Abstract
Purpose: The associations between hypoxia-inducible factor-1 alpha (HIF-1alpha) and clinicopathological characteristics of cancers have been evaluated in various studies, with the conflicting results. The common rs11549465 (1772C/T) genetic polymorphism has been reported to be functional and may contribute to genetic susceptibility to cancers. However, the association between rs11549465 (1772C/T) and cancer risk remains inconclusive. Methods: To better understand the role of rs11549465 (1772C/T) polymorphism in global cancer, we conducted this comprehensive meta-analysis encompassing 7807 cases and 8633 controls. Results: Overall, the rs11549465 (1772C/T) genetic polymorphism was associated with higher cancer risk, especially exists in Asians. In the stratified analysis, significant associations were found between the HIF-1 rs11549465 polymorphism and gynecologic cancer among Caucasian population. We observed that the TT genotype might modulate gynecologic cancer (OR=9.92 [2.15-45.66]) risk comparing with the CC genotype. Moreover, a significantly increased lung and breast cancer risk was found among Asian population comparing with Caucasian population. When stratified by study design, significantly elevated susceptibility to cancer was found among hospital -based studies. Conclusions: Our meta-analysis suggested that the HIF-1 rs11549465 (1772C/T) genetic polymorphism is significantly associated with higher risk among Asian population and lower risk among Caucasian population in breast and lung cancer, and this SNP was significantly associated with the gynecologic cancer among Caucasian population. The effect of the rs11549465 polymorphism on cancer especially exists in Asians.
Keywords: Hif-1, rs11549465, cancer, genetic polymorphism, meta-analysis
Introduction
Cancer is one of the leading causes of death in the world. It has become a worldwide public health problem [1]. The exact mechanism of carcinogenesis is not yet fully elucidated [2]. Recently, it has become clear that genetic variation contributes to the development and progression of cancer [2,3]. However, due to various reasons, including considerable heterogeneity of the disease, the identification of susceptibility genes is difficult and most associations have not been replicated.
One of the most important features of tumors is hypoxia. Intratumoral hypoxia occurs when cells are located further from a functional blood vessel than is required for adequate diffusion of oxygen, as a result of rapid tumor cell proliferation and abnormal blood vessels [4]. Hypoxia conditions in tumor tissues induce a molecular response, which drives the activation of transcription factors. Among these, hypoxia-inducible factor-1 (HIF-1) plays an essential role in adaptive responses to reduced oxygen levels [5,6].
HIF-1 is a dimeric protein complex, consisting of α and β subunits. The activity of HIF-1 is predominantly regulated through the stability of the subunit [7]. Koshiji et al. demonstrated that hif-1α (PASD8) inhibits the DNA mismatch repair system (MSH2 and MSH6), which is responsible for genetic instability [8]. Other researchers have also reported that hypoxia down regulates the expression of DNA double-stranded break repair genes [9-12]. These data support the concept that defective DNA repair pathways cause genomic instability within the tumor microenvironment. PASD8 (Hif-1α) is overexpressed in >90% of colon, lung and prostate cancers, whereas no expression was detected in corresponding normal tissues [13], indicating a role of hif-1α in cancer. It is over expressed in several human cancers, such as head-neck, colon, breast, stomach, pancreas, prostate, kidney, esophagus, endometrial, and non-small-cell lung cancer [14-20]. The target genes of hif-1α are particularly relevant to cancer, encoding angiogenic factors, proliferation/survival factors, glucose transporters and glycolytic enzymes [21]. As such, variability in this protein is likely to influence individual risk to this pathology.
A number of investigators have studied the possible association between the hif-1 polymorphisms and cancer risk, but the results have been conflicting [20,22-39]. Thus, the association between the HIF-1 polymorphisms and cancers requires further investigation. In an attempt to clarify this inconsistency, we have combined all the published studies of hospital and population up to July 2014 in a meta-analysis to give a comprehensive picture of the role of HIF-1α gene using multiple research methods and models.
In this study, a comprehensive meta-analysis was performed on previous reports to investigate the association of hif-1α rs11549465 (1772C/T) polymorphisms with all cancers, different kinds of cancers, different kinds of detection method, and different kinds of populations.
Methods
Search strategy and data extraction
In this meta-analysis, a comprehensive literature research of the US National Library of Medicine’s PubMed database, ISI Web of Knowledge, Medline, Embase and Google Scholar Search (update to July 2014) was conducted using the search terms including “hif-1α” or “hypoxia-inducible factor-1” or “rs11549465” or “1772C/T” or “P582S”, ”polymorphisms” or “variation” or “mutation” or “SNP”, “tumour” or “tumor” or “cancer” or “neoplasm” or “phyma” or “oncoma” or “knub” or “carcinoma” or “malignancy”, and the combined phrases in order to obtain all genetic studies on the relationship of rs11549465 polymorphism and cancers. We also used a hand search of references of original studies or reviewed articles on this topic to identify additional studies. Eligible studies were selected according to the following explicit inclusion criteria: (1) a case control study on the association between rs11549465 polymorphism and cancer risk, (2) detailed number of different genotypes for estimating an odds ratio (OR) with 95% confidence interval (CI), (3) when several publications reported on the same population data, the largest or most complete study was chosen, (4) cases with carcinomas were diagnosed by histopathology, (5) animal studies, case reports, review articles, abstracts, editorials, reports with incomplete data, and studies based on pedigree data were excluded (Figure 1). For each eligible study, the following information was recorded: the first author’s name, the year of publication, ethnicity, genotyping methods, sources of control, racial descent of the study population, genotype and allele distributions and main results of each study.
Figure 1.
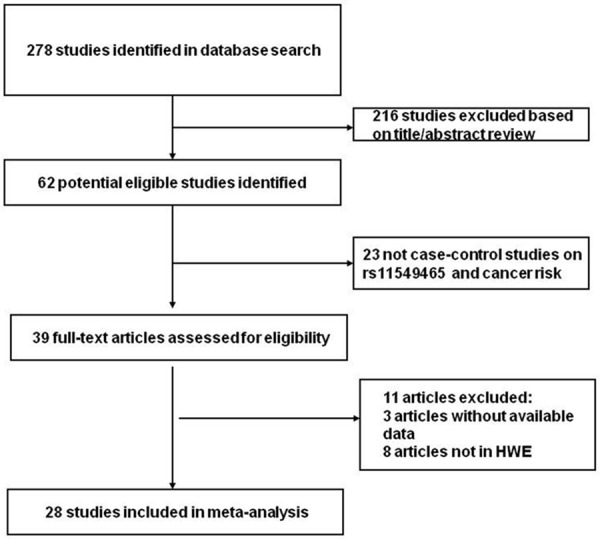
Flow diagram of study identification.
Statistics
The strength of relationship between rs11549465 polymorphism and cancer was assessed by using crude OR with 95% CI. We examined the association between the rs11549465 polymorphism and cancer risk using the following genetic models: homozygote comparison (TT vs. CC), heterozygote comparison (TC vs. CC), dominant genetic model (TT/TC vs. CC), recessive genetic model (TT vs. TC/CC) and additive model (T vs. C). Firstly, we checked the Hardy-Weinberg equilibrium (HWE) in controls for each study. Then we performed Q-test for evaluating the heterogeneity [40]. Fixed effects model was used to pool the data when the P-value of Q-test ≥0.05; otherwise, random effects model was selected [41]. I2 was also used to assess the heterogeneity in this meta-analysis. If I2 >50%, the heterogeneity exists [42]. We also performed sensitivity analysis and subgroup analysis to explore the reason of heterogeneity. Both funnel plot and Egger’s test were used to assess the publication bias (P<0.05 was representative of statistical significance) [43]. All statistical analysis were performed using STATA 12.0 software (Stata Corp., College Station, Texas, USA) and Review Manager 5.2 (The Cochrane Collaboration, http://ims.cochrane.org/revman).
Results
Eligible studies
Overall, 28 relevant studies involving 7807 cases and 8633 controls were selected in this meta-analysis [20,24,27,28,30,31,33,34,39,44-62]. The main characteristics of these studies were shown in Table 1. Genotype and allele distributions of rs11549465 polymorphism among cancer cases and controls and P value of HWE in controls were shown in Tables 1 and 2. All studies were case-control studies, including four prostate cancer studies [20,30,45,52], three colorectal cancer studies [24,27,54], two gynecologic carcinoma studies [28,55] , five breast cancer studies [31,33,34,51,61], two oral squamous cell carcinoma (OSCC) studies [47,62], three lung cancer studies [48,56,59], two renal cell carcinoma studies [50,60] and the others (including esophageal squamous cell carcinoma (ESCC) [39], head and neck squamous cell carcinoma (HNSCC) [44], transitional cell carcinoma of the bladder [46], gastric cancer [49], hepatocellular carcinoma [53], pancreatic cancer [57] and glioma [58]). Cancers were diagnosed histopathologically in most studies. There were nineteen studies [20,24,30,33,34,39,44,46,47,49,51,53-60,62] of Asian descent, nine studies [27,28,30,31,45,48,50,52,61] of Caucasian descent. Population-based controls were carried out in 11 studies, while hospital-based controls were carried out in 17 studies. All studies were reported in English and the genotyping methods contained the classic polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) assay, PCR-sequencing, SNP-ITTM, PCR-LDR, SnaPShot and Taqman. The genotype distributions of controls were all in agreement with HWE.
Table 1.
Main characteristics of included studies in the meta-analysis
| Studies (cancer type) | Country | Ethnicity | Genotype assay | Source of control | Case/control | P |
|---|---|---|---|---|---|---|
| Tanimoto 2003 HNSCC | Japan | Asian | PCR-Sequencing | Population | 55/110 | 0.545 |
| Foley 2009 prostate cancer | Dublin | Caucasian | PCR-Sequencing | Population | 95/188 | 0.623 |
| Li 2007 prostate cancer | USA | Caucasian | PCR-RFLP | Population | 1041/1234 | 0.159 |
| Orr-Urtreger 2007 prostate cancer | Israel | Caucasian | PCR-RFLP | Population | 402/300 | 0.137 |
| Lee 2008 breast cancer | Korean | Asian | SNP-ITTM | Population | 1332/1369 | 0.250 |
| Apaydin 2008 breast cancer | Turkey | Caucasian | PCR-RFLP | Population | 102/102 | 0.415 |
| Kim 2008 breast cancer | Korea | Asian | PCR-Sequencing | Hospital | 90/102 | 0.641 |
| Konac 2007 gynecologic cancer | Turkey | Caucasian | PCR-RFLP | Hospital | 102/107 | 0.229 |
| Li 2012 prostate cancer | China | Asian | Taqman | Population | 662/716 | 0.267 |
| Fransen 2006 colorectal cancer | Sweden | Caucasian | PCR-RFLP | Hospital | 198/258 | 0.916 |
| Kuwai 2004 colorectal cancer | Japan | Asian | PCR-Sequencing | Population | 100/100 | 0.561 |
| Ling 2005 ESCC | China | Asian | PCR-RFLP | Population | 95/104 | 0.569 |
| Naidu 2009 breast cancer | Malaysia | Asian | PCR-RFLP | Hospital | 410/275 | 0.922 |
| Zagouri 2012 breast cancer | Greece | Caucasian | PCR-RFLP | Hospital | 113/124 | 0.413 |
| Kuo 2012 lung cancer | China | Asian | PCR-RFLP | Hospital | 285/300 | 0.132 |
| Wang 2011 pancreatic cancer | China | Asian | PCR-Sequencing | Hospital | 263/271 | 0.352 |
| Kang 2011 colorectal cancer | Korea | Asian | PCR-RFLP | Hospital | 50/50 | 0.335 |
| Xu 2011 Glioma | China | Asian | PCR-RFLP | Hospital | 150/150 | 0.354 |
| Hsiao 2010 hepatocellular carcinoma | China | Asian | PCR-RFLP | Hospital | 102/347 | 0.722 |
| Chen 2009 OSCC | China | Asian | PCR-RFLP | Population | 174/347 | 0.722 |
| Konac 2009 lung cancer | Turkey | Caucasian | PCR-RFLP | Hospital | 141/156 | 0.335 |
| Li 2009 gastric cancer | Tibetan | Asian | PCR-LDR | Hospital | 87/106 | 0.501 |
| Nadaoka 2008 bladder cancer | Japan | Asian | PCR-RFLP | Hospital | 219/461 | 0.305 |
| Kim 2011 cervical cancer | Korea | Asian | SNaPShot | Hospital | 199/214 | 0.325 |
| Qin 2012 renal cell carcinoma | China | Asian | Taqman | Hospital | 620/623 | 0.219 |
| Morris 2009 renal cell carcinoma | Poland | Caucasian | Taqman | Population | 332/313 | 0.083 |
| Putra 2011 lung cancer | Japan | Asian | PCR-Sequencing | Hospital | 83/110 | 0.545 |
| Shieh 2010 OSCC | China | Asian | PCR-Sequencing | Hospital | 305/96 | 0.711 |
P Value of Hardy-Weinberg equilibrium in controls.
Table 2.
Distribution of rs11549465 polymorphism and the main results of eligible studies
| Stuies (cancer type) | Case (TT/TC/CC) | Control (TT/TC/CC) | OR (95% CI) | ||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| TT vs. CC | TC vs. CC | TT/TC vs. CC | TT vs. TC/CC | T vs. C | |||
| Tanimoto 2003 HNSCC | 55 (0/10/45) | 110 (0/12/98) | - | 1.81 (0.73-4.51) | 1.81 (0.73-4.51) | - | 1.73 (0.72-4.15) |
| Foley 2009 prostate cancer | 95 (0/30/65) | 188 (0/13/175) | - | 6.21 (3.05-12.64) | 6.21 (3.05-12.64) | - | 5.24 (2.66-10.30) |
| Li 2007 prostate cancer | 1041 (14/209/818) | 1234 (18/221/995) | 0.95 (0.47-1.91) | 1.15 (0.93-1.42) | 1.13 (0.92-1.39) | 0.92 (0.46-1.86) | 1.11 (0.92-1.33) |
| Orr-Urtreger 2007 prostate cancer | 402 (16/99/287) | 300 (3/80/217) | 4.03 (1.16-14.01) | 0.94 (0.66-1.32) | 1.05 (0.75-1.46) | 4.10 (1.18-14.21) | 1.16 (0.87-1.56) |
| Lee 2008 breast cancer | 1332 (6/119/1207) | 1369 (1/123/1245) | 6.19 (0.74-51.48) | 1.00 (0.77-1.30) | 1.04 (0.80-1.35) | 6.19 (0.74-51.49) | 1.08 (0.84-1.39) |
| Apaydin 2008 breast cancer | 102 (2/21/79) | 102 (5/29/68) | 0.34 (0.06-1.83) | 0.62 (0.33-1.19) | 0.58 (0.31-1.08) | 0.39 (0.07-2.05) | 0.59 (0.34-1.02) |
| Kim 2008 breast cancer | 90 (1/8/81) | 102 (0/9/93) | 3.44 (0.14-85.66) | 1.02 (0.38-2.77) | 1.15 (0.43-3.03) | 3.44 (0.14-85.40) | 1.27 (0.51-3.21) |
| Konac 2007 gynecologic cancer | 102 (14/40/48) | 107 (2/37/68) | 9.92 (2.15-45.66) | 1.53 (0.86-2.74) | 1.96 (1.13-3.41) | 8.35 (1.85-37.75) | 2.11 (1.35-3.30) |
| Li 2012 prostate cancer | 662 (2/48/612) | 716 (0/57/659) | 5.38 (0.26-112.36) | 0.91 (0.61-1.35) | 0.94 (0.64-1.40) | 5.42 (0.26-113.18) | 0.99 (0.67-1.45) |
| Fransen 2006 colorectal cancer | 198 (3/28/167) | 258 (2/43/213) | 1.91 (0.32-11.58) | 0.83 (0.50-1.39) | 0.88 (0.53-1.45) | 1.97 (0.33-11.90) | 0.94 (0.59-1.49) |
| Kuwai 2004 colorectal cancer | 100 (0/0/100) | 100 (0/11/89) | - | 0.04 (0.00-0.67) | 0.04 (0.00-0.67) | - | 0.04 (0.00-0.70) |
| Ling 2005 ESCC | 95 (0/11/84) | 104 (0/11/93) | - | 1.11 (0.46-2.69) | 1.11 (0.46-2.69) | - | 1.10 (0.47-2.60) |
| Naidu 2009 breast cancer | 410 (16/100/294) | 275 (3/50/222) | 4.03 (1.16-13.99) | 1.51 (1.03-2.21) | 1.65 (1.14-2.39) | 3.68 (1.06-12.76) | 1.69 (1.21-2.36) |
| Zagouri 2012 breast cancer | 113 (0/15/98) | 124 (0/17/107) | - | 0.96 (0.46-2.03) | 0.96 (0.46-2.03) | - | 0.97 (0.47-1.98) |
| Kuo 2012 non–small-cell lung cancer | 285 (38/94/153) | 300 (11/73/216) | 4.88 (2.42-9.84) | 1.82 (1.26-2.63) | 2.22 (1.57-3.13) | 4.04 (2.02-8.08) | 2.26 (1.70-3.00) |
| Wang 2011 pancreatic cancer | 263 (0/54/209) | 271 (0/29/242) | - | 2.16 (1.32-3.51) | 2.16 (1.32-3.51) | - | 2.02 (1.27-3.23) |
| Kang 2011 colorectal cancer | 50 (0/4/46) | 50 (0/12/38) | - | 0.28 (0.08-0.92) | 0.28 (0.08-0.92) | - | 0.31 (0.10-0.98) |
| Xu 2011 Glioma | 150 (2/27/121) | 150 (1/14/135) | 2.23 (0.20-24.92) | 2.15 (1.08-4.29) | 2.16 (1.10-4.21) | 2.01 (0.18-22.45) | 2.05 (1.09-3.83) |
| Hsiao 2010 Hepatocellular carcinoma | 102 (0/8/94) | 347 (0/13/334) | - | 2.19 (0.88-5.43) | 2.19 (0.88-5.43) | - | 2.14 (0.87-5.23) |
| Chen 2009 OSCC | 174 (1/10/163) | 347 (0/13/334) | 6.14 (0.25-151.49) | 1.58 (0.68-3.67) | 1.73 (0.76-3.95) | 6.01 (0.24-148.26) | 1.87 (0.84-4.14) |
| Konac 2009 Lung cancer | 141 (0/31/110) | 156 (2/43/111) | 0.20 (0.01-4.25) | 0.73 (0.43-1.24) | 0.70 (0.41-1.18) | 0.22 (0.01-4.59) | 0.70 (0.43-1.13) |
| Li 2009 Gastric cancer | 87 (0/4/83) | 106 (0/13/93) | - | 0.34 (0.11-1.10) | 0.34 (0.11-1.10) | - | 0.36 (0.12-1.13) |
| Nadaoka 2008 TCC | 219 (0/22/197) | 461 (0/42/419) | - | 1.11 (0.65-1.92) | 1.11 (0.65-1.92) | - | 1.11 (0.65-1.88) |
| Kim 2011 Cervical cancer | 199 (0/22/177) | 214 (0/27/187) | - | 0.86 (0.47-1.57) | 0.86 (0.47-1.57) | - | 0.87 (0.49-1.55) |
| Qin 2012 renal cell carcinoma | 620 (2/46/572) | 623 (2/43/578) | 1.01 (0.14-7.20) | 1.08 (0.70-1.66) | 1.08 (0.71-1.65) | 1.00 (0.14-7.16) | 1.07 (0.71-1.61) |
| Morris 2009 renal cell carcinoma | 332 (3/39/290) | 313 (5/46/262) | 0.54 (0.13-2.29) | 0.77 (0.48-1.21) | 0.74 (0.48-1.16) | 0.56 (0.13-2.37) | 0.74 (0.49-1.11) |
| Putra 2011 lung cancer | 83 (0/9/74) | 110 (0/12/98) | - | 0.99 (0.40-2.48) | 0.99 (0.40-2.48) | - | 0.99 (0.41-2.42) |
| Shieh 2010 OSCC | 305 (0/23/282) | 96 (0/7/89) | - | 1.04 (0.43-2.50) | 1.04 (0.43-2.50) | - | 1.04 (0.44-2.45) |
The numbers in parentheses represent 95% confidence interval [CI].
Meta-analysis
Overall, as shown in Table 3, we observed that the rs11549465 (1772C/T) polymorphism increased the cancer risk in the homozygote (TT vs. CC, OR=2.15 [1.19-3.88]) (Figure 2), heterozygote model (TC vs. CC, OR=1.15 [0.96-1.36]) (Figure 3), dominant genetic model (OR=1.19 [0.99-1.42]) (Figure 4), recessive model (OR=2.21 [1.60-3.05]) (Figure 5) and additive model (T vs. C, OR=1.20 [1.01-1.44]) (Figure 6) when all the eligible studies were pooled into the meta-analysis. In the homozygote comparison, heterozygote comparison, dominant genetic, recessive genetic and additive models, all the P values of Q-test were lower than 0.05 and I2 values were higher than 50%. So we performed the sensitive analysis by deleting one single study from overall pooled analysis each time to check the influence of the removed data. However, the results revealed that no extreme sensitive study changed the between-study heterogeneities.
Table 3.
Results of meta-analysis for rs11549465 polymorphism and cancer risk
| Study Groups | NO.of studies | Case (TT/TC/CC) | Control (TT/TC/CC) | TT vs. CC | TC vs. CC | TT/TC vs. CC | TT vs. TC/CC | T vs. C | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||
| OR (95% CI) | Pa; Pb; I2 (%) | OR (95% CI) | Pa; Pb; I2 (%) | OR (95% CI) | Pa; Pb; I2 (%) | OR (95% CI) | Pa; Pb; I2 (%) | OR (95% CI) | Pa; Pb; I2 (%) | ||||
| All population | 28 | 7807 (120/1131/6556) | 8633 (55/1100/7478) | 2.15 (1.19-3.88) | 0.011; 0.010; 52.0% | 1.15 (0.96-1.36) | 0.127; 0.000; 63.8% | 1.19 (0.99-1.42) | 0.071; 0.000; 69.1% | 2.21 (1.60-3.05) | 0.010; 0.028; 45.5% | 1.20 (1.01-1.44) | 0.043; 0.000; 71.8% |
| Ethnicity | |||||||||||||
| Asian | 19 | 5281 (68/619/4594) | 5851 (18/571/5262) | 4.17 (2.48-7.01) | 0.000; 0.913; 0.0% | 1.19 (0.97-1.47) | 0.097; 0.003; 53.7% | 1.24 (0.99-1.55) | 0.063; 0.000; 60.7% | 3.70 (2.21-6.19) | 0.000; 0.936; 0.0% | 1.26 (1.01-1.57) | 0.041; 0.000; 63.2% |
| Caucasian | 9 | 2526 (52/512/1962) | 2782 (37/529/2216) | 1.34 (0.55-3.31) | 0.521; 0.012; 63.5% | 1.09 (0.79-1.50) | 0.613; 0.000; 76.3% | 1.12 (0.80-1.56) | 0.503; 0.000; 78.9% | 1.36 (0.57-3.21) | 0.489; 0.019; 60.4% | 1.13 (0.83-1.54) | 0.432; 0.000; 80.6% |
| Source of control | |||||||||||||
| Population | 11 | 4390 (44/596/3750) | 4883 (32/616/4235) | 1.39 (0.88-2.20) | 0.158; 0.067; 49.0% | 1.12 (0.84-1.50) | 0.430; 0.000; 73.4% | 1.14 (0.86-1.52) | 0.360; 0.000; 74.0% | 1.40 (0.89-2.22) | 0.148; 0.073; 47.9% | 1.15 (0.88-1.49) | 0.302; 0.000; 73.3% |
| Hospital | 17 | 3417 (76/535/2806) | 3750 (23/484/3243) | 3.75 (2.34-6.01) | 0.000; 0.326; 13.3% | 1.17 (0.94-1.46) | 0.164; 0.005; 53.3% | 1.21 (0.95-1.55) | 0.121; 0.000; 63.1% | 3.36 (2.10-5.37) | 0.000; 0.455; 0.0% | 1.23 (0.97-1.57) | 0.090; 0.000; 68.0% |
| Detection method | |||||||||||||
| PCR-Sequencing | 7 | 991 (1/134/856) | 977 (0/93/884) | 3.44 (0.14-85.66) | 0.451; -; - | 1.51 (0.78-2.94) | 0.000; 0.001; 75.0% | 1.54 (0.80-2.97) | 0.198; 0.001; 74.5% | 3.44 (0.14-85.40) | 0.452; -; - | 1.53 (0.84-2.79) | 0.000; 0.002; 71.3% |
| PCR-RFLP | 15 | 3584 (106/719/2759) | 4315 (47/698/3570) | 2.31 (1.12-4.73) | 0.000; 0.005; 61.6% | 1.15 (0.95-1.40) | 0.012; 0.010; 51.9% | 1.21 (0.97-1.52) | 0.098; 0.000; 66.0% | 2.21 (1.13-4.30) | 0.020; 0.016; 55.8% | 1.24 (0.98-1.56) | 0.000; 0.000; 73.2% |
| SNP-ITTM | 1 | 1332 (6/119/1207) | 1369 (1/123/1245) | 6.19 (0.74-51.48) | 0.092; -; - | 1.00 (0.77-1.30) | 0.988; -; - | 1.04 (0.80-1.35) | 0.769; -; - | 6.19 (0.74-51.49) | 0.092; -; - | 1.08 (0.84-1.39) | 0.543; -; - |
| Taqman | 3 | 1614 (7/133/1474) | 1652 (7/146/1499) | 0.97 (0.35-2.66) | 0.950; 0.397; 0.0% | 0.91 (0.71-1.17) | 0.477; 0.562; 0.0% | 0.92 (0.72-1.17) | 0.488; 0.486; 0.0% | 0.99 (0.36-2.71) | 0.842; 0.407; 0.0% | 0.92 (0.73-1.16) | 0.502; 0.415; 0.0% |
| PCR-LDR | 1 | 87 (0/4/83) | 106 (0/13/93) | - | - | 0.34 (0.11-1.10) | 0.072; -; - | 0.34 (0.11-1.10) | 0.072; -; - | - | - | 0.36 (0.12-1.13) | 0.079; -; - |
| SNaPShot | 1 | 199 (0/22/177) | 214 (0/27/187) | - | - | 0.86 (0.47-1.57) | 0.624; -; - | 0.86 (0.47-1.57) | 0.624; -; - | - | - | 0.87 (0.49-1.55) | 0.635; -; - |
| Cancer type | |||||||||||||
| HNSCC | 1 | 55 (0/10/45) | 110 (0/12/98) | - | - | 1.81 (0.73-4.51) | 0.199; -; - | 1.81 (0.73-4.51) | 0.199; -; - | - | - | 1.73 (0.72-4.15) | 0.217; -; - |
| Prostate | 4 | 2200 (32/386/1782) | 2438 (21/371/2046) | 2.02 (0.60-6.83) | 0.117; 0.090; 58.5% | 1.42 (0.84-2.40) | 0.062; 0.000; 87.7% | 1.46 (0.89-2.40) | 0.031; 0.000; 86.9% | 2.03 (0.58-7.16) | 0.124; 0.077; 60.9% | 1.43 (0.93-2.21) | 0.017; 0.000; 85.0% |
| Prostate in Asian | 1 | 662 (2/48/612) | 716 (0/57/659) | 5.38 (0.26-112.36) | 0.278; -; - | 0.91 (0.61-1.35) | 0.631; -; - | 0.94 (0.64-1.40) | 0.777; -; - | 5.42 (0.26-113.18) | 0.275; -; - | 0.99 (0.67-1.45) | 0.943; -; - |
| Prostate in Caucasian | 3 | 1538 (30/338/1170) | 1722 (21/313/1387) | 1.78 (0.43-7.40) | 0.427; 0.045; 75.2% | 1.71 (0.83-3.51) | 0.144; 0.000; 91.2% | 1.75 (0.89-3.47) | 0.107; 0.000; 90.7% | 1.78 (0.41-7.74) | 0.443; 0.038; 76.8% | 1.68 (0.94-3.02) | 0.081; 0.000; 89.5% |
| Breast | 5 | 2047 (25/263/1759) | 1972 (9/228/1735) | 2.16 (0.52-8.85) | 0.031; 0.084; 54.8% | 1.07 (0.88-1.29) | 0.516; 0.188; 35.0% | 1.07 (0.76-1.50) | 0.254; 0.061; 55.6% | 2.27 (1.06-4.87) | 0.035; 0.120; 48.6% | 1.09 (0.76-1.55) | 0.106; 0.022; 64.9% |
| Breast in Asian | 3 | 1832 (23/227/1582) | 1746 (4/182/1560) | 4.38 (1.58-12.12) | 0.004; 0.932; 0.0% | 1.14 (0.92-1.41) | 0.228; 0.211; 35.6% | 1.26 (0.89-1.79) | 0.198; 0.132; 50.7% | 4.16 (1.51-11.48) | 0.006; 0.911; 0.0% | 1.32 (0.93-1.86) | 0.115; 0.109; 54.9% |
| Breast in Caucasian | 2 | 215 (2/36/177) | 226 (5/46/175) | 0.34 (0.06-1.83) | 0.211; -; - | 0.75 (0.46-1.22) | 0.251; 0.388; 0.0% | 0.72 (0.44-1.16) | 0.178; 0.309; 3.2% | 0.39 (0.07-2.05) | 0.265; -; - | 0.71 (0.45-1.14) | 0.156; 0.286; 12.3% |
| Gynecologicc | 2 | 301 (14/62/225) | 321 (2/64/255) | 9.92 (2.15-45.66) | 0.003; -; - | 1.16 (0.77-1.75) | 0.488; 0.176; 45.4% | 1.31 (0.58-2.94) | 0.152; 0.048; 74.5% | 8.35 (1.85-37.75) | 0.006; -; - | 1.38 (0.58-3.29) | 0.020; 0.018; 82.2% |
| Gynecologic in Asian | 1 | 199 (0/22/177) | 214 (0/27/187) | - | - | 0.86 (0.47-1.57) | 0.624; -; - | 0.86 (0.47-1.57) | 0.624; -; - | - | - | 0.87 (0.49-1.55) | 0.635; -; - |
| Gynecologic in Caucasian | 1 | 102 (14/40/48) | 107 (2/37/68) | 9.92 (2.15-45.66) | 0.003; -; - | 1.53 (0.86-2.74) | 0.150; -; - | 1.96 (1.13-3.41) | 0.017; -; - | 8.35 (1.85-37.75) | 0.006; -; - | 2.11 (1.35-3.30) | 0.001; -; - |
| Colorectal | 3 | 348 (3/32/313) | 408 (2/66/340) | 1.91 (0.32-11.58) | 0.480; -; - | 0.34 (0.09-1.34) | 0.009; 0.030; 71.5% | 0.34 (0.08-1.41) | 0.016; 0.023; 73.4% | 1.97 (0.33-11.90) | 0.460; -; - | 0.38 (0.09-1.50) | 0.035; 0.021; 74.0% |
| Colorectal in Asian | 2 | 150 (0/4/146) | 150 (0/13/127) | - | - | 0.15 (0.02-1.01) | 0.051; 0.182; 43.8% | 0.15 (0.02-1.01) | 0.051; 0.182; 43.8% | - | - | 0.16 (0.02-1.15) | 0.069; 0.169; 47.1% |
| Colorectal in Caucasian | 1 | 198 (3/28/167) | 258 (2/43/213) | 1.91 (0.32-11.58) | 0.480; -; - | 0.83 (0.50-1.39) | 0.482; -; - | 0.88 (0.53-1.45) | 0.612; -; - | 1.97 (0.33-11.90) | 0.460; -; - | 0.94 (0.59-1.49) | 0.783; -; - |
| ESCC | 1 | 95 (0/11/84) | 104 (0/11/93) | - | - | 1.11 (0.46-2.69) | 0.822; -; - | 1.11 (0.46-2.69) | 0.822; -; - | - | - | 1.10 (0.47-2.60) | 0.827; -; - |
| Lung | 3 | 509 (38/134/337) | 566 (13/128/425) | 1.41 (0.07-30.44) | 0.000; 0.044; 75.3% | 1.13 (0.59-2.19) | 0.067; 0.018; 75.2% | 1.19 (0.51-2.76) | 0.003; 0.001; 85.6 | 1.38 (0.09-22.18) | 0.000; 0.065; 70.6% | 1.19 (0.50-2.86) | 0.000; 0.000; 88.9% |
| Lung in Asian | 2 | 368 (38/103/227) | 410 (11/85/314) | 4.88 (2.42-9.84) | 0.000; -; - | 1.56 (0.94-2.61) | 0.088; 0.230; 30.6% | 1.67 (0.79-3.54) | 0.183; 0.107; 61.5% | 4.04 (2.02-8.08) | 0.000; -; - | 1.68 (0.77-3.64) | 0.191; 0.084; 66.4% |
| Lung in Caucasian | 1 | 141 (0/31/110) | 156 (2/43/111) | 0.20 (0.01-4.25) | 0.303; -; - | 0.73 (0.43-1.24) | 0.241; -; - | 0.70 (0.41-1.18) | 0.177; -; - | 0.22 (0.01-4.59) | 0.327; -; - | 0.70 (0.43-1.13) | 0.144; -; - |
| Pancreatic | 1 | 263 (0/54/209) | 271 (0/29/242) | - | - | 2.16 (1.32-3.51) | 0.002; -; - | 2.16 (1.32-3.51) | 0.002; -; - | - | - | 2.02 (1.27-3.23) | 0.003; -; - |
| Glioma | 1 | 150 (2/27/121) | 150 (1/14/135) | 2.23 (0.20-24.92) | 0.514; -; - | 2.15 (1.08-4.29) | 0.030; -; - | 2.16 (1.10-4.21) | 0.025; -; - | 2.01 (0.18-22.45) | 0.569; -; - | 2.05 (1.09-3.83) | 0.025; -; - |
| Hepatocellular | 1 | 102 (0/8/94) | 347 (0/13/334) | - | - | 2.19 (0.88-5.43) | 0.092; -; - | 2.19 (0.88-5.43) | 0.092; -; - | - | - | 2.14 (0.87-5.23) | 0.096; -; - |
| OSCC | 2 | 479 (1/33/445) | 443 (0/20/423) | 6.14 (0.25-151.49) | 0.267; -; - | 1.28 (0.69-2.38) | 0.432; 0.501; 0.0% | 1.35 (0.73-2.49) | 0.334; 0.403; 0.0% | 6.01 (0.24-148.26) | 0.273; -; - | 1.41 (0.78-2.56) | 0.257; 0.323; 0.0% |
| Gastric | 1 | 87 (0/4/83) | 106 (0/13/93) | - | - | 0.34 (0.11-1.10) | 0.072; -; - | 0.34 (0.11-1.10) | 0.072; -; - | - | - | 0.36 (0.12-1.13) | 0.079; -; - |
| Bladder | 1 | 219 (0/22/197) | 461 (0/2/419) | - | - | 1.11 (0.65-1.92) | 0.697; -; - | 1.11 (0.65-1.92) | 0.697; -; - | - | - | 1.11 (0.65-1.88) | 0.704; -; - |
| RCC | 2 | 952 (5/85/862) | 936 (7/89/840) | 0.67 (0.21-2.15) | 0.498; 0.616; 0.0% | 0.92 (0.67-1.26) | 0.599; 0.283; 13.1% | 0.90 (0.67-1.22) | 0.509; 0.235; 29.2% | 0.69 (0.22-2.17) | 0.521; 0.640; 0.0% | 0.89 (0.67-1.19) | 0.432; 0.207; 37.1% |
| RCC in Asian | 1 | 620 (2/46/572) | 623 (2/43/578) | 1.01 (0.14-7.20) | 0.992; -; - | 1.08 (0.70-1.66) | 0.724; -; - | 1.08 (0.71-1.65) | 0.728; -; - | 1.00 (0.14-7.16) | 0.996; -; - | 1.07 (0.71-1.61) | 0.738; -; - |
| RCC in Caucasian | 1 | 332 (3/39/290) | 313 (5/46/262) | 0.54 (0.13-2.29) | 0.405; -; - | 0.77 (0.48-1.21) | 0.254; -; - | 0.74 (0.48-1.16) | 0.189; -; - | 0.56 (0.13-2.37) | 0.432; -; - | 0.74 (0.49-1.11) | 0.149; -; - |
P value for Z test.
P value for Q test for between-study heterogeneity.
Ovarian, cervical and endometrial cancer.
The numbers in parentheses represent 95% confidence interval [CI]. The bold numbers mean that the OR values for the contrast models are significant.
Figure 2.
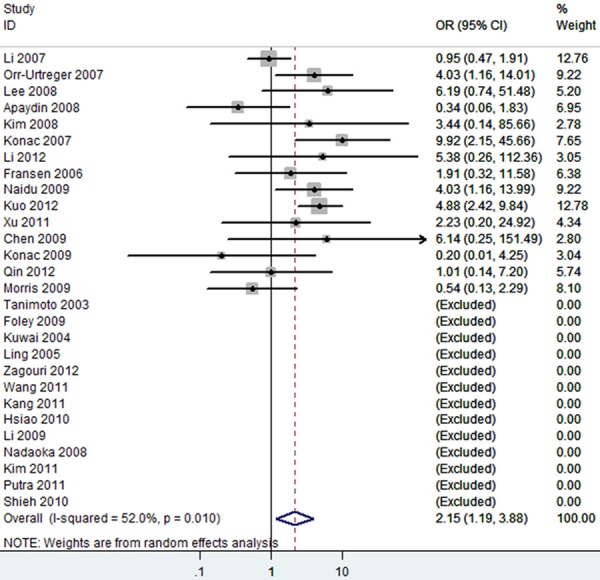
The forest plot of TT vs. CC of rs11549465 polymorphism and overall cancer risk (Random model). The overall OR is shown. The OR of each study is marked with a black dot. The overall OR is indicated by blue diamond.
Figure 3.
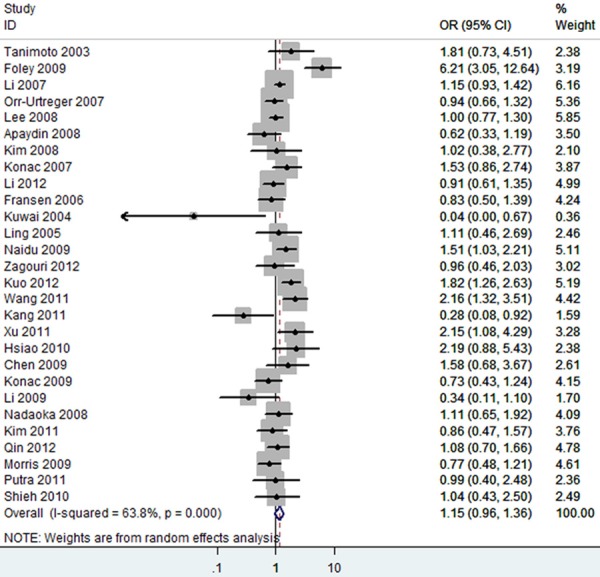
The forest plot of TC vs. CC of rs11549465 polymorphism and overall cancer risk (Random model). The overall OR is shown. The OR of each study is marked with a black dot. The overall OR is indicated by blue diamond.
Figure 4.
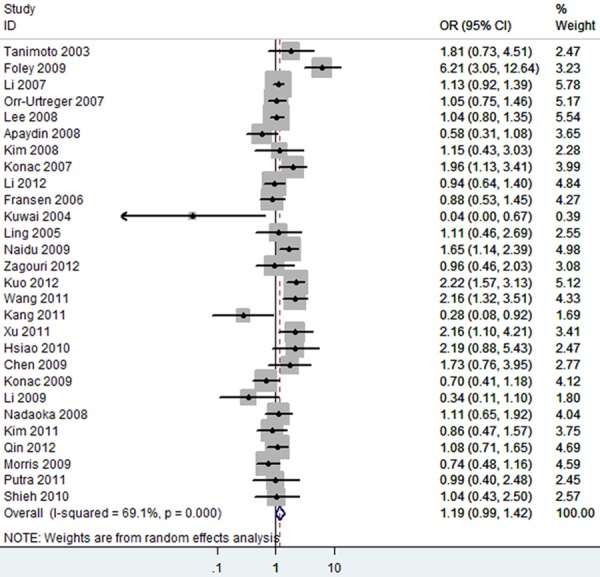
The forest plot of TT/TC vs. CC of rs11549465 polymorphism and overall cancer risk (Random model). The overall OR is shown. The OR of each study is marked with a black dot. The overall OR is indicated by blue diamond.
Figure 5.
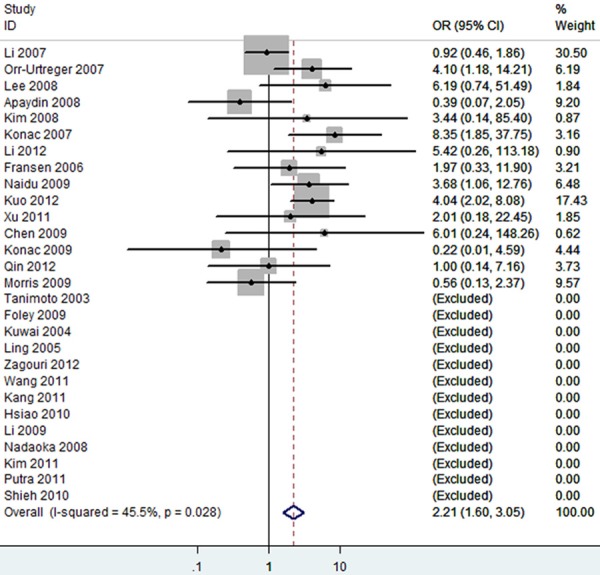
The forest plot of TT vs. TC/CC of rs11549465 polymorphism and overall cancer risk (Fixed model). The overall OR is shown. The OR of each study is marked with a black dot. The overall OR is indicated by blue diamond.
Figure 6.
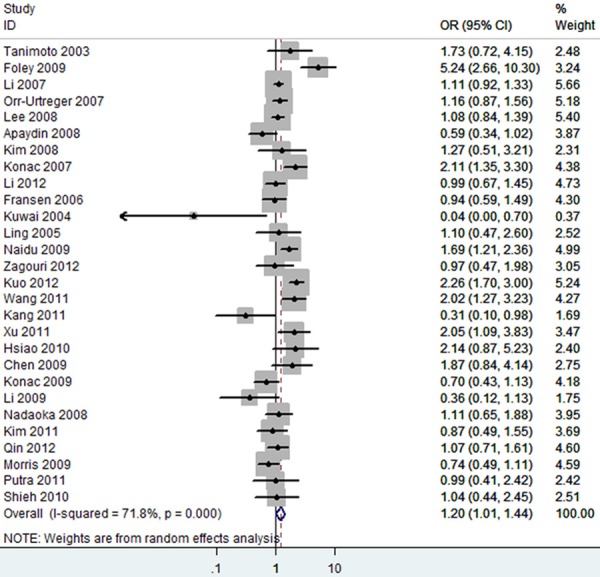
The forest plot of T vs. C of rs11549465 polymorphism and overall cancer risk (Random model). The overall OR is shown. The OR of each study is marked with a black dot. The overall OR is indicated by blue diamond.
We then evaluated the effects of the rs11549465 (1772C/T) polymorphism according to specific cancer types, different ethnicities, different detection methods and different sources of control. The results of stratified analyses were listed out in Table 3. Subgroup analyses for cancer types indicated that the pooled ORs for the homozygote (TT vs. CC, OR=9.92 [2.15-45.66]), heterozygote model (TC vs. CC, OR=1.53 [0.86-2.74]), dominant genetic model (OR=1.96 [1.13-3.41]), recessive model (OR=8.35 [1.85-37.75]) and additive model (T vs. C, OR=2.11 [1.35-3.30]) (Table 3) suggested the rs11549465 polymorphism was significantly associated with an increased gynecologic cancer risk in Caucasian. A marginal significant association between the rs11549465 polymorphism and increased lung cancer risk was detected in Asians under homozygote comparison (TT vs. CC, OR=4.88 [2.42-9.84]) and recessive genetic model (TT vs. TC/CC, OR=4.04 [2.02-8.08]) (Table 3) and the pooled ORs for all genetic models tested suggested that rs11549465 polymorphism was significantly associated with a decreased lung cancer risk in Caucasian (Table 3). A marginal significant association between the rs11549465 polymorphism and increased breast cancer risk was detected in Asians under homozygote comparison (TT vs. CC, OR=4.38 [1.58-12.12]) (Figure 7) and recessive genetic model (TT vs. TC/CC, OR=4.16 [1.51-11.48]) (Figure 8) and the pooled ORs for all genetic models tested suggested that rs11549465 polymorphism was significantly associated with a decreased breast cancer risk in Caucasian (Table 3). For pancreatic cancer and glioma, significant associations were observed in heterozygote comparison (TC vs. CC), dominant genetic model (TT/TC vs. CC) and additive model (T vs. C) (Table 3). Significant association was not observed for head and neck squamous cell carcinoma (HNSCC), prostate cancer, colorectal cancer, esophageal squamous cell carcinoma (ESCC), hepatocellular carcinoma, oral squamous cell carcinoma (OSCC), Gastric cancer, transitional cell carcinoma of the bladder and renal cell carcinoma in all genetic models tested. In the stratified analysis by ethnicity, significantly increased risks were found in Asian in almost all genetic models tested (Table 3). The remaining polled ORs from this analysis were not significant (Table 3). Significant association was not observed for different detection methods. According to the source of controls, signification effects in two genetic models were observed in hospital-based studies; while in population-based studies, significant association was not observed in any genetic model.
Figure 7.
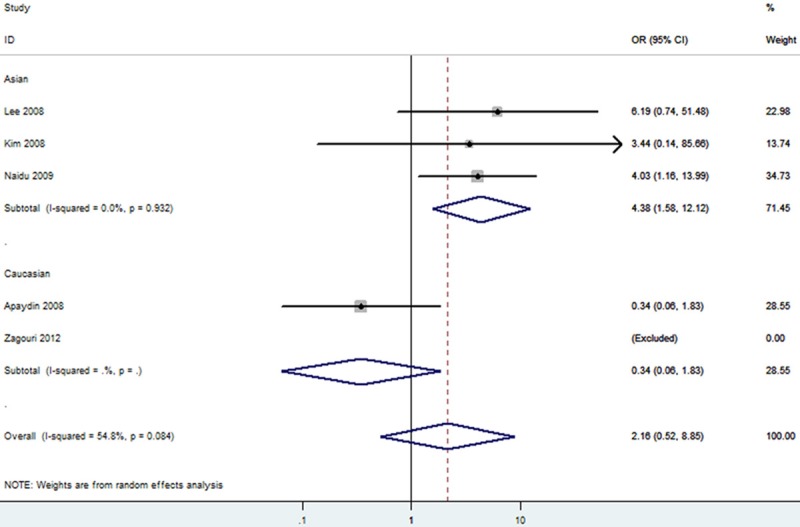
The forest plot of T vs. C of rs11549465 polymorphism and breast cancer risk (Random model). The overall OR is shown. The OR of each study is marked with a black dot. The overall OR is indicated by blue diamond.
Figure 8.
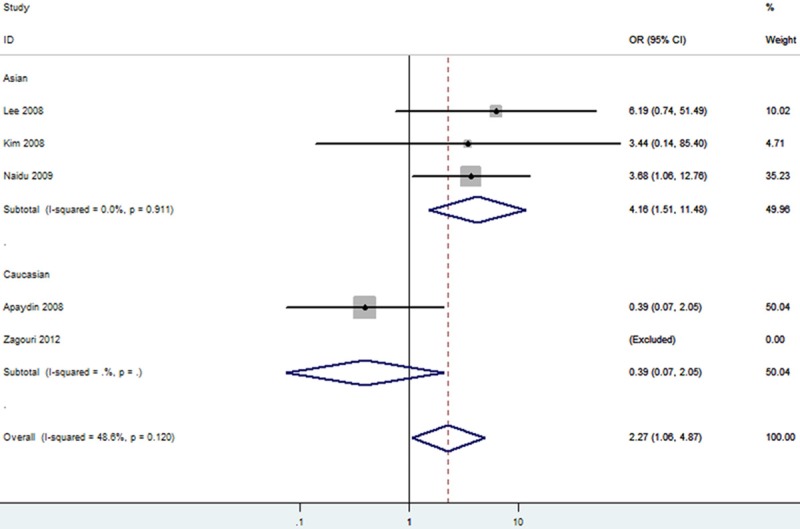
The forest plot of TT vs. TC/CC of rs11549465 polymorphism and breast cancer risk (Fixed model). The overall OR is shown. The OR of each study is marked with a black dot. The overall OR is indicated by blue diamond.
Publication bias
Both Begg’s funnel plot and Egger’s test were performed to assess the publication bias. The shape of the funnel plots did not reveal any evidence of obvious asymmetry in the overall meta-analysis. Then, Egger’s test was used to provide statistical evidence of funnel plot symmetry. The results still did not present any obvious evidence of publication bias (TT vs. CC. P=0.908; TC vs. CC. P=0.660; TT/TC vs. CC, P=0.627; TT vs. TC/CC, P=0.992; T vs. C. P=0.516).
Discussion
This meta-analysis of 28 studies involving 7807 cases and 8633 controls was conducted in order to yield a valid conclusion concerning the potential association between rs11549465 (1772C/T) polymorphism and cancer risk. HIF-1 plays a major role in cancer progression and metastasis through activation of various genes that are linked to regulation of angiogenesis, cell survival, and energy metabolism [63,64]. The HIF-1 was previously found to be implicated in the development and progression of cancer [63,64]. In 2009, Zhao T et al. [65] have done a meta-analysis on the relationship between HIF-1 and cancers, but their study only referred to the case-control studies before 2009. The polymorphisms analyzed in the present study consist of G to A nucleotide substitutions at positions 1772 of the exon 12 of the HIF-1. Because a study by Tanimoto [64] showed both of the substitutions displayed an increased transactivation capacity of HIF-1α in vitro, the presence of the variant alleles might be associated with increased cancer susceptibility. However, studies focusing on the association of the HIF-1 polymorphism with cancer susceptibility had controversial conclusions [20,22,27,28,30,31,33,44-51,53,55-57,59,60,62,66,67]. The lack of concordance across many of these studies reflects limitation in the studies, such as small sample sizes, ethnic difference and research methodology and so on. Meta-analysis is a powerful tool for summarizing the results from different studies by producing a single estimate of the major effect with enhanced precision.
In our analysis, there was significant association between this polymorphism and increased gynecologic cancer risk in Caucasian. Patients carrying the T allele at position 1772 of the exon 12 of the HIF-1 had more cancer risk than did patients homozygous for the C allele. A marginal significant association between the rs11549465 polymorphism and increased lung and breast cancer risk was detected in Asians under homozygote comparison and recessive genetic model. The pooled effects for all genetic models tested suggested a significant association between the rs11549465 (1772C/T) polymorphism and a decreased lung and breast cancer risk in Caucasian. Furthermore, We found that Asians with TT genotype had higher risk of cancer compared to Caucasians under the homozygote, recessive and additive models. Inconsistency between the two ethnicities can be explained by the possibility that different ethnic groups live with multiple life styles and environmental factors. And different populations carry different genotype and/or allele frequencies of this locus polymorphism may lead to various degrees of cancer susceptibility. In our meta-analysis, we also observed inconsistent results between hospital-based studies and population-based studies. Controls in hospital-based studies are more representative of general population than controls from population-based studies. Several factors such as environmental factors and genetic backgrounds might contribute to the discrepancy.
There were some limitations in our meta-analysis. First, sample size in any given cancer was not sufficiently large, which could increase the probability of false positive or false negative. It might be difficult to get a concrete conclusion if the number of included studies in subgroup was few. Besides, the sample size was not large enough, studies involved in different ethnicities were warranted to estimate the effects of this functional polymorphism on cancer risk. Second, due to the original data of the eligible studies was unavailable, it was difficult for us to evaluate the roles of some special environmental factors and lifestyles such as diet, alcohol consumption, and smoking status in developing cancer. Third, the influence of bias in the present analysis could not be completely excluded because positive results are supposed to be published much more quickly than articles with “negatives” results.
Conclusions
Our meta-analysis suggested that the rs11549465 (1772C/T) genetic polymorphism is significantly associated with higher breast and lung cancer risk among Asian population, and this SNP is significantly associated with decreased breast and lung cancer risk among Caucasian population, but this SNP was significantly associated with the gynecologic cancer among Caucasian population. The effect of the rs11549465 polymorphism on cancer especially exists in Asians. Large well designed epidemiological studies are needed to validate our findings.
Disclosure of conflict of interest
None for all authors.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Wright ME, Peters U, Gunter MJ, Moore SC, Lawson KA, Yeager M, Weinstein SJ, Snyder K, Virtamo J, Albanes D. Association of variants in two vitamin e transport genes with circulating vitamin e concentrations and prostate cancer risk. Cancer Res. 2009;69:1429–38. doi: 10.1158/0008-5472.CAN-08-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheung WY, Liu G. Genetic variations in esophageal cancer risk and prognosis. Gastroenterol Clin North Am. 2009;38:75–91. doi: 10.1016/j.gtc.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 4.Ruan K, Song G, Ouyang G. Role of hypoxia in the hallmarks of human cancer. J Cell Biochem. 2009;107:1053–62. doi: 10.1002/jcb.22214. [DOI] [PubMed] [Google Scholar]
- 5.Brahimi-Horn MC, Pouyssegur J. HIF at a glance. J Cell Sci. 2009;122:1055–7. doi: 10.1242/jcs.035022. [DOI] [PubMed] [Google Scholar]
- 6.Brahimi-Horn MC, Chiche J, Pouyssegur J. Hypoxia and cancer. J Mol Med (Berl) 2007;85:1301–7. doi: 10.1007/s00109-007-0281-3. [DOI] [PubMed] [Google Scholar]
- 7.Schmid T, Zhou J, Brune B. HIF-1 and p53: communication of transcription factors under hypoxia. J Cell Mol Med. 2004;8:423–31. doi: 10.1111/j.1582-4934.2004.tb00467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koshiji M, To KK, Hammer S, Kumamoto K, Harris AL, Modrich P, Huang LE. HIF-1alpha induces genetic instability by transcriptionally downregulating MutSalpha expression. Mol Cell. 2005;17:793–803. doi: 10.1016/j.molcel.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 9.Chan N, Koch CJ, Bristow RG. Tumor hypoxia as a modifier of DNA strand break and cross-link repair. Curr Mol Med. 2009;9:401–10. doi: 10.2174/156652409788167050. [DOI] [PubMed] [Google Scholar]
- 10.Lee YM, Lim JH, Chun YS, Moon HE, Lee MK, Huang LE, Park JW. Nutlin-3, an Hdm2 antagonist, inhibits tumor adaptation to hypoxia by stimulating the FIH-mediated inactivation of HIF-1alpha. Carcinogenesis. 2009;30:1768–75. doi: 10.1093/carcin/bgp196. [DOI] [PubMed] [Google Scholar]
- 11.Huang LE, Bindra RS, Glazer PM, Harris AL. Hypoxia-induced genetic instability--a calculated mechanism underlying tumor progression. J Mol Med (Berl) 2007;85:139–48. doi: 10.1007/s00109-006-0133-6. [DOI] [PubMed] [Google Scholar]
- 12.Meng AX, Jalali F, Cuddihy A, Chan N, Bindra RS, Glazer PM, Bristow RG. Hypoxia down-regulates DNA double strand break repair gene expression in prostate cancer cells. Radiother Oncol. 2005;76:168–76. doi: 10.1016/j.radonc.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 13.Semenza GL. Expression of hypoxia-inducible factor 1: mechanisms and consequences. Biochem Pharmacol. 2000;59:47–53. doi: 10.1016/s0006-2952(99)00292-0. [DOI] [PubMed] [Google Scholar]
- 14.Zhong H, De Marzo AM, Laughner E, Lim M, Hilton DA, Zagzag D, Buechler P, Isaacs WB, Semenza GL, Simons JW. Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res. 1999;59:5830–5. [PubMed] [Google Scholar]
- 15.Talks KL, Turley H, Gatter KC, Maxwell PH, Pugh CW, Ratcliffe PJ, Harris AL. The expression and distribution of the hypoxia-inducible factors HIF-1alpha and HIF-2alpha in normal human tissues, cancers, and tumor-associated macrophages. Am J Pathol. 2000;157:411–21. doi: 10.1016/s0002-9440(10)64554-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bos R, Zhong H, Hanrahan CF, Mommers EC, Semenza GL, Pinedo HM, Abeloff MD, Simons JW, van Diest PJ, van der Wall E. Levels of hypoxia-inducible factor-1 alpha during breast carcinogenesis. J Natl Cancer Inst. 2001;93:309–14. doi: 10.1093/jnci/93.4.309. [DOI] [PubMed] [Google Scholar]
- 17.Koukourakis MI, Papazoglou D, Giatromanolaki A, Panagopoulos I, Maltezos E, Harris AL, Gatter KC, Sivridis E. C2028T polymorphism in exon 12 and dinucleotide repeat polymorphism in intron 13 of the HIF-1alpha gene define HIF-1alpha protein expression in non-small cell lung cancer. Lung Cancer-J Iaslc. 2006;53:257–62. doi: 10.1016/j.lungcan.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 18.Tzao C, Lee SC, Tung HJ, Hsu HS, Hsu WH, Sun GH, Yu CP, Jin JS, Cheng YL. Expression of hypoxia-inducible factor (HIF)-1alpha and vascular endothelial growth factor (VEGF)-D as outcome predictors in resected esophageal squamous cell carcinoma. Dis Markers. 2008;25:141–8. doi: 10.1155/2008/468323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koukourakis MI, Giatromanolaki A, Skarlatos J, Corti L, Blandamura S, Piazza M, Gatter KC, Harris AL. Hypoxia inducible factor (HIF-1α and HIF-2α) expression in early esophageal cancer and response to photodynamic therapy and radiotherapy. Cancer Res. 2001;61:1830–2. [PubMed] [Google Scholar]
- 20.Li P, Cao Q, Shao PF, Cai HZ, Zhou H, Chen JW, Qin C, Zhang ZD, Ju XB, Yin CJ. Genetic polymorphisms in HIF1A are associated with prostate cancer risk in a Chinese population. Asian J Androl. 2012;14:864–9. doi: 10.1038/aja.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–32. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 22.Clifford SC, Astuti D, Hooper L, Maxwell PH, Ratcliffe PJ, Maher ER. The pVHL-associated SCF ubiquitin ligase complex: molecular genetic analysis of elongin B and C, Rbx1 and HIF 1alpha in renal cell carcinoma. Oncogene. 2001;20:5067–74. doi: 10.1038/sj.onc.1204602. [DOI] [PubMed] [Google Scholar]
- 23.Tanimoto K, Yoshiga K, Eguchi H, Kaneyasu M, Ukon K, Kumazaki T, Oue N, Yasui W, Imai K, Nakachi K, Poellinger L, Nishiyama M. Hypoxia-inducible factor-1alpha polymorphisms associated with enhanced transactivation capacity, implying clinical significance. Carcinogenesis. 2003;24:1779–83. doi: 10.1093/carcin/bgg132. [DOI] [PubMed] [Google Scholar]
- 24.Kuwai T, Kitadai Y, Tanaka S, Kuroda T, Ochiumi T, Matsumura S, Oue N, Yasui W, Kaneyasu M, Tanimoto K, Nishiyama M, Chayama K. Single nucleotide polymorphism in the hypoxia-inducible factor-1alpha gene in colorectal carcinoma. Oncol Rep. 2004;12:1033–7. [PubMed] [Google Scholar]
- 25.Ollerenshaw M, Page T, Hammonds J, Demaine A. Polymorphisms in the hypoxia inducible factor-1alpha gene (HIF1A) are associated with the renal cell carcinoma phenotype. Cancer Genet Cytogenet. 2004;153:122–6. doi: 10.1016/j.cancergencyto.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 26.Chau CH, Permenter MG, Steinberg SM, Retter AS, Dahut WL, Price DK, Figg WD. Polymorphism in the hypoxia-inducible factor 1alpha gene may confer susceptibility to androgen-independent prostate cancer. Cancer Biol Ther. 2005;4:1222–5. doi: 10.4161/cbt.4.11.2091. [DOI] [PubMed] [Google Scholar]
- 27.Fransen K, Fenech M, Fredrikson M, Dabrosin C, Soderkvist P. Association between ulcerative growth and hypoxia inducible factor-1alpha polymorphisms in colorectal cancer patients. Mol Carcinog. 2006;45:833–40. doi: 10.1002/mc.20209. [DOI] [PubMed] [Google Scholar]
- 28.Konac E, Onen HI, Metindir J, Alp E, Biri AA, Ekmekci A. An investigation of relationships between hypoxia-inducible factor-1 alpha gene polymorphisms and ovarian, cervical and endometrial cancers. Cancer Detect Prev. 2007;31:102–9. doi: 10.1016/j.cdp.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 29.Li H, Bubley GJ, Balk SP, Gaziano JM, Pollak M, Stampfer MJ, Ma J. Hypoxia-inducible factor-1alpha (HIF-1alpha) gene polymorphisms, circulating insulin-like growth factor binding protein (IGFBP)-3 levels and prostate cancer. Prostate. 2007;67:1354–61. doi: 10.1002/pros.20589. [DOI] [PubMed] [Google Scholar]
- 30.Orr-Urtreger A, Bar-Shira A, Matzkin H, Mabjeesh NJ. The homozygous P582S mutation in the oxygen-dependent degradation domain of HIF-1 alpha is associated with increased risk for prostate cancer. Prostate. 2007;67:8–13. doi: 10.1002/pros.20433. [DOI] [PubMed] [Google Scholar]
- 31.Apaydin I, Konac E, Onen HI, Akbaba M, Tekin E, Ekmekci A. Single nucleotide polymorphisms in the hypoxia-inducible factor-1alpha (HIF-1alpha) gene in human sporadic breast cancer. Arch Med Res. 2008;39:338–45. doi: 10.1016/j.arcmed.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 32.Horree N, Groot AJ, van Hattem WA, Heintz AP, Vooijs M, van Diest PJ. HIF-1A gene mutations associated with higher microvessel density in endometrial carcinomas. Histopathology. 2008;52:637–9. doi: 10.1111/j.1365-2559.2008.02991.x. [DOI] [PubMed] [Google Scholar]
- 33.Kim HO, Jo YH, Lee J, Lee SS, Yoon KS. The C1772T genetic polymorphism in human HIF-1alpha gene associates with expression of HIF-1alpha protein in breast cancer. Oncol Rep. 2008;20:1181–7. [PubMed] [Google Scholar]
- 34.Lee JY, Choi JY, Lee KM, Park SK, Han SH, Noh DY, Ahn SH, Kim DH, Hong YC, Ha E, Yoo KY, Ambrosone CB, Kang D. Rare variant of hypoxia-inducible factor-1alpha (HIF-1A) and breast cancer risk in Korean women. Clin Chim Acta. 2008;389:167–70. doi: 10.1016/j.cca.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 35.Foley R, Marignol L, Thomas AZ, Cullen IM, Perry AS, Tewari P, O’Grady A, Kay E, Dunne B, Loftus B, Watson WR, Fitzpatrick JM, Woodson K, Lehman T, Hollywood D, Lynch TH, Lawler M. The HIF-1alpha C1772T polymorphism may be associated with susceptibility to clinically localised prostate cancer but not with elevated expression of hypoxic biomarkers. Cancer Biol Ther. 2009;8:118–24. doi: 10.4161/cbt.8.2.7086. [DOI] [PubMed] [Google Scholar]
- 36.Marignol L, Foley R, Southgate TD, Coffey M, Hollywood D, Lawler M. Hypoxia response element-driven cytosine deaminase/5-fluorocytosine gene therapy system: a highly effective approach to overcome the dynamics of tumour hypoxia and enhance the radiosensitivity of prostate cancer cells in vitro. J Gene Med. 2009;11:169–79. doi: 10.1002/jgm.1281. [DOI] [PubMed] [Google Scholar]
- 37.Munoz-Guerra MF, Fernandez-Contreras ME, Moreno AL, Martin ID, Herraez B, Gamallo C. Polymorphisms in the hypoxia inducible factor 1-alpha and the impact on the prognosis of early stages of oral cancer. Ann Surg Oncol. 2009;16:2351–8. doi: 10.1245/s10434-009-0503-8. [DOI] [PubMed] [Google Scholar]
- 38.Smaldone MC, Maranchie JK. Clinical implications of hypoxia inducible factor in renal cell carcinoma. Urol Oncol. 2009;27:238–45. doi: 10.1016/j.urolonc.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 39.Ling TS, Shi RH, Zhang GX, Zhu H, Yu LZ, Ding XF. Common single nucleotide polymorphism of hypoxia-inducible factor-1alpha and its impact on the clinicopathological features of esophageal squamous cell carcinoma. Chin J Dig Dis. 2005;6:155–8. doi: 10.1111/j.1443-9573.2005.00223.x. [DOI] [PubMed] [Google Scholar]
- 40.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–88. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 41.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–48. [PubMed] [Google Scholar]
- 42.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Egger M, Davey SG, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanimoto K, Yoshiga K, Eguchi H, Kaneyasu M, Ukon K, Kumazaki T, Oue N, Yasui W, Imai K, Nakachi K, Poellinger L, Nishiyama M. Hypoxia-inducible factor-1alpha polymorphisms associated with enhanced transactivation capacity, implying clinical significance. Carcinogenesis. 2003;24:1779–83. doi: 10.1093/carcin/bgg132. [DOI] [PubMed] [Google Scholar]
- 45.Li H, Bubley GJ, Balk SP, Gaziano JM, Pollak M, Stampfer MJ, Ma J. Hypoxia-inducible factor-1alpha (HIF-1alpha) gene polymorphisms, circulating insulin-like growth factor binding protein (IGFBP)-3 levels and prostate cancer. Prostate. 2007;67:1354–61. doi: 10.1002/pros.20589. [DOI] [PubMed] [Google Scholar]
- 46.Nadaoka J, Horikawa Y, Saito M, Kumazawa T, Inoue T, Narita S, Yuasa T, Satoh S, Nishiyama H, Ogawa O, Tsuchiya N, Habuchi T. Prognostic significance of HIF-1 alpha polymorphisms in transitional cell carcinoma of the bladder. Int J Cancer. 2008;122:1297–302. doi: 10.1002/ijc.23256. [DOI] [PubMed] [Google Scholar]
- 47.Chen MK, Chiou HL, Su SC, Chung TT, Tseng HC, Tsai HT, Yang SF. The association between hypoxia inducible factor-1alpha gene polymorphisms and increased susceptibility to oral cancer. Oral Oncol. 2009;45:e222–6. doi: 10.1016/j.oraloncology.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 48.Konac E, Dogan I, Onen HI, Yurdakul AS, Ozturk C, Varol A, Ekmecki A. Genetic variations in the hypoxia-inducible factor-1alpha gene and lung cancer. Exp Biol Med (Maywood) 2009;234:1109–16. doi: 10.3181/0902-RM-49. [DOI] [PubMed] [Google Scholar]
- 49.Li K, Zhang Y, Dan Z, Wang Y, Ren ZC. Association of the hypoxia inducible factor-1alpha gene polymorphisms with gastric cancer in Tibetans. Biochem Genet. 2009;47:625–34. doi: 10.1007/s10528-009-9254-2. [DOI] [PubMed] [Google Scholar]
- 50.Morris MR, Hughes DJ, Tian YM, Ricketts CJ, Lau KW, Gentle D, Shuib S, Serrano-Fernandez P, Lubinski J, Wiesener MS, Pugh CW, Latif F, Ratcliffe PJ, Maher ER. Mutation analysis of hypoxia-inducible factors HIF1A and HIF2A in renal cell carcinoma. Anticancer Res. 2009;29:4337–43. [PubMed] [Google Scholar]
- 51.Naidu R, Har YC, Taib NA. Associations between hypoxia-inducible factor-1alpha (HIF-1alpha) gene polymorphisms and risk of developing breast cancer. Neoplasma. 2009;56:441–7. doi: 10.4149/neo_2009_05_441. [DOI] [PubMed] [Google Scholar]
- 52.Foley R, Marignol L, Thomas AZ, Cullen IM, Perry AS, Tewari P, O’Grady A, Kay E, Dunne B, Loftus B, Watson WR, Fitzpatrick JM, Woodson K, Lehman T, Hollywood D, Lynch TH, Lawler M. The HIF-1alpha C1772T polymorphism may be associated with susceptibility to clinically localised prostate cancer but not with elevated expression of hypoxic biomarkers. Cancer Biol Ther. 2009;8:118–24. doi: 10.4161/cbt.8.2.7086. [DOI] [PubMed] [Google Scholar]
- 53.Hsiao PC, Chen MK, Su SC, Ueng KC, Chen YC, Hsieh YH, Liu YF, Tsai HT, Yang SF. Hypoxia inducible factor-1alpha gene polymorphism G1790A and its interaction with tobacco and alcohol consumptions increase susceptibility to hepatocellular carcinoma. J Surg Oncol. 2010;102:163–9. doi: 10.1002/jso.21539. [DOI] [PubMed] [Google Scholar]
- 54.Kang MJ, Jung SA, Jung JM, Kim SE, Jung HK, Kim TH, Shim KN, Yi SY, Yoo K, Moon IH. Associations between single nucleotide polymorphisms of MMP2, VEGF, and HIF1A genes and the risk of developing colorectal cancer. Anticancer Res. 2011;31:575–84. [PubMed] [Google Scholar]
- 55.Kim YH, Park IA, Park WY, Kim JW, Kim SC, Park NH, Song YS, Kang SB. Hypoxia-inducible factor 1alpha polymorphisms and early-stage cervical cancer. Int J Gynecol Cancer. 2011;21:2–7. doi: 10.1097/IGC.0b013e318204f6e6. [DOI] [PubMed] [Google Scholar]
- 56.Putra AC, Tanimoto K, Arifin M, Hiyama K. Hypoxia-inducible factor-1alpha polymorphisms are associated with genetic aberrations in lung cancer. Respirology. 2011;16:796–802. doi: 10.1111/j.1440-1843.2011.01972.x. [DOI] [PubMed] [Google Scholar]
- 57.Wang X, Liu Y, Ren H, Yuan Z, Li S, Sheng J, Zhao T, Chen Y, Liu F, Wang F, Huang H, Hao J. Polymorphisms in the hypoxia-inducible factor-1alpha gene confer susceptibility to pancreatic cancer. Cancer Biol Ther. 2011;12:383–7. doi: 10.4161/cbt.12.5.15982. [DOI] [PubMed] [Google Scholar]
- 58.Xu G, Wang M, Xie W, Bai X. Hypoxia-inducible factor-1 alpha C1772T gene polymorphism and glioma risk: a hospital-based case-control study from China. Genet Test Mol Biomarkers. 2011;15:461–4. doi: 10.1089/gtmb.2010.0265. [DOI] [PubMed] [Google Scholar]
- 59.Kuo WH, Shih CM, Lin CW, Cheng WE, Chen SC, Chen W, Lee YL. Association of hypoxia inducible factor-1alpha polymorphisms with susceptibility to non-small-cell lung cancer. Transl Res. 2012;159:42–50. doi: 10.1016/j.trsl.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 60.Qin C, Cao Q, Ju X, Wang M, Meng X, Zhu J, Yan F, Li P, Ding Q, Chen J, Gu M, Zhang W, Yin C, Zhang Z. The polymorphisms in the VHL and HIF1A genes are associated with the prognosis but not the development of renal cell carcinoma. Ann Oncol. 2012;23:981–9. doi: 10.1093/annonc/mdr325. [DOI] [PubMed] [Google Scholar]
- 61.Zagouri F, Sergentanis TN, Gazouli M, Tsigginou A, Dimitrakakis C, Papaspyrou I, Eleutherakis-Papaiakovou E, Chrysikos D, Theodoropoulos G, Zografos GC, Antsaklis A, Dimopoulos AM, Papadimitriou CA. HSP90, HSPA8, HIF-1 alpha and HSP70-2 polymorphisms in breast cancer: a case-control study. Mol Biol Rep. 2012;39:10873–9. doi: 10.1007/s11033-012-1984-2. [DOI] [PubMed] [Google Scholar]
- 62.Shieh TM, Chang KW, Tu HF, Shih YH, Ko SY, Chen YC, Liu CJ. Association between the polymorphisms in exon 12 of hypoxia-inducible factor-1alpha and the clinicopathological features of oral squamous cell carcinoma. Oral Oncol. 2010;46:e47–53. doi: 10.1016/j.oraloncology.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 63.Smaldone MC, Maranchie JK. Clinical implications of hypoxia inducible factor in renal cell carcinoma. Urol Oncol. 2009;27:238–45. doi: 10.1016/j.urolonc.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 64.Tanimoto K, Yoshiga K, Eguchi H, Kaneyasu M, Ukon K, Kumazaki T, Oue N, Yasui W, Imai K, Nakachi K, Poellinger L, Nishiyama M. Hypoxia-inducible factor-1alpha polymorphisms associated with enhanced transactivation capacity, implying clinical significance. Carcinogenesis. 2003;24:1779–83. doi: 10.1093/carcin/bgg132. [DOI] [PubMed] [Google Scholar]
- 65.Zhao T, Lv J, Zhao J, Nzekebaloudou M. Hypoxia-inducible factor-1alpha gene polymorphisms and cancer risk: a meta-analysis. J Exp Clin Cancer Res. 2009;28:159. doi: 10.1186/1756-9966-28-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Munoz-Guerra MF, Fernandez-Contreras ME, Moreno AL, Martin ID, Herraez B, Gamallo C. Polymorphisms in the hypoxia inducible factor 1-alpha and the impact on the prognosis of early stages of oral cancer. Ann Surg Oncol. 2009;16:2351–8. doi: 10.1245/s10434-009-0503-8. [DOI] [PubMed] [Google Scholar]
- 67.Ruiz-Tovar J, Fernandez-Contreras ME, Martin-Perez E, Gamallo C. Association of thymidylate synthase and hypoxia inducible factor-1alpha DNA polymorphisms with pancreatic cancer. Tumori. 2012;98:364–9. doi: 10.1177/030089161209800314. [DOI] [PubMed] [Google Scholar]


