Abstract
The mammalian intestine harbors a community of trillions of microbes, collectively known as the gut microbiota, which co-evolved with the host in a mutually beneficial relationship. Among the numerous gut microbial species, certain commensal bacteria are known to provide health benefits to the host when administered in adequate amounts, and as such are labeled “probiotics”. Here we review some of the mechanisms by which probiotics and other beneficial commensals provide colonization resistance to pathogens. The battle for similar nutrients and the bacterial secretion of antimicrobials provide a direct means of competition between beneficial and harmful microbes. Beneficial microbes can also indirectly diminish pathogen colonization by stimulating the development of innate and adaptive immunity as well as the function of the mucosal barrier. Altogether, we gather and present evidence that beneficial microbes cooperate with host immunity in an effort to shut out pathogens.
Introduction
The mammalian gastrointestinal (GI) tract is home to a community of trillions of microorganisms commonly known as the microbiota. The long co-existence of the microbiota and the host intestinal mucosa has established a mutual beneficial relationship: On the one hand, the microbiota protects the host from infection with pathogenic microorganisms and contributes to both nutrient metabolism as well as to the development and function of the GI immune system; on the other hand, the host provides nutrient-rich niches to ensure the survival of its resident bacterial communities [1].
In the past decade, studies in germ-free (GF) mice and the advent of metagenomics have tremendously contributed to elucidate the complexity of the intestinal microbiota and its contribution to health and disease [2, 3]. In healthy subjects, at least 1,000 different bacterial species contribute to intestinal homeostasis, with Firmicutes and Bacteroidetes representing the most common intestinal phyla, followed by Actinobacteria and Proteobacteria [4, 5]. Within these phyla, some bacterial species including Gram-positive Lactobacillus spp. (phylum Firmicutes) and Bifidobacterium spp. (phylum Actinobacteria) as well as certain Gram-negative bacteria such as Escherichia coli Nissle 1917 (phylum Proteobacteria) have been shown to benefit the host by blocking harmful microorganisms; for this reason, they are referred to as “probiotics.”
The first observation that certain commensal bacteria have beneficial properties dates back to 1907, when Elie Mechnikoff proposed that lactic acid-producing strains are beneficial to the host by inhibiting the growth of other species within the colon [6]. Today, probiotics are defined by the World Health Organization as “live bacterial species that confer a health benefit when administered in adequate amounts” [7]. In addition to the few known probiotics, the microbiota in general has beneficial effects on the host; for example, the absence of the microbiota renders GF mice more susceptible to infection in comparison to conventionally raised mice [1, 8]. Moreover, the use of antibiotics has been shown to enhance intestinal colonization of enteric pathogens, as alterations to the composition of the gut flora track with increased susceptibility to infection with pathogens such as Salmonella enterica serovar Typhimurium (Salmonella Typhimurium) and Clostridium difficile [9–12].
More recently, several studies have begun to elucidate the molecular mechanisms behind the beneficial role of commensal and probiotic strains. It is now becoming clear that beneficial bacteria provide colonization resistance to pathogens by two major mechanisms [13, 14]. The first mechanism involves the direct competition between certain commensals and pathogens for nutrients or niche establishment. The second mechanism comprises indirect effects on pathogen colonization, deriving from the stimulation of the innate and adaptive immune system by commensal bacteria. In this review, we will summarize some of the mechanisms by which commensal bacteria, including certain probiotic species, contribute to colonization resistance against pathogens, both by direct competition with pathogenic bacteria and by stimulation of host immunity.
Direct competition with pathogens
One of the mechanisms by which commensal and probiotic bacteria provide colonization resistance to pathogens is by directly competing for the same niche. Some beneficial microbes acquire similar nutrients as pathogens, often more efficiently, thus hindering the replication and colonization of infectious agents. In addition, some microbes produce antimicrobial proteins that can target pathogens. Below, we will discuss these two aspects of direct competition between beneficial and harmful bacteria (Figure 1).
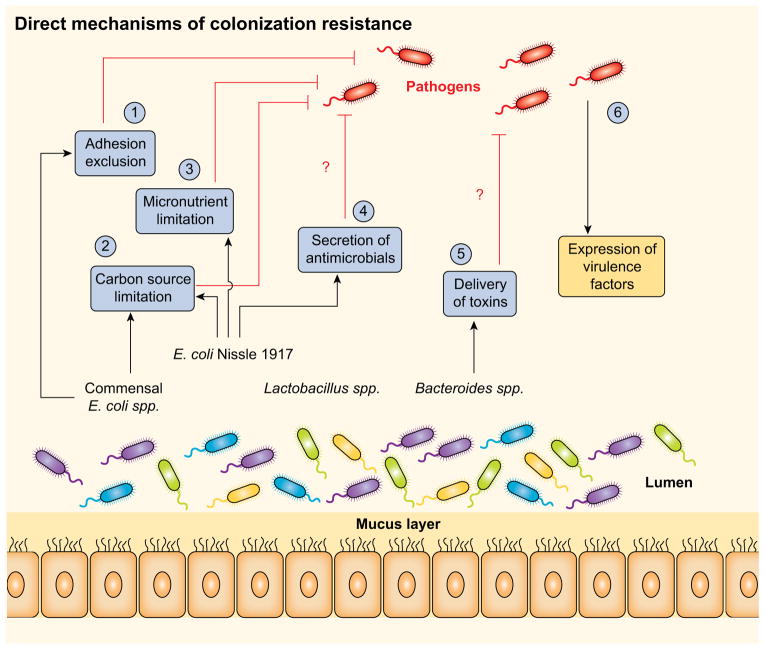
Figure 1. Direct mechanisms of colonization resistance against enteropathogens.
The microbiota provide a barrier against incoming enteric pathogens via multiple mechanisms. (1) Adhesion exclusion: Certain commensals reduce pathogen adherence to the intestinal mucosa. (2) Carbon source limitation: Human commensal Escherichia coli strain HS and probiotic strain Escherichia coli Nissle 1917 both metabolize multiple sugar molecules, which limits the availability of nutrients in the gut to certain pathogens. (3) Micronutrient limitation: The probiotic strain E. coli Nissle 1917 can uptake iron via several mechanisms, limiting its availability to pathogens such as Salmonella Typhimurium. (4) Secretion of antimicrobials: Commensals act against pathogens not only by limiting nutrients, but also by the production antimicrobial compounds, such as bacteriocins and microcins. (5) Direct delivery of toxins: Although not yet demonstrated in vivo, commensals can express type 6-secretion systems (T6SSs) and contact-dependent inhibition (CDI) systems, means by which to deliver growth-inhibiting toxins to close competitors. (6) Circumventing colonization resistance: Pathogens employ a variety of virulence factors to colonize the host and cause disease.
Competition for nutrients
Numerous studies on the battle between commensals and pathogens belonging to the Enterobacteriaceae family have put forward the idea that competition for nutrients in the gut appears to occur primarily between metabolically related bacteria. For example, in GF mice, certain commensal Escherichia coli strains reduce cecal colonization of the enteric pathogen enterohemorrhagic E. coli (EHEC), a leading cause of bloody diarrhea in humans, by competing for the amino acid proline [15]. Similarly, pre-colonization of streptomycin-treated mice with specific human commensal E. coli strains prevented the growth of EHEC [16]. This latter effect was later reported to be linked to the capacity of the human commensal E. coli strain HS and of the probiotic strain E. coli Nissle 1917 (discussed below) to occupy a unique nutritional niche in the mouse gut. Indeed, utilization of multiple sugar molecules (6 by E. coli HS and 7 by E. coli Nissle 1917) limited the nutrient availability to pathogenic strains of E. coli, thereby impeding their successful colonization of the gut [17]. It is worth noting that the competition for nutrients is not limited to bacteria within the same species. For instance, a commensal strain of E. coli was able to delay the intestinal colonization and translocation of the pathogen Salmonella Typhimurium in GF mice, possibly due to competition for nutrients [18]. More recently, it has been shown that the intestinal microbiota provides colonization resistance to infection with Citrobacter rodentium, a mouse pathogen used to model infection with diarrheagenic E. coli strains including enteropathogenic E. coli (EPEC) and EHEC, by competing for similar carbohydrates [19]. Similarly, consumption of fucose during infection supports the growth of the microbiota and protects mice from infection with C. rodentium [20].
As competition for nutrients is one of the mechanisms by which commensals provide colonization resistance, it is not surprising that certain pathogens have evolved to evade this mechanism. This is, for instance, the case for certain pathogenic E. coli strains, which can utilize sugars that are not used by commensal E. coli [21]. Moreover, pathogens like EHEC, Salmonella Typhimurium, and C. difficile can catabolize sugars liberated by the microbiota [22, 23]. Additionally, the types of nutrient sources available to both commensals and pathogens can vary if the intestinal environment is altered; e.g. because of inflammation or antibiotic treatment [12, 22, 24]. In such conditions, changes in nutrient type and availability enhance the growth of specific strains capable of exploiting the new nutrient sources; for example, the pathogen Salmonella Typhimurium, but not the microbiota, can utilize ethanolamine and fructose-asparagine in the inflamed intestine [25]. Similarly, antibiotic treatment of mice induces substantial changes in the gut metabolome, resulting in an increase of specific carbon sources that support the germination and growth of C. difficile [12]. Taken together, these observations indicate that commensal bacteria are best equipped to provide colonization resistance to pathogens in the healthy, unperturbed intestine. Nevertheless, it may be feasible to design probiotics by engineering commensal strains to better compete with pathogens for nutrients in more hostile environments like the inflamed gut. Of note, there is at least one example of an E. coli strain that is able to compete with pathogenic Enterobacteriaceae in such conditions: The widely studied and utilized probiotic E. coli Nissle 1917 [26].
E. coli Nissle 1917 (hereafter referred to as E. coli Nissle) was isolated in 1917 by Dr. Alfred Nissle from the stool sample of a soldier who did not develop diarrhea during an outbreak of shigellosis [27]. Today, E. coli Nissle is the active component of a probiotic preparation used for the treatment of both infectious diarrheal diseases and inflammatory bowel disease (IBD) [26]. Although the mechanisms by which E. coli Nissle exerts its beneficial effects are not completely understood, new pieces of evidence suggest that this probiotic strain competes with related bacteria and modulates the immune system (reviewed in [28]).
E. coli Nissle possesses multiple features that might contribute to its ability to colonize the intestine, including Curli, Type 1 and F1C fimbriae, which increase its adherence to the mucosa and can prevent the adhesion of other pathogens [29]. In addition, the E. coli Nissle genome encodes several redundant mechanisms that might contribute to its fitness, one of which is a large number of iron acquisition systems, including genes for the synthesis and uptake of small iron chelators known as siderophores [30–32]. Such systems allow for growth in environments where the essential metal ion iron is limited, including the intestinal lumen during inflammation [33]. As reported by our lab in 2013, the presence of multiple iron acquisition systems allows E. coli Nissle to successfully compete with the pathogen Salmonella Typhimurium for colonization of the inflamed gut [34]. As other commensal E. coli strains were unable to reduce Salmonella Typhimurium gut colonization [34, 35], our work suggests that E. coli Nissle exhibits the uncommon ability to compete with Salmonella Typhimurium, and possibly with other enteric pathogens, for iron. Nevertheless, it is feasible that other E. coli strains which colonize the gut but cause extraintestinal infections, such as those which cause urinary tract infections, may also compete with Salmonella Typhimurium and/or with other bacteria via similar mechanisms. Overall, E. coli Nissle is a prime example of how a commensal strain can provide colonization resistance against pathogens by better competing for the limited nutritional resources available in the inflamed intestine.
Production of antimicrobial peptides and toxins
Another means of direct competition between commensals and pathogens within the gut involves the secretion of toxins and antimicrobial peptides. Though originally described as a virulence trait of pathogens to kill their commensal competitors [36], new evidence suggests that the secretion of antibacterial toxins via phage-like machinery known as a “type VI secretion system” (T6SS) is also employed by commensals to attack competitors vying for the same ecological niche [37]. To this end, it has been postulated that the abundance of Bacteroidetes in the intestine could be attributed to their T6SSs [38].
Other mechanisms employed by bacteria to deliver toxins to their competitors (for instance, the contact-dependent growth inhibition system [39]) could also promote inter and intra-species competition as well as colonization resistance to pathogens. Moreover, some commensal Enterobacteriaceae, including the probiotic E. coli Nissle, secrete small antimicrobial peptides called bacteriocins or microcins, which specifically target and kill related competitors, including pathogenic organisms [40, 41]. Some microcins target competitors expressing the same nutrient receptors as the microcin producers, and are internalized by hijacking these transporters via a so-called “Trojan horse” mechanism [41, 42]. Other probiotic strains such as Bifidobacterium spp. also secrete bacteriocins, which can exhibit either a narrow or broad spectrum of activity [43]. As most of the evidence for the activity of bacteriocins and toxins derives from in vitro studies, future studies should address the role of antimicrobial peptides and toxins secreted by commensal and probiotic bacteria to compete with pathogens in the host. Moreover, as most studies on human commensals and probiotics have analyzed their effects on the murine gut microbiota, their relevance in the human host is largely unknown. To this end, broader utilization of humanized gnotobiotic mice (i.e., gnotobiotic mice colonized with human microbiota [3]) could begin to unravel mechanisms by which human commensals and probiotics compete with the human microbiota.
Indirect effects against pathogens
Studies of GF mice have unequivocally shown the importance of the microbiota for the development of a normal gut mucosa and gut-associated lymphoid tissue (GALT) [44]. It thus follows that the ability of the microbiota and of certain probiotics to provide colonization resistance to pathogens is also mediated by their enhancement of the gut mucosal barrier and of the innate and adaptive immune systems. Examples of such mechanisms are described in detail below (Figure 2).
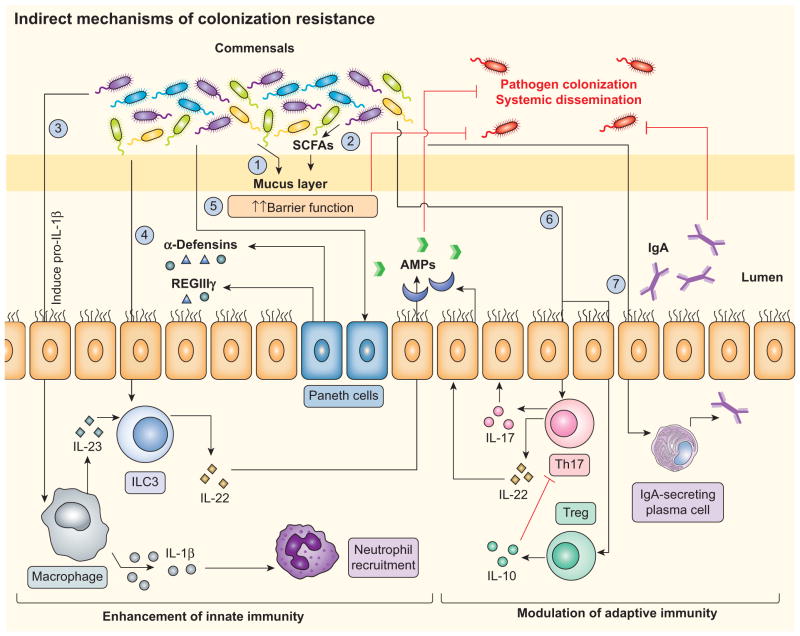
Figure 2. Microbiota-stimulated host immunity provides colonization resistance.
Commensal bacteria can indirectly control pathogen colonization by a variety of means. (1) Barrier function: The commensal microbiota up-regulates host barrier function by contributing to the development of the mucus layer. (2) Short-chain fatty acid (SCFA) production: Members of the microbiota such as Bifidobacterium spp. can enhance epithelial barrier function by producing SCFAs such as acetate. (3) IL-1β-mediated neutrophil recruitment: Commensals can promote protection against certain pathogens by stimulating IL-1β processing and secretion, resulting in the recruitment of neutrophils to the site of the infection. (4) IL-22-dependent release of antimicrobials: Certain commensals (such as Lactobacillus reuteri) induce the secretion of IL-22 by innate lymphoid cells (ILCs), which can in turn protect against some pathogens via the induction of antimicrobial release by epithelial cells. (5) Direct stimulation of antimicrobial production by the host: Secretion of antimicrobial proteins (AMPs), including α-defensins and REG3γ, is a key component in controlling pathogen growth, and is in part mediated by commensal-dependent mechanisms. (6) Induction of T cell differentiation: Commensals can promote adaptive immunity by inducing the differentiation of T cells, such as by stimulating Th17 and Treg cell differentiation and activation. (7) Secretory IgA (sIgA): Commensals can facilitate host-barrier function by inducing B cells and by regulating the secretion of IgA.
Enhancement of epithelial barrier function
An important first step for gut colonization by pathogens is the adherence to mucosal surfaces. This step, however, is hindered by the thick mucus layer that covers the intestinal epithelium, providing a first layer of defense against bacterial colonization [45]. The optimal development of the mucus layer is dependent on the microbiota; indeed, GF mice develop a much thinner epithelial mucus layer in comparison to conventionally-raised mice. In line with this, the administration of bacterial components such as LPS or peptidoglycan to GF mice restores their mucus layer formation and reduces their susceptibility to bacterial infections [46]. Of particular importance is the highly glycosylated mucin protein MUC2, which is densely packed and insoluble in the inner mucus layer, but loose and soluble in the outer layer, thereby providing a barrier to the colonization and translocation of both commensal and pathogenic bacteria [47, 48].
Although the development of the mucosal barrier primarily results from the generally cooperative interactions between host and microbiota, some specific bacterial species likely contribute to this process. For instance, certain commensal bacteria can enhance the epithelial barrier function through the production of specific metabolites. One example is Bifidobacterium longum subspecies Infantis (a probiotic bacterium that constitutes up to 90% of the microbiota of healthy infants [49]), which secretes peptides that normalize intestinal permeability and reduce intestinal pathology in a mouse model of colitis [50]. Additionally, production of the short-chain fatty acid (SCFA) acetate by Bifidobacterium improved intestinal defenses mediated by epithelial cells as well as induced protection against lethal infection with EHEC [51]. Tissue culture studies have also shown that certain probiotics modulate the immune response through a direct interaction with intestinal epithelial cells. One such example is E. coli Nissle, whose K5 capsule was shown to stimulate the production of chemokines from Caco-2 cells and from ex vivo mouse intestine [52], although it is not known whether the K5 capsule plays an immunomodulatory role in vivo.
Enhancement of innate immunity
Commensal organisms also employ several mechanisms to boost the immune response against pathogens at epithelial mucosal surfaces. Although pathogens and commensals can exhibit similarity in terms of surface molecules and antigens, mucosal secretion of pro-inflammatory cytokines is typically dependent on whether the host encounters commensal or pathogenic microbes. In general, macrophages and dendritic cells in the lamina propria of the intestine are hypo-responsive to commensal microbial ligands, whereas their interaction with a pathogen like Salmonella Typhimurium results in the secretion of mature IL-1β and in the recruitment of neutrophils to the site of infection [53]. Nevertheless, during infection with certain pathogens, the intestinal microbiota is also able to promote immune defenses by triggering specific responses, which ultimately lead to secretion of host pro-inflammatory and antimicrobial proteins. To this end, Hasegawa et al. have shown that commensal bacteria are able to stimulate the innate immune system, and thus protect the host from infection [54]. Mice lacking the adaptor protein ASC, an essential mediator of IL-1β and IL-18 processing, are highly susceptible to C. difficile infection, mostly because of impaired recruitment of neutrophils to the intestine [54]. Surprisingly, translocation of commensal bacteria was essential to promote IL-1β secretion and protection against C. difficile intestinal colonization, indicating that stimulation of pro-inflammatory responses by commensal organisms can have a protective function [54]. Nevertheless, the particular mechanisms by which commensals induce IL-1β processing and secretion remain to be elucidated.
An additional mechanism by which commensals enhance the host immune response to pathogens is by triggering the secretion of host antimicrobial peptides. In the small intestine, Paneth cells are the main source of these peptides (predominantly C-type lectins and α-defensins), whose primary function is to protect the host against enteric pathogens [55]. Paneth cell secretion of the C-type lectins REG3γ and REG3β requires the stimulation of the MyD88 pathway, underlining the role of Toll-like receptor (TLR)-mediated recognition of the microbiota for this process [56, 57]. The importance of REG3γ and REG3β is highlighted by studies showing that these two antimicrobial peptides provide protection to infection with some bacterial pathogens, including Enterococcus faecalis, Yersinia pseudotuberculosis, and Listeria monocytogenes [58–60].
The α-defensins are also key components for the control of enteric pathogens [61]. Similar to C-type lectins, the microbiota plays an important role in the induction of α-defensin expression, which in turn controls pathogen growth and contains commensals within the intestine [62–65]. In particular, both mice lacking the MHC class I-related protein CD1D as well as Crohn’s disease patients with mutations in the peptidoglycan sensor NOD2 show a reduced secretion of α-defensins, suggesting that recognition of commensal ligands enhances α-defensin expression [66, 67]. However, Nod2 mutations in mice do not affect the expression of α-defensins or the composition of the microbiota, which could possibly reflect differences between humans and mice [68, 69]. With regard to β-defensins, some bacteria such as the probiotic strain E. coli Nissle 1917 were shown to induce β-defensin expression in cell culture through TLR5-mediated recognition of flagellin [70]. Still, in vivo studies are required to determine if the ability of E. coli Nissle to induce β-defensins subsequently reduces the colonization of enteropathogens and perhaps controls gut homeostasis by inhibiting bacterial translocation.
Another example of commensal-mediated gut immunity enhancement is the induction of IL-22, a cytokine that enhances the mucosal barrier against pathogens by inducing the secretion of chemokines and antimicrobials by epithelial cells [71]. Mice that lack IL-22 were shown to be more susceptible to gut infection with C. rodentium, [72, 73]. Secretors of IL-22 include innate lymphoid cells (ILCs), in which the production of this cytokine is enhanced by activation of the aryl hydrocarbon receptor (Ahr) via specific bacterially-derived molecules [74, 75]. Recently, Zelante et al. showed that a subset of Lactobacillus species (specifically, L. reuteri in the gastrointestinal tract), utilize tryptophan as an energy source and produce a metabolite, indole-3-aldehyde (IAld), which in turns activates Ahr on ILCs. Once activated, ILCs secrete IL-22, which protects the host against the opportunistic pathogen Candida albicans by reducing its colonization [76]. This host-commensal interplay is a prime example of how beneficial bacteria might enhance the immune response in order to protect the host from pathogens. In addition, the observation that only a subset of Lactobacilli produces IAld and activates Ahr-mediated secretion of IL-22 highlights the importance of how differences in genomic content and gene expression, even within the same genus, might account for significant differences in host immune regulation.
Enhancement of adaptive immunity
Commensal and probiotic bacteria are not only implicated in the activation of innate immune responses, but are also able to promote adaptive immune responses. In recent years, a few studies have provided evidence that specific commensal bacteria are able to preferentially drive the differentiation of individual T cell subsets. A seminal study by Ivanov et al. demonstrated that bacteria of the Clostridiales family, Segmented Filamentous Bacteria (SFB), specifically induce the differentiation of mucosal T helper 17 (Th17) cells in the lamina propria of the intestine, which in turn secrete the pro-inflammatory cytokines IL-17 and IL-22 [77]. As IL-22 induces the production of antimicrobial proteins, mice administered with SFB were more resistant to infection with C. rodentium [77].
In addition to Th17 cells, the differentiation of the T regulatory (Treg) cell lineage is also impacted by the microbiota. Atarashi et al. demonstrated that colonization of GF mice with Clostridium spp. generates an environment rich in TGF-β and colonic Treg cells [78]. Moreover, oral administration of a mixture of 17 Clostridia strains attenuated the pathogenesis of colitis by shaping an anti-inflammatory environment rich in Treg cells and IL-10 [79]. In some cases, a single component of a commensal bacterium is sufficient to induce a specific cell subset. For example, the protective effect of Bacteroides fragilis against experimental colitis induced by Helicobacter hepaticus was shown to be dependent on the expression of a specific capsule, termed polysaccharide A (PSA). In this model, administration of PSA alone was able to protect mice from colitis by inducing anti-inflammatory, IL-10-producing CD4+ T cells [80]. In other cases, certain metabolites produced by the microbiota can impact Treg lineage homeostasis. For instance, SCFAs generated by the intestinal microbiota contribute to the regulation of Treg cell size and function by interacting with the G-protein-coupled free fatty acid receptor 43 (GPR43) [81].
Another important arm of the adaptive immune response modulated by the microbiota involves the differentiation and activation of B cells, along with mucosal secretory IgA (sIgA) [82, 83]. Because sIgA plays an essential role in promoting intestinal barrier function and in determining the composition of the gut microbiota, the presence of certain bacterial species is essential to regulate this important aspect of gut homeostasis [84]. In particular, dendritic cells which phagocytize small numbers of commensal microbes in the intestine can selectively induce sIgA and protect the host from its own commensal bacteria [85, 86]. Akin to the development of T cell subsets, specific commensal microbes can contribute to the development of B cells. For instance, SFB but not commensal E. coli were shown to shown to contribute to the development of intestinal lymphoid tissue, including Peyer’s patches, and to the development of intestinal sIgA [87]. Some human in vivo studies have also reported the importance of the microbiota for B cell maturation, such as with early life colonization by E. coli and Bifidobacterium longum subspecies Infantis being associated with higher numbers of mature CD20+ B cells [88]. As most of these studies were aimed at investigating the development of the immune response rather than how the microbiota contributes to colonization resistance to pathogens, future studies should address whether the development of specific immune responses are linked to colonization resistance.
Conclusions
Beneficial microbes provide colonization resistance against harmful microorganisms by stimulating the immune response and by directly inhibiting pathogen growth. Although some such mechanisms have now been described, future studies are needed to identify and/or characterize probiotics and their modes of action against specific pathogens. For instance, IL-22-mediated induction of antimicrobial responses, stimulated by Lactobacillus reuteri, could be advantageous against pathogens that are susceptible to these effects, such as C. albicans [76]. However, the same probiotic bacterium would likely be ineffective against pathogens like Salmonella Typhimurium, which evades and exploits IL-22-mediated antimicrobial responses [35]. In contrast, as Salmonella requires iron to grow in the inflamed intestine, administration of E. coli Nissle, which can compete with the pathogen for iron, would be a better therapeutic and preventive strategy [34]. In light of this, the identification of vulnerabilities within target pathogens is an essential first step to discover and design probiotics.
Another promising strategy is to screen the intestinal microbiota for natural competitors to specific pathogens. This approach has recently proven successful in a seminal study by Buffie et al, who discovered that a specific intestinal commensal bacterium, Clostridium scindens, provides colonization resistance to C. difficile infection by producing inhibitory metabolites derived from bile salts [89]. With the constant emergence of antibiotic resistance, as well as new knowledge concerning the detrimental effects antibiotics have on the microbiota, the discovery of new probiotics and their mechanisms of action could provide a strong foundation for developing novel therapeutics against infection.
Acknowledgments
The authors would like to thank Alborz Karimzadeh and Sean-Paul Nuccio for help with editing the manuscript.
Footnotes
Work in MR lab is supported by Public Health Service Grants AI083663, AI101784, AI105374, and by Burroughs Wellcome Fund.
References
- 1.Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- 2.Kamada N, Seo SU, Chen GY, Nuñez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 3.Faith JJ, Rey FE, O’Donnell D, Karlsson M, McNulty NP, Kallstrom G, Goodman AL, Gordon JI. Creating and characterizing communities of human gut microbes in gnotobiotic mice. ISME J. 2010;4:1094–1098. doi: 10.1038/ismej.2010.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Podolsky S. Cultural divergence: Elie Metchnikoff’s Bacillus bulgaricus therapy and his underlying concept of health. Bull Hist Med. 1998;72:1–27. doi: 10.1353/bhm.1998.0056. [DOI] [PubMed] [Google Scholar]
- 7.FAO/WHO. Expert consultation on evaluation of health and nutritional properties of probiotics in food including milk powder with live lactic acid bacteria. Food and Agriculture Organization/World Health Organization; 2001. [Google Scholar]
- 8.Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 9.Croswell A, Amir E, Teggatz P, Barman M, Salzman NH. Prolonged impact of antibiotics on intestinal microbial ecology and susceptibility to enteric Salmonella infection. Infect Immun. 2009;77:2741–2753. doi: 10.1128/IAI.00006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawley TD, Clare S, Walker AW, Goulding D, Stabler RA, Croucher N, Mastroeni P, Scott P, Raisen C, Mottram L, Fairweather NF, Wren BW, Parkhill J, Dougan G. Antibiotic treatment of Clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect Immun. 2009;77:3661–3669. doi: 10.1128/IAI.00558-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sekirov I, Tam NM, Jogova M, Robertson ML, Li Y, Lupp C, Finlay BB. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect Immun. 2008;76:4726–4736. doi: 10.1128/IAI.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Theriot CM, Koenigsknecht MJ, Carlson PE, Jr, Hatton GE, Nelson AM, Li B, Huffnagle GB, ZLJ, Young VB. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun. 2014;5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawley TD, Walker AW. Intestinal colonization resistance. Immunology. 2013;138:1–11. doi: 10.1111/j.1365-2567.2012.03616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013;13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Momose Y, Hirayama K, Itoh K. Competition for proline between indigenous Escherichia coli and E. coli O157:H7 in gnotobiotic mice associated with infant intestinal microbiota and its contribution to the colonization resistance against E. coli O157:H7. Antonie Van Leeuwenhoek. 2008;94:165–171. doi: 10.1007/s10482-008-9222-6. [DOI] [PubMed] [Google Scholar]
- 16.Leatham MP, Banerjee S, Autieri SM, Mercado-Lubo R, Conway T, Cohen PS. Precolonized human commensal Escherichia coli strains serve as a barrier to E. coli O157:H7 growth in the streptomycin-treated mouse intestine. Infect Immun. 2009;77:2876–2886. doi: 10.1128/IAI.00059-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maltby R, Leatham-Jensen MP, Gibson T, Cohen PS, Conway T. Nutritional basis for colonization resistance by human commensal Escherichia coli strains HS and Nissle 1917 against E. coli O157:H7 in the mouse intestine. PLoS One. 2013;8:e53957. doi: 10.1371/journal.pone.0053957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hudault S, Guignot J, Servin AL. Escherichia coli strains colonising the gastrointestinal tract protect germfree mice against Salmonella typhimurium infection. Gut. 2001;49:47–55. doi: 10.1136/gut.49.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamada N, Kim YG, Sham HP, Vallance BA, Puente JL, Martens EC, Nuñez G. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science. 2012;336:1325–1329. doi: 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pickard JM, Maurice CF, Kinnebrew MA, Abt MC, Schenten D, Golovkina TV, Bogatyrev SR, Ismagilov RF, Pamer EG, Turnbaugh PJ, Chervonsky AV. Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature. 2014;514:638–641. doi: 10.1038/nature13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fabich AJ, Jones SA, Chowdhury FZ, Cernosek A, Anderson A, Smalley D, McHargue JW, Hightower GA, Smith JT, Autieri SM, Leatham MP, Lins JJ, Allen RL, Laux DC, Cohen PS, Conway T. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect Immun. 2008;76:1143–1152. doi: 10.1128/IAI.01386-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, Sonnenburg JL. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, Moreira CG, Sperandio V. Fucose sensing regulates bacterial intestinal colonization. Nature. 2012;492:113–117. doi: 10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schumann S, Alpert C, Engst W, Loh G, Blaut M. Dextran sodium sulfate-induced inflammation alters the expression of proteins by intestinal Escherichia coli strains in a gnotobiotic mouse model. Appl Environ Microbiol. 2012;78:1513–1522. doi: 10.1128/AEM.07340-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Bäumler AJ. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci U S A. 2011;108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobi CA, Malfertheiner P. Escherichia coli Nissle 1917 (Mutaflor): new insights into an old probiotic bacterium. Dig Dis. 2011;29:600–607. doi: 10.1159/000333307. [DOI] [PubMed] [Google Scholar]
- 27.Nissle A. Old and new experiences on therapeutic successes by restoration of the colonic flora with mutaflor in gastrointestinal diseases. Med Welt. 1961;29–30:1519–1523. [PubMed] [Google Scholar]
- 28.Behnsen J, Deriu E, Sassone-Corsi M, Raffatellu M. Probiotics: properties, examples, and specific applications. Cold Spring Harb Perspect Med. 2013;3:a010074. doi: 10.1101/cshperspect.a010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lasaro MA, Salinger N, Zhang J, Wang Y, Zhong Z, Goulian M, Zhu J. F1C fimbriae play an important role in biofilm formation and intestinal colonization by the Escherichia coli commensal strain Nissle 1917. Appl Environ Microbiol. 2009;75:246–251. doi: 10.1128/AEM.01144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grozdanov L, Raasch C, Schulze J, Sonnenborn U, Gottschalk G, Hacker J, Dobrindt U. Analysis of the genome structure of the nonpathogenic probiotic Escherichia coli strain Nissle 1917. J Bacteriol. 2004;186:5432–5441. doi: 10.1128/JB.186.16.5432-5441.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grosse C, Scherer J, Koch D, Otto M, Taudte N, Grass G. A new ferrous iron-uptake transporter, EfeU (YcdN), from Escherichia coli. Mol Microbiol. 2006;62:120–131. doi: 10.1111/j.1365-2958.2006.05326.x. [DOI] [PubMed] [Google Scholar]
- 32.Valdebenito M, Crumbliss AL, Winkelmann G, Hantke K. Environmental factors influence the production of enterobactin, salmochelin, aerobactin, and yersiniabactin in Escherichia coli strain Nissle 1917. Int J Med Microbiol. 2006;296:513–520. doi: 10.1016/j.ijmm.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 33.Diaz-Ochoa VE, Jellbauer S, Klaus S, Raffatellu M. Transition metal ions at the crossroads of mucosal immunity and microbial pathogenesis. Front Cell Infect Microbiol. 2014;4:2. doi: 10.3389/fcimb.2014.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deriu E, Liu JZ, Pezeshki M, Edwards RA, Ochoa RJ, Contreras H, Libby SJ, Fang FC, Raffatellu M. Probiotic bacteria reduce salmonella typhimurium intestinal colonization by competing for iron. Cell Host Microbe. 2013;14:26–37. doi: 10.1016/j.chom.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Behnsen J, Jellbauer S, Wong CP, Edwards RA, George MD, Ouyang W, Raffatellu M. The cytokine IL-22 promotes pathogen colonization by suppressing related commensal bacteria. Immunity. 2014;40:262–273. doi: 10.1016/j.immuni.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Unterweger D, Miyata ST, Bachmann V, Brooks TM, Mullins T, Kostiuk B, Provenzano D, Pukatzki S. The Vibrio cholerae type VI secretion system employs diverse effector modules for intraspecific competition. Nat Commun. 2014;5:3549. doi: 10.1038/ncomms4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jani AJ, Cotter PA. Type VI secretion: not just for pathogenesis anymore. Cell Host Microbe. 2010;8:2–6. doi: 10.1016/j.chom.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russell AB, Wexler AG, Harding BN, Whitney JC, Bohn AJ, Goo YA, Tran BQ, Barry NA, Zheng H, Peterson SB, Chou S, Gonen T, Goodlett DR, Goodman AL, Mougous JD. A Type VI Secretion-Related Pathway in Bacteroidetes Mediates Interbacterial Antagonism. Cell Host Microbe. 2014;16:227–236. doi: 10.1016/j.chom.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruhe ZC, Low DA, Hayes CS. Bacterial contact-dependent growth inhibition. Trends Microbiol. 2013;21:230–237. doi: 10.1016/j.tim.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patzer SI, Baquero MR, Bravo D, Moreno F, Hantke K. The colicin G, H and X determinants encode microcins M and H47, which might utilize the catecholate siderophore receptors FepA, Cir, Fiu and IroN. Microbiology. 2003;149:2557–2570. doi: 10.1099/mic.0.26396-0. [DOI] [PubMed] [Google Scholar]
- 41.Rebuffat S. Microcins in action: amazing defence strategies of Enterobacteria. Biochem Soc Trans. 2012;40:1456–1462. doi: 10.1042/BST20120183. [DOI] [PubMed] [Google Scholar]
- 42.Nolan EM, Walsh CT. Investigations of the MceIJ-catalyzed posttranslational modification of the microcin E492 C-terminus: linkage of ribosomal and nonribosomal peptides to form “trojan horse” antibiotics. Biochemistry. 2008;47:9289–9299. doi: 10.1021/bi800826j. [DOI] [PubMed] [Google Scholar]
- 43.Martinez FA, Balciunas EM, Converti A, Cotter PD, de Souza Oliveira RP. Bacteriocin production by Bifidobacterium spp. A review. Biotechnol Adv. 2013;31:482–488. doi: 10.1016/j.biotechadv.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 44.Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, Reading NC, Villablanca EJ, Wang S, Mora JR, Umesaki Y, Mathis D, Benoist C, Relman DA, Kasper DL. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johansson ME, Ambort D, Pelaseyed T, Schutte A, Gustafsson JK, Ermund A, Subramani DB, Holmen-Larsson JM, Thomsson KA, Bergstrom JH, van der Post S, Rodriguez-Pineiro AM, Sjovall H, Backstrom M, Hansson GC. Composition and functional role of the mucus layers in the intestine. Cell Mol Life Sci. 2011;68:3635–3641. doi: 10.1007/s00018-011-0822-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petersson J, Schreiber O, Hansson GC, Gendler SJ, Velcich A, Lundberg JO, Roos S, Holm L, Phillipson M. Importance and regulation of the colonic mucus barrier in a mouse model of colitis. Am J Physiol Gastrointest Liver Physiol. 2011;300:G327–333. doi: 10.1152/ajpgi.00422.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johansson ME, Larsson JM, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garrido D, Barile D, Mills DA. A molecular basis for bifidobacterial enrichment in the infant gastrointestinal tract. Adv Nutr. 2012;3:415S–421S. doi: 10.3945/an.111.001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ewaschuk JB, Diaz H, Meddings L, Diederichs B, Dmytrash A, Backer J, Looijer-van Langen M, Madsen KL. Secreted bioactive factors from Bifidobacterium infantis enhance epithelial cell barrier function. Am J Physiol Gastrointest Liver Physiol. 2008;295:G1025–1034. doi: 10.1152/ajpgi.90227.2008. [DOI] [PubMed] [Google Scholar]
- 51.Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, Taylor TD, Itoh K, Kikuchi J, Morita H, Hattori M, Ohno H. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 52.Hafez M, Hayes K, Goldrick M, Warhurst G, Grencis R, Roberts IS. The K5 capsule of Escherichia coli strain Nissle 1917 is important in mediating interactions with intestinal epithelial cells and chemokine induction. Infect Immun. 2009;77:2995–3003. doi: 10.1128/IAI.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Franchi L, Kamada N, Nakamura Y, Burberry A, Kuffa P, Suzuki S, Shaw MH, Kim YG, Nuñez G. NLRC4-driven production of IL-1beta discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nat Immunol. 2012;13:449–456. doi: 10.1038/ni.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hasegawa M, Kamada N, Jiao Y, Liu MZ, Nuñez G, Inohara N. Protective role of commensals against Clostridium difficile infection via an IL-1beta-mediated positive-feedback loop. J Immunol. 2012;189:3085–3091. doi: 10.4049/jimmunol.1200821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol. 2011;9:356–368. doi: 10.1038/nrmicro2546. [DOI] [PubMed] [Google Scholar]
- 56.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brandl K, Plitas G, Mihu CN, Ubeda C, Jia T, Fleisher M, Schnabl B, DeMatteo RP, Pamer EG. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455:804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brandl K, Plitas G, Schnabl B, DeMatteo RP, Pamer EG. MyD88-mediated signals induce the bactericidal lectin RegIII gamma and protect mice against intestinal Listeria monocytogenes infection. J Exp Med. 2007;204:1891–1900. doi: 10.1084/jem.20070563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dessein R, Gironella M, Vignal C, Peyrin-Biroulet L, Sokol H, Secher T, Lacas-Gervais S, Gratadoux JJ, Lafont F, Dagorn JC, Ryffel B, Akira S, Langella P, Nunez G, Sirard JC, Iovanna J, Simonet M, Chamaillard M. Toll-like receptor 2 is critical for induction of Reg3 beta expression and intestinal clearance of Yersinia pseudotuberculosis. Gut. 2009;58:771–776. doi: 10.1136/gut.2008.168443. [DOI] [PubMed] [Google Scholar]
- 61.Ayabe T, Satchell DP, Wilson CL, Parks WC, Selsted ME, Ouellette AJ. Secretion of microbicidal alpha-defensins by intestinal Paneth cells in response to bacteria. Nat Immunol. 2000;1:113–118. doi: 10.1038/77783. [DOI] [PubMed] [Google Scholar]
- 62.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci U S A. 2008;105:20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chu H, Pazgier M, Jung G, Nuccio SP, Castillo PA, de Jong MF, Winter MG, Winter SE, Wehkamp J, Shen B, Salzman NH, Underwood MA, Tsolis RM, Young GM, Lu W, Lehrer RI, Bäumler AJ, Bevins CL. Human alpha-defensin 6 promotes mucosal innate immunity through self-assembled peptide nanonets. Science. 2012;337:477–481. doi: 10.1126/science.1218831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Salzman NH, Ghosh D, Huttner KM, Paterson Y, Bevins CL. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature. 2003;422:522–526. doi: 10.1038/nature01520. [DOI] [PubMed] [Google Scholar]
- 65.Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D, Stoel M, Zhou Y, Sodergren E, Weinstock GM, Bevins CL, Williams CB, Bos NA. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wehkamp J, Harder J, Weichenthal M, Schwab M, Schaffeler E, Schlee M, Herrlinger KR, Stallmach A, Noack F, Fritz P, Schroder JM, Bevins CL, Fellermann K, Stange EF. NOD2 (CARD15) mutations in Crohn’s disease are associated with diminished mucosal alpha-defensin expression. Gut. 2004;53:1658–1664. doi: 10.1136/gut.2003.032805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nieuwenhuis EE, Matsumoto T, Lindenbergh D, Willemsen R, Kaser A, Simons-Oosterhuis Y, Brugman S, Yamaguchi K, Ishikawa H, Aiba Y, Koga Y, Samsom JN, Oshima K, Kikuchi M, Escher JC, Hattori M, Onderdonk AB, Blumberg RS. Cd1d-dependent regulation of bacterial colonization in the intestine of mice. J Clin Invest. 2009;119:1241–1250. doi: 10.1172/JCI36509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Robertson SJ, Zhou JY, Geddes K, Rubino SJ, Cho JH, Girardin SE, Philpott DJ. Nod1 and Nod2 signaling does not alter the composition of intestinal bacterial communities at homeostasis. Gut Microbes. 2013;4:222–231. doi: 10.4161/gmic.24373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shanahan MT, Carroll IM, Grossniklaus E, White A, von Furstenberg RJ, Barner R, Fodor AA, Henning SJ, Sartor RB, Gulati AS. Mouse Paneth cell antimicrobial function is independent of Nod2. Gut. 2014;63:903–910. doi: 10.1136/gutjnl-2012-304190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schlee M, Wehkamp J, Altenhoefer A, Oelschlaeger TA, Stange EF, Fellermann K. Induction of human beta-defensin 2 by the probiotic Escherichia coli Nissle 1917 is mediated through flagellin. Infect Immun. 2007;75:2399–2407. doi: 10.1128/IAI.01563-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eidenschenk C, Rutz S, Liesenfeld O, Ouyang W. Role of IL-22 in microbial host defense. Curr Top Microbiol Immunol. 2014;380:213–236. doi: 10.1007/978-3-662-43492-5_10. [DOI] [PubMed] [Google Scholar]
- 72.Satoh-Takayama N, Vosshenrich CA, Lesjean-Pottier S, Sawa S, Lochner M, Rattis F, Mention JJ, Thiam K, Cerf-Bensussan N, Mandelboim O, Eberl G, Di Santo JP. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29:958–970. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 73.Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, Ouyang W. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 74.Spits H, Di Santo JP. The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nat Immunol. 2010;12:21–27. doi: 10.1038/ni.1962. [DOI] [PubMed] [Google Scholar]
- 75.Qiu J, Guo X, Chen ZM, He L, Sonnenberg GF, Artis D, Fu YX, Zhou L. Group 3 innate lymphoid cells inhibit T-cell-mediated intestinal inflammation through aryl hydrocarbon receptor signaling and regulation of microflora. Immunity. 2013;39:386–399. doi: 10.1016/j.immuni.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, Zecchi R, D’Angelo C, Massi-Benedetti C, Fallarino F, Carvalho A, Puccetti P, Romani L. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 77.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov, Umesaki Y, Itoh K, Honda K. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, Kim S, Fritz JV, Wilmes P, Ueha S, Matsushima K, Ohno H, Olle B, Sakaguchi S, Taniguchi T, Morita H, Hattori M, Honda K. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 80.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 81.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cerutti A, Rescigno M. The biology of intestinal immunoglobulin A responses. Immunity. 2008;28:740–750. doi: 10.1016/j.immuni.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cerutti A, Cols M, Puga I. Marginal zone B cells: virtues of innate-like antibody-producing lymphocytes. Nat Rev Immunol. 2013;13:118–132. doi: 10.1038/nri3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Macpherson AJ. IgA adaptation to the presence of commensal bacteria in the intestine. Curr Top Microbiol Immunol. 2006;308:117–136. doi: 10.1007/3-540-30657-9_5. [DOI] [PubMed] [Google Scholar]
- 85.Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 86.Macpherson AJ, Gatto D, Sainsbury E, Harriman GR, Hengartner H, Zinkernagel RM. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science. 2000;288:2222–2226. doi: 10.1126/science.288.5474.2222. [DOI] [PubMed] [Google Scholar]
- 87.Lecuyer E, Rakotobe S, Lengline-Garnier H, Lebreton C, Picard M, Juste C, Fritzen R, Eberl G, McCoy KD, Macpherson AJ, Reynaud CA, Cerf-Bensussan N, Gaboriau-Routhiau V. Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity. 2014;40:608–620. doi: 10.1016/j.immuni.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 88.Lundell AC, Bjornsson V, Ljung A, Ceder M, Johansen S, Lindhagen G, Tornhage CJ, Adlerberth I, Wold AE, Rudin A. Infant B cell memory differentiation and early gut bacterial colonization. J Immunol. 2012;188:4315–4322. doi: 10.4049/jimmunol.1103223. [DOI] [PubMed] [Google Scholar]
- 89.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, van den Brink MR, Jenq RR, Taur Y, Sander C, Cross J, Toussaint NC, Xavier JB, Pamer EG. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2014 doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]