Abstract
Multiple primary malignant tumours (MPMT) are frequently taken as an indicator of potential inherited cancer susceptibility and occur at appreciable frequency both among unselected cancer patients and, particularly, among referrals to cancer genetics services. However, there is a paucity of information on the clinical genetic evaluation of cohorts of MPMT patients representing a variety of tumour types. We ascertained a referral-based series of MPMT cases and describe the patterns of tumours observed. Service-based molecular genetic testing had demonstrated a pathogenic germline variant in an inherited cancer gene in fewer than one in four unselected referrals. To assess for evidence of thus far unidentified variants in those who tested negative, comparisons were made with those who tested positive. This revealed considerable overlap between the two groups with respect to clinical characteristics indicative of an inherited cancer syndrome. We therefore proceeded to test a subset of unexplained MPMT cases (n=62) for pathogenic germline variants in TP53 and PTEN but none were detected. Individuals with MPMT may receive negative genetic test results for a number of reasons, which are discussed. Many of these may be addressed by the increasing application of next generation sequencing techniques such as inherited cancer gene panels.
Introduction
Multiple primary malignant tumours (MPMT) describes a scenario whereby two or more histologically distinct malignant tumours not due to metastasis, recurrence or local spread are diagnosed in the same individual. These may be diagnosed at the same time (synchronous) or separated by months to years (metachronous). The first description of MPMT is attributed to Billroth in 18891 and it initially appeared to be a rare phenomenon. However, with improved survival from many forms of cancer,2 MPMT is increasingly recognised as an important medical problem.3 Indeed, a review of 69 European cancer registries revealed that 6.3% of registered tumours were part of an MPMT clinical picture.4 Furthermore, registry-based evidence suggests that the incidence of cancer in previously diagnosed individuals is greater than the expected population incidence with an increased risk of a wide variety of concordant and discordant tumours after an initial primary malignancy.5
Multiple factors may contribute to the occurrence of MPMT. Thus, increased clinical surveillance following an initial diagnosis may lead to increased detection of second malignancies through lead-time bias or may identify cancers that would not present otherwise in the individual's lifetime. Alternatively, radiotherapy or cytotoxic chemotherapy regimens for the initial tumour may predispose to second primary tumours. Even non-cytotoxic drug treatment may increase cancer risk as is seen for endometrial cancer after tamoxifen treatment for breast cancer.6 Two or more tumours may also result from carcinogenic environmental exposures relevant to both cancer types.7 In addition, it is widely recognised that genetic susceptibility can be a major cause of MPMT and many monogenic familial cancer syndromes are associated with a high frequency of this phenomenon.8, 9, 10, 11, 12 Indeed, particular combinations of multiple tumours may suggest specific cancer syndromes (eg haemangioblastomas and renal cancers in von Hippel-Lindau disease). Accordingly, many patients with MPMT will be referred for clinical genetics evaluation because of a suspicion of such a syndrome. However, the outcome of such evaluation is not well described and although there are often large published series of individuals with a specific familial cancer syndrome, to our knowledge, there are no large studies of individuals with MPMT referred for clinical genetics assessment. In particular, it is highly relevant to know whether individuals with MPMT who test negative for a suspected familial cancer syndrome are likely to represent phenocopies or whether there is evidence to indicate a need for more extensive genetic testing. To address these questions, we undertook a retrospective review of referrals for MPMT to two regional genetics centres. We hypothesized that a group of patients with MPMT might harbour germline pathogenic variants in TP53 or PTEN and consequently initiated analysis of these genes in a subset of individuals.
Materials and methods
Ascertainment of cases
To identify MPMT cases referred for genetic assessment (directly or through a family member), we undertook a records-based interrogation of two UK Regional Genetics Services covering a combined population of >10 million.13, 14 Firstly, the West Midlands Regional Genetics Service database was used to identify individuals with two or more malignant tumours diagnosed before the age of 60. Referrals and genetic analysis had taken place between February 1993 and February 2013. Medical and pathology records were then inspected to confirm the inclusion criteria. Those individuals with benign histology, metastases, recurrence of the primary tumour or tumours of the same site and histological type were excluded from further analysis if those tumour characteristics led to non-fulfilment of the criteria. Multicentric or multifocal cancers were counted as a single malignancy. Thus, the definition of MPMT was made according to international guidelines.15 Additionally, two databases of individuals referred to the North West regional genetics service in Manchester with a suspected diagnosis of hereditary colorectal cancer or familial breast/ovarian cancer were interrogated to identify additional cases of MPMT satisfying the same criteria specified above.
Assessment of clinical indicators
To provide an indicator, in a broad range of clinical scenarios, of the strength of the clinical evidence for an inherited cancer syndrome, we developed a ‘multiple tumour score' (MTS). To record this score for individuals within the series, clinical genetics case records were inspected to extract details of family history, age at diagnosis and histopathological type of cancer. The MTS was designed to be analogous to the Manchester score for prioritising genetic testing in familial breast/ovarian cancer kindreds.16 This system assigns a score for each cancer of a particular type that occurs in a single lineage, with higher scores for tumours more characteristic of pathogenic BRCA1/BRCA2 variants (Supplementary Table 1). The sum of scores is incorporated into clinical guidelines, with ≥15 providing a threshold for testing in breast/ovarian cancer cases.17 Thus, within a single family lineage, the MTS was calculated according to whether a common or rare cancer had occurred and the age at which it had occurred (Table 1). Within the scoring system, rarer cancers were weighted more heavily than common cancers (as phenocopies should be less likely for the former). Cervical cancer is not a common cancer but as it is mostly attributable to human papilloma virus (HPV) infection18, 19 and it was therefore scored lower than other uncommon cancers. The development of this malignancy may involve a degree of genetic predisposition interacting with HPV infection. However, additional analyses were also performed excluding cases within the series with cervical cancer as part of their MPMT clinical presentation (results and Supplementary Information). For individuals with a diagnosis of breast or ovarian cancer, the Manchester score was calculated and fulfilment of the revised Bethesda criteria was noted if colorectal or endometrial cancer had been diagnosed. The latter criteria are designed to prompt microsatellite instability (MSI) analysis of colorectal tumours where one or more of five clinical indicators are suggestive of Lynch syndrome (Supplementary Table 2).20 Records of genetic testing were interrogated to obtain details of assays performed and variants identified (including whether deemed causative by the laboratory/clinical team). These are listed in Supplementary Table 3 and were uploaded to public databases (MMR/APC/MUTYH – chromium.liacs.nl/LOVD2/colon_cancer/home.php, BRCA1/BRCA2–research.nhgri.nih.gov/bic, PTEN/RB1–ncbi.nlm.nih.gov/clinvar). Statistical significance testing was performed using two-tailed z-tests.
Table 1. Multiple tumour score.
| Malignant tumour | Age at diagnosis | Score |
|---|---|---|
| Breast, lung, colorectal, prostate, non-melanoma skin, cervical | <30 | 5 |
| 30–39 | 4 | |
| 40–49 | 3 | |
| 50–59 | 2 | |
| >59 | 1 | |
| Any other malignant tumour | <50 | 5 |
| 50–59 | 3 | |
| >59 | 1 |
Sequencing of candidate genes
DNA samples from 62 patients with no detected causative variant and who had given consent for diagnostic genetic testing were subjected to whole genome amplification (REPLI-g Mini Kit, Qiagen, Hilden, Germany) according to the manufacturer's protocol, and targeted PCR amplification was then performed to amplify PTEN exons 1–9 and TP53 exons 2–11. A 25 μl reaction volume was used with 5 μl template DNA and 20 μl BioMix Red (Bioline, London, UK). Primers used are listed in Supplementary Table 4. PCR reaction products were cleaned with an equal volume of microCLEAN (Microzone, Haywards Heath, UK) and subsequently added to a sequencing reaction incorporating (per reaction) 0.5 μl BigDye Terminator v3.1 Ready Reaction Mix and 2 μl 5X sequencing buffer from the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Austin TX, USA). Also added were 2 μl of 2 mM primer solution and 5.5 μl water. Following an ethanol-based precipitation, products were suspended in 10 μl Hi-Di Formamide sequenced with an ABI 3730 automated sequencer (Applied Biosystems). Obtained chromatograms were analysed with the assistance of the Mutation Surveyor software package (Softgenetics, State College, PA, USA) using reference sequences generated from the GRCh37 assembly.
Results
Evaluation and analysis of a series of MPMT cases – West Midlands series
A total of 212 individuals with an MPMT phenotype (441 tumours) satisfied the eligibility criteria for inclusion. Most individuals (179, 84.4%) were female and breast cancer was the most frequent tumour type (Table 2). The most frequent combination of tumour types was breast with ovarian followed by breast with non-melanoma skin (NMSC), endometrial and colorectal cancers (Supplementary Table 5).
Table 2. Tumour frequency among MPMT subgroups.
| Tumour type | All tumours (n=441) | Testing sent (n=234) | No testing sent (n=208) | Test+ve group (n =96) | Test−ve group (n=138) |
|---|---|---|---|---|---|
| Breast | 128 (29%) | 69 (29.5%) | 59 (28.3%) | 21 (21.9%) | 48 (34.8%) |
| Ovarian | 54 (12.2%) | 35 (14.9%) | 18 (8.6%) | 14 (14.6%) | 21 (15.2%) |
| Colorectal | 71 (16.1%) | 34 (14.5%) | 37 (17.8%) | 21 (21.9%) | 13 (9.4%) |
| Endometrial | 43 (9.7%) | 28 (12%) | 16 (7.7%) | 12 (12.5%) | 16 (11.6%) |
| Non-melanoma skin | 42 (9.5%) | 22 (9.4%) | 21 (10.1%) | 7 (7.3%) | 15 (10.9%) |
| Other tumours individually comprising <5% total | 103 (23.3%) | 46 (19.6%) | 57 (27.4%) | 21 (21.9%) | 25 (18.1%) |
At least one gene relevant to inherited cancer had been analysed in 111 of the 212 (52.3%) individuals. Tests were performed in BRCA1 (n=71 tests), BRCA2 (n=72), MSH2 (n=32), MLH1 (n=29), MSH6 (n=11), PMS2 (n=2), APC (n=3), MUTYH (n=5), PTEN (n=4), TP53 (n=3) and RB1 (n=2). Eight of these were tests for known familial variants suspected to be present in the consultand with MPMT and two cases were obligate carriers. Of BRCA1 and BRCA2 analyses, 89% had sequenced the entire coding region with MLPA being conducted in the same proportion. Other strategies included sequencing of selected exons and heteroduplex analysis. Of the 66 (excluding familial tests) mismatch repair gene analyses performed, 48% sequenced the coding region and 33% were heteroduplex or single-strand conformation-based. MLPA was performed as part of 63% of tests. MSI and/or immunohistochemistry (IHC) analyses had been undertaken in 36 cases, with subsequent germline analysis following 10 positive and 4 negative results. Seven analyses (9%) were as part of a next generation sequencing (NGS) gene panel (bowel cancer) approach. Of the other 17 analyses performed, 3 also used this panel. All NGS analyses were among the last eight cases to be referred. Nineteen of 30 initially non-direct sequencing based approaches (PTT, SSCP, heteroduplex) were among the first 30 referred cases, whereas no such analyses were undertaken for the last 30 patients.
Comparison of tumour types between the 111 tested patients and the 101 individuals who were not tested revealed similar tumour frequencies (Table 2) with only thyroid cancer (higher P=<0.05) and ovarian cancer (lower P=<0.05) being significantly different in the untested group. There were no differences in gender distribution between tested and untested groups (94/111, 84.7% female in tested group vs 86/101, 85.1% female in untested group) but the mean age at diagnosis of first (41.5 vs 43.2), second/synchronous (48.8 vs 50.7) and all (45.2 vs 47.3) tumours was lower in those who had undergone testing. Where a particular tumour occurred and a genetic test was performed, a pathogenic variant was identified in between 27.3% (NMSC) and 61.8% (colorectal) of patients (Table 3). The relatively high diagnostic rates for colorectal and endometrial tumours are likely due to Lynch syndrome investigations prior to germline testing.
Table 3. Testing sent group – Genetic testing results by cancer type.
| Cancer type | No. of occurrences where patient had causative variant demonstrated |
|---|---|
| Breast | 21 (30.4%) |
| Ovarian | 14 (40%) |
| Colorectal | 21 (61.8%) |
| Endometrial | 11 (39.3%) |
| Non melanoma skin | 6 (27.3%) |
| Other tumours individually comprising <5% total | 22 (47.8%) |
Overall, 44 of the 111 (39.6%) patients who underwent germline genetic testing in one or more genes had a pathogenic variant demonstrated (designated Test+ve group) and 67 did not (designated Test−ve group). Of the cases, 22 had negative MSI and/or IHC analysis for suspected Lynch syndrome and did not proceed to germline testing. These cases were not assigned Test−ve status because of the fact that the sensitivity of these investigations is quoted as between 55 and 83% depending on the gene involved and the assay. Similarly, four cases who had positive MSI/IHC but did not have a germline test (because of death in three cases) were not assigned Test+ve status because of high but incomplete specificity.21 These individuals are nevertheless likely to have a mismatch repair deficit and further calculations were performed (see below) where these investigations were taken as negative or positive genetic tests, respectively.
Pathogenic variants were most frequent in mismatch repair genes (n=21, MSH2 (n=9), MLH1 (n=9), MSH6 (n=3)) or BRCA1/BRCA2 (n=16), BRCA1 (n=12) and BRCA2 (n=4). Seven patients harboured pathogenic variants in genes relevant to rarer syndromes (PTEN (n=3), RB1 (n=2), APC (n=1), MUTYH (homozygous) (n=1)). Comparison of the Test+ve and Test−ve groups revealed that the mean MTS was significantly higher (21 vs 14.1, P= <0.01) and the mean age at tumour diagnosis lower, though not statistically significant, (44 vs 46, P=0.3271) (Table 4) in the Test+ve group. The proportion of cases with a concordant tumour in a first-degree relative was also higher in the Test+ve group (63.6 vs 56.7% P=0.4654) though again, this was not statistically significant.
Table 4. Comparison of clinical indicators observed in West Midlands series.
| Clinical indicator | All cases (n=212) | Causative variant identified (Test+ve n=44) | No causative variant identified and ≥ 1 germline test (Test−ve n=67) | No causative variant identified (n=168) |
|---|---|---|---|---|
| Mean age at tumour diagnosis | 46.2 SD 10.2 | 44 SD 10.9 | 46 (SD 9.8, P=0.3271) | 46.8 (SD 9.9 1.85, P=0.0643) |
| Mean age diagnosis first tumour | 42.5 SD 11.3 | 39.4 SD 12.3 | 42.9 (SD 10, P=0.1141) | 43.1 (SD 10.7, P=0.3681) |
| Mean age diagnosis second or synchronous tumour | 49.6 SD 7.4 | 48.1 SD 6.8 | 49.2 (SD 8.4, P=0.4473) | 50.1 (SD 7.2, P=0.0891) |
| Mean individual age at tumour diagnosis | 46.2 SD 8.4 | 44.2 SD 8.6 | 46 (SD 8.6, P=0.2801) | 46.7 (SD 8.3, P=0.0819) |
| % cases with individual mean age at tumour diagnosis ≤ Test+ve group median (46.25) | N/A | N/A | 41.8 (28) | 39.9 (67) |
| % cases with concordant tumour in first degree relative | 52.3 (111) | 63.6 (28) | 56.7 (38) (P=0.4654) | 49.4 (83) (P=0.09296) |
| Mean multiple tumour score | 14.6 SD 8.7 | 21 SD 14.3 | 14.1 (SD 5.4, P= <0.01) | 12.9 (SD 5.6, P= <0.01) |
| % cases with multiple tumour score ≥ Test+ve group median (17) | N/A | N/A | 28.3 (19) | 21.4 (36) |
Mean individual age at diagnosis=Combined age of tumours/number of tumours in individual.
P-values describe two-tailed comparison with Mut+ group (H0=μ1=μ2).
There was, however, significant overlap between the MTS in the Test+ve and Test−ve groups such that ∼28% of Test−ve patients had a value equal to or higher than the median seen in the Test+ve group (Figure 1) and 88% had a score equal to or higher than the lowest value in the Test+ve group. Of all cases without a causative variant demonstrated, 21.4% (including those who had no germline genetic testing performed and were therefore of unknown status) had a MTS equal to or higher than the Test+ve group median. The individual mean age at tumour diagnosis (combined age at diagnosis/no. of primary tumours) was also calculated and 41.8% of Test−ve cases had a value at or below the median value observed in the Test+ve group (Figure 2). This figure was 39.9% if individuals who had not undergone genetic testing were also included in analysis. If the results of MSI/IHC analysis were given equal status to that of germline genetic testing, then similar figures were observed. Of the revised Test−ve group, 28.1% had an MTS at or above and 38.6% had an individual mean age of diagnosis below the revised Test+ve medians; 55% had a first degree relative with a concordant tumour compared with 62.5% in the revised Test+ve group.
Figure 1.

Individual case scores. Black indicates score ≥ Test+ve group median.
Figure 2.
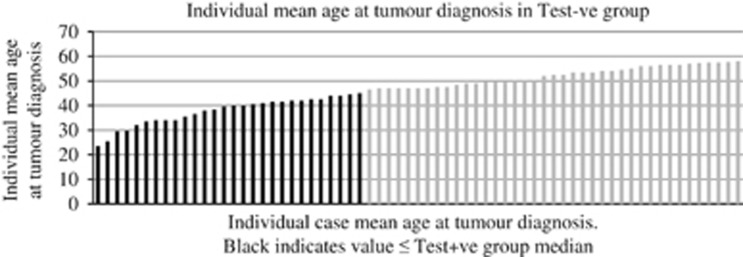
Individual case mean age at tumour diagnosis. Black indicates value ≤ Test+ve group median.
Analysis was also performed without the cervical cancers present in the patients within the series. This led to the exclusion of nine individuals, five of whom had received genetic testing with a causative variant identified in four BRCA1, BRCA2 (n=2) and APC. Results are shown in Supplementary Table 6. No significant differences in results were shown between the two analyses both in terms of tumour profile and clinical indicators of a causative variant.
Further evaluation of MPMT cases – North West series
To further evaluate MPMT referrals to cancer genetics services, we analysed the characteristics of 240 further cases referred to the North West region genetics service and entered into database of individuals referred for suspected hereditary colorectal cancer or familial breast cancer. Of them, 230 (95.8%) cases had at least one diagnosis of breast, ovarian, colorectal or endometrial cancer; 166 cases had received a diagnosis of breast and/or ovarian cancer and 144 had previously had colorectal and/or endometrial malignancy. Details of the tumour combinations are shown in Supplementary Table 7.
Comparison of MTS with existing scoring systems – combined series
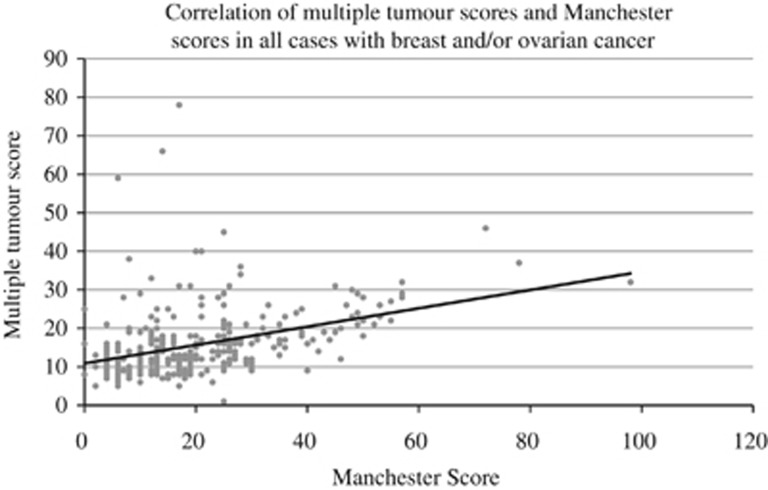
To consider the MTS compared with existing clinical evaluation systems, the data from North West cases were combined with that from those West Midlands patients who had breast and/or ovarian cancer (combined total n=320) or colorectal and/or endometrial cancer (combined total n=244). We then investigated the relationships between MTS and Manchester score or fulfilment of revised Bethesda criteria in the two cancer phenotype subgroups. The relationships are shown in Figures 3 and 4, respectively. Manchester score showed a positive correlation with MTS. Among the colorectal/endometrial cases, 58 (23.8%) had a MTS higher than 20. Four out of the 59 (6.8%) individuals not fulfilling the Revised Bethesda criteria had scores above this level. Therefore, the proportion of patients with the highest MTSs were less likely to fall short of criteria fulfilment
Figure 3.
Correlation of multiple tumour scores and Manchester scores in all cases with breast and/or ovarian cancer.
Figure 4.
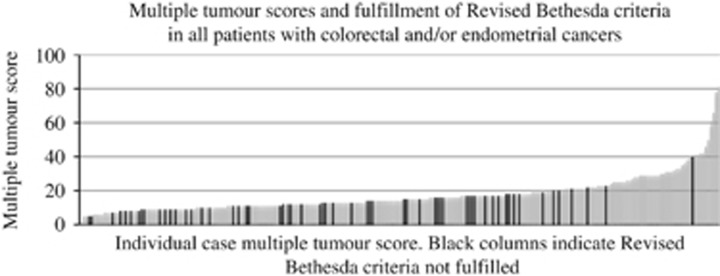
Multiple tumour scores and fulfilment of revised Bethesda criteria in all patients with colorectal and/or endometrial cancers.
Sequencing analysis of PTEN and TP53 in multiple primary tumour cases
The overlap between Test+ve and Test−ve groups in MTS distributions and age at diagnosis suggested that some of the Test−ve group might harbour a pathogenic variant in an unanalysed gene. To test this, sequencing of the coding regions of PTEN and TP53 was performed in 62 patients not previously tested for variants in these genes. The details of the tumour phenotypes of the 62 patients are shown in Supplementary Table 8. Of the 62 patients, 56 (90.3%) had been diagnosed with a tumour previously associated with variants in PTEN and/or TP5322, 23 (breast cancer in 42 patients, endometrial carcinoma (n=10), papillary thyroid cancer (n=2), sarcoma (n=1) and gliomas (n=1)). Of the 62 patients, 41 (66.1%) had previously undergone testing for BRCA1/BRCA2 and/or mismatch repair genes. No germline test had been performed in 21 cases, 8 of whom had negative MSI and/or IHC analysis. The 13 remaining untested patients all had been diagnosed with breast cancer in combination with another tumour but there were no colorectal cancers among this group. No pathogenic variants were detected in PTEN or TP53. The common TP53 p.Pro72Arg variant was observed in the samples although the genotype distribution (C;C∼10%, C;G ∼30%, G;G ∼60%) among the series was similar to that expected in a European population24 and no phenotypic differences were observed between the different genotype groups.
Discussion
We investigated the clinical features of a series of MPMT patients referred to clinical genetics services. Half of the (West Midlands) referrals that satisfied the inclusion criteria were referred in the last 7 years of the 20-year period, suggesting an increasing awareness among non-genetic health-care professionals of the relevance of inherited cancer syndromes for MPMT patients.
Clinical observations of MPMT patients have previously been made at a population level. A series based on data from the Surveillance, Epidemiology and End Results (SEER) programme collected between 1975 and 2001 identified 557 685 patients who had developed two tumours at different sites. Only 10.1% of these cases had both cancers diagnosed before the age of 60. Among this group, the most common first tumours were breast (19.7%), lymphoma (11.6%), female genital tract (10.9%), melanoma (9.5%) and colorectal (6.8%).3 The high proportion of lymphoma cases may, in part, be accounted for by earlier carcinogenic treatments for this condition. In particular, the risk of breast cancer post mantle radiotherapy in lymphoma cases diagnosed under 30 years is known to be very high.25 Analysis of the most frequent first tumours observed in our series reveals a similar picture albeit with higher proportions of BRCA1/BRCA2 or Lynch syndrome associated cancers with breast tumours making up 32.5% of first diagnoses, female genital tract 18.9%, colorectal 17% and melanoma 5.2%. Of the first cancers, 9% were NMSCs, but the incidence of these was not quoted in the SEER data. There were no cases of lymphoma and few cases with common cancers lung (5) or prostate (3). The most frequent tumours observed in our series, therefore, reflect both incidence in the general population and the clinical genetics referral-based nature of the series. Breast, ovarian and colorectal cancer referrals made up over 90% of cancer in a descriptive study of referrals to cancer genetics services in the UK26 with a figure of 82.2% in a similar analysis of French services.27 In both reports, breast and bowel cancer accounted for around 60% and 20% of referrals, respectively. Breast, colorectal and NMSC were among the five most frequent tumours, an observation that is also seen in the UK population.28 Germline genetic cancer predisposition may be suggested by rarer tumours but there is not a simple relationship between the incidence of specific cancer types and a genetic or environmental origin. Thus, assessment of the proportion of cancer cases attributable to 14 common preventable environmental exposures has shown relatively low figures for many of the most frequent tumours in the series including breast (26.8%), colorectal (54.4%), ovarian (20.7%) and uterine (36.9%).18 The remaining proportion would include genetic factors and environmental exposures not considered.
Only a minority of cases were ultimately identified with a constitutional variant conferring cancer risk. Diagnostic rates are influenced by clinical techniques to target testing, with more specific criteria not only producing higher variant detection rates but also excluding some potential carriers from analysis. Of the 71 (66.2%) individuals undergoing BRCA1/BRCA2 testing, 47 had a Manchester score at or above the commonly used threshold of 15 and 34% of these had a pathogenic variant, contrasting with none of the 24 cases with a score under 15. Of the 34 (88.2%) cases having mismatch repair gene analysis, 30 fulfilled revised Bethesda criteria with 21 (70%) causative variants identified and none in the remaining 4 patients. The clinical techniques used in the series, therefore, generally showed specificity. The sensitivity of services in detection of causative variants is likely to improve with more widespread testing with NGS gene panels, two of which were utilized in the series.
We compared clinical aspects of Test+ve and Test−ve cases to assess the clinical evidence for underlying causative variants in cancer susceptibility genes in individuals not demonstrated to harbour one by clinical investigation. The latter group reflects those individuals where the testing pathway was completed (ie consent and blood sample/s given, clinical suspicion present) and represents those who may be candidates for more extensive analysis. Comparison of most frequent tumours between the two groups revealed similar patterns for cancer subtypes though a higher proportion of breast cancers was observed in the Test−ve group, likely reflecting the fact that colorectal cases are less likely to require germline testing following MSI/IHC analysis. Although, on average, comparison between Test+ve and Test−ve groups revealed a lower age at diagnosis, higher MTS and more familial (first degree relative affected) cases in the former group, our analysis also revealed a significant number of Test−ve individuals with clinical indicators equally or more indicative of a causative germline variant than many Test+ve group cases. This was also the case if all individuals without a genetic diagnosis were analysed, including those who had not had any genetic testing performed. This suggests that there may be an appreciable, as yet unidentified genetic aetiology contributing to tumourigenesis in these individuals.
Additional analysis revealed that individuals with a higher MTS were more likely to have an elevated Manchester score or fulfil revised Bethesda criteria, implying that MTS is also likely to be higher in patients harbouring a variant predisposing to tumour development. This in turn suggests that the development of validated scores along similar lines incorporating all tumours may be useful in the assessment of individuals referred to cancer genetics and that further genotyping may be particularly likely to reveal causative variants in cases within our series with an MTS at the higher end of the range.
What might be the possible reasons for an MPMT case, even with other indicators suggestive of a constitutional genetic aetiology, testing negative after genetic analysis? Multiple tumours may occur by chance alone. A rough estimate of how frequent this could be arrived at by considering the lifetime risk of developing cancer under the age of 60 years,29, 30 multiplying that probability and considering it in the context of the observed age distribution in a population MPMT cohort.3 For each age group, an expected number of cases in a given time period could be generated. Applied to the West Midlands population, the sum of these figures suggests 16.8 cases per year, but the lifetime risk figure notably does not include NMSC. Discounting NMSC, the last 7 years of referral in our series produced 12.6 cases per year. Clearly, not all MPMT patients will be referred for genetic assessment. Registry-based evidence from the same population (1995–1999) showed 1425 new MPMT cases per year including NMSC,2 which would be estimated to be around 200 per year without NMSC if it is considered that these cancers makes up around 23.6% of diagnoses in the UK31 and assumed that these had a typical age distribution and occurred evenly among first and second cancers. On this basis, observed MPMT cases appear to be more frequent than that expected by chance alone.
Environmental exposures relevant to the development of two or more tumour types account for a proportion of MPMT cases and are important to consider in the assessment of such patients. These may be easily identifiable clinically such as a smoking history where, for example, an increased risk of oral and pharyngeal cancers are observed after an initial lung adenocarcinoma.32 Alternatively, they may be less obvious and/or shared between family members so as to mimic constitutional genetic factors leading to cancer risk. A meta-analysis of risk factors contributing to an early onset cancer Wilms tumour identified some relevant in utero exposures (eg pesticides).33 One pregnancy affected by these exposures could mimic a de novo variant conferring cancer risk whereas further affected pregnancies could produce a family history and falsely suggest inherited genetic factors.
A causative variant might not be detected because the relevant gene is overlooked. Possible reasons include a de novo mutation leading to insufficient family history or a phenotype not highly characteristic of a particular syndrome. To investigate this possibility, we undertook analysis of TP53 and PTEN, variants in which are associated with a wide range of cancers.22, 23 Although many individuals in the Test−ve group had cancers that are associated with germline aberrations in these genes, we did not identify any in our series. These findings add support to existing diagnostic criteria relating to these genes and in particular are consistent with the finding that Cowden stigmata are detectable in all individuals with a pathogenic PTEN variant34 (none of our tested cases had such findings). Previous studies have assessed the frequency of deleterious variants in these genes within an MPMT cohort. Shiseki et al35 found a pathogenic TP53 variant in one out of five patients diagnosed with three primary cancers, though that individual did conform to the Li Fraumeni syndrome (LFS) phenotype according to classical diagnostic criteria. Another analysis of TP53 in 59 cases diagnosed with a second primary cancer revealed four individuals with pathogenic variants.36 None of these individuals fulfilled classical diagnostic criteria for LFS though all met the Chompret criteria for TP53 testing.37 Deleterious variants in TP53 were identified in only 1 case of 21 (5%) breast/sarcoma double primaries not fulfilling classical criteria for LFS38 and not identified in a series of 88 breast cancer cases with a personal or family history of MPMT.39 De Vivo et al40 identified 5 individuals harbouring germline PTEN variants (p.Val119Leu x3 and p.Val158Leu x2) from a series of 103 MPMT cases from a cohort of 32 826 nurses. There was no evidence of a diagnosis of Cowden syndrome among the variant carriers but all had had a cancer associated with that condition. Functional studies of the identified variants via PTEN-null cell line transfection assays suggested an increase in cell size and number, but lack of further clinical information regarding them renders it difficult to interpret these findings as evidence that PTEN variants are significant within the context of an MPMT cohort.
If a variant conferring cancer risk is mosaic, it may elude detection by clinical tests because of relatively low sensitivity of Sanger sequencing techniques to this phenomenon. Instances of mosaicism, such as the detection of activating HIF2A variants in multiple tumours but not in blood from patients with multiple paragangliomas,41 may account for a proportion of individuals with MPMT and no relevant family history. Further genetic studies in such cases may still be revealing even without a pedigree consistent with inherited predisposition.
Finally, variants in gene not previously associated with cancer risk may be significant in a proportion of MPMT cases. Recently identified inherited cancer genes (eg POLD1, POLE) associated with colorectal and endometrial cancers42 had not been tested in our cohort and might explain some cases.
We identified evidence in favour of the hypothesis that many MPMT patients referred to clinical genetics services may harbour causative constitutional variants in cancer predisposition genes but a limited candidate gene approach to identify them did not do so. The development and increased availability of massively parallel sequencing platforms in clinical genetics service laboratories will enable individuals to be tested simultaneously for variants in multiple inherited cancer genes and also provide better sensitivity for mosaicism.
Acknowledgments
JW is supported by the Birmingham Women's Hospital Springboard Fellowship. EM is supported by an ERC Advanced Researcher Award and the Cambridge NIHR BRC. DGE and FL are supported by the Manchester NIHR BRC.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Billroth T. Die Allgemeine Chirurgische Pathologie and Therapie. 51 Vorlesungen. Ein Handbuch fur Studierende and Artze. Berlin: G Reimer; 1889. pp. 934–953. [Google Scholar]
- Berrino F, De Angelis R, Sant M, et al. Survival for eight major cancers and all cancers combined for European adults diagnosed in 1995-99: results of the EUROCARE-4 study. Lancet Oncol. 2007;8:773–783. doi: 10.1016/S1470-2045(07)70245-0. [DOI] [PubMed] [Google Scholar]
- Mariotto AB, Rowland JH, Ries LaG, Scoppa S, Feuer EJ. Multiple cancer prevalence: a growing challenge in long-term survivorship. Cancer Epidemiol Biomarkers Prev. 2007;16:566–571. doi: 10.1158/1055-9965.EPI-06-0782. [DOI] [PubMed] [Google Scholar]
- Rosso S, De Angelis R, Ciccolallo L, et al. Multiple tumours in survival estimates. Eur J Cancer. 2009;45:1080–1094. doi: 10.1016/j.ejca.2008.11.030. [DOI] [PubMed] [Google Scholar]
- Dong C, Hemminki K. Second primary neoplasms in 633,964 cancer patients in Sweden, 1958-1996. Int J Cancer. 2001;93:155–161. doi: 10.1002/ijc.1317. [DOI] [PubMed] [Google Scholar]
- Abe O, Abe R, Enomoto K, Kikuchi K. Tamoxifen for early breast cancer: an overview of the randomised trials. Lancet. 1998;351:1451–1467. [PubMed] [Google Scholar]
- Hemminki K, Boffetta P. Multiple primary cancers as clues to environmental and heritable causes of cancer and mechanisms of carcinogenesis. IARC Sci Publ. 2004. pp. 289–297. [PubMed]
- Gonzalez KD, Noltner KA, Buzin CH, et al. Beyond Li Fraumeni Syndrome: clinical characteristics of families with p53 germline mutations. J Clin Oncol. 2009;27:1250–1256. doi: 10.1200/JCO.2008.16.6959. [DOI] [PubMed] [Google Scholar]
- Kleinerman R, Yu C-L, Little MP, et al. Variation of second cancer risk by family history of retinoblastoma among long-term survivors. J Clin Oncol. 2012;30:950–957. doi: 10.1200/JCO.2011.37.0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim W, Olschwang S, Keller JJ, et al. Relative frequency and morphology of cancers in STK11 mutation carriers1 ⋆. Gastroenterology. 2004;126:1788–1794. doi: 10.1053/j.gastro.2004.03.014. [DOI] [PubMed] [Google Scholar]
- Metcalfe Ka, Lynch HT, Ghadirian P, et al. The risk of ovarian cancer after breast cancer in BRCA1 and BRCA2 carriers. Gynecol Oncol. 2005;96:222–226. doi: 10.1016/j.ygyno.2004.09.039. [DOI] [PubMed] [Google Scholar]
- Win AK, Lindor NM, Winship I, et al. Risks of colorectal and other cancers after endometrial cancer for women with Lynch syndrome. J Natl Cancer Inst. 2013;105:274–279. doi: 10.1093/jnci/djs525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birmingham Women's Hospital NHS Foundation Trust West Midlands Family Cancer Service - The West Midlands Region2013. At < < http://www.bwhct.nhs.uk/west-midlands-family-cancer-service/the-west-midlands-region >.
- Manchester Centre for Genomic Medicine Manchester Centre for Genomic Medicine2013. At < < http://www.manchesterbrc.org/OurFacilities/ManchesterCentreforGenomicMedicine.php >.
- International Agency for Research on Cancer International Rules for Multiple Primary Cancers (ICD-O Third Edition)2004
- Evans DGR. A new scoring system for the chances of identifying a BRCA1/2 mutation outperforms existing models including BRCAPRO. J Med Genet. 2004;41:474–480. doi: 10.1136/jmg.2003.017996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Institute of Cancer Research Protocol 2 BRCA1 and BRCA2 mutation testing guidelines Woman with Woman with Male with breast cancer who Individual unaffected. 2013 (2013). At < < http://www.icr.ac.uk/research/team_leaders/Rahman_Nazneen/Rahman_Nazneen_Protocols/Protocols/22778.pdf >.
- Boyd L, Parkin DM. The fraction of cancer attributable to lifestyle and environmental factors in the UK in 2010. J Epidemiol Community Heal. 2011;65:A143–A143. doi: 10.1038/bjc.2011.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baseman JG, Koutsky LA. The epidemiology of human papillomavirus infections. J Clin Virol. 2005;32 (Suppl 1:S16–S24. doi: 10.1016/j.jcv.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group Recommendations from the EGAPP Working Group: genetic testing strategies in newly diagnosed individuals with colorectal cancer aimed at reducing morbidity and mortality from Lynch syndrome in relatives. Genet Med. 2009;11:35–41. doi: 10.1097/GIM.0b013e31818fa2ff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng C.PTEN Hamartoma Tumor Syndrome (PHTS) University of Washington: Seattle; 2012. At < < http://www.ncbi.nlm.nih.gov/books/NBK1488/ >. [PubMed] [Google Scholar]
- Schneider K, Zelley K, Nichols K, Garber J.Li-Fraumeni Syndrome. GeneReviewsTM [Internet] University of Washington: Seattle; 2013. at < < http://www.ncbi.nlm.nih.gov/books/NBK1311/ >. [PubMed] [Google Scholar]
- SNPedia. Rs1042522 - SNPedia2013. At < < http://snpedia.com/index.php/Rs1042522 >.
- Yahalom J, Petrek J. Breast cancer in patients irradiated for Hodgkin's disease: a clinical and pathologic analysis of 45 events in 37 patients. J Clin. 1992;10:1674–1681. doi: 10.1200/JCO.1992.10.11.1674. [DOI] [PubMed] [Google Scholar]
- Wonderling D, Hopwood P, Cull A, et al. A descriptive study of UK cancer genetics services: an emerging clinical response to the new genetics. Br J Cancer. 2001;85:166–170. doi: 10.1054/bjoc.2001.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol P, Lyonnet DS, Frebourg T, et al. Lack of referral for genetic counseling and testing in BRCA1/2 and Lynch syndromes: a nationwide study based on 240,134 consultations and 134,652 genetic tests. Breast Cancer Res Treat. 2013;141:135–144. doi: 10.1007/s10549-013-2669-9. [DOI] [PubMed] [Google Scholar]
- Cancer Research UK Cancer incidence for common cancers2013. At < < http://www.cancerresearchuk.org/cancer-info/cancerstats/incidence/commoncancers/ >.
- UK, C. R. Lifetime risk of cancer2014. at < < http://www.cancerresearchuk.org/cancer-info/cancerstats/incidence/risk/ >.
- UK, C. R. Cancer incidence by age. at < < http://www.cancerresearchuk.org/cancer-info/cancerstats/incidence/age/ >.
- UK, C. R. Skin cancer incidence statistics2014. at < < http://www.cancerresearchuk.org/cancer-info/cancerstats/types/skin/incidence/ >.
- Levi F, Randimbison L, Te VC, La Vecchia C. Second primary cancers in patients with lung carcinoma. Cancer. 1999;86:186–190. doi: 10.1002/(sici)1097-0142(19990701)86:1<186::aid-cncr25>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Chu A, Heck JE, Ribeiro KB, et al. Wilms' tumour: a systematic review of risk factors and meta-analysis. Paediatr Perinat Epidemiol. 2010;24:449–469. doi: 10.1111/j.1365-3016.2010.01133.x. [DOI] [PubMed] [Google Scholar]
- Rustad CF, Bjørnslett M, Heimdal KR, Mæhle L, Apold J, Møller P. Germline PTEN mutations are rare and highly penetrant. Hered Cancer Clin Pract. 2006;4:177–185. doi: 10.1186/1897-4287-4-4-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiseki M, Nishikawa R, Yamamoto H, et al. Germ-line p53 mutation is uncommon in patients with triple primary cancers. Cancer Lett. 1993;73:51–57. doi: 10.1016/0304-3835(93)90187-e. [DOI] [PubMed] [Google Scholar]
- Malkin D, Jolly KW, Barbier N, et al. Germline mutations of the p53 tumor-suppressor gene in children and young adults with second malignant neoplasms. N Engl J Med. 1992;326:1309–1315. doi: 10.1056/NEJM199205143262002. [DOI] [PubMed] [Google Scholar]
- Chompret A, Abel A, Stoppa-lyonnet D, et al. Sensitivity and predictive value of criteria for p53 germline mutation. J Med Genet. 2001;38:43–47. doi: 10.1136/jmg.38.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D, Birch J, Thorneycroft M, McGown G, Lalloo F, Varley J. Low rate of TP53 germline mutations in breast cancer/sarcoma families not fulfilling classical criteria for Li-Fraumeni syndrome. J Med Genet. 2002;39:941–944. doi: 10.1136/jmg.39.12.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Kanestsky P, Amirimani B, et al. Germline TP53 mutation in breast cancer families with multiple primary cancers: is Tp53 a modifier of BRCA1. J Med Genet. 2003;40:e34. doi: 10.1136/jmg.40.4.e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vivo I, Gertig DM, Nagase S, et al. Novel germline mutations in the PTEN tumour suppressor gene found in women with multiple cancers. J Med Genet. 2000;37:336–341. doi: 10.1136/jmg.37.5.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Z, Yang C, Lorenzo F, et al. Somatic HIF2A gain-of-function mutations in paraganglioma with polycythemia. N Engl J Med. 2012;367:922–930. doi: 10.1056/NEJMoa1205119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palles C, Cazier J-B, Howarth KM, et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet. 2013;45:136–144. doi: 10.1038/ng.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.