Abstract
The type and the amount of dietary fat have a significant influence on the metabolic pathways involved in the development of obesity, metabolic syndrome, diabetes type 2 and cardiovascular diseases. However, it is unknown to what extent this modulation is achieved through DNA methylation. We assessed the effects of cholesterol intake, the proportion of energy intake derived from fat, the ratio of polyunsaturated fatty acids (PUFA) to saturated fatty acids (SFA), the ratio of monounsaturated fatty acids (MUFA) to SFA, and the ratio of MUFA+PUFA to SFA on genome-wide DNA methylation patterns in normal-weight and obese children. We determined the genome-wide methylation profile in the blood of 69 Greek preadolescents (∼10 years old) as well as their dietary intake for two consecutive weekdays and one weekend day. The methylation levels of one CpG island shore and four sites were significantly correlated with total fat intake. The methylation levels of 2 islands, 11 island shores and 16 sites were significantly correlated with PUFA/SFA; of 9 islands, 26 island shores and 158 sites with MUFA/SFA; and of 10 islands, 40 island shores and 130 sites with (MUFA+PUFA)/SFA. We found significant gene enrichment in 34 pathways for PUFA/SFA, including the leptin pathway, and a significant enrichment in 5 pathways for (MUFA+PUFA)/SFA. Our results suggest that specific changes in DNA methylation may have an important role in the mechanisms involved in the physiological responses to different types of dietary fat.
Introduction
According to the World Health Organization1, worldwide obesity has nearly doubled since 1980, resulting in an increase in cardiovascular diseases and diabetes type 2. One of the possible causes to this negative development is the increase of consumption of energy-dense foods that are high in fat. Dietary guidelines do not only recommend to eat a moderate amount of fat, but they also recommend to consume the right type of fat.2 Fatty acids include saturated fatty acids (SFA), monounsaturated fatty acids (MUFA) or polyunsaturated fatty acids (PUFA), and their structural differences explain why they have different biological effects.3 Consuming PUFA or MUFA instead of SFA is known to improve the blood lipid profile.4 Moreover, consumption of SFA in place of MUFA may worsen glucose-insulin homeostasis.5 Finally, replacing SFA with PUFA has been reported to lower coronary heart disease risk.6
Some of the effects of the qualitative and quantitative aspects of fat intake have been imputed to a modification of the transcription of key genes involved in pathways related to lipid and glucose metabolism, and/or inflammation.7 The regulation of gene expression can be achieved by mechanisms other than changes in the nucleotide sequence, namely epigenetic processes. Such processes are responsible for the establishment, maintenance, and reversal of metastable transcriptional states.8 One major example of such processes is the methylation of cytosine, usually at CpG dinucleotides, called DNA methylation. Regions rich in CpGs are called ‘CpG islands' and are mostly unmethylated when located in the promoter of active genes. Conversely, methylated promoters are associated with reduced gene expression.9
Five studies have investigated the link between DNA methylation and fat intake in humans, but the methylation assays in those studies were limited to only few key genes. One study found a significantly higher methylation in the peroxisome proliferator-activated receptor coactivator-1 gene (PPARGC1A) in high-fat overfed men.10 Another study found that the clock circadian regulator gene (CLOCK) methylation was negatively associated with MUFA intake, but positively associated with PUFA intake.11 A third study showed that higher n-6 PUFA intake was associated with lower methylation in the promoter of tumor necrosis factor-α (TNFα).12 A fourth study found no significant correlation between a diet rich in fat and sucrose, and methylation of hydroxyacyl-coenzyme A dehydrogenase (HADH) and glucokinase (GCK) genes.13 The fifth paper reported a lack of correlation between four diets enriched in different types of fat and the methylation levels of leptin (LEP), leptin receptor (LEPR), and pro-opiomelanocortin (POMC) genes.14
Here we explore the genome-wide DNA methylation profiles of Greek preadolescents with respect to parameters related to dietary fat quantity, and dietary fat quality. To our knowledge, this is the first time that parameters related to both quantitative and qualitative aspects of fat intake with respect to DNA methylation are investigated at a genome-wide scale. Moreover, no such studies have been performed in children.
Materials and Methods
Genome-wide changes of DNA methylation pattern associated with parameters related to fat intake were assessed. Two variables related to dietary fat quantity (proportion of energy intake derived from fat, cholesterol intake) and three related to dietary fat quality (MUFA/SFA, PUFA/SFA and (MUFA+PUFA)/SFA) were analyzed. A linear model that explains the methylation level for each CpG site/island corrected for gender, weight category, Tanner stage (an estimation of physical development), and white blood cell count was utilized. The ratios between the unsaturated and saturated fatty acid intakes were preferred to their individual values, as they have been reported to have antagonistic effects. A higher fatty acids ratio would account for a ‘healthier' fatty acid intake profile, while a lower ratio would account for an ‘unhealthier' fatty acid intake profile.
Ethics
All participants and their guardians gave informed written consent and the study was approved by the Greek Ministry of National Education (7055/C7-Athens, 19-01-2007) and the Ethical Committee of Harokopio University (16/ Athens, 19-12-2006).
Subjects
The ‘Healthy Growth Study' was a cross-sectional epidemiological study initiated in May 2007. Approval to conduct the study was granted by the Greek Ministry of National Education (7055/C7-Athens, 19-01-2007) and the Ethics Committee of Harokopio University of Athens (16/Athens, 19-12-2006). The study population comprised school children attending the fifth and sixth grades of primary schools located in the regions of Attica, Etoloakarnania, Thessaloniki and Heraklion. The sampling procedure is fully described elsewhere.15 For the purpose of the current analysis, a subsample of 24 obese and 23 normal-weight preadolescent girls, as well as 11 obese and 11 normal-weight preadolescent boys (Table 1) was selected. This subsample was initially used to investigate the effect of polymorphism in the FTO gene on genome-wide DNA methylation patterns.16
Table 1. Demographic data stratified for weight category and gender.
| Gender | Normal-weight | Obese | P-valuea |
|---|---|---|---|
| Male | |||
| N | 11 | 11 | |
| Age (years) | 10.34±0.25 | 10.82±0.56 | 0.03 |
| Height (z-score)b | −1.0±0.20 | 0.44±0.21 | <0.001 |
| Weight (z-score)b | −0.94±0.093 | 1.5±0.15 | <0.001 |
| BMI (z-score)b | −0.71±0.11 | 1.6±0.19 | <0.001 |
| White blood cell count (103/mm3) | 8.56±3.85 | 6.93±1.27 | n.s. |
| Tanner stagec (z-score)b | −1.5±0.27 | −1.5±0.32 | n.s. |
| Total fat intake (% of total energy intake) | 39.92±8.96 | 45.82±9.54 | n.s. |
| MUFA intake (% of total energy intake) | 19.00±5.97 | 21.74±5.91 | n.s. |
| PUFA intake (% of total energy intake) | 7.83±7.99 | 4.77±0.92 | n.s. |
| SFA intake (% of total energy intake) | 13.32±3.62 | 16.29±3.54 | n.s. |
| Cholesterol (g/day) | 188.90±101.84 | 304.49±137.54 | n.s. |
| Female | |||
| N | 23 | 24 | |
| Age (years) | 10.54±0.46 | 10.94±0.71 | 0.05 |
| Height (z-score)b | −0.76±0.16 | 0.30±0.24 | 0.001 |
| Weight (z-score)b | −0.86±0.088 | 1.5±0.19 | <0.001 |
| BMI (z-score)b | −0.72±0.079 | 1.8±0.13 | <0.001 |
| White blood cell count (103/mm3) | 7.26±2.07 | 7.28±1.70 | n.s. |
| Tanner stagec (z-score)b | −1.45±0.14 | −0.70±0.21 | n.s. |
| Total fat intake (% of total energy intake) | 42.22±6.59 | 38.42±7.66 | n.s. |
| MUFA intake (% of total energy intake) | 18.83±4.85 | 17.96±±5.15 | n.s. |
| PUFA intake (% of total energy intake) | 7.56±11.42 | 4.30±2.13 | 0.04 |
| SFA intake (% of total energy intake) | 15.55±2.98 | 13.67±4.28 | 0.04 |
| Cholesterol (g/day) | 216.25±89.86 | 211.04±125.58 | n.s. |
Abbreviations: BMI, body mass index; MUFA, monounsaturated fatty acid; n.s., nonsignificant; PUFA, polyunsaturated fatty acid; SFA, saturated fatty acid.
Indicates P-value for significant or n.s. differences between obese and normal-weight individuals. All values are means±SEs.
z-Scores were calculated using all samples from the Healthy Growth Study as a reference population.
Describes pubertal development.
Dietary assessment
Dietary intake data was obtained for two consecutive weekdays and one weekend day, via morning interviews with the children at the school site using the 24-h recall technique. More specifically, all study participants were asked to describe the type and amount of foods and beverages consumed during the previous day, provided that it was a usual day according to the participant's perception. To improve the accuracy of food descriptions, standard household measures (cups, tablespoons, etc) and food models were used to define amounts. At the end of each interview, the interviewers, who were dietitians rigorously trained to minimize interviewer's effect, reviewed the collected food intake data with the respondent to clarify entries, servings and possible forgotten foods. Food intake data were analyzed using the Nutritionist V diet analysis software (version 2.1, 1999, First Databank, San Bruno, CA, USA), which was extensively amended to include traditional Greek recipes, as described in Food Composition Tables of Greek Cooked Foods and Dishes. Furthermore, the database was updated with nutritional information of processed foods provided by independent research institutes, food companies and fast-food chains.
DNA methylation profiling
The genome-wide Illumina Infinium HumanMethylation27 BeadChip (Illumina, San Diego, CA, USA), which allows interrogation of 27 578 CpG dinucleotides covering 14 495 genes was applied to determine the methylation profile of genomic DNA isolated and purified from the peripheral whole blood. This chip has been shown to give a reliable and reproducible estimation of the methylation profile on a genomic scale.17 First, bisulfite conversion of genomic DNA was performed using the EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA, USA) according to the manufacturer's protocol. Briefly, 500 ng of DNA was sodium bisulfite-treated, denatured at 98 °C for 10 min, and bisulfite converted at 64 °C for 2.5 h. After conversion, samples were desulfonated and purified using column preparation. Approximately 200 ng of bisulfate-converted DNA was processed according to the Illumina Infinium Methylation Assay protocol. This assay is based on the conversion of unmethylated cytosine (C) nucleotides into uracil/thymine (T) nucleotides by the bisulfite treatment. The DNA was whole-genome amplified, enzymatically fragmented, precipitated, resuspended, and hybridized overnight at 48 °C to locus-specific oligonucleotide primers on the BeadChip. After hybridization, the C or T nucleotides were detected by single-base primer extension. The fluorescence signals corresponding to the C or T nucleotides were measured from the BeadChips using the Illumina iScan scanner. Phenotypes, raw data and background-corrected normalized DNA methylation data are available through the GEO database (http://www.ncbi.nlm.nih.gov/geo/) with accession numbers GSE27860 for the girls and GSE57484 for the boys.
Data processing
All downstream data processing and statistical analyses were performed with the statistical software R (www.r-project.org) together with the lumi,18 limma19 and IMA20 packages of the Bioconductor project.
Data preprocessing
The fluorescence data were preprocessed using the GenomeStudio 2009.2 (Illumina) software. We used the log2 ratio of the intensities of methylated probe versus unmethylated probe, also called M-value, which is more statistically valid for the differential analysis of methylation levels.21
Quality control
The data were imported and submitted to quality control using a modified version of the IMA.methy450PP function of the IMA package. The following CpG sites and samples were removed: the sites with missing β-values, the sites with detection P-value>0.05, the sites having <75% of samples with detection P-value<10−5, the samples with missing β-values, the samples with detection P-value>10−5 and the samples having <75% of sites with detection P-value<10−5. A total of 26 168 probes were included in the analysis, after discarding 328 probes that did not reach the quality control together with 1082 probes from the sex chromosomes.
Normalization
Quantile normalization was performed on the M-values of all the 26 168 CpG sites using the lumiMethyN function of the lumi package.
Annotation
For better interpretation of the genome-wide methylation patterns, we chose to use the expanded annotation table for the Illumina Infinium HumanMethylation450 BeadChip array generated by Price et al.22 There are a total of 27 578 loci for 27k array, and 1600 of them are not mapped to 450k array. For those unmapped loci, we kept their original annotation from the 27k array. The expanded annotation file was used to determine the average methylation value of CpG sites belonging to the same island or island shores (all sites with the same name in the ‘HIL_CpG_Island_Name' column of the annotation file were averaged). We obtained the average methylation value of 5980 islands/island shores, which reduced the number of interrogated locations to 19 437 sites/islands. The CpG island classification developed by Price et al22 provides good enrichment discrimination of CpG islands. This classification is a combination of Weber et al's classification23 where CpG islands are defined according to the GC content, the Obs/Exp CpG ratio and the island length, and Illumina's classification, where CpG islands are defined according to their physical position (islands, island shores, and shelves). The location within a CpG island or shore are suggested to be relevant,24 and Price et al's definition of CpG islands allowed to distinguish different methylation distribution between probes, which remained undetectable with the Illumina CpG island classification.22 Besides, their classification demonstrated a more extreme DNA methylation profile and a larger proportion of differentially methylated regions between different tissues.
The expanded annotation file was also used to determine which gene each interrogated CpG site/island may be associated with (‘Closest_TSS_gene_name' column of the annotation file), the distance of each interrogated CpG site/island to the closest TSS (transcription start site) (‘Distance_closest_TSS' column of the annotation file) and the CpG density surrounding each interrogated CpG site/island (‘HIL_CpG_class' column of the annotation file). Each site can either be located in a high-density CpG island, an intermediate-density CpG island, a region of intermediate-density CpG island that borders HCs, or a non-island. Indeed, the local CpG density has been shown to influence the role of methylated cytosines, with methylation having more transcriptional effect in high-density CpG island and less at non-islands.25
The Illumina-provided MAPINFO GenomeStudio column was used to determine the genomic location of each interrogated CpG site. For CpG islands, the name of the island was used to determine its genomic location (eg the island ‘chr19_IC:17905037-17906698' would be a CpG island of intermediate density located on chromosome 19, between 17 905 037 and 17 906 698).
Statistics
Linear model
We developed the following linear model for each CpG site k, using limma's robust regression method, with a maximum number of iteration equal to 10 000:
where Mk is the M-value of CpG site/island k, G is the dichotomized gender (female=1 and male=0), T is the Tanner stage, B is the white blood cell count, W is the dichotomized weight category (normal-weight=0 and obese=1), ɛk is the unexplained variability, and V is one of the following variables: proportion of energy intake derived from total fat intake, cholesterol intake (g/day), MUFA/SFA, PUFA/SFA, or (MUFA+PUFA)/SFA.
The coefficients bkx summarize the correlation between the methylation level and the variables of interest. Moderated t-statistics for each contrast and CpG site/island were created using an empirical Bayes model, to rank genes in order of evidence for differential methylation.19 To control the proportion of false positives, P-values were adjusted for multiple comparisons as proposed by Benjamini and Hochberg (BH).26 An adjusted P-value>0.05 was considered nonsignificant.
Three children from the cohort had a MUFA/SFA, a PUFA/SFA, and a (MUFA+PUFA)/SFA higher than the mean±3 × SD. Thus, they were excluded from the linear models developed for MUFA/SFA, PUFA/SFA, and (MUFA+PUFA)/SFA.
Functional enrichment analysis
The unique Entrez Gene ID associated with each significant gene-based site/island was identified. Three gene lists were generated for MUFA/SFA, PUFA/SFA, and (MUFA+PUFA)/SFA, respectively.
We used the web-based ConsensusPathDB-human (CPDB)27, 28 to determine the significant pathways each gene list may be involved in. On the basis of the reference gene set (all Entrez Gene IDs from the 27k BeadChip annotation file were used as a background), the expected number of genes in each pathway of the CPDB database is compared with the actual number of genes found for this pathway. For each pathway, a P-value and a q-value are calculated according to the hypergeometric test. The pathways with a raw P-value<0.05 together with a q-value<0.05 were selected. As CPDB includes information from 30 databases, the pathways often overlap with each other to some extent. Thus, to show the relationships between the different pathways, we constructed a heatmap of the proportion of shared input genes between the significant pathways. For instance, if P1 is a given pathway containing genes A, B, and C from the input gene list, and P2 is a given pathway containing genes B, C, D, and E from the input gene list, the proportion of shared genes between P1 and P2 is:
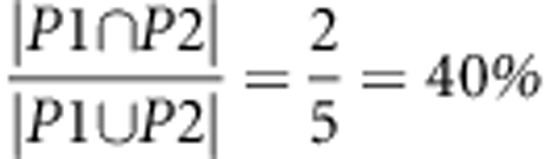 |
We also used the web-based g:Profiler29, 30 as an alternative method for pathway analysis, to confirm the significant results obtained with CPDB. The g:GOSt tool was used for enrichment analysis, with the same background gene list, and the g:GOSt native method g:SCS for multiple testing correction. The pathways with an adjusted P-value<0.05 were selected. It is important to note that g:Profiler only includes pathways from two databases: KEGG and Reactome.
Results
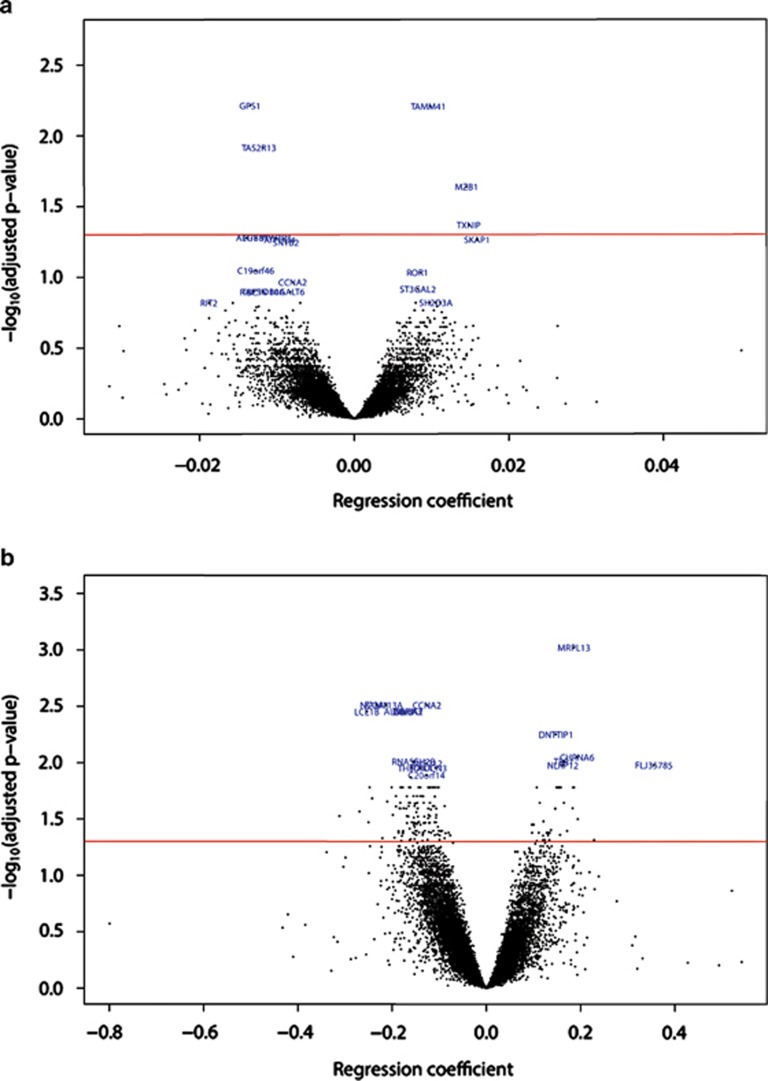
Four CpG sites and one CpG island were found to be significantly associated with the proportion of overall fat intake (Figure 1a), while no significance was found for cholesterol intake. The methylation levels of 2 islands, 11 island shores, and 16 sites were significantly correlated with PUFA/SFA; 9 islands, 26 island shores, and 158 sites for MUFA/SFA; 10 islands, 40 island shores, and 130 sites for (MUFA+PUFA)/SFA (Figure 1b and Supplementary Tables 1–3).
Figure 1.
Volcano plots for proportion of total energy intake derived from fat (a) and (MUFA+PUFA)/SFA (b). The regression coefficient refers to the coefficient of the linear model and each point represents a CpG site or a CpG island. The red horizontal line is the significance threshold (P-value=0.05) and all points above this line are significant. (a) proportion of total energy intake derived from fat (positive coefficients refer to an increased methylation in children for whom fat represents a higher proportion of total energy intake). (b) (MUFA+PUFA)/SFA (positive coefficients refer to an increased methylation in children having a higher (MUFA+PUFA)/SFA).
What genes are associated with the significant CpG sites/islands?
To determine which gene may be regulated by each CpG site and island, we identified the gene whose TSS is closest to each CpG site and island. Each significant site, island or island shore can show either a positive fold change if its methylation is higher in children having an elevated dietary variable (eg, a higher cholesterol intake), or a negative fold change if its methylation is lower in children having an elevated dietary variable.
Regarding the proportion of fat intake, one CpG site associated with taste receptor, type 2, member 13 (TAS2R13) that may have a role in the perception of bitterness, while another site associated with thioredoxin interacting protein (TXNIP), a regulator of cellular oxidative stress downregulated by SFA uptake31 (Table 2).
Table 2. Information on the significant CpG sites/island found for proportion of energy intake derived from fat and the top 10 most significant CpG sites/islands found for MUFA/SFA, PUFA/SFA, and (MUFA+PUFA)/SFA.
| Gene | Entrez Gene ID | Genomic location of the probe/island (hg19) | HIL classa | Genomic location of the closest TSS (hg19) | Coefficientb | Adjusted P-valuec |
|---|---|---|---|---|---|---|
| Proportion of energy intake derived from fat | ||||||
| GPS1 | 2873 | chr17:80009807 | HC | 80 009 762 | −0.0135 | 0.00612 |
| TAMM41 | 13 2001 | chr3_HCshore:11887600_11888782; chr3_ICshore:11887684_11888691 | HC | 11 888 351 | 0.00987 | 0.00621 |
| TAS2R13 | 50 838 | chr12:11061985 | LC | 11 062 160 | −0.0118 | 0.0121 |
| MZB1 | 51 237 | chr5:138725350 | LC | 138 725 604 | 0.0145 | 0.023 |
| TXNIP | 10 628 | chr1:145438031 | IC | 145 438 461 | 0.0148 | 0.043 |
| MUFA/SFA | ||||||
| ALDH3A2 | 224 | chr17:19552343 | HC | 19 552 063 | −0.289 | 0.00097 |
| MYLK3 | 91 807 | chr16:46782176 | LC | 46 782 220 | −0.238 | 0.00363 |
| LOC642852 | 257 103 | chr21:46716835 | LC | 46 707 966 | −0.317 | 0.00364 |
| TPPP2 | 122 664 | chr14:21498837 | IC | 21 498 344 | −0.309 | 0.00364 |
| RXFP2 | 122 042 | chr13:32313824 | NA | 32 313 679 | −0.262 | 0.00364 |
| TMEM80 | 283 232 | chr11_HCshore:694282_696564; chr11_ICshore:694282_697179 | HC | 695 615 | −0.245 | 0.00364 |
| SEMA3G | 56 920 | chr3:52478874 | HC | 52 479 042 | 0.28 | 0.00388 |
| VCAM1 | 7412 | chr1:101185020 | LC | 101 185 195 | −0.259 | 0.00482 |
| KRT73 | 319 101 | chr12:53013281 | LC | 53 012 342 | −0.245 | 0.00496 |
| KRTCAP2 | 200 185 | chr1:155145737 | HC | 155 145 803 | −0.301 | 0.0051 |
| PUFA/SFA | ||||||
| CBR1 | 873 | chr21_HCshore:37441920_37443032; chr21_ICshore:37442016_37442892 | HC | 37 442 284 | 1.28 | 4.02e–06 |
| RBCK1 | 10 616 | chr20:388351 | HC | 388 708 | 0.687 | 2.3e–05 |
| ABHD16A | 7920 | chr6_HCshore:31670422_31671462; chr6_ICshore:31670279_31671902 | HC | 31 671 136 | −0.302 | 7.18e–05 |
| KRT23 | 25 984 | chr17:39095141 | LC | 39 093 835 | −0.326 | 0.00536 |
| PDE3A | 5139 | chr12_HCshore:20521268_20523183; chr12_ICshore:20520944_20523341 | HC | 20 522 178 | −0.274 | 0.0066 |
| NCOA1 | 8648 | chr2:24806720 | LC | 24 807 344 | −0.42 | 0.00722 |
| PCED1A | 64 773 | chr20:2822804 | LC | 2 821 796 | −0.412 | 0.00914 |
| MRPL13 | 27 085 | chr8:121457500 | HC | 121 457 646 | 0.308 | 0.0193 |
| AKR7A2 | 54 896 | chr1_HCshore:19638013_19639253; chr1_ICshore:19637904_19639606; | HC | 19 638 639 | 0.237 | 0.0193 |
| FAM154A | 158 297 | chr9_IC:19032509_19033364 | IC | 19 033 255 | −0.357 | 0.0193 |
| (MUFA+PUFA)/SFA | ||||||
| MRPL13 | 27 085 | chr8:121457500 | HC | 121 457 646 | 0.186 | 0.000952 |
| NCOA1 | 8648 | chr2:24806720 | LC | 24 807 344 | −0.233 | 0.00308 |
| PCED1A | 64773 | chr20:2822804 | LC | 2821796 | −0.213 | 0.00308 |
| CCNA2 | 890 | chr4_HCshore:122744257_122745486; chr4_ICshore:122744093_122745437 | HC | 122 745 087 | −0.126 | 0.00308 |
| LCE1B | 353 132 | chr1:152783674 | LC | 152 784 446 | −0.254 | 0.00352 |
| ALDH3A2 | 224 | chr17:19552343 | HC | 19 552 063 | −0.176 | 0.00352 |
| MYLK3 | 91 807 | chr16:46782176 | LC | 46 782 220 | −0.166 | 0.00352 |
| GBP7 | 388 646 | chr1:89641121 | LC | 89 641 722 | −0.175 | 0.00352 |
| DGKI | 9162 | chr7_HCshore:137530917_137532628; chr7_ICshore:137530976_137532560 | HC | 137 531 608 | −0.178 | 0.00352 |
| DNTTIP1 | 140 686 | chr20:44421526 | LC | 44 420 575 | 0.148 | 0.00561 |
Abbreviations: HC, high-density CpG island; IC, intermediate-density CpG island; LC, non-island; MUFA, monounsaturated fatty acid; PUFA, polyunsaturated fatty acid; SFA, saturated fatty acid.
CpG density surrounding each interrogated CpG site/island.
Value of the coefficient of the linear model associated with (MUFA+PUFA)/SFA.
P-value calculated by moderated t-statistics and adjusted for multiple comparisons according to Benjamini and Hochberg.
The 10 most significant sites/islands/island shores found for MUFA/SFA included one CpG site associated with aldehyde dehydrogenase 3 family, member A2 (ALDH3A2) (P=0.00097), whose expression is reduced in insulin-resistant murine models.32 It also included a CpG site associated with sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3G (SEMA3G) (P=0.0039), whose expression increases during adipogenesis.33 Among the top 10 found for PUFA/SFA, there was 1 CpG site associated with nuclear receptor coactivator 1 (NCOA1) (P=0.0072) and another 1 associated with PC-esterase domain containing 1A (PCED1A) (P=0.0091), as well as an island shore associated with phosphodiesterase 3A, cGMP-inhibited (PDE3A) (P=0.0066; Table 2).
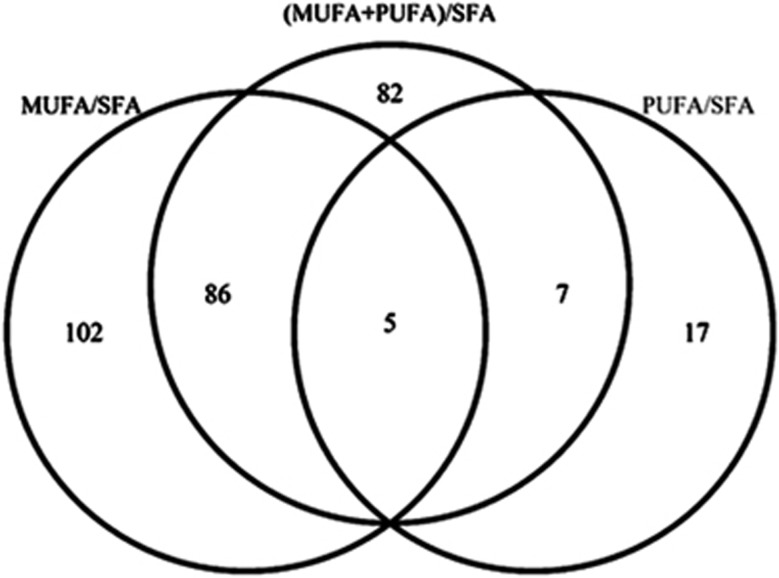
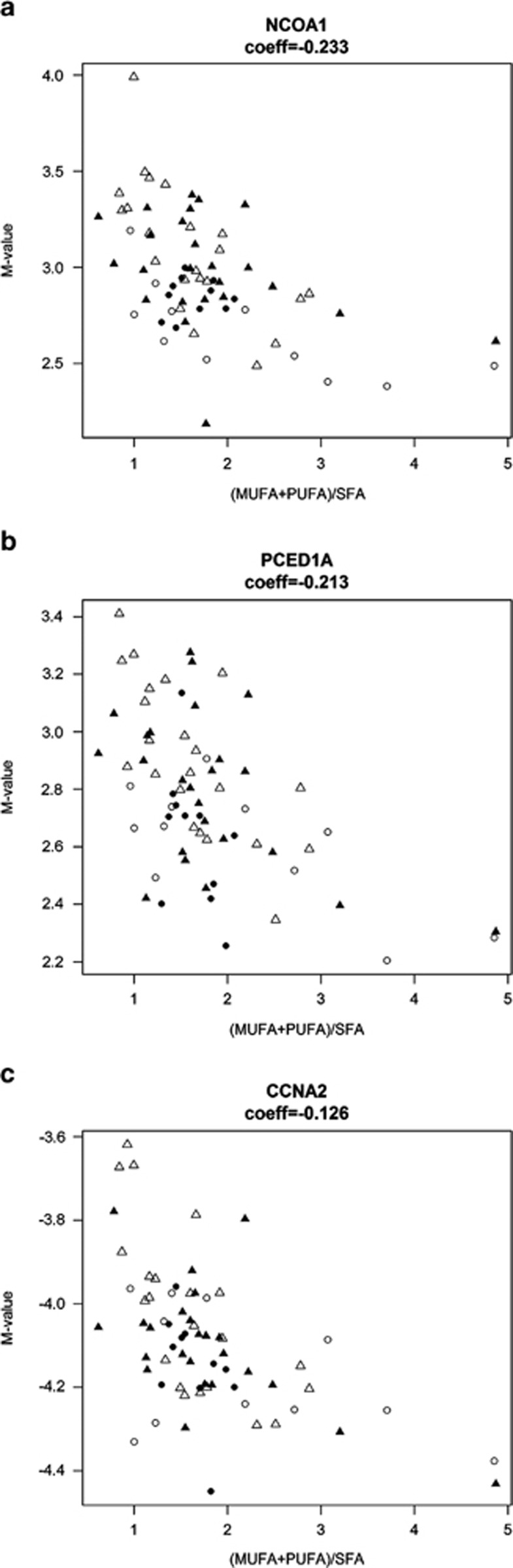
There were only 4 sites and 1 island shore found significant for all fatty acid ratios, but 86 sites/islands/island shores in common between MUFA/SFA and (MUFA+PUFA)/SFA, and 7 in common between PUFA/SFA and (MUFA+PUFA)/SFA (Figure 2). Notably, the four sites found significant for all fatty acid ratios contained some of previously mentioned sites (Table 2), for example, the ones associated with NCOA1 (P=0.0031) (Figure 3a) and PCED1A (P=0.0031) (Figure 3b). It also included an island shore associated with CCNA2 (Figure 3c), a gene recently shown to be associated with serum phosphatidylcholine concentration in mice.34
Figure 2.
Venn diagram of the significant CpG sites and islands found for MUFA/SFA, PUFA/SFA, and (MUFA+PUFA)/SFA.
Figure 3.
Correlation between methylation of three sites associated with NCOA1 (a), PCED1A (b), CCNA2 (c), and (MUFA+PUFA)/SFA. Coeff (coefficient) of the linear model associated with (MUFA+PUFA)/SFA; full triangles, obese girls (n=23); full circles, obese boys (n=11); empty triangles, normal-weight girls (n=22); empty circles, normal-weight boys (n=10).
In which pathways are the significant genes involved?
Instead of going through all the genes associated with the significant sites found for MUFA/SFA, PUFA/SFA, and (MUFA+PUFA)/SFA, it was preferred to perform a gene enrichment analysis. Using CPDP,27 we identified the significant pathways for each of the fatty acid ratios. We considered a pathway significant if the significant CpG sites/island/island shores were associated with a high proportion of genes involved in this particular pathway.
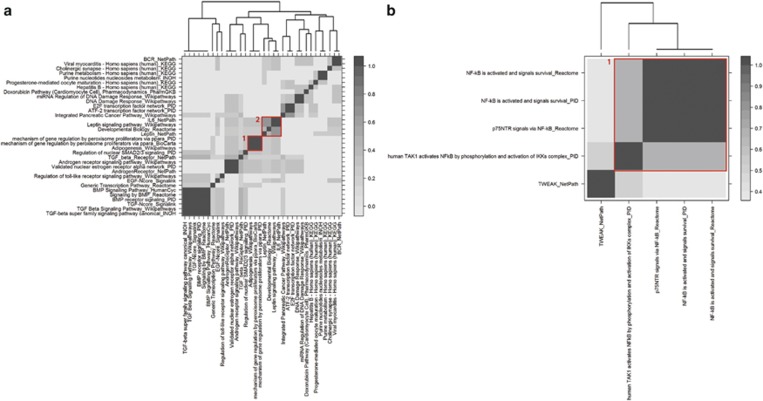
Neither CPDB nor g:Profiler identified significant pathways for MUFA/SFA, but CPDB found 34 significant pathways for PUFA/SFA (Supplementary Table 4), including 1 group of pathways related to adipogenesis and mechanism of gene regulation by peroxisome proliferators via PPARα (Group 1, Figure 4a), and another group of pathways related to leptin and IL6 (Group 2, Figure 4a). Five significant pathways were identified for (MUFA+PUFA)/SFA using CPDB (Supplementary Table 4), including one group of pathways linked to NF-κB (Group 1, Figure 4b). g:Profiler identified only one significant pathway for (MUFA+PUFA)/SFA, also linked to NF-κB (IKKβ phosphorylates IkB causing NF-κB to dissociate, P-value=0.041).
Figure 4.
Heatmap representation of the proportion of shared genes between the significant pathways found for PUFA/SFA (a) and (MUFA+PUFA)/SFA (b). Each significant pathway retrieved from CPDB (P-value<0.05 and q-value<0.05) is represented on the graph, along with the database it comes from. A stronger color indicates a higher proportion of shared genes between two pathways. (a) Red rectangle 1: group of pathways related to adipogenesis and mechanism of gene regulation by peroxisome proliferators via PPARα; red rectangle 2: group of pathways related to leptin and IL6. (b) Red rectangle 1: group of pathways related to NF-κB. The full colour version of this figure is available at European Journal of Human Genetics online.
Discussion
In the present study of Greek preadolescents, we found a large number of CpG sites and regions significantly associated with variables related to the quality of fat intake and few sites significantly associated with variables related to the quantity of fat intake.
Our findings suggest that fat quality is likely to influence DNA methylation on a large genomic scale. NCOA1, one of the most significant gene found for all fatty acids ratios, is involved in the mechanism of gene regulation by peroxisome proliferators via PPARα, a master gene whose regulation is altered in obesity.35 NCOA1 is a transcriptional coactivator whose ablation confers susceptibility to diet-induced obesity.36 Interestingly, various fatty acids, but especially PUFAs, act as ligands for PPARα. Moreover, along with PDE3A, the fifth most significant gene found for PUFA/SFA, NCOA1, is part of the leptin pathway. Leptin is an adipokine that has a key role in regulating energy intake by inhibiting the sensation of hunger.37 Fish oil has been reported to increase plasma leptin concentrations,38 and leptin induces the expression of NCOA1 in human cells.39 Besides, PDE3A's expression is enhanced in cows fed with a diet enriched in fish oil or in SFA.40 Interestingly, an island shore located near the TSS of PDE3A was less methylated in children with a higher PUFA/SFA. All this information is consistent with the negative fold change observed for NCOA1 in our cohort.
There was substantial overlap between the significant sites/islands/island shores found for the different fatty acid ratios, but little overlap between all fatty acid ratios. This may reflect how MUFA and PUFA affect DNA methylation in a different way. Interestingly, the site associated with NCOA1 was more significant for (MUFA+PUFA)/SFA than for PUFA/SFA or MUFA/SFA, suggesting that PUFA and MUFA affect the methylation of this gene in an identical way. A similar observation can be made for PCED1A and CCNA2 that were more significant for (MUFA+PUFA)/SFA than for PUFA/SFA or MUFA/SFA. However, this may also be due to differences in power to detect significant correlations, as the fatty acids ratios distributions were quite different (Supplementary Figure 1).
It should be noted that two of the four individual CpG sites found to be significantly associated with the proportion of energy intake derived from fat might be relevant to obesity. It has been hypothesized that individuals with increased bitter taste sensitivity avoid antioxidant-rich vegetables because of their perceived bitterness, consuming instead sweet, fatty foods.41 The site associated with TAS2R13 was more methylated in children for whom fat represents a higher proportion of the total energy intake. In addition, children with a higher proportion of energy intake derived from fat had a higher methylation at a site located in an island shore near TXNIP, which is consistent with the observed downregulation of TXNIP by SFA uptake.31 None of these genes were previously reported to be differentially methylated depending on fat intake, probably because the methylation assays of previous studies were limited in scope only addressing key genes.
The present work was not devoid of limitations. First of all, our sample size is limited (n=69) and therefore replication is needed to confirm our findings and to allow generalization to larger populations. Second, the fatty acid ratios investigated herein are among the most interesting to compare with respect to health, as their roles are heavily debated and researched. However, other fatty acids not examined in this study may reflect other aspects of the quality of fat intake. For example, unsaturated fatty acids includes trans unsaturated fatty acids, which have been demonstrated to have adverse effects on health.42 In addition, we did not separate n-3 and n-6 PUFA in our study, but these two fatty acids do not have the same effects; while both n-3 and n-6 PUFA have beneficial effects, an excess of n-6 PUFA can cause health disorders.43 DNA methylation was assessed in whole peripheral blood, which is the case for most epigenetic studies focused on nutrition, as peripheral changes may occur in relation to overall energy balance.44 However, the methylation pattern observed in blood may not always reflect the pattern in other tissues.45 The other weakness of this approach is that DNA methylation can vary by blood cell type, and thus the methylation changes associated with the variables investigated in this study may represent an alteration in blood cell composition, rather than a change in methylation. However, no correlation was found between any of the investigated variables and the relative proportions of granulocytes, lymphocytes, or mid cells (P-value>0.05 on Spearman's correlation test). Finally, an increasing number of human studies suggest that parental BMI impacts DNA methylation in the offspring, especially at imprinted genes.46, 47, 48 However, evidences in humans are still scarce and limited to two available tissues at birth: umbilical cord and/or placenta; thus, we did not take parental BMI into account in our analysis.
In conclusion, this study is the first to demonstrate the roles of fat quantity and quality in DNA methylation patterns at a genome-wide scale. Our results suggest that specific changes in DNA methylation may have an important role in the mechanisms involved in the physiological responses to different types of dietary fat. Future studies could reveal other potential impacts of dietary fat quality on DNA methylation in controlled, randomized designs, and perhaps investigate further the downstream effects of this process.
Acknowledgments
The methylation array was performed at the Genotyping Technology Platform, Uppsala, Sweden (http://www.genotyping.se), with support from Uppsala University and the Knut and Alice Wallenberg foundation, and at the Uppsala Genome Centre. The studies were supported by the Swedish Research Council (VR, medicine), Ecole Normale Superieure de Lyon and the Novo Nordisk Foundation. We thank Miguel Xavier for his guidance, advice and support.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- World Health Organization Obesity and overweight. Media Cent. Fact sheet N 311 (2013). Available at http://www.who.int/mediacentre/factsheets/fs311/en/ .
- UN Food and Agriculture Organization. Fats and Fatty Acids in Human Nutrition. Rome, Italy: Publishing Policy and Support Branch, Office of Knowledge Exchange, Research and Extension, FAO, 2010.
- Tvrzicka E, Kremmyda L-S, Stankova B, Zak A. Fatty acids as biocompounds: their role in human metabolism, health and disease—a review. Part 1: classification, dietary sources and biological functions. Biomed Pap Med Fac Univ Palacky Olomouc Czechoslov. 2011;155:117–130. doi: 10.5507/bp.2011.038. [DOI] [PubMed] [Google Scholar]
- Micha R, Mozaffarian D. Trans fatty acids: effects on metabolic syndrome, heart disease and diabetes. Nat Rev Endocrinol. 2009;5:335–344. doi: 10.1038/nrendo.2009.79. [DOI] [PubMed] [Google Scholar]
- Vessby B, Uusitupa M, Hermansen K, et al. Substituting dietary saturated for monounsaturated fat impairs insulin sensitivity in healthy men and women: The KANWU Study. Diabetologia. 2001;44:312–319. doi: 10.1007/s001250051620. [DOI] [PubMed] [Google Scholar]
- Jakobsen MU, Reilly EJO, Heitmann BL, et al. Major types of dietary fat and risk of coronary heart disease: a pooled analysis of 11 cohort studies 1–3. Am J Clin Nutr. 2009;89:1425–1432. doi: 10.3945/ajcn.2008.27124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremmyda L-S, Tvrzicka E, Stankova B, Zak A. Fatty acids as biocompounds: their role in human metabolism, health and disease - a review. Part 2: Fatty acid physiological roles and applications in human health and disease. Biomed Pap. 2011;155:195–218. doi: 10.5507/bp.2011.052. [DOI] [PubMed] [Google Scholar]
- Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science (80-) 2010;330:612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimshony T, Zhang J, Keshet I, Bustin M, Cedar H. The role of DNA methylation in setting up chromatin structure during development. Nat Genet. 2003;34:187–192. doi: 10.1038/ng1158. [DOI] [PubMed] [Google Scholar]
- Brøns C, Jacobsen S, Nilsson E, et al. Deoxyribonucleic acid methylation and gene expression of PPARGC1A in human muscle is influenced by high-fat overfeeding in a birth-weight-dependent manner. J Clin Endocrinol Metab. 2010;95:3048–3056. doi: 10.1210/jc.2009-2413. [DOI] [PubMed] [Google Scholar]
- Milagro FI, Gómez-Abellán P, Campión J, et al. CLOCK, PER2 and BMAL1 DNA methylation: association with obesity and metabolic syndrome characteristics and monounsaturated fat intake. Chronobiol Int. 2012;29:1–15. doi: 10.3109/07420528.2012.719967. [DOI] [PubMed] [Google Scholar]
- Hermsdorff HH, Mansego ML, Campión J, et al. TNF-alpha promoter methylation in peripheral white blood cells: Relationship with circulating TNFα, truncal fat and n-6 PUFA intake in young women. Cytokine. 2013;64:265–271. doi: 10.1016/j.cyto.2013.05.028. [DOI] [PubMed] [Google Scholar]
- Lomba A, Milagro FI, García-Díaz DF, et al. Obesity induced by a pair-fed high fat sucrose diet: methylation and expression pattern of genes related to energy homeostasis. Lipids Health Dis. 2010;9:60. doi: 10.1186/1476-511X-9-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Liu X, Shen W, Deckelbaum RJ, Qi K. The regulation of leptin, leptin receptor and pro-opiomelanocortin expression by n-3 PUFAs in diet-induced obese mice is not related to the methylation of their promoters. Nutr Metab (Lond) 2011;8:31. doi: 10.1186/1743-7075-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moschonis G, Tanagra S, Vandorou A, et al. Social, economic and demographic correlates of overweight and obesity in primary-school children: preliminary data from the Healthy Growth Study. Public Health Nutr. 2010;13:1693–1700. doi: 10.1017/S1368980010002247. [DOI] [PubMed] [Google Scholar]
- Almén MS, Schiöth HB, Fredriksson R, et al. Genome wide analysis reveals association of a FTO gene variant with epigenetic changes. Genomics. 2012;99:132–137. doi: 10.1016/j.ygeno.2011.12.007. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Le J, Barnes B, et al. Genome-wide DNA methylation profiling using Infinium®® assay. Epigenomics. 2009;1:24. doi: 10.2217/epi.09.14. [DOI] [PubMed] [Google Scholar]
- Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- Smyth GK.Linear models and empirical bayes methods for assessing differential expression in microarray experiments Stat Appl Genet Mol Biol 20043Article 3. [DOI] [PubMed] [Google Scholar]
- Wang D, Yan L, Hu Q, et al. IMA: an R package for high-throughput analysis of Illumina's 450K Infinium methylation data. Bioinformatics. 2012;28:729–730. doi: 10.1093/bioinformatics/bts013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P, Zhang X, Huang C, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price ME, Cotton AM, Lam LL, et al. Additional annotation enhances potential for biologically-relevant analysis of the Illumina Infinium HumanMethylation450 BeadChip array. Epigenetics Chromatin. 2013;6:4. doi: 10.1186/1756-8935-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- Michels KB, Binder AM, Dedeurwaerder S, et al. Recommendations for the design and analysis of epigenome-wide association studies. Nat Methods. 2013;10:949–955. doi: 10.1038/nmeth.2632. [DOI] [PubMed] [Google Scholar]
- Hackett JA, Surani MA. DNA methylation dynamics during the mammalian life cycle. Philos Trans R Soc B Biol Sci. 2013;368:20110328. doi: 10.1098/rstb.2011.0328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57:289–300. [Google Scholar]
- Kamburov A, Wierling C, Lehrach H, Herwig R. ConsensusPathDB—a database for integrating human functional interaction networks. Nucleic Acids Res. 2009;37:D623–D628. doi: 10.1093/nar/gkn698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamburov A, Stelzl U, Lehrach H, Herwig R. The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res. 2013;41:D793–D800. doi: 10.1093/nar/gks1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimand J, Arak T, Vilo J. g:Profiler—a web server for functional interpretation of gene lists (2011 update) Nucleic Acids Res. 2011;39:W307–W315. doi: 10.1093/nar/gkr378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimand J, Kull M, Peterson H, Hansen J, Vilo J. g:Profiler—a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007;35:W193–W200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaked M, Ketzinel-Gilad M, Cerasi E, Kaiser N, Leibowitz G. AMP-activated protein kinase (AMPK) mediates nutrient regulation of thioredoxin-interacting protein (TXNIP) in pancreatic beta-cells. PLoS One. 2011;6:e28804. doi: 10.1371/journal.pone.0028804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demozay D, Rocchi S, Mas J, et al. Fatty aldehyde dehydrogenase: potential role in oxidative stress protection and regulation of its gene expression by insulin. J Biol Chem. 2004;279:6261–6270. doi: 10.1074/jbc.M312062200. [DOI] [PubMed] [Google Scholar]
- Ullah M, Stich S, Häupl T, et al. Reverse differentiation as a gene filtering tool in genome expression profiling of adipogenesis for fat marker gene selection and their analysis. PLoS One. 2013;8:e69754. doi: 10.1371/journal.pone.0069754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer N, Yu Z, Wagener A, et al. Changes in metabolite profiles caused by genetically determined obesity in mice. Metabolomics. 2014;10:461–472. doi: 10.1007/s11306-013-0590-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway GP, Bonen A, Spriet LL. Regulation of skeletal muscle mitochondrial fatty acid metabolism in lean and obese individuals. Am J Clin Nutr. 2009;89:455S–462S. doi: 10.3945/ajcn.2008.26717B. [DOI] [PubMed] [Google Scholar]
- Picard F, Gehin M, Anicotte J, et al. SRC-1 and TIF2 control energy balance between white and brown adipose tissues. Cell. 2002;111:931–941. doi: 10.1016/s0092-8674(02)01169-8. [DOI] [PubMed] [Google Scholar]
- Brennan AM, Mantzoros CS. Drug Insight: the role of leptin in human physiology and pathophysiology[mdash]emerging clinical applications. Nat Clin Pr End Met. 2006;2:318–327. doi: 10.1038/ncpendmet0196. [DOI] [PubMed] [Google Scholar]
- Puglisi MJ, Hasty AH, Saraswathi V. The role of adipose tissue in mediating the beneficial effects of dietary fish oil. J Nutr Biochem. 2011;22:101–108. doi: 10.1016/j.jnutbio.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin N, Wang D, Zhang H, et al. Molecular mechanisms involved in the growth stimulation of breast cancer cells by leptin. Cancer Res. 2004;64:5870–5875. doi: 10.1158/0008-5472.CAN-04-0655. [DOI] [PubMed] [Google Scholar]
- Schmitt E, Ballou MA, Correa MN, et al. Dietary lipid during the transition period to manipulate subcutaneous adipose tissue peroxisome proliferator-activated receptor-gamma co-regulator and target gene expression. J Dairy Sci. 2011;94:5913–5925. doi: 10.3168/jds.2011-4230. [DOI] [PubMed] [Google Scholar]
- Garcia-Bailo B, Toguri C, Eny KM, El-Sohemy A. Genetic variation in taste and its influence on food selection. Omi J Integr Biol. 2009;13:69–80. doi: 10.1089/omi.2008.0031. [DOI] [PubMed] [Google Scholar]
- White B. Dietary fatty acids. Am Fam Phys. 2009;80:345–350. [PubMed] [Google Scholar]
- Lands B. Consequences of essential fatty acids. Nutrients. 2012;4:1338–1357. doi: 10.3390/nu4091338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pjetri E, Schmidt U, Kas MJ, Campbell IC. Epigenetics and eating disorders. Curr Opin Clin Nutr Metab Care. 2012;15:330–335. doi: 10.1097/MCO.0b013e3283546fd3. [DOI] [PubMed] [Google Scholar]
- McKay JA, Xie L, Harris S, et al. Blood as a surrogate marker for tissue-specific DNA methylation and changes due to folate depletion in post-partum female mice. Mol Nutr Food Res. 2011;55:1026–1035. doi: 10.1002/mnfr.201100008. [DOI] [PubMed] [Google Scholar]
- Soubry A, Murphy SK, Wang F, et al. Newborns of obese parents have altered DNA methylation patterns at imprinted genes Int J Obes (Lond) 2013. e-pub ahead of print 25 October 2013;doi: 10.1038/ijo.2013.193 [DOI] [PMC free article] [PubMed]
- Soubry A, Schildkraut JM, Murtha A, et al. Paternal obesity is associated with IGF2 hypomethylation in newborns: results from a Newborn Epigenetics Study (NEST) cohort. BMC Med. 2013;11:29. doi: 10.1186/1741-7015-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Chen Q, Tsai H, Wang G. Maternal preconception body mass index and offspring cord blood DNA methylation: exploration of early life origins of disease. Environ Mol Mutagen. 2013;55:223–230. doi: 10.1002/em.21827. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.