Abstract
Myotonic dystrophy type 1 (DM1) is caused by the expansion of an unstable CTG repeat (g.17294_17296(45_1000)) with more repeats associated with increased disease severity and reduced age at onset. Expanded disease-associated alleles are highly unstable in both the germline and soma. Germline instability is expansion biased, providing a molecular explanation for anticipation. Somatic instability is expansion biased, size- and age-dependent, features that have compromised genotype–phenotype correlations and intergenerational studies. We corrected these confounding factors by estimating the progenitor allele length in 54 father–offspring and 52 mother–offspring pairs in Costa Rican DM1 families. Not surprisingly, we found major parental allele length effects on the size of the allele transmitted, the magnitude of the intergenerational length change, the age at onset in the next generation and the degree of anticipation in both male and female transmissions. We also detected, for the first time, an age-of-parent effect for both male and female transmission. Interestingly, we found no evidence for an intrauterine effect in the transmission of congenital DM1, suggesting previous reports may have been an artefact of age-dependent somatic instability and sampling bias. These data provide new insights into the germline dynamics of the CTG repeat and opportunities for providing additional advice and more accurate risk assessments to prospective parents in DM1 families.
Introduction
Myotonic dystrophy type 1 (DM1) is a highly variable progressive multisystem disorder characterised by myotonia, muscle weakness, cardiac defects and cataracts.1 DM1 is an autosomal dominant disorder, affects individuals of all ages and both sexes, and presents with striking anticipation.2 The DM1 mutation is the expansion of a polymorphic CTG repeat (5 to ∼37 repeats (g.17294_17296(5_37)) in the general population) located in the 3′-untranslated region of the DMPK gene.3, 4, 5, 6, 7, 8 Affected patients carry from ∼45 to several thousand repeats (g.17294_17296(45_3000)).4, 6, 8 Repeat number correlates positively with disease severity and negatively with age of onset.9, 10, 11, 12 The expanded repeat is highly unstable and frequent germline expansions explain anticipation.10, 11, 12, 13 However, many apparent intergenerational contractions are associated with clinical anticipation.14 Proto-mutations 50 to 79 repeats (g.17294_17296(50_79)) appear to be particularly unstable and liable to relatively large expansions in the male germline, providing an explanation for the excess of transmitting grandfathers relative to congenitally affected grandchildren.10, 13, 15, 16, 17, 18, 19 Conversely, full mutations >79 repeats (g.17294_17296(80_3000)) appear to be prone to larger expansions when transmitted by females, providing an apparent explanation for the maternal transmission bias for congenital DM (CDM).10, 13, 20, 21 However, it has been revealed that approximately 25% of DM1 cases inherited from affected fathers appear to have inherited alleles equal to or greater in size than those found in maternally inherited CDM cases,19 supporting the suggestion that CDM is at least partially mediated by an intrauterine effect in affected mothers.19, 22, 23
Traditionally, DM1 patients have been genotyped using Southern blot hybridisation of restriction digested genomic DNA. However, as the DM1 CTG repeat is also highly unstable in the soma,24, 25, 26, 27, 28 this approach frequently yields a diffuse smear for the expanded allele, from which it is only possible to estimate the modal allele length.4, 6, 8, 10, 11, 12, 13, 14, 15, 16, 20, 29 As somatic mosaicism is expansion biased and age dependent,24, 25, 26, 27, 28 the modal allele length thus measured also increases with age.26 By performing multiple reactions with reduced amounts of input DNA, it is possible to use small pool PCR (SP-PCR) to resolve the diffuse smear into its component alleles25 and estimate the progenitor allele length (PAL) transmitted from the affected parent.25 Accounting for age-dependent somatic mosaicism by estimating PAL dramatically improves genotype–phenotype relationships.28 Nearly all previous analyses of intergenerational transmission have compared modal allele length in the blood DNA of parent and child and have not accounted for age-dependent somatic mosaicism. Combined SP-PCR analysis of sperm DNA in a small number of affected males with estimation of PAL in themselves and their offspring has suggested that some intergenerational transmissions have been mis-interpreted using the traditional approach.25, 30 We hypothesise that failure to take into account age-dependent somatic mosaicism has compromised the interpretation of previous pedigree analyses in DM1. To test this hypothesis, we recruited a large cohort of Costa Rican DM1 families and investigated intergenerational dynamics by estimating PAL in parents and offspring.
Materials and methods
Genealogical studies in Costa Rican DM1 families
The study included all 41 families that have been referred to Universidad de Costa Rica since 1998, in which at least one individual has a confirmed molecular diagnosis of DM1. Pedigrees of the families were constructed during the visit of the research team to their home and included all affected and unaffected family members known by the proband or their parents. The research team included a neurologist and each individual in each family was examined for clinical signs of DM1. Generation one was defined as the generation presenting the late-onset form of the disease; generation two as that presenting classic adult onset DM1; and generation three as that in which juvenile or CDM cases were observed. The majority of patients in these families have been confirmed to carry the DM1 mutation and this information was used to complement the genealogical study and to determine obligate mutation carriers. As expected,2 we observed high levels of anticipation with the disease typically progressing from a mild late-onset form in generation one to congenital or juvenile disease in generation three. We also observed the expected10, 13, 15, 16, 17, 18, 19 excess of transmitting males in generation one and the exclusive maternal transmission of CDM (Supplementary Information).22, 31
Molecular analyses
Molecular analyses of the CTG repeat was undertaken in 153 patients from 32 Costa Rican DM1 families, all of the families in which DNA was available from at least one parent-offspring pair. Patients were recruited with informed consent and the study was approved by the Institutional Review Board of the University of Costa Rica. Genomic DNA was purified from peripheral blood using proteinase K digestion and phenol–chloroform extraction. The PAL was estimated from the lower boundary of the allele length distribution obtained from SP-PCR analysis of five replicate reactions with ∼180 to 300 pg of genomic DNA as described previously.25, 28, 32 Modal allele length was estimated as the point of highest band density. Alleles are described using Human Genome Variation Society nomenclature (http://www.hgvs.org/mutnomen/recs-DNA.html#var) based on the DMPK RefSeq: NG_009784.1. Data have been deposited in the NCBI ClinVar database (http://www.ncbi.nlm.nih.gov/clinvar/), accession numbers: SCV000120188 and SCV000120189.
Statistical analysis
Statistical analyses were conducted using SPSS statistics (IBM, v21, New York, NY, USA) and STATA package (StataCorp, v12, College Station, TX, USA). For linear and multivariate regression, all squared coefficients of correlation have been adjusted for the number of parameters. For proportions, we calculated 95% confidence intervals (CIs) for the point estimate.
Results
Intergenerational transmission of the CTG repeat expansion
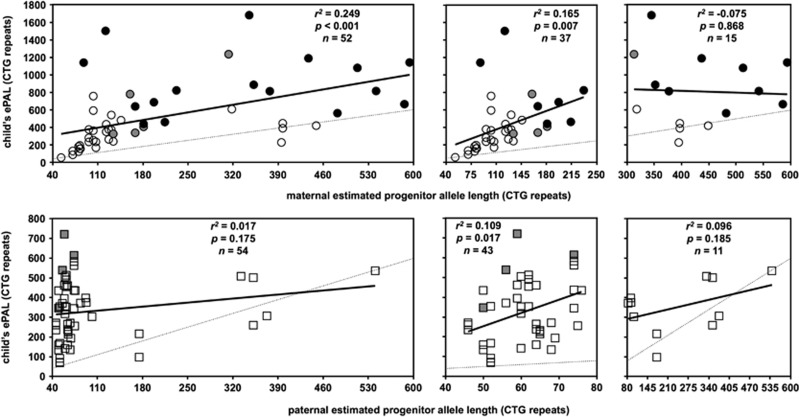
To correct the confounding effects of somatic mosaicism and biased age at sampling in assessing intergenerational transmissions when comparing the blood DNA of parents and offspring,25 we used SP-PCR to estimate PAL (ePAL) in 106 parent–offspring pairs from 32 Costa Rican DM1 families: 54 father–offspring pairs (from 24 different fathers) and 52 mother–offspring pairs (from 36 different mothers), including 16 mother–CDM pairs. Surprisingly, paternal ePAL was not significantly correlated with the child's ePAL (r2=0.017, P=0.175, n=54; Figure 1). Previously, a highly significant effect of male ePAL with mean allele length in sperm was reported.33 However, this was largely driven by a strong effect for males carrying proto-mutations <80 repeats (g.17294_17296(50_79)).33 Similarly, when male transmissions were split according to paternal ePAL, a significant correlation with the child's ePAL was observed for fathers carrying pre- and proto-mutations (38–79 repeats (g.17294_17296(38_79)); r2=0.109, P=0.017, n=43), but no length effect towards the transmission of larger alleles was observed for fathers with full mutations (>80 repeats (g.17294_17296(80_3000)); r2=0.096, P=0.185, n=11; Figure 1). The ePAL in the mother was correlated with the ePAL in the child (r2=0.249, P<0.001, n=52; Figure 1). However, this correlation also appeared to be driven by mothers with smaller ePALs and when cases were split by maternal ePAL, the correlation remained significant for mothers with ePAL <250 repeats (g.17294_17296(50_249); r2=0.165, P=0.007, n=37), but was not significant for mothers with larger ePALs (>249 repeats (g.17294_17296(250_3000)), r2=−0.075, P=0.868, n=15; Figure 1). When considering the actual intergenerational allele length change for paternal transmissions, an inverse relationship was detected across the whole data set (r=−0.419, r2=0.159, P=0.002, n=54; Supplementary Figure 2). However, when paternal transmissions were split dependent on the father's ePAL, a positive allele length effect was observed for pre- and proto-mutations (r=0.313, r2=0.076, P=0.041, n=43) and a negative effect for full mutations (r=−0.623, r2=0.320, P=0.041, n=11; Supplementary Figure 2). Across the whole data set, no allele length effect was observed for female transmissions (r2=−0.006, P=0.406, n=52; Supplementary Figure 2). However, there appeared to be a trend towards the transmission of larger intergenerational expansions for mothers with smaller ePALs. Indeed, a marginally significant allele length effect was detected for alleles <250 repeats (r2=0.069, P=0.064, n=37; Supplementary Figure 2).
Figure 1.
The child's progenitor allele length is dependent on the sex of the parent and correlated with the parent's progenitor allele length. The scatter plots show the relationship between parental ePAL and the child's ePAL for female (circles) and male transmission (squares). Cases with congenital (black) and juvenile (grey) onset in the child are indicated. The faint dotted line shows zero length change. The line of best fit under linear regression (solid line) and relevant statistics are shown. Data are split according to sex of parent (females top, males bottom). Left panels show data over the whole range, whereas the right panels show the data split according to parental ePAL (males </> 80 repeats, females </> 250 repeats).
Sex-of-parent effects on intra-sibship variability
To gain insight into the predictive power once we know there is an affected offspring in a DM1 family, we investigated the intraclass correlation between parent–full sibling pairs. Intraclass correlation is commonly used to quantify the degree to which individuals with a fixed degree of relatedness (eg, full siblings) resemble each other in terms of a quantitative trait.34 In father–full sibling pairs, the intraclass correlation for ePAL was very low (r=0.04, P=0.44, n=43). However, in mother–full sibling pairs a significant intraclass correlation was detected (r=0.49, P=0.04, n=29).
Intergenerational expansions and anticipation
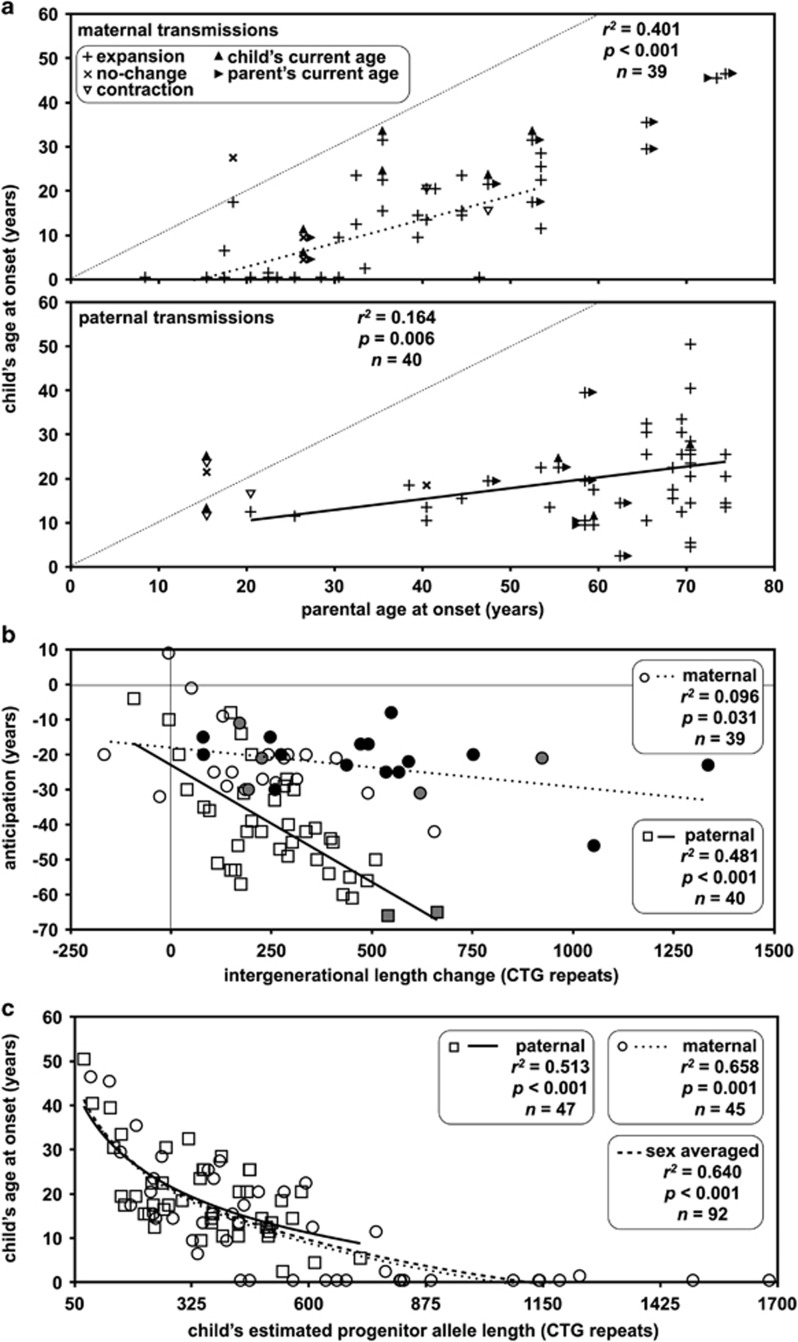
Clinical anticipation was observed in most transmissions (maternal transmission: ∼85% (n=44/52, CI=0.72–0.92), paternal transmission: ∼87% (n=47/54, CI=0.76–0.94); Figure 2a), and was accompanied by intergenerational expansion in the vast majority of these (maternal transmission: ∼95% (n=42/44, CI=0.85–0.99), paternal transmission: ∼96% (n=45/47, CI=0.86–0.99)). Details of the 6 out of 94 cases where the expected relationship between repeat length transmitted and clinical anticipation were not observed are described in the Supplementary Information.
Figure 2.
Sex-of-parent effects on anticipation and age at onset. (a) Sex-of-parent and parental age at onset dependent effects on the child's age at onset. The scatter plots show the relationship between parental age at onset and the child's age at onset. Cases associated with an intergenerational repeat expansion are indicated with a vertical cross, contractions with an open downward triangle and cases with no length change as diagonal crosses. In most cases, age at onset in parent and offspring are known. Where one or more individual remains asymptomatic, the point is placed at the age at sampling. Cases with asymptomatic parent are marked with a right facing filled triangle. Cases with an asymptomatic child are indicated with a closed upward facing triangle. The faint dotted line shows the zero age at onset change line. (b) The degree of anticipation is correlated with the sex of the parent and the intergenerational length change. Cases with congenital (black) and juvenile (grey) onset in the child are indicated. (c) Age at onset in the child is dependent on the length of the allele transmitted, but not on the sex of the parent. In all graphs, the line of best fit under linear regression (a, b) or logarithmic regression (c) (dotted line female, solid line male, dashed line sex-independent transmissions) and relevant statistics are shown.
Overall, the mother's age at onset was highly correlated with the age at onset in the child (r2=0.401, P<0.001, n=39; Figure 2a) and the degree of anticipation (r2=0.436, P<0.001, n=39; Supplementary Figure 3). Interestingly, relative to maternal transmissions, the correlation for the father's age at onset and the age at onset in the child was much lower (r2=0.164, P=0.006, n=40; Figure 2a), but the correlation between the father's age at onset and the degree of anticipation was much greater (r2=0.642, P<0.001, n=40; Supplementary Figure 3). Similarly, the correlation between the degree of anticipation and the intergenerational repeat length change was much greater for paternal transmission (r2=0.481, P<0.001, n=40) than maternal (r2=0.096, P=0.031, n=39; Figure 2b).
As is typical, the Costa Rican cohort contained no examples of paternal transmission of CDM. Although this appears to be largely driven by the failure of affected men to transmit very large alleles to their offspring,10, 13, 20, 21 it has been suggested that there may be a role for an intrauterine effect in affected mothers.19 To test for a sex-of-parent effect on age at onset over and above repeat length, we investigated the correlation between age at onset and ePAL (Figure 2c) using the logarithmic model we previously defined.28 The child's allele length versus age at onset models for male parents (β0=97.5, β1=−31.0, r2=0.513, P<0.001, n=47) and female parents (β0=106.5, β1=−35.1, r2=0. 658, P<0.001, n=45) were remarkably similar (Figure 2c). Moreover, a t-test revealed no difference in the mean residuals (t=0.924, P=0.817) under the null sex-of-parent-independent model (β0=105.2, β1=−34.3, r2=0.640, P<0.001, n=92; Figure 2c).
Age effects on the transmission of the CTG repeat expansion
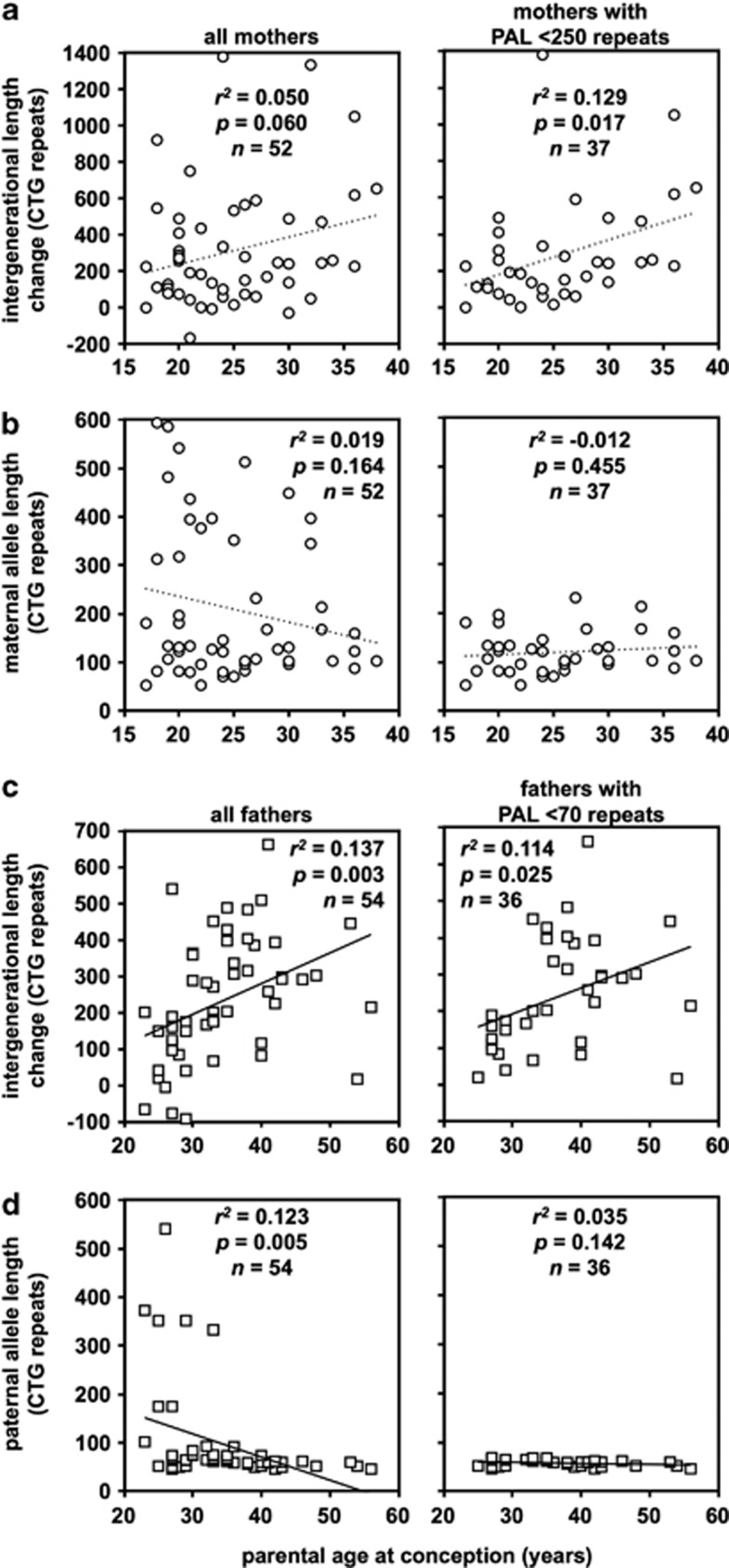
Somatic expansion in DM1 is highly age dependent24, 25, 26, 28 and it might be expected that germline expansion would be also. However, there are no reports of a parental age effect in pedigree analyses, and even direct sperm DNA analyses have proven inconclusive.33 In female transmission, a modest marginally significant increase in the size of intergenerational expansions with age was observed (r2=0.050, P=0.060, n=52; Figure 3a). In addition to the inherent variability in transmissions from a given individual, a possible confounding factor in detecting age effects are possible allele length-mediated sampling biases. Indeed, analysis of the correlation between age at conception and maternal allele length suggested a slight deficit of transmission from older mothers with large alleles (Figure 3b), consistent with an allele length-dependent decrease in fecundity. Thus, we also analysed female transmissions after excluding mothers with an ePAL >249 repeats. In this range (g.17294_17296(50_249)), there was no obvious maternal allele length effect on fecundity (Figure 3b), and a more obvious maternal age effect (r2=0.129, P=0.017, n=37; Figure 3a). These data were supported by multivariate linear regression analysis in which maternal age at conception was the only parameter that added significant value to the model (Table 1, model 1 and model 2). In male transmission, a highly significant paternal age effect was detected in the whole data set (r2=0.137, P=0.003, n=54; Figure 3c). However, regression analysis between age at conception and paternal allele length also revealed a clear effect on male fecundity (r2=0.123, P=0.005, n=54; Figure 3d). Such an effect is an expected consequence of the well-documented male infertility in DM1.1 Thus, we examined the relationship between age at conception and paternal ePAL for males with pre- and proto-mutations (<80 repeats (g.17294_17296(38_79))). However, even in this range there was still a statistically significant correlation between paternal age at conception and paternal allele length (r2=0.073, P=0.045, n=43). Examining males with <70 repeats (g.17294_17296(38_69)) revealed no effect of paternal allele length on fecundity (r2=0.035, P=0.142, n=36; Figure 3d), but a significant paternal age effect on intergenerational length change was still evident (r2=0.114, P=0.025, n=36; Figure 3c). This effect was replicated in multivariate linear regression analysis in which paternal age at conception and paternal ePAL both added significant value to the model when considering the whole data set (Table 1, model 3). Indeed, when considering only those fathers with <70 repeats, the effect of paternal allele length was no longer statistically significant, but the effect of paternal age at conception was statistically significant (P=0.012, Table 1, model 4).
Figure 3.
Sex- and age-of-parent-dependent effects on intergenerational length change, and sex and parental progenitor allele length effects on fecundity. (a, c) Age-of-parent-dependent effects on intergenerational length changes. The scatter plots show the relationship between parental age at conception and the intergenerational length change. (b, d) Age-of-parent-dependent effects on fecundity. The scatter plots show the relationship between parental age at conception and parental ePAL. The line of best fit under linear regression (dotted line female, solid line male transmissions) and relevant statistics are shown. Data are split according to sex of the parent (females a, b (circles), males c, d (squares)). Left panels show data over the whole range, whereas the right panels show the data split according to parental ePAL (males <70 repeats, females <250 repeats).
Table 1. Regression models of the relationship between the Int, estimated PAL and AgC.
| Model | r2 | P | Parameter | Coefficient | Standard error | t-Statistic | P | ||
|---|---|---|---|---|---|---|---|---|---|
| 1 | Int=β0+β1PAL+β2AgC | 0.062 | 0.079 | Intercept | β0 | −184.5 | 220.4 | −0.8 | 0.406 |
| All maternal transmission (n=52) | PAL | β1 | 0.3 | 0.3 | 1.3 | 0.210 | |||
| AgC | β2 | 16.9 | 7.9 | 2.1 | 0.037 | ||||
| 2 | Int=β0+β1PAL+β2AgC | 0.175 | 0.015 | Intercept | β0 | −364.5 | 215.2 | −1.7 | 0.099 |
| Maternal transmissions with PAL <250 repeats (n=37) | PAL | β1 | 1.7 | 1.0 | 1.7 | 0.095 | |||
| AgC | β2 | 17.3 | 7.4 | 2.3 | 0.025 | ||||
| 3 | Int=β0+β1PAL+β2AgC | 0.210 | 0.001 | Intercept | β0 | 82.1 | 109.3 | 0.8 | 0.456 |
| All paternal transmission (n=54) | PAL | β1 | −0.535 | 0.2 | 2.4 | 0.020 | |||
| AgC | β2 | 5.9 | 2.8 | 2.1 | 0.043 | ||||
| 4 | Int=β0+β1PAL+β2AgC | 0.136 | 0.034 | Intercept | β0 | −351.5 | 266.5 | −1.3 | 0.196 |
| Paternal transmissions with PAL <70 repeats (n=36) | PAL | β1 | 5.1 | 3.7 | 1.4 | 0.178 | |||
| AgC | β2 | 8.1 | 3.1 | 2.6 | 0.012 |
Abbreviations: AgC, age at conception; Int, intergenerational length change; PAL, progenitor allele length.
The table shows the squared coefficient of correlation (r2), and statistical significance (P) for each model, and the coefficient, standard error, t-statistic and statistical significance (P), associated with each parameter in the model. The coefficient provides an indication of the relative weight of the contribution of each parameter to the model and its associated standard error. The t-statistic and corresponding P-value provide an indication of the statistical significance that the parameter is adding explanatory power to the model.
Estimated risk for the transmission of juvenile/CDM from an affected mother
To determine if defining ePAL in the mother facilitated a more accurate risk assessment of giving birth to a congenitally affected child, we used the receiver operating characteristics curve.35 The accuracy of the test was ∼84% (CI=0.73–0.96, n=52) with a cutoff of 164 repeats with mothers, and with ePALs exceeding this threshold at ∼64% risk of transmitting CDM to an affected child (Supplementary Figure 4). The cutoff for juvenile or CDM predictions using maternal ePAL was lower at 153 repeats with a similar accuracy of ∼85% (CI=0.74–0.96, n=50; Supplementary Figure 4). We have not measured modal repeat length using the traditional Southern blot analysis of restriction digested genomic DNA with which to compare these data. However, estimating the mothers mode from the SP-PCR analyses revealed a similar level of accuracy (∼83%, CI=0.71–0.96, n=52), although with a higher cutoff of 284 repeats, which was also the same as that defined for juvenile or CDM (∼83%, CI=0.71–0.95, n=50; Supplementary Figure 4). In addition, we also performed logistic regression revealing that a one repeat increase in maternal ePAL increases the odds of transmitting CDM by 1.009 times (Supplementary Information).
Discussion
Extreme variability in disease severity, coupled with unusual sex-of-parent effects, grossly complicate genetic counseling in DM1. Much of the difficulty derives from the complex relationship between allele length and disease severity, and extreme germline instability. Nonetheless, previous pedigree analyses have yielded considerable insight into this process and the broad sex-dependent dynamics of DM1 transmission have been established.10, 11, 12, 13, 15 However, these studies have not taken into account ongoing somatic expansion.25 Here, we corrected for the confounding effects of age at sampling by using SP-PCR to estimate PAL in parent and offspring. Our analyses have confirmed that intergenerational transmissions are highly biased towards expansions and that the main driver of ePAL in offspring is parental ePAL. This effect was much more pronounced for females, and was only apparent for males carrying pre- and proto-mutations (<80 repeats). These observations are consistent with the intraclass correlation between siblings, which was significant for female transmissions, but was not detectable for paternal transmissions; consistent with the very high levels of intra-individual variation observed by direct sperm analysis25, 30, 33 and suggesting that inter-egg variation is less pronounced. Our analyses have also confirmed that the main driver of the size of the intergenerational length change is also parental ePAL. However, these effects are nonlinear and complex. Pre- and proto-mutations (<80 repeats) are much more unstable in the male germline and biased towards large expansions, consistent with the previously reported excess of transmitting grandfathers.10, 13, 15, 16, 17, 18, 19 Full mutations (>79 repeats) in males are highly unstable, but appear to be more likely to contract; consistent with direct sperm analyses.25, 30, 33 In females, pre- and proto-mutations are relatively stable, but full mutations are much more unstable and biased towards much larger expansion than in males, consistent with the excess of transmitting mothers of CDM.10, 13, 20, 21 However, transmissions from mothers with very long alleles (>249 repeats) are less well explained by maternal ePAL and, as in males, appear to be more likely to contract. The increased risk of contraction with longer alleles parallels similar observations made by Ashizawa et al in traditional pedigree analyses.14 In contrast to Ashizawa et al however, some of the contractions we identified resulted in clear reverse anticipation, as would be expected, suggesting that some of the apparent contractions observed by Ashizawa et al were artefacts of age-dependent somatic instability.25 Nonetheless, we also detected several cases that appeared to be genuine intergenerational contractions, but that were still accompanied by anticipation. These examples highlight the existence of additional modifiers of disease severity in DM1. Recent data have revealed that promoter polymorphisms in one of the key mediators of downstream pathology, MBNL1, account for some of the residual variation in age at onset not accounted for by modal allele length.36 In addition, disease severity is also affected by genetic modifiers that alter the rate of somatic expansion,28 which, by analogy to mice, are likely to include polymorphisms in DNA mismatch repair genes.37 Defining the influence of modifiers would be expected to account for some of the atypical inheritance patterns and provide the basis for offering even more accurate risk estimates.
Reflecting the complex relationships between parental age at onset, parental ePAL and the allele length transmitted to the child, the relationships between parental age at onset, parental ePAL and the child's ePAL and the degree of anticipation were similarly complex. The child's age at onset was highly correlated with parental age at onset, particularly for females, but less so for males, again likely reflecting the high degree of intra-individual variability in sperm. In contrast, the degree of anticipation was much more strongly correlated with the intergenerational length change and parental age at onset in male transmission. However, rather than reflecting any sex of parent difference in the absolute relationship between repeat length and disease onset, this most likely reflects the relative position of transmitting males and females in pedigrees and the nonlinear relationship between repeat length and disease severity: transmitting males are over-represented in the mildly affected first generation with relatively small expansions from whom the transmission of relatively modest increases in repeat length result in a dramatic decrease in the age at onset. Whereas most female transmissions are observed in generation two from women who already have adult onset disease and for whom the absolute degree of anticipation is bounded by their own age at onset.
Given the strong age dependence of somatic mosaicism in DM1,24, 25, 26, 28 it is surprising that an age effect for intergenerational transmission has not been observed. Here, we provide evidence for an age effect on the size of the intergenerational length change for males with alleles <70 repeats and females with <250 repeats. However, our analyses highlight the confounding factors that can obfuscate such relationships. In both sexes, but most dramatically in males, we found evidence for an allele length effect on fecundity that leads to an over-representation of individuals with smaller alleles among older parents. The age effect revealed here for paternal transmissions from males with alleles <70 repeats is consistent with the effect observed in direct sperm DNA analyses in males with pre- and proto-mutations.33 The allele length-dependent age effect on fecundity limits our ability to detect any possible age effect in males carrying larger expansions. Previous direct sperm analyses in such males did not reveal any evidence for changes in repeat length over time periods in which clear differences in somatic mosaicism were detectable.33 A possible explanation for these apparently conflicting results may reside in the complex dynamics of the expanded CTG repeat in the male germline. Expanded DM1 alleles are highly unstable in the male germline, with a major bias towards expansion.25, 30, 33 However, in most DM1 men, even those carrying large expanded alleles, the absolute range of variation appears to be limited.25, 30, 33, 38 Moreover, the pattern of variation is distinct from that observed in the soma with distributions far more normally dispersed, with a measurable frequency of large contractions, including occasional reversions.25, 30, 33 These data suggest that the male germline mutational pathway is much more bidirectional than in the soma, with the possibility that an equilibrium may be reached whereby ongoing expansions and contractions cancel each other out. It is possible that this equilibrium is reached at a relatively young age in DM1 males with larger expansions, negating any age effect in the reproductive window, but that males with pre- and proto-mutations take much longer to reach such an equilibrium and consequently display age-dependent germline mutations. Variation in the female germline appears to mirror more closely somatic variation with pre- and proto-mutations being relatively stable and a lower frequency of contractions and an age dependence in arrested eggs that may mimic the instability observed in post-mitotic muscle39, 40, 41 and brain.42
The final step in the anticipatory cascade in DM1 is the transmission of many hundreds or even thousands of repeats and the birth of a congenitally affected child. As is usually observed,22, 31 all of the congenitally affected children identified in the Costa Rican DM1 population were born to affected mothers. This phenomenon was initially attributed to a maternal intrauterine effect on fetal development.22 However, it is clear that this effect can now partly be explained by transmission of the very largest expansions almost exclusively through the maternal germline,10, 13, 20, 21 as we have also observed here in the Costa Rican DM1 population. Nonetheless, it has also been suggested that the paternal transmission of alleles of a similar magnitude to those observed in maternally transmitted congenital DM1 do not precipitate congenital DM1.19 Although these observations may support an intrauterine effect, an alternative explanation is that the apparent discrepancy in sex-of-parent-dependent genotype–phenotype relationships is an artefact of age at sampling biases within DM1 families. Many CDM patients are sampled shortly after birth at which point levels of somatic mosaicism in blood DNA are very low26, 43 and measured allele length should be close to the PAL. In contrast, individuals without congenital symptoms are usually not sampled until later in life, by which time their measured allele length would have increased because of age-dependent expansion-biased somatic instability. Here, we have corrected for any such bias by estimating PAL in all individuals and we find the genotype–phenotype relationship in offspring of parents of either sex is indistinguishable. These data strongly argue against an intrauterine effect and suggest that the previously reported discrepancy between paternally transmitted expansions and CDM,19 may be another artefact of age-dependent somatic instability.
An important consideration in providing genetic counseling to DM1 families is assessing the relative risk of CDM. Here, by using receiver operating characteristics analysis, we have revealed a maternal allele length cutoff of 164 CTG repeats, above which the risk of CDM is ∼64%. The cutoff we have identified is lower than that predicted using modal allele length and that previously reported cutoff of 300 repeats,20 as using ePAL corrects for age-dependent somatic expansion in the mother. However, the use of ePAL in this study did not result in a significant increase in the accuracy of the predictions, probably reflecting the relatively small sample size and highlighting the contribution of additional modifiers in precipitating CDM. Our analyses have also allowed us to calculate the relative risk of CDM associated with each CTG repeat in the mother revealing an odds ratio of 1.009 per CTG repeat. This translates to a doubling in the risk of CDM for an increase in the maternal allele of ∼110 repeats.
In the absence of effective therapies, one of the most important applications of a positive molecular diagnosis is to provide accurate reproductive risk assessments to prospective parents in DM1 families. Here, we have corrected for the confounding effect of somatic instability to confirm the importance of parental allele length, reveal new insights into the role of parental age and argue against a major role for sex-of-parent effects beyond transmitted repeat length. These data provide additional insights into repeat biology and may lead to the provision of more accurate prognostic information to prospective parents.
Acknowledgments
We thank our colleagues in the DGM group for productive discussions and helpful comments on the manuscript. We also thank the myotonic dystrophy patients and their families for their assistance. This work was supported by awards from: the Muscular Dystrophy Association; the Universidad de Costa Rica; the Ministerio de Ciencia y Tecnologia of Costa Rica; and the National Council for Scientific and Technological Research in Costa Rica (CONICIT).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Harper PS. Myotonic Dystrophy. London: WB Saunders Co; 2001. [Google Scholar]
- Höweler CJ, Busch HFM, Geraedts JPM, Niermeijer MF, Staal A. Anticipation in myotonic dystrophy: fact or fiction. Brain. 1989;112:779–797. doi: 10.1093/brain/112.3.779. [DOI] [PubMed] [Google Scholar]
- Aslanidis C, Jansen G, Amemiya C, et al. Cloning of the essential myotonic dystrophy region and mapping of the putative defect. Nature. 1992;355:548–550. doi: 10.1038/355548a0. [DOI] [PubMed] [Google Scholar]
- Brook JD, McCurrach ME, Harley HG, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- Buxton J, Shelbourne P, Davies J, et al. Detection of an unstable fragment of DNA specific to individuals with myotonic dystrophy. Nature. 1992;355:547–548. doi: 10.1038/355547a0. [DOI] [PubMed] [Google Scholar]
- Fu YH, Pizzuti A, Fenwick RG, et al. An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science. 1992;255:1256–1258. doi: 10.1126/science.1546326. [DOI] [PubMed] [Google Scholar]
- Harley HG, Brook JD, Rundle SA, et al. Expansion of an unstable DNA region and phenotypic variation in myotonic dystrophy. Nature. 1992;355:545–546. doi: 10.1038/355545a0. [DOI] [PubMed] [Google Scholar]
- Mahadevan M, Tsilfidis C, Sabourin L, et al. Myotonic dystrophy mutation: an unstable CTG repeat in the 3' untranslated region of the gene. Science. 1992;255:1253–1255. doi: 10.1126/science.1546325. [DOI] [PubMed] [Google Scholar]
- Hunter A, Tsilfidis C, Mettler G, et al. The correlation of age of onset with CTG trinucleotide repeat amplification in myotonic dystrophy. J Med Genet. 1992;29:774–779. doi: 10.1136/jmg.29.11.774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley HG, Rundle SA, MacMillan JC, et al. Size of the unstable CTG repeat sequence in relation to phenotype and parental transmission in myotonic dystrophy. Am J Hum Genet. 1993;52:1164–1174. [PMC free article] [PubMed] [Google Scholar]
- Redman JB, Fenwick RG, Fu Y-H, Pizzuti A, Caskey CT. Relationship between parental trinucleotide GCT repeat length and severity of myotonic dystrophy in offspring. J Am Med Assoc. 1993;269:1960–1965. [PubMed] [Google Scholar]
- Ashizawa T, Dubel JR, Dunne PW, et al. Anticipation in myotonic dystrophy. II. Complex relationships between clinical findings and structure of the GCT repeat. Neurology. 1992;42:1877–1883. doi: 10.1212/wnl.42.10.1877. [DOI] [PubMed] [Google Scholar]
- Lavedan C, Hofmann-Radvanyi H, Shelbourne P, et al. Myotonic dystrophy: size- and sex-dependent dynamics of CTG meiotic instability, and somatic mosaicism. Am J Hum Genet. 1993;52:875–883. [PMC free article] [PubMed] [Google Scholar]
- Ashizawa T, Anvret M, Baiget M, et al. Characteristics of intergenerational contractions of the CTG repeat in myotonic dystrophy. Am J Hum Genet. 1994;54:414–423. [PMC free article] [PubMed] [Google Scholar]
- Brunner HG, Bruggenwirth HT, Nillesen W, et al. Influence of sex of the transmitting parent as well as of parental allele size on the CTG expansion in myotonic dystrophy (DM) Am J Hum Genet. 1993;53:1016–1023. [PMC free article] [PubMed] [Google Scholar]
- Ashizawa T, Dunne PW, Ward PA, Seltzer WK, Richards CS. Effects of sex of myotonic dystrophy patients on the unstable triplet repeat in their affected offspring. Neurology. 1994;44:120–122. doi: 10.1212/wnl.44.1.120. [DOI] [PubMed] [Google Scholar]
- Lopez de Munain A, Cobo AM, Poza JJ, et al. Influence of the sex of the transmitting grandparent in congenital myotonic dystrophy. J Med Genet. 1995;32:689–691. doi: 10.1136/jmg.32.9.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barceló JM, Mahadevan MS, Tsilfidis C, Mackenzie AE, Korneluk RG. Intergenerational stability of the myotonic dystrophy protomutation. Hum Mol Genet. 1993;2:705–709. doi: 10.1093/hmg/2.6.705. [DOI] [PubMed] [Google Scholar]
- Barceló JM, Pluscauskas M, MacKenzie AE, Tsilfidis C, Narang M, Korneluk RG. Additive influence of maternal and offspring DM-kinase gene CTG repeat lengths in the genesis of congenital myotonic dystrophy. Am J Hum Genet. 1994;54:1124–1125. [PMC free article] [PubMed] [Google Scholar]
- Cobo AM, Poza JJ, Martorell L, Lopez de Munain A, Emparanza JI, Baiget M. Contribution of molecular analyses to the estimation of the risk of congenital myotonic dystrophy. J Med Genet. 1995;32:105–108. doi: 10.1136/jmg.32.2.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsilfidis C, MacKenzie AE, Mettler G, Barcelo J, Korneluk RG. Correlation between CTG trinucleotide repeat length and frequency of severe congenital myotonic dystrophy. Nat Genet. 1992;1:192–195. doi: 10.1038/ng0692-192. [DOI] [PubMed] [Google Scholar]
- Harper PS, Dyken PR. Early-onset dystrophia myotonica. Evidence supporting a maternal environmental factor. Lancet. 1972;2:53–55. doi: 10.1016/s0140-6736(72)91548-6. [DOI] [PubMed] [Google Scholar]
- Koch MC, Grimm T, Harley HG, Harper PS. Genetic risks for children of women with myotonic dystrophy. Am J Hum Genet. 1991;48:1084–1091. [PMC free article] [PubMed] [Google Scholar]
- Martorell L, Monckton DG, Gamez J, et al. Progression of somatic CTG repeat length heterogeneity in the blood cells of myotonic dystrophy patients. Hum Mol Genet. 1998;7:307–312. doi: 10.1093/hmg/7.2.307. [DOI] [PubMed] [Google Scholar]
- Monckton DG, Wong LJ, Ashizawa T, Caskey CT. Somatic mosaicism, germline expansions, germline reversions and intergenerational reductions in myotonic dystrophy males: small pool PCR analyses. Hum Mol Genet. 1995;4:1–8. doi: 10.1093/hmg/4.1.1. [DOI] [PubMed] [Google Scholar]
- Wong LJ, Ashizawa T, Monckton DG, Caskey CT, Richards CS. Somatic heterogeneity of the CTG repeat in myotonic dystrophy is age and size dependent. Am J Hum Genet. 1995;56:114–122. [PMC free article] [PubMed] [Google Scholar]
- Higham CF, Morales F, Cobbold CA, Haydon DT, Monckton DG. High levels of somatic DNA diversity at the myotonic dystrophy type 1 locus are driven by ultra frequent expansion and contraction mutations. Hum Mol Genet. 2012;21:2450–2463. doi: 10.1093/hmg/dds059. [DOI] [PubMed] [Google Scholar]
- Morales F, Couto JM, Higham CF, et al. Somatic instability of the expanded CTG triplet repeat in myotonic dystrophy type 1 is a heritable quantitative trait and modifier of disease severity. Hum Mol Genet. 2012;21:3558–3567. doi: 10.1093/hmg/dds185. [DOI] [PubMed] [Google Scholar]
- Lopez de Munain A, Blanco A, Emparanza JI, et al. Anticipation in myotonic dystrophy: a parental-sex-related phenomenon. Neuroepidemiology. 1994;13:75–78. doi: 10.1159/000110362. [DOI] [PubMed] [Google Scholar]
- Martorell L, Monckton DG, Gamez J, Baiget M. Complex patterns of male germline instability and somatic mosaicism in myotonic dystrophy type 1. Eur J Hum Genet. 2000;8:423–430. doi: 10.1038/sj.ejhg.5200478. [DOI] [PubMed] [Google Scholar]
- Vanier TM. Dystrophia myotonica in childhood. BMJ. 1960;2:1284–1288. doi: 10.1136/bmj.2.5208.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes-Pereira M, Bidichandani SI, Monckton DG. Analysis of unstable triplet repeats using small-pool polymerase chain reaction. Methods Mol Biol. 2004;277:61–76. doi: 10.1385/1-59259-804-8:061. [DOI] [PubMed] [Google Scholar]
- Martorell L, Gamez J, Cayuela ML, et al. Germline mutational dynamics in myotonic dystrophy type 1 males: allele length and age effects. Neurology. 2004;62:269–274. doi: 10.1212/wnl.62.2.269. [DOI] [PubMed] [Google Scholar]
- Fisher RA. Statistical Methods for Research Workers. Edinburgh: Oliver and Boyd; 1954. [Google Scholar]
- Wray NR, Yang J, Goddard ME, Visscher PM. The genetic interpretation of area under the ROC curve in genomic profiling. PLoS Genet. 2010;6:e1000864. doi: 10.1371/journal.pgen.1000864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huin V, Vasseur F, Schraen-Maschke S, et al. MBNL1 gene variants as modifiers of disease severity in myotonic dystrophy type 1. J Neurol. 2013;260:998–1003. doi: 10.1007/s00415-012-6740-y. [DOI] [PubMed] [Google Scholar]
- Tome S, Manley K, Simard JP, et al. MSH3 polymorphisms and protein levels affect CAG repeat instability in Huntington's disease mice. PLoS Genet. 2013;9:e1003280. doi: 10.1371/journal.pgen.1003280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen G, Willems P, Coerwinkel M, et al. Gonosomal mosaicism in myotonic dystrophy patients: Involvement of mitotic events in (CTG)n variation and selection against extreme expansion in sperm. Am J Hum Genet. 1994;54:575–585. [PMC free article] [PubMed] [Google Scholar]
- Ashizawa T, Dubel JR, Harati Y. Somatic instability of CTG repeat in myotonic dystrophy. Neurology. 1993;43:2674–2678. doi: 10.1212/wnl.43.12.2674. [DOI] [PubMed] [Google Scholar]
- Anvret M, Ahlberg G, Grandell U, Hedberg B, Johnson K, Edstrom L. Larger expansions of the CTG repeat in muscle compared to lymphocytes from patients with myotonic dystrophy. Hum Mol Genet. 1993;2:1397–1400. doi: 10.1093/hmg/2.9.1397. [DOI] [PubMed] [Google Scholar]
- Thornton CA, Johnson KJ, Moxley RT. Myotonic dystrophy patients have larger CTG expansions in skeletal muscle than in leukocytes. Ann Neurol. 1994;35:104–107. doi: 10.1002/ana.410350116. [DOI] [PubMed] [Google Scholar]
- Ishii S, Nishio T, Sunohara N, et al. Small increase in triplet repeat length of cerebellum from patients with myotonic dystrophy. Hum Genet. 1996;98:138–140. doi: 10.1007/s004390050176. [DOI] [PubMed] [Google Scholar]
- Tachi N, Ohya K, Chiba S, Sato T, Kikuchi K. Minimal somatic instability of CTG repeat in congenital myotonic dystrophy. Pediatr Neurol. 1995;12:81–83. doi: 10.1016/0887-8994(94)00112-f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.