Abstract
Background: RAD51 interacting with BRCA1 and BRCA2 could modulate the penetrance of BRCA1/BRCA2 mutations, which may increase susceptibility for breast cancer by inhibiting DNA repair and genome stability. The purpose of this study was to provide refined statistical evidence for the association between RAD51 polymorphism and breast cancer risk. Design and results: We conducted a meta-analysis of 15 publications with a total of 11,766 cancer cases and 11,227 controls. We summarized the data on the association of RAD51 polymorphism with breast cancer risk and performed subgroup analyses by ethnicity and control source. The pooled ORs based on fixed-effects model did not indicate a modified risk of breast cancer associated with RAD51 polymorphism in the overall population. Nor did we find a significant association in any stratified analysis. Conclusions: This meta-analysis suggested that RAD51 polymorphism did not appear to represent a significant risk factor for breast cancer.
Keywords: RAD51, breast cancer, polymorphism, susceptibility
Introduction
Breast cancer incidences have increased steadily worldwide in recent years and it remains the major cause of cancer-related deaths among women [1,2]. Several lines of evidence implicate that exposure to radiation is a risk factor for breast cancer, due to its capability of inducing double-strand DNA damage that may consequently contribute to the occurrence of this disease [3-5]. The mutations of BRCA1 and BRCA2 genes involved in double-strand break repair should be responsible for approximately 45% to 65% of all breast cancer cases [6]. DNA double-strand breaks repair gene RAD51 is able to modulate cancer risk through interaction with BRCA1 and BRCA2, two critical genes in response to ionizing radiation and genome stability [7,8]. Genetic polymorphisms of DNA repair genes have been reported to play an important role in DNA damage repair [9]. The most commonly studied has been a polymorphism in the 5’ untranslated region of the RAD51 gene.
A number of previous investigations have focused on the role of RAD51 polymorphism in the susceptibility to breast cancer [10-24]. The results, however, are highly controversial. This controversy stimulated great interest of several investigators to carry out a meta-analysis. The initial study by Wang et al. [25] suggested that the RAD51 polymorphism may contribute to breast cancer susceptibility. In the following meta-analyses, the reported associations are also inconsistent: RAD51 polymorphism as a protective factor [26], as a cancer promoter [27,28], and even no association [29]. These results are probably biased on account of the inclusion of repeated data and failure to identify all usable data [30].
In this study, we rigorously reviewed the eligibility of studies included in previous analyses and identified newly published articles to provide compelling statistical evidence for the association between RAD51 polymorphism and breast cancer risk.
Materials and methods
Search strategy
Potentially relevant studies were identified by searching the electronic databases of PubMed, EMBASE and CNKI from March 2008 to March 2014 using the following key words: “RAD51”, “polymorphism” and “breast cancer”. To obtain additional articles that may have been missed in the electronic search, we scanned the references cited in all extracted publications. If the same case series was included in multiple studies published in the name of the same authors, the most informative study with the largest number of subjects was finally selected.
Inclusion criteria
We defined the following criteria to select the studies eligible for the current meta-analysis: (1) the case population was composed of breast cancer patients and cancer-free healthy individuals were used as controls, (2) the association of RAD51 G135C polymorphism with breast cancer risk was investigated, and (3) published as a full-length article with detailed genotyping data that could help to estimate the odds ratios (ORs) with 95% confidence intervals (CIs).
Data extraction
Two authors independently collected data on the following items for each study: first author’s surname, publication date, location where the study was conducted, ethnicity of study population, total numbers of cases and controls, allele and genotype frequency in cancer cases and control subjects. When several ethnic groups were investigated in a single article, they were classified into Asian, European or American category and the data were separately extracted. Discrepancies were handled by consensus involving a third author.
Statistical methods
The association between RAD51 polymorphism and breast cancer risk was assessed by calculating ORs with 95% CIs using five genetic models (GG vs. CC, GG + GC vs. CC, GG vs. GC + CC, G vs. C, GC vs. CC). Significance of the summary ORs was assessed by the Z-test, and P<0.05 was considered significant. Subgroup analyses were performed by ethnicity and source of controls.
A Chi square based ‘Q’ test defined by Cochran was applied to evaluate the between-study heterogeneity in the meta-analysis [31]. A P value lower than 0.05 was deemed statistically significant. Combined effect sizes were measured by a fixed-effects model (the Mantel-Haenszel method) [32] when there was no indication of substantial heterogeneity (P>0.05). Otherwise, a random-effects model (the DerSimonian and Laird method) that includes assumptions on potential variance across studies was used [33] . Violation of Hardy-Weinberg equilibrium (HWE) was determined by x2 test using genotype data in control groups.
Publication bias was determined by Begg’s funnel plots and Egger’s test [34], which uses a weighted regression method to investigate the relationship between outcome effects (log odds ratio) and its standard error in each study. We considered a p value less than 0.05 as statistically significant. All statistical analyses were performed with STATA 12.0 (StataCorp, College Station, TX).
Results
Selection of studies
We initially identified a total of 61 relevant articles by a systematic literature search. Among them, 26 studies appeared to meet the pre-designed includion criteria and were singled out for further examination. After screening the full texts, 11 articles were excluded for the following reasons: (a) using the same patient population as the studies included [8,35-42]; (b) presenting no or insufficient data [43,44]. Therefore, our final data set consisted of 15 studies, providing 11,766 cancer cases and 11,227 controls (Figure 1).
Figure 1.

Flow diagram of included studies for this meta-analysis.
Characteristics of included studies
The main characteristics of the eligible studies are summarized in Table 1. All studies were based on a case-control design, of which three were conducted among Americans [13,19,22], four among Asians [10,18,21,24] and eight among Europeans [11,12,14-17,20,23]. For genotyping method, most studies used the typical polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) assay. Genotype distribution in all control groups did not deviate from values predicted by HWE except for four studies [12,13,19,21].
Table 1.
Characteristics of studies included in the meta-analysis
| First author | Year | Source of control | Genotyping method | Sample size | Country of study | Ethnicity | |
|---|---|---|---|---|---|---|---|
|
| |||||||
| Cases | controls | ||||||
| Lee | 2005 | HB | MassARRAY | 782 | 587 | Korea | Asian |
| Sliwinski | 2005 | NA | PCR-RFLP | 150 | 150 | Poland | European |
| Webb | 2005 | PB | TaqMan | 1295 | 660 | Australia | European |
| Dufloth | 2005 | PB | PCR-RFLP | 78 | 119 | Brazil | American |
| Tarasov | 2006 | PB | PCR-RFLP | 151 | 191 | Russia | European |
| Costa | 2007 | PB | PCR-RFLP | 265 | 435 | Portugal | European |
| Antoniou | 2007 | NA | TaqMan | 4443 | 4069 | UK | European |
| Pharoah | 2007 | NA | TaqMan | 2160 | 2266 | UK | European |
| Hu | 2008 | NA | PCR | 71 | 85 | China | Asian |
| Brooks | 2008 | PB | PCR-RFLP | 606 | 611 | USA | American |
| Jakubowska | 2009 | PB | Simple Probe | 1007 | 1069 | Poland | European |
| Akisik | 2010 | NA | PCR-RFLP | 147 | 120 | Turkey | Asian |
| Jara | 2010 | HB | PCR-RFLP | 267 | 500 | Chili | American |
| Hosseini | 2012 | PB | PCR-RFLP | 294 | 315 | Poland | European |
| Smolarz | 2013 | NA | PCR-RFLP | 50 | 50 | Iran | Asian |
PCR: polymerase chain reaction; PCR-RFLP: PCR-restriction fragment length polymorphism; NA: not available; HB: hospital-based; PB: population-based; HWE: Hardy-Weinberg equilibrium.
Quantitative synthesis
In the meta-analysis including a total of 11,766 cancer cases and 11,227 controls, we examined the association of RAD51 polymorphism with breast cancer risk. As shown in Table 2, overall, the RAD51 polymorphism was not found to be associated with either an increased or a decreased risk of breast cancer (GG vs. CC: OR = 1.00, 95% CI = 0.96-1.04, P for heterogeneity = 0.987; GG + GC vs. CC: OR = 1.00, 95% CI = 0.96-1.03, P for heterogeneity = 0.946; GG vs. GC + CC: OR = 1.00, 95% CI = 0.96-1.04, P for heterogeneity = 0.769; G vs. C: OR = 1.00, 95% CI = 0.97-1.03, P for heterogeneity = 0.656; GC vs. CC: OR = 0.98, 95% CI = 0.89-1.07, P for heterogeneity = 0.778). Similarly, no major effects were revealed in subsequent stratification analyses by ethnicity and source of controls. To test reliability of the obtained results, we excluded the studies that disobeyed HWE. The primary pooled ORs were not significantly altered (Figure 2).
Table 2.
Meta-analysis of the association between RAD51 G135C polymorphism and breast cancer risk
| Subgroups (No. Of studies) | GG vs. CC | GG + GC vs. CC | GG vs. GC + CC | G vs. C | GC vs. CC | |||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| OR (95% CI) | P-het | OR (95% CI) | P-het | OR (95% CI) | P-het | OR (95% CI) | P-het | OR (95% CI) | P-het | |
| Ethnicity | ||||||||||
| Asian (n = 4) | 0.96 (0.83, 1.12) | 0.203 | 0.96 (0.84, 1.09) | 0.101 | 1.07 (0.93, 1.24) | 0.080 | 1.01 (0.91, 1.11) | 0.015 | 0.83 (0.64, 1.08) | 0.075 |
| European (n = 8) | 1.00 (0.96, 1.04) | 1.000 | 1.00 (0.96, 1.04) | 1.000 | 1.00 (0.96, 1.04) | 0.952 | 1.00 (0.97, 1.03) | 0.998 | 1.00 (0.90, 1.11) | 0.990 |
| American (n = 3) | 1.01 (0.88, 1.15) | 0.988 | 1.01 (0.89, 1.14) | 0.990 | 1.00 (0.89, 1.14) | 0.979 | 1.01 (0.92, 1.10) | 0.970 | 1.05 (0.76, 1.45) | 0.939 |
| Source of control | ||||||||||
| Hospital (n = 2) | 0.99 (0.86, 1.13) | 0.956 | 0.99 (0.88, 1.12) | 0.959 | 1.01 (0.88, 1.15) | 0.806 | 1.00 (0.91, 1.09) | 0.904 | 0.94 (0.70, 1.25) | 0.933 |
| Population (n = 7) | 1.00 (0.93, 1.08) | 1.000 | 1.00 (0.94, 1.07) | 1.000 | 0.98 (0.91, 1.05) | 0.943 | 0.99 (0.95, 1.04) | 0.994 | 1.04 (0.89, 1.22) | 0.984 |
| Total (n = 15) | 1.00 (0.96, 1.04) | 0.987 | 1.00 (0.96, 1.03) | 0.946 | 1.00 (0.96, 1.04) | 0.769 | 1.00 (0.97, 1.03) | 0.656 | 0.98 (0.89, 1.07) | 0.778 |
CI: confidence interval; OR, odds ratio.
Figure 2.
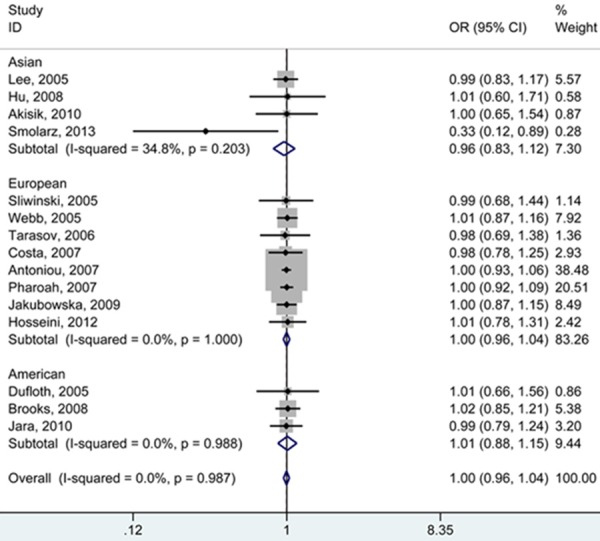
Forest plot (fixed-effects model) describing the association of the RAD51 G135C polymorphism with risk of breast cancer. The RAD51 G135C polymorphism was not associated with breast cancer under GG vs. CC.
Sensitivity analysis
Sensitivity analysis was performed to assess the influence conferred by the independent studies on the association of RAD51 polymorphism with risk of breast cancer. The combined ORs were not obviously affected by excluding each study. Therefore, our results were reliable.
Bias diagnostics
Begg’s funnel plots and Egger’s test were performed to determine the publication bias in the meta-analysis. The symmetrical shape of each funnel plot and Egger’s test did not reveal any evidence of publication bias (Egger’s test: P = 0.364 for GG vs. GC + CC) (Figure 3).
Figure 3.
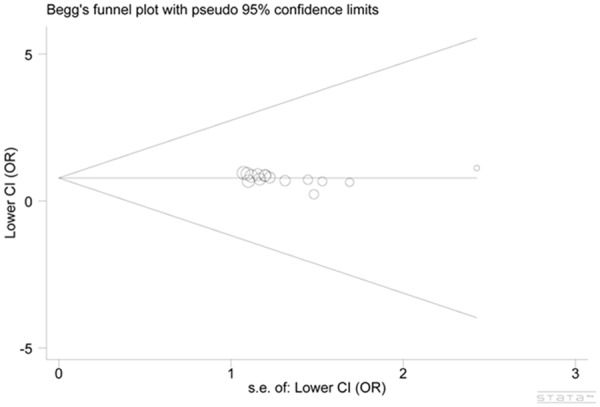
Funnel plot analysis to detect publication bias. Each point represents an individual study for the indicated association (Egger’s test: P = 0.364 for GG vs. GC + CC).
Discussion
Genetic instability and reduced DNA repair capacity may result in the breast carcinogenesis [45]. Harmful mutations in BRCA1 and BRCA2 prevent reconstruction of damaged DNA and thereby increase susceptibility for breast cancer. RAD51 is a homologue of bacterial RecA protein, playing a key role in meiotic and mitotic recombination and homology-dependent recombinational repair of DNA double-strand breaks. RAD51 could modulate the penetrance of BRCA1/BRCA2 mutations by interacting with BRCA1 and BRCA2 [46]. These results suggested that the RAD51 gene is a potential susceptibility locus for breast cancer. Therefore, investigating and establishing the role of single nucleotide polymorphisms within the region are substantially important to identify the populations at higher risk of the malignancy.
The G135C polymorphism in the RAD51 gene has been investigated in a number of genetic association studies in different ethnic groups with conflicting findings, making it important to perform a meta-analysis, a statistical method that is different from a single study tending to achieve a less precise measure of interest and contributes to a higher statistical power for the measure. In the present study, we failed to find statistical evidence for an increased risk of breast cancer associated with RAD51 polymorphism, an observation supported by Yu et al. [29], who identified 12 studies involving 7,065 cases and 6,981 controls, showing that the G135C polymorphism is not a risk factor for breast cancer. Inconsistent with former findings, Zhou et al. [27] demonstrated that RAD51 G135C polymorphism is associated with an elevated risk of breast cancer. While data from the study by Sun et al. [26] suggested that the polymorphism of interest is a low-penetrant risk factor for developing breast cancer. Lack of accuracy in data and inclusion of studies with overlapped information may result in decreases in the precision of reported associations and thus make the results less credible [30].
In the subgroup analysis by ethnicity, none of the ethnic groups showed a significant association with breast cancer, even though we excluded all repeated data and included new subjects into the meta-analysis. Interestingly, Gao et al. found an increased breast cancer risk in European populations [28] and Sun et al. reported a statistically decreased risk in Asians [26]. Ethnicity is a crucial host-related factor that may modify the association between polymorphism and cancer, because different genetic backgrounds may result in potential gene/gene and gene/environment interactions. A second possibility is that the RAD51 polymorphism is common among Europeans and Asians and functions biologically in both ethnic groups, thus modulating the risk of developing this cancer. In addition to the aforementioned explanations, another reason may again relate to data inaccuracy, a cause of false positive and false negative findings.
Although we attempted to avoid the shortcomings mentioned by He et al. [30], our results need to be interpreted with caution because of some limitations. First, significant HWE deviation was tested in several studies and this deviation may have more or less influenced the results despite no change in combined results was observed when the outlier were excluded. Second, RAD51 G135C polymorphism might be a low-penetrance risk factor for breast cancer, and the exact genetic association merits further investigations. Third, gene/gene and gene/environment interactions were not considered in this work. Further, the real association for Americans and Asians may have been masked as a result of the relatively sample sizes.
In conclusion, our study provided some evidence for lack of an association between RAD51 G135C polymorphism and risk of breast cancer. Subgroup analysis by ethnicity likewise did not implicate a statistically significant association. Further studies with a much larger number are needed to establish the RAD51-breast cancer relationship. As human diseases are a result of both environmental and genetic factors, the effects of exogenous and endogenous mutagens are expected to be considered in future.
Disclosure of conflict of interest
None.
References
- 1.Pedraza AM, Pollan M, Pastor-Barriuso R, Cabanes A. Disparities in breast cancer mortality trends in a middle income country. Breast Cancer Res Treat. 2012;134:1199–1207. doi: 10.1007/s10549-012-2026-4. [DOI] [PubMed] [Google Scholar]
- 2.Hortobagyi GN, de la Garza Salazar J, Pritchard K, Amadori D, Haidinger R, Hudis CA, Khaled H, Liu MC, Martin M, Namer M, O’Shaughnessy JA, Shen ZZ, Albain KS. The global breast cancer burden: variations in epidemiology and survival. Clin Breast Cancer. 2005;6:391–401. doi: 10.3816/cbc.2005.n.043. [DOI] [PubMed] [Google Scholar]
- 3.Hoffman DA, Lonstein JE, Morin MM, Visscher W, Harris BS 3rd, Boice JD Jr. Breast cancer in women with scoliosis exposed to multiple diagnostic x rays. J Natl Cancer Inst. 1989;81:1307–1312. doi: 10.1093/jnci/81.17.1307. [DOI] [PubMed] [Google Scholar]
- 4.Holmberg E, Holm LE, Lundell M, Mattsson A, Wallgren A, Karlsson P. Excess breast cancer risk and the role of parity, age at first childbirth and exposure to radiation in infancy. Br J Cancer. 2001;85:362–366. doi: 10.1054/bjoc.2001.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gietko M. [Hiatal hernia in children with diseases and injuries of the central nervous system] . Pediatr Pol. 1977;52:177–181. [PubMed] [Google Scholar]
- 6.Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, Loman N, Olsson H, Johannsson O, Borg A, Pasini B, Radice P, Manoukian S, Eccles DM, Tang N, Olah E, Anton-Culver H, Warner E, Lubinski J, Gronwald J, Gorski B, Tulinius H, Thorlacius S, Eerola H, Nevanlinna H, Syrjakoski K, Kallioniemi OP, Thompson D, Evans C, Peto J, Lalloo F, Evans DG, Easton DF. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72:1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tarsounas M, Davies D, West SC. BRCA2-dependent and independent formation of RAD51 nuclear foci. Oncogene. 2003;22:1115–1123. doi: 10.1038/sj.onc.1206263. [DOI] [PubMed] [Google Scholar]
- 8.Kadouri L, Kote-Jarai Z, Hubert A, Durocher F, Abeliovich D, Glaser B, Hamburger T, Eeles RA, Peretz T. A single-nucleotide polymorphism in the RAD51 gene modifies breast cancer risk in BRCA2 carriers, but not in BRCA1 carriers or noncarriers. Br J Cancer. 2004;90:2002–2005. doi: 10.1038/sj.bjc.6601837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiao Y, Spitz MR, Guo Z, Hadeyati M, Grossman L, Kraemer KH, Wei Q. Rapid assessment of repair of ultraviolet DNA damage with a modified host-cell reactivation assay using a luciferase reporter gene and correlation with polymorphisms of DNA repair genes in normal human lymphocytes. Mutat Res. 2002;509:165–174. doi: 10.1016/s0027-5107(02)00219-1. [DOI] [PubMed] [Google Scholar]
- 10.Lee KM, Choi JY, Kang C, Kang CP, Park SK, Cho H, Cho DY, Yoo KY, Noh DY, Ahn SH, Park CG, Wei Q, Kang D. Genetic polymorphisms of selected DNA repair genes, estrogen and progesterone receptor status, and breast cancer risk. Clin Cancer Res. 2005;11:4620–4626. doi: 10.1158/1078-0432.CCR-04-2534. [DOI] [PubMed] [Google Scholar]
- 11.Sliwinski T, Krupa R, Majsterek I, Rykala J, Kolacinska A, Morawiec Z, Drzewoski J, Zadrozny M, Blasiak J. Polymorphisms of the BRCA2 and RAD51 genes in breast cancer. Breast Cancer Res Treat. 2005;94:105–109. doi: 10.1007/s10549-005-0672-5. [DOI] [PubMed] [Google Scholar]
- 12.Webb PM, Hopper JL, Newman B, Chen X, Kelemen L, Giles GG, Southey MC, Chenevix-Trench G, Spurdle AB. Double-strand break repair gene polymorphisms and risk of breast or ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:319–323. doi: 10.1158/1055-9965.EPI-04-0335. [DOI] [PubMed] [Google Scholar]
- 13.Dufloth RM, Costa S, Schmitt F, Zeferino LC. DNA repair gene polymorphisms and susceptibility to familial breast cancer in a group of patients from Campinas, Brazil. Genet Mol Res. 2005;4:771–782. [PubMed] [Google Scholar]
- 14.Tarasov VA, Aslanyan MM, Tsyrendorzhiyeva ES, Litvinov SS, Gar’kavtseva RF, Altukhov YP. Genetically determined subdivision of human populations with respect to the risk of breast cancer in women. Dokl Biol Sci. 2006;406:66–69. doi: 10.1134/s0012496606010182. [DOI] [PubMed] [Google Scholar]
- 15.Costa S, Pinto D, Pereira D, Rodrigues H, Cameselle-Teijeiro J, Medeiros R, Schmitt F. DNA repair polymorphisms might contribute differentially on familial and sporadic breast cancer susceptibility: a study on a Portuguese population. Breast Cancer Res Treat. 2007;103:209–217. doi: 10.1007/s10549-006-9364-z. [DOI] [PubMed] [Google Scholar]
- 16.Antoniou AC, Sinilnikova OM, Simard J, Leone M, Dumont M, Neuhausen SL, Struewing JP, Stoppa-Lyonnet D, Barjhoux L, Hughes DJ, Coupier I, Belotti M, Lasset C, Bonadona V, Bignon YJ, Rebbeck TR, Wagner T, Lynch HT, Domchek SM, Nathanson KL, Garber JE, Weitzel J, Narod SA, Tomlinson G, Olopade OI, Godwin A, Isaacs C, Jakubowska A, Lubinski J, Gronwald J, Gorski B, Byrski T, Huzarski T, Peock S, Cook M, Baynes C, Murray A, Rogers M, Daly PA, Dorkins H, Schmutzler RK, Versmold B, Engel C, Meindl A, Arnold N, Niederacher D, Deissler H, Spurdle AB, Chen X, Waddell N, Cloonan N, Kirchhoff T, Offit K, Friedman E, Kaufmann B, Laitman Y, Galore G, Rennert G, Lejbkowicz F, Raskin L, Andrulis IL, Ilyushik E, Ozcelik H, Devilee P, Vreeswijk MP, Greene MH, Prindiville SA, Osorio A, Benitez J, Zikan M, Szabo CI, Kilpivaara O, Nevanlinna H, Hamann U, Durocher F, Arason A, Couch FJ, Easton DF, Chenevix-Trench G. RAD51 135G-->C modifies breast cancer risk among BRCA2 mutation carriers: results from a combined analysis of 19 studies. Am J Hum Genet. 2007;81:1186–1200. doi: 10.1086/522611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pharoah PD, Tyrer J, Dunning AM, Easton DF, Ponder BA. Association between common variation in 120 candidate genes and breast cancer risk. PLoS Genet. 2007;3:e42. doi: 10.1371/journal.pgen.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu R, Wei Y, Jiang WJ, Yao WX, Long QM, Zhang JH, Liang Y, Tang XL. [Association of polymorphisms of N372H in BRCA2 gene and 135G/C in RAD51 gene and breast cancers] . Sichuan Da Xue Xue Bao Yi Xue Ban. 2008;39:973–975. [PubMed] [Google Scholar]
- 19.Brooks J, Shore RE, Zeleniuch-Jacquotte A, Currie D, Afanasyeva Y, Koenig KL, Arslan AA, Toniolo P, Wirgin I. Polymorphisms in RAD51, XRCC2, and XRCC3 are not related to breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2008;17:1016–1019. doi: 10.1158/1055-9965.EPI-08-0065. [DOI] [PubMed] [Google Scholar]
- 20.Jakubowska A, Jaworska K, Cybulski C, Janicka A, Szymanska-Pasternak J, Lener M, Narod SA, Lubinski J. Do BRCA1 modifiers also affect the risk of breast cancer in non-carriers? Eur J Cancer. 2009;45:837–842. doi: 10.1016/j.ejca.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 21.Akisik E, Yazici H, Dalay N. ARLTS1, MDM2 and RAD51 gene variations are associated with familial breast cancer. Mol Biol Rep. 2011;38:343–348. doi: 10.1007/s11033-010-0113-3. [DOI] [PubMed] [Google Scholar]
- 22.Jara L, Dubois K, Gaete D, de Mayo T, Ratk evicius N, Bravo T, Margarit S, Blanco R, Gomez F, Waugh E, Peralta O, Reyes JM, Ibanez G, Gonzalez-Hormazabal P. Variants in DNA double-strand break repair genes and risk of familial breast cancer in a South American population. Breast Cancer Res Treat. 2010;122:813–822. doi: 10.1007/s10549-009-0709-2. [DOI] [PubMed] [Google Scholar]
- 23.Hosseini M, Houshmand M, Ebrahimi A. RAD51 polymorphisms and breast cancer risk. Mol Biol Rep. 2013;40:665–668. doi: 10.1007/s11033-012-2105-y. [DOI] [PubMed] [Google Scholar]
- 24.Smolarz B, Zadrozny M, Duda-Szymanska J, Makowska M, Samulak D, Michalska MM, Mojs E, Brys M, Forma E, Romanowicz-Makowska H. RAD51 genotype and triple-negative breast cancer (TNBC) risk in Polish women. Pol J Pathol. 2013;64:39–43. doi: 10.5114/pjp.2013.34602. [DOI] [PubMed] [Google Scholar]
- 25.Wang Z, Dong H, Fu Y, Ding H. RAD51 135G>C polymorphism contributes to breast cancer susceptibility: a meta-analysis involving 26,444 subjects. Breast Cancer Res Treat. 2010;124:765–769. doi: 10.1007/s10549-010-0885-0. [DOI] [PubMed] [Google Scholar]
- 26.Sun H, Bai J, Chen F, Jin Y, Yu Y, Jin L, Fu S. RAD51 G135C polymorphism is associated with breast cancer susceptibility: a meta-analysis involving 22,399 subjects. Breast Cancer Res Treat. 2011;125:157–161. doi: 10.1007/s10549-010-0922-z. [DOI] [PubMed] [Google Scholar]
- 27.Zhou GW, Hu J, Peng XD, Li Q. RAD51 135G>C polymorphism and breast cancer risk: a meta-analysis. Breast Cancer Res Treat. 2011;125:529–535. doi: 10.1007/s10549-010-1031-8. [DOI] [PubMed] [Google Scholar]
- 28.Gao LB, Pan XM, Li LJ, Liang WB, Zhu Y, Zhang LS, Wei YG, Tang M, Zhang L. RAD51 135G/C polymorphism and breast cancer risk: a meta-analysis from 21 studies. Breast Cancer Res Treat. 2011;125:827–835. doi: 10.1007/s10549-010-0995-8. [DOI] [PubMed] [Google Scholar]
- 29.Yu KD, Yang C, Fan L, Chen AX, Shao ZM. RAD51 135G>C does not modify breast cancer risk in non-BRCA1/2 mutation carriers: evidence from a meta-analysis of 12 studies. Breast Cancer Res Treat. 2011;126:365–371. doi: 10.1007/s10549-010-0937-5. [DOI] [PubMed] [Google Scholar]
- 30.He XF, Su J, Zhang Y, Ding DP, Wang W, Liu Y. Need for clarification of data in the recent meta-analysis about RAD51 135G>C polymorphism and breast cancer risk. Breast Cancer Res Treat. 2011;129:649–651. doi: 10.1007/s10549-011-1537-8. author reply 652-643. [DOI] [PubMed] [Google Scholar]
- 31.Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127:820–826. doi: 10.7326/0003-4819-127-9-199711010-00008. [DOI] [PubMed] [Google Scholar]
- 32.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 33.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 34.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jakubowska A, Gronwald J, Menkiszak J, Gorski B, Huzarski T, Byrski T, Edler L, Lubinski J, Scott RJ, Hamann U. The RAD51 135 G>C polymorphism modifies breast cancer and ovarian cancer risk in Polish BRCA1 mutation carriers. Cancer Epidemiol Biomarkers Prev. 2007;16:270–275. doi: 10.1158/1055-9965.EPI-06-0562. [DOI] [PubMed] [Google Scholar]
- 36.Krupa R, Synowiec E, Pawlowska E, Morawiec Z, Sobczuk A, Zadrozny M, Wozniak K, Blasiak J. Polymorphism of the homologous recombination repair genes RAD51 and XRCC3 in breast cancer. Exp Mol Pathol. 2009;87:32–35. doi: 10.1016/j.yexmp.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 37.Kuschel B, Auranen A, McBride S, Novik KL, Antoniou A, Lipscombe JM, Day NE, Easton DF, Ponder BA, Pharoah PD, Dunning A. Variants in DNA double-strand break repair genes and breast cancer susceptibility. Hum Mol Genet. 2002;11:1399–1407. doi: 10.1093/hmg/11.12.1399. [DOI] [PubMed] [Google Scholar]
- 38.Levy-Lahad E, Lahad A, Eisenberg S, Dagan E, Paperna T, Kasinetz L, Catane R, Kaufman B, Beller U, Renbaum P, Gershoni-Baruch R. A single nucleotide polymorphism in the RAD51 gene modifies cancer risk in BRCA2 but not BRCA1 carriers. Proc Natl Acad Sci U S A. 2001;98:3232–3236. doi: 10.1073/pnas.051624098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Romanowicz-Makowska H, Smolarz B, Kulig A. Germline BRCA1 mutations and G/C polymorphism in the 5’-untranslated region of the RAD51 gene in Polish women with breast cancer. Pol J Pathol. 2005;56:161–165. [PubMed] [Google Scholar]
- 40.Romanowicz-Makowska H, Smolarz B, Zadrozny M, Kulig A. Analysis of RAD51 polymorphism and BRCA1 mutations in Polish women with breast cancer. Exp Oncol. 2006;28:156–159. [PubMed] [Google Scholar]
- 41.Synowiec E, Stefanska J, Morawiec Z, Blasiak J, Wozniak K. Association between DNA damage, DNA repair genes variability and clinical characteristics in breast cancer patients. Mutat Res. 2008;648:65–72. doi: 10.1016/j.mrfmmm.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 42.Wang WW, Spurdle AB, Kolachana P, Bove B, Modan B, Ebbers SM, Suthers G, Tucker MA, Kaufman DJ, Doody MM, Tarone RE, Daly M, Levavi H, Pierce H, Chetrit A, Yechezkel GH, Chenevix-Trench G, Offit K, Godwin AK, Struewing JP. A single nucleotide polymorphism in the 5’ untranslated region of RAD51 and risk of cancer among BRCA1/2 mutation carriers. Cancer Epidemiol Biomarkers Prev. 2001;10:955–960. [PubMed] [Google Scholar]
- 43.Blasiak J, Przybylowska K, Czechowska A, Zadrozny M, Pertynski T, Rykala J, Kolacinska A, Morawiec Z, Drzewoski J. Analysis of the G/C polymorphism in the 5’-untranslated region of the RAD51 gene in breast cancer. Acta Biochim Pol. 2003;50:249–253. [PubMed] [Google Scholar]
- 44.Chang TW, Wang SM, Guo YL, Tsai PC, Huang CJ, Huang W. Glutathione S-transferase polymorphisms associated with risk of breast cancer in southern Taiwan. Breast. 2006;15:754–761. doi: 10.1016/j.breast.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 45.Parshad R, Price FM, Bohr VA, Cowans KH, Zujewski JA, Sanford KK. Deficient DNA repair capacity, a predisposing factor in breast cancer. Br J Cancer. 1996;74:1–5. doi: 10.1038/bjc.1996.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen PL, Chen CF, Chen Y, Xiao J, Sharp ZD, Lee WH. The BRC repeats in BRCA2 are critical for RAD51 binding and resistance to methyl methanesulfonate treatment. Proc Natl Acad Sci U S A. 1998;95:5287–5292. doi: 10.1073/pnas.95.9.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]


