598 homodimer (Crystal 1, PDB code 4WIJ; chain A, gray surface; chain B, cartoon). Domains that are discussed throughout this work are labeled. (e) Crystal structure of SFPQ-369–598 (Crystal 2, PDB code 4WIK; chain A, gray surface; chain B, cartoon). The distance between the Cα of residue 597 in chain A and that of residue 594 in chain B is displayed. (f) Crystal packing of SFPQ (Crystal 1; asymmetric unit, red).
However, the two structures of longer SFPQ proteins (Crystal 1: PDB code 4WIJ and Crystal 2: PDB code 4WIK), display remarkable extended α-helical regions of over 110 amino acids in a single helical span. As a result of the α-helix beginning in the dimerization domain, where it forms an unusual right-handed antiparallel coiled-coil, the C-termini of the two molecules in the dimer extend over 260 Å apart (Figure 1d and e). Each of the crystal structures contains two independent SFPQ molecules (as a homodimer) in the asymmetric unit and the extended α-helix, which we term the polymerization domain, participates in numerous intermolecular contacts within the crystal (e.g. Figure 1f). Evaluation of the potential biological relevance of these interactions with PISA (30) indicates that the majority are typical low affinity polar crystal contacts. However, in addition to the dimer interface that is common to all DBHS structures (ΔiG = −42.7 kcal/mol; P-value = 0.05), two novel interactions are identified as stable with a ‘complex significance score’ of 1. While one of these potential interfaces (ΔiG = −7.1 kcal/mol; P-value = 0.44) is only observed in Crystal 2, the other one is, notably, observed in both Crystal 1 and Crystal 2 (ΔiG = −7.6 kcal/mol; P-value = 0.29), giving us further confidence of its significance (Figure 2). The residues involved in this latter high-confidence interface, 528 to 555, belong to a region of high sequence-conservation (Figure 2a) and inspection of the structure reveals a coiled-coil motif immediately C-terminal to the dimerization domain of the structures (Figure 2b and c). This motif interacts with the next dimer in the unit cell in a specific, homotypic antiparallel left-handed coiled-coil that is readily detected from the structure by the program SOCKET (31). The result is a linear polymer of SFPQ running through the crystals. We hereafter refer to this 528 to 555 region as the coiled-coil interaction motif (Figures 1d and 2). Although we note that the use of crystal contacts to infer biologically-relevant macromolecular interactions is open to question, the observation of identical sequence-conserved interfaces in two unrelated crystal forms from two differently-truncated proteins provides considerable validation that this interaction is likely to be biologically relevant.
Figure 2.
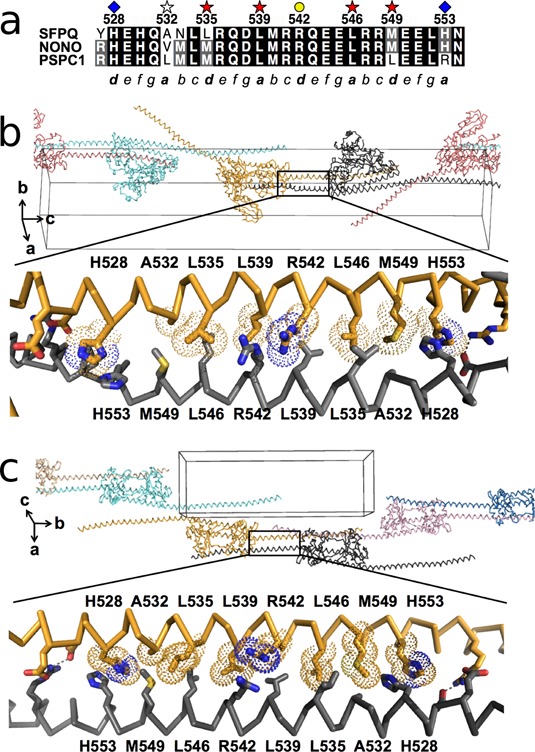
A novel conserved coiled-coil interaction motif amongst DBHS proteins. (a) Sequence alignment of the coiled-coil interaction motifs from SFPQ, NONO and PSPC1. Key residues are marked. (b and c.) Molecular details of the coiled-coil interaction motif in Crystal 1 (PDB code: 4WIJ) and Crystal 2 (PDB code: 4WIK), respectively.
Solution studies reveal a reversible coiled-coil mediated interaction at moderate concentrations
While the observation of coiled-coil based polymerization is consistent with SFPQ being an essential ‘building’ component of nuclear bodies such as paraspeckles, it raises questions regarding additional SFPQ function: the roles of SFPQ in the nucleus are diverse and have been shown to be dynamic, involving mobility between paraspeckles, nucleoli and sites of action, such as double strand breaks in DNA and sites of transcription (12,32). To address the question of whether SFPQ proteins can reversibly associate and dissociate, we measured a small-angle X-ray solution scattering dilution series on a dimeric protein sample where only one of the monomers includes the complete polymerization domain, while the other only includes the dimerization domain (Figure 3, Supplementary Figure S1, and Supplementary Table S1). This was achieved by exploiting the apparent preference of SFPQ and NONO to heterodimerize. Thus co-expressing the same SFPQ protein used for Crystal 1 along with the dimerization domain of NONO results in formation of SFPQ-276–598/NONO-53–312 heterodimers. We note that the SFPQ/NONO heterodimer may in fact be more physiologically relevant than the SFPQ homodimer, as SFPQ was first discovered as a heterodimer with NONO (33) and many studies since have observed SFPQ and NONO colocalized and co-purified (12,34).
Figure 3.
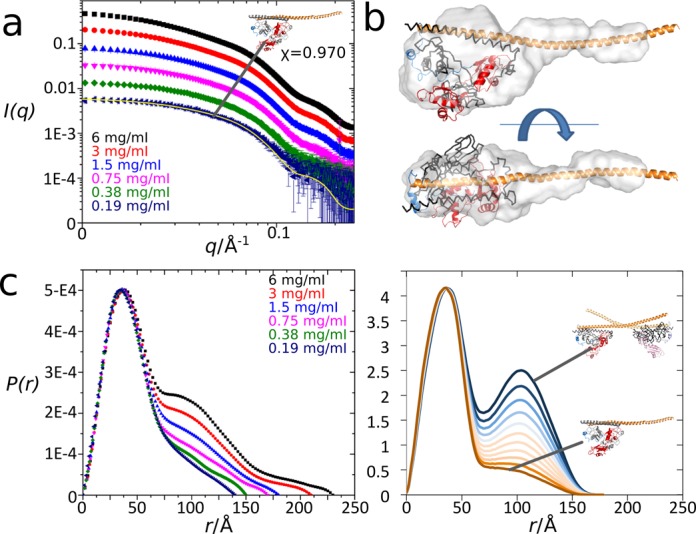
Small angle X-ray scattering studies of an SFPQ-276–598/NONO-53–312 heterodimer. (a) Scattering profiles (log(I) vs log(q)) for a dilution series (6.0–0.19 mg/ml), including fit of a model derived from the Crystal 1 structure (inset) to the 0.19 mg/ml concentration sample. See Supplementary Table S1 and Supplementary Figure S3 for additional details. (b) Ab initio model calculated from 0.38 mg/ml data and superimposed on an atomic model derived from the Crystal 1 structure. (c) Experimental P(r) distribution calculated from the scattering data (left panel) is comparable with a calculated P(r) distribution (right panel) for atomic heterodimer and tetramer models (and mixtures thereof) derived from the Crystal 1 structure. The lowest concentration sample contains no tetramer, but a 100 Å feature consistent with increasing tetramer content appears with rising concentration.
At lower concentrations, the solutions display scattering consistent with pure heterodimers, having a characteristic shape of a mostly globular molecule with an extended tail (Figure 3a and b), with excellent fit to an appropriate atomic model derived from the SFPQ-276–598 structure. At higher concentrations, protein association occurs: the appearance and increasing intensity of a peak in the pair distribution function around 100 Å is consistent with the formation of an increasing population of extended tetramers generated by interaction of the coiled-coil interaction motif, as observed in our crystal structures. At the highest concentrations measured, further higher order associations are observed. These associations are not surprising as there are many polar interactions observed in both crystal structures and an additional region of potentially relevant coiled-coil in Crystal 2 which could allow non-specific aggregation into larger particles at higher concentrations (Figure 3a and c). The presence of these multiple species in the higher concentration solutions convolutes the X-ray scattering profile and so prohibits more detailed modeling of the tetramer species.
Nevertheless, as the lowest concentration samples were obtained by dilution from the highest, the associations observed are demonstrated to be reversible. A potential basis for this reversibility can be found in the amino acid bias in this region of DBHS proteins compared to other coiled-coil proteins (35): over-representation of relatively polar glutamine and methionine residues and under-representation of apolar alanine and leucine may explain the ability of the unpaired coiled-coil to remain in aqueous solution. This study paints a picture of reversible dynamic equilibrium between soluble dimeric protein and coiled-coil associated tetramers, dependent on concentration.
The coiled-coil interaction motif is important for optimal DNA-binding in vitro
SFPQ, reported to bind DNA as well as RNA, acts in transcriptional regulation of genes—both repression and activation—by direct interaction with promoter regions (7,36,37). In this regard, a key defining structural feature of SFPQ, different with respect to the related proteins NONO and PSPC1, is a unique region immediately N-terminal to the RRM1 that encodes a putative DBD (12,32). Our attempts to crystallize a protein containing this complete DBD, while unsuccessful, allowed us to determine the structure of Crystal 3 encompassing residues 276–535, lacking the polymerization domain but including 10 amino acids of the DBD (Figure 1b). Unlike in the structure of SFPQ-276–598, where the first 16 amino acids (276–291) prior to the RRM1 are not observed in the electron density, this N-terminal region in SFPQ-276–535 revealed a short α-helical structure. It is notable that this short stretch of sequence in SFPQ is conspicuously divergent from NONO and PSPC1 (Supplementary Figure S4).
To examine the significance of the intact DBD and the polymerization domain for DNA binding, we next tested the ability of a number of the SFPQ constructs to bind to promoter DNA using an EMSA with the GAGE6 promoter—a reported SFPQ target gene (7)—as a probe (Figure 4). In order to limit extensive unworkable polymerization, we again used heteromeric SFPQ/NONO proteins, where NONO is unchanged (dimerization domain, 53–312) and SFPQ is varied: SFPQ-214–598 comprises DBD, dimerization domain and polymerization domain, SFPQ-214–535 comprises DBD and dimerization domain, and SFPQ-276–535 matches Crystal 3 (dimerization domain) (Figure 1b). To specifically address the role of the coiled-coil interaction motif in the polymerization domain, a mutated variant SFPQ-214–598mut was also generated, where the four key residues of the motif—L535, L539, L546 and M549 (Figure 2a)—were substituted to alanine. These four hydrophobic residues are the ‘knobs’ that stick into ‘holes’ that stabilize coiled-coil interactions. The alanine mutation was designed to maintain α-helical structure while disrupting the ability to form the coiled-coil interaction.
Figure 4.
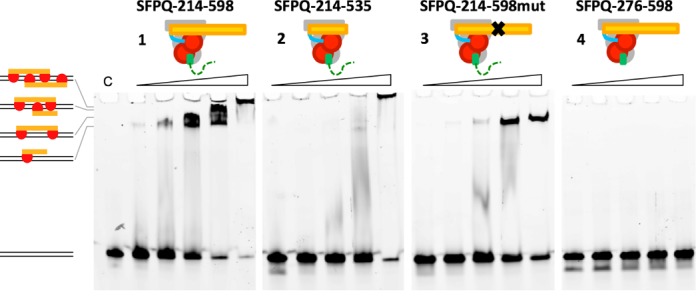
The coiled-coil interaction motif is critical for DNA binding of SFPQ. Electrophoretic mobility shift assay using a 61-bp double-strand DNA probe (GAGE6 promoter). Increasing concentrations of proteins (0.1, 0.2, 0.4, 0.8 and 1.6 μM) were incubated with the probe. Maximum DNA-binding activity is achieved when both DBD and the coiled-coil polymerization domain are present.
We observed maximal DNA binding with SFPQ protein that contains both the full DBD and the polymerization domain (SFPQ-214–598; Figures 1–4). Interestingly, increasing protein concentration of SFPQ-214–598 resulted in multiple bands in the EMSA, indicating that more than one SFPQ-214–598 molecule is capable of interacting with the 61-bp probe. These higher order species are likely to be no more than two heterotetramers due to steric considerations for this size protein on a 61-bp probe (Supplementary Figure S2). Removal of most of the DBD but including the short stretch of α-helix (SFPQ-276–598) abolished DNA binding (Figure 4-4).
Most interestingly, deletion of the polymerization domain significantly impaired DNA binding, despite the inclusion of the full DBD (SFPQ-214–535; Figures 2–4). We further validated this observation by showing that SFPQ-214–598mut (Figures 3 and 4) also has significantly impaired DNA-binding ability: only a single protein:DNA complex is observed at higher concentrations of the mutant, presumably corresponding to a single heterodimer bound to the 61-bp probe. We also note that at higher concentrations, the presence of the impaired polymerization domain in SFPQ-214–598mut results in less aggregation (protein/DNA retained in the well of the gel) than when it is completely absent (SFPQ-276–535). The disfunctional polymerization domain could be acting as a steric barrier to non-specific protein:protein interactions.
Noting that the EMSA shows multiple binding events and considering that SFPQ is enriched over thousands of bases around transcription start sites (38), and that SFPQ accumulates at double-strand breaks where no sequence conservation can be expected, we sought to visualize the interaction of SFPQ with bulk double-stranded DNA using negative-stain transmission electron microscopy (Figure 5). SFPQ containing the full DBD as well as the polymerization domain (SFPQ-214–598, the optimal DNA-binding protein in the EMSA assay) clearly forms a thick contiguous coating of protein around linearized plasmid DNA (Figure 5a). The protein coat is 17 nm thick and typically 350 nm or longer in length. In remarkable contrast, the equivalent coiled-coil quadruple mutant of this protein exists as individual protein particles of 20 nm, occasionally associated with DNA (Figure 5b), demonstrating the importance of the coiled-coil interaction motif to this DNA coating property of SFPQ. In the absence of DNA, SFPQ-214–598 forms amorphous aggregates composed of protein particles at 10 nm separation (Figure 5c), While it is hard to judge the concentration-dependent effects on protein interactions in the dehydrated environment of the electron microscopy grid, it is clear that wild-type protein alone is unique in demonstrating regular, reproducible, coating of DNA. The thickness of the fiber (17 nm) is approximately as would be expected for a sleeve of protein, a single dimer thick (7 nm) surrounding DNA duplex (2 nm).
Figure 5.
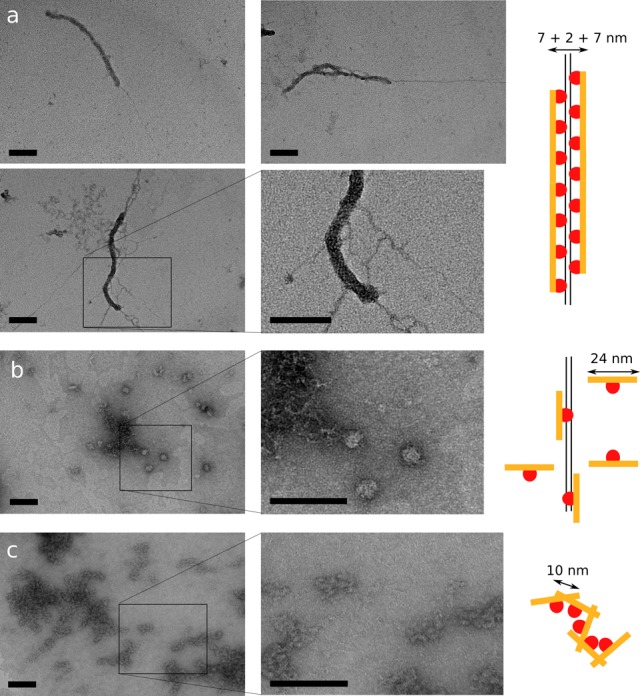
The coiled-coil interaction motif in SFPQ promotes the regular coating of double-stranded DNA, as revealed by transmission electron microscopy. Scale bars are 100 nm throughout. Images are representative of a homogeneous and numerous fields of view. (a) SFPQ-214–598, containing both DBD and coiled-coil polymerization domain coats double-stranded DNA in a single layer of protein, forming 17 nm think fibers for contiguous regions of around 400 nm. Naked DNA can be seen protruding from the end. (b) SFPQ-214–598mut, where the coiled-coil interaction motif is impaired by mutation, is unable to form fibres, instead existing as individual protein dimers (∼20 nm) which occasionally associate with DNA. (c) In the absence of DNA, SFPQ-214-598 forms poorly ordered aggregates.
Overall these in vitro results illustrate a synergistic dependency between the DBD and coiled-coil interaction motif for optimal DNA-binding. This can be interpreted as the coiled-coil interaction promoting a more avid interaction by colocalizing multiple DBDs along a piece of DNA. Extrapolation of this notion to native proteins capable of forming polymers and thus highly avid multivalent interactions, is commensurate with their role in forming large nucleoprotein complexes in the cell nucleus.
Polymerization of SFPQ is important for optimal transcriptional activation in cell culture
We next tested the functional significance of the coiled-coil interaction motif on the ability of SFPQ to mediate transcriptional regulation in cells. We used the RNA levels of the SFPQ target gene, ADARB2 (37), in HeLa cells as a readout of SFPQ transcriptional activation in a series of knockdown-rescue experiments (Figure 6). ADARB2 is an enzymatically-impaired adenosine deaminase protein, whose expression is reduced by SFPQ depletion or sub-nuclear sequestration and whose functional depletion is associated with ALS (8,39).
Figure 6.
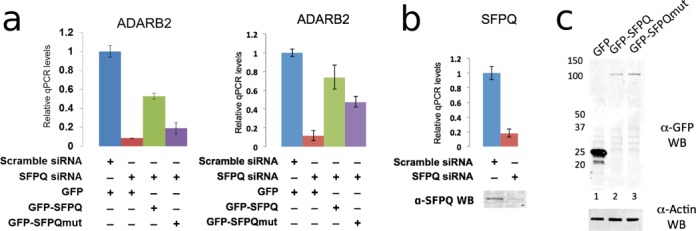
The coiled-coil interaction motif is critical for transcriptional regulation activity of SFPQ. (a) Relative qPCR levels of the SFPQ target gene, ADARB2, following knockdown of endogenous SFPQ and rescue with transiently expressed GFP-SFPQ or GFP-SFPQmut. Wild-type SFPQ can rescue ADARB2 expression to a greater extent than mutant SFPQ (two independent biological replicates shown). Error bars represent standard error of three technical replicates. (b) RT-qPCR and western blot showing endogenous SFPQ levels in HeLa cells treated with either scramble (control) or SFPQ siRNA. (c) Anti-GFP and anti-β-actin western blot of protein extracts from HeLa cells transiently expressing GFP (lane 1), GFP-SFPQ (lane 2) or GFP-SFPQmut (lane 3).
We first showed that knockdown of SFPQ resulted in a five-fold reduction in ADARB2 levels (Figure 6a and b), and then tested the ability of transiently expressed full-length SFPQ and quadruple mutant full-length SFPQ (L535A/L539A/L546A/M549A), to rescue ADARB2 expression. Both constructs contain silent mutations in the siRNA target region to render them siRNA resistant, allowing complementation of the knockdown. We found wild-type SFPQ restored ADARB2 mRNA levels to over 60% of endogenous levels, whereas the quadruple mutant SFPQ has significantly impaired capacity to recover the activity (Figure 6a, compare lanes 3 and 4).
Despite both SFPQ variants being over-expressed at similar protein levels (Figure 6c), the polymerization-deficient mutant was unable to restore ADARB2 at a level similar to wild-type protein. SFPQ binds directly to the ADARB2 promoter, as demonstrated by chromatin immunoprecipitation (37). Beyond this binding, the precise mechanism of SFPQ activation of the ADARB2 gene is unknown. One possibility suggested by our data is that the polymerization of SFPQ at the ADARB2 promoter may facilitate recruitment of other proteins involved in transcriptional activation, however this is yet to be tested.
Localization of SFPQ to existing paraspeckles requires both a functional coiled-coil interaction motif and a full-length polymerization domain
In previous studies we showed that YFP-tagged C-terminally truncated versions of PSPC1 require additional sequence beyond the dimerization domain in order to be localized to paraspeckles (18). This shortest paraspeckle-targeting construct of PSPC1 (PSPC1-61-340, schematically marked as protein ‘1’ in Figure 7a) contains the dimerization domain as well as the equivalent residues to the SFPQ coiled-coil interaction motif (Figure 2a). We therefore introduced the equivalent quadruple substitution of key leucine and methionine residues to alanine within PSPC1-61-340 and observed that, when tagged with YFP and overexpressed in HeLa cells, this construct is no longer able to be targeted to paraspeckles (Figure 7a). Clearly, the coiled-coil interaction motif is absolutely essential for PSPC1 localization to paraspeckles. Fully expecting a similar observation for SFPQ, we made the same length YFP-tagged SFPQ (the protein ‘1’ version of SFPQ; SFPQ-276–565; Figure 7a and Supplementary Figure S3), but were surprised to observe that this length of SFPQ construct does not localize to paraspeckles (Figure 7a), despite including the coiled-coil interaction motif. This led us to speculate that additional sequence within the polymerization domain may be playing a role in paraspeckle targeting by SFPQ. Indeed, adding more and more coiled-coil, by increasing the length of polymerization domain improves paraspeckle localization progressively (proteins ‘2’-‘4’, Figure 7a) until a native-like distribution is observed for SFPQ-267–598 (protein ‘5’; complete polymerization domain, Figure 7a). It is possible that the additional potential coiled-coil based interaction observed only in the Crystal 2 structure around residues 565–585 may be playing a role in paraspeckle localization. However, given its relatively low P-value from PISA analysis and the fact that its detection as a coiled-coil by SOCKET is marginal (distance cut-off 8.5 Å required), an alternative possibility is that this region could interact with other proteins in the cell, enabling paraspeckle targeting. Nevertheless, once robust paraspeckle targeting by SFPQ could be achieved (protein ‘5’), the importance of the coiled-coil interaction motif was again highlighted, as the protein ‘5’ SFPQ quadruple mutant was unable to be targeted to paraspeckles (Figure 7a). Thus paraspeckle targeting by SFPQ is underpinned by extensive coiled-coil interactions mediating polymerization.
Figure 7.
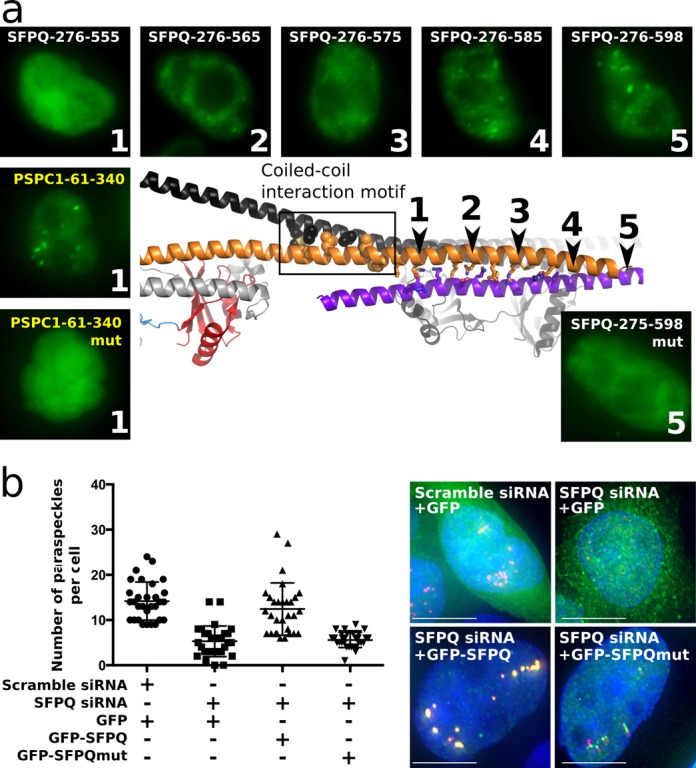
The coiled-coil interaction motif within the polymerization domain is essential for both localization of SFPQ to exiting paraspeckles and formation of new paraspeckles. (a) Truncated variants of SFPQ and PSPC1 have differing coiled-coil requirements for localization to paraspeckles. For both proteins, the coiled-coil interaction motif is essential, but for SFPQ, additional potential coiled-coil is required (purple helix indicates potential coiled-coil interaction with a neighboring molecule from another asymmetric unit in Crystal 2). Protein ‘1’ corresponds to the minimum paraspeckle-targeted PSPC1 while protein ‘5’ corresponds to the optimal paraspeckle-targeted SFPQ. Quadruple mutation of the coiled-coil interaction motif abolishes paraspeckle localization. Typical fluorescent micrographs of representative HeLa cells transiently expressing YFP-NLS (nuclear localization sequence)-fusions of the proteins indicated in each panel are shown: GFP, green; DAPI blue. Scale bars, 10 μm. (b) Number of paraspeckles per cell (n = 30) under a number of conditions, measured by counting NEAT1 FISH foci: scramble siRNA control (endogenous SFPQ present), SFPQ siRNA knockdown, wild-type GFP-SFPQ rescue of SFPQ knockdown and mutant GFP-SFPQ rescue of SFPQ knockdown. Whille wild-type GFP-SFPQ can rescue paraspeckle formation, mutant GFP-SFPQ is no more effective than GFP alone. Typical fluorescent micrographs of representative HeLa cells are shown: GFP, green; NEAT1 red, DAPI blue. Significantly reduced numbers of NEAT1 foci are accompanied by reduced colocalization with SFPQ. Scale bars are 5 μm. All samples are significantly different (P < 0.0001, unpaired t-test).
Polymerization of SFPQ is essential to the formation of paraspeckles
As SFPQ has been shown to be essential for paraspeckle formation (40), we next chose to use our knockdown-rescue assay to investigate the importance of the coiled-coil interaction motif for polymerization of SFPQ in the formation of paraspeckles. Fluorescent in situ hybridization (FISH) using probes against NEAT1 lncRNA is the gold-standard marker for paraspeckle localization and can be used to count the number of paraspeckles per cell. Against a background of siRNA knocked-down endogenous SFPQ, we transfected HeLa cells with siRNA-resistant GFP-fusion proteins and counted the numbers of paraspeckles (NEAT1 foci) per cell. Cells lacking SFPQ and overexpressing GFP alone had significantly reduced numbers of paraspeckles, commensurate with no rescue, while rescue with recombinant full-length SFPQ restored paraspeckle numbers to wild-type levels (P < 0.0001, Figure 7b). Full-length mutant SFPQ is, however, unable to restore paraspeckle numbers, yielding a similar number of NEAT1 foci to the GFP-only case (P < 0.0001, Figure 7b). Thus, the presence of an intact coiled-coil interaction motif is absolutely necessary for the essential role of SFPQ in paraspeckle formation.
DISCUSSION
Our studies suggest that the coiled-coil interaction motif in the SFPQ polymerization domain is critical for paraspeckle formation, targeting SFPQ to paraspeckles, binding DNA and mediating transcriptional regulation. Our SAXS data further support the notion that this polymerization is dynamic and reversible, suggesting a mechanism for regulation via equilibrium into and out of the polymerized state. The polymerized protein–DNA complex is readily observed under the electron microscope.
The structural studies described here have revealed a remarkable and highly unusual protein arrangement where extensive coiled-coil structures link functional globular domains. Examination of secondary structure element length in the complete PDB reveals that the SFPQ α-helix is the longest single α-helix found in the structure of a nuclear protein: other long helices are found in numerous myosin-related proteins, viral spike proteins and proteins implicated in extracellular roles such as cell adhesion, clotting and chemotaxis. Only two known structures with equally long α-helices have any relevance to nucleic acid metabolism—the enigmatic vault ribonucleoprotein compartment (PDB code: 2ZUO) (41) and the E3 ligase protein TRIM25 (PDB code: 4LTB) (42). The observation of a protein that can easily span 25 nm, that is involved in nuclear bodies of minimum dimension 300 nm (43) points to the exciting possibility in the near future of reconciling atomic structure with microscopic ultrastructure.
It is known that paraspeckle formation is initiated with transcription of NEAT1 lncRNA (23 kb), which acts as a structural scaffold, nucleating the bodies by recruiting SFPQ and other essential paraspeckle proteins (13,44). Paraspeckles have recently been shown to play a role in the cellular response to viral infection and other stresses, primarily via the sub-nuclear sequestration of SFPQ. Under these conditions, paraspeckle formation and subsequent SFPQ sequestration prevents SFPQ from acting at chromatin to regulate important target genes such as IL-8 and ADARB2 (9,37). Interestingly, through overexpression of portions of NEAT1, Imamura et al. showed that SFPQ sequestration only occurs when NEAT1 is of sufficient length (9). Our data, added to this, raises the possibility of involvement of multiple binding sites on NEAT1 to recruit multiple polymerizing SFPQ molecules as critical for paraspeckle formation and sequestration of SFPQ.
The polymerization of SFPQ is underpinned by the coiled-coil interaction motif. This region is linked to SFPQ's important roles in cancer. Chromosomal translocations of SFPQ resulting in a fusion protein are well documented; SFPQ fused to the transcription factor TFE3 in papillary renal cell carcinoma (45) and fused to ABL kinase in acute lymphoid leukaemia (46). Both resulting fusion proteins consist of the N-terminal 662 amino acids of SFPQ followed by the partner protein (TFE3 or ABL kinase) at the C-terminus. In particular, it has been speculated that constitutive activation of fusion ABL kinase, essential for transforming activity, is mediated by an oligomerization domain within SFPQ (46). This fits with our findings of polymerization of SFPQ via the coiled-coil interaction motif that is contained within the fusions. Given this new-found structural knowledge of SFPQ, the door is now open for future structure-based development of an inhibitor of SFPQ polymerization as a potentially useful therapeutic.
Interestingly, there already exists an animal model demonstrating the functional importance of the coiled-coil interaction motif to DBHS protein function. Drosophila nonA is the invertebrate ortholog of DBHS proteins. The nonAdiss allele encodes an arginine to cysteine mutation at a position equivalent to SFPQ R542 (Figure 2a, yellow circle) exhibiting a severe neurological phenotype with global defects in viability and courtship song (47). This point mutation could potentially result in destabilization of the coiled-coil as cysteine is heavily under-represented in coiled-coils (only proline is less prevalent). It is also possible that constitutive polymerization of nonA may result from this mutation, as C542 residues of neighboring units in the polymer would be within 5 Å and could potentially form a disulfide bond in a conducively oxidative environment.
Further, alterations in the functional aggregation propensity of SFPQ, potentially via mutations within the polymerization domain, may also play a role in human neurodegeneration. In addition to the drosophila nonAdiss neurological phenotype described above, SFPQ has strong expression in the developing zebrafish brain, particularly in regions enriched for neuronal progenitors and ablation of SFPQ in the zebrafish embryo shows a brain phenotype within 28 h (2). In human disease, changes in the subcellular localization of SFPQ and its heterodimeric partner NONO have been observed in neurons of Alzheimer's disease and ALS patients (48,49). In regard to ALS, it is also intriguing to note that a significant number of paraspeckle proteins have known ALS-causing mutations (6 of ∼40 proteins) (40). These six proteins (FUS, EWSR1, TDP43, TAF15, SS18L1 and HNRNPA1) are prone to aggregation when mutated or mislocalized to the cytoplasm (19). As SFPQ shares similar traits of functional aggregation and paraspeckle enrichment with these proteins, it is possible that SFPQ may also play a role in the pathways perturbed in ALS, and studies are underway to investigate this possibility.
The dynamic and functional aggregation exhibited by SFPQ thus explains its ability to participate in many cellular processes, modulated by local protein and nucleic acid concentrations, thereby providing temporal and spatial regulation and potentially reducing biological noise. A general model for DBHS participation in these processes could involve a seeding event, such as binding to a hotspot in a nucleic acid (e.g. a specific DNA promoter sequence, a DNA double-strand break or a binding site on a long non-coding RNA), followed by condensation of additional DBHS dimers via protein–protein and protein–nucleic acid interactions (Figure 8). Such an assembly, as observed by electron microscopy (Figure 5a), would provide a large local concentration of other peripheral domains of DBHS proteins thus allowing the bulk recruitment of additional factors which could be crucial in mounting a rapid response to DNA damage or a transcriptional event. Similar themes are emerging to explain the involvement of higher order assemblies in signaling complexes (50). Interestingly, the network of coiled-coil interactions observed in the crystals, is redolent of a fibrillar gel. This fibrillar potential is reminiscent of recent discoveries that several other paraspeckle proteins, namely FUS, EWS and TAF15, harbor low complexity domains that form reversible, fibrillar functional aggregates in vivo and in vitro (51,52). These fibrillar mesh networks with liquid like and phase-transition properties mediate the recruitment of the C-terminal domain of RNA polymerase II to promoters, resulting in strong transcriptional activation (52). Analogous to these low-complexity domains, the findings presented in this study offer a framework how a common coiled-coil domain, in combination with nucleic acid binding domains, can form reversible functional aggregates. This framework provides new insight into the multifunctional biological implications of SFPQ in the organization of dynamic sub-cellular structures and gene regulation.
Figure 8.
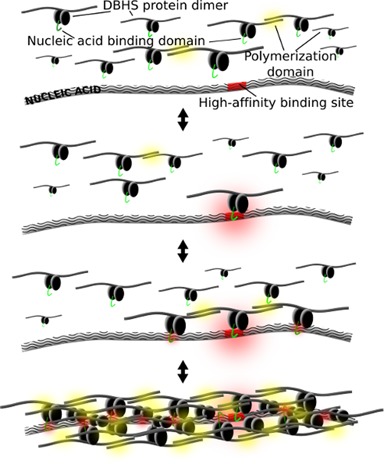
A general model for cooperative nucleic acid-templated functional polymerization of DBHS proteins.
ACCESSION NUMBERS
Crystal structure coordinates and structure factors have been deposited with the PDB with accession codes 4WII, 4WIJ and 4WIK. The raw X-ray diffraction data used in this experiment have been made available at https://store.synchrotron.org.au/ (53).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
Aspects of this research were undertaken on the Macromolecular Crystallography and SAXS/WAXS beamlines at the Australian Synchrotron (Victoria, Australia) and we thank the beamline staff for their enthusiastic and professional support. The authors acknowledge the facilites of the Centre for Microscopy, Characterisation & Analysis, The University of Western Australia, a facility funded by the University, State and Commonwealth Governments.
Author Contributions: M.L., A.H.F., K.S.I. and C.S.B. designed the experiments; M.L. prepared and crystallized proteins, collected X-ray data, solved structures, performed EMSA. A.S. performed the paraspeckle targeting experiments and siRNA experiments. I.B. and D.H. performed electron microscopy experiments. B.G. collected small-angle X-ray scattering data. M.L., Y.L., J.T., A.H.F. and C.S.B. analyzed the data. M.L., J.T., A.H.F. and C.S.B. wrote the manuscript.
Footnotes
Present address: Mihwa Lee, La Trobe Institute for Molecular Science, La Trobe Unviersity, Melbourne, Victoria 3086, Australia.
FUNDING
National Health and Medical Research Council of Australia Project Grants [513880, 1048659, 1050585 to C.S.B. and A.H.F.]; National Health and Medical Research Council of Australia, Postdoctoral Training Fellowship [513935 to M.L.]; Cancer Council of Western Australia Project Grant (to C.S.B.); Cancer Council of Western Australia Fellowship (to A.H.F.); The University of Western Australia, Research Development Award (to M.L.). Funding for open access charge: The University of Western Australia.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kowalska E., Ripperger J.A., Muheim C., Maier B., Kurihara Y., Fox A.H., Kramer A., Brown S.A. Distinct roles of DBHS family members in the circadian transcriptional feedback loop. Mol. Cell. Biol. 2012;32:4585–4594. doi: 10.1128/MCB.00334-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lowery L.A., Rubin J., Sive H. Whitesnake/sfpq is required for cell survival and neuronal development in the zebrafish. Dev. Dyn. 2007;236:1347–1357. doi: 10.1002/dvdy.21132. [DOI] [PubMed] [Google Scholar]
- 3.Duong H.A., Robles M.S., Knutti D., Weitz C.J. A molecular mechanism for circadian clock negative feedback. Science. 2011;332:1436–1439. doi: 10.1126/science.1196766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chia N.Y., Chan Y.S., Feng B., Lu X., Orlov Y.L., Moreau D., Kumar P., Yang L., Jiang J., Lau M.S., et al. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010;468:316–320. doi: 10.1038/nature09531. [DOI] [PubMed] [Google Scholar]
- 5.Patton J.G., Porro E.B., Galceran J., Tempst P., Nadal-Ginard B. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 1993;7:393–406. doi: 10.1101/gad.7.3.393. [DOI] [PubMed] [Google Scholar]
- 6.Shav-Tal Y., Zipori D. PSF and p54(nrb)/NonO–multi-functional nuclear proteins. FEBS Lett. 2002;531:109–114. doi: 10.1016/s0014-5793(02)03447-6. [DOI] [PubMed] [Google Scholar]
- 7.Song X., Sun Y., Garen A. Roles of PSF protein and VL30 RNA in reversible gene regulation. Proc. Natl. Acad. Sci. U.S.A. 2005;102:12189–12193. doi: 10.1073/pnas.0505179102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirose T., Virnicchi G., Tanigawa A., Naganuma T., Li R., Kimura H., Yokoi T., Nakagawa S., Benard M., Fox A.H., et al. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol. Biol. Cell. 2014;25:169–183. doi: 10.1091/mbc.E13-09-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imamura K., Imamachi N., Akizuki G., Kumakura M., Kawaguchi A., Nagata K., Kato A., Kawaguchi Y., Sato H., Yoneda M., et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell. 2014;53:393–406. doi: 10.1016/j.molcel.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 10.Morozumi Y., Takizawa Y., Takaku M., Kurumizaka H. Human PSF binds to RAD51 and modulates its homologous-pairing and strand-exchange activities. Nucleic Acids Res. 2009;37:4296–4307. doi: 10.1093/nar/gkp298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajesh C., Baker D.K., Pierce A.J., Pittman D.L. The splicing-factor related protein SFPQ/PSF interacts with RAD51D and is necessary for homology-directed repair and sister chromatid cohesion. Nucleic Acids Res. 2011;39:132–145. doi: 10.1093/nar/gkq738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ha K., Takeda Y., Dynan W.S. Sequences in PSF/SFPQ mediate radioresistance and recruitment of PSF/SFPQ-containing complexes to DNA damage sites in human cells. DNA Rep. 2011;10:252–259. doi: 10.1016/j.dnarep.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bond C.S., Fox A.H. Paraspeckles: nuclear bodies built on long noncoding RNA. J. Cell Biol. 2009;186:637–644. doi: 10.1083/jcb.200906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takayama K., Horie-Inoue K., Katayama S., Suzuki T., Tsutsumi S., Ikeda K., Urano T., Fujimura T., Takagi K., Takahashi S., et al. Androgen-responsive long noncoding RNA CTBP1-AS promotes prostate cancer. EMBO J. 2013;32:1665–1680. doi: 10.1038/emboj.2013.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji Q., Zhang L., Liu X., Zhou L., Wang W., Han Z., Sui H., Tang Y., Wang Y., Liu N., et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br. J Cancer. 2014;111:736–748. doi: 10.1038/bjc.2014.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fox A.H., Bond C.S., Lamond A.I. P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol. Biol. Cell. 2005;16:5304–5315. doi: 10.1091/mbc.E05-06-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee M., Passon D.M., Hennig S., Fox A.H., Bond C.S. Construct optimization for studying protein complexes: obtaining diffraction-quality crystals of the pseudosymmetric PSPC1-NONO heterodimer. Acta Crystallogr. D Biol. Crystallogr. 2011;67:981–987. doi: 10.1107/S0907444911039606. [DOI] [PubMed] [Google Scholar]
- 18.Passon D.M., Lee M., Rackham O., Stanley W.A., Sadowska A., Filipovska A., Fox A.H., Bond C.S. Structure of the heterodimer of human NONO and paraspeckle protein component 1 and analysis of its role in subnuclear body formation. Proc. Natl. Acad. Sci. U.S.A. 2012;109:4846–4850. doi: 10.1073/pnas.1120792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y.R., King O.D., Shorter J., Gitler A.D. Stress granules as crucibles of ALS pathogenesis. J. Cell Biol. 2013;201:361–372. doi: 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramaswami M., Taylor J.P., Parker R. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell. 2013;154:727–736. doi: 10.1016/j.cell.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kabsch W. Xds. Acta crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evans P. Scaling and assessment of data quality. Acta crystallogr. D Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 23.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blanc E., Roversi P., Vonrhein C., Flensburg C., Lea S.M., Bricogne G. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 26.Chen V.B., Arendall W.B., 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bond C.S. Easy editing of Protein Data Bank formatted files with EMACS. J. Appl. Crystallogr. 2003;36:350–351. [Google Scholar]
- 28.Bond C.S., Schuttelkopf A.W. ALINE: a WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr. D. 2009;65:510–512. doi: 10.1107/S0907444909007835. [DOI] [PubMed] [Google Scholar]
- 29.Petoukhov M.V., Franke D., Shkumatov A.V., Tria G., Kikhney A.G., Gajda M., Gorba C., Mertens H.D.T., Konarev P.V., Svergun D.I. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 31.Walshaw J., Woolfson D.N. Socket: a program for identifying and analysing coiled-coil motifs within protein structures. J. Mol. Biol. 2001;307:1427–1450. doi: 10.1006/jmbi.2001.4545. [DOI] [PubMed] [Google Scholar]
- 32.Urban R.J., Bodenburg Y.H., Wood T.G. NH2 terminus of PTB-associated splicing factor binds to the porcine P450scc IGF-I response element. Am. J. Physiol. Endocrinol. Metab. 2002;283:E423–E427. doi: 10.1152/ajpendo.00057.2002. [DOI] [PubMed] [Google Scholar]
- 33.Zhang W.W., Zhang L.X., Busch R.K., Farres J., Busch H. Purification and characterization of a DNA-binding heterodimer of 52 and 100 Kda from Hela-cells. Biochem. J. 1993;290:267–272. doi: 10.1042/bj2900267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyamoto K., Sakurai H., Sugiura T. Proteomic identification of a PSF/p54nrb heterodimer as RNF43 oncoprotein-interacting proteins. Proteomics. 2008;8:2907–2910. doi: 10.1002/pmic.200800083. [DOI] [PubMed] [Google Scholar]
- 35.Gromiha M.M., Parry D.A. Characteristic features of amino acid residues in coiled-coil protein structures. Biophys. Chem. 2004;111:95–103. doi: 10.1016/j.bpc.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 36.Mathur M., Tucker P.W., Samuels H.H. PSF is a novel corepressor that mediates its effect through Sin3A and the DNA binding domain of nuclear hormone receptors. Mol. Cell. Biol. 2001;21:2298–2311. doi: 10.1128/MCB.21.7.2298-2311.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hirose T., Virnicchi G., Tanigawa A., Naganuma T., Li R., Kimura H., Yokoi T., Nakagawa S., Benard M., Fox A.H., et al. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol. Biol. Cell. 2013;25:169–183. doi: 10.1091/mbc.E13-09-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.West J.A., Davis C.P., Sunwoo H., Simon M.D., Sadreyev R.I., Wang P.I., Tolstorukov M.Y., Kingston R.E. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell. 2014;55:791–802. doi: 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stepto A., Gallo J.M., Shaw C.E., Hirth F. Modelling C9ORF72 hexanucleotide repeat expansion in amyotrophic lateral sclerosis and frontotemporal dementia. Acta Neuropathol. 2014;127:377–389. doi: 10.1007/s00401-013-1235-1. [DOI] [PubMed] [Google Scholar]
- 40.Naganuma T., Nakagawa S., Tanigawa A., Sasaki Y.F., Goshima N., Hirose T. Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 2012;31:4020–4034. doi: 10.1038/emboj.2012.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka H., Kato K., Yamashita E., Sumizawa T., Zhou Y., Yao M., Iwasaki K., Yoshimura M., Tsukihara T. The structure of rat liver vault at 3.5 angstrom resolution. Science. 2009;323:384–388. doi: 10.1126/science.1164975. [DOI] [PubMed] [Google Scholar]
- 42.Sanchez J.G., Okreglicka K., Chandrasekaran V., Welker J.M., Sundquist W.I., Pornillos O. The tripartite motif coiled-coil is an elongated antiparallel hairpin dimer. Proc. Natl. Acad. Sci. U.S.A. 2014;111:2494–2499. doi: 10.1073/pnas.1318962111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Souquere S., Beauclair G., Harper F., Fox A., Pierron G. Highly ordered spatial organization of the structural long noncoding NEAT1 RNAs within paraspeckle nuclear bodies. Mol. Biol. Cell. 2010;21:4020–4027. doi: 10.1091/mbc.E10-08-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mao Y.S., Sunwoo H., Zhang B., Spector D.L. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat. Cell Biol. 2011;13:95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuroda N., Mikami S., Pan C.C., Cohen R.J., Hes O., Michal M., Nagashima Y., Tanaka Y., Inoue K., Shuin T., et al. Review of renal carcinoma associated with Xp11.2 translocations/TFE3 gene fusions with focus on pathobiological aspect. Histol. Histopathol. 2012;27:133–140. doi: 10.14670/HH-27.133. [DOI] [PubMed] [Google Scholar]
- 46.Hidalgo-Curtis C., Chase A., Drachenberg M., Roberts M.W., Finkelstein J.Z., Mould S., Oscier D., Cross N.C., Grand F.H. The t(1;9)(p34;q34) and t(8;12)(p11;q15) fuse pre-mRNA processing proteins SFPQ (PSF) and CPSF6 to ABL and FGFR1. Genes Chromosomes Cancer. 2008;47:379–385. doi: 10.1002/gcc.20541. [DOI] [PubMed] [Google Scholar]
- 47.Rendahl K.G., Gaukhshteyn N., Wheeler D.A., Fry T.A., Hall J.C. Defects in courtship and vision caused by amino acid substitutions in a putative RNA-binding protein encoded by the no-on-transient A (nonA) gene of Drosophila. J. Neurosci. 1996;16:1511–1522. doi: 10.1523/JNEUROSCI.16-04-01511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ke Y.D., Dramiga J., Schutz U., Kril J.J., Ittner L.M., Schroder H., Gotz J. Tau-mediated nuclear depletion and cytoplasmic accumulation of SFPQ in Alzheimer's and Pick's disease. PLoS One. 2012;7:e35678. doi: 10.1371/journal.pone.0035678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shelkovnikova T.A., Robinson H.K., Troakes C., Ninkina N., Buchman V.L. Compromised paraspeckle formation as a pathogenic factor in FUSopathies. Hum. Mol. Genet. 2014;23:2298–2312. doi: 10.1093/hmg/ddt622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu H. Higher-order assemblies in a new paradigm of signal transduction. Cell. 2013;153:287–292. doi: 10.1016/j.cell.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kato M., Han T.W., Xie S., Shi K., Du X., Wu L.C., Mirzaei H., Goldsmith E.J., Longgood J., Pei J., et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kwon I., Kato M., Xiang S., Wu L., Theodoropoulos P., Mirzaei H., Han T., Xie S., Corden J.L., McKnight S.L. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell. 2013;155:1049–1060. doi: 10.1016/j.cell.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meyer G.R., Aragao D., Mudie N.J., Caradoc-Davies T.T., McGowan S., Bertling P.J., Groenewegen D., Quenette S.M., Bond C.S., Buckle A.M., et al. Operation of the Australian Store.Synchrotron for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2014;70:2510–2519. doi: 10.1107/S1399004714016174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


