Abstract
The prognosis of gastric cancer is mainly linked to tumor recurrence. MicroRNA-34a (miR-34a) is a direct transcriptional target of p53 and links tumor suppressor function and the oncogenic pathways in some cancers. However, the role of miR-34a in predicting prognosis of gastric cancer has not been fully elucidated. In this study, we aimed to investigate the expression level of miR-34a and its prognostic value in gastric cancer. A total of 137 consecutive gastric cancer patients who underwent gastrectomy with D2 lymph node dissection were included in this study. Quantitative real-time polymerase chain reaction (qRT-PCR) was utilized to detect miR-34a expression in gastric cancer tissues and adjacent normal tissues. The results showed that the levels of miR-34a expression were significantly decreased in the tumor tissues compared with the adjacent normal tissues (P<0.001). Low miR-34a expression level was associated with lymph node involvement (P=0.004), advanced TNM stage (P=0.006), poor tumor differentiation (P=0.024), high tumor recurrence rate (P=0.008), and poor five-year survival (P<0.001). The median time to recurrence and median overall survival time were significantly shorter in patients with low miR-34a levels compared with those with high miR-34a levels (P=0.028 and P=0.021, respectively). Furthermore, when analyzed with a multivariate Cox regression model, a low miR-34a level was significantly correlated with high recurrence rate and poor overall survival. Taken together, our results suggest that downregulation of miR-34a in gastric cancer is associated with high recurrence rate and poor overall survival and that miR-34a may be served as a prognostic marker for gastric cancer.
Keywords: microRNA-34a, gastric cancer, tumor recurrence, overall survival
Introduction
Gastric cancer is the fifth leading cause of cancers and the third leading cause of cancer-related death in the world [1]. A total of 950,000 new gastric cancer cases and 723,000 deaths have occurred in 2012, accounting for 7% of the total cancer cases and 9% of total cancer-related deaths, respectively [1]. Although treatment approaches including surgery, chemotherapy, and radiotherapy have been proven to be effective in the past two decades, the prognosis of gastric cancer remains poor with the 5-year survival rate to be less than 10%, due to the fact that gastric cancer has often metastasized by the time of discovery and the fact that most people with this disease condition are elderly at diagnosis [2]. To improve the survival of gastric cancer patients, a better understanding of the underlying molecular mechanisms of gastric cancer, and their application towards helping determine the prognosis and guiding clinicians in designing personalized treatment strategies, is urgently needed.
MicroRNAs (miRNAs) are small non-coding RNA molecules (about 22 nucleotides in length), which function in RNA silencing and post-transcriptional regulation of gene expression [3,4]. Currently, more than several hundred unique mature human miRNAs are known, and many are involved in tumorigenesis, acting either as oncogenes [5] or tumor suppressors [6,7]. Aberrant expression of miRNAs has also been linked to gastric cancer, suggesting that miRNAs may serve as prognostic factors for gastric cancer [8]. The microRNA-34 (miR-34) family is consisted of miR-34a, miR-34b, and miR-34c, with miR-34a being a part of the p53 tumor suppressor network [9]. It has been demonstrated that the miR-34 family directly links tumor suppressor function and the oncogenic pathways in some cancers [10]. In p53-deficient human gastric cancer cells, restoration of functional miR-34 inhibits cell growth and induces chemosensitization and apoptosis, indicating that miR-34 may restore p53 function [11]. Previous studies have demonstrated that miR-34a expression was downregulated in gastric cancer cells and tissues [12-14]. However, the study by Osawa et al. examining miRNA expression profile from formalin-fixed, paraffin embedded gastric cancer samples showed an upregulated miR-34a level in gastric cancers compared with normal gastric tissues [8]. Furthermore, in that study, miR34a was the only independent prognostic factor for overall survival. In another study examining miRNA profile of gastric cancer, 22 miRNAs including miR-34a were significantly upregulated [15]. Therefore, the expression level of miR-34a in gastric cancer remains controversial. Thus, aiming to investigate the exact expression level of miR-34a and its prognostic value in gastric cancer, we assessed the expression of miR-34a in gastric cancer tissue samples from operative resection.
Materials and methods
Patients and tissue specimens
A total of 137 consecutive gastric cancer patients who underwent gastrectomy with D2 lymph node dissection at Yantai Yuhuangding Hospital between January 2007 and December 2019 were included in this study. None of the patients had received chemotherapy or radiotherapy prior to surgery. Fresh tissues including gastric cancer tissues and adjacent normal tissues were collected and immediately snap-frozen in liquid nitrogen after surgery and were stored at -196°C until used. Patient preoperative demographic and clinical data, including age, gender, details of pathological diagnosis, serum carcino-embryonic antigen (CEA) and carbohydrate antigen (CA 72-4) levels, follow-up period, tumor recurrence, and overall survival were collected prospectively. Patients with stage II, IIIA, or IIIB gastric cancer were given postoperative adjuvant chemotherapy every three weeks for six months (Oxaliplatin 130 mg/m2 d1 + Capecitabine 1000 mg/m2 d1-d14). The study has been conducted in accordance with the ethical standards and according to the principles of the Declaration of Helsinki and has been approved by the Institutional Review Board of Yuhuangding Hospital. Written informed consent was obtained from all of the patients.
RNA isolation and qRT-PCR
Quantitative real-time polymerase chain reaction (qRT-PCR) was utilized to detect miR-34a expression in gastric cancer tissues. Briefly, RNA was extracted using the TaqMan miRNA Isolation kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. U6 small nuclear RNA (snRNA) was used as an internal control. The reverse transcriptase (RT) reaction contained 10 ng of total RNAs, 50 nmol/l stem-loop RT primer, 1×RT buffer, 0.25 mmol/l each of deoxynucleotide triphosphate (dNTP), 3.33 U/μl MultiScribe reverse transcriptase, and 0.25 U/μl RNase Inhibitor. The 20 μl reaction volumes were incubated at 16°C for 30 min, 40°C for 30 min, and 85°C for 5 min. Real-time PCR was then performed on a StepOnePlus real-time PCR system (Applied Biosystems, Foster City, CA, USA). The following PCR parameters were used: 95°C for 30 sec, followed by 40 cycles of 95°C for 10 sec and 60°C for 30 sec. All reactions were performed in triplicate and the cycle threshold (CT) value in each reaction well was recorded. The relative quantification method was used to determine the changes in the expression of miR-34a. The change in amplification was normalized to the expression of the U6 snRNA. The fold change in expression was calculated using 2-∆∆CT, where ∆∆CT= (CTmiR-34a-CTU6)gastric cancer tissue-(CTmiR-34a-CTU6)adjacent normal tissue.
Statistical analysis
The main end points were tumor recurrence and overall survival. Tumor recurrence was defined as evidence of a new lesion in the remaining stomach or elsewhere outside the stomach and was classified as local recurrence and distant recurrence. Time to recurrence was defined as the interval between the time of surgery and the discovery of the recurrence. Overall survival was measured from the time of surgery to death. Continuous variables were presented as means ± standard deviations (SD). Categorical variables were expressed as frequencies (percentages). Point-biserial correlation coefficients were calculated to estimate the correlation between tumor recurrence and the miR-34a level. The Kaplan-Meier method was used to estimate recurrence and survival rates, and the log-rank test was used to assess the differences between groups. The Cox proportional hazards model for multivariate survival analysis was used to assess predictors related to overall survival. All statistical analyses were performed using SPSS software (SPSS 19.0, Chicago, IL, USA) and the difference was considered statistically significant when the P value was less than 0.05.
Results
One hundred and thirty-seven consecutive patients with gastric cancer treated with surgical resection were enrolled in this study. The baseline characteristics of patients are summarized in Table 1. To explore the roles of miR-34a in human gastric cancer development, we detected its expression levels in these cases of gastric cancer tissues and adjacent normal tissues. According to the qRT-PCR analysis, the levels of miR-34a expression were significantly decreased in the tumor tissues compared with the adjacent normal tissues (P<0.001, Figure 1). The median miR-34a expression level in tumor tissue was 2.44 (range 0.12-29.83). By adopting cut-off value according to the median miR-34a level, patients were sorted into two categories: 64 patients with high miR-34a levels and 73 patients with low miR-34a levels. To determine the clinical revelance of miR-34a expressionclinical relevancethe, we further assessed its correlation with patient clinicopathological factors. As shown in Table 1, low miR-34a expression level was associated with lymph node involvement (P=0.004), advanced TNM stage (P=0.006), poor tumor differentiation (P=0.024), high tumor recurrence rate (P=0.008), and poor five-year survival (P<0.001).
Table 1.
Patient characteristics according to the expression of miR-34a
| Characteristics | All patients (n=137) | High miR-34a level (n=64) | Low miR-34a level (n=73) | P-value |
|---|---|---|---|---|
| Age (years) | 58.3±12.4 | 56.3±11.9 | 59.6±14.1 | 0.538 |
| Gender | 0.681 | |||
| Male | 75 (54.7%) | 36 (56.3%) | 39 (53.4%) | |
| Female | 62 (45.3%) | 28 (43.7%) | 34 (46.6%) | |
| Tumor size (cm) | 0.097 | |||
| ≤5 | 81 (59.1%) | 39 (60.9%) | 42 (57.5%) | |
| >5 | 56 (40.9%) | 25 (39.1%) | 31 (42.5%) | |
| Depth of wall invasion | 0.065 | |||
| T1~T2 | 79 (57.7%) | 33 (51.6%) | 46 (63.0%) | |
| T3~T4 | 58 (42.3%) | 31 (48.4%) | 27 (37.0%) | |
| Lymph node involvement | 0.004 | |||
| Negative | 67 (48.9%) | 40 (62.5%) | 27 (37.0%) | |
| Positive | 70 (51.1%) | 24 (37.5%) | 46 (63.0%) | |
| Distant metastasis | 0.079 | |||
| No | 134 (97.9%) | 63(98.4%) | 71 (97.3%) | |
| Yes | 3 (2.1%) | 1 (1.6%) | 2 (2.7%) | |
| TNM stagea | 0.006 | |||
| IA~IB | 22 (16.1%) | 14 (21.9%) | 8 (11.0%) | |
| IIA~IIB | 59 (43.1%) | 27 (42.2%) | 32 (43.8%) | |
| IIIA~IIIB | 51 (37.2%) | 22 (34.4%) | 29 (39.7%) | |
| IIIC~IV | 5 (3.6%) | 1 (1.5%) | 4 (5.5%) | |
| Histologic types | 0.285 | |||
| Adenocarcinoma | 104 (75.9%) | 49 (76.6%) | 55 (75.3%) | |
| Othersb | 33 (24.1%) | 15 (23.4%) | 18 (24.7%) | |
| Differentiation | 0.024 | |||
| Well | 38 (27.7%) | 23 (35.9%) | 15 (20.6%) | |
| Moderate | 42 (30.7%) | 18 (28.1%) | 24 (32.9%) | |
| Poor | 49 (35.8%) | 20 (31.3%) | 29 (39.7%) | |
| Undifferentiate | 8 (5.8%) | 3 (4.7%) | 5 (6.8%) | |
| Adjuvant chemotherapy | 0.077 | |||
| No | 24 (17.5%) | 10 (15.6%) | 14 (19.2%) | |
| Yes | 113 (82.5%) | 54 (84.4%) | 59 (80.8%) | |
| Tumor recurrence | 0.008 | |||
| No | 77 (56.2%) | 42 (65.6%) | 35 (47.9%) | |
| Yes | 60 (43.8%) | 22 (34.4%) | 38 (52.1%) | |
| Five-year survival | <0.001 | |||
| No | 66 (48.2%) | 24 (37.5%) | 42 (57.5%) | |
| Yes | 71 (51.8%) | 40 (62.5%) | 31 (42.5%) |
TNM staging was classified according to the 7th edition of the AJCC cancer staging manual.
Other cell types included signet ring cell carcinoma (n=15), adenosquamous carcinoma (n=11), and squamous cell carcinoma (n=7).
Figure 1.
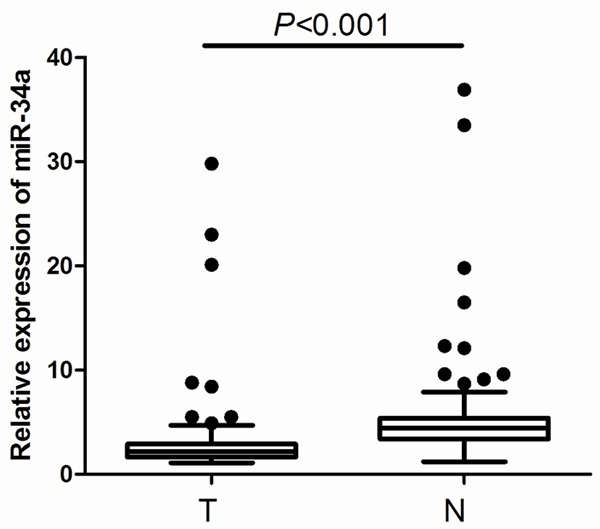
Relative expression of miR-34a in 137 pairs of gastric cancer tissues (T) and adjacent normal tissues (N).
Univariate analysis was carried out to identify those factors significantly associated with tumor recurrence. As shown in Table 2, tumor size, lymph node involvement, TNM stage, and tumor differentiation were associated with recurrence. In addition, low miR-34a level was associated with high recurrence rate, while high CEA and CA 72-4 levels were not. Multivariate analysis indicated that lymph node involvement, TNM stage, tumor differentiation, and miR-34a expression were independent prognostic factors of tumor recurrence (Table 2). In the univariate analysis of overall survival, the significant factors were lymph node involvement, TNM stage, and miR-34a expression. In the multivariate analysis including all the variables from the univariate analysis, only advanced TNM stage and low miR-34a level were independent prognostic factors of poor overall survival (Table 3).
Table 2.
Univariate and multivariate Cox analyses of clinical variables for time to recurrence
| Variables | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
|
| ||||
| HR (95% CI) | P-value | HR (95% CI) | P-value | |
| Age (>50 vs. ≤50 years) | 1.14 (0.83-1.45) | 0.547 | ||
| Gender (Male vs. Female) | 0.97 (0.78-1.21) | 0.821 | ||
| Tumor size (>5 vs. ≤5 cm) | 1.34 (1.12-1.56) | 0.003 | 1.17 (0.99-1.35) | 0.052 |
| Depth of wall invasion | 1.19 (0.98-1.43) | 0.058 | ||
| Lymph node involvement | 1.29 (1.09-1.51) | 0.017 | 1.20 (1.02-1.37) | 0.044 |
| Distant metastasis | 1.05 (0.66-1.49) | 0.732 | ||
| TNM stage | ||||
| IA~IB | 1.0 | 1.0 | ||
| IIA~IIB | 1.16 (1.08-1.25) | 0.022 | 1.09 (1.01-1.22) | 0.047 |
| IIIA~IIIB | 1.37 (1.12-1.64) | 0.006 | 1.28 (1.07-1.46) | 0.018 |
| IIIC~IV | 1.39 (1.11-1.72) | 0.004 | 1.31 (1.13-1.52) | 0.009 |
| Differentiation | ||||
| Well/Moderate | 1.0 | 1.0 | ||
| Poor/Undifferentiate | 1.35 (1.08-1.56) | 0.009 | 1.28 (1.04-1.56) | 0.016 |
| Adjuvant chemotherapy (No vs. Yes) | 1.09 (0.80-1.44) | 0.652 | ||
| Serum CEA level (high vs. low) | 1.14 (0.92-1.36) | 0.216 | ||
| Serum CA 72-4 level (high vs. low) | 1.06 (0.85-1.29) | 0.579 | ||
| MiR-34a level (low vs. high) | 1.23 (1.11-1.34) | 0.003 | 1.17 (1.05-1.30) | 0.015 |
HR hazard ratio, CI confidence interval.
Table 3.
Univariate and multivariate Cox analyses of clinical variables for overall survival
| Variables | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
|
| ||||
| HR (95% CI) | P-value | HR (95% CI) | P-value | |
| Age (>50 vs. ≤50 years) | 1.21 (0.78-1.65) | 0.692 | ||
| Gender (Male vs. Female) | 1.01 (0.85-1.19) | 0.546 | ||
| Tumor size (>5 vs. ≤5 cm) | 1.17 (0.98-1.38) | 0.054 | ||
| Depth of wall invasion | 1.26 (0.93-1.57) | 0.069 | ||
| Lymph node involvement | 1.33 (1.18-1.51) | 0.022 | 1.19 (1.00-1.40) | 0.050 |
| Distant metastasis | 1.07 (0.76-1.38) | 0.439 | ||
| TNM stage | ||||
| IA~IB | 1.0 | 1.0 | ||
| IIA~IIB | 1.28 (1.14-1.45) | 0.008 | 1.19 (1.10-1.28) | 0.010 |
| IIIA~IIIB | 1.44 (1.20-1.71) | 0.002 | 1.35 (1.16-1.65) | 0.007 |
| IIIC~IV | 1.42 (1.09-1.68) | 0.014 | 1.29 (1.05-1.54) | 0.019 |
| Differentiation | ||||
| Well/Moderate | 1.0 | 1.0 | ||
| Poor/Undifferentiate | 1.29 (1.05-1.54) | 0.023 | 1.18 (0.99-1.44) | 0.053 |
| Adjuvant chemotherapy (No vs. Yes) | 1.02 (0.77-1.41) | 0.548 | ||
| Serum CEA level (high vs. low) | 1.23 (0.97-1.47) | 0.068 | ||
| Serum CA 72-4 level (high vs. low) | 1.12 (0.89-1.38) | 0.431 | ||
| MiR-34a level (low vs. high) | 1.46 (1.23-1.70) | <0.001 | 1.33 (1.14-1.61) | 0.008 |
HR hazard ratio, CI confidence interval.
The Kaplan-Meier curves for the effect of miR-34a expression on tumor recurrence and overall survival are shown in Figure 2. The median times to recurrence were 41.3 months and 29.7 months for the patients with high miR-34a levels and those with low miR-34a levels, respectively, and there was a significant difference between the two groups (P=0.028; Figure 2A). The median survival times were 56.9 months and 37.4 months for the patients with high miR-34a levels and those with low miR-34a levels, respectively, and there was also a significant difference between the two groups (P=0.021; Figure 2B).
Figure 2.
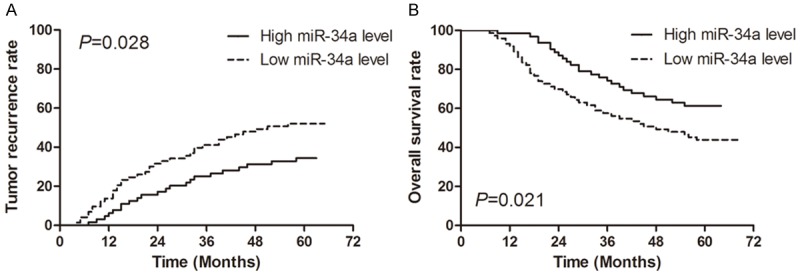
Kaplan-Meier estimates of tumor recurrence (A) and overall survival (B) for low miR-34a expression level versus high miR-34a expression level in patients with gastric cancer.
Discussion
MiR-34a is a direct proapoptotic transcriptional target of p53 that can mediate some of p53’s biological effects [16]. It is commonly deleted in various types of cancers, such as breast cancer, lung cancer, pancreatic cancer, and bladder cancer [17]. The downregulated expression of miR-34a genes may occur due to mutations that inactivate p53 in tumor cells [18]. In previous studies [8,12-15], there was a dispute about the expression level of miR-34a in gastric cancer. Therefore, we conducted this study to investigate the exact expression level of miR-34a. We found that the expression level of miR-34a was significantly downregulated in gastric cancer. We also demonstrate herein that miR-34a has an independent predictive value of tumor recurrence and overall survival of gastric cancer. These results could improve decision-making in the gastric cancer therapeutic area, as stratification according to miR-34a expression level would allow a better selection of patients for adjuvant therapy after tumor resection, help determine the prognosis, and guide clinicians in designing personalized treatment strategies.
In our current study, we found that miR-34a expression level was significantly associated with lymph node involvement, TNM stage, tumor differentiation, tumor recurrence, and five-year survival. This is in line with previous reports [19], indicating that miR-34a expression may be complementary to other clinical relevant prognostic indicators in patients with gastric cancer. A previous study has also found that miR-34a overexpression could improve the sensitivity of gastric cancer cells against cisplatin-based chemotherapies [13]. However, we didn’t identify any significant correlation between miR-34a expression level and adjuvant chemotherapy. This may be due to the different chemotherapy regimens used and the fact that the patients in our study were given chemotherapy after tumor resection. In addition, low miR-34a level predicted high tumor recurrence rate and poor overall survival in patients with gastric cancer. As miR-34a expression level is measured by qRT-PCR using tumor tissue specimens, it may be widely used in clinical practice and may prove beneficial in prognostic stratification of patients with gastric cancer in clinical trials and improve the outcomes of patients with gastric cancer.
The already identified targets of miR-34a include Bcl-2, E2F3, Met, SATB2, CD44, AXIN2, and PDGFR [20]. Besides, miR-34a can inhibit breast cancer proliferation by targeting LMTK3 (lemur tyrosine kinase 3), inhibit colonic cancer cell proliferation by targeting PAR2 (proteinase activated receptor 2), inhibit pancreatic cancer cell proliferation by targeting Notch1 [21-23]. In gastric cancer, miR-34a can inhibit gastric cancer tumourigenesis by targeting PDGFR and Met through the PI3K/Akt pathway [13]. In addition, Cao et al. has found miR-34a can negatively regulate the expression of the survivin protein and inhibit gastric caner cell proliferation and invasion [12]. Furthermore, a recent study has also found that miR-34a-YY1 (Yin Yang 1) axis plays an important role in the control of gastric carcinogenesis [9]. These results indicate that the effect of miR-34a on the development and progression of gastric caner may be based on multiple targets and pathways. Figuring out the exact mechanisms of miR-34a may contribute to the development of new therapeutic strategies for gastric cancer.
In conclusion, we have reported the differential expression of miR-34a in gastric cancer and adjacent normal tissues. Our results suggest that downregulation of miR-34a in gastric cancer is associated with high recurrence rate and poor overall survival and that miR-34a may serve as a prognostic marker for gastric cancer.
Acknowledgements
We thank the LetPub language editing company for their excellant language editing work for this manuscript.
Disclosure of conflict of interest
None.
References
- 1.World Health Organization: Globocan 2012 Stomach Cancer: Estimated Incidence, Mortality and Prevalence Worldwide in 2012. http://globocan.iarc.fr/old/FactSheets/cancers/stomach-new.asp. Accessed October 14, 2014.
- 2.Orditura M, Galizia G, Sforza V, Gambardella V, Fabozzi A, Laterza MM, Andreozzi F, Ventriglia J, Savastano B, Mabilia A, Lieto E, Ciardiello F, De Vita F. Treatment of gastric cancer. World J Gastroenterol. 2014;20:1635–1649. doi: 10.3748/wjg.v20.i7.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Yu SL, Chen HY, Chang GC, Chen CY, Chen HW, Singh S, Cheng CL, Yu CJ, Lee YC, Chen HS, Su TJ, Chiang CC, Li HN, Hong QS, Su HY, Chen CC, Chen WJ, Liu CC, Chan WK, Chen WJ, Li KC, Chen JJ, Yang PC. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13:48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 6.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A, Zlotorynski E, Yabuta N, De Vita G, Nojima H, Looijenga LH, Agami R. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 8.Osawa S, Shimada Y, Sekine S, Okumura T, Nagata T, Fukuoka J, Tsukada K. MicroRNA profiling of gastric cancer patients from formalin-fixed paraffin-embedded samples. Oncol Lett. 2011;2:613–619. doi: 10.3892/ol.2011.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang AM, Huang TT, Hsu KW, Huang KH, Fang WL, Yang MH, Lo SS, Chi CW, Lin JJ, Yeh TS. Yin Yang 1 is a target of microRNA-34 family and contributes to gastric carcinogenesis. Oncotarget. 2014;5:5002–5016. doi: 10.18632/oncotarget.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cha YH, Kim NH, Park C, Lee I, Kim HS, Yook JI. MiRNA-34 intrinsically links p53 tumor suppressor and Wnt signaling. Cell Cycle. 2012;11:1273–1281. doi: 10.4161/cc.19618. [DOI] [PubMed] [Google Scholar]
- 11.Ji Q, Hao X, Meng Y, Zhang M, Desano J, Fan D, Xu L. Restoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumorspheres. BMC Cancer. 2008;8:266. doi: 10.1186/1471-2407-8-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao W, Fan R, Wang L, Cheng S, Li H, Jiang J, Geng M, Jin Y, Wu Y. Expression and regulatory function of miRNA-34a in targeting survivin in gastric cancer cells. Tumour Biol. 2013;34:963–971. doi: 10.1007/s13277-012-0632-8. [DOI] [PubMed] [Google Scholar]
- 13.Cao W, Yang W, Fan R, Li H, Jiang J, Geng M, Jin Y, Wu Y. miR-34a regulates cisplatin-induce gastric cancer cell death by modulating PI3K/AKT/survivin pathway. Tumour Biol. 2014;35:1287–1295. doi: 10.1007/s13277-013-1171-7. [DOI] [PubMed] [Google Scholar]
- 14.Peng Y, Guo JJ, Liu YM, Wu XL. MicroRNA-34A inhibits the growth, invasion and metastasis of gastric cancer by targeting PDGFR and MET expression. Biosci Rep. 2014;34:e00112. doi: 10.1042/BSR20140020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Yao Y, Suo AL, Li ZF, Liu LY, Tian T, Ni L, Zhang WG, Nan KJ, Song TS, Huang C. MicroRNA profiling of human gastric cancer. Mol Med Rep. 2009;2:963–970. doi: 10.3892/mmr_00000199. [DOI] [PubMed] [Google Scholar]
- 16.Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 17.Bader AG. miR-34 - a microRNA replacement therapy is headed to the clinic. Front Genet. 2012;3:120. doi: 10.3389/fgene.2012.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balca-Silva J, Sousa Neves S, Goncalves AC, Abrantes AM, Casalta-Lopes J, Botelho MF, Sarmento-Ribeiro AB, Silva HC. Effect of miR-34b overexpression on the radiosensitivity of non-small cell lung cancer cell lines. Anticancer Res. 2012;32:1603–1609. [PubMed] [Google Scholar]
- 19.Stanitz E, Juhasz K, Toth C, Gombos K, Natali PG, Ember I. Evaluation of MicroRNA expression pattern of gastric adenocarcinoma associated with socioeconomic, environmental and lifestyle factors in northwestern Hungary. Anticancer Res. 2013;33:3195–3200. [PubMed] [Google Scholar]
- 20.Agostini M, Knight RA. miR-34: from bench to bedside. Oncotarget. 2014;5:872–881. doi: 10.18632/oncotarget.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao G, Guo J, Li D, Jia C, Yin W, Sun R, Lv Z, Cong X. MicroRNA-34a suppresses cell proliferation by targeting LMTK3 in human breast cancer mcf-7 cell line. DNA Cell Biol. 2013;32:699–707. doi: 10.1089/dna.2013.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma Y, Bao-Han W, Lv X, Su Y, Zhao X, Yin Y, Zhang X, Zhou Z, MacNaughton WK, Wang H. MicroRNA-34a mediates the autocrine signaling of PAR2-activating proteinase and its role in colonic cancer cell proliferation. PLoS One. 2013;8:e72383. doi: 10.1371/journal.pone.0072383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia J, Duan Q, Ahmad A, Bao B, Banerjee S, Shi Y, Ma J, Geng J, Chen Z, Rahman KM, Miele L, Sarkar FH, Wang Z. Genistein inhibits cell growth and induces apoptosis through up-regulation of miR-34a in pancreatic cancer cells. Curr Drug Targets. 2012;13:1750–1756. doi: 10.2174/138945012804545597. [DOI] [PubMed] [Google Scholar]


