Abstract
Previous studies have investigated the associations between the two polymorphisms (prostate stem cell antigen (PSCA) rs2294008 C/T and c-MYC rs9642880 G/T) and bladder cancer (BC) risk. However, the results are inconsistent. We therefore carried out a meta-analysis to estimate the relationship between PSCA/c-MYC polymorphisms and BC risk. We searched PubMed up to November 2014 to identify potentially eligible literatures. Odds ratios (ORs) and 95% confidence intervals (CIs) were used to estimate the strength of the associations, the data were further stratified by ethnicity. Heterogeneity was evaluated by Q test and I2 statistics. Begg’s funnel plot and Egger’s test were used to assess the publication bias. 11 studies from 9 articles were identified, including a total of 16,814 cancer cases and 52,868 case-free controls. We found a significant association between PSCA rs2294008 polymorphism and BC risk (the allele contrast model: OR = 1.14, 95% CI = 1.11-1.18; homozygote comparison: OR = 1.28, 95% CI = 1.20-1.37; heterozygote comparison: OR = 1.23, 95% CI = 1.17-1.30; dominant model: OR = 1.25, 95% CI = 1.19-1.31 and recessive model: OR = 1.13, 95% CI = 1.07-1.20). Moreover, a significant increased risk of BC was confirmed both in Caucasian and in Asians. For c-MYC rs9642880 polymorphism, significant increased BC risk was detected under the following genetic models (the allele contrast model: OR = 1.20, 95% CI = 1.13-1.27; homozygote comparison: OR = 1.37, 95% CI = 1.21-1.55; heterozygote comparison: OR = 1.20, 95% CI = 1.09-1.32; dominant model: OR = 1.25, 95% CI = 1.14-1.37 and recessive model: OR = 1.26, 95% CI = 1.13-1.40). Further stratified analysis by ethnicity also observed the same results. This meta-analysis suggested that PSCA rs2294008 and c-MYC rs9642880 polymorphisms may increase the BC risk. Further studies are needed to clarify the effects.
Keywords: PSCA, c-MYC, polymorphism, bladder cancer, meta-analysis
Introduction
Bladder cancer (BC) is one of the most common malignant cancer worldwide and the eighth cause of death in male cancer. As the second most frequent malignancy of the genitourinary tract, in 2014, the estimated number of new cases is 74,690 and result in 15,580 deaths in the US [1]. In china, BC is the 10th most common cancer, the incidence and mortality of BC is significantly increased from 1991 to 2005 [2]. The known risk factors are smoking and occupational exposures [3]. Furthermore, genetic factors also play an important role in BC susceptibility [4,5].
Recently, genome-wide association studies (GWAS) show certain number of new BC-associated single-nucleotide polymorphisms (SNPs). Among them, rs2294008 (C/T) within the prostate stem cell antigen (PSCA) gene on 8q24.3 and rs9642880 (G/T) within c-MYC gene on 8q24.1 are most widely discussed in BC [6-9,14-15].
PSCA gene is locate on chromosome 8q24.2, consists of 3 exons and 2 introns, encodes a 123-amino acid glycoprotein, which is belong to the LY-6/Thy-1 family of cell surface antigens [16]. PSCA was initially identified as a prostate-specific antigen, which is overexpressed in most of prostate cancer, and influence cell adhesion, proliferation, and survival [17]. However, it is also expressed in other solid tumors, such as pancreas cancer, bladder cancer, esophagus cancer and gastric cancer [14,18-20]. c-MYC gene is a member of the MYC gene family, locate on band q24.1 of chromosome 8 and consists of 3 exons and 2 introns. c-MYC plays an essential role in the regulation of various physiological processes such as cell cycle, cell adhesion, apoptosis and protein synthesis [21]. Aberrant expression of c-MYC is very likely to attribute to direct gene alteration, which can lead to tumorigenesis and maintain tumor growth [22]. The inhibition of c-MYC is expected to become a therapeutic strategy of human cancer [23].
However, because of ethnic diversity and various backgrounds of the studies, there are controversial results regarding the association of PSCA/c-MYC polymorphisms with the risk of BC [6,8,9,14,15]. So we performed a meta-analysis to clarify the relationship between the PSCA rs2294008 (C/T) and c-MYC rs9642880 (G/T) polymorphisms and BC risk.
Materials and methods
Literature searching strategy
We searched for relevant literatures in PubMed up to November 2014 using the following terms: “PSCA rs2294008 (C/T)” “c-MYC rs9642880 (G/T)” “polymorphism” and “bladder cancer”. Only studies published in English were included. To search for more potentially relevant studies, reference lists from studies included were reviewed to identify additional relevant publications.
Selection criteria
The inclusion criteria of this meta-analysis were as follows: (1) case-control studies; (2) the studies evaluated the relationship between the PSCA rs2294008 (C/T) or c-MYC rs9642880 (G/T) polymorphisms and BC risk; (3) the studies included detailed genotyping data.
The exclusion criteria were: (1) not case-control studies; (2) the source of cases and controls, and other essential information were not provided; (3) no available genotype frequency (4) reviews and duplicated publications.
Data extraction
For each study, the following data were collected: first author’s surname, year of publication, country of origin, ethnicity, source of control, genotyping method, total numbers of cases and controls as well as numbers of cases and controls with CC (GG), CT (GT) and TT (TT) genotypes. Disagreement was resolved by discussion between all authors until a consensus was reached. The non-cancer controls had no history of any malignant disease. When studies included, subjects of ethnicity and genotype data were extracted separately according to ethnicities for subgroup analyses. We did not define any minimum number of patients for inclusion in our meta-analysis.
Statistical analysis
The association of PSCA rs2294008 (C/T) and c-MYC rs9642880 (G/T) polymorphisms with BC were measured by odds ratio (OR) with 95% confidence interval (CI). The statistical significance for each OR value was evaluated by the Z test. Statistical heterogeneity was measured by using the Q test and I2 statistics. The Q test and I2 were claimed to test the variation which was due to heterogeneity or by random error, when P value of heterogeneity tests was no more than 0.1 (P ≤ 0.1), we used random effects model. On the contrary (P > 0.1), the fixed effects model was performed. Additionally, subgroup analysis was conducted on the basis of ethnicity. Funnel plots were used to assess publication bias. All of the calculations were performed using the review manager version 5.3 (Revman; The Cochrane Collaboration, Oxford, UK), and all statistical tests were two-sided, P value less than 0.05 was considered significant.
Results
Characteristics of studies
As shown in Figure 1, we preliminarily identified 42 studies about the relationship between the PSCA rs2294008 and c-MYC rs9642880 polymorphisms and BC risk. Following the above inclusion and exclusion criteria, we excluded 33 studies (17 were not case-control studies, 2 were duplicate literature, 5 were not for bladder cancer, 4 were review or meta-analysis articles, 5 did not report detailed allele frequency data). Finally, 11 studies from 9 articles were included in this meta-analysis [7-15], including a total of 15,138 BC cases and 22,594 case-free controls. The characteristics of the included studies are listed in Table 1. Detailed genotypes data for PSCA rs2294008 (C/T) and c-MYC rs9642880 (G/T) polymorphism are list in Table 2.
Figure 1.
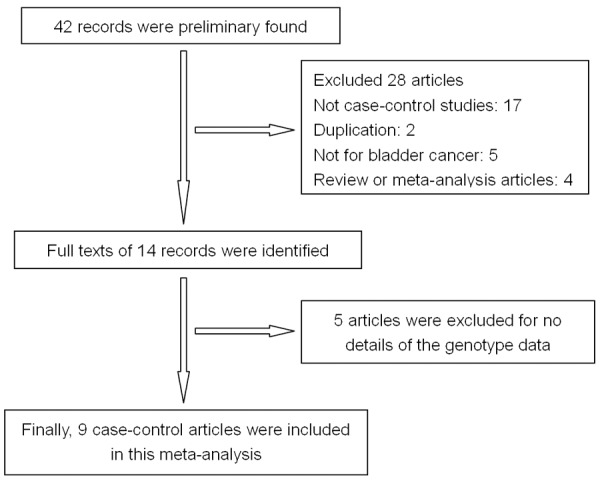
Flow chart showing the detailed steps for study selection.
Table 1.
Characteristics of the included studies in the meta-analysis
| First author | Year | Country | Ethnicity | Genotyping medthod | Source of control | Total sample size (case/control) | SNP No. | HWE (P) |
|---|---|---|---|---|---|---|---|---|
| Wang [8] | 2014 | China | Asian | TaqMan | Population | 1210/1008 | 1,2 | > 0.05 |
| Ma [11] | 2013 | China | Asian | MassARRAY | Population | 184/962 | 1,2 | > 0.05 |
| Fu [12] | 2012 | Europe | Caucasian | GWAS | Population | 5416/7349 | 2 | > 0.05 |
| Yates DR [13] | 2013 | France | Caucasian | TaqMan | Hospital | 231/261 | 1 | > 0.05 |
| Schwender H [7] | 2012 | Europe | Caucasian | TaqMan | Hospital | 1595/1760 | 1 | > 0.05 |
| Golka K [9] | 2009 | Germany | Caucasian | TaqMan | Hospital | 212/194 | 1 | > 0.05 |
| Golka K [9] | 2009 | Germany | Caucasian | TaqMan | Hospital | 303/699 | 1 | > 0.05 |
| Wang [10] | 2009 | China | Asian | PCR-RFLP | Hospital | 230/255 | 1 | > 0.05 |
| Wang [10] | 2009 | China | Asian | PCR-RFLP | Hospital | 185/210 | 1 | > 0.05 |
| Wu [14] | 2009 | U.S.A | Caucasian | GWAS | Population | 6667/39590 | 2 | > 0.05 |
| Wang [15] | 2010 | China | Asian | PCR-RFLP | Hospital | 581/580 | 2 | > 0.05 |
CC: case-control; PCR: polymerase chain reaction; RFLP: restriction fragment length polymorphism; GWAS: Genome-wide association studies; HWE: Hardy-Weinberg equilibrium; SNP: single-nucleotide polymorphisms; SNP No. 1: c-MYC rs9642880 (G/T); 2: PSCA rs2294008 (C/T).
Table 2.
PSCA rs2294008 and c-MYC rs9642880 polymorphisms genotype distribution and allele frequency in cases and controls
| First author | Genotype (N) | Allele frequency (N) | MAF | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||
| Case | Control | Case | Control | ||||||||||
|
| |||||||||||||
| total | AA | AB | BB | total | AA | AB | BB | A | B | A | B | ||
| rs9642880 | |||||||||||||
| Wang 2014 | 1210 | 550 | 536 | 124 | 1008 | 514 | 389 | 105 | 1636 | 784 | 1417 | 599 | 0.32 |
| Ma 2013 | 171 | 74 | 74 | 23 | 962 | 489 | 371 | 102 | 222 | 120 | 1349 | 575 | 0.35 |
| Yates DR 2013 | 231 | 64 | 114 | 53 | 261 | 81 | 130 | 50 | 242 | 220 | 292 | 230 | 0.48 |
| Schwender H 2012 | 1584 | 391 | 767 | 426 | 1738 | 486 | 876 | 376 | 1549 | 1619 | 1848 | 1628 | 0.51 |
| Golka K 2009 | 212 | 47 | 93 | 72 | 194 | 52 | 97 | 45 | 187 | 237 | 201 | 187 | 0.56 |
| Golka K 2009 | 303 | 74 | 151 | 78 | 699 | 178 | 364 | 157 | 299 | 307 | 720 | 678 | 0.51 |
| Wang 2009 | 230 | 81 | 114 | 35 | 255 | 120 | 109 | 26 | 276 | 184 | 349 | 161 | 0.4 |
| Wang 2009 | 185 | 68 | 89 | 28 | 210 | 103 | 83 | 24 | 225 | 145 | 289 | 131 | 0.39 |
| rs2294008 | |||||||||||||
| Wang 2014 | 1210 | 604 | 509 | 97 | 1008 | 566 | 376 | 66 | 1717 | 703 | 1508 | 508 | 0.29 |
| Ma 2013 | 175 | 84 | 80 | 11 | 962 | 543 | 355 | 64 | 248 | 102 | 1441 | 483 | 0.29 |
| Fu 2012 | 5393 | 1363 | 2804 | 1226 | 7324 | 2107 | 3645 | 1572 | 5530 | 5256 | 7859 | 6789 | 0.49 |
| Wu 2009 | 5038 | 1288 | 2613 | 1137 | 9363 | 2842 | 4668 | 1853 | 5189 | 4887 | 10352 | 8374 | 0.49 |
| Wang 2010 | 581 | 272 | 259 | 50 | 580 | 316 | 220 | 44 | 803 | 359 | 852 | 308 | 0.31 |
A represents the major allele, B represents the minor allele. MAF: minor allele frequencies.
Meta-analysis results
The main results of this meta-analysis were listed in Table 3. There were 8 studies including 4,126 cases and 5,327 controls used to evaluate the relationship between PSCA rs2294008 (C/T) polymorphism and BC susceptibility, and 5 studies including 12,397 cases and 19,237 controls were performed to assess the effect of c-MYC rs9642880 (G/T) polymorphism and BC susceptibility.
Table 3.
Meta-analysis results
| Comparisons | OR | 95% CI | P value | Heterogeneity | Effects model | |
|---|---|---|---|---|---|---|
|
| ||||||
| I2 | P value | |||||
| B vs A | ||||||
| rs9642880 | 1.20 | 1.13–1.27 | < 0.00001 | 0% | 0.53 | F |
| Caucasian | 1.18 | 1.09-1.28 | < 0.0001 | 0% | 0.63 | F |
| Asian | 1.22 | 1.11-1.35 | < 0.0001 | 25% | 0.26 | F |
| rs2294008 | 1.14 | 1.11-1.18 | < 0.00001 | 14% | 0.32 | F |
| Caucasian | 1.13 | 1.09-1.17 | < 0.00001 | 69% | 0.07 | F |
| Asian | 1.22 | 1.11-1.35 | < 0.0001 | 0% | 0.99 | F |
| BB vs AA | ||||||
| rs9642880 | 1.37 | 1.21–1.55 | < 0.00001 | 0% | 0.54 | F |
| Caucasian | 1.39 | 1.19-1.62 | < 0.0001 | 0% | 0.71 | F |
| Asian | 1.33 | 1.07-1.65 | 0.009 | 33% | 0.21 | F |
| rs2294008 | 1.28 | 1.20-1.37 | < 0.00001 | 0% | 0.57 | F |
| Caucasian | 1.28 | 1.19-1.37 | < 0.00001 | 61% | 0.11 | F |
| Asian | 1.32 | 1.03-1.69 | 0.03 | 0% | 0.86 | F |
| AB vs AA | ||||||
| rs9642880 | 1.20 | 1.09–1.32 | 0.0002 | 6% | 0.39 | F |
| Caucasian | 1.07 | 0.94-1.23 | 0.30 | 0% | 0.97 | F |
| Asian | 1.36 | 1.18-1.54 | < 0.0001 | 0% | 0.68 | F |
| rs2294008 | 1.23 | 1.17-1.30 | < 0.00001 | 0% | 0.65 | F |
| Caucasian | 1.21 | 1.14-1.29 | < 0.00001 | 0% | 0.53 | F |
| Asian | 1.32 | 1.16-1.51 | < 0.0001 | 0% | 0.74 | F |
| AB + BB vs AA | ||||||
| rs9642880 | 1.25 | 1.14–1.37 | < 0.00001 | 0% | 0.56 | F |
| Caucasian | 1.17 | 1.03-1.33 | 0.01 | 0% | 0.90 | F |
| Asian | 1.35 | 1.18-1.54 | < 0.00001 | 0% | 0.41 | F |
| rs2294008 | 1.25 | 1.19-1.31 | < 0.00001 | 0% | 0.64 | F |
| Caucasian | 1.23 | 1.17-1.30 | < 0.00001 | 13% | 0.28 | F |
| Asian | 1.32 | 1.17-1.50 | < 0.0001 | 0% | 0.86 | F |
| BB vs AA + AB | ||||||
| rs9642880 | 1.26 | 1.13-1.40 | < 0.0001 | 0% | 0.54 | F |
| Caucasian | 1.33 | 1.17-1.51 | < 0.0001 | 0% | 0.63 | F |
| Asian | 1.09 | 0.89-1.34 | 0.40 | 0% | 0.63 | F |
| rs2294008 | 1.13 | 1.07-1.20 | < 0.0001 | 0% | 0.56 | F |
| Caucasian | 1.13 | 1.06-1.20 | < 0.0001 | 57% | 0.13 | F |
| Asian | 1.17 | 0.92-1.48 | 0.20 | 0% | 0.76 | F |
A represents the major allele, B represents the minor allele, F: fixed effects model, R: random effects model.
As shown in Table 3 and Figure 2, the individuals carrying variant genotypes of PSCA rs2294008 had an increased risk of BC in all genetic models (the allele contrast model: OR = 1.14, 95% CI = 1.11-1.18; homozygote comparison: OR = 1.28, 95% CI = 1.20-1.37; heterozygote comparison: OR = 1.23, 95% CI = 1.17-1.30; dominant model: OR = 1.25, 95% CI = 1.19-1.31 and recessive model: OR = 1.13, 95% CI = 1.07-1.20). In the stratified analysis by ethnicity, significant increased BC risk were detected both in Caucasians and in Asians in the following genetic models (the allele contrast model: OR =1.13, 95% CI = 1.09-1.17; homozygote comparison: OR = 1.28, 95% CI = 1.19-1.37; heterozygote comparison: OR = 1.21, 95% CI = 1.14-1.29; dominant model: OR = 1.23, 95% CI = 1.17-1.30 and recessive model: OR = 1.13, 95% CI = 1.06-1.20 for Caucasians. the allele contrast model: OR = 1.22, 95% CI = 1.11-1.35; homozygote comparison: OR = 1.32, 95% CI = 1.03-1.69; heterozygote comparison: OR = 1.32, 95% CI = 1.16-1.51 and dominant model: OR = 1.32, 95% CI = 1.17-1.50 for Asians).
Figure 2.
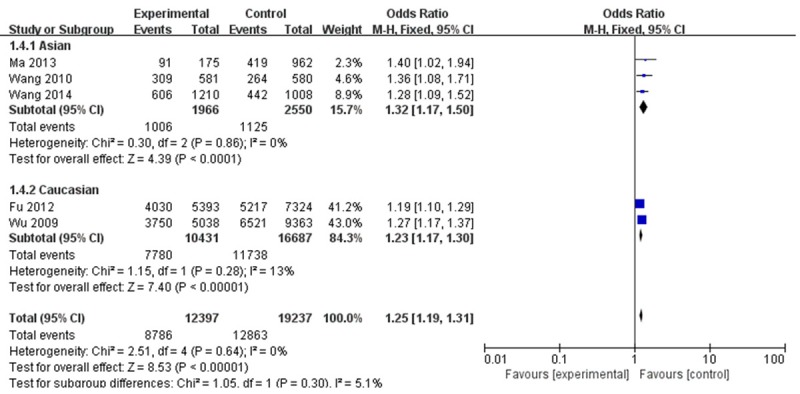
Forest Plot for the association between PSCA rs2294008 polymorphism and BC risk (AB + BB vs AA; subgroup analysis base on ethnicity). CI: confidence interval; OR: odds ratio; M-H: Mantel-Haenszel.
For c-MYC rs9642880 (G/T), from Table 3 and Figure 3, we also observed an increased risk of bladder cancer in all genetic models (the allele contrast model: OR = 1.20, 95% CI = 1.13-1.27; homozygote comparison: OR = 1.37, 95% CI = 1.21-1.55; heterozygote comparison: OR = 1.20, 95% CI = 1.09-1.32; dominant model: OR = 1.25, 95% CI = 1.14-1.37 and recessive model: OR = 1.26, 95% CI = 1.13-1.40). When stratified by ethnicity, an increased BC risk was found in Caucasians under the allele contrast model (OR = 1.18, 95% CI = 1.09-1.28), homozygote comparison (OR = 1.39, 95% CI = 1.19-1.62), dominant model (OR = 1.17, 95% CI = 1.03-1.33) and recessive model (OR = 1.13, 95% CI = 1.17-1.51), but not the heterozygote comparison (OR = 1.07, 95% CI = 0.94-1.23). For Asian, the same effects were observed in the allele contrast model (OR = 1.22, 95% CI = 1.11-1.35), homozygote comparison (OR = 1.33, 95% CI = 1.07-1.65), heterozygote comparison (OR = 1.36, 95% CI = 1.18-1.54) and dominant model (OR = 1.35, 95% CI = 1.18-1.54), except the recessive model (OR = 1.09, 95% CI = 0.89-1.34).
Figure 3.
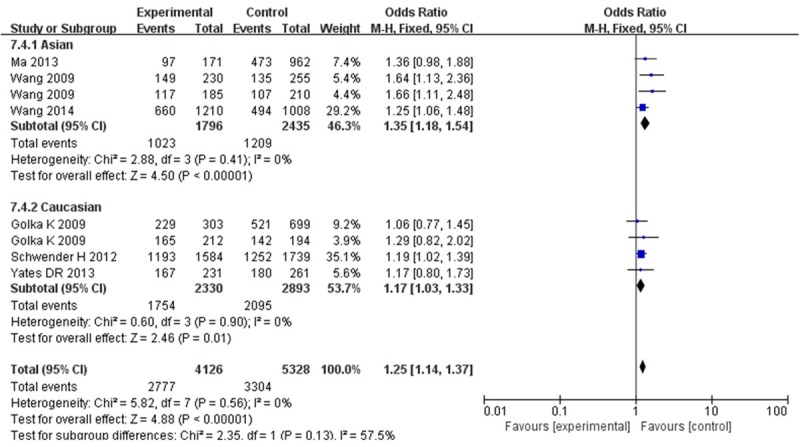
Forest Plot for the association between c-MYC rs9642880 polymorphism and BC risk (AB + BB vs AA; subgroup analysis base on ethnicity). CI: confidence interval; OR: odds ratio; M-H: Mantel-Haenszel.
Tests of heterogeneity
Statistically significant heterogeneity was observed between trials of the following analyses by using Q statistic. As shown in Table 3, there was no significant heterogeneity in any genetic models, so the fixed-effects model was performed in all analysis.
Publication bias
Begg’s funnel plot and Egger’s test were performed to assess the publication bias. As show in Figure 4, the shape of the funnel plots was symmetrical in all comparison models. The statistical results did not suggest any evidence of publication bias in this meta-analysis (P > 0.05).
Figure 4.
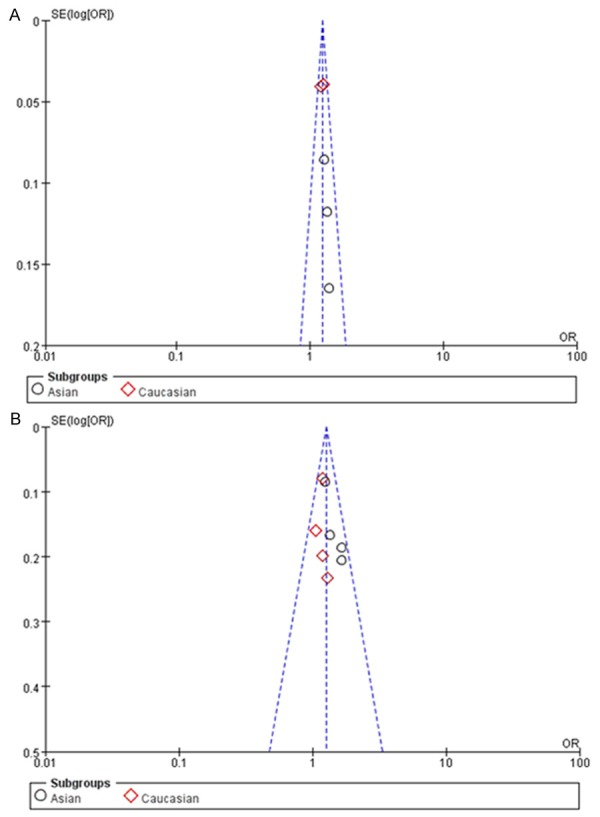
Funnel plot assessing evidence of publication bias (A: PSCA rs2294008; B: c-MYC rs9642880).
Discussion
Although there had great progress in the treatment of BC, the prognosis of BC patients was still poor. To the best of our knowledge, the most important risk factors are smoking and occupational exposures, but there also have significant differences within the same Lifestyle. Therefore, we need to explore other new biomarkers, which have a great relationship of BC risk. By genome-wide association studies (GWAS), we observed that PSCA rs2294008 (C/T) and c-MYC rs9642880 (G/T) polymorphisms were closely associated with the risk and survival of BC [6-9,14,15].
PSCA was initially identified as a prostate-specific cell-surface antigen, and not only expressed in normal prostate and prostate cancer but also in other non-prostatic malignancies [24]. Cheng et al. reported immunocytochemical analysis of PSCA on archived voided urine samples may provided a complementary marker for cytological diagnosis of urothelial carcinoma [25]. The PSCA expression level could be a valuable prognostic marker of recurrence in superficial transitional cell carcinoma (TCC) of the bladder [26]. c-MYC also expressed in a wide range of malignancies, which was involved in cell apoptosis, senescence, and DNA damage responses [27]. C-MYC plays an important role in tumor progression and maintenance [28]. Thus, c-MYC become feasible target for novel therapies of human malignancies, recently, some low-molecular weight compounds were found have a potential to be developed into therapeutic drugs in cancer therapy, which target the transcription of c-MYC gene directly or the c-MYC downstream pathway [29].
The relationships between the PSCA/c-MYC polymorphisms and BC are inconsistent [6,8,9,14,15]. In this meta-analysis, we pooled 11 eligible case-control studies to estimate the effects of PSCA rs2294008 and c-MYC rs9642880 on BC risk. We found that the two polymorphisms had significant increased risk of BC in the overall population. Kiemeney et al. observed c-MYC rs9642880 polymorphism was susceptibility to urinary bladder cancer in hospital-based case-control groups, if there was scarce specific occupational exposure [6]. Ma et al. obtained the same result [11]. In addition, by stratified analysis in non-muscle invasive cases, PSCA rs2294008 had potential effect on non-muscle invasive bladder cancer, but not on muscle invasive BC [11]. The results suggested the pathological stage and environment factors may influence the relationship between PSCA or c-MYC polymorphism and BC. Therefore, association studies with detailed pathological stage and individual data need to be performed in large samples in the future to validate the relationship between PSCA/c-MYC and BC.
In the subgroup analysis based on ethnicity, we also found this result, except the recessive model in Asians for PSCA, heterozygote comparison in Caucasian for c-MYC and recessive mode in Asian for c-MYC. However, there were no studies to evaluate the ethnic differences between the susceptibility gene and BC, especially on differences between Caucasians and Asians. Further investigations with large scale on Caucasian and Asian populations were needed to verify this result.
Some limitations of this meta-analysis should be noted. Firstly, this meta-analysis was based on pooled data and no individual data was available, thus, we could not assess the risk of cancer according to stratification of age, gender, pathological stage, environment factors or other risk factors of bladder cancer. Secondly, small study effect, in which effects reported in small studies were larger, could not be avoided in that some studies were of a relative small size. Moreover, further large scale multicenter studies with more detailed individual data, and different environmental background are needed to further validate gene-gene and gene-environment interactions on PSCA/c-MYC polymorphism and BC risk.
Conclusion
In summary, this meta-analysis provides evidence of the association between PSCA/c-MYC polymorphisms and BC risk. PSCA rs2294008 and c-MYC rs9642880 are both significantly associated with the increased BC risk. Further studies based on different ethnicity are warranted to verify our findings.
Acknowledgements
We acknowledge the Grants from the National Natural Science Foundation of China, No. 81471670; the International Cooperative Project of Shaanxi province, China, No. 2013KW-32-01; the Fundamental Research Funds for the Central Universities, China and Specialized Research Fund of the Second Affiliated Hospital of Xi’an Jiaotong University, China, No. RC (GG) 201203.
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Yang L, Parkin DM, Li LD, Chen YD, Bray F. Estimation and projection of the national profile of cancer mortality in China: 1991-2005. Br J Cancer. 2004;90:2157–2166. doi: 10.1038/sj.bjc.6601813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freedman ND, Silverman DT, Hollenbeck AR, Schatzkin A, Abnet CC. Association between smoking and risk of bladder cancer among men and women. JAMA. 2011;306:737–745. doi: 10.1001/jama.2011.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K. Environmental and heritable factors in the causation of cancer-analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 5.Czene K, Lichtenstein P, Hemminki K. Environmental and heritable causes of cancer among 9.6 million individuals in the Swedish Family-Cancer Database 2002. Int J Cancer. 2002;99:260–266. doi: 10.1002/ijc.10332. [DOI] [PubMed] [Google Scholar]
- 6.Kiemeney LA, Thorlacius S, Sulem P, Geller F, Aben KK, Stacey SN, Gudmundsson J, Jakobsdottir M, Bergthorsson JT, Sigurdsson A, Blondal T, Witjes JA, Vermeulen SH, Hulsbergen-van de Kaa CA, Swinkels DW, Ploeg M, Cornel EB, Vergunst H, Thorgeirsson TE, Gudbjartsson D, Gudjonsson SA, Thorleifsson G, Kristinsson KT, Mouy M, Snorradottir S, Placidi D, Campagna M, Arici C, Koppova K, Gurzau E, Rudnai P, Kellen E, Polidoro S, Guarrera S, Sacerdote C, Sanchez M, Saez B, Valdivia G, Ryk C, de Verdier P, Lindblom A, Golka K, Bishop DT, Knowles MA, Nikulasson S, Petursdottir V, Jonsson E, Geirsson G, Kristjansson B, Mayordomo JI, Steineck G, Porru S, Buntinx F, Zeegers MP, Fletcher T, Kumar R, Matullo G, Vineis P, Kiltie AE, Gulcher JR, Thorsteinsdottir U, Kong A, Rafnar T, Stefansson K. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat Genet. 2008;40:1307–1312. doi: 10.1038/ng.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwender H, Selinski S, Blaszkewicz M, Marchan R, Ickstadt K, Golka K, Hengstler JG. Distinct SNP combinations confer susceptibility to urinary bladder cancer in smokers and non-smokers. PLoS One. 2012;7:e51880. doi: 10.1371/journal.pone.0051880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang P, Ye D, Guo J, Liu F, Jiang H, Gong J, Gu C, Shao Q, Sun J, Zheng SL, Yu H, Lin X, Xia G, Fang Z, Zhu Y, Ding Q, Xu J. Genetic score of multiple risk-associated single nucleotide polymorphisms is a marker for genetic susceptibility to bladder cancer. Genes Chromosomes Cancer. 2014;53:98–105. doi: 10.1002/gcc.22121. [DOI] [PubMed] [Google Scholar]
- 9.Golka K, Hermes M, Selinski S, Blaszkewicz M, Bolt HM, Roth G, Dietrich H, Prager HM, Ickstadt K, Hengstler JG. Susceptibility to urinary bladder cancer: relevance of rs9642880 [T] , GSTM1 and occupational exposure. Pharmacogenet Genomics. 2009;19:903–6. doi: 10.1097/FPC.0b013e328331b554. [DOI] [PubMed] [Google Scholar]
- 10.Wang M, Wang M, Zhang W, Yuan L, Fu G, Wei Q, Zhang Z. Common genetic variants on 8q24 contribute to susceptibility to bladder cancer in a Chinese population. Carcinogenesis. 2009;30:991–6. doi: 10.1093/carcin/bgp091. [DOI] [PubMed] [Google Scholar]
- 11.Ma Z, Hu Q, Chen Z, Tao S, Macnamara L, Kim ST, Tian L, Xu K, Ding Q, Zheng SL, Sun J, Xia G, Xu J. Systematic evaluation of bladder cancer risk-associated single-nucleotide polymorphisms in a Chinese population. Mol Carcinog. 2013;52:916–21. doi: 10.1002/mc.21932. [DOI] [PubMed] [Google Scholar]
- 12.Fu YP, Kohaar I, Rothman N, Earl J, Figueroa JD, Ye Y, Malats N, Tang W, Liu L, Garcia-Closas M, Muchmore B, Chatterjee N, Tarway M, Kogevinas M, Porter-Gill P, Baris D, Mumy A, Albanes D, Purdue MP, Hutchinson A, Carrato A, Tardón A, Serra C, García-Closas R, Lloreta J, Johnson A, Schwenn M, Karagas MR, Schned A, Diver WR, Gapstur SM, Thun MJ, Virtamo J, Chanock SJ, Fraumeni JF Jr, Silverman DT, Wu X, Real FX, Prokunina-Olsson L. Common genetic variants in the PSCA gene influence gene expression and bladder cancer risk. Proc Natl Acad Sci U S A. 2012;109:4974–9. doi: 10.1073/pnas.1202189109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yates DR, Rouprêt M, Drouin SJ, Audouin M, Cancel-Tassin G, Comperat E, Bitker MO, Cussenot O. Genetic polymorphisms on 8q24.1 and 4p16.3 are not linked with urothelial carcinoma of the bladder in contrast to their association with aggressive upper urinary tract tumours. World J Urol. 2013;31:53–9. doi: 10.1007/s00345-012-0954-6. [DOI] [PubMed] [Google Scholar]
- 14.Wu X, Ye Y, Kiemeney LA, Sulem P, Rafnar T, Matullo G, Seminara D, Yoshida T, Saeki N, Andrew AS, Dinney CP, Czerniak B, Zhang ZF, Kiltie AE, Bishop DT, Vineis P, Porru S, Buntinx F, Kellen E, Zeegers MP, Kumar R, Rudnai P, Gurzau E, Koppova K, Mayordomo JI, Sanchez M, Saez B, Lindblom A, de Verdier P, Steineck G, Mills GB, Schned A, Guarrera S, Polidoro S, Chang SC, Lin J, Chang DW, Hale KS, Majewski T, Grossman HB, Thorlacius S, Thorsteinsdottir U, Aben KK, Witjes JA, Stefansson K, Amos CI, Karagas MR, Gu J. Genetic variation in the prostate stem cell antigen gene PSCA confers susceptibility to urinary bladder cancer. Nat Genet. 2009;41:991–5. doi: 10.1038/ng.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang S, Tang J, Wang M, Yuan L, Zhang Z. Genetic variation in PSCA and bladder cancer susceptibility in a Chinese population. Carcinogenesis. 2010;31:621–4. doi: 10.1093/carcin/bgp323. [DOI] [PubMed] [Google Scholar]
- 16.Reiter RE, Gu Z, Watabe T, Thomas G, Szigeti K, Davis E, Wahl M, Nisitani S, Yamashiro J, Le Beau MM, Loda M, Witte ON. Prostate stem cell antigen: a cell surface marker overexpressed in prostate cancer. Proc Natl Acad Sci U S A. 1998;95:1735–40. doi: 10.1073/pnas.95.4.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eshel R, Zanin A, Kapon D, Sagi-Assif O, Brakenhoff R, van Dongen G, Witz IP. Human Ly-6 antigen E48 (Ly-6D) regulates important interaction parameters between endothelial cells and head-and-neck squamous carcinoma cells. Int J Cancer. 2002;98:803–10. doi: 10.1002/ijc.10301. [DOI] [PubMed] [Google Scholar]
- 18.Argani P, Rosty C, Reiter RE, Wilentz RE, Murugesan SR, Leach SD, Ryu B, Skinner HG, Goggins M, Jaffee EM, Yeo CJ, Cameron JL, Kern SE, Hruban RH. Discovery of new markers of cancer through serial analysis of gene expression: prostate stem cell antigen is overexpressed in pancreatic adenocarcinoma. Cancer Res. 2001;61:4320–4. [PubMed] [Google Scholar]
- 19.Shi Y, Hu Z, Wu C, Dai J, Li H, Dong J, Wang M, Miao X, Zhou Y, Lu F, Zhang H, Hu L, Jiang Y, Li Z, Chu M, Ma H, Chen J, Jin G, Tan W, Wu T, Zhang Z, Lin D, Shen H. A genome-wide association study identifies new susceptibility loci for non-cardia gastric cancer at 3q13.31 and 5p13.1. Nat Genet. 2011;43:1215–8. doi: 10.1038/ng.978. [DOI] [PubMed] [Google Scholar]
- 20.Bahrenberg G, Brauers A, Joost HG, Jakse G. Reduced expression of PSCA, a member of the LY-6 family of cell surface antigens, in bladder, esophagus, and stomach tumors. Biochem Biophys Res Commun. 2000;275:783–8. doi: 10.1006/bbrc.2000.3393. [DOI] [PubMed] [Google Scholar]
- 21.Dang CV. MYC on the Path to Cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin CY, Lovén J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, Zhao X, Fiskus W, Lin J, Lwin T, Rao R, Zhang Y, Chan JC, Fu K, Marquez VE, Chen-Kiang S, Moscinski LC, Seto E, Dalton WS, Wright KL, Sotomayor E, Bhalla K, Tao J. Coordinated silencing of MYC-mediated miR-29 by HDAC3 and EZH2 as a therapeutic target of histone modification in aggressive B-cell lymphomas. Cancer Cell. 2012;22:506–23. doi: 10.1016/j.ccr.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Elsamman EM, Fukumori T, Tanimoto S, Nakanishi R, Takahashi M, Toida K, Kanayama HO. The expression of prostate stem cell antigen in human clear cell renal cell carcinoma: a quantitative reverse transcriptasepolymerase chain reaction analysis. BJU Int. 2006;3:668–73. doi: 10.1111/j.1464-410X.2006.06350.x. [DOI] [PubMed] [Google Scholar]
- 25.Cheng L, Reiter RE, Jin Y, Sharon H, Wieder J, Lane TF, Rao J. Immunocytochemical analysis of prostate stem cell antigen as adjunct marker for detection of urothelial transitional cell carcinoma in voided urine specimens. J Urol. 2003;169:2094–100. doi: 10.1097/01.ju.0000064929.43602.17. [DOI] [PubMed] [Google Scholar]
- 26.Elsamman E, Fukumori T, Kasai T, Nakatsuji H, Nishitani MA, Toida K, Ali N, Kanayama HO. Prostate stem cell antigen predicts tumour recurrence in superficial transitional cell carcinoma of the urinary bladder. BJU Int. 2006;97:1202–7. doi: 10.1111/j.1464-410X.2006.06153.x. [DOI] [PubMed] [Google Scholar]
- 27.Zhang W, Kater AP, Widhopf GF 2nd, Chuang HY, Enzler T, James DF, Poustovoitov M, Tseng PH, Janz S, Hoh C, Herschman H, Karin M, Kipps TJ. B-cell activating factor and v-Myc myelocytomatosis viral oncogene homolog (c-Myc) influence progression of chronic lymphocytic leukemia. PNAS. 2010;107:18956–60. doi: 10.1073/pnas.1013420107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seenisamy J, Bashyam S, Gokhale V, Vankayalapati H, Sun D, Siddiqui-Jain A, Streiner N, Shin-Ya K, White E, Wilson WD, Hurley LH. Design and synthesis of an ex-panded porphyrin that has selectivity for the c-MYC G-quadruplex structure. J Am Chem Soc. 2005;127:2944–59. doi: 10.1021/ja0444482. [DOI] [PubMed] [Google Scholar]
- 29.Chen BJ, Wu YL, Tanaka Y, Zhang W. Small Molecules Targeting c-Myc Oncogene: Promising Anti-Cancer Therapeutics. Int J Biol Sci. 2014;10:1084–96. doi: 10.7150/ijbs.10190. [DOI] [PMC free article] [PubMed] [Google Scholar]


