Abstract
MicroRNAs are increasingly important in the study of cancer because of their ability to down regulate the expression of tumor suppressors and promote tumorigenesis. Here, miR-221, which is dysregulated in various tumors, was investigated for its expression in colon cancer tissues and its correlation with patient prognosis. Colon cancer tissue samples were obtained from 182 individuals who underwent surgical resection in our hospital from June 2008 to September 2009. Real-time PCR was used to detect the expression of miR-221 in these tissues. Patient survival was determined by telephone interview, and survival curves were plotted by using the Kaplan-Meier method and compared by the Log-rank test. Statistical methods also included X2 test and Cox proportional hazard regression model. Differences in the expression of miR-221 by gender, pathology, and pathological staging were not statistically significant (P>0.05), but differences in the expression of miR-221 among age groups were statistically significant (P<0.05). A survival analysis indicated that high expression of miR-221 was closely associated with a shorter survival time (P<0.05). Further, later p-TNM (hazard ratio, HR=2.973, 95% confidence interval, CI: 1.329-6.519, P=0.003) and high expression of miR-211 (HR=2.394, 95% CI: 1.210-4.910, P=0.006) were identified as risk factors for colon cancer prognosis. Thus, high miRNA-221 expression might be a prognostic marker of colon cancer patients. The high expression of miRNA-221 was associated with poor prognosis of patients with colon cancer.
Keywords: miRNA-221, colon cancer, prognosis
Introduction
Colon cancer occurs in the digestive tract, typically at the junction of the rectum and the sigmoid colon. One of the most common malignancies in China, the overall incidence has displayed a marked upward trend in recent years [1]. The current surgical and chemo- or radio-therapy treatment approaches enable approximately 60% of colon cancer patients to survive 5 years after diagnosis, but many colon cancers remain undiagnosed until the disease has progressed to a late stage. Therefore, identifying new diagnostic and prognostic markers may enable earlier detection and better treatment of the disease.
MicroRNAs (miRNAs) have emerging importance as potential cancer biomarkers [3]. These small, non-protein-coding, single-stranded, short RNAs bind to the 3’-untranslated region of target mRNAs to regulate their expression. This regulation leads to downstream effects on diverse biological processes including early embryonic development and cell proliferation, differentiation, and apoptosis [4-6]. Further, miRNAs regulate the expression of various genes involved in tumorigenesis, including oncogenes and tumor suppressors, and tumors often display aberrant miRNA expression profiles [7-10]. miRNA-221 has demonstrated higher expression in multiple tumors including colon cancer [11,12]. It is hypothesized that upregulation of this miRNA can contribute to disease initiation or progression. To better understand the contribution of miR-221 to colon cancer, in this study, the clinical data of 182 colon cancer patients were analyzed retrospectively to investigate the correlation between patient survival time and miRNA-221 expression.
Participants and methods
Participants
Paraffin-embedded colon cancer specimens were collected and stored from 182 patients who underwent surgical resection between June 2008 and September 2009 and were confirmed by pathologists to have colon cancer. Before surgery, none of the patients received anti-cancer treatment involving chemotherapy, radiotherapy, or biologicals. The pathological classification and staging of colon cancer was in accordance with the AJCC TNM Staging System (7th edition, 2010). This study was approved by the Hospital Ethics Committee, and informed consent was obtained from all participants.
Polymerase chain reaction (PCR)
The human pre-miR-221 sequence was obtained from the miRBase (www.mirbase.org), which was 5’-AGCUACAUUGUCUG CUGGGUUUC-3’.
RNA extraction
Total RNA of paraffin-embedded colon cancer tissues was extracted according to the instructions in the Recover All Total Nucleic Acid Isolation Kit (Applied Biosystems). The extracted RNA was diluted with 50 μL of DEPC-treated water and was then stored at -80°C for use. A NanoDrop 1000 Micro-volume UV-Vis Spectrophotometer (Tripbiotech, Shanghai, China) was used to measure the purity of total RNA, i.e., the ratio of OD260/OD280.
Reverse transcription
Reverse transcription was performed strictly according to the instructions in the Real-time Reverse Transcription Kit (Invitrogen). In brief, 0.5 μg of RNA was added with the following reagents: 1 μL of total RNA, 1 μL of PrimeScript RT Enzyme Mix I, 5 mL of 5 × PrimeScript Buffer, 2 μL of stem-loop RT primer, and 20 μL of DEPC-treated water. The reaction conditions were: 42°C for 15 min, 85°C for 5 s and 4°C for 15 min. After the reaction, the mixture was stored at -80°C.
Real-time PCR
The PCR reaction system comprised a total volume of 20 μL, containing 10 μL of SYBR Green Real time PCR Master Mix (Mingyangkehua Bio Technology Co. Ltd., Beijing), 1 μL of downstream primer and 1 μL of upstream primer [RNU6B primer and has-miRNA-221 primer (Generay Biotech, Shanghai) as internal controls], 2 μL of cDNA and added DEPC-treated water. The 7300 Real-time PCR System was used under following conditions in a 9700 Thermal Cycler: denaturation at 95°C for 10 min, 95°C for 15 s and 1min for 60°C, for 40 cycles. The process above was repeated three times.
Follow-up
To determine patient survival time, telephone follow-up was performed for all patients from whom a paraffin-embedded tissue sample was used. The deadline for completing the interviews was Dec. 2013, and the median follow-up time was 30 months (ranging from 5 to 6 months).
Statistical analysis
Relative expression of miR-221 was determined using the 2-ΔΔCT method [13]. The median of 2-ΔΔCT was used as the demarcation value to divide all patients into groups with highly-expressed miRNA-221 or lowly-expressed miRNA-221. Statistical methods included a chi-squared test, Kaplan-Meier method, Log-rank test, and Cox proportional hazards model. P<0.05 was considered to indicate the difference was statistically significant.
Results
miR-221 expression is associated with clinicopathological features in patients with colon cancer
Of the 182 colon cancer patients, 73 were male and 109 were female (mean age, 52 ± 6.25 years). Pathological assessment indicated that112 cases had adenocarcinoma, and 70 cases had squamous cell carcinoma; additionally, 67 cases had stage I cancer, and 115 cases had stage II-IIIa.
miR-221 expression was detected in paraffin-embedded tissues by qRT-PCR. Patients were then stratified by the level of relative expression (high or low) of miR-221 in their colon cancer tissue samples (Table 1). Differences in miR-221 expression were not statistically significant by sex, pathological type, or tumor staging. In contrast, differences in miR-221 expression were statistically significant by patient age (P<0.05).
Table 1.
Relation between miRNA-221 expression and clinical characteristics in patients with colon cancer
| Variables | miRNA-221 expression* | X2 | P | |
|---|---|---|---|---|
|
| ||||
| Low [n (%)] | High [n (%)] | |||
| Gender | 0.111 | 0.740 | ||
| Male (73) | 35 (47.95) | 38 (52.05) | ||
| Female (109) | 55 (50.46) | 54 (49.54) | ||
| Age (years) | 8.423 | 0.004 | ||
| <45 (81) | 29 (35.80) | 52 (64.20) | ||
| ≥45 (101) | 58 (57.43) | 43 (42.57) | ||
| Histology | 0.712 | 0.399 | ||
| SCC (70) | 37 (52.86) | 33 (47.14) | ||
| ADC (112) | 52 (46.43) | 60 (53.57) | ||
| Stage -TNM | 0.145 | 0.704 | ||
| I (67) | 34 (50.75) | 33 (49.25) | ||
| II-IIIa (115) | 55 (47.83) | 60 (52.17) | ||
Relative expression of miR-221 was determined using the 2-ΔΔCT method, and the median of 2-ΔΔCT was used to divide all patients into groups based on high expression of miRNA-221 or low expression of miRNA-221.
SCC = squamous cell carcinoma; ADC = adenocarcinoma.
miR-221 expression is associated with the survival time of colon cancer patients
A Kaplan-Meier survival analysis was performed on the 182 colon cancer patients with stratification by miR-221 expression. In the group with highly-expressed miR-211, the median survival time of colon cancer patients was 39.6 months; in the group with low miR-211 expression had a median survival of 53.9 months (Figure 1). The difference between these groups was statistically significant (Log-rank test: P=0.004). A Cox proportional hazards model for single factor analysis showed that both late (II-IIIa) TNM staging for colon cancer [hazard ratio (HR)=3.014, 95% confidence interval (CI): 1.389-6.421, P=0.002)] and the high expression of miR-211 (HR=2.185, 95% CI: 1.108-4.430, P=0.009) were correlated with poorer prognosis of colon cancer patients (Table 2). Similarly, a multivariate Cox regression analysis showed that late TNM staging (HR=2.973, 95% CI: 1.329-6.519, P=0.003) and the high expression of miR-211 (HR=2.394, 95% CI: 1.210-4.910, P=0.006) were risk factors for the prognosis of colon cancer patients.
Figure 1.
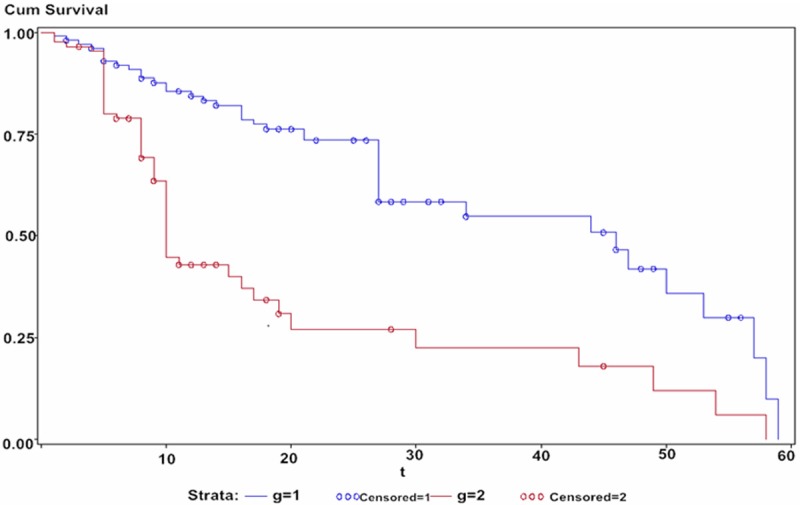
Survival analysis of colon cancer patients based on miRNA-221 expression. Note: g=1: Low expression; Censored =1: Low expression-censored; g=2: High expression; Censored=2: High expression-censored.
Table 2.
Univariate and multivariate analyses of overall survival (OS) of colon cancer patients
| variable | Univariate analysis | Mutivariate analysis | ||
|---|---|---|---|---|
|
| ||||
| HR (95% CI) | P | HR (95% CI) | P | |
| Age | ||||
| <45 | 1 | 1 | 1 | 1 |
| ≥45 | 1.164 (0.435-2.011) | 0.620 | 1.219 (0.397-2.103) | 0.607 |
| Gender | ||||
| Male | 1 | 1 | 1 | 1 |
| Female | 0.852 (0.283-1.726) | 0.538 | 0.981 (0.374-2.206) | 0.841 |
| Histology | ||||
| SCC | 1 | 1 | 1 | 1 |
| ADC | 0.895 (0.399-1.507) | 0.611 | 0.742 (0.320-1.497) | 0.540 |
| p-TNM stage | ||||
| I | 1 | 1 | 1 | 1 |
| II-IIIa | 3.014 (1.389-6.421) | 0.002 | 2.973 (1.329-6.519) | 0.003 |
| MiR221 expression | ||||
| Low | ||||
| High | 2.185 (1.108-4.430) | 0.009 | 2.394 (1.210-4.910) | 0.006 |
Discussion
With the third highest cancer incidence, colon cancer poses a significant health threat [14]. The investigation of miRNAs as contributors to colon cancer pathogenesis offers the potential to uncover new therapeutic targets [15,16]. miR-221 plays a role similar to that of oncogenes in tumor initiation and progression, and is correlated with multiple cancers including ovarian, breast, lung, and papillary thyroid cancer [17-20]. In tumor cells, miR-221 tends to be highly expressed, and overexpressed miR-221 can target tumor suppressor genes such as P57, BIM, PUMA, TIMP3, and PTEN. Inhibition of these target genes thereby activates the AKT-induced bypass, initiates the cell cycle, inhibits related apoptosis-inducing ligands, and contributes to tumor proliferation. Similarly, high expression of miR-211 in colon cancer can inhibit TIMP3, thereby activating the AKT-induced bypass and promoting the proliferation and infiltration of colon cancer cells [12,15]. Thus, miR-221 may play a key role in colon cancer.
The results of the current study support the hypothesis that miR-221 may contribute to the etiology of colon cancer. Interestingly, although differences among patients in the level of miR-221 expression in colon cancer samples were not meaningful by patient gender or by tumor type or staging, miR-221 expression did differ by patient age.
Further, the difference in miR-221 expression was significantly associated with survival time. The Cox multivariate analysis indicated that highly-expressed miR-221 is a risk factor for poorer prognosis of colon cancer patients. Indeed, the miR-221 expression showed a negative correlation with the overall survival time of patients, i.e., the lower the miR-221 expression level, the longer the overall survival time, and vice versa.
In summary, in colon cancer patients with low expression of miR-221, the survival time was longer; in contrast, patients with high expression of miR-221 had shorter survival. Thus, miR-221 could serve as a molecular marker for the prognosis of colon cancer patients. Because this study was a retrospective study with inadequate clinical data, further prospective clinical studies should be conducted in future to further confirm the reliability of the findings.
Disclosure of conflict of interest
None.
References
- 1.Xu AG, Yu ZJ, Jiang B, Wang XY, Zhong XH, Liu JH, Lou QY, Gan AH. Colorectal cancer in Guangdong Province of China: A demographic and anatomic survey. World J Gastroenterol. 2010;16:960–965. doi: 10.3748/wjg.v16.i8.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moghimi-Dehkordi B, Safaee A. An overview of colorectalcancer survival ratesandprognosis in Asia. World J Gastrointest Oncol. 2012;4:71–75. doi: 10.4251/wjgo.v4.i4.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Osman A. MicroRNAs in health and disease--basic science and clinical applications. Clin Lab. 2012;58:393–402. [PubMed] [Google Scholar]
- 4.Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223:102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Brit J Cancer. 2006;94:776–780. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 7.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, Calin GA, Ménard S, Croce CM. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 9.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 10.Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29:903–906. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- 11.Garofalo M, Quintavalle C, Romano G, Croce CM, Condorelli G. miR221/222 in cancer: their role in tumor progression and response to therapy. Curr Mol Med. 2012;12:27–33. doi: 10.2174/156652412798376170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun K, Zeng JJ, Wang W. MicroRNA-221 controls CDKN1C/P57 expression in human colorectal carcinoma. Acta Pharmacol Sin. 2011;14:279–283. doi: 10.1038/aps.2010.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barness R, Watson P, Allen-Vercoe E, Moore RA, Holt RA. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, Poon TC, Ng SS, Sung JJ. Differential expression of microRNAs in plasma of colorectal cancer patients: a potential marker for colorectal cancer screening. Gut. 2009;58:1375–1381. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- 16.Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127:118–126. doi: 10.1002/ijc.25007. [DOI] [PubMed] [Google Scholar]
- 17.Hong F, Li Y, Xu Y, Zhu L. Prognostic significance of serum microRNA-221 expression in human epithelial ovarian cancer. J Int Med Res. 2013;41:64–71. doi: 10.1177/0300060513475759. [DOI] [PubMed] [Google Scholar]
- 18.Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, Jacob S, Majumder S. MicroRNA-221/222 conferstamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem. 2008;283:29897–29903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 20.Visone R, Russo L, Pallante P, De Martino I, Ferraro A, Leone V, Borbone E, Petrocca F, Alder H, Croce CM, Fusco A. MicroRNAs (miR)-221 and miR-222,both overexpressed in human thyroid papillary carcinomas, regulatep27Kip1 protein levels and cell cycle. Endocr Relat Cancer. 2007;14:791–798. doi: 10.1677/ERC-07-0129. [DOI] [PubMed] [Google Scholar]


