Abstract
Objective: Our study aims to measure the expression level of serum microRNA-205 (miRNA-205, miR-205) in breast cancer, and evaluate the diagnostic value of miR-205 as a noninvasive biomarker for the early detection of breast cancer and other human cancers through a meta-analysis. Methods: In the first phase, an experiment based on 58 breast cancer patients and 93 healthy controls was carried out to figure out the diagnostic performance of serum miR-205 for breast cancer. The study was conducted by using Reverse Transcription and quantitative Real-time polymerase Cycle Reaction (RT-qPCR). In the second phase, a meta-analysis was performed to evaluate the diagnostic value of miR-205 for various cancers. Results: The test showed that the normalized expression levels of miR-205 in serum were evidently higher in healthy control than in breast cancer with a sensitivity and specificity at the optimal cutoff of 86.2% and 82.8%, respectively. The meta-analysis received a sensitivity of 75% and a specificity of 84%, and an AUC of 0.87. Conclusion: Our study determined that as a noninvasive and accessible biomarker, miR-205 not only has high clinical diagnostic value in the detection of breast cancer, but also plays an important role in clinical utilization of diagnosing various cancers. Further researches based on larger-scale subjects and additional improvement should be carried out to confirm our results.
Keywords: miR-205, breast cancer, serum, diagnostic value, meta-analysis, various cancers
Introduction
Breast cancer is the leading cause of death in women, contributing to 23% (1.38 million) of all new cancer cases and 14% (458,400) of all deaths in 2008 in females worldwide [1]. Early detection is an effective way to decline the mortality rate since the overall five-year survival rate is higher than 90% if it was diagnosed at an early stage in contrast to about 20% at a late stage when the malignant cells have already spread to other organs [2]. As a heterogeneous disease, breast cancer has evident tumor phenotypes, which indicates a wide range of potential molecular alterations and originally motivating events [3]. Therefore, breast cancer theoretically can be easily diagnosed in the early stage, but still many patients every year, which mostly accounts for the lack of specific diagnostic tests.
Currently, mammography serves as a gold diagnostic tool which has been widely applied to the detection of breast cancer in early stage, but it is strongly not recommended to young women because of its unpredictable hazards to body in ionizing radiation [4,5]. Ultrasound is also a useful way for detection, but it largely depends on the testing environment [6]. Over the past decades, scientists have made progress to find noninvasive and stable biomarkers, and thereby some protein-based circulating tumor biomarkers were discovered, which include carbohydrate antigen 15-3 (CA 15-3), carcinoembryonic antigen (CEA) and tissue polypeptide specific antigen (TPS) [7,8]. However, these promising biomarkers were non-specific for breast cancer and suffered from a low sensitivity and specificity [9]. Therefore, novel minimally invasive or even noninvasive diagnostic biomarkers with high sensitivity and specificity are urgently needed to the early detection of breast cancer [10].
Recently, new biomarkers, microRNAs (miRNAs), were found as important diagnostic tools for the detection of breast cancer. Composed of about 19-25 nucleotides, miRNAs serve as an important role in the regulation and development of cancer, which regulate the expression of a large number of genes based on the 3’untranslated regions of target mRNA in a sequence-specific process, causing the degradation or suppression of translation in mRNA [11-13]. Besides, miRNAs have the advantages of easy extraction and strong stability, since they exist in tissue and body fluid, such as urine, plasma, tissue, etc, and are resistant to extended storage, RNase activity, boiling, extremes of PH, and multiple freeze-thaw cycles [14]. The evidences mentioned above show miRNAs can be noninvasive biomarkers for cancer with a high accuracy.
Among these tumor-specific miRNAs, microRNA-205 (miR-205) is likely to be a new biomarker in the early detection of breast cancer, according to previous studies [15-18]. MiR-205, a miRNA of high conservation, was initially discovered by computational methods, whose expression mechanism in vivo was figured out subsequently, which indicated miR-205 can be a tumor suppressor [19-21]. Evidences suggested that miR-205 would down-regulate ErbB2, and meanwhile the expressions of cyclin E, cyclin D1 and cyclin-dependent kinase 6 (CDK6) go up in breast cancer [22]. Combined with the evidences of miRNAs mentioned above, miR-205 was estimated to be qualified to be a breast tumor-specific biomarker along with the advantages of noninvasiveness, convenience, cost-effectiveness, and high accuracy.
However, there were insufficient data to confirm the diagnostic value of miR-205 to the detection of breast cancer. According to the statistics we have gathered, we found that the majority of studies which concern both miR-205 and breast cancer only focused on the exploration of mechanism or the association between miR-205 and breast cancer [16,23,24], and few of them performed a research based on experiment of patients and healthy controls to confirm the diagnostic value of miR-205 for breast cancer. So far, only one study carried out by Shahram Savad et al., who performed a population-based research of the expression of miR-205 on breast cancer, showing miR-205 was under-expressed in breast cancer patients, and only had diagnostic proficiency in triple negative breast cancer (estrogen receptor/progesterone receptor/Her-2 negative) [17]. Therefore, a fundamental research using breast cancer patients and healthy controls as experiment objects was conducted to comprehensively identify whether miR-205 can be a useful biomarker for the early-stage diagnosis of breast cancer with high sensitivity and specificity. Besides, a meta-analysis based on the study and other studies with respect to miR-205 as a biomarker to other cancers were carried out to evaluate the diagnostic value of miR-205 [25-34] for various cancers.
Materials and methods
Ethics statement
The present study was conducted in conformity to the declaration of the ethics committee of the Affiliated Hospital of Hebei Engineering University, with abidance by the international ethical guidelines for biomedical research in which human subjects are involved. In this double- blind design, we have obtained informed consents from all participant subjects approved by the local institutional review board.
Study design and patients
The study was divided into two phases. In the first phase, we appraised the diagnostic performance of serum miR-205 for breast cancer. Patient blood samples were collected from 58 breast cancer patients with various stages and different clinicopathological characteristics. In total, 25 had stage I breast cancer, 33 with stage II. Histopathological characteristics and tumor stages of patients were identified by using breast biopsy specimens and imaging techniques in accordance with the World Health Organization (WHO) categories. Blood samples of healthy control were collected from 93 healthy women with no history of malignant gynecological diseases, no blood donations within the latest 3 years and no inflammation during the blood-drawing process. All the subjects are women, and no significant differences of age and ethnicity exist between the breast cancer patients and healthy controls. Table 1 summarized all the relative information of patients and healthy controls.
Table 1.
Clinicopathological characteristics of patients with breast cancer and healthy control
| Clinical characteristic | Breast cancer (n = 58) | Healthy control (n = 93) |
|---|---|---|
| Age, years | ||
| Median | 47 | 48 |
| Range | 21-78 | 27-74 |
| Menstrual status | ||
| Premenopausal | 24 (41.4%) | 42 (45.2%) |
| Postmenopausal | 34 (58.6%) | 51 (54.8%) |
| Tumor size (mean ± SD) (range) (cm) | ||
| > 2 | 32 (55.1%) | |
| q 2 | 26 (44.8%) | |
| TNM stage | ||
| I | 25 (43.1%) | |
| II | 33 (56.9%) | |
| Histology subtype | ||
| Ductal | 39 (67.2%) | |
| Lobular | 12 (20.7%) | |
| Other | 7 (12.1%) | |
| Lymph node metastasis | ||
| Yes | 31 (53.4%) | |
| No | 27 (46.4%) | |
| Estrogen receptor (ER) | ||
| Positive | 34 (58.6%) | |
| Negative | 24 (41.4%) | |
| Progesterone receptor (PR) | ||
| Positive | 36 (62.1%) | |
| Negative | 22 (37.9%) | |
| Proliferation index (Ki 67) | ||
| Positive | 30 (51.7%) | |
| Negative | 28 (48.3%) | |
| Her-2 | ||
| Positive | 23 (39.7%) | |
| Negative | 35 (60.3%) |
Data were expressed as N (%).
In the second phase of the study, a meta-analysis was carried out to further evaluate the diagnostic value of miR-205 in the detection of various cancers, covering this study and other 10 relative studies gathered. PubMed, Embase, Sinomed electronic databases, Chinese National Knowledge Infrastructure (CNKI), and Chinese Biomedical Literature Database (CBM) were searched up to July 15, 2014 without language restrictions to find out all the articles concerning the diagnostic proficiency of miR-205 for cancer. To be qualified for inclusion in this meta-analysis, studies should fulfill the following criteria: (1) concerning the diagnostic potential of miR-145 for cancers based on at least ten patients as subjects; (2) using the diagnostic gold standard to confirm cancer patients; and (3) providing sufficient data to work out relevant parameters, including true positive (TP), false positive (FP), true negative (TN) and false negative (FN).
Samples processing and miRNA extraction
To identify the discrimination of miR-205 expression between breast cancer patients and healthy controls, blood (10 ml) were drawn from subjects into PAXgene Blood RNA Tubes (PreAnalytiX GmbH, Switzerland) before surgery or adjuvant therapy. Each blood sample was then centrifuged at 3,000 rpm for 5 min at 4°C so as to separate the serum from cellular components. Serum was then immediately frozen and stored at -75°C into a fresh tube until RNA extraction. In all 10 μL RNA was isolated from each serum sample in 10 μL reactions including reverse transcription (RT) mixture and primers, using miRNAeasy kit (Qiagen, Valencia, CA, USA) on the basis of the manufacturer’s protocol. Quantitative PCR was conducted on Mx3005P qPCR System (Agilent, Santa Clara, CA, USA). Comparative cycle threshold (Ct) method is used to calculate the expression level of the miR-205. Since levels of miR-16 was found to be relatively stable in the test environment, so it was used as interval standard substance to normalize the expression of miR-205. The expression level of miR-205 was calculated and assessed using this equation: DCt = Ct (reference miR-16)-Ct (miR-205), the relative expression equal to 2-ΔΔCt. Each RT-PCR was performed in triplicate, including no-template controls and repeated three times.
Statistical analysis
In the first stage, all statistical data were analyzed by SPSS 20.0 software (SPSS, Inc., Chicago, IL, USA). The significance of serum miR-205 levels of different stages was determined with Kruskal-Wallis test, while the expression of miR-205 between breast cancer patients and healthy controls was performed with Mann-Whitney test. Receiver operating characteristic (ROC) curves was established to evaluate the proficiency of miR-205 as a biomarker in differentiating patients from healthy control, using the following parameters: area under curve, diagnostic sensitivity and specificity, positive and negative cutoff, positive and negative predictive values. The sensitivity and specificity were calculated using the various cutoff points based on the standard formulas, among which, the optimal sensitivity and specificity from ROC curves were decided by a pre-test probability and cost ratio. A P value less than 0.05 was considered as highly statistical significance.
In the meta-analysis, the bivariate meta-analysis model was employed to calculate the following pooled parameters: sensitivity, specificity, positive likelihood (PLR) and negative likelihood ratio (NLR), and diagnostic odds ratio (DOR), with corresponding 95% confidence intervals (CIs). The summary receiver operator characteristic (SROC) was constructed and the area under the SROC curve (AUC) was calculated by using the sensitivity and specificity offered by all the included studies. Furthermore, the heterogeneity of all studies (Table 2) was quantified by the Q test and the I 2 to figure out whether there is inconformity. The Fagan’s nomogram was conducted to dig out the clinical diagnostic value of miR-205 in the detection of cancer. Finally, the Deek’s funnel plot was performed so as to explore the potential sources of publication bias [35]. All statistical analyses were performed on Stata 12.0 software, when with a P < 0.05 shows statistical significance.
Table 2.
Summary of articles evaluating the diagnostic value of miR-205 for human cancers
| First author | Year | Country | Ethnicity | Case | Control | Cancer | Specimen | Diagnostic power | QUADAS-2 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||||
| No. | Age | No. | Age | TP | FP | FN | TN | |||||||
| Schaefer et al. | 2010 | German | Caucasian | 76 | 63 | 76 | 63 | Prostate cancer | Tissue | 54 | 14 | 22 | 62 | 4 |
| Xing et al. | 2010 | USA | Caucasian | 48 | 68 | 48 | 65 | Lung cancer | Sputum | 31 | 4 | 17 | 44 | 5 |
| Le et al. | 2012 | China | Asian | 82 | n.a. | 50 | n.a. | Lung cancer | Blood | 70 | 14 | 12 | 36 | 5 |
| Lee et al. | 2012 | Korea | Asian | 22 | 47 | 31 | n.a. | Uterine cancer | Tissue | 17 | 4 | 5 | 27 | 4 |
| 22 | 47 | 22 | n.a. | Uterine cancer | Tissue | 17 | 5 | 5 | 17 | |||||
| Srivastava et al. | 2013 | USA | Caucasian/African | 40 | 59.3 | 40 | 59.3 | Prostate cancer | Tissue | 29 | 7 | 11 | 33 | 4 |
| 36 | 66.5 | 12 | 66.8 | Prostate cancer | Urine | 26 | 4 | 10 | 8 | |||||
| Torres et al. | 2013 | Poland | Caucasian | 73 | 62.8 | 31 | 44.8 | Uterine cancer | Tissue | 61 | 4 | 12 | 27 | 4 |
| Zheng et al. | 2013 | China | Asian | 134 | 53.8 | 70 | 53.8 | Ovarian cancer | Plasma | 40 | 4 | 94 | 66 | 5 |
| Orang et al. | 2014 | Iran | Caucasian | 36 | n.a. | 36 | n.a. | Colorectal cancer | Tissue | 31 | 3 | 5 | 33 | 6 |
| Tsukamoto et al. | 2014 | Japan | Asian | 28 | n.a. | 14 | n.a. | Uterine cancer | Tissue | 22 | 3 | 6 | 11 | 5 |
| 12 | n.a. | 12 | n.a. | Uterine cancer | Plasma | 12 | 0 | 0 | 12 | |||||
| Shen et al. | 2014 | USA | Caucasian | 66 | 64 | 8 | 65 | Lung cancer | Sputum | 36 | 3 | 30 | 5 | 5 |
| Current study | 2014 | China | Asian | 58 | 47 | 93 | 48 | Breast cancer | Serum | 50 | 16 | 8 | 77 | 5 |
n.a. not available, QUADAS-2 quality assessment of diagnostic accuracy studies-2.
Results
Clinicopathological characteristics patients involved in the study
A total of 58 breast cancer patients and 93 healthy controls were recruited in this test. No significant difference of age, ethnicity and gender (all subjects are women) existed between breast cancer patients and healthy controls. Moreover, we investigated clinicopathological characteristics of breast cancer patients, including tumor stage, menstrual status, histology subtype, lymph node metastasis, status of ER, PR, Ki 67, Her-2, which were shown in Table 1.
Dysregulated expression of miR-205 in serum of breast cancer
We further investigated whether the regulation of miR-205 in serum become abnormal in breast cancer patients. Figure 1A indicated the concentrations of normalized levels of miR-205 in serum were evidently higher in healthy control than in breast cancer, which suggested that miR-205 was under-expressed in breast cancer patients. The P value of between healthy controls and breast cancer was less than 0.001, which indicated significant difference in the expression levels of these two types of subjects in serum. The P value of between stage I and healthy control was less than 0.01, suggesting that miR-205 can differentiate those with early-stage breast cancer from healthy controls, while the P value of between stage I and stage II of breast cancer was higher than 0.05, showing no statistical significance in expression levels between these two stages.
Figure 1.
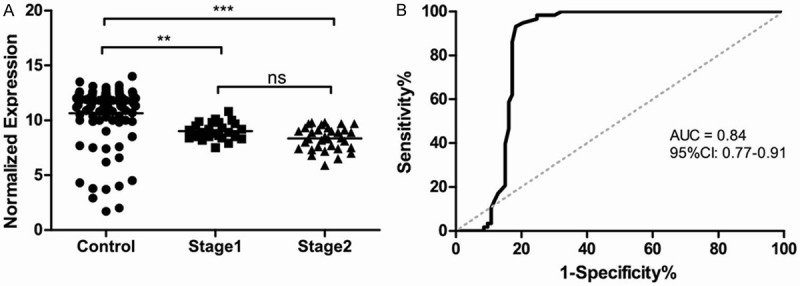
A: Different expression levels of serum miR-205 in healthy controls and stage I & II breast cancer; B: ROC curve to evaluate the application value of miR-205 for diagnosing breast cancer.
High sensitivity and specificity of miR-205 for diagnosing breast cancer
To further evaluate the diagnostic proficiency of miR-205 for breast cancer, a ROC curve was performed, which can be seen at Figure 1B. The AUC and confidence interval (CI) for miR-205 were as follows: 0.84, (0.77-0.91). The sensitivity and specificity at the optimal cutoff were 86.2% and 82.8%, respectively. The statistics described above indicated miR-205 had a moderately high accuracy in discriminating the breast cancer patients from healthy controls. Combined with the fact that miR-205 showed dysregulated expression in breast cancer, we determined that miR-205 revealed high diagnostic value as a biomarker for the detection of breast cancer.
Basic characteristics of included studies
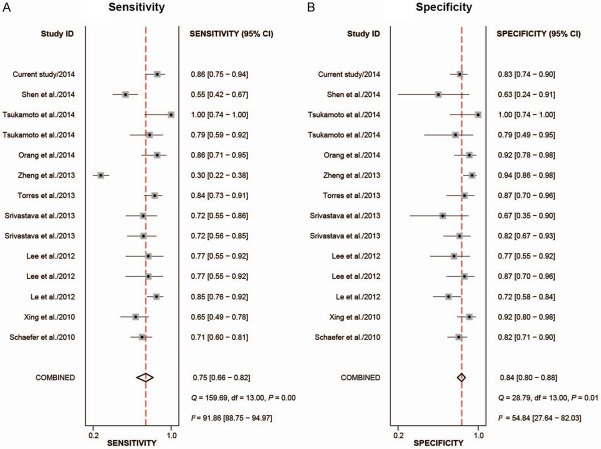
In total 13 studies from 10 articles and 1 study from our current research were included in this meta-analysis (Table 2), with 733 patients and 543 healthy controls involved in [25-34]. Among these 13 included studies from ten articles studying the diagnostic value of miR-205, 1focused on colorectal cancer, 1 on ovarian cancer, 3 on prostate cancer, 3 on lung cancer, 5 on uterine cancer. The related characteristics of the studies and subjects were shown in Table 2. To appraise the publication bias of all included studies, Deeks’ funnel plot asymmetry test was performed (Figure 2), with a P value of 0.51 higher 0.10, which indicated no statistically significant publication bias between these studies.
Figure 2.
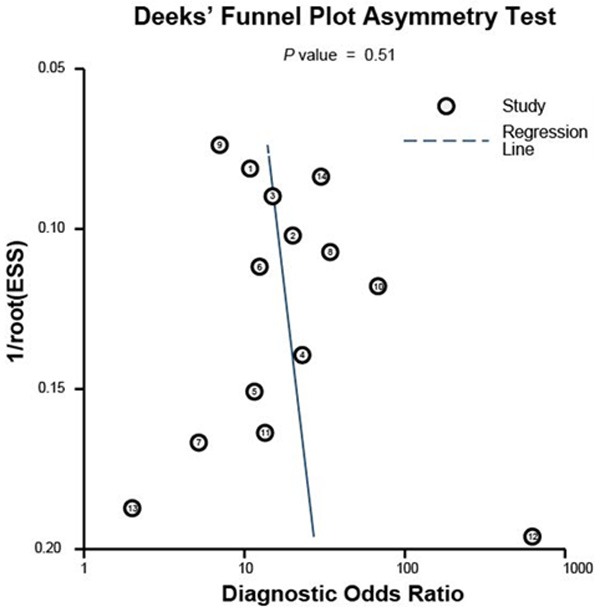
The Deeks’ test of the meta-analysis to analyze the publication bias of included studies.
Diagnostic accuracy and clinical value of miR-205 for cancers
A forest plot was performed for the overall studies to evaluate the diagnostic proficiency of miR-205 for cancers (Figure 3). We conducted a random-effect model, since the I 2 for sensitivity and specificity were 91.86% and 54.84% respectively. Other relative parameters are as follows: sensitivity, 0.75 (95% CI: 0.66-0.82); specificity, 0.84 (95% CI: 0.80-0.88); PLR, 4.8 (95% CI: 3.9-6.0); NLR, 0.30 (95% CI: 0.22-0.41); and DOR, 16 (95% CI: 11-24). The AUC of the overall SROC curve (Figure 4) is of 0.87 (95% CI: 0.84-0.90), demonstrating that miR-205 as a biomarker has highly accuracy in the detection of cancer. Furthermore, a Fagan’s nomogram was carried out to explore the clinical utilization of miR-205 for diagnosing cancers (Figure 5). For any people with a pre-test probability of 25% of suffering from cancer, the results showed that the positive results would raise the post-test probability of correctly diagnosing cancer to 62% with miR-205 as a screen tool, while negative post-test probability will lower to 7%. This test implied that miR-205 can be a moderately persuasive and useful biomarker in differentiating the patients from healthy controls.
Figure 3.
Forest plots to estimate the sensitivity and specificity of the meta-analysis.
Figure 4.
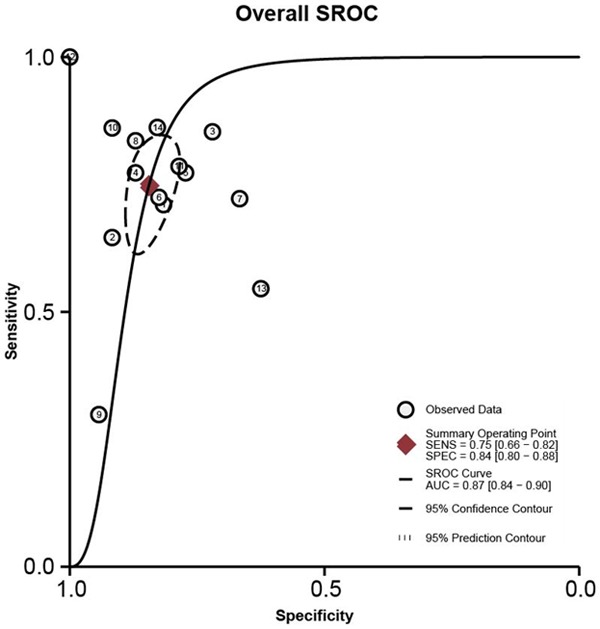
Summary receiver operator characteristic (SROC) curves to appraise the diagnostic value of miR-205 as biomarker for various cancers.
Figure 5.
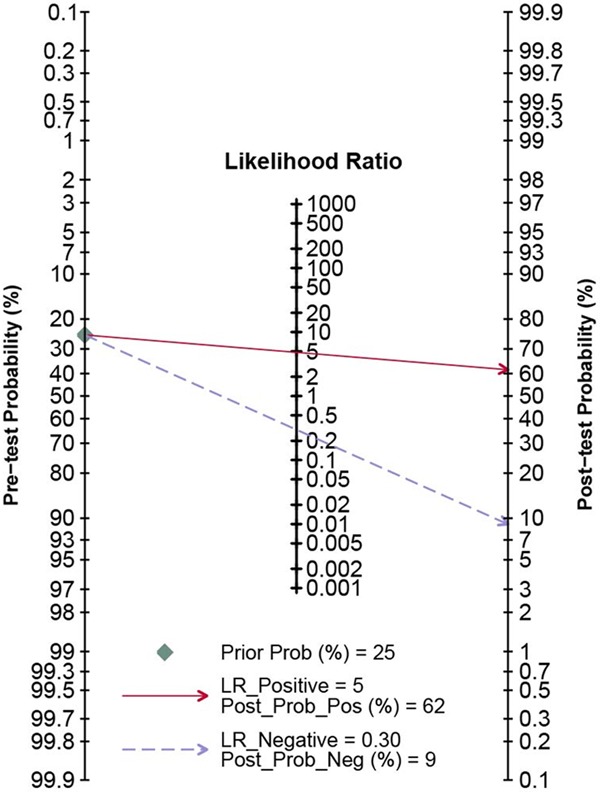
Nomogram of Fagan of all included studies to estimate the post-test probabilities for positive and negative findings.
Discussion
One of the effective methods to reduce the high mortality of breast cancer is early detection as the five-year survival rate of early stage is much higher than late stage [36]. To date, the standard screening tools for breast cancer are mammography, ultrasound and some useful biomarkers, which have been applied to the early detection of breast cancer successfully for decades, but still a lot of limitations showed up to hamper their further application. Thus, novel minimally invasive or even noninvasive and easily accessible diagnostic methods are urgently needed to improve or replace the current methods for early diagnosis of breast cancer.
Recently, the research of tumor-specific miRNAs become more and more popular as they can be serve as biomarkers noninvasively with easy accessibility and high accuracy [37-39]. In our study, we made a research to identify the correlation between miR-205 expression level and breast cancer based on 58 breast cancer patients (25 for stage I and 33 for stage II) and 93 healthy controls by RT-qPCR. There was no significant difference of age, gender and ethnicity between breast cancer patients and healthy controls. As we know, the present study is the first study using serum samples to comprehensively assess the diagnostic value of miR-205 as a screening tool for breast cancer. Results were presented in Figure 1. The data indicated that the expression levels of miR-205 in healthy controls were markedly higher than that of breast cancer patients. However, there was no outstanding significance between stage I breast cancer and stage II breast cancer, which suggested miR205 has potential diagnostic value to be a serum-based biomarker to differentiate the healthy controls from breast cancer patients. A ROC curve was further performed to investigate the accuracy of our results, which showed that the sensitivity and specificity and AUC are 86.2%, 82.8% and 0.84, respectively, indicating the highly accuracy of miR-205 as a biomarker for breast cancer detection.
MiR-205 was shown to be a breast cancer suppressor as it was identified to be down-regulated in serum in this study. We suspected that the dysregulated expression of miR-205 has an impact on breast cells or related proteins based on some relevant researches. One of the related study by Hailong Wu et al. in 2009 suggested that miR-205 is evidently under-expressed in malignant cells of breast tissue through suppressing anchorage independent growth, cellular proliferation and cellular invasion [40]. The other study by Marilena V. Iorio et al. in 2009 has proven that the under-expression of miR-205 targets Her-3 receptor, inhibiting the activation of Akt, which is a kind of downstream mediator proteins [41]. Although these underlying mechanisms have been figured out to investigate why miR-205 can be diagnostic target for breast cancer, more research are still needed to be done for comprehensive explanation based on larger subject sample.
Additionally, miR-205 has been proven to function as regulator for many different types of cancers. From the studies we included in our meta-analysis, miR-205 was up-expressed in lung cancer, uterine cancer and ovarian cancer, while under-expression of miR-205 was found in breast cancer, prostate cancer and colorectal cancer, compared with the healthy controls, but they were based on different specimens. Even so, these facts suggested that miR-205 also can serve as a biomarker for various cancers on the basis of dysregulated expression. However, inconsistency on accuracy existed among these studies. Therefore, we performed a meta-analysis to systematically evaluate the diagnostic value of miR-205 for various cancers.
In total, 13 clinical diagnostic studies and this study based on breast cancer were included in the analysis. All of the studies satisfied the QUADAS-2 standards (Table 2) and showed no publication bias (Figure 2), thus ensuring the quality of these included studies. The pooled parameters of meta-analysis are as follows: sensitivity, 0.75 (95% CI: 0.66-0.82); specificity, 0.84 (95% CI: 0.80-0.88), AUC of the overall SROC curve, 0.87 (95% CI: 0.84-0.90) (Figures 3, 4), indicating miR-205 as a promising biomarker for cancer detection with moderately high accuracy. Furthermore, we performed the Fagan’ nomogram to evaluate the clinical utilization of miR-205 for diagnosing cancers, and found a moderately high post-test probability of 62% to correct cancer detection and a negative post-test probability of 6% under the circumstance of 25% pre-test possibility of carrying cancer, which suggested assays based on miR-205 can serve as confirmatory test to complement current screening methods for cancer due to the high accuracy on clinical diagnosis.
However, there are some undeniable limitations in this study, expect for the deficiency on exploring mechanisms we mentioned above. We divided the limitations into two parts, one part for our fundamental research on the diagnostic value of miR-205 as a biomarker for breast cancer, the other part for the meta-analysis. In the first part, the limitations are summarized as follows: (1) our sample size was relatively small, and the long-term follow-up studies haven’t been carried out; (2) we only focused on the expression levels of serum miR-205 on the diagnosis of breast cancer. It is necessary to perform tests on other body fluids or tissues to find out the most sensitive specimen for extracting miR-205 persuasively [42]. The second part of limitations are listed below: firstly, we may miss some relevant studies during the screening process; secondly, only few of African population are involved in this meta-analysis compared with Asian and Caucasian population, making it hard to fully evaluate the diagnostic value of miR-205 for cancer. Despite these deficiencies mentioned above, our study was still the first one to systematically confirm the diagnostic application of miR-205 for breast cancer based on moderately sufficient experimental data and fully evaluate the diagnostic value of miR-205 as a biomarker in the detection of various cancers by meta-analysis.
In conclusion, our study shows that as a noninvasive and accessible biomarker, miR-205 not only has high clinical diagnostic value in the detection of early-stage breast cancer, but also plays an important role in clinical diagnosis of various cancers. However, more complementary researches with large scale samples should be considered in the future study.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Howlader N, Noone A, Krapcho M, Neyman N, Aminou R, Waldron W, Altekruse S, Kosary C, Ruhl J, Tatalovich Z. SEER cancer statistics review. Bethesda, MD: National Cancer Institute; 1975-2008. p. 19. [Google Scholar]
- 3.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taplin S, Abraham L, Barlow WE, Fenton JJ, Berns EA, Carney PA, Cutter GR, Sickles EA, Carl D, Elmore JG. Mammography facility characteristics associated with interpretive accuracy of screening mammography. J Natl Cancer Inst. 2008;100:876–887. doi: 10.1093/jnci/djn172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mandelblatt JS, Cronin KA, Bailey S, Berry DA, de Koning HJ, Draisma G, Huang H, Lee SJ, Munsell M, Plevritis SK, Ravdin P, Schechter CB, Sigal B, Stoto MA, Stout NK, van Ravesteyn NT, Venier J, Zelen M, Feuer EJ. Effects of mammography screening under different screening schedules: model estimates of potential benefits and harms. Ann Intern Med. 2009;151:738–747. doi: 10.1059/0003-4819-151-10-200911170-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kopans DB, Kopans D. Breast imaging. Philadelphia, PA: Lippincott Williams & Wilkins; 2007. [Google Scholar]
- 7.Duffy MJ. CA 15-3 and related mucins as circulating markers in breast cancer. Ann Clin Biochem. 1999;36:579–586. doi: 10.1177/000456329903600503. [DOI] [PubMed] [Google Scholar]
- 8.O’Hanlon DM, Kerin MJ, Kent P, Maher D, Grimes H, Given HF. An evaluation of preoperative CA 15-3 measurement in primary breast carcinoma. Br J Cancer. 1995;71:1288–1291. doi: 10.1038/bjc.1995.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uehara M, Kinoshita T, Hojo T, Akashi-Tanaka S, Iwamoto E, Fukutomi T. Long-term prognostic study of carcinoembryonic antigen (CEA) and carbohydrate antigen 15-3 (CA 15-3) in breast cancer. Int J Clin Oncol. 2008;13:447–451. doi: 10.1007/s10147-008-0773-3. [DOI] [PubMed] [Google Scholar]
- 10.Zeng RC, Zhang W, Yan XQ, Ye ZQ, Chen ED, Huang DP, Zhang XH, Huang GL. Down-regulation of miRNA-30a in human plasma is a novel marker for breast cancer. Med Oncol. 2013;30:477. doi: 10.1007/s12032-013-0477-z. [DOI] [PubMed] [Google Scholar]
- 11.Ng EK, Li R, Shin VY, Jin HC, Leung CP, Ma ES, Pang R, Chua D, Chu KM, Law WL, Law SY, Poon RT, Kwong A. Circulating microRNAs as specific biomarkers for breast cancer detection. PLoS One. 2013;8:e53141. doi: 10.1371/journal.pone.0053141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fiorucci G, Chiantore MV, Mangino G, Percario ZA, Affabris E, Romeo G. Cancer regulator microRNA: potential relevance in diagnosis, prognosis and treatment of cancer. Curr Med Chem. 2012;19:461–474. doi: 10.2174/092986712798918798. [DOI] [PubMed] [Google Scholar]
- 13.Anindo MI, Yaqinuddin A. Insights into the potential use of microRNAs as biomarker in cancer. Int J Surg. 2012;10:443–449. doi: 10.1016/j.ijsu.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 14.Brase JC, Wuttig D, Kuner R, Sultmann H. Serum microRNAs as non-invasive biomarkers for cancer. Mol Cancer. 2010;9:306. doi: 10.1186/1476-4598-9-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo D, Wilson JM, Harvel N, Liu J, Pei L, Huang S, Hawthorn L, Shi H. A systematic evaluation of miRNA:mRNA interactions involved in the migration and invasion of breast cancer cells. J Transl Med. 2013;11:57. doi: 10.1186/1479-5876-11-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, Schwartz G, Wells W, Kauppinen S, Cole CN. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67:11612–11620. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- 17.Savad S, Mehdipour P, Miryounesi M, Shirkoohi R, Fereidooni F, Mansouri F, Modarressi MH. Expression analysis of MiR-21, MiR-205, and MiR-342 in breast cancer in Iran. Asian Pac J Cancer Prev. 2012;13:873–877. doi: 10.7314/apjcp.2012.13.3.873. [DOI] [PubMed] [Google Scholar]
- 18.Wang Z, Liao H, Deng Z, Yang P, Du N, Zhanng Y, Ren H. miRNA-205 affects infiltration and metastasis of breast cancer. Biochem Biophys Res Commun. 2013;441:139–143. doi: 10.1016/j.bbrc.2013.10.025. [DOI] [PubMed] [Google Scholar]
- 19.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540–1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 20.Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 21.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adachi R, Horiuchi S, Sakurazawa Y, Hasegawa T, Sato K, Sakamaki T. ErbB2 down-regulates microRNA-205 in breast cancer. Biochem Biophys Res Commun. 2011;411:804–808. doi: 10.1016/j.bbrc.2011.07.033. [DOI] [PubMed] [Google Scholar]
- 23.Qin AY, Zhang XW, Liu L, Yu JP, Li H, Wang SZ, Ren XB, Cao S. MiR-205 in cancer: an angel or a devil? Eur J Cell Biol. 2013;92:54–60. doi: 10.1016/j.ejcb.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 24.Quesne JL, Jones J, Warren J, Dawson SJ, Ali HR, Bardwell H, Blows F, Pharoah P, Caldas C. Biological and prognostic associations of miR-205 and let-7b in breast cancer revealed by in situ hybridization analysis of micro-RNA expression in arrays of archival tumour tissue. J Pathol. 2012;227:306–314. doi: 10.1002/path.3983. [DOI] [PubMed] [Google Scholar]
- 25.Le HB, Zhu WY, Chen DD, He JY, Huang YY, Liu XG, Zhang YK. Evaluation of dynamic change of serum miR-21 and miR-24 in pre- and post-operative lung carcinoma patients. Med Oncol. 2012;29:3190–3197. doi: 10.1007/s12032-012-0303-z. [DOI] [PubMed] [Google Scholar]
- 26.Schaefer A, Jung M, Mollenkopf HJ, Wagner I, Stephan C, Jentzmik F, Miller K, Lein M, Kristiansen G, Jung K. Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int J Cancer. 2010;126:1166–1176. doi: 10.1002/ijc.24827. [DOI] [PubMed] [Google Scholar]
- 27.Xing L, Todd NW, Yu L, Fang H, Jiang F. Early detection of squamous cell lung cancer in sputum by a panel of microRNA markers. Mod Pathol. 2010;23:1157–1164. doi: 10.1038/modpathol.2010.111. [DOI] [PubMed] [Google Scholar]
- 28.Lee H, Choi HJ, Kang CS, Lee HJ, Lee WS, Park CS. Expression of miRNAs and PTEN in endometrial specimens ranging from histologically normal to hyperplasia and endometrial adenocarcinoma. Mod Pathol. 2012;25:1508–1515. doi: 10.1038/modpathol.2012.111. [DOI] [PubMed] [Google Scholar]
- 29.Srivastava A, Goldberger H, Dimtchev A, Ramalinga M, Chijioke J, Marian C, Oermann EK, Uhm S, Kim JS, Chen LN, Li X, Berry DL, Kallakury BV, Chauhan SC, Collins SP, Suy S, Kumar D. MicroRNA profiling in prostate cancer--the diagnostic potential of urinary miR-205 and miR-214. PLoS One. 2013;8:e76994. doi: 10.1371/journal.pone.0076994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torres A, Torres K, Pesci A, Ceccaroni M, Paszkowski T, Cassandrini P, Zamboni G, MacIejewski R. Diagnostic and prognostic significance of miRNA signatures in tissues and plasma of endometrioid endometrial carcinoma patients. Int J Cancer. 2013;132:1633–1645. doi: 10.1002/ijc.27840. [DOI] [PubMed] [Google Scholar]
- 31.Zheng H, Zhang L, Zhao Y, Yang D, Song F, Wen Y, Hao Q, Hu Z, Zhang W, Chen K. Plasma miRNAs as diagnostic and prognostic biomarkers for ovarian cancer. PLoS One. 2013;8:e77853. doi: 10.1371/journal.pone.0077853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orang AV, Safaralizadeh R, Hosseinpour Feizi MA, Somi MH. Diagnostic and prognostic value of miR-205 in colorectal cancer. Asian Pac J Cancer Prev. 2014;15:4033–4037. doi: 10.7314/apjcp.2014.15.9.4033. [DOI] [PubMed] [Google Scholar]
- 33.Shen J, Liao J, Guarnera MA, Fang H, Cai L, Stass SA, Jiang F. Analysis of MicroRNAs in Sputum to Improve Computed Tomography for Lung Cancer Diagnosis. J Thorac Oncol. 2014;9:33–40. doi: 10.1097/JTO.0000000000000025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsukamoto O, Miura K, Mishima H, Abe S, Kaneuchi M, Higashijima A, Miura S, Kinoshita A, Yoshiura K, Masuzaki H. Identification of endometrioid endometrial carcinoma-associated microRNAs in tissue and plasma. Gynecol Oncol. 2014;132:715–721. doi: 10.1016/j.ygyno.2014.01.029. [DOI] [PubMed] [Google Scholar]
- 35.Deeks JJ. Systematic reviews in health care: Systematic reviews of evaluations of diagnostic and screening tests. BMJ. 2001;323:157–162. doi: 10.1136/bmj.323.7305.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carter CL, Allen C, Henson DE. Relation of tumor size, lymph node status, and survival in 24,740 breast cancer cases. Cancer. 1989;63:181–187. doi: 10.1002/1097-0142(19890101)63:1<181::aid-cncr2820630129>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 37.Duffy MJ. Role of tumor markers in patients with solid cancers: a critical review. Eur J Int Med. 2007;18:175–184. doi: 10.1016/j.ejim.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 38.Ha TY. MicroRNAs in Human Diseases: From Cancer to Cardiovascular Disease. Immune Netw. 2011;11:135–154. doi: 10.4110/in.2011.11.3.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patel N, Sauter ER. Body fluid micro(mi)RNAs as biomarkers for human cancer. Journal of Nucleic Acids Investigation; 2:1–4. [Google Scholar]
- 40.Wu H, Zhu S, Mo YY. Suppression of cell growth and invasion by miR-205 in breast cancer. Cell research. 2009;19:439–448. doi: 10.1038/cr.2009.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iorio MV, Casalini P, Piovan C, Di Leva G, Merlo A, Triulzi T, Ménard S, Croce CM, Tagliabue E. microRNA-205 regulates HER3 in human breast cancer. Cancer Res. 2009;69:2195–2200. doi: 10.1158/0008-5472.CAN-08-2920. [DOI] [PubMed] [Google Scholar]
- 42.Hu Z, Chen X, Zhao Y, Tian T, Jin G, Shu Y, Chen Y, Xu L, Zen K, Zhang C, Shen H. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J. Clin. Oncol. 2010;28:1721–1726. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]