Abstract
MicroRNA (miRNA-221) has been reported to be a regulator of cell proliferation. Here we intended to investigate the role of miRNA-221 in regulating the growth of human non-small cell lung cancer cell line H460. H460 cells were transfected with miRNA-221 mimics/inhibitors or their respective negative controls. Real-time quantitative PCRs (qRT-PCRs) were used to confirm the effects of miRNA-221 mimics and inhibitors in H460 cells while Cell Counting Kit 8 (CCK-8) and 5-Ethynyl-2’-deoxyuridine (EdU) assay were used to access the cell viability and proliferation. P27 and P57, as putative targets of miRNA-221, were determined by qRT-PCRs in H460 cells. We found that overexpression of miRNA-221 led to increased proliferative rate and cell viability in H460 cells while down-regulation of miRNA-221 decreased those effects. P27 but not P57 was identified as a potential target gene of miRNA-221 in H460 as P27 was negatively regulated by miRNA-221 in the protein level. In conclusion, this study suggests that miRNA-221 controls human non-small cell lung cancer cell H460 growth potentially by targeting P57. Inhibition of miRNA-221 represents a novel potential treatment for human non-small cell lung cancer.
Keywords: MicroRNA, human non-small cell lung cancer, growth
Introduction
Non small-cell lung cancer (NSCLC), a common type of lung cancer, is among the most frequently diagnosed types of cancer and is also a leading cause of mortality worldwide [1]. Great efforts in the early diagnosis have been made. However, despite emerging technologies and newly developed chemotherapy that improve treatment responses, only 15% of patients diagnosed with NSCLC could survive over 5 years and the recurrence rate is extremely high as well, even receiving treatment in early-stage [2]. Therefore, emerging targeted therapies directed against specific cellular alterations might give alternative strategies for NSCLC treatment. Moreover, accumulating knowledge gained from genomic medicine also provides the possibility of unravelling the remaining mysteries of NSCLC. In other words, molecular targeted therapies based on gene expression profiles and microRNA (miRNA) signatures are promising in developing novel therapies for NSCLC [3].
MicroRNAs (miRNAs) are a group of small non-coding RNAs containing about 20~ nucleotides identified in plants, animals, and even some viruses. miRNAs can function in RNA silencing and post-transcriptional regulation of gene expression [4,5]. Generally, miRNAs can negatively regulate genes expression by binding to the 3’-untranslated region (3’-UTR) of their target genes. One miRNA can regulate several or even up to hundreds of target genes while one gene can also be regulated by multiple miRNAs [6]. Therefore, more than 60% of all human genes have been predicted to be regulated by miRNAs [6]. miRNAs are considered to be involved in many physiological and pathological processes such as development, proliferation, cancers and inflammation response, being the center players of gene regulations [7-12].
MiRNA-221 is a member of miRNA-221/222 family, and has been reported to critically participate in multiple cancers [13-15]. Moreover, it has also been indicated that down-regulation of miRNA-221 was associated with poor prognosis of patients with NSCLC [16]. In addition, miRNA-221 regulation has also been related to drug resistance in NSCLC [17]. As miRNA-221 has been reported to be a regulator of cell growth, here we intended to investigate the role of miRNA-221 in regulating the growth of human non-small cell lung cancer cell line H460.
Methods
Cell culture
Human non-small cell lung cancer cell line (NSCLC) H460 was purchased from Cell Bank of Chinese Academy of Sciences (Shanghai, China). In general, H460 cells were cultured at 37°C under 5% CO2 in 10 cm plastic dishes containing 10 ml of RPMI 1640 (Hyclone, USA), supplemented with 10% FBS (Hyclone, USA), 100 U/ml penicillin and 100 mg/ml Streptomycin (KeyGen, China).
Cell transient transfection
miRNA-221 mimics, inhibitors and their negative controls (nc-mimics and nc-inhibitors) were purchased from RiboBio (Guangzhou, China). H460 cells were staved with serum free medium for 6 h, and then were transfected with miRNA-221 mimics (50 nM), inhibitors (100 nM) or their negative controls for 48 h using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s instructions.
Cell viability
The effects of miRNA-221 mimics and inhibitors in H460 cell viability were determined using a Cell Counting Kit 8 (CCK-8) assay. Cells (2×103/well) were seeded in 96-well plates and adhered overnight. After 48 h of miRNA-221 mimics, inhibitors and their negative controls transfection, CCK-8 solution was added to each well and incubated for 1 h at 37°C. Absorbance was then measured at 450 nm using a spectrophotometer.
Cell proliferative assay
H460 cells were planted into 24-well dishes at a density of 2×104 per milliliter and were allowed to adhere overnight. After transfection with miRNA-221 mimics, inhibitors and their negative controls, H460 cells were cultured with 5-Ethyny-2’-deoxyuridine (EdU) for 8 h before detection. The proliferative rate of H460 was then evaluated using a Cell-Light™ EdU Cell Proliferation Detection kit (RiboBio, China) following the manufacturer’s instructions.
Real-time quantitative PCR
Total RNA was isolated using the TRIzol RNA extraction kit (Invitrogen, USA). For miRNA analysis, cDNA was synthesized using Bulge-Loop™ miRNA qRT-PCR Primer Set (Riobio, China) according to the manufacturer’s instructions, normalized to that of U6 of the same RNA. For quantitative analysis of P27 and P57, real-time PCRs using primers as indicated were performed. Briefly, 400 ng of RNA was subjected to reverse transcription-PCR using Bio-Rad iScripTM cDNA Synthesis Kit (Bio-Rad, USA), and then cDNA was subjected into 40 cycles of quantitative PCR with Takara SYBR Premix Ex TaqTM (Takara, Japan) in a CFX96TM Real-Time PCR Detection System (Bio-Rad, USA). GAPDH was used as a house-keeping gene for mRNA analysis. The sequence of p27 primer: FOR AACGTGCGAGTGTCTAACGG (5’-3’), REV CCCTCTAGGGGTTTGTGATTCT (5’-3’). The sequence of p57 primer: FOR GCGGCGATCAAGAAGCTGT (5’-3’), REV GCTTGGCGAAGAAATCGGAGA (5’-3’). The sequence of GAPDH primer: FOR TGTGGGCATCAATGGATTTGG (5’-3’), REV ACACCATGTATTCCGGGT CAAT (5’-3’).
Western blot
Total proteins from H460 cells were lysed with RIPA buffer (Beyotime Biotechnology, China). Extracted proteins were separated with SDS-PAGE gels and were transferred onto PVDF membranes (Millipore, Bedford, MA, USA) for western blotting analysis. The following primary antibodies were used: p27 at a dilution of 1:1000 (Bioworld, USA), β-actin at a dilution of 1:10000 (Bioworld, USA), then were detected using HRP-linked secondary Antibodies and the ECL System.
Statistical analysis
Data are presented as the mean ± SEM. An independent-samples t-test or one-way ANOVA was conducted with a Bonferroni’s post-hoc test. P value of <0.05 was thought as statistically significant. Statistical analysis was performed with IBM SPSS 19.0 for Windows.
Results
MiRNA-221 controls H460 cells’ cell viability
To understand whether miR-221 could regulate the cell viability of H460 cells, we first transfected miRNA-221 mimics, inhibitors or their negative controls to human NSCLC cell line H460. As indicated in Figure 1, the transfection rate was at least 70%. In addition, as demonstrated by real time qPCRs, 48 h after transfection of miRNA-221 mimics, the miRNA-221 level was significantly up-regulated in H460 cells, while miRNA-221 inhibitors decreased the miRNA-221 level (Figure 2), indicating that miRNA-221 mimics and inhibitors used in this study took effects in increasing or decreasing miRNA-221 levels. Based on that, overexpression of miRNA-221 with miRNA-221 mimics in H460 cells increased cell vitality and down-regulation of miRNA-221 with miRNA-221 inhibitors decreased H460 cell vitality (Figure 3). These results suggest miRNA-221 may contribute to H460 tumor properties by regulating cell vitality.
Figure 1.
The transfection rate in this study is over 70%.
Figure 2.
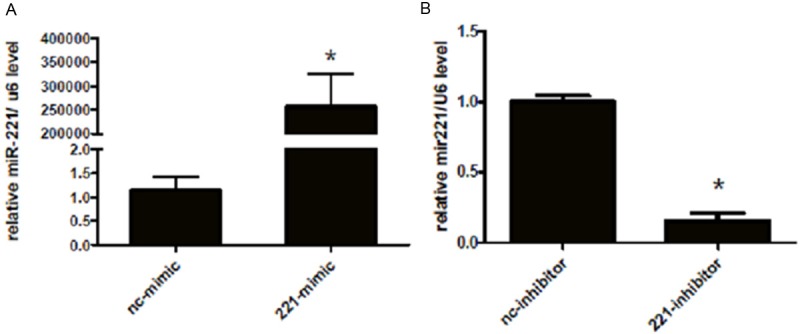
miRNA-221 mimics and inhibitors take effects in H460 cells. A miRNA-221 mimics increase miRNA-221 levels in H460 cells. *, P<0.05. B miRNA-221 inhibitors decrease miRNA-221 levels in H460 cells. *, P<0.05.
Figure 3.
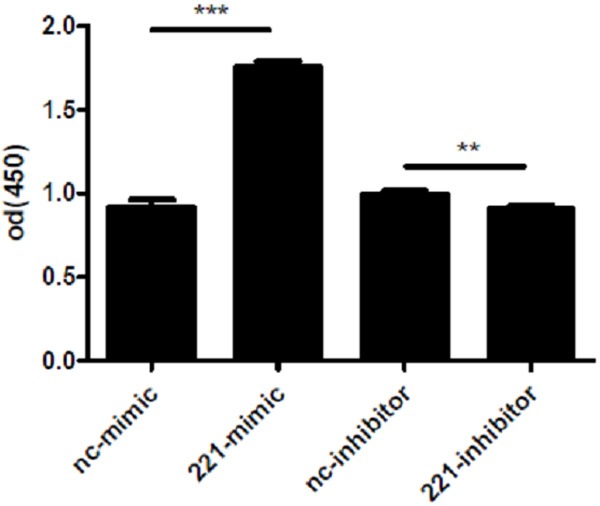
miRNA-221 regulates cell viability of H460 cells.CCK-8 assay shows that miRNA-221 mimics lead to an increased cell vitality of H460 (A) while miRNA-221 inhibitors decrease cell viability (B). **, P<0.01; ***, P<0.001.
MiRNA-221 induces H460 cell proliferation
To determine the effects of miRNA-221 in regulating cell proliferation, EdU assays were used in the present study. We found that overexpression of miRNA-221 with miRNA-221 mimics led to higher EdU positive cells percentage, indicating a higher proliferative level of H460 cells. Conversely, down-regulation of miRNA-221 with miRNA-221 inhibitors deceased the proliferative level of H460 cells (Figure 4). These data suggest that miRNA-221 may contribute to H460 tumor properties by promoting cell proliferation.
Figure 4.
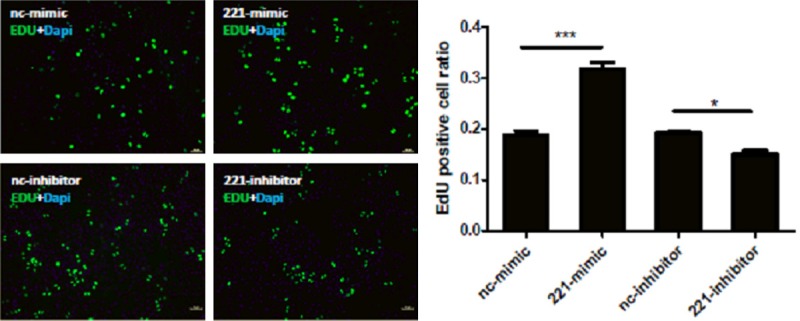
miRNA-221 controls cell proliferation of H460 cells. Edu staining indicates that miRNA-221 mimics increase the proliferation of H460 cells (A) while miRNA-221 inhibitors decrease that (B). *, P<0.05; ***, P<0.001.
P27 is a potential target gene of miRNA-221 in H460 cells
P27, as well as p57, are two well-known targets of miRNA-221 in multiple types of cells including cancer cells. They are members of the Cip/Kip family of cyclin-dependent kinase inhibitors and function to negatively control cell cycle progression and cell proliferation. To identify whether p27 and p57 are the potential targets of miRNA-221 in H460 cells, we firstly assessed the effects of miRNA-221 on endogenous expressions of p27 and p57 in h460 cells by real time quantitative PCR. As shown in Figure 5A, the p27 mRNA level was regulated by both overexpression and down-regulation of miRNA-221, but p57 mRNA level remained unchanged. To determine whether p27 is the putative target of miRNA-221 in H460 cells, we next detected the protein level of p27 by Western blotting and found that the p27 protein level was down-regulated by overexpressing miRNA-221, and was increased when silencing miRNA-221 in H460 cells (Figure 5B). The results showed that p27 may be a putative target gene of miRNA-221 in H460 cells.
Figure 5.
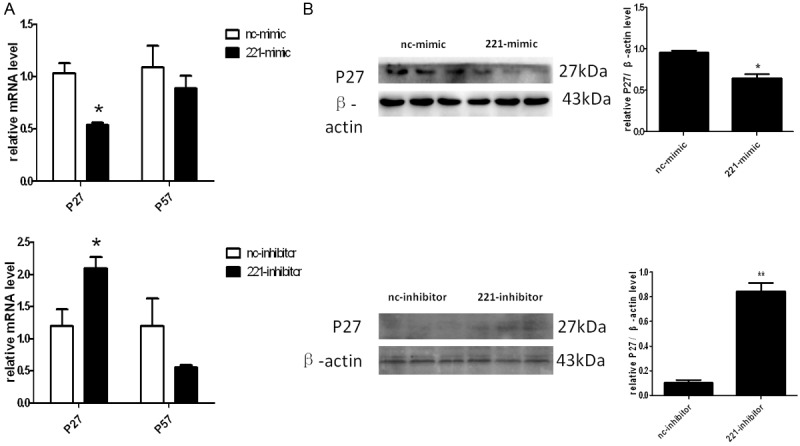
P27 is a potential target gene of miRNA-221 in H460 cells. A. miRNA-221 negatively regulates p27 but not P57 at the mRNA level. *, P<0.05. B. miRNA-221 negatively regulates p27 at the protein levels. *, P<0.05.
Discussion
NSCLC is among the leading causes of death caused by cancer worldwide [1]. With the help of modern genetic techniques for understanding candidate gene and genome wide, increasing evidence show that lung cancers accumulate numerous epigenetic alterations during a multistep process. These alterations include the inactivation of tumor suppressor genes and activation of oncogenes that promote growth or survival. MiRNAs comprise a novel class of regulatory molecules with the ability to negatively control gene expression. The complex interactions between miRNAs and their target genes form a gene regulatory network that control lots of physiological and pathological processes in human. Recent evidence indicates that miRNAs can work as tumor-suppression factors or they can promote tumorigenesis and may play crucial roles in the prominent cell proliferation, dys-regulated cell cycle and invasion ability of human cancers [18-22]. The differential expressions of miRNAs suggest that miRNAs may be involved in the genesis and development of tumor. Moreover, in malignant diseases, the alteration of miRNAs level in the circulation shows great values in diagnosis, providing a novel strategy for tumor early stage detection [23-25]. Here we show that miRNA-221 controls human non-small cell lung cancer cell H460 growth by targeting P57. Inhibition of miRNA-221 represents a novel potential treatment for human non-small cell lung cancer.
Several studies have indicated that miR-221 could enhance cellular migration through the activation of the AKT pathway and metallopeptidases in NSCLC [26]. In the current study, our data provide further evidence that miRNA-221 can regulate human NSCLC cell line H460 proliferation rate and cell vitality. By overexpressing miRNA-221, H460 cells showed higher rate of EdU positive cells and cell vitality than NC-mimics. Conversely, down-regulation of miRNA-221 led to lower rate of EdU positive cells and cell vitality. These data provide more insights of the function of miRNA-221 in NSCLC progression.
P27 is a well established target of miRNA-221. By targeting p27, miRNA-221 was demonstrated to mediate the effects of PDGF-BB on migration, proliferation, and the epithelial-mesenchymal transition of pancreatic cancer cells. Our data here shows that p27 is regulated by miRNA-221 in H460 cells, indicating it might mediate the effects of miRNA-221 in this study, though this needs to be further confirmed with reverse evidence in the future, which is a limitation of the present study.
In conclusion, our study provided evidence that miR-221 controls H460 cells growth potentially by targeting P27. Our data demonstrate that changes in miR-221/p27 signaling may contribute to cell proliferation in NSCLC.
Acknowledgements
This work was supported by the grants from National Clinical Key Subject Construction Project and Jiangsu Province’s Key Discipline/Laboratory of Medicine (XK201118).
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Chansky K, Sculier JP, Crowley JJ, Giroux D, Van Meerbeeck J, Goldstraw P International Staging Committee and Participating Institutions. The International Association for the Study of Lung Cancer Staging Project: prognostic factors and pathologic TNM stage in surgically managed non-small cell lung cancer. J Thorac Oncol. 2009;4:792–801. doi: 10.1097/JTO.0b013e3181a7716e. [DOI] [PubMed] [Google Scholar]
- 3.Lin PY, Yu SL, Yang PC. MicroRNA in lung cancer. Br J Cancer. 2010;103:1144–1148. doi: 10.1038/sj.bjc.6605901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 7.Schetter AJ, Heegaard NH, Harris CC. Inflammation and cancer: interweaving microRNA, free radical, cytokine and p53 pathways. Carcinogenesis. 2010;31:37–49. doi: 10.1093/carcin/bgp272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao J, Shen B, Li J, Lv D, Zhao Y, Wang F, Xu J. Serum microRNA-499 and microRNA-208a as biomarkers of acute myocardial infarction. Int J Clin Exp Med. 2014;7:136–141. [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, Xu J, Cheng Y, Wang F, Song Y, Xiao J. Circulating microRNAs as mirrors of acute coronary syndromes: MiRacle or quagMire. J Cell Mol Med. 2013;17:1362–1270. doi: 10.1111/jcmm.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang YF, Wang F, Xiao JJ, Song Y, Zhao YY, Cao Y, Bei YH, Yang CQ. MiR-222 overexpression promotes proliferation of human hepatocellular carcinoma HePG2 cells by downregulating p27. Int J Clin Exp Med. 2014;7:893–902. [PMC free article] [PubMed] [Google Scholar]
- 11.Shenoy A, Blelloch RH. Regulation of microRNA function in somatic stem cell proliferation and differentiation. Nat Rev Mol Cell Biol. 2014;15:565–576. doi: 10.1038/nrm3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang R, Ma J, Wu Q, Xia J, Miele L, Sarkar FH, Wang Z. Functional role of miR-34 family in human cancer. Curr Drug Targets. 2013;13:1185–1191. doi: 10.2174/13894501113149990191. [DOI] [PubMed] [Google Scholar]
- 13.Liu S, Sun X, Wang M, Hou Y, Zhan Y, Jiang Y, Liu Z, Chen X, Tao Y, Xu C, Mao J, Cheng C, Li C, Hu Y, Wang L, Chin YE, Shi Y, Siebenlist U, Zhang X. A microRNA 221- and 222-mediated feedback loop maintains constitutive activation of NFkappaB and STAT3 in colorectal cancer cells. Gastroenterology. 2014;147:847–859. doi: 10.1053/j.gastro.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Liang C, Ma H, Zhao Q, Lu Y, Xiang Z, Li L, Qin J, Chen Y, Cho WC, Pestell RG, Liang L, Yu Z. miR-221/222 promotes S-phase entry and cellular migration in control of basal-like breast cancer. Molecules. 2014;19:7122–7137. doi: 10.3390/molecules19067122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lv J, Xu L, Xu Y, Qiu M, Yang X, Wang J, Yin R, Xu L. Expression of MiRNA-221 in non-small cell lung cancer tissues and correlation with prognosis. Zhongguo Fei Ai Za Zhi. 2014;17:221–225. doi: 10.3779/j.issn.1009-3419.2014.03.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Acunzo M, Visone R, Romano G, Veronese A, Lovat F, Palmieri D, Bottoni A, Garofalo M, Gasparini P, Condorelli G, Chiariello M, Croce CM. miR-130a targets MET and induces TRAIL-sensitivity in NSCLC by downregulating miR-221 and 222. Oncogene. 2012;31:634–642. doi: 10.1038/onc.2011.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 19.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 20.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 21.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 22.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 23.Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, Xiao T, Schafer J, Lee ML, Schmittgen TD, Nana-Sinkam SP, Jarjoura D, Marsh CB. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 25.Garofalo M, Di Leva G, Romano G, Nuovo G, Suh SS, Ngankeu A, Taccioli C, Pichiorri F, Alder H, Secchiero P, Gasparini P, Gonelli A, Costinean S, Acunzo M, Condorelli G, Croce CM. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16:498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Su A, He S, Tian B, Hu W, Zhang Z. MicroRNA-221 mediates the effects of PDGF-BB on migration, proliferation, and the epithelial-mesenchymal transition in pancreatic cancer cells. PLoS One. 2013;8:e71309. doi: 10.1371/journal.pone.0071309. [DOI] [PMC free article] [PubMed] [Google Scholar]


