Abstract
The genes RAD51 and XRCC2 encode proteins that are important for the repair of double-strand DNA breaks by recombination. Therefore, genetic variability in these genes may contribute to the occurrence and progression of carcinoma. We investigated the association of polymorphisms in the DNA repair genes XRCC2-A/G and RAD51-135G/C with the colorectal cancer risk. Genotypes were determined by PCR-RFLP assays in 71 patients with colorectal cancer and 86 age-matched healthy controls. After amplification, we used a restriction enzyme (RAD51; MvaI and XRCC2; HphI) and digested the PCR product. Then, this DNA fragments were passed through gel electrophoresis. By examining these images, we identified changes in the nucleotides in these specific regions. To clarify fragments polymorphisms, the PCR products were sequenced with an Applied Biosystems Automated Sequencer. We observed the Arg188His polymorphism of XRCC2 genes in 42.2%, as shown in 30 of the 71 cancer patients. Only 21 out of 86 controls showed this polymorphism (24.2%). We also observed that 21 of the 71 patients (29.5%) carried the RAD51135G/C polymorphism of this gene. The same polymorphism was observed in 11 of the 86 controls (12.7 %; p < 0.05). The obtained results indicate that the polymorphism of RAD51 and XRCC2 genes may be associated with the incidence of colon cancer in the Turkish population. Further studies, including those on a larger group of patients, are required to further clarify this point.
Keywords: Colon cancer, RAD51, XRCC2 genes, polymorphism, Turkey
Introduction
Colorectal cancer (CRC) is a common and lethal disease affecting individuals worldwide. According to the World Health Organization GLOBAN database CRC is the third most common cancer in men and the second in women worldwide [1,2]. Likewise, according to the Turkish Ministry of Health cancer registry data, CRC was the third most common cancer in women and the fourth in men in Turkey between years 2007 and 2008 [3]. The incidence of CRC is approximately 7 per 100,000, with approximately 5,000 new cases and 3,200 deaths annually [4].
Environmental, genetic, and epigenetic factors are the related risk factors for developing CRC [5]. Nowadays, an intense effort has been made to discover the genetic pathways and molecular mechanisms of CRC formation and progression. Thus, it is now generally accepted that CRC tumorigenesis is a multistep process from normal colonic epithelium to cancer following adenoma-carcinoma sequence, involving both inactivation of tumor-suppressor genes and/or activation of oncogenes [6,7].
Another research area is DNA repair mechanisms, which work to prevent the unintended changes during the replication of DNA that might cause malformations or cancer development. DNA repair genes and the enzyme products of these genes decrease cell death rate, mutations occurring in DNA, replication errors, and genomic instability. In this context, any change in these genes and products may cause the distortion of cell death and thus a formation of cancer [8].
Previous research on DNA repair genes showed that the X-ray repair cross complementing (XRCC) 2 and RAD51 genes might have a vital importance in DNA repair and chromosome arrangements and the lack of appropriate processes of these repairing genes can cause cancer formation including colorectal cancer [9,10].
In this study, we have investigated the polymorphism variations in DNA repair genes XRCC2 and RAD51 in patients suffering from colon cancer and in healthy control group.
Material and methods
Patients
Histologically proven 71 colon cancer patients (35 females and 36 males) were included in this study. All patients had histologically confirmed invasive adenocarcinoma of the colon. None of them were associated with an inherited cancer syndrome. The histopathological reports were reviewed retrospectively from the hospital medical records. Eighty-six volunteer healthy individuals (45 females and 41 males) were selected as the control group.
For genotyping, 10-ml blood samples were taken from each patient and the control group into tubes with EDTA. All of the genomic analyses were conducted in the laboratory of molecular genetics in the Department of Bioen-gineering, Kafkas University.
Determination of the XRCC2 and RAD51 Genotypes
DNA was extracted using commercially available Invitrogen™ genomic DNA extraction mini kits (CS11010, London, UK) and purified using DNA purification kit, according to the manufacturer’s instructions. The isolated DNA samples were measured using the Nanodrop Spectrophotometer (Thermo ND1000) and kept at -20°C. RAD51 and XRCC2 gene polymorphisms, primers, PCR product sizes, restriction enzymes, and annealing temperatures are given in Table 1 [10]. PCR protocol for a final volume of 25 μl, including 2.5-μl 10× Taq polymerase buffer solution, 2-μl magnesium chloride (2 mM), 2-μl dNTP mix (0.2 mM), 1-μl forward primer (10 pmol), 1-μl reverse primer (10 pmol), 2-μl genomic DNA (100 ng/μl), 0.5-μl DNA taq polymerase enzyme (5 u/μl), and 14-μl distilled water, was followed [11]. PCR conditions were as follows: an initial denaturation at 94°C for 4 min, then 35 cycles at 94°C for 30 s, at 60°C for 30 s, at 72°C for 40 s, and a final extension cycle at 72°C for 7 min. The PCR products were detected by agarose gel electrophoresis (at 90 V, 300 A for 1.5 h) on 2% agarose gel containing ethidium bromide, and the fluorescence intensity of each band was evaluated using a UV transilluminator (Gel Logic Pro 2200, Montreal, Canada). The PCR amplification bands were observed as 290 bp and 157 bp for XRCC2 (Arg188His) polymorphism and RAD51 polymorphism, respectively. Amplified products were digested: XRCC2 (Arg188His) with 3U Haemophilus parahaemolyticus (HphI) and RAD51 with 3 U Micrococcus varians (MvaI) (New England Biolabs, INC UK). Digestion products were visualized, and the resulting fragments were separated on 2.5% agarose gels and with ethidium bromide staining under ultraviolet illumination (Gel Logic Pro 2200, Canada). The single amplicon of 290 bp as a result of the section of the XRCC2 (Arg188His) polymorphism with the restriction enzyme was separated into two DNA fragments of 148 bp and 142 bp. In the evaluation, XRCC2 (Arg188His) was analyzed as having the HphI enzyme (290-bp bands Arg/Arg [homozygote wild tip] genotype; 148-bp + 142-bp bands as having the Arg/His [heterozygote] genotype; 290-bp + 148-bp + 142-bp bands as having the His/His [homozygote mutant genotype]). As a result of the section of RAD51 with MvaI enzyme, the 157-bp bands were observed as having 86 bp + 71 bp. The PCR products were then isolated using agarose gel electrophoresis (Bigdye Cycle Sequencing kit v.3.1, Applied Biosystems). Approximately 5 ul of PCR products were sequenced with an Applied Biosystems Automated Sequencer (ABI 3130 XL Genetic Analyzer, Foster City, CA 94404 USA). Restriction maps and bioinformatics analysis of SNPs were performed using Vector NTI software (Life Technologies).
Table 1.
RAD51 and XRCC2 gene polymorphisms, primers, PCR product sizes, restriction enzymes, and annealing temperatures
| Genes | Primer Sequences (5’ → 3’) | Expected size of the PCR product (bp) | Restriction enzymes | Annealing temperature (°C) |
|---|---|---|---|---|
| RAD51 | F: GGGAACTGCAACTCATCTGG | 157 | MvaI | 62.5 |
| R: GCGCTCCTCTCTCCAGCAG | ||||
| XRCC2 | F: TGTAGTCACCCATCTCTCTGC | 290 | HphI | 61 |
| R: AGTTGCTGCCATGCCTTACA |
F: Forward, R: Reverse.
Statistical analysis
For each polymorphism, deviation of the genotype frequencies in the controls from those expected under Hardy-Weinberg equilibrium was assessed using the standard χ2 test. Genotypic frequencies in cases and controls were compared by χ2 tests. The genotypic specific risks were estimated as odds ratios (ORs) with associated 95% confidence intervals (CIs) by unconditional logistic regression. P-values < 0.05 were considered to be significant (GraphPad Prism software package [v. 6; GraphPad Software Inc, La Jolla, CA, USA] or SPSS 16.0 software [SPSS, Chicago, IL, USA]).
Results
As a result of the genomic analysis, RAD51 gene polymorphic changes were seen in 21 of 71 patients (29.5%) in the patient group, whereas they were observed in 11 of 86 individuals (12.7%) in the control group. The RAD51 gene polymorphic changes were two-fold in the patient group compared with the control group; and the difference was statistically significant (p < 0.05).
According to the genomic analyses for XRCC2 gene, the polymorphic changes were seen in 30 of 71 patients (42.2%) in the patient group and in 11 of 86 individuals (12.7%) in the control group. XRCC2 gene polymorphic changes were also increased in the patient group and this increase was found to be statistically significant between the two groups (p < 0.05).
Demographical data of the patient and the control group are given in Tables 2 and 3, respectively. There was no statistically difference between the two groups in terms of the demographic data. The findings of the genomic analysis for RAD51 and XRCC2 genes in the patient and control groups are given in Table 4. Restriction sites for restriction enzyme (MvaI) in RAD51 gene sequences are shown in Figure 1. SNP could not be analyzed due to the small sample size of sequenced RAD51 gene. SNP regions were detected in XRRC2 gene sequences and are shown in Figure 2.
Table 2.
Demographical data of the colon cancer patients
| Ages | Patients | Females (n = 35) | % | Males (n = 36) | % | SD |
|---|---|---|---|---|---|---|
| 25-40 | 8 | 3 | 0.38 | 5 | 0.63 | 1.41 |
| 41-60 | 31 | 15 | 0.48 | 16 | 0.52 | 0.707 |
| 61-82 | 32 | 17 | 0.53 | 15 | 0.47 | 1.41 |
| Total | 71 | 35 | 0.49 | 36 | 0.51 | 0.707 |
Table 3.
Demographical data of the controls
| Ages | Controls | Females (n = 45) | % | Males (n = 41) | % | SD |
|---|---|---|---|---|---|---|
| 25-40 | 19 | 10 | 0.53 | 9 | 0.47 | 0.707 |
| 41-60 | 46 | 24 | 0.52 | 22 | 0.48 | 1.41 |
| 61-72 | 21 | 11 | 0.53 | 10 | 0.47 | 0.707 |
| Total | 86 | 45 | 0.53 | 41 | 0.47 | 2.82 |
Table 4.
Genotypic and allelic results of RAD51 and XRCC2 genes polymorphisms in the patients and controls
| RAD51 | Colon Cancer (n = 71) | Control Group (n = 86) | ||||||||||||
|
| ||||||||||||||
| Genotypes | GG | % | CC | % | GC | % | P | GG | % | CC | % | GC | % | P |
|
| ||||||||||||||
| Frequencies of Genotypes | 39 | 0.55 | 11 | 0.15 | 21 | 0.30 | 1.26 | 21 | 0.24 | 54 | 0.63 | 11 | 0.13 | 0.0053 |
| Frequencies of Alleles | G 0.37 | C 0.63 | G 0.25 | C 0.75 | ||||||||||
|
| ||||||||||||||
| XRCC2 | ||||||||||||||
|
| ||||||||||||||
| Genotypes | GG | % | AA | % | GA | % | P | GG | % | AA | % | GA | % | P |
|
| ||||||||||||||
| Frequencies of Genotypes | 32 | 0.45 | 9 | 0.13 | 30 | 0.42 | 0.90 | 54 | 0.63 | 11 | 0.13 | 21 | 0.24 | 0.0053 |
| Frequencies of Alleles | G 0.66 | A 0.34 | G 0.75 | A 0.25 | ||||||||||
XRCC2 Gene: GG(Arg/Arg); AA(His/His); GA(Arg188His).
Figure 1.
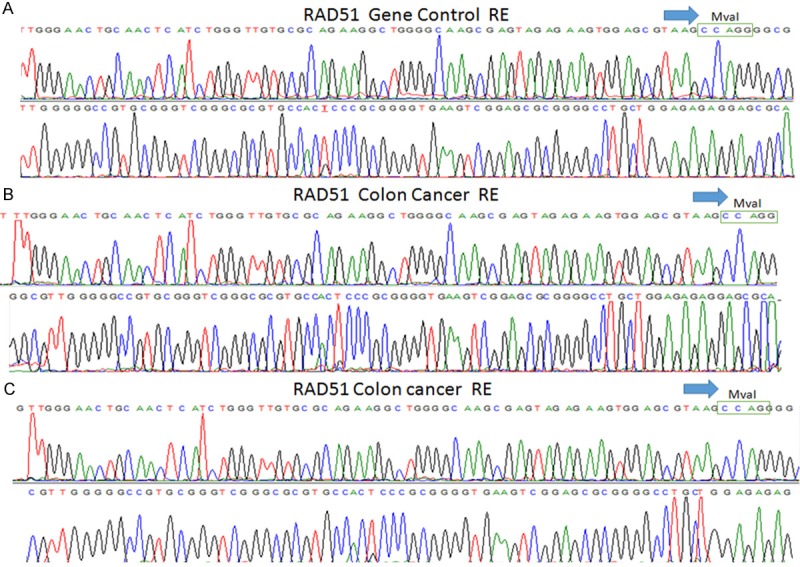
RAD51 gene sequencing results. A. Restriction sites for restriction enzyme (MvaI) in control subjects. B and C. Restriction sites for restriction enzyme (MvaI) in colon cancer patients.
Figure 2.
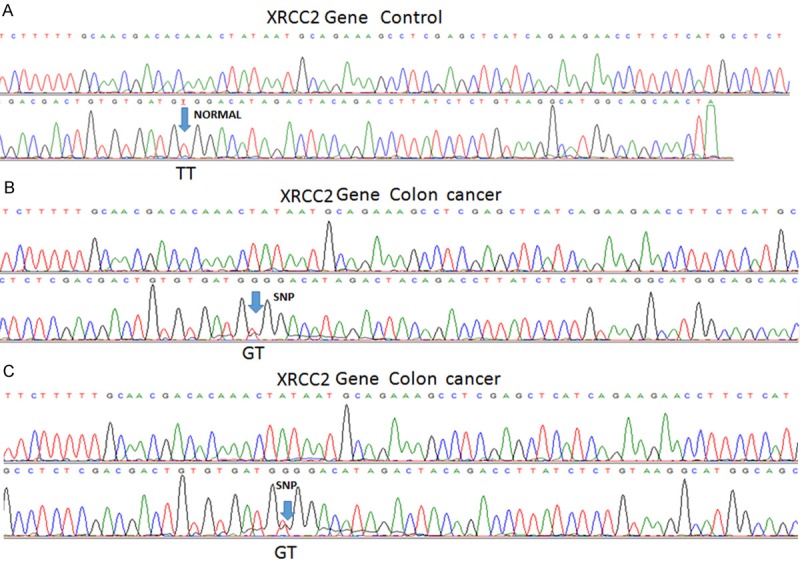
XRCC2 gene sequencing results. A. In control subjects the SNP region is shown as normal (TT). B and C. Sequence results for SNP site difference (GT) in colon cancer patients.
Discussion
Colon cancer is a widely seen and life threatening disease worldwide. Despite improvements in different kinds of treatment modalities, early diagnosis and treatment are the key points in both better prognosis and long survival.
In the present study, we investigated the differences in the polymorphism rates of XRCC2 and RAD51 genes in colon cancer patients in Turkey. To the best of our knowledge, this study is the first to investigate these genes in colon cancer development in the Turkish population.
RAD51 gene and the X-ray repair complementing defective repair in Chinese hamster cells 2 (XRCC2) genes are essential in DNA repair pathways, and functional disorders of these genes may lead to tumor development and progression [12-14].
DNA double-strand breaks (DSBs), in which the double helix of the DNA is severed, the RAD51 and XRCC2 genes participate in the DNA damage response pathway associated with the activation of homologous recombination and double-strand break repair [13,15]. Inaccurate repair of DSBs leads to genomic instability, which consequently causes chromosomal instability, mutations, apoptosis, tumor predisposition, and carcinogenesis [12,13,16,17].
Studies on the polymorphisms of XRCC2 and RAD51 genes have been conducted for various cancer types including CRC [12,18-22].
On the contrary, a reverse association as a protective effect and no association between XRCC2 polymorphism and cancer risk have been reported in several studies [23-26]. The different results of XRCC2 polymorphism association with cancer risk may depend on the ethnicity of the study groups, environmental differences (particularly smoking and alcohol consumption), and sample size or the selection bias of the studies.
In this study, we found that the frequency of the XRCC2 gene polymorphism was two-fold higher in colon cancer patients than in healthy control individuals. This finding is consistent with the results of several studies [12,27].
This study has several limitations. First, the study group was small. Second, we only investigated Turkish population; maybe comparing different ethnicities may reveal different results. Third, we could not conduct an analysis between the histopathological prognostic factors, such as tumor size, differentiation, and gene polymorphism. The strong association between XRCC2 and RAD51 gene polymorphisms and colon cancer suggest that these polymorphisms may be prognostic factors. Despite these lacks, this is the first study conducted in Turkish population that investigates the association between colon cancer and XRCC2 and RAD51 gene polymorphisms.
The strong association between RAD51 and XRCC2 gene polymorphism and colon cancer encouraged us that the investigation of these polymorphisms may be used in several areas in colon cancer management, such as early detection of the carcinogenesis, prediction of tumor biologic behavior, and being a candidate therapeutic target for colon cancer.
In short, our study provides evidence that RAD51 and XRCC2 gene polymorphisms may be risk factors for colon cancer development. These findings should be further evaluated with large-scale population-based, prospective and collaborative studies.
Conclusion
Due to the positive impact of early diagnosis in the prognosis and survival of the colon cancer, nowadays new investigations have been focused on this topic. As a result of our study RAD51 and XRCC2 genes polymorphisms increase the risk of colon cancer and might be an early diagnostic marker for colon cancer.
Acknowledgements
We are grateful to the Kafkas University Scientific Research Project Unit (Kars, Turkey, Grant No: MMF: 2011-60) for financially supporting this study.
Disclosure of conflict of interest
The authors declare that there are no conflicts of interest.
References
- 1.Bray F, Ren JS, Masuyer E, Ferlay J. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer. 2013;132:1133–1145. doi: 10.1002/ijc.27711. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F, editors. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 [Internet] 09.10.2014; Available from: http://globocan.iarc.fr.
- 3.Department of Cancer, P.H.I.o.T.T.M.o.H., Turkey. 09.10.2014; Available from: http://kanser.gov.tr/Dosya/ca_istatistik/2009kanseraporu.pdf.
- 4.Tatar M, Tatar F. Colorectal cancer in Turkey: current situation and challenges for the future. Eur J Health Econ. 2010;10(Suppl 1):S99–105. doi: 10.1007/s10198-009-0197-7. [DOI] [PubMed] [Google Scholar]
- 5.Chan AT, Giovannucci EL. Primary prevention of colorectal cancer. Gastroenterology. 2010;138:2029–2043. e2010. doi: 10.1053/j.gastro.2010.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zoratto F, Rossi L, Verrico M, Papa A, Basso E, Zullo A, Tomao L, Romiti A, Lo Russo G, Tomao S. Focus on genetic and epigenetic events of colorectal cancer pathogenesis: implications for molecular diagnosis. Tumour Biol. 2014;35:6195–6206. doi: 10.1007/s13277-014-1845-9. [DOI] [PubMed] [Google Scholar]
- 7.Armaghany T, Wilson JD, Chu Q, Mills G. Genetic alterations in colorectal cancer. Gastrointest Cancer Res. 2012;5:19–27. [PMC free article] [PubMed] [Google Scholar]
- 8.Gok I, Baday M, Cetinkunar S, Kilic K, Bilgin BC. Polymorphism in DNA repair genes XRCC2 and XRCC3 risk of gastric cancer in Turkey. Bosn J Basic Med Sci. 2014;14:214–218. doi: 10.17305/bjbms.2014.4.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li XB, Luo H, Huang J, Zhang JD, Yang ZX, Sun XW. XRCC2 gene polymorphisms and its protein are associated with colorectal cancer susceptibility in Chinese Han population. Med Oncol. 2014;31:245. doi: 10.1007/s12032-014-0245-8. [DOI] [PubMed] [Google Scholar]
- 10.Krupa R, Sliwinski T, Wisniewska-Jarosinska M, Chojnacki J, Wasylecka M, Dziki L, Morawiec J, Blasiak J. Polymorphisms in RAD51, XRCC2 and XRCC3 genes of the homologous recombination repair in colorectal cancer--a case control study. Mol Biol Rep. 2011;38:2849–2854. doi: 10.1007/s11033-010-0430-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sliwinski T, Krupa R, Majsterek I, Rykala J, Kolacinska A, Morawiec Z, Drzewoski J, Zadrozny M, Blasiak J. Polymorphisms of the BRCA2 and RAD51 genes in breast cancer. Breast Cancer Res Treat. 2005;94:105–109. doi: 10.1007/s10549-005-0672-5. [DOI] [PubMed] [Google Scholar]
- 12.Curtin K, Lin WY, George R, Katory M, Shorto J, Cannon-Albright LA, Smith G, Bishop DT, Cox A, Camp NJ. Genetic variants in XRCC2: new insights into colorectal cancer tumorigenesis. Cancer Epidemiol Biomarkers Prev. 2009;18:2476–2484. doi: 10.1158/1055-9965.EPI-09-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffin CS, Simpson PJ, Wilson CR, Thacker J. Mammalian recombination-repair genes XRCC2 and XRCC3 promote correct chromosome segregation. Nat Cell Biol. 2000;2:757–761. doi: 10.1038/35036399. [DOI] [PubMed] [Google Scholar]
- 14.Thacker J, Zdzienicka MZ. The XRCC genes: expanding roles in DNA double-strand break repair. DNA Repair (Amst) 2004;3:1081–1090. doi: 10.1016/j.dnarep.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 15.Johnson RD, Liu N, Jasin M. Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature. 1999;401:397–399. doi: 10.1038/43932. [DOI] [PubMed] [Google Scholar]
- 16.Liu N, Lamerdin JE, Tebbs RS, Schild D, Tucker JD, Shen MR, Brookman KW, Siciliano MJ, Walter CA, Fan W, Narayana LS, Zhou ZQ, Adamson AW, Sorensen KJ, Chen DJ, Jones NJ, Thompson LH. XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol Cell. 1998;1:783–793. doi: 10.1016/s1097-2765(00)80078-7. [DOI] [PubMed] [Google Scholar]
- 17.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 18.Silva SN, Tomar M, Paulo C, Gomes BC, Azevedo AP, Teixeira V, Pina JE, Rueff J, Gaspar JF. Breast cancer risk and common single nucleotide polymorphisms in homologous recombination DNA repair pathway genes XRCC2, XRCC3, NBS1 and RAD51. Cancer Epidemiol. 2010;34:85–92. doi: 10.1016/j.canep.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 19.Jiao L, Hassan MM, Bondy ML, Wolff RA, Evans DB, Abbruzzese JL, Li D. XRCC2 and XRCC3 gene polymorphism and risk of pancreatic cancer. Am J Gastroenterol. 2008;103:360–367. doi: 10.1111/j.1572-0241.2007.01615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benhamou S, Tuimala J, Bouchardy C, Dayer P, Sarasin A, Hirvonen A. DNA repair gene XRCC2 and XRCC3 polymorphisms and susceptibility to cancers of the upper aerodigestive tract. Int J Cancer. 2004;112:901–904. doi: 10.1002/ijc.20474. [DOI] [PubMed] [Google Scholar]
- 21.Han J, Hankinson SE, Ranu H, De Vivo I, Hunter DJ. Polymorphisms in DNA double-strand break repair genes and breast cancer risk in the Nurses’ Health Study. Carcinogenesis. 2004;25:189–195. doi: 10.1093/carcin/bgh002. [DOI] [PubMed] [Google Scholar]
- 22.Vineis P, Manuguerra M, Kavvoura FK, Guarrera S, Allione A, Rosa F, Di Gregorio A, Polidoro S, Saletta F, Ioannidis JP, Matullo G. A field synopsis on low-penetrance variants in DNA repair genes and cancer susceptibility. J Natl Cancer Inst. 2009;101:24–36. doi: 10.1093/jnci/djn437. [DOI] [PubMed] [Google Scholar]
- 23.Figueroa JD, Malats N, Rothman N, Real FX, Silverman D, Kogevinas M, Chanock S, Yeager M, Welch R, Dosemeci M, Tardon A, Serra C, Carrato A, Garcia-Closas R, Castano-Vinyals G, Garcia-Closas M. Evaluation of genetic variation in the double-strand break repair pathway and bladder cancer risk. Carcinogenesis. 2007;28:1788–1793. doi: 10.1093/carcin/bgm132. [DOI] [PubMed] [Google Scholar]
- 24.Auranen A, Song H, Waterfall C, Dicioccio RA, Kuschel B, Kjaer SK, Hogdall E, Hogdall C, Stratton J, Whittemore AS, Easton DF, Ponder BA, Novik KL, Dunning AM, Gayther S, Pharoah PD. Polymorphisms in DNA repair genes and epithelial ovarian cancer risk. Int J Cancer. 2005;117:611–618. doi: 10.1002/ijc.21047. [DOI] [PubMed] [Google Scholar]
- 25.Pooley KA, Baynes C, Driver KE, Tyrer J, Azzato EM, Pharoah PD, Easton DF, Ponder BA, Dunning AM. Common single-nucleotide polymorphisms in DNA double-strand break repair genes and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2008;17:3482–3489. doi: 10.1158/1055-9965.EPI-08-0594. [DOI] [PubMed] [Google Scholar]
- 26.Moreno V, Gemignani F, Landi S, Gioia-Patricola L, Chabrier A, Blanco I, Gonzalez S, Guino E, Capella G, Canzian F. Polymorphisms in genes of nucleotide and base excision repair: risk and prognosis of colorectal cancer. Clin Cancer Res. 2006;12:2101–2108. doi: 10.1158/1078-0432.CCR-05-1363. [DOI] [PubMed] [Google Scholar]
- 27.Xu K, Chen Z, Qin C, Song X. miR-7 inhibits colorectal cancer cell proliferation and induces apoptosis by targeting XRCC2. Onco Targets Ther. 2014;7:325–332. doi: 10.2147/OTT.S59364. [DOI] [PMC free article] [PubMed] [Google Scholar]


