SUMMARY
Leishmania is an infectious protozoan parasite related to African and American trypanosomes. All Leishmania species that are pathogenic to humans can cause dermal disease. When one is confronted with cutaneous leishmaniasis, identification of the causative species is relevant in both clinical and epidemiological studies, case management, and control. This review gives an overview of the currently existing and most used assays for species discrimination, with a critical appraisal of the limitations of each technique. The consensus taxonomy for the genus is outlined, including debatable species designations. Finally, a numerical literature analysis is presented that describes which methods are most used in various countries and regions in the world, and for which purposes.
INTRODUCTION
Leishmania species are digenetic parasitic protozoans of the order Kinetoplastida and the family Trypanosomatidae. They are transmitted by phlebotomine sand flies to wild and domestic animals and to humans. Cutaneous leishmaniasis (CL) is one of the main clinical manifestations of human infection (reviewed by Reithinger et al. [1]). In contrast to visceral leishmaniasis (VL), it is not lethal but is often traumatic and associated with social stigmatization. According to a recent estimate (2), the annual incidence of the disease is between 0.7 and 1.2 million cases worldwide. It is endemic in 87 countries, 10 of which (Afghanistan, Algeria, Colombia, Brazil, Iran, Syria, Ethiopia, Sudan, Costa Rica, and Peru) bear 70 to 75% of the burden (2). All species of Leishmania that are pathogenic to humans can cause cutaneous disease, albeit with various severities.
On one side of the spectrum is Leishmania major, which generally inflicts self-healing localized skin lesions at the site of the infection (localized CL [LCL]) and is found in the Mediterranean region, Africa, the Middle East, and further, as far as India. On the opposite side one finds L. braziliensis, which is endemic in South America and is able to cause severe mutilating mucocutaneous leishmaniasis (MCL) even years after cure of the initial local lesion. Between these extremes are species inflicting difficult-to-treat localized lesions and those often giving rise to diffuse cutaneous leishmaniasis (DCL), whereby several lesions occur distant from the site of infection (1).
To complete the picture, L. infantum (syn., L. chagasi), found in Mediterranean countries and Brazil, can lead to both CL and VL in humans and to canine leishmaniasis in dogs. VL (also known as kala-azar) is a lethal condition if untreated, with parasites affecting the liver, spleen, and bone marrow. Also, L. donovani causes VL, whereby cured patients sometimes develop a particular cutaneous complication known as post-kala-azar dermal leishmaniasis (PKDL), which is not seen with L. infantum and manifests as nodules covering large parts of the body (3). Especially in Sri Lanka, L. donovani infection often leads to CL (4, 5), making dermal leishmaniasis a disease that can be provoked by all Leishmania species that are infectious to humans.
The association between different clinical forms of CL and specific Leishmania species is, however, not absolute. The best example is probably mucosal leishmaniasis (ML), which is generally encountered in patients infected with L. braziliensis but has also been reported for other species of the Leishmania (Viannia) subgenus, such as L. guyanensis (6). Especially with HIV coinfection and in immunosuppressed individuals, unexpected clinical presentations may be encountered (e.g., see reference 7). Also, the traditional geographical connotation of clinical forms should be taken with caution, because expansion or movement of transmission cycles (e.g., see references 8 to 11) may cause unexpected species to circulate in unexpected regions or habitats.
It is thus clear from the above information, and also because species display specific transmission patterns, affect disease prognosis, and may differentially react to certain drugs or treatment regimens, that efficient control of dermal leishmaniasis requires species typing (e.g., see references 12 to 19). Besides parasite identity, other factors that guide the choice of treatment are the clinical presentation, the host's genetic background and immunity, and additional confounding conditions (1). The need to determine the infecting Leishmania species depends largely on the specific context. For clinical and epidemiological studies, confirmation of the infecting species is definitely recommended in all cases (19), preferably using a globally applicable technique that has been validated on all species. In these studies, such an approach is certainly feasible.
For day-to-day clinical management, on the other hand, individual species typing is not always achievable, for either technical, logistic, or financial reasons. The most straightforward situation is presented by a primary health center located in a region of endemicity where only one species circulates. In such cases, accurate diagnosis by genus detection is all that is needed, provided that the epidemiology of the area is sufficiently monitored. Such genus detection can be simple and based on clinical or microscopic examinations of lesions, even though these are not the most sensitive or specific methods. Serology is not very sensitive because of the strong Th1 bias, and molecular methods are superior when the parasite load is low (20). A more complicated setting is that of a primary health center located in a region where two or more species or variants are transmitted, each requiring a different treatment approach. In such a scenario, experienced physicians may separate species based on the lesion morphology and clinical syndrome. If this is not possible, then the species can be identified using techniques separating the local parasite variants. Furthermore, a regional or national reference center typically deals with patients infected in various environments, which makes individual species identification more relevant, as the exact geographical origin of infection and the local epidemiology are often difficult to assess. Finally, the most complicated setting is that of a travel clinic seeing patients who have visited several regions or countries of endemicity. Generally, the exact region of infection is unknown, and treating physicians have less knowledge of or access to epidemiological information, in addition to often having less experience with diagnosis and treatment. Such an environment hence requires a globally applicable species typing strategy.
This review aims to give a state-of-the-art overview of the currently deployed and available Leishmania species typing assays in the context of clinical and epidemiological studies and disease management. The focus lies on methods that are globally applicable rather than on those validated locally. The limitations of each technique are discussed. In addition, current problems with the taxonomic framework of the genus are treated from a practical point of view, and suggestions toward a more solid approach are formulated.
LEISHMANIA TAXONOMY
A Bird's-Eye View
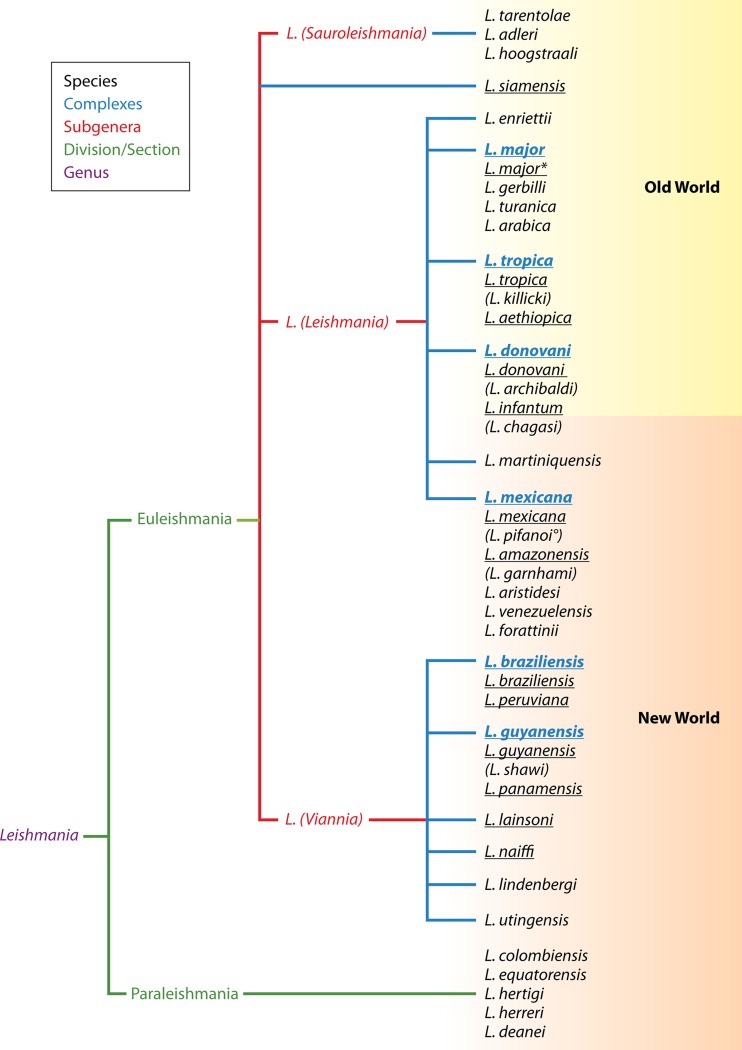
The taxonomy of the Leishmania genus is outlined in Fig. 1, essentially as compiled by Schönian et al. (21), but with the addition of L. siamensis, recently described from Thailand (22); L. martiniquensis, from the French West Indies (23); and L. adleri and L. hoogstraali (24). This classification of Leishmania is by no means undisputed, but neither is any other proposed scheme. The different layers between the classical Linnaean levels of genus and species and the growing number of described species are illustrative of the troublesome classification of the genus (25). Several species have been grouped into so-called “species complexes,” further referred to as “complexes,” whereby each complex is named after one of the constituting species. Complexes and species were assigned to subgenera, which in turn were grouped into sections or divisions (Fig. 1).
FIG 1.
MLEE-based taxonomy of the Leishmania genus as listed by Schönian et al. (21), but with the addition of L. siamensis (22), L. martiniquensis (23), L. adleri, and L. hoogstraali (24). The various levels are indicated by their respective colors. Several species (black) are grouped into “species complexes,” or “complexes” (blue), whereby the complex is named after one of its species. The underlined species are those documented in the studies analyzed in Currently Applied Methods, which are those relevant for human and domestic animal diseases. Species names between brackets are not recognized as separate entities by most authors and in fact are part of the species listed above them. L. chagasi is a synonym of L. infantum of the New World. °, some L. pifanoi strains are more related to L. amazonensis than to L. mexicana (61, 62); *, several authors have reported L. major-like parasites from the New World (62, 79–83). The figure does not represent a dendrogram with evolutionary relationships but a practical classification system.
The lack of an unequivocal classification scheme is not unique to Leishmania and is essentially the result of using different markers to study phylogeny and of the lack of a universally applicable species definition for unicellular organisms. Multilocus enzyme electrophoresis (MLEE) (26) laid the basis of current Leishmania taxonomy, but species were often defined when only minimal differences in isoenzyme characteristics were observed, while these really concerned intraspecies variability or convergent evolution (27). With the advent of molecular techniques to study the variability of genera, it became possible to test the MLEE-derived classifications with more informative genetic methods, and this gave rise to various adaptations. Some of these have now been generally accepted, but others remain subjects of debate (28).
When it comes to identifying species for human or veterinary clinical practice, the situation is not dramatic, however, and almost all authors classify parasites as one of the species underlined in Fig. 1. Generally the species complexes are easily discriminated by many techniques, but often identification of the individual species poses more problems (see “Clinically Relevant Taxa: Certainties and Doubts”). The reason behind this is that species belonging to the same complex are often very closely related, which not only complicates their identification but also hampers an unambiguous species definition. Sometimes more variable markers are needed to distinguish species within the same complex than to distinguish different complexes.
Evaluation of Current Taxonomy
One may ask how species should be defined from a clinical perspective. Some would argue that a species is a set of organisms that impose identical clinical presentations on humans or animals. However, this is not a workable criterion for several reasons. First, what is regarded as clinically identical may change over time. New drugs may become available that specifically affect one group of parasites, while another group shows intrinsic tolerance. Also, vaccines targeting one particular group of parasites may be deployed. Second, clinical presentations may differ according to the geographical region or human population, for instance, in travelers versus individuals with endemic cases (18). Third, for prevention and control, different species can have different transmission cycles, requiring other control measures.
Conversely, some species are clinically pleomorphic. For instance, L. braziliensis causes MCL in some patients after cure of a CL lesion. Currently, no difference has been identified between parasites from CL or MCL lesions, and the clinical presentation seems to be governed by the immune system of the host or other confounding factors rather than by the parasite. In such cases, it is impossible to separate isolates into different species.
Consequently, the only logical definition of species relies on parasite identity, which is best studied based on molecular characteristics, such as those revealed by zymodeme and DNA analyses, as these reflect the parasites' evolution. This is also known as the phylogenetic species concept (28) and forms the basis for the current Leishmania taxonomy. Inevitably, the next question to address is how different two groups need to be for them to be assigned to different species. It was one of the pioneers of modern evolutionary theory, Charles Darwin, who already observed the difficulty of defining species in higher sexual organisms (29), and it is even much more challenging in single-celled protozoa, such as Leishmania. In such cases, defining the species boundary is a matter of convention once the evolutionary relationships have been established, and what may be considered distinct populations in one genus may be regarded as species in another group of organisms.
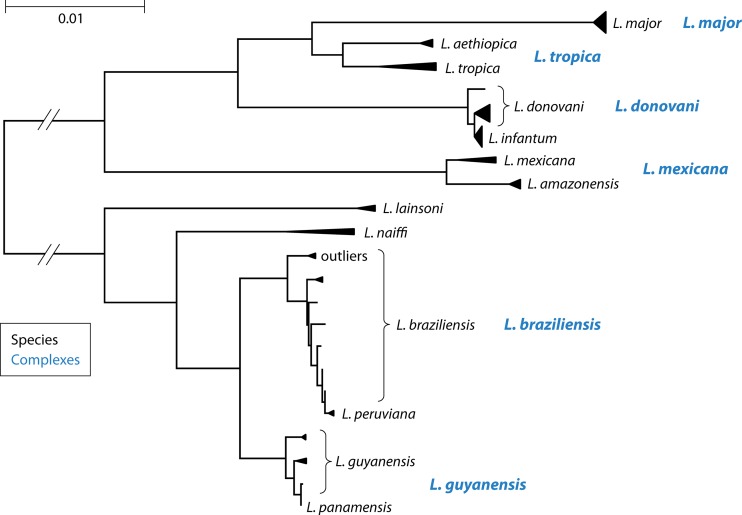
In this respect, the current Leishmania classification is not entirely consistent. As can be observed in Fig. 2, some species are so closely related to each other that one can question their validity as distinct species, even though discriminating them is clinically and epidemiologically relevant. In particular, L. infantum, L. peruviana, and L. panamensis are subgroups of L. donovani, L. braziliensis, and L. guyanensis, respectively, as further detailed in “Clinically Relevant Taxa: Certainties and Doubts.”
FIG 2.
Multilocus sequence analysis of Van der Auwera et al. (60), based on sequences of 7 housekeeping genes. Each species and species complex are indicated, as in Fig. 1. L. braziliensis outliers are discussed in “Clinically Relevant Taxa: Certainties and Doubts.” L. shawi was not included in this analysis, and L. panamensis was represented by a single strain. The dissimilarity scale is depicted in the top left corner, in substitutions per nucleotide.
A further taxonomic complication is presented by the occasional occurrence of natural interspecies hybrids (30–38), although these are quite rare. At the intraspecies or population level, sexual recombination might be frequent (39–44), but in general, a clonal mode of propagation is observed when looking at the greater picture (45). In theory, interspecies hybrids may give rise to new species, which develop their own characteristics by combining those of both parents. This happened for two discrete typing units (DTUs) in Trypanosoma cruzi (46), but as far as we know, it has not occurred in Leishmania, where interspecies hybrids have been confined in space and time. Nevertheless, they do seem to exist as isolated populations but have not gained any formal taxonomic status. In a specific epidemiological or clinical context, it may, however, be relevant to identify them (see “Detection of Interspecies Hybrids”). Finally, taxonomy is troubled by intermediate forms between closely related parasites, such as those between L. braziliensis and L. peruviana (33). Some of these may represent remnant transition forms of the speciation process or may represent interspecies hybrids (38, 47). Such forms complicate the clear distinction between species, also leading to difficulties in the species typing process.
Clinically Relevant Taxa: Certainties and Doubts
Several species and species complexes form separate entities that are clearly different from all others. In limiting the list to clinically relevant groups, this applies to L. siamensis, L. major, the L. tropica complex, the L. donovani complex, the L. mexicana complex, the L. braziliensis complex, the L. guyanensis complex, L. lainsoni, and L. naiffi (Fig. 1 and 2). Within the 5 complexes, several typing and species definition problems are observed.
L. donovani complex.
L. infantum strains are a clear subgroup within the L. donovani complex, but other equally valid subgroups identified from microsatellites, amplified fragment length polymorphisms (AFLP), and multilocus sequence typing (MLST) have not been assigned separate species names (48–52). L. chagasi was formerly separated from L. infantum because of its New World endemicity, but it is now regarded as the same species, which was imported from Europe to Latin America in the 16th century (53, 54). However, Marcili et al. (24) could discriminate them. L. archibaldi, which is an MLEE-defined species of the complex (55), was not validated as a monophyletic subgroup by DNA analyses and is no longer used by most authors (27, 49, 51). The species L. donovani is thus defined by exclusion and comprises all strains of the complex that are not L. infantum. Nevertheless, L. donovani strains have some common characteristics, such as their presumed anthroponotic transmission and the PKDL complication. Because both species are close, methods that are fit for typing to the species complex level are often not able to distinguish them.
L. tropica complex.
L. aethiopica and L. tropica belong to the L. tropica complex, but these species seem quite well defined genetically, as evidenced by MLST (56). Consequently, most methods can distinguish them. L. killicki, on the other hand, behaves more like a local L. tropica variant and is better not regarded as a separate species (56–59).
L. mexicana complex.
In clinical reports, only two species of the L. mexicana complex have been described: L. amazonensis and L. mexicana. The complex seems to be understudied, because most reports dealing with the Leishmania (Leishmania) subgenus focus on the Old World, and conversely, most studies of the New World focus on the Leishmania (Viannia) subgenus. This often leaves the L. mexicana complex in an orphaned position, as it is a Leishmania (Leishmania) subgenus complex of the New World. No comprehensive studies covering the full genetic and geographic variability of the complex are available.
Multilocus sequence analysis identified L. amazonensis (including L. garnhami) as a clearly defined group in the complex (Fig. 2) (60), which agrees with other markers (61). Also, Schönian et al. (21) considered L. garnhami to be a synonym of L. amazonensis. L. pifanoi, another species of the complex, seems to form a heterogeneous group and is not a valid species. Some strains have been found to be more related to L. mexicana, and others to L. amazonensis (61, 62).
L. braziliensis complex.
L. peruviana and L. braziliensis have been described as the only species in the L. braziliensis complex, based on a minor difference in MLEE profiles (47, 63). If L. peruviana is considered a different species, then the situation is comparable to that of the L. donovani complex, with L. braziliensis being defined by exclusion of L. peruviana (Fig. 2). The situation is more complicated, however, as molecular analyses revealed the existence of several intermediate strains that may be hybrids or transitional forms between both species (33, 38, 64).
Current data hence do not support a clear-cut dichotomy between L. peruviana and L. braziliensis, leading to frequent problems in identifying both species. Nevertheless, in contrast to infection with L. peruviana, infection with L. braziliensis can result in mucocutaneous complications after the initial cure (65), which is why identification of these species has a pronounced clinical relevance. Currently, full-genome sequences are being determined for several members of the complex, which may help to better define both species (unpublished results).
Some studies identified a cluster of L. braziliensis strains that are genetically distinct from the main L. braziliensis-L. peruviana group (“outliers” in Fig. 2), comprising parasites from different countries (33, 60). This group has been given various names, such as “outlier L. braziliensis” (66), “atypical L. braziliensis” (67), and “L. braziliensis type 2” (60). It has so far not been widely recognized, even though it is discriminated much more easily than L. peruviana. Most assays probably type the group as L. braziliensis. Other authors reported an atypical L. braziliensis group on the basis of MLEE and DNA sequence analysis (68), but it is unclear whether this concerns the same variant. A more detailed phylogenetic study is needed to decide the taxonomic status of the L. braziliensis outliers, as some markers place them at the same distance from L. braziliensis as other species complexes (33).
L. guyanensis complex.
The L. guyanensis complex unites the species L. guyanensis, L. panamensis, and L. shawi (Fig. 1). Members of the complex are widespread in Latin America and have been reported from Brazil, Venezuela, Colombia, Peru, Ecuador, Surinam, French Guiana, Guyana, Argentina, and Central American countries, such as Panama and Costa Rica. So far, studies have been limited to analyzing strains from a particular geographical origin or country, and the entire genetic variability of the complex has not been assessed. Nevertheless, even based on a limited number of strains, several reports have questioned the species status of L. panamensis (69) and L. shawi (41). Given the current evidence, it can be extrapolated that L. panamensis and L. shawi may be local variants of L. guyanensis, which would then be defined by exclusion of the two subgroups (Fig. 2).
Looking for Answers and Finding Pragmatic Solutions
A common denominator underlies all the above-mentioned problems with current taxonomy, namely, the lack of comprehensive studies that include the entire variability observed in each complex (28). Analyzing a globally representative set of strains is the only possibility for determining the exact status of each entity as either a complex, species, or subspecies. Adequate sampling not only relates to geographical coverage but also to including strains from asymptomatically infected individuals, animal reservoirs, and vectors; otherwise, one risks looking just at the tip of the iceberg. However, for clinical practice, it may be more convenient to discriminate disease-causing parasites, i.e., those isolated from actual lesions, even though this is not recommended for establishing a general taxonomy of the genus.
After gathering all samples, the next question is how to analyze them. Studying the population structure of each complex seems to be the way to go in order to define well-characterized groups, but on which data should this be based? MLEE has the best coverage of genus variation, at least for cultured strains, and remains advocated as the gold standard by the World Health Organization (65). Unfortunately, this method is not able to characterize uncultured parasites and is based on a limited number of mutations affecting physicochemical enzyme properties. The advent of next-generation sequencing might be taken as an opportunity to exploit the entire genome (70), certainly as new developments are in the pipeline that may allow whole-genome sequencing even from clinical samples (71). Taking into account the vast information content from a full-genome sequence compared to that obtained from MLEE, this would be the method of choice. Multilocus sequence analysis is the next best option, as it uses sequence information from various genes distributed over different chromosomes (72).
Taken together, the data indicate that characterizing a global panel with the best techniques available is the ideal scenario in the quest for defining a useful and unbiased reference framework for the classification of Leishmania. This requires a global effort, however, which is not likely to happen in the near future, for financial or political rather than technical reasons. A profound study of each species complex is more realistic, but even that requires extensive additional sampling. At present, studies documenting parasite diversity in-depth focus on particular local issues only (e.g., see reference 73).
For both human and veterinary clinical practice, the species underlined in Fig. 1 provide an adequate framework despite the open questions for some species complexes. The following sections present an overview of how these species can be identified by using the methods at hand in whatever setting one works in. These methods are a practical approach allowing one to conduct clinical and epidemiological studies and also enabling optimization of patient care on a day-to-day basis.
METHODS FOR SPECIES TYPING
Levels of Typing and Focus of This Review
The current review primarily discusses typing methods that have the ability to discriminate many different species or complexes in a clinical context. Each technique looking at parasite variability is designed for a particular purpose, and three steps can be discriminated in a diagnostic workflow.
The first stage usually targets detection of the Leishmania genus rather than a specific species (20, 74–77). Assays focusing on this level are not covered here, except when species typing relies on downstream analysis of PCR amplification products generated in the detection process. Methods designed for detection of the genus should be sensitive and Leishmania specific.
After Leishmania infection has been confirmed, the subsequent step involves identification below the genus level, down to the subgenus, species complex, or species level. In contrast to the Leishmania detection tools described above, the most useful typing assays were designed to discriminate many different species, with less emphasis on sensitivity. These tests are the subject of this paper. Unless otherwise specified, we focus specifically on discriminating clinically relevant species, i.e., those underlined in Fig. 1, without relying on parasite isolation in culture. Methods applicable to clinical samples are prioritized over those used for sand flies, as insects can harbor additional species and genera that are not encountered in humans. We highlight Leishmania species typing methods that are applicable for everyday use, discriminate many species, and have been validated in a large geographical area.
In particular cases, i.e., population or epidemiological studies, it may be relevant to discriminate isolates to below the species level, or even to the strain level. This involves methods with higher discriminative potential, which are outside the scope of this review unless such methods are also useful for species typing.
The distinction between these three diagnostic levels is not always clear-cut. In some settings, the genus detection step is replaced by a sensitive assay that targets a particular complex, species, or subgenus rather than the entire genus. Such methods are able to identify a particular group of Leishmania parasites, but they are useful only when no other variants of the parasite are expected. This can be difficult to establish in the current context of human mobility, urbanization, and changing climate and ecology. On the other hand, some species typing methods also allow characterization below the species level.
Criteria for Species Typing Tools
A multitude of different methods have been designed for discrimination of Leishmania species, a search that has been going on for many decades, as evidenced in early publications (e.g., see reference 78). They can roughly be categorized by function of the technology involved or by function of the biological feature evaluated. Regardless, in our opinion, an ideal typing tool for clinical purposes should adhere to the criteria listed here.
First, a species typing tool must be able to discriminate species. But which species should it discriminate? The absolute minimal requirement is that it can identify species circulating in the studied area. In order to assess this, one needs to take into account the entire Leishmania variability encountered in the region where the test is to be applied, both between and within species. In many studies describing a new technique or marker, however, validation is performed on only a few reference type strains, often from a region or country that is different from that where the test is used. Given the tremendous variability of single-cell parasites, this is a major flaw, and tests not validated with adequate reference strains must not be regarded as reliable.
Second, the tool is preferably globally applicable. Especially in times of climate change and rapidly changing ecology, evaluating whether a test can discriminate species in a particular area entails a certain risk. It assumes that we accurately know the epidemiology of the area and ignores the possibility of other species invading new territories. For example, a test designed to discriminate L. major from L. tropica in Kenya may mistakenly identify an L. aethiopica sample as L. tropica if the test does not distinguish these species. Because up-to-date epidemiological data on a particular study area are often scanty, the safest methods are those validated on a global scale, taking into account Leishmania's worldwide variability. The larger the region where the test can be applied, the better the result will be. For instance, a global test is better than one for the New World, which in turn outperforms one for Colombia. And even in large areas, such as the entire New World, unexpected parasite variants may turn up, such as occasionally documented L. major-like parasites, which are expected to be found exclusively in the Old World (62, 79–83).
Third, the assay must be sensitive. For typing of cultured parasite isolates, sensitivity is not an issue, as plenty of starting material is available for processing. However, clinical and epidemiological studies often require culture-independent and high-throughput applications, which must be able to analyze a minimal amount of parasites, even more so when asymptomatic infections are studied. Also, in the diagnostic pipeline, parasite isolation is often replaced by fast and sensitive methods that are directly applicable to clinical samples.
Fourth, the test must be Leishmania specific. Leishmania parasites are typically present in a high background of human or animal cells, sometimes infected by other organisms. Any cross-reaction with non-Leishmania biomolecules must not prevent parasite identification. Therefore, typing methods need to be targeted to Leishmania, or at least must be able to discriminate Leishmania from other pathogens in the sample. This is a major issue with HIV patients, in whom several trypanosomatids can be encountered (84) but also other coinfections have been documented, such as mycobacteria and Schistosoma (28). These problems are less of an issue for analyzing cultured isolates, where Leishmania promastigotes form most of the harvested biomass. In cultures, however, other problems may arise, such as other pathogens from the sample outgrowing Leishmania (85).
Fifth, standardization is an issue. In order to compare species typing results across different studies, the technology employed should be reproducible in various settings. Some methods are easier to standardize than others. PCR followed by amplicon sequencing, for instance, is quite easily standardized, as (good-quality) sequences leave little room for interpretation. Restriction fragment length polymorphism (RFLP) analysis is already more difficult, as small fragment size differences and incomplete digests may interfere with a correct reading of the patterns. Even more problematic are species-specific PCRs, hybridization assays, and melting assays, where even small experimental changes to the reaction conditions (buffer, pH, ionic strength, thermocycling parameters, optical detection, and amount of sample or parasite DNA) can result in an altered specificity. Currently, no commercial standard tests are available for Leishmania species typing.
Sixth, a given tool must be applicable in the setting where it is needed. This is probably what hampers most development of a generic typing technology. What springs to mind when talking of feasibility are limited-resource labs, which often have no access to common technology available in more advanced settings, for example, resources for DNA extraction and PCR. But equally so, large-scale clinical and epidemiological studies may benefit from high-throughput applications not generally of use in a patient management setting, where speed may be a key requirement. Nevertheless, different technologies can be based on the most informative biological information available for species discrimination, such as a given gene target.
Seventh, any assay needs proper validation. Before venturing into species typing for a particular purpose and context, one has to prove that the assay can, with a certain degree of confidence, assign the correct species. This context can be defined geographically but is equally dependent on the kind of sample involved, e.g., samples from humans, animal reservoirs, or vectors, where different Leishmania and non-Leishmania parasite species may be encountered. Validation is a critical point in any assay development and implementation but is often overlooked, especially for in-house assays. It requires testing the technology with all parasite variants that are circulating, both Leishmania and non-Leishmania parasites. Participation in an external quality control program can be beneficial in this regard. Validation can be highly simplified by selecting a standardized test that has already been evaluated extensively in various settings. In this respect, using a well-established assay is generally preferred over use of an in-house test.
As illustrated by various examples below, only a minority of currently available species discrimination assays comply with these, in our view, logical test requirements.
Species Typing Assays
Many methods have been described for species typing of Leishmania in clinical, environmental, or cultured samples. The following sections give an overview of the different methods and targets currently available, with an emphasis on assays applicable to clinical samples in a large geographical area and on those that are widely used. Some sections deal with a particular method, and others report on the biological target, whichever is more convenient for clarity. Undoubtedly, the list is not exhaustive, and other locally deployed methods can be found in the literature. Nevertheless, the techniques that best abide by the criteria listed above are included. For each method, an appreciation is given of the pros and cons of the technique, and Table 1 presents a comparative overview of the best-validated and most used tests.
TABLE 1.
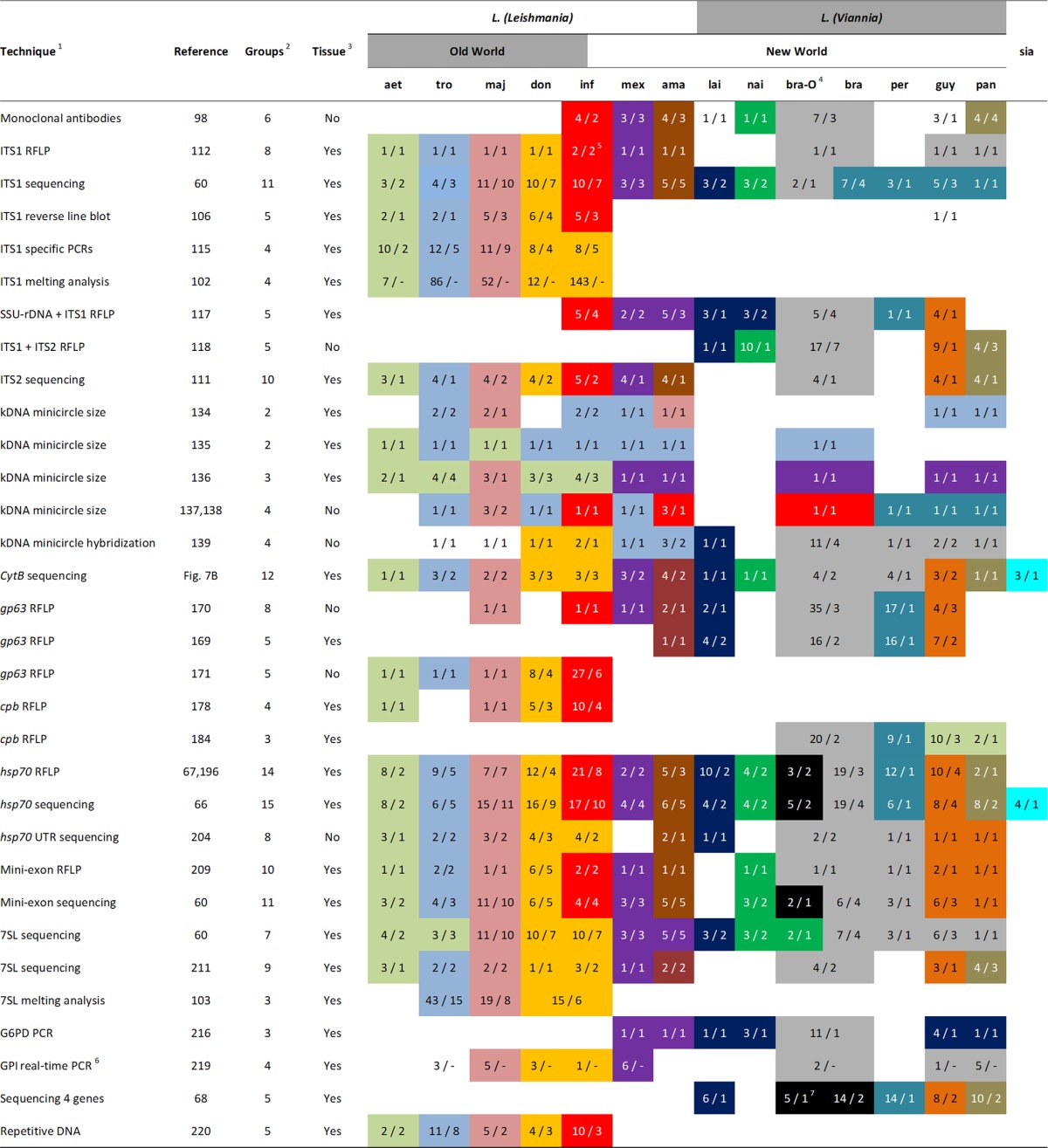
For each assay, groups of species that could be discriminated are indicated by different color patterns, and those not shaded could not be identified. The table is indicative, as each assay was tested on a different set of strains. Species abbreviations: aet, L. aethiopica; tro, L. tropica (includes L. killicki); maj, L. major; don, L. donovani (includes L. archibaldi); inf, L. infantum (includes L. chagasi); mex, L. mexicana (excludes L. pifanoi); ama, L. amazonensis (includes L. garnhami, excludes L. pifanoi); lai, L. lainsoni; nai, L. naiffi; bra-O, L. braziliensis outliers (Fig. 2); bra, L. braziliensis; per, L. peruviana; guy, L. guyanensis (includes L. shawi); pan, L. panamensis; sia, L. siamensis. For each assay and species, the first number indicates the number of type strains tested, and the second number gives the number of different countries from which these originated, giving an indication of the intraspecies variability that was evaluated. The number of countries of origin may sometimes be slightly higher, as it was not always mentioned for each strain in the publications.
1, Techniques are listed in the order that they appear in the text. 2, Number of species or groups of species that could be discriminated. 3, Whether the method, to our knowledge, was used on sand fly or clinical samples. Application in such samples was not necessarily done in the referenced paper and refers to application of the PCR assay rather than the downstream PCR analysis. 4, Most studies did not recognize L. braziliensis outliers (Fig. 2) as a separate entity. In such cases, the columns were taken together to represent all L. braziliensis strains as one category. 5, In silico analysis showed that L. infantum could not be distinguished as a separate group when intraspecies variability was taken into account (see “rDNA array” in the text). 6, Countries where the reference strains originated were not mentioned in the referenced paper. 7, Tsukayama et al. (68) described atypical L. braziliensis strains, but these might be different from the L. braziliensis outliers shown in Fig. 2.
MLEE
MLEE is still considered by many (including the World Health Organization [65]) to be the gold standard in parasite typing, even though the method is cumbersome, time-consuming, only applicable to cultured parasites, and exclusively applied in a few labs across the globe. Because it has been the reference test for so long, it is the only technique that has been evaluated for almost all currently identified Leishmania species.
MLEE entails a biochemical characterization based on the pH-dependent electrophoretic mobility of a predefined set of proteins (usually around 10 to 15) in a gel. The combined pattern of all these proteins constitutes a so-called zymodeme, which serves as the basis for species assignment as well as for classification below the species level (30, 55, 58, 86–88). Nevertheless, different labs make use of other enzymes, leading to several identification systems, such as the MON (Montpellier, France) (26), LON (London, United Kingdom) (89), and IOC (Instituto Oswaldo Cruz, Rio de Janeiro, Brazil) (90) classifications. As only a few labs nowadays persist in performing these analyses, and the method relies on parasite isolation and culture, MLEE is not suitable as a typing method for everyday use (91).
Other Culture-Dependent Methods
Several authors have described the use of monoclonal antibodies to identify Leishmania species of the Leishmania (Viannia) (92–95) or Leishmania (Leishmania) (96, 97) subgenus, or both (62, 98–100). These antibodies to some extent specifically recognize cultured promastigotes of different species, or they are genus specific. As such, all assays require parasite isolation and are unfit for analyzing clinical or environmental samples. Grimaldi and McMahon-Pratt (98) described the most extensive validation on nearly all New World species, of both the Leishmania (Leishmania) and Leishmania (Viannia) subgenera, whereby most species could be discriminated (Table 1). No extensive antibody panel has been designed for identifying Old World species. At present, antibodies are rarely used, as more reliable PCR techniques have become available.
Recently, mass spectrometry was suggested for typing of Leishmania parasites (23, 101), providing a promising means of rapid identification to the species level. Nevertheless, it is suitable only for identifying cultured parasites, and spectral data are not comparable across different laboratories. Therefore, each center must develop its own validated library of reference spectra.
PCR-Based Methods
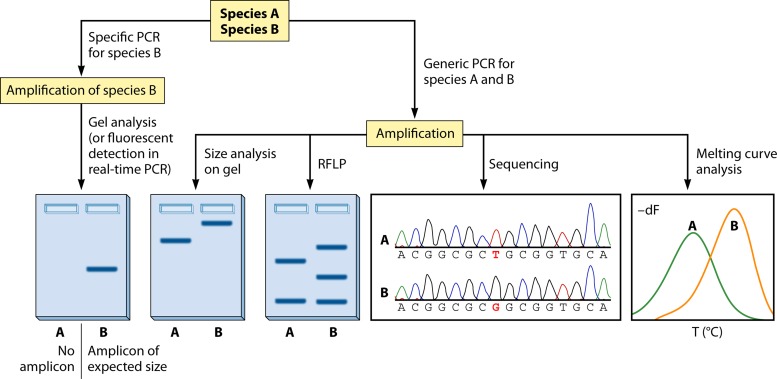
The vast majority of methods for species typing, in either a clinical study or an epidemiological, environmental, experimental, or patient management context, involve the use of PCR. Because PCR allows a massive specific amplification of Leishmania DNA, it is applicable directly on clinical samples, without the need for parasite isolation. The technique uses either a generic PCR that amplifies any Leishmania species or a specific PCR that amplifies a single or multiple species, species complexes, or subgenera (Fig. 3). In using a specific PCR, detection of the PCR product can be achieved either on a conventional agarose gel or in a real-time PCR format using fluorescence detection. In case of a generic PCR, downstream analysis is required in order to determine the parasite species on the basis of size or sequence information in the PCR amplicon.
FIG 3.
Schematic overview of common techniques to discriminate between two species (A and B). See “PCR-Based Methods” for details.
The most widely accessible and most used sequence-dependent technique is RFLP analysis, whereby the PCR product is digested with one or several restriction endonucleases. Depending on the presence or absence of the enzyme's recognition site, differently sized DNA fragments are generated. Gel-based analysis of the resulting DNA fragment mixture subsequently allows classification of the parasite at hand (Fig. 3). As RFLP is a simple technique that requires minimal lab infrastructure, it is available in each lab where PCR can be done. A more informative and equally straightforward method is sequence analysis of the PCR amplicon. By identification of single-nucleotide polymorphisms (SNPs) or comparison of the obtained sequence with available reference sequences in a dendrogram, such as in Fig. 2, the species can be determined. It should be noted that dendrograms are also constructed for studying evolution, but these analyses require mathematical models that may differ from those applied for typing.
Other techniques have been described but have rarely been used for everyday applications outside the lab where they were developed. Such techniques entail the use of probe hybridization and melting curve assays, in various formats. For example, some authors have described species-specific melting points of double-stranded DNA fragments or bound oligonucleotide probes (Fig. 3) (102–105). Nasereddin et al. (106) used a reverse line blot assay on a conventional membrane, whereby the similarity between the PCR amplicon and species-specific oligonucleotides was assessed.
The following sections deal with various targets that have been used in PCR-based technology. Some of the enzyme-encoding loci had previously been included in MLEE analysis, and DNA sequence-based approaches were initially developed to study the mutations underlying different zymodemes.
rDNA Array
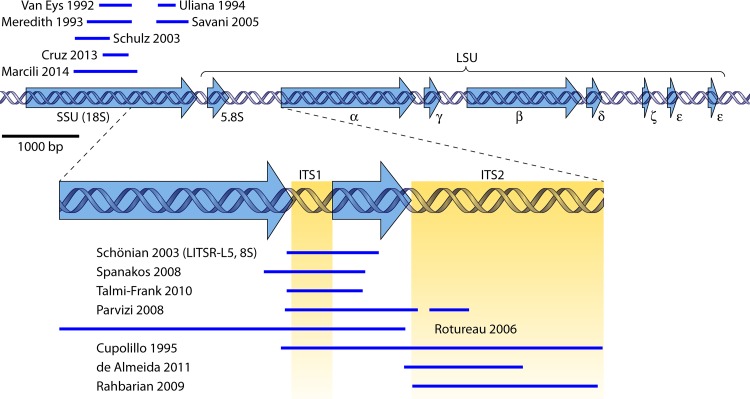
As detailed in Currently Applied Methods, below, the ribosomal DNA (rDNA) array is one of the most popular species typing targets. In Leishmania, an estimated 10 to 20 copies are tandemly repeated per haploid genome, rendering sufficient sensitivity for analyzing clinical sample DNA. Each repeat unit consists of several genes and spacers (Fig. 4) (107, 108).
FIG 4.
rRNA gene array of L. major strain MHOM/IL/81/Friedlin. The sequence and annotations were taken from www.tritrypdb.org (LmjF chromosome 27, nucleotides 989640 to 998595; accessed on 10 August 2014). Arrows in blue indicate the 5′-to-3′ direction of the genes. The lower panel represents a more detailed view of the most relevant fragment for species typing. Regions used by different authors are indicated above (top) or below (bottom) each panel (24, 77, 102, 111, 112, 114, 116–118, 120–124, 126). The scale of the upper panel is given on the left. SSU, small ribosomal subunit rRNA gene (18S rRNA); LSU, large ribosomal subunit rRNA gene; ITS, internal transcribed spacer.
Within the array, primarily the spacer regions contain sufficient variability for species discrimination, even though they have also been used to discriminate isolates at the subspecies level (57, 109, 110). Some minor sequence variation between the different copies has been observed, which occasionally complicates interpretation of sequence reads (60, 111). It is not clear, however, whether these differences are biological or caused by enzymes in PCR or sequencing reaction mixtures. The following paragraphs give an overview of the fragments and techniques that have been described (Fig. 4).
(i) LITSR-L5,8S.
The primers for the LITSR-L5,8S assay were originally reported by el Tai et al. (109). They were applied for species discrimination by RFLP analysis with the enzyme HaeIII by Schönian et al. (112), who evaluated the technique in clinical samples. It allowed separation of species from the Leishmania (Leishmania) subgenus but did not discriminate Leishmania (Viannia) species (77, 113). The authors evaluated their method using only one type strain of each investigated species (Table 1), thereby overlooking possible intraspecies variation that could lead to erroneous results.
The amplified fragment was also used for identification by sequencing, allowing a better assessment of the discrimination power obtained with this target. The most extensive analysis was recently carried out by Van der Auwera et al. (60), who evaluated the sequences of a globally representative panel of all medically relevant species (Table 1). Their findings corroborated the RFLP analysis in that the complete sequences could also best discriminate the Leishmania (Leishmania) subgenus. The sequence analysis included the additional species L. naiffi, L. lainsoni, and L. braziliensis outliers (Fig. 2), also allowing their identification (Table 1).
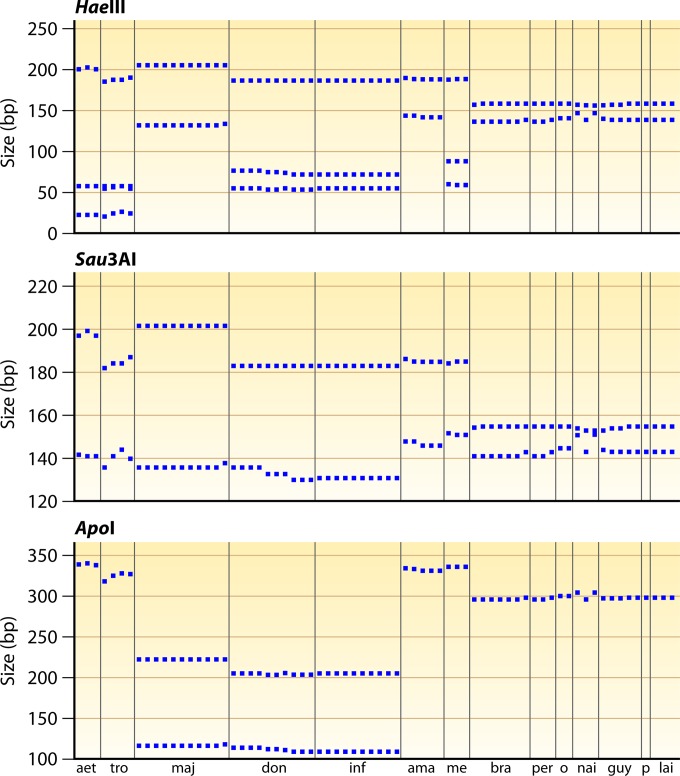
As sequence variability was observed in nearly all distinguished groups, the question of whether the observed sequence variation hampers correct identification by RFLP analysis using HaeIII remained. To assess this, we undertook an in silico RFLP analysis of the data set of Van der Auwera et al. (60), and the results are depicted in Fig. 5. This largely confirmed the previously published RFLP data (112), even though the separation of L. donovani and L. infantum was no longer possible. This was also observed by Cruz et al. (77) and is caused by the higher level of coverage of intraspecies variation.
FIG 5.
In silico RFLP analysis of the ITS1 sequences of Van der Auwera et al. (60), covering the LITSR-L5,8S fragment used by Schönian et al. (Fig. 4) (112). For each of the 3 enzymes, the expected fragments are plotted as a function of size in base pairs, whereby each data point on the abscissa represents a different strain. Species abbreviations: aet, L. aethiopica; tro, L. tropica; maj, L. major; don, L. donovani; inf, L. infantum; ama, L. amazonensis; me, L. mexicana; bra, L. braziliensis; per, L. peruviana; o, L. braziliensis outliers (Fig. 2); nai, L. naiffi; guy, L. guyanensis; p, L. panamensis; lai, L. lainsoni.
Tojal da Silva et al. (30) proposed using Sau3AI for RFLP analysis of this target to allow separation of species of the Leishmania (Viannia) subgenus. Nevertheless, in silico analysis of the global data set of Van der Auwera et al. (60) disproved the possibility of identifying the individual species or species complexes (Fig. 5). Spanakos et al. (114) used an alternative fragment also covering the ITS1 region (Fig. 4), but with ApoI as the restriction endonuclease. Figure 5 includes an in silico analysis of the LITSR-L5,8S fragment digested with ApoI, which was less efficient than HaeIII and Sau3AI at discriminating Leishmania (Leishmania) species.
Besides sequencing and RFLP analysis, a reverse line blot hybridization assay was developed based on the LITSR-L5,8S fragment (106). In this setup, genus- and species-specific probes are immobilized on a membrane, and reactivity of each of these probes against the PCR product is scored visually. This assay was evaluated on Old World Leishmania parasites by use of a few type strains of each species and was able to discriminate all five species (Table 1). It was also evaluated on L. tropica and L. major samples from patients in Israel, where a perfect correlation with RFLP analysis was demonstrated. Even though hybridization experiments in general are difficult to standardize, the assay uses the probes for all species simultaneously, and therefore a relative measure of hybridization to the various species-specific probes is obtained for each sample, rendering the assay quite robust. In addition, the sensitivity was significantly higher than that of PCR, which is explained by the additional colorimetric signal amplification. With the genus-specific probes, some nonspecific results were observed, but this did not interfere with correct species typing. Despite its promising characteristics, the method has, to our knowledge, only been used in the lab where it was developed.
(ii) Species-specific PCRs.
Odiwuor et al. (115) developed a set of species-specific PCRs to discriminate the Old World Leishmania complexes. These PCRs are nested within the LITSR-L5,8S fragment described above, which opens possibilities for nested PCR to increase the sensitivity. As Odiwuor et al. pointed out, the 4 species-specific PCRs are best used in parallel to avoid misclassification due to cross-reactions, which would be apparent if a sample is positive for more than one species at a time. This strategy was also used in the above-mentioned hybridization assay (106). Species-specific PCRs need proper validation due to their dependence on minor technical variations, but the authors in this case showed that the technique is transferable to another lab with minimal effort. The assays were validated on various reference strains from each species and from different geographic origins and were able to identify three species and the L. donovani complex (Table 1). As for the hybridization assays, these species-specific PCRs were also used only in the labs where they were originally developed.
(iii) High-resolution melting analysis.
Talmi-Frank et al. (102) developed a melting assay based on a 265- to 288-bp fragment within ITS1 (Fig. 4). After PCR, the temperature at which both DNA strands separate is recorded with high resolution, and this was found to be species specific. As with RFLP analysis, the Old World species could all be distinguished, with the exception of L. infantum from L. donovani. The assay was tested on a large panel of isolates from different geographic origins (Table 1) but so far has not been used outside the lab where it was optimized.
(iv) Other ITS1 typing methods.
Spanakos et al. (114) described the use of sequencing, RFLP, and single-strand conformation polymorphisms for species discrimination in the Old World, based on an ITS1 fragment that largely overlaps the LITSR-L5,8S target (Fig. 4). These methods showed the same species discrimination as that obtained with the LITSR-L5,8S region but were not as extensively validated. Other examples based on ITS1 sequencing include the work of Parvizi and Ready (116), who used a PCR region covering the LITSR-L5,8S target. Their fragment extended into the 5.8S rRNA gene (Fig. 4), but this did not show any advantage. Rotureau et al. (117) further extended the analyzed fragment into the small-subunit (SSU) rRNA to achieve typing in the New World by RFLP analysis (Fig. 4), but this strategy also failed to discriminate all species, or even all complexes (Table 1).
(v) ITS2 typing.
In contrast to the above-described ITS1-based methods, Cupolillo et al. (118) could discriminate all tested species of the Leishmania (Viannia) subgenus by PCR-RFLP analysis of a fragment covering both ITS1 and ITS2 (Fig. 4; Table 1). However, 10 enzymes were needed, while sequencing of this fragment was found to be complicated because of microsatellite regions in ITS2 (116). Davila and Momen (119) separated most Leishmania (Leishmania) species on the basis of ITS1-5.8S-ITS2 rDNA sequences, but this showed no added value compared to the use of the LITSR-L5,8S fragment.
Some authors have used exclusively the ITS2 region for typing purposes (Fig. 4). A quite recently published ITS2-based assay was performed by de Almeida et al. (111). They compared typing results from single-nucleotide polymorphism analysis and MLEE, which proved largely congruent, and they could separate the three tested Leishmania (Viannia) species (Table 1). Other authors (116, 120) identified Old World species based on the size or sequence of various ITS2 fragments (Fig. 4), but these assays were not tested on a variety of type strains. Because for the Leishmania (Leishmania) subgenus ITS2 does not show any advantage over ITS1 and has been less used and evaluated, its potential lies in complementing ITS1 for discriminating species of the Leishmania (Viannia) subgenus, even though a more extensive validation is required.
(vi) SSU rDNA.
The 18S or SSU rDNA has limited use for discriminating Leishmania species due to its conserved nature within the genus. Single-nucleotide polymorphisms have been exploited to identify the L. mexicana and L. donovani complexes and the Leishmania (Viannia) subgenus. Methods (Fig. 4) included probe hybridization (121, 122), specific PCRs (123), RFLP analysis (124), and sequencing (24, 77, 124–126). The SSU rDNA region, however, has many merits for first-line diagnostic purposes that need to detect the genus (e.g., see references 124, 127, and 128).
kDNA
Minicircles.
Several kinetoplast DNA (kDNA) assays have been described for both the Old World and the New World. In contrast to most other targets used for species typing, the minicircles are not encoded on the parasite's chromosomes but are part of a dense extrachromosomal DNA network called the kinetoplast. This organelle contains the mitochondrial DNA and is found at the base of the flagellum in the order Kinetoplastida to which Trypanosoma also belongs (129). The kinetoplast is composed of mini- and maxicircles.
Minicircles are small circular molecules of around 800 bp, containing a conserved region of 120 nucleotides and a highly variable region coding for guide RNAs involved in editing maxicircle genes (130). The general advantage of using minicircles in diagnostics is their high copy number (10,000 copies per parasite), which results in a high sensitivity of assays based on this target. However, minicircles within one strain are not all identical, and they can be divided into minicircle classes (131–133). Because this variability complicates sequencing, most assays for identifying Leishmania subgenera and species are based on size discrimination, hybridization with specific probes, or species-specific PCRs.
(i) Size discrimination.
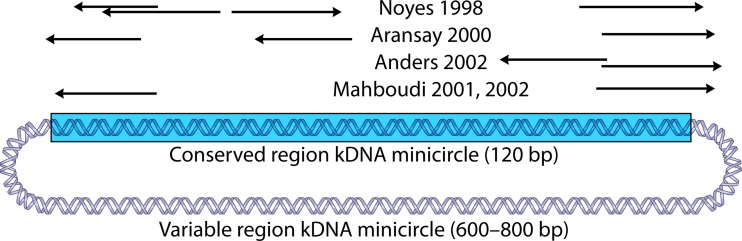
Several authors have used size differences of the variable region as a basis for typing of species in the Old World (Fig. 6). The assay of Noyes et al. (134) is widely used, together with those of Aransay et al. (135) and Anders et al. (136). These PCR assays amplify the variable part of the Leishmania minicircles (Fig. 6), but many species have minicircles of the same size (Table 1). In all studies, however, L. major can be separated from L. tropica-L. infantum-L. donovani. Whether L. aethiopica can be distinguished based on size alone remains to be established. Aransay et al. (135) complemented size analysis with sequencing of the obtained PCR products, but given the variability of minicircles in a single parasite, and even more so within all strains of the same species, sequence analysis of the variable region is difficult.
FIG 6.
Primers used for species discrimination by size differences of the variable region of kDNA minicircles (134–138). The primers are mapped to a composite sequence obtained from GenBank accession numbers AF308685 and JF831926 (L. major). The arrows indicate the 5′-to-3′ orientation. The variable region is not drawn to scale.
Other authors used different primers essentially amplifying the same region (Fig. 6), and included a more extensive panel of New World isolates (137, 138), but, again, only some groups could be separated (Table 1). Even though all PCR primers used in these studies can be mapped to the same minicircle sequence (Fig. 6), it is striking that not all authors could discriminate the same groups on the basis of size (Table 1). This can be explained by the fact that different strains and gel resolutions were tested in each analysis, but it may also be due to amplification of other minicircle classes in different PCRs, as the primers do not have perfect sequence identity with the depicted L. major minicircle. Hence, no standardized assays are yet available. In short, kDNA size variation has no global use for separating isolates to the species level and can only be applied in a regional context when certain groups of species need to be discriminated.
(ii) Hybridization.
Both partial and complete minicircles and oligonucleotide probes have been used in hybridization assays, either to discriminate the Leishmania (Leishmania) and Leishmania (Viannia) subgenera or to distinguish particular species (e.g., see references 132 and 139 to 143). Most assays rely on hybridization of PCR-amplified minicircle products, even though some have applied the method directly on clinical samples (141), on pelleted Leishmania cells (140), or on total Leishmania DNA (142). Only the assay of Brenière et al. (139) has been tested on a larger panel of New World parasites and could separate some species complexes (Table 1). The probes of Fernandes et al. (140) could identify the subgenus Leishmania (Viannia) but were tested on only 8 strains representing 7 species. Nevertheless, they were used by several authors without further validation (e.g., see references 144 and 145). None of the available minicircle hybridization assays has been validated properly to allow separation of all species in either the Old or the New World. Assays often discriminate only to the subgenus level or, at best, identify a few species or species complexes.
(iii) Species-specific PCRs.
Sequence polymorphisms in the minicircle kDNA have been exploited to design PCR primers that specifically amplify a species or group of species (e.g., see references 104, 123, 146, and 147). Only the specific primers of Meredith et al. (123) discriminated 4 Old World species, but that assay was validated on one strain of each species only. Several authors have observed cross-reactions between different species (104, 147), which is probably caused by the use of slightly different reaction conditions across labs and the variability of the minicircle population in each species. De Bruijn and Barker (148) reported a widely used Leishmania (Viannia)-specific PCR, validated on one strain each of 10 species, but cross-reaction of one primer with human and mouse DNAs was found (149), which could lead to false-positive results. Also, Lopez et al. (150) reported a PCR that amplifies several Leishmania (Viannia) species but which was evaluated on only 5 strains.
(iv) RFLP analysis.
Several reports have described RFLP analysis of the variable minicircle region as an aid in species typing. The obtained fragment patterns form the basis of so-called schizodeme identification (132, 134, 151). In our experience, such patterns are extremely difficult to reproduce, even within the same lab (152), and they have more use in strain tracking or studying intraspecies variability (e.g., see references 62, 153, and 154). Volpini et al. (155) described an RFLP-based species discrimination test based on the conserved region of the minicircles, but only to discriminate L. braziliensis from L. amazonensis. Rocha et al. (147) further evaluated this RFLP method and demonstrated that L. lainsoni and L. infantum could also be identified.
(v) Melting curve analysis.
Nicolas et al. (156) used differences in melting temperature of PCR-amplified variable and conserved minicircle regions to differentiate some species in the Old World. Their assay was validated on only one or a few type strains from four species and could not separate L. tropica from L. donovani. The conserved region allowed Pita-Pereira et al. (157) to differentiate a few type strains of Leishmania (Leishmania) from those of Leishmania (Viannia), but species identification was not possible. Weirather et al. (104) evaluated several primer sets that could discriminate groups of species.
(vi) LAMP.
Even though it is not a PCR procedure, loop-mediated isothermal amplification (LAMP) also constitutes an amplification technology (158). Unlike PCR, it does not require a thermocycler, and the amplified product is visualized in the reaction tube and seen by the naked eye, not requiring gels or fluorescence detection. Even though the technique has been used primarily in first-line diagnostics for parasite detection (159, 160), an L. donovani-specific assay based on kDNA minicircles was developed (161, 162). It was tested on only a few type strains for some species, and hence it currently has limited validity.
In conclusion, minicircle kDNA assays show poor performance for separating Leishmania species, mainly due to the lack of proper validation and the problem of extensive variability within single strains, and even more so within an entire species. Current methods are not standardized and not usable on a global scale for discriminating to the complex or species level. kDNA minicircle methods seem to be restricted primarily to separating the Leishmania (Viannia) and Leishmania (Leishmania) subgenera, and some assays can be used in a regional context for discriminating between species.
Cytochrome b (maxicircles).
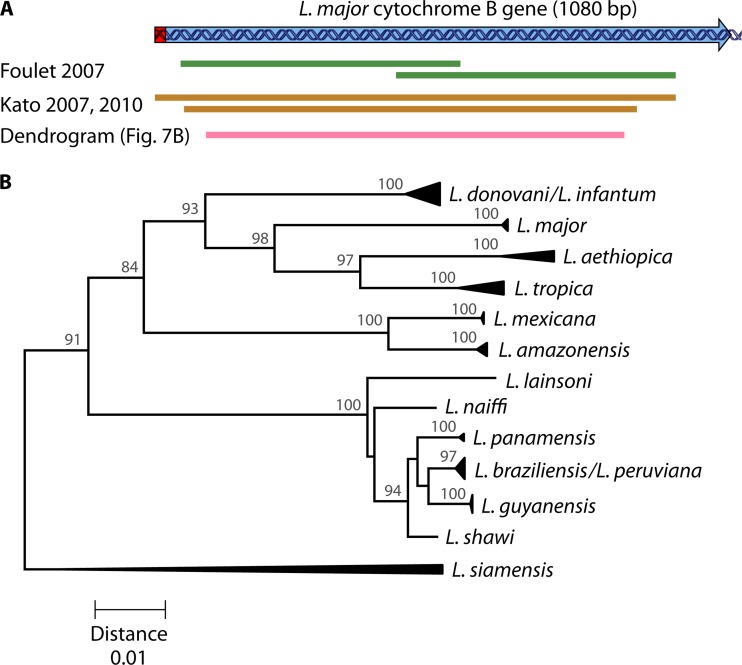
The Leishmania cytochrome b gene (cytB) is encoded on the kDNA maxicircles, of which an estimated 25 to 50 copies are present in each cell, making it a sensitive target for analysis of clinical samples without the need for culturing. Sequencing of the cytB coding region has successfully separated most tested species (82, 163), and PCRs were applied directly to clinical and sand fly samples (Fig. 7A) (164–167). Figure 7B shows a dendrogram for a fragment covered by these PCRs, by which most species can be discriminated (Table 1). L. donovani and L. infantum cannot be separated, nor can L. braziliensis and L. peruviana. L. guyanensis can be separated from L. shawi. Sequence analysis of the gene is straightforward, as no size differences were observed, which facilitates sequence alignment. Some cytB real-time PCRs were evaluated by Weirather et al. (104), but they could identify only L. tropica.
FIG 7.
(A) Cytochrome b coding sequence of L. major (GenBank accession number AB095961) (163). The editing region is shown in red, and the arrow indicates the sense direction. The two overlapping fragments sequenced for typing of clinical samples, covering 887 bp in total, are depicted in green (164). Other fragments amplified for typing of both clinical samples and sand flies are shown in brown, the smallest of which (817 bp) is sequenced (165). The larger brown fragment is the outer PCR amplicon in a nested PCR approach needed for human sample analysis (166). (B) Dendrogram constructed on the basis of cytochrome b gene sequences of reference isolates reported by Luyo-Acero et al. (163), Asato et al. (82), Foulet et al. (164), and Leelayova et al. (22), complemented with the sequences under GenBank accession numbers AB433279, AB433280, AB433282, AB566382, AB566381, and AB566380. The analyzed fragment is indicated in panel A (pink fragment). The dendrogram was constructed by the neighbor-joining method and is based on uncorrected p-distances (the scale is shown below, in substitutions per nucleotide). Bootstrap values from a 2,000-replicate analysis are depicted in percentages at the internodes, when higher than 70%. The dendrogram was constructed with the software package MEGA5 (253).
cytB is one of the few genes of L. siamensis that have been sequenced, and the species is readily identified from sequence dendrograms (Fig. 7B) (22). In all, cytB is a good typing target, rendering a good resolution for all tested species across the globe, is sensitive enough for use on clinical and environmental samples, and is easily analyzed. Sequence validation on a more extended strain panel would further increase its reliability.
Antigen Genes
GP63.
The metalloprotease glycoprotein 63 (GP63) is the major surface glycoprotein of Leishmania and is considered an important virulence factor and a strong immunogen (168). The gene is arranged as a tandemly repeated unit, and both the intra- and intergenic regions have been used for typing by RFLP analysis. Because of its multicopy nature, assays based on this gene array have been found to be sensitive enough for application directly on clinical samples, without the need for culturing. One report describes a sensitivity of 85% compared with that of kDNA minicircle amplification (169).
For the New World, different fragments were evaluated (169, 170), whereby all tested species could be separated (Table 1). For the Old World, intra- and intergenic gp63 regions have been used primarily for looking at intraspecies variability (171–174), even though all species could be distinguished (Table 1). Nevertheless, a proper validation with several strains of each species is lacking. Although RFLP analysis seems to be successful for species identification, the obtained patterns can be complicated and difficult to interpret because of so-called isogenes. These are variations between copies within the same parasite (169, 175), which also render sequencing of the gp63 target impractical. Also, because antigen genes are under constant pressure from the immune system (176, 177), they are primarily suited for exploring clinical pleomorphism at the intraspecies level rather than for typing species.
CPB.
Cysteine proteinase B (CPB) is another antigenic protein, and several copies of the gene are present in the Leishmania genome, arranged in a tandem array (178). The copies are not all identical and are classified into various subgroups (179–182). Like GP63, it is an important factor in the host-parasite relationship (183) and is therefore especially fit for looking at population structure in a clinical context.
Both the coding sequence and the intergenic regions between the gene copies have been used for typing purposes, primarily using PCR-RFLP approaches to distinguish strains below the species level (e.g., see references 174, 178, and 184). Quispe-Tintaya et al. (178) included in a study of the L. donovani species complex a few outgroups that resulted in discriminant RFLP patterns, but L. tropica was not tested, nor was the intraspecies variability in L. aethiopica and L. major (Table 1). PCR-RFLP analysis was also used in the New World (184), but not all species could be amplified, and only L. braziliensis, L. peruviana, and the L. guyanensis complex were identified (Table 1). These PCR-RFLP assays have been used on clinical samples, without culturing (64, 185). In summary, however, none of the cpb PCR-RFLP methods have been well validated for species typing. Because the cpb array is made up of nonidentical isogenes (178), RFLP patterns are often complicated and vary within the same species.
Some authors have used species-specific cpb PCRs that amplify one or a few species (179, 181, 186). Lopez et al. (187) developed a more general assay, based on specific PCR amplification of certain groups followed by RFLP analysis to discriminate species within each group, but the assay was developed in silico and was not validated on type strains or compared with other methods. Given the complexity of the cpb locus, this is a major flaw. Because all these specific assays target only one particular copy of the cpb gene array, their sensitivity is reduced, and the PCRs have been applied only for typing of cultured parasites, not directly on environmental or clinical material. A more sensitive LAMP assay was developed by Chaouch et al. (188) for the detection of the L. donovani complex, but its specificity was validated on a limited set of type strains.
HSP70.
Heat shock protein 70 (HSP70) is also a Leishmania antigen, playing a role as a molecular chaperone in protein folding and transport (189). Between 5 and 10 copies of the gene are present in the Leishmania genome (190), and minor differences between them may exist in a particular strain (66). Fraga et al. (191) studied Leishmania evolution based on hsp70. Because many studies have illustrated the value of the heat shock protein 70 gene for species discrimination, it has been used by various researchers (9, 22, 64, 192, 193).
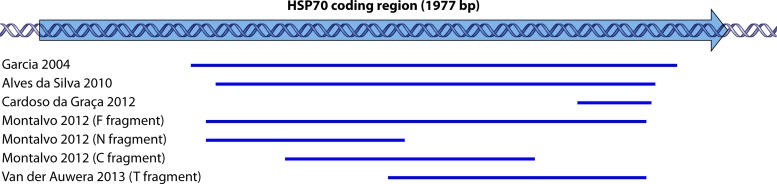
Garcia et al. (Fig. 8) (194) were the first to exploit the variability of the gene for species discrimination in the Leishmania (Viannia) subgenus on the basis of RFLP analysis. Several authors (67, 195, 196) increased the number of restriction enzymes to allow typing of all Leishmania species, irrespective of origin or subgenus (Table 1). Alves da Silva et al. (Fig. 8) (197) evaluated the target on all species circulating in Brazil, using both RFLP analysis and sequencing. This study was further complemented by Van der Auwera et al. (60, 66), who reported over 200 sequences from across the globe and from all medically relevant species (Table 1). All studies showed a nearly perfect congruence with MLEE typing, which makes hsp70 the most widely validated species typing target to date.
FIG 8.
Mapping of hsp70 fragments used in different studies to the complete coding region of the HSP70 gene (GenBank accession number XM001684512.1) (the arrow indicates the sense direction). These regions were used in either RFLP or sequence analysis (66, 194, 196, 197, 200).
Even though the original PCR of Garcia et al. (194) was used on both clinical and sand fly samples (198, 199), more sensitive PCRs have been developed, some of which amplify shorter fragments (Fig. 8). These were applied in Brazil (200), Peru (201), and several Old World countries (202) and on various clinical sample types. The sensitivities of these assays have not been compared systematically to those of other species typing PCRs but were generally found to be 60 to 90% compared to those of higher-copy-number targets, such as rRNA genes or kDNA minicircles, which are used for diagnostic Leishmania genus detection.
Compared to some other targets, hsp70 combines the possibility of accurate typing to the species level over the entire genus with the ease of sequence or RFLP analysis, as hardly any size variation is observed and almost no intraspecies variation is present in currently used RFLP assays. Some authors have developed species-specific PCRs (203). Finally, Requena et al. (204) used the 3′-untranslated region of the gene to distinguish species complexes based on size differences, RFLP analysis, or sequencing (Table 1), but this was not tested on clinical samples.
Miniexon
The miniexon or spliced leader (Fig. 9) is tandemly repeated in the Leishmania genome. The exon encodes an RNA that is added to the 5′ end of each protein-encoding RNA during maturation. Even though the exon and intron are conserved in all Leishmania species, nontranscribed spacers are variable in both size and sequence (205). Around 100 to 200 copies are present in each genome, which makes the target sufficiently sensitive for analyzing tissue samples (e.g., see references 113 and 206 to 208).
FIG 9.
Miniexon repeat of L. major (GenBank accession number X69449). The nontranscribed spacer varies in length and sequence between species, from 51 to 341 bp (209). The PCR product amplified in the assay of Marfurt et al. (209) is indicated, whereby primers are shown with arrows in the 5′-to-3′ direction. All regions are drawn to scale.
Even though some species groups can be discriminated based on size only (60, 132, 147), RFLP analysis is needed for discriminating species. The PCR (Fig. 9) and RFLP scheme of Marfurt et al. (209) was validated on 12 species, 8 of which could be typed to the species level, with the remaining 4 identified to the complex level (Table 1). Nevertheless, RFLP patterns varied within the same species, and up to 5 restriction endonucleases were required. The assay is therefore quite complicated and can be cumbersome (113). In addition, most species were analyzed based on a single strain, which precludes assessment of the full range of intraspecies variability.
Recently, Van der Auwera et al. (60) carried out a sequence analysis of the same PCR-amplified region on a larger global panel of strains (Table 1). Based on that analysis, the miniexon could discriminate all species from both the Old and the New World, 4 of which were identified only to the complex level. Roelfsema et al. (113) found the sequencing approach to be more practical than RFLP analysis for typing of clinical samples. Nevertheless, sequencing of the miniexon was sometimes problematic due to differences between copies in the same genome and to sequence features, such as homopolymer stretches, causing technical difficulties (60).
7SL-RNA
The 7SL-RNA is an RNA molecule of 250 to 300 nucleotides that plays a role in the translocation process of proteins across the endoplasmic reticulum. Because of its variability and abundance, Zelazny et al. (210) proposed sequencing of a 140-bp PCR-amplified product for typing purposes. They tested the approach on 30 strains, and their analysis was complemented by Van der Auwera et al. (60), using a larger panel of 71 isolates (Table 1). These sequences allowed separation of species complexes rather than individual species, except for L. major and L. lainsoni, which could be typed individually.
Stevenson et al. (211) extended the region to obtain a 385-bp fragment allowing better discrimination for both subgenera, even though this improved method was validated on fewer strains (Table 1). Nasereddin and Jaffe (103) designed a high-resolution melting assay for the Old World by using a 119-bp fragment of the gene. The assay was tested on a large panel of strains and could differentiate them to the complex level, but L. aethiopica was not evaluated (Table 1). Both methods have been used on environmental and clinical samples (212–214).
Carbohydrate Metabolism Enzymes
Glucose-6-phosphate dehydrogenase (G6PDH or G6PD) has been exploited to discriminate between the subgenera Leishmania (Viannia) and Leishmania (Leishmania), and within Leishmania (Viannia) to distinguish the L. braziliensis complex, including L. peruviana (215, 216). These conventional and real-time specific PCR assays were validated mainly on Brazilian strains, and intraspecies variability was only moderately taken into account (Table 1). They therefore have limited general use. Since they are based on single-copy genes, their sensitivity is lower than that of other methods based on multicopy targets, such as ITS1, hsp70, and kDNA minicircles (200), but they have been applied to clinical specimens (30).
Mannose-phosphate isomerase (MPI) has been used in PCR assays to separate L. peruviana from L. braziliensis, either with a specific PCR (217) or by RFLP analysis (167), even though it is unclear whether the latter was checked for intraspecies variability. These assays can thus be deployed as a second line of testing when the L. braziliensis species complex is identified by G6PDH analysis, and they have been applied to clinical samples (64). Another enzyme downstream in the glycolysis pathway, 6-phosphogluconate dehydrogenase (6PGDH or 6PGD), was also exploited for Leishmania typing (5, 218), but only a few Old World species were tested.
Glucose phosphate isomerase (GPI) was applied to discriminate the Leishmania (Viannia) subgenus and some species complexes in the Leishmania (Leishmania) subgenus (219). Real-time PCR could be used on clinical samples and was based on specific amplification. It was not evaluated on all species, however, and whether intraspecies variability was properly evaluated is not clear (Table 1). In addition, Weirather et al. (104) used the target in an evaluation of real-time PCR and melting assays.
A combination of carbohydrate metabolism genes was deployed by Tsukayama et al. (68), who combined 6PGD, MPI, and glucose-6-phosphate isomerase with malate dehydrogenase to identify species of the Leishmania (Viannia) subgenus (Table 1). Single-nucleotide polymorphisms were found for each of the species, but other authors have questioned their general validity (72). They further based two real-time PCRs on melting analysis of fluorescent probes for typing of clinical samples, using mpi and 6pgd. Their assay was evaluated on an extensive reference panel of 64 strains, almost exclusively from Peru. Validation on more strains from other countries would further increase the validity of this technique.
Miscellaneous Targets
Some additional assays have been applied to clinical samples. Piarroux et al. (220) separated all Old World species on the basis of a 222-bp repetitive DNA sequence. This assay was validated on a large sample set (Table 1) and has been applied to clinical samples by using sequence analysis and RFLP analysis (221). Despite these promising results, the method has not been widely applied. Haouas et al. (222) designed a PCR-RFLP scheme based on the topoisomerase II gene for the Old World species, but only 3 reference strains were evaluated, and hence it was not properly validated. Mimori et al. (223) discriminated New World species on the basis of two subgenus- and five species-specific primer sets derived from sequenced randomly amplified polymorphic DNA (RAPD) fragments, but a limited number of strains were evaluated. Weirather et al. (104) designed real-time PCR and melting assays based on a set of genes and could discriminate several species when analyzed on a panel of reference strains. The assays were successfully tested in some clinical samples, but the authors presented no clear and straightforward strategy for general and global use.
Several authors have used a variety of other genomic targets for species typing purposes, but these were not further explored for analyzing clinical samples and were applied to reference strains and cultured isolates only. Strategies include histone 2B RFLP analysis (184); sequencing and RFLP analysis of N-acetylglucosamine-1-phosphate transferase (181, 224); PCR and RFLP analysis of hydrophilic acylated surface protein B, also known as K26 (9, 225); glycosomal glyceraldehyde 3-phosphate dehydrogenase sequencing (24); meta2 RFLP analysis (226); chitinase sequencing (27); sequence analysis of DNA and RNA polymerase genes (227); pteridine reductase 1 sequencing (228); β-tubulin analysis (229); heat shock protein 20 sequencing (230); and sequence analysis of a calmodulin intergenic spacer (231).
Multilocus Typing
Combining different genomic targets has an apparent advantage: one uses information from various loci to gather evidence for a certain species. Such an approach can use either a targeted strategy, whereby several predefined loci are characterized, or a nontargeted approach that randomly documents genome variation. These methods often have limited practical use for species typing, as they provide too much information, which is either redundant or better fit for population-based studies documenting intraspecies variation. When it comes to species typing, multilocus methods are primarily useful for discriminating species belonging to the same species complex, such as L. infantum-L. donovani (49–51), L. guyanensis-L. panamensis (69, 232), and L. braziliensis-L. peruviana (33, 47).
Widely used targeted approaches include MLEE, MLST, and multilocus satellite typing (MLMT). MLEE has already been discussed at length. For MLST, typically several single-copy housekeeping genes are sequenced, and the combined sequence set is used for typing (41, 56, 60, 68, 193). As such, MLST can be considered an upgraded version of MLEE, whereby each enzyme is evaluated by analyzing its coding sequence instead of on the basis of a single gel mobility assessment. Thereby variations that do not affect this mobility are also charted. Even though an MLST outcome is highly robust, the method has no immediate applicability for routine use but rather for establishing the genus's phylogeny (72). So far, it has not been applied to clinical samples: since it is based on mostly single-copy genes, the success rate is expected to be low because of the limited sensitivity. Also, routinely amplifying several genes in parallel and then sequencing them is too costly and time-consuming.
For MLMT (e.g., see references 27, 28, 50, and 233 to 237), several microsatellite regions across the genome are amplified. Microsatellites are repeat regions of di- or trinucleotides, and because the number of repeats is highly variable, the size of the microsatellites makes up a fingerprint pattern. This pattern is used as the basis for typing and studying populations. MLMT has been used on clinical samples (238), but the method requires several PCR amplifications and accurate size analysis. In addition, the method is mainly suited to study variability within one complex, as the markers evolve too rapidly for typing at the species complex level, and markers are complex dependent (28, 72).
RAPD and amplified fragment length polymorphism (AFLP) analyses are both nontargeted strategies. In RAPD analysis, PCR conditions allow permissive primers to hybridize to various nondefined genomic loci, which generates a fingerprint pattern used for species typing (e.g., see references 185 and 239). Due to its nature of permissive priming, the outcome is highly dependent on the exact PCR conditions, making the method unfit for standardization. Variants of the technique have been described for species discrimination, with one specific primer in combination with random priming (240). For AFLP analysis (241, 242), a selection of restriction endonuclease-generated genome fragments is amplified, again resulting in fingerprint patterns that allow discrimination of species (33, 52, 232, 243). However, untargeted methods, such as RAPD and AFLP analyses, are of no use for typing of clinical or environmental samples. As they rely on amplification of random DNA fragments, they are not Leishmania specific and would primarily amplify the abundant host DNA. Hence, they can be applied only to axenically cultured parasites.
Even though the use of multilocus approaches offers the theoretical advantage of sampling information across the genome, in practice single-locus markers render equally valid species typing results, except maybe in the case of simple binary assays (see “Interpreting Species Typing Results.” Van der Auwera et al. (60) compared typing outcomes from MLST with those from 4 single-locus methods and found hardly any inconsistencies. Despite the fact that sexual recombination most probably occurs in Leishmania, it does not seem to break the clonal population structure observed at the species level (45). As a result, typing from different targets is expected to yield consistent results. This is not the same as saying that typing can be achieved using any locus, as a good typing target needs to fulfill the criteria outlined in “Criteria for Species Typing Tools.”
Some authors have used combinations of the single-locus assays described above (e.g., see references 9, 22, 166, 181, 184, and 244). Not all such methods are true multilocus approaches, as they are sometimes deployed in a sequential manner. Veland et al. (64), for instance, discriminated various species based on a predefined algorithm incorporating mpi, cpb, and hsp70, but they used only one locus in each step. Once typing was achieved, the result was not confirmed with a second locus, which in fact makes the method a single-locus approach. In a true multilocus assay, all methods are used in parallel to confirm the detected species.
Detection of Interspecies Hybrids
One possible exception to the general rule that multilocus typing has few advantages over single-locus species determination lies in the identification of hybrid parasites (e.g., see references 30 to 36). Even though recent hybridization can be observed in many loci, alleles from one hybrid parent can be lost over time, and this risk is increased by the highly variable ploidy in natural populations (73). Nevertheless, several examples exist where hybrids were evidenced even in looking at a single locus. For instance, in natural L. donovani-L. aethiopica hybrids, both species alleles were present in all genes investigated, including ITS1 and hsp70 (32, 112), even though this was not recognized at the time of the ITS1 study, which classified the hybrid as an L. aethiopica variant. In an L. infantum-L. major hybrid, both genomes were present (34), enabling typing of the parasite based on a single-gene assay.
Other examples also exist: for instance, MHOM/PE/2003/LH2538, which was shown to be a hybrid by AFLP analysis, was not recognized as such by hsp70 analysis (33). As this isolate derived a much smaller proportion of its genome from the undetected parent, such classification is not necessarily problematic. The problem of missing out on hybrids may be an academic one. When a certain hybrid strain has few remnants of the genome of parent 1 in comparison to that of parent 2, typing techniques are more likely to pick up only parent 2 purely by chance. As this parent constitutes the major part of the genome, the parasite likely behaves more like parent 2 than like parent 1, making the typing outcome equally valid. In practice, such a case would be classified as intraspecies variability.
Even though the chance of hybrid detection increases with the number of loci investigated and their locations on different chromosomes, it equally depends on the technology deployed to analyze these loci. SNP analysis, such as RFLP analysis, looks at only a few nucleotides and has a smaller chance of picking up hybrids than does sequencing. In all, given the fact that intraspecies hybrids are rarely documented for Leishmania, they do not seem to present a major problem in current typing practices.
Choosing a Tree in the Forest: Which Method To Use?
Selecting the most appropriate species typing technology in a given context can be a challenging task. As illustrated in Currently Applied Methods, below, many different assays have been used, but the question of the basis on which a particular assay is chosen for a given application remains, and this is project rather than lab dependent. Logical factors that can guide the choice of the best technique include technology (what is available in the lab), speed (importance of a short turnaround time), throughput (how many samples need processing in total and simultaneously), personnel (how many hands and brains are available), cost, host (which species are thought to appear in the studied host), location (which species are thought to occur in the region examined), and sample type (cultures, symptomatic infections, asymptomatic carriers, and low or high parasite loads). Especially regarding the presumed species in a particular host or location, one should always keep in mind that this may not be fully understood. In addition, the specifications in “Criteria for Species Typing Tools” should be considered.
One of the items not listed is “history.” Researchers often stick with their known territory when species typing is not the main focus of a given project. Often a method is used that was once introduced in the lab but may not be the best choice for the question at hand. Granted, sometimes it can be beneficial to stick with an assay or target with which one has experience, even if it is not optimal in a given situation, but in such a case, at least a critical evaluation should ensure valid results.
In our lab, we have found it a clear advantage to adhere to one particular target for species typing: hsp70. Based on this gene, we developed and evaluated both sequencing and RFLP strategies, and this allowed us to build a wide range of experience and a vast range of different methods. We have optimized PCRs for sensitivity and specificity, which makes the target applicable in various settings (66, 67, 191, 194–196, 201, 202). On top of this, several leading labs have developed supplementary hsp70 assays (9, 197, 200). Confining oneself to one target allows an appreciation of what it can and—at least as importantly—cannot do. When hsp70 is not able to answer particular questions, other targets are deployed.
In comparing the assays in Table 1, hsp70 seems to be the best marker for global use, for both New and Old World species and for both subgenera. It discriminates all tested species, including L. braziliensis outliers (Fig. 2) and the little-studied L. siamensis, and has been tested on more strains from a wide geographical distribution than the case for any other method, for both RFLP analysis and sequencing. In addition, different PCR primers were developed and validated for typing in clinical samples, and sequence analysis is straightforward. Unfortunately, it is not the most sensitive PCR target. Cytochrome b has a comparable discrimination efficiency but so far has been evaluated on fewer species and strains, and it relies on sequencing only. Other assays have more restricted use and were designed for either the Old or the New World, or they distinguish fewer species.
Ideally, each lab in the world would base its species typing on the same target, as this would ensure maximal comparability of results from different studies. Because many factors come into play in selecting the best technique, this will never be possible. Fortunately, outcomes from various methods seem to be largely congruent (60), which is a welcomed consequence of the clonal population structure of Leishmania.
Nevertheless, we argue against reinventing the wheel when there is no good reason to do so. New and improved methods should be looked for only when they can give a real added value. In cases where a properly validated and standardized technique is available to implement in a setting or study, there is no need to create yet additional assays.
Interpreting Species Typing Results
In interpreting a species typing result, it is important to keep the limitations of the technique in mind (see also “Criteria for Species Typing Tools”). As further detailed in the next section, many recently published clinical reports specify that species were determined but do not mention the method that was used or mention only that they were determined “by PCR.” These papers list no reference or any other information that allows tracing of the applied methodology, illustrating that a species typing result is taken for granted by many. Furthermore, one should always bear in mind that unexpected species may be encountered, such as the L. major-like parasites in the New World (Fig. 1), or that patients may be infected by more than one Leishmania species (28).
In particular, methods that give a simple binary outcome should be interpreted with caution. Take as an example the species-specific mpi PCRs developed by Zhang et al. (217). One PCR specifically amplifies L. braziliensis and the other L. peruviana, and each test gives a clear outcome depending on whether a product is amplified. However, in many analyses, the dichotomy between both species is not clear-cut, and a large gray zone exists (see “Clinically Relevant Taxa: Certainties and Doubts”). Yet even in this gray zone, each mpi assay gives a clear black-or-white result, which can be misleading.
Methods such as sequencing analyze many polymorphisms simultaneously, thereby allowing a more balanced typing outcome. However, in comparative sequence analysis, caution is needed with regard to the selection of a reference sequence set, as several species designations in the public databases have been shown to be erroneous (e.g., see reference 66). Also, a combination of several binary assays gives a smaller chance of errors, as more data are evaluated in such cases. Methods using only a single feature to discriminate species should thus be interpreted with caution, whether the feature is a restriction endonuclease recognition site for RFLP analysis, a specific PCR, a single SNP, a hybridization result, or an antibody binding assay. The chance of misclassification is increased because uncharted mutations may exist, new mutations may arise, or species that were not validated with the method may be taken for other species.
CURRENTLY APPLIED METHODS
General Overview of PubMed Records
Relatively few of the published assays have made it to the field, and the majority have been used primarily in the lab where they were first developed. Such practice makes it difficult to assess their general validity. In order to get an idea of the currently used Leishmania species typing methods, we undertook an extensive numerical literature survey based on PubMed records filed between 1 January 2012 and 31 January 2014, with a focus on typing at the subgenus to species levels as explained in “Levels of Typing and Focus of This Review.” Papers were filtered based on the title word “leishmaniasis” in conjunction with any of the following search terms: “cutaneous,” “mucocutaneous,” “mucosal,” “tegumentary,” “PKDL,” or “dermal.” The search was performed on 3 February 2014 and returned 410 publications. Of these, 320 were original research papers, both prospective and retrospective. A detailed listing with a summary of each individual analysis can be obtained from the authors upon request. These articles could be categorized into the following 4 groups.
The first group consists of clinical research papers. These 108 papers focus primarily on evaluations of small and large groups of patients and contain reports of clinical, observational, and preclinical studies but not case reports (see below). Of these, 82 were prospective studies. Topics ranged from assessing therapies to evaluation of diagnostic and typing tools, vaccine trials, and descriptive clinical symptomatology, for both humans and canines. In these papers, we looked at the methods used to determine parasite species or subgenus in the patient samples. Many studies also used cultured promastigotes for serologic or protective purposes, but typing of these cultures was not taken into consideration.
The second group, comprising 79 papers, deals with patient management. These are primarily case reports and give an idea of how species typing is performed in actual diagnostic settings. A few papers in this category describe guidelines for management. The third group of publications, 98 in total, are on epidemiology. These papers focus on epidemiological aspects of transmission, reservoirs, vectors, infection, or disease. Not all papers in this category intended to study the parasite or infection of the hosts. Finally, 35 papers dealt with in vivo or in vitro studies (35 papers), reporting on laboratory analyses of parasites in promastigote cultures, cell lines, or animal models. Such studies used well-characterized strains or clinical isolates, and we assessed whether the reported species were accurately determined to exclude contaminations or culture mix-ups.
Half of the 320 studies did not report any method of typing Leishmania at the species level or below the genus level (Table 2). These papers either did not mention any species (34%), relied exclusively on epidemiological data (7%), or used previously documented cultured type strains (9%). The remaining 161 papers reported at least one laboratory method for parasite identification. They also often relied partly on epidemiological information, either because only a subset of samples was analyzed or because the methods discriminated only locally circulating species. In the latter case, it was assumed that no other species had invaded the region under study, implying that the local epidemiology is exactly known.
TABLE 2.
Overview of typing methods stratified by study type
| Typing method or target | % of studies |
||||
|---|---|---|---|---|---|
| Clinicalg | Patient management | Epidemiology | In vivo and in vitro | Total | |
| No species reporteda | 42 (35) | 35 | 37 | 0 | 34 |
| Epidemiology-based studiesb | 14 (16) | 1 | 5 | 3 | 7 |
| Type strain-based studiesc | NA | NA | NA | 80 | 9 |
| rDNA arrayd | 7 (7) | 11 | 28 | 6 | 14 |
| kDNA minicirclesd | 7 (9) | 9 | 21 | 0 | 11 |
| Other genomic targetsd | 14 (16) | 11 | 11 | 6 | 12 |
| Unspecified PCRd | 6 (9) | 28 | 1 | 0 | 9 |
| Isoenzymes (MLEE)d | 7 (6) | 4 | 4 | 11 | 6 |
| Antibodiesd | 5 (4) | 5 | 3 | 9 | 5 |
| Unspecifiedd,e | 6 (9) | 6 | 4 | 0 | 5 |
| Totalf | 108 (82) | 79 | 98 | 35 | 320 |
Studies not mentioning any species.
Studies relying only on epidemiology, not using any laboratory-based methods for typing.
Studies relying on previously obtained typing outcomes for cultured strains but not describing any method for confirmation. This category applies only to in vitro and in vivo studies. NA, not applicable.
Studies using each method for at least one sample. Studies often combined different methods.
Studies using at least one method that was not properly described or referenced.
Total number of publications for each study category. The category in which each individual study was classified can be obtained upon request (see “General Overview of PubMed Records” in the text).
Numbers are for all clinical reports, and numbers in parentheses relate to prospective studies.
In the 161 studies that used at least one laboratory typing technique, the different methods and targets were distributed as shown in Fig. 10, whereby several papers reported more than one method. Genetic identification was used in nearly three quarters of all these cases, among which rRNA genes and kDNA minicircles accounted for more than half of the identifications. In 30 reports, the authors used a PCR without specifying a literature reference, technique, or genomic locus, making it difficult to assess whether the typing outcome is valid and illustrating researchers' perception that PCR is a magical technique able to determine a species unequivocally. MLEE, which is still advocated by the World Health Organization as the reference method (65), was used in 12% of studies where typing was done. Antibodies were used in 9% of cases, and 10% of studies did not specify any details other than that the species was determined.
FIG 10.
Relative use of different species typing methods and targets in the 161 studies where parasite typing was done. The numbers give the percentages of studies using the different methods. The total percentage is more than 100, as many studies used more than one technique.
Methods in Different Study Types
Table 2 gives an overview of the methods for species typing that were used in the different study types. It is noteworthy that in 42% of clinical studies, the causative species was not reported, and in another 14% of these studies, the authors relied exclusively on epidemiological data. Hence, in fewer than half of the clinical studies (44%) was the identity of the species checked to some extent. This was slightly better in prospective studies, even though typing was done in only 49% of these reports. Based on extensive reviews in the Old World and the New World, Gonzalez et al. (19) found similar numbers, with about 50% of clinical studies actually determining the causative species.
Unfortunately, reports that do not mention or check the Leishmania species are of limited use outside the area where data were gathered, and in that sense they contribute little to our global understanding of the behavior of species in patients. In recent recommendations on designing good clinical studies, species typing was generally advised because of the possible differential drug efficacy (76, 245). This was also concluded in several meta-analyses of clinical trials of CL and MCL (19, 246). For patient management, 64% of studies indicated the use of species identification methods, even though in such cases it is also considered relevant (16, 247).
In vitro and in vivo studies always reported the species of the isolates used in the assays, but rarely did the authors specify whether they included some verification of whether the parasites they used actually corresponded to the reported species. Such practice may result in describing a species different from the one actually studied as a result of undetected culture contamination. Van der Auwera et al. (66) listed several examples of erroneous Leishmania species designations in GenBank, with some sequences belonging to a different species complex from the one on file, and even with some being more related to Leptomonas than to Leishmania. These erroneous classifications are likely the result of culture switches or contamination, which can be avoided by applying adequate typing. As for epidemiological studies, 58% reported Leishmania species identification, while the remaining studies focused on different aspects of epidemiology.
With regard to the chosen method, clinical studies relied on a variety of genomic targets, while this was less the case for patient management and epidemiology. The latter made more use of kDNA minicircles and rDNA, which may be related to the routine nature of these labs, where parasite identification has a practical use and less effort is spent on precise (intra)species characterization. Also, labs where clinical trials are carried out often have a more biology-oriented character, and, frequently, different aspects of the parasite are investigated. This may include the study of various genomic loci not commonly deployed in routine settings.
Methods in Different Geographical Areas
Table 3 stratifies the methods used by the locality where they were carried out. The sites where the studies were performed rather than where the samples originated from were considered, which were generally the same for areas of endemicity. From these data, it is apparent that Old World countries relied mostly on rDNA-based methods for species typing, except for the Middle East, where kDNA minicircles were equally popular. Given the fact that studies in these regions focused on the Leishmania (Leishmania) subgenus, where variability in the rDNA sequences is sufficient to determine the species or species complexes, this is logical. Minicircle kDNA was used particularly in Iran, where 16 of the 22 Old World minicircle-based studies were performed.
TABLE 3.
Overview of typing methods stratified by region
| Typing method or target | % of studies |
||||||
|---|---|---|---|---|---|---|---|
| Mediterranean basin | Middle East (Iran)g,h | Far East (India)i | Africag | Latin America (except Brazil)j | Brazil | Countries where leishmaniasis is not endemic | |
| No species reporteda | 34 | 30 (26) | 44 (43) | 36 | 26 | 46 | 13 |
| Epidemiology-based studiesb | 4 | 3 (3) | 8 (7) | 18 | 3 | 15 | 0 |
| Type strain-based studiesc | 6 | 5 (5) | 5 (7) | 0 | 3 | 13 | 23 |
| rDNA arrayd | 21 | 27 (28) | 18 (17) | 36 | 6 | 4 | 5 |
| kDNA minicirclesd | 2 | 27 (28) | 10 (7) | 0 | 6 | 11 | 8 |
| Other genome targetsd | 13 | 3 (3) | 3 (3) | 18 | 31 | 8 | 21 |
| Unspecified PCRd | 11 | 5 (5) | 13 (17) | 0 | 9 | 3 | 28 |
| Isoenzymes (MLEE)d | 6 | 2 (2) | 0 (0) | 0 | 14 | 9 | 8 |
| Antibodiesd | 2 | 2 (2) | 10 (10) | 0 | 11 | 6 | 0 |
| Unspecifiedd,e | 11 | 5 (5) | 3 (3) | 0 | 6 | 1 | 8 |
| Totalf | 53 | 63 (58) | 39 (30) | 11 | 35 | 80 | 39 |
Studies not mentioning any species.
Studies relying only on epidemiology, not using any laboratory-based methods for typing.
Studies relying on previously obtained typing outcomes for cultured strains but not describing any method for confirmation. This category applies only to in vitro and in vivo studies.
Studies using each method for at least one sample. Studies often combined different methods.
Studies using at least one method that was not properly described or referenced.
Total number of publications for each study category. The category in which each individual study was classified can be obtained upon request (see “General Overview of PubMed Records” in the text).
Countries bordering the Mediterranean Sea are excluded here and are listed in the category “Mediterranean basin.”
The first numbers include studies from Iran, and numbers in parentheses are for Iran only.
The Far East category includes the Indian subcontinent. The total values are given, and numbers for India alone are given in parentheses.
Mexico and Central and South America, excluding Brazil.
In Latin America, the picture is more diverse, and a variety of targets were used. Only in Brazil did kDNA minicircle assays slightly outnumber other techniques, probably for historical reasons, as many minicircle-based assays were developed and validated there. In other countries, only cpb and hsp70 assays were used in more than one lab, illustrating the scattered typing landscape. This relates to the difficulties in efficiently discriminating Leishmania (Viannia) species, leading to many assay development efforts. In addition, rDNA offers limited possibilities for this subgenus, so it was not frequently deployed.
In countries where leishmaniasis is not endemic but was investigated, no studies relied only on epidemiology, and only 13% did not mention parasite species. A variety of methods were used. In clinical practice, these countries deal with import leishmaniasis (15, 16, 248), and as the site of infection in such cases is not always known, globally applicable typing strategies are needed. Neither minicircles nor rDNA provides such a possibility, so other methods must be sought. In addition, labs in these countries frequently participate in clinical or in vitro and in vivo studies. For these studies, a detailed parasite identification is frequently needed to correlate with clinical and biological data, relying on several targets or even the entire parasite's genome (e.g., see references 73, 249, and 250).
It is striking that Brazil is among the countries where the fewest efforts for parasite typing were reported, as parasite identification was done in merely 21 of 80 studies. Also, India and the Far East countries scored low, but they have far less parasite diversity. In India, visceral leishmaniasis caused by L. donovani is the prominent disease, and diagnosis of PKDL can be established on clinical grounds and patient history. Brazil, on the other hand, is home to 8 Leishmania species and an entire spectrum of diseases (2), yet confirmation of the infecting agent is apparently not considered important by several authors, or they rely blindly on epidemiological data.
Limitations of PubMed Analysis
This overview of currently applied Leishmania typing strategies is by no means complete. First, it is based only on PubMed records gathered over 2 years, whereby routinely applied techniques in various types of health centers remain hidden because they often are not published in indexed journals. Two countries were responsible for 43% of the original research papers published in the analyzed period: Brazil and Iran. This is not a coincidence, as both are in the top 4 countries with the highest estimated incidences of cutaneous leishmaniasis (2). Only Afghanistan and Algeria have higher estimated burdens, but each of them published merely one paper in the studied period.
Nevertheless, this literature review illustrates the scattered use of various typing methods and genetic targets. It also gives an idea of how often scientists and physicians rely on epidemiological information, even though species distribution may change over time and is mostly not systematically monitored. Our overview underscores the need to use more standardized and uniform species typing tools, as this would allow a better evaluation of the assays on a global scale and facilitate comparison between different studies. It also highlights the need for accessible but accurate assays in various settings, allowing continuous monitoring of an ever-changing epidemiology.
PARASITE TYPING BEYOND THE SPECIES LEVEL
Parasite identification should not be limited to species determination. Even though particular species can be associated with certain forms of the disease or can affect prognosis and cure, two cases can be envisioned where additional typing is clinically relevant and the development of additional typing techniques is recommended.
First, within each species, different populations are expected to exist. Such populations can have specific characteristics, such as virulence or tolerance to certain drugs, which make their identification relevant. This concept relates to identification of different discrete typing units (DTUs) within a given species or species complex (28). Studying population dynamics in a given geographical location requires methods that are able to expose a higher resolution than those of the methods focused on here. MLMT, MLST, AFLP, MLEE, and RAPD analyses have been used successfully to uncover intraspecies differences. Of these, MLMT has the most potential for use on clinical samples without the need for culturing, because it is sensitive and targeted. Nevertheless, in cases of recent outbreaks, in which strains are genetically very similar, this method is not applicable (251). The most informative method to date is sequencing of the entire genome (e.g., see reference 73), even though this too still requires parasite isolation. A major hurdle when it comes to studying populations lies in the difficulty of examining parasites in wild animal reservoirs, because of their poor accessibility, and in asymptomatically infected individuals, because few parasites can be found. Also, parasite loads can be low in symptomatic disease. These problems all lead to a potential bias in uncovering the complete population structure (28).
Second, certain biological traits may be conferred by genetic mechanisms that surpass the population boundary (1, 28). For instance, strains belonging to different populations or even species may share pathways that render them drug resistant or that enable them to colonize certain cell types or sand fly species. In such cases, if the biological pathway is known or markers correlated with these traits have been identified, characterizing parasites by such features may be more relevant than looking at the species.
RECOMMENDATIONS
Verification of the Leishmania species is considered a must in any report describing clinical data and is a valuable aid in day-to-day patient management (16, 19, 76, 245–247). To this end, single-gene sequencing is the most practical method in a clinical (human or animal) context. Several genes render good resolution at the species level, but ITS1 (Old World) and hsp70 (global) seem to be the best choices in terms of sensitivity and validation, possibly along with the cytochrome b gene. The miniexon offers good resolution globally, but the sequencing process is cumbersome. If sequencing is not possible, RFLP analyses of hsp70, ITS1 (Old World), and the miniexon may be considered, as these have been used and validated successfully. However, the miniexon RFLP scheme is complicated and suffers from a high degree of intraspecies variability. For species typing in epidemiological and environmental studies, only MLEE has been applied to all currently described species. No other single method has been used on the entire genus. Consequently, additional validation is needed to include species exclusively found in sand fly vectors or animals.
Wherever possible, we advise investigators to adhere to one target, preferably one that others use as well, as this allows one to gain a good sense of what it can or cannot do in a given context. “Reinventing the wheel” is discouraged unless a marked improvement of the existing range of assays can be realized.
In the absence of a clear species border within certain complexes, we advise adoption of the “better sure than sorry” strategy. When sequencing places a parasite somewhere halfway between different species in the complex, it is better to report at the complex than the species level. This rule is difficult to adhere to when using RFLP- or other SNP-based methods, which inherently give a black-or-white answer (see “Interpreting Species Typing Results”).
These are our opinions, taking into consideration the current knowledge and flaws. Often practical limitations stand in the way of using the best available technique, and one is forced to find a pragmatic compromise. Ideally, everyone would use the same target for all experiments, but this is not realistic. Full-genome sequencing is the ultimate target for establishing a consensus reference taxonomy, to be mirrored by simpler field-applicable assays. While awaiting such comprehensive analysis, we will have to cope with assays that best match the existing species reference framework for many years to come. This may not be ideal, but it seems to be quite manageable and fairly accurate when it comes to species typing for dermal leishmaniasis.
ACKNOWLEDGMENTS
We express our sincere gratitude to all colleagues with whom we have worked and discussed species typing of Leishmania and who helped to shape our views. The LeishMan consortium (www.leishman.eu) is acknowledged for valuable input from a clinical perspective. We thank the anonymous reviewers for their constructive comments and Patrick Lane from ScEYEnce Studios for figure editing.
Gert Van der Auwera is financially supported by the Third Framework agreement between the Belgian Directorate General for Development and the Institute of Tropical Medicine.
Biographies

Gert Van der Auwera obtained his Ph.D. in molecular biology at the University of Antwerp (Belgium), studying the evolutionary relationships of protists. Subsequently he studied the genetic variability of HIV and was involved in the design of HIV subtyping kits for the NIH, while affiliated with the Institute of Tropical Medicine (ITM), Antwerp, Belgium. During a second postdoctoral position at the University of Ghent and the Flanders Interuniversity Institute for Biotechnology (Belgium), he worked on gene silencing and site-directed mutagenesis in Arabidopsis thaliana, after which he returned to ITM to focus on the epidemiology, diagnosis, and identification of Leishmania and other trypanosomatids. He has been a research associate at ITM for 9 years, and during this time, he has published several papers on Leishmania species typing. He plays a leading role in a European network on clinical and diagnostic management of imported leishmaniasis (LeishMan).

Jean-Claude Dujardin is a biologist. He obtained his doctorate in sciences at the Free University of Brussels (Belgium), studying the adaptive significance of karyotype variability in Leishmania. He has worked at the Institute of Tropical Medicine in Antwerp (Belgium) since 1986, and he is now Head of the Department of Biomedical Sciences and the Unit of Molecular Parasitology. He dedicated his career to the exploration of molecular diversity in natural populations of Leishmania in both clinical and epidemiological contexts, especially in Latin America and the Indian subcontinent. He is a pioneer in the application of next-generation sequencing methods for Leishmania typing. He has been coordinating several European consortia, more recently in the domain of drug resistance (Kaladrug-R).
APPENDIX
GLOSSARY
- AFLP
Amplified fragment length polymorphism. Fingerprinting technique whereby genomic DNA is digested with a frequent and a rare-cutting restriction endonuclease, after which a selection of the obtained fragments are amplified by PCR and separated based on size (241).
- amplicon
Amplified PCR product.
- CL
Cutaneous leishmaniasis. In this review used to refer to MCL, ML, LCL, and DCL.
- DCL
Diffuse cutaneous leishmaniasis. Characterized by multiple lesions resulting from dissemination of the infection from the inoculation site.
- DTU
Discrete typing unit.
- HRM
High-resolution melting. Real-time PCR technique whereby the melting temperature of a double-stranded DNA fragment or bound oligonucleotide probe is determined.
- isoenzymes
Variations of the same enzyme across different strains. These variations migrate differently in a pH-gradient gel, and the collective pattern of all isoenzymes is called a zymodeme.
- ITS
Internal transcribed spacer of the rDNA array (Fig. 4).
- kDNA
Kinetoplast DNA. Extrachromosomal mitochondrial DNA in kinetoplastid organisms, such as Leishmania. kDNA consists of a dense network of mini- and maxicircles.
- LCL
Localized cutaneous leishmaniasis. Characterized by a single nodular or ulcerative lesion at the site of parasite inoculation by the sand fly vector.
- MCL
Mucocutaneous leishmaniasis. Typical of the New World and generally produced by mucosal metastasis from a primary local cutaneous lesion, often after healing of the latter, even years later.
- microsatellite
Short nucleotide repeat that varies in size between different strains.
- ML
Mucosal leishmaniasis. A variant of MCL in which only mucosae are affected, possibly by extension of contiguous facial skin lesions or direct mucosal inoculation by an insect (252).
- MLEE
Multilocus enzyme electrophoresis. Technique to determine the zymodeme of strains, long used as the gold standard for Leishmania species typing.
- MLMT
Multilocus microsatellite typing. In this technique, several microsatellites in the genome are PCR amplified, and the combination of their lengths allows definition of parasites and parasite populations.
- MLST
Multilocus sequence typing. Technique whereby several genes are sequenced and the species is determined on the basis of this sequence information. Usually, single-copy housekeeping genes are selected.
- PKDL
Post-kala-azar dermal leishmaniasis, a dermal manifestation characterized by multiple lesions and following successful treatment of visceral leishmaniasis caused by L. donovani (3).
- RAPD
Randomly amplified polymorphic DNA. Also called permissively primed PCR. Technique whereby primers are allowed to hybridize to nonspecified fragments in a DNA sample, leading to a collection of fragments that together define a fingerprint pattern.
- rDNA
rRNA gene array (Fig. 4).
- restriction endonuclease
Enzyme cutting DNA in a sequence-dependent manner. Upon recognizing a specific DNA sequence, the enzyme cuts both strands of the DNA. Each restriction enzyme recognizes a particular sequence, most frequently of 4 or 6 nucleotides.
- RFLP
Restriction fragment length polymorphism. Method whereby a DNA fragment (in the case of typing; usually a PCR product) is cut by one or a combination of restriction endonucleases. The obtained fragments are separated based on length, resulting in an RFLP pattern that is sequence dependent (Fig. 3).
- SNP
Single-nucleotide polymorphism. DNA base change in the genome of a particular species or parasite variant.
- species complex
Group of related Leishmania species, also called a “complex.” The level of species complex is situated between subgenera and individual species (Fig. 1).
- VL
Visceral leishmaniasis (kala-azar).
- zymodeme
Collection of Leishmania strains sharing the same isoenzyme profile.
REFERENCES
- 1.Reithinger R, Dujardin JC, Louzir H, Pirmez C, Alexander B, Brooker S. 2007. Cutaneous leishmaniasis. Lancet Infect Dis 7:581–596. doi: 10.1016/S1473-3099(07)70209-8. [DOI] [PubMed] [Google Scholar]
- 2.Alvar J, Velez ID, Bern C, Herrero M, Desjeux P, Cano J, Jannin J, den Boer M, WHO Leishmaniasis Control Team . 2012. Leishmaniasis worldwide and global estimates of its incidence. PLoS One 7:e35671. doi: 10.1371/journal.pone.0035671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh S, Sharma U, Mishra J. 2011. Post-kala-azar dermal leishmaniasis: recent developments. Int J Dermatol 50:1099–1108. doi: 10.1111/j.1365-4632.2011.04925.x. [DOI] [PubMed] [Google Scholar]
- 4.Karunaweera ND. 2009. Leishmania donovani causing cutaneous leishmaniasis in Sri Lanka: a wolf in sheep's clothing? Trends Parasitol 25:458–463. doi: 10.1016/j.pt.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Siriwardana HV, Noyes HA, Beeching NJ, Chance ML, Karunaweera ND, Bates PA. 2007. Leishmania donovani and cutaneous leishmaniasis, Sri Lanka. Emerg Infect Dis 13:476–478. doi: 10.3201/eid1303.060242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guerra JA, Prestes SR, Silveira H, Coelho LI, Gama P, Moura A, Amato V, Barbosa M, Ferreira LC. 2011. Mucosal leishmaniasis caused by Leishmania (Viannia) braziliensis and Leishmania (Viannia) guyanensis in the Brazilian Amazon. PLoS Negl Trop Dis 5:e980. doi: 10.1371/journal.pntd.0000980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Madeddu G, Fiori ML, Ena P, Riu F, Lovigu C, Nunnari G, Bagella P, Maida I, Babudieri S, Mura MS. 2014. Mucocutaneous leishmaniasis as presentation of HIV infection in Sardinia, insular Italy. Parasitol Int 63:35–36. doi: 10.1016/j.parint.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 8.Nawaratna SS, Weilgama DJ, Wijekoon CJ, Dissanayake M, Rajapaksha K. 2007. Cutaneous leishmaniasis, Sri Lanka. Emerg Infect Dis 13:1068–1070. doi: 10.3201/eid1307.060773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chicharro C, Llanes-Acevedo IP, Garcia E, Nieto J, Moreno J, Cruz I. 2013. Molecular typing of Leishmania infantum isolates from a leishmaniasis outbreak in Madrid, Spain, 2009 to 2012. Euro Surveill 18:20545 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20545. [DOI] [PubMed] [Google Scholar]
- 10.Dujardin JC. 2006. Risk factors in the spread of leishmaniases: towards integrated monitoring? Trends Parasitol 22:4–6. doi: 10.1016/j.pt.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Salomon OD, Quintana MG, Zaidenberg M. 2008. Urban distribution of Phlebotominae in a cutaneous leishmaniasis focus, Argentina. Mem Inst Oswaldo Cruz 103:282–287. doi: 10.1590/S0074-02762008005000016. [DOI] [PubMed] [Google Scholar]
- 12.Arévalo J, Ramirez L, Adaui V, Zimic M, Tulliano G, Miranda-Verastegui C, Lazo M, Loayza-Muro R, De Doncker S, Maurer A, Chappuis F, Dujardin JC, Llanos-Cuentas A. 2007. Influence of Leishmania (Viannia) species on the response to antimonial treatment in patients with American tegumentary leishmaniasis. J Infect Dis 195:1846–1851. doi: 10.1086/518041. [DOI] [PubMed] [Google Scholar]
- 13.Romero GA, Guerra MV, Paes MG, Macedo VO. 2001. Comparison of cutaneous leishmaniasis due to Leishmania (Viannia) braziliensis and L. (V.) guyanensis in Brazil: therapeutic response to meglumine antimoniate. Am J Trop Med Hyg 65:456–465. [DOI] [PubMed] [Google Scholar]
- 14.Modabber F, Buffet PA, Torreele E, Milon G, Croft SL. 2007. Consultative meeting to develop a strategy for treatment of cutaneous leishmaniasis. Institute Pasteur, Paris, 13 to 15 June 2006. Kinetoplastid Biol Dis 6:3. doi: 10.1186/1475-9292-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buffet P. 2010. Leishmaniasis—a guide for disease management. Sanofi Aventis, Paris, France. [Google Scholar]
- 16.Blum J, Buffet P, Visser L, Harms G, Bailey MS, Caumes E, Clerinx J, van Thiel PP, Morizot G, Hatz C, Dorlo TP, Lockwood DN. 2014. LeishMan recommendations for treatment of cutaneous and mucosal leishmaniasis in travelers, 2014. J Travel Med 21:116–129. doi: 10.1111/jtm.12089. [DOI] [PubMed] [Google Scholar]
- 17.World Health Organization. 2002. Control of Chagas disease, WHO technical report series 905. World Health Organization, Geneva, Switzerland. [PubMed] [Google Scholar]
- 18.Blum J, Lockwood DN, Visser L, Harms G, Bailey MS, Caumes E, Clerinx J, van Thiel PP, Morizot G, Hatz C, Buffet P. 2012. Local or systemic treatment for New World cutaneous leishmaniasis? Re-evaluating the evidence for the risk of mucosal leishmaniasis. Int Health 4:153–163. doi: 10.1016/j.inhe.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez U, Pinart M, Reveiz L, Rengifo-Pardo M, Tweed J, Macaya A, Alvar J. 2010. Designing and reporting clinical trials on treatments for cutaneous leishmaniasis. Clin Infect Dis 51:409–419. doi: 10.1086/655134. [DOI] [PubMed] [Google Scholar]
- 20.Reithinger R, Dujardin JC. 2007. Molecular diagnosis of leishmaniasis: current status and future applications. J Clin Microbiol 45:21–25. doi: 10.1128/JCM.02029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schönian G, Mauricio I, Cupolillo E. 2010. Is it time to revise the nomenclature of Leishmania? Trends Parasitol 26:466–469. doi: 10.1016/j.pt.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 22.Leelayoova S, Siripattanapipong S, Hitakarun A, Kato H, Tan-ariya P, Siriyasatien P, Osatakul S, Mungthin M. 2013. Multilocus characterization and phylogenetic analysis of Leishmania siamensis isolated from autochthonous visceral leishmaniasis cases, southern Thailand. BMC Microbiol 13:60. doi: 10.1186/1471-2180-13-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mouri O, Morizot G, Van der Auwera G, Ravel C, Passet M, Chartrel N, Joly I, Thellier M, Jaureguiberry S, Caumes E, Mazier D, Marinach-Patrice C, Buffet P. 2014. Easy identification of Leishmania species by mass spectrometry. PLoS Negl Trop Dis 8:e2841. doi: 10.1371/journal.pntd.0002841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marcili A, Speranca MA, Da Costa AP, Madeira MF, Soares HS, Sanches CO, Acosta IC, Girotto A, Minervino AH, Horta MC, Shaw JJ, Gennari SM. 2014. Phylogenetic relationships of Leishmania species based on trypanosomatid barcode (SSU rDNA) and gGAPDH genes: taxonomic revision of Leishmania (L.) infantum chagasi in South America. Infect Genet Evol 25:44–51. doi: 10.1016/j.meegid.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 25.Dedet J, Pratlong F. 2000. Taxonomy of Leishmania and geographical distribution of leishmaniasis. Ann Dermatol Venereol 127:421–424. [PubMed] [Google Scholar]
- 26.Rioux JA, Lanotte G, Serres E, Pratlong F, Bastien P, Perieres J. 1990. Taxonomy of Leishmania. Use of isoenzymes. Suggestions for a new classification. Ann Parasitol Hum Comp 65:111–125. [DOI] [PubMed] [Google Scholar]
- 27.Jamjoom MB, Ashford RW, Bates PA, Chance ML, Kemp SJ, Watts PC, Noyes HA. 2004. Leishmania donovani is the only cause of visceral leishmaniasis in East Africa; previous descriptions of L. infantum and “L. archibaldi” from this region are a consequence of convergent evolution in the isoenzyme data. Parasitology 129:399–409. doi: 10.1017/S0031182004005955. [DOI] [PubMed] [Google Scholar]
- 28.Bañuls AL, Hide M, Prugnolle F. 2007. Leishmania and the leishmaniases: a parasite genetic update and advances in taxonomy, epidemiology and pathogenicity in humans. Adv Parasitol 64:1–109. doi: 10.1016/S0065-308X(06)64001-3. [DOI] [PubMed] [Google Scholar]
- 29.Darwin C. 1859. On the origin of species by means of natural selection. John Murray, London, United Kingdom. [Google Scholar]
- 30.Tojal da Silva AC, Cupolillo E, Volpini AC, Almeida R, Romero GA. 2006. Species diversity causing human cutaneous leishmaniasis in Rio Branco, State of Acre, Brazil. Trop Med Int Health 11:1388–1398. doi: 10.1111/j.1365-3156.2006.01695.x. [DOI] [PubMed] [Google Scholar]
- 31.Nolder D, Roncal N, Davies CR, Llanos-Cuentas A, Miles MA. 2007. Multiple hybrid genotypes of Leishmania (Viannia) in a focus of mucocutaneous leishmaniasis. Am J Trop Med Hyg 76:573–578. [PubMed] [Google Scholar]
- 32.Odiwuor S, De Doncker S, Maes I, Dujardin JC, Van der Auwera G. 2011. Natural Leishmania donovani/Leishmania aethiopica hybrids identified from Ethiopia. Infect Genet Evol 11:2113–2118. doi: 10.1016/j.meegid.2011.04.026. [DOI] [PubMed] [Google Scholar]
- 33.Odiwuor S, Veland N, Maes I, Arévalo J, Dujardin JC, Van der Auwera G. 2012. Evolution of the Leishmania braziliensis species complex from amplified fragment length polymorphisms, and clinical implications. Infect Genet Evol 12:1994–2002. doi: 10.1016/j.meegid.2012.03.028. [DOI] [PubMed] [Google Scholar]
- 34.Ravel C, Cortes S, Pratlong F, Morio F, Dedet JP, Campino L. 2006. First report of genetic hybrids between two very divergent Leishmania species: Leishmania infantum and Leishmania major. Int J Parasitol 36:1383–1388. doi: 10.1016/j.ijpara.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 35.Belli AA, Miles MA, Kelly JM. 1994. A putative Leishmania panamensis/Leishmania braziliensis hybrid is a causative agent of human cutaneous leishmaniasis in Nicaragua. Parasitology 109:435–442. doi: 10.1017/S0031182000080689. [DOI] [PubMed] [Google Scholar]
- 36.Delgado O, Cupolillo E, Bonfante-Garrido R, Silva S, Belfort E, Grimaldi JG, Momen H. 1997. Cutaneous leishmaniasis in Venezuela caused by infection with a new hybrid between Leishmania (Viannia) braziliensis and L. (V.) guyanensis. Mem Inst Oswaldo Cruz 92:581–582. doi: 10.1590/S0074-02761997000500002. [DOI] [PubMed] [Google Scholar]
- 37.Bañuls AL, Guerrini F, Le Pont F, Barrera C, Espinel I, Guderian R, Echeverria R, Tibayrenc M. 1997. Evidence for hybridization by multilocus enzyme electrophoresis and random amplified polymorphic DNA between Leishmania braziliensis and Leishmania panamensis/guyanensis in Ecuador. J Eukaryot Microbiol 44:408–411. doi: 10.1111/j.1550-7408.1997.tb05716.x. [DOI] [PubMed] [Google Scholar]
- 38.Dujardin JC, Bañuls AL, Llanos-Cuentas A, Alvarez E, De Doncker S, Jacquet D, Le Ray D, Arévalo J, Tibayrenc M. 1995. Putative Leishmania hybrids in the Eastern Andean valley of Huanuco, Peru. Acta Trop 59:293–307. doi: 10.1016/0001-706X(95)00094-U. [DOI] [PubMed] [Google Scholar]
- 39.Sadlova J, Yeo M, Seblova V, Lewis MD, Mauricio I, Volf P, Miles MA. 2011. Visualisation of Leishmania donovani fluorescent hybrids during early stage development in the sand fly vector. PLoS One 6:e19851. doi: 10.1371/journal.pone.0019851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuhls K, Cupolillo E, Silva SO, Schweynoch C, Boite MC, Mello MN, Mauricio I, Miles M, Wirth T, Schönian G. 2013. Population structure and evidence for both clonality and recombination among Brazilian strains of the subgenus Leishmania (Viannia). PLoS Negl Trop Dis 7:e2490. doi: 10.1371/journal.pntd.0002490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boité MC, Mauricio IL, Miles MA, Cupolillo E. 2012. New insights on taxonomy, phylogeny and population genetics of Leishmania (Viannia) parasites based on multilocus sequence analysis. PLoS Negl Trop Dis 6:e1888. doi: 10.1371/journal.pntd.0001888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rougeron V, De Meeus T, Hide M, Le Falher G, Bucheton B, Dereure J, El-Safi SH, Dessein A, Bañuls AL. 2011. Multifaceted population structure and reproductive strategy in Leishmania donovani complex in one Sudanese village. PLoS Negl Trop Dis 5:e1448. doi: 10.1371/journal.pntd.0001448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rougeron V, Bañuls AL, Carme B, Simon S, Couppie P, Nacher M, Hide M, De Meeus T. 2011. Reproductive strategies and population structure in Leishmania: substantial amount of sex in Leishmania Viannia guyanensis. Mol Ecol 20:3116–3127. doi: 10.1111/j.1365-294X.2011.05162.x. [DOI] [PubMed] [Google Scholar]
- 44.Rougeron V, De Meeus T, Hide M, Waleckx E, Bermudez H, Arévalo J, Llanos-Cuentas A, Dujardin JC, De Doncker S, Le Ray D, Ayala FJ, Bañuls AL. 2009. Extreme inbreeding in Leishmania braziliensis. Proc Natl Acad Sci U S A 106:10224–10229. doi: 10.1073/pnas.0904420106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tibayrenc M, Ayala FJ. 2013. How clonal are Trypanosoma and Leishmania? Trends Parasitol 29:264–269. doi: 10.1016/j.pt.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 46.Westenberger SJ, Barnabé C, Campbell DA, Sturm NR. 2005. Two hybridization events define the population structure of Trypanosoma cruzi. Genetics 171:527–543. doi: 10.1534/genetics.104.038745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bañuls AL, Dujardin JC, Guerrini F, De Doncker S, Jacquet D, Arévalo J, Noel S, Le Ray D, Tibayrenc M. 2000. Is Leishmania (Viannia) peruviana a distinct species? A MLEE/RAPD evolutionary genetics answer. J Eukaryot Microbiol 47:197–207. doi: 10.1111/j.1550-7408.2000.tb00039.x. [DOI] [PubMed] [Google Scholar]
- 48.Mauricio IL, Yeo M, Baghaei M, Doto D, Pratlong F, Zemanová E, Dedet JP, Lukeš J, Miles MA. 2006. Towards multilocus sequence typing of the Leishmania donovani complex: resolving genotypes and haplotypes for five polymorphic metabolic enzymes (ASAT, GPI, NH1, NH2, PGD). Int J Parasitol 36:757–769. doi: 10.1016/j.ijpara.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 49.Zemanová E, Jirku M, Mauricio IL, Horák A, Miles MA, Lukeš J. 2007. The Leishmania donovani complex: genotypes of five metabolic enzymes (ICD, ME, MPI, G6PDH, and FH), new targets for multilocus sequence typing. Int J Parasitol 37:149–160. doi: 10.1016/j.ijpara.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 50.Kuhls K, Keilonat L, Ochsenreither S, Schaar M, Schweynoch C, Presber W, Schönian G. 2007. Multilocus microsatellite typing (MLMT) reveals genetically isolated populations between and within the main endemic regions of visceral leishmaniasis. Microbes Infect 9:334–343. doi: 10.1016/j.micinf.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 51.Lukeš J, Mauricio IL, Schönian G, Dujardin JC, Soteriadou K, Dedet JP, Kuhls K, Quispe-Tintaya KW, Jirku M, Chocholová E, Haralambous C, Pratlong F, Oborník M, Horák A, Ayala FJ, Miles MA. 2007. Evolutionary and geographical history of the Leishmania donovani complex with a revision of current taxonomy. Proc Natl Acad Sci U S A 104:9375–9380. doi: 10.1073/pnas.0703678104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Odiwuor S, Vuylsteke M, De Doncker S, Maes I, Mbuchi M, Dujardin JC, Van der Auwera G. 2011. Leishmania AFLP: paving the way towards improved molecular assays and markers of diversity. Infect Genet Evol 11:960–967. doi: 10.1016/j.meegid.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 53.Leblois R, Kuhls K, Francois O, Schönian G, Wirth T. 2011. Guns, germs and dogs: on the origin of Leishmania chagasi. Infect Genet Evol 11:1091–1095. doi: 10.1016/j.meegid.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 54.Mauricio IL, Stothard JR, Miles MA. 2000. The strange case of Leishmania chagasi. Parasitol Today 16:188–189. doi: 10.1016/S0169-4758(00)01637-9. [DOI] [PubMed] [Google Scholar]
- 55.Pratlong F, Lami P, Ravel C, Balard Y, Dereure J, Serres G, Baidouri FE, Dedet JP. 2013. Geographical distribution and epidemiological features of Old World Leishmania infantum and Leishmania donovani foci, based on the isoenzyme analysis of 2277 strains. Parasitology 140:423–434. doi: 10.1017/S0031182012001825. [DOI] [PubMed] [Google Scholar]
- 56.El Baidouri F, Diancourt L, Berry V, Chevenet F, Pratlong F, Marty P, Ravel C. 2013. Genetic structure and evolution of the Leishmania genus in Africa and Eurasia: what does MLSA tell us. PLoS Negl Trop Dis 7:e2255. doi: 10.1371/journal.pntd.0002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schönian G, Schnur L, el Fari M, Oskam L, Kolesnikov AA, Sokolowska-Kohler W, Presber W. 2001. Genetic heterogeneity in the species Leishmania tropica revealed by different PCR-based methods. Trans R Soc Trop Med Hyg 95:217–224. doi: 10.1016/S0035-9203(01)90173-7. [DOI] [PubMed] [Google Scholar]
- 58.Pratlong F, Dereure J, Ravel C, Lami P, Balard Y, Serres G, Lanotte G, Rioux JA, Dedet JP. 2009. Geographical distribution and epidemiological features of Old World cutaneous leishmaniasis foci, based on the isoenzyme analysis of 1048 strains. Trop Med Int Health 14:1071–1085. doi: 10.1111/j.1365-3156.2009.02336.x. [DOI] [PubMed] [Google Scholar]
- 59.Schwenkenbecher JM, Wirth T, Schnur LF, Jaffe CL, Schallig H, Al-Jawabreh A, Hamarsheh O, Azmi K, Pratlong F, Schönian G. 2006. Microsatellite analysis reveals genetic structure of Leishmania tropica. Int J Parasitol 36:237–246. doi: 10.1016/j.ijpara.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 60.Van der Auwera G, Ravel C, Verweij JJ, Bart A, Schönian G, Felger I. 2014. Evaluation of four single-locus markers for Leishmania species discrimination by sequencing. J Clin Microbiol 52:1098–1104. doi: 10.1128/JCM.02936-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Momen H, Grimaldi G Jr. 1984. On the identity of Leishmania mexicana pifanoi and L. mexicana garnhami. Trans R Soc Trop Med Hyg 78:701–702. [DOI] [PubMed] [Google Scholar]
- 62.Hashiguchi Y, Gomez EA, de Coronel VV, Mimori T, Kawabata M, Furuya M, Nonaka S, Takaoka H, Alexander JB, Quizhpe AM, Kreutzer RD, Tesh RB. 1991. Andean leishmaniasis in Ecuador caused by infection with Leishmania mexicana and L. major-like parasites. Am J Trop Med Hyg 44:205–217. [DOI] [PubMed] [Google Scholar]
- 63.Arana M, Evans DA, Zolessi A, Cuentas AL, Arévalo J. 1990. Biochemical characterization of Leishmania (Viannia) braziliensis and Leishmania (Viannia) peruviana by isoenzyme electrophoresis. Trans R Soc Trop Med Hyg 84:526–529. doi: 10.1016/0035-9203(90)90025-A. [DOI] [PubMed] [Google Scholar]
- 64.Veland N, Boggild AK, Valencia C, Valencia BM, Llanos-Cuentas A, Van der Auwera G, Dujardin JC, Arévalo J. 2012. Leishmania (Viannia) species identification on clinical samples from cutaneous leishmaniasis patients in Peru: assessment of a molecular stepwise approach. J Clin Microbiol 50:495–498. doi: 10.1128/JCM.05061-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.World Health Organization. 2010. Control of the leishmaniasis: report of a meeting of the WHO Expert Committee on the Control of Leishmaniases, Geneva, 22 to 26 March 2010. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 66.Van der Auwera G, Maes I, De Doncker S, Ravel C, Cnops L, Van Esbroeck M, Van Gompel A, Clerinx J, Dujardin JC. 2013. Heat-shock protein 70 gene sequencing for Leishmania species typing in European tropical infectious disease clinics. Euro Surveill 18:20543 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20543. [DOI] [PubMed] [Google Scholar]
- 67.Fraga J, Montalvo AM, Maes L, Dujardin JC, Van der Auwera G. 2013. HindII and SduI digests of heat-shock protein 70 PCR for Leishmania typing. Diagn Microbiol Infect Dis 77:245–247. doi: 10.1016/j.diagmicrobio.2013.07.023. [DOI] [PubMed] [Google Scholar]
- 68.Tsukayama P, Lucas C, Bacon DJ. 2009. Typing of four genetic loci discriminates among closely related species of New World Leishmania. Int J Parasitol 39:355–362. doi: 10.1016/j.ijpara.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 69.Bañuls AL, Jonquieres R, Guerrini F, Le Pont F, Barrera C, Espinel I, Guderian R, Echeverria R, Tibayrenc M. 1999. Genetic analysis of Leishmania parasites in Ecuador: are Leishmania (Viannia) panamensis and Leishmania (V.) guyanensis distinct taxa? Am J Trop Med Hyg 61:838–845. [DOI] [PubMed] [Google Scholar]
- 70.Van der Auwera G, Fraga J, Montalvo AM, Dujardin JC. 2011. Leishmania taxonomy up for promotion? Trends Parasitol 27:49–50. doi: 10.1016/j.pt.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 71.Macaulay IC, Voet T. 2014. Single cell genomics: advances and future perspectives. PLoS Genet 10:e1004126. doi: 10.1371/journal.pgen.1004126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schönian G, Kuhls K, Mauricio IL. 2011. Molecular approaches for a better understanding of the epidemiology and population genetics of Leishmania. Parasitology 138:405–425. doi: 10.1017/S0031182010001538. [DOI] [PubMed] [Google Scholar]
- 73.Downing T, Imamura H, Decuypere S, Clark TG, Coombs GH, Cotton JA, Hilley JD, De Doncker S, Maes I, Mottram JC, Quail MA, Rijal S, Sanders M, Schönian G, Stark O, Sundar S, Vanaerschot M, Hertz-Fowler C, Dujardin JC, Berriman M. 2011. Whole genome sequencing of multiple Leishmania donovani clinical isolates provides insights into population structure and mechanisms of drug resistance. Genome Res 21:2143–2156. doi: 10.1101/gr.123430.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Masmoudi A, Hariz W, Marrekchi S, Amouri M, Turki H. 2013. Old World cutaneous leishmaniasis: diagnosis and treatment. J Dermatol Case Rep 7:31–41. doi: 10.1186/1752-1947-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bensoussan E, Nasereddin A, Jonas F, Schnur LF, Jaffe CL. 2006. Comparison of PCR assays for diagnosis of cutaneous leishmaniasis. J Clin Microbiol 44:1435–1439. doi: 10.1128/JCM.44.4.1435-1439.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Goto H, Lindoso JA. 2010. Current diagnosis and treatment of cutaneous and mucocutaneous leishmaniasis. Expert Rev Anti Infect Ther 8:419–433. doi: 10.1586/eri.10.19. [DOI] [PubMed] [Google Scholar]
- 77.Cruz I, Millet A, Carrillo E, Chenik M, Salotra P, Verma S, Veland N, Jara M, Adaui V, Castrillon C, Arévalo J, Moreno J, Canavate C. 2013. An approach for interlaboratory comparison of conventional and real-time PCR assays for diagnosis of human leishmaniasis. Exp Parasitol 134:281–289. doi: 10.1016/j.exppara.2013.03.026. [DOI] [PubMed] [Google Scholar]
- 78.Kirk R. 1949. The differentiation and nomenclature of Leishmania. Parasitology 39:263–273. doi: 10.1017/S0031182000083839. [DOI] [PubMed] [Google Scholar]
- 79.Katakura K, Matsumoto Y, Gomez EA, Furuya M, Hashiguchi Y. 1993. Molecular karyotype characterization of Leishmania panamensis, Leishmania mexicana, and Leishmania major-like parasites: agents of cutaneous leishmaniasis in Ecuador. Am J Trop Med Hyg 48:707–715. [DOI] [PubMed] [Google Scholar]
- 80.de Oliveira Silva S, Wu AA, Evans DA, Vieira LQ, Melo MN. 2009. Leishmania sp. isolated from human cases of cutaneous leishmaniasis in Brazil characterized as Leishmania major-like. Acta Trop 112:239–248. doi: 10.1016/j.actatropica.2009.07.026. [DOI] [PubMed] [Google Scholar]
- 81.Yamasaki H, Agatsuma T, Pavon B, Moran M, Furuya M, Aoki T. 1994. Leishmania major-like parasite, a pathogenic agent of cutaneous leishmaniasis in Paraguay. Am J Trop Med Hyg 51:749–757. [DOI] [PubMed] [Google Scholar]
- 82.Asato Y, Oshiro M, Myint CK, Yamamoto Y, Kato H, Marco JD, Mimori T, Gomez EA, Hashiguchi Y, Uezato H. 2009. Phylogenic analysis of the genus Leishmania by cytochrome b gene sequencing. Exp Parasitol 121:352–361. doi: 10.1016/j.exppara.2008.12.013. [DOI] [PubMed] [Google Scholar]
- 83.Momen H, Grimaldi G Jr, Pacheco RS, Jaffe CL, McMahon-Pratt D, Marzochi MC. 1985. Brazilian Leishmania stocks phenotypically similar to Leishmania major. Am J Trop Med Hyg 34:1076–1084. [DOI] [PubMed] [Google Scholar]
- 84.Chicharro C, Alvar J. 2003. Lower trypanosomatids in HIV/AIDS patients. Ann Trop Med Parasitol 97(Suppl 1):S75–S78. doi: 10.1179/000349803225002552. [DOI] [PubMed] [Google Scholar]
- 85.Srivastava P, Prajapati VK, Vanaerschot M, Van der Auwera G, Dujardin JC, Sundar S. 2010. Detection of Leptomonas sp. parasites in clinical isolates of kala-azar patients from India. Infect Genet Evol 10:1145–1150. doi: 10.1016/j.meegid.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hide M, Bañuls AL, Tibayrenc M. 2001. Genetic heterogeneity and phylogenetic status of Leishmania (Leishmania) infantum zymodeme MON-1: epidemiological implications. Parasitology 123:425–432. doi: 10.1017/S003118200100871X. [DOI] [PubMed] [Google Scholar]
- 87.Saravia NG, Segura I, Holguin AF, Santrich C, Valderrama L, Ocampo C. 1998. Epidemiologic, genetic, and clinical associations among phenotypically distinct populations of Leishmania (Viannia) in Colombia. Am J Trop Med Hyg 59:86–94. [DOI] [PubMed] [Google Scholar]
- 88.Pratlong F, Rioux JA, Marty P, Faraut-Gambarelli F, Dereure J, Lanotte G, Dedet JP. 2004. Isoenzymatic analysis of 712 strains of Leishmania infantum in the south of France and relationship of enzymatic polymorphism to clinical and epidemiological features. J Clin Microbiol 42:4077–4082. doi: 10.1128/JCM.42.9.4077-4082.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Evans DA, Lanham SM, Baldwin CI, Peters W. 1984. The isolation and isoenzyme characterization of Leishmania braziliensis subsp. from patients with cutaneous leishmaniasis acquired in Belize. Trans R Soc Trop Med Hyg 78:35–42. doi: 10.1016/0035-9203(84)90168-8. [DOI] [PubMed] [Google Scholar]
- 90.Cupolillo E, Grimaldi G Jr, Momen H. 1994. A general classification of New World Leishmania using numerical zymotaxonomy. Am J Trop Med Hyg 50:296–311. [DOI] [PubMed] [Google Scholar]
- 91.Schönian G, Mauricio I, Gramiccia M, Canavate C, Boelaert M, Dujardin JC. 2008. Leishmaniases in the Mediterranean in the era of molecular epidemiology. Trends Parasitol 24:135–142. doi: 10.1016/j.pt.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 92.McMahon-Pratt D, Bennett E, David JR. 1982. Monoclonal antibodies that distinguish subspecies of Leishmania braziliensis. J Immunol 129:926–927. [PubMed] [Google Scholar]
- 93.Saravia NG, Weigle K, Navas C, Segura I, Valderrama L, Valencia AZ, Escorcia B, McMahon-Pratt D. 2002. Heterogeneity, geographic distribution, and pathogenicity of serodemes of Leishmania Viannia in Colombia. Am J Trop Med Hyg 66:738–744. [DOI] [PubMed] [Google Scholar]
- 94.Shaw JJ, De Faria DL, Basano SA, Corbett CE, Rodrigues CJ, Ishikawa EA, Camargo LM. 2007. The aetiological agents of American cutaneous leishmaniasis in the municipality of Monte Negro, Rondonia State, western Amazonia, Brazil. Ann Trop Med Parasitol 101:681–688. doi: 10.1179/136485907X229103. [DOI] [PubMed] [Google Scholar]
- 95.Brito ME, Andrade MS, Mendonca MG, Silva CJ, Almeida EL, Lima BS, Felix SM, Abath FG, da Graca GC, Porrozzi R, Ishikawa EA, Shaw JJ, Cupolillo E, Brandao-Filho SP. 2009. Species diversity of Leishmania (Viannia) parasites circulating in an endemic area for cutaneous leishmaniasis located in the Atlantic rainforest region of northeastern Brazil. Trop Med Int Health 14:1278–1286. doi: 10.1111/j.1365-3156.2009.02361.x. [DOI] [PubMed] [Google Scholar]
- 96.Jaffe CL, Bennett E, Grimaldi G Jr, McMahon-Pratt D. 1984. Production and characterization of species-specific monoclonal antibodies against Leishmania donovani for immunodiagnosis. J Immunol 133:440–447. [PubMed] [Google Scholar]
- 97.McMahon-Pratt D, Bennett E, Grimaldi G, Jaffe CL. 1985. Subspecies- and species-specific antigens of Leishmania mexicana characterized by monoclonal antibodies. J Immunol 134:1935–1940. [PubMed] [Google Scholar]
- 98.Grimaldi G, McMahon-Pratt D. 1996. Monoclonal antibodies for the identification of New World Leishmania species. Mem Inst Oswaldo Cruz 91:37–42. [DOI] [PubMed] [Google Scholar]
- 99.Hanham CA, Zhao FS, Shaw JJ, Lainson R. 1991. Monoclonal antibodies for the identification of New World Leishmania. Trans R Soc Trop Med Hyg 85:220. doi: 10.1016/0035-9203(91)90029-X. [DOI] [PubMed] [Google Scholar]
- 100.Canto-Lara SB, Van Wynsberghe NR, Vargas-Gonzalez A, Ojeda-Farfan FF, Andrade-Narvaez FJ. 1999. Use of monoclonal antibodies for the identification of Leishmania spp. isolated from humans and wild rodents in the State of Campeche, Mexico. Mem Inst Oswaldo Cruz 94:305–309. [DOI] [PubMed] [Google Scholar]
- 101.Cassagne C, Pratlong F, Jeddi F, Benikhlef R, Aoun K, Normand AC, Faraut F, Bastien P, Piarroux R. 2014. Identification of Leishmania at the species level with matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Microbiol Infect 20:551–557. doi: 10.1111/1469-0691.12387. [DOI] [PubMed] [Google Scholar]
- 102.Talmi-Frank D, Nasereddin A, Schnur LF, Schönian G, Özensoy Töz S, Jaffe CL, Baneth G. 2010. Detection and identification of Old World Leishmania by high resolution melt analysis. PLoS Negl Trop Dis 4:e581. doi: 10.1371/journal.pntd.0000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nasereddin A, Jaffe CL. 2010. Rapid diagnosis of Old World leishmaniasis by high-resolution melting analysis of the 7SL RNA gene. J Clin Microbiol 48:2240–2242. doi: 10.1128/JCM.00553-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Weirather JL, Jeronimo SM, Gautam S, Sundar S, Kang M, Kurtz MA, Haque R, Schriefer A, Talhari S, Carvalho EM, Donelson JE, Wilson ME. 2011. Serial quantitative PCR assay for detection, species discrimination, and quantification of Leishmania spp. in human samples. J Clin Microbiol 49:3892–3904. doi: 10.1128/JCM.r00764-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tsukayama P, Nunez JH, De Los Santos M, Soberon V, Lucas CM, Matlashewski G, Llanos-Cuentas A, Ore M, Baldeviano GC, Edgel KA, Lescano AG, Graf PC, Bacon DJ. 2013. A FRET-based real-time PCR assay to identify the main causal agents of New World tegumentary leishmaniasis. PLoS Negl Trop Dis 7:e1956. doi: 10.1371/journal.pntd.0001956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nasereddin A, Bensoussan-Hermano E, Schönian G, Baneth G, Jaffe CL. 2008. Molecular diagnosis of Old World cutaneous leishmaniasis and species identification by use of a reverse line blot hybridization assay. J Clin Microbiol 46:2848–2855. doi: 10.1128/JCM.00951-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Martinez-Calvillo S, Sunkin SM, Yan S, Fox M, Stuart K, Myler PJ. 2001. Genomic organization and functional characterization of the Leishmania major Friedlin ribosomal RNA gene locus. Mol Biochem Parasitol 116:147–157. doi: 10.1016/S0166-6851(01)00310-3. [DOI] [PubMed] [Google Scholar]
- 108.Inga R, De Doncker S, Gomez J, Lopez M, Garcia R, Le Ray D, Arévalo J, Dujardin JC. 1998. Relation between variation in copy number of ribosomal RNA encoding genes and size of harbouring chromosomes in Leishmania of subgenus Viannia. Mol Biochem Parasitol 92:219–228. doi: 10.1016/S0166-6851(98)00009-7. [DOI] [PubMed] [Google Scholar]
- 109.el Tai NO, Osman OF, El Fari M, Presber W, Schönian G. 2000. Genetic heterogeneity of ribosomal internal transcribed spacer in clinical samples of Leishmania donovani spotted on filter paper as revealed by single-strand conformation polymorphisms and sequencing. Trans R Soc Trop Med Hyg 94:575–579. doi: 10.1016/S0035-9203(00)90093-2. [DOI] [PubMed] [Google Scholar]
- 110.Schönian G, Akuffo H, Lewin S, Maasho K, Nylen S, Pratlong F, Eisenberger CL, Schnur LF, Presber W. 2000. Genetic variability within the species Leishmania aethiopica does not correlate with clinical variations of cutaneous leishmaniasis. Mol Biochem Parasitol 106:239–248. doi: 10.1016/S0166-6851(99)00216-9. [DOI] [PubMed] [Google Scholar]
- 111.de Almeida ME, Steurer FJ, Koru O, Herwaldt BL, Pieniazek NJ, da Silva AJ. 2011. Identification of Leishmania spp. by molecular amplification and DNA sequencing analysis of a fragment of rRNA internal transcribed spacer 2. J Clin Microbiol 49:3143–3149. doi: 10.1128/JCM.01177-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schönian G, Nasereddin A, Dinse N, Schweynoch C, Schallig HD, Presber W, Jaffe CL. 2003. PCR diagnosis and characterization of Leishmania in local and imported clinical samples. Diagn Microbiol Infect Dis 47:349–358. doi: 10.1016/S0732-8893(03)00093-2. [DOI] [PubMed] [Google Scholar]
- 113.Roelfsema JH, Nozari N, Herremans T, Kortbeek LM, Pinelli E. 2011. Evaluation and improvement of two PCR targets in molecular typing of clinical samples of Leishmania patients. Exp Parasitol 127:36–41. doi: 10.1016/j.exppara.2010.06.024. [DOI] [PubMed] [Google Scholar]
- 114.Spanakos G, Piperaki ET, Menounos PG, Tegos N, Flemetakis A, Vakalis NC. 2008. Detection and species identification of Old World Leishmania in clinical samples using a PCR-based method. Trans R Soc Trop Med Hyg 102:46–53. doi: 10.1016/j.trstmh.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 115.Odiwuor SO, Saad AA, De Doncker S, Maes I, Laurent T, El Safi S, Mbuchi M, Buscher P, Dujardin JC, Van der Auwera G. 2011. Universal PCR assays for the differential detection of all Old World Leishmania species. Eur J Clin Microbiol Infect Dis 30:209–218. doi: 10.1007/s10096-010-1071-3. [DOI] [PubMed] [Google Scholar]
- 116.Parvizi P, Ready PD. 2008. Nested PCRs and sequencing of nuclear ITS-rDNA fragments detect three Leishmania species of gerbils in sandflies from Iranian foci of zoonotic cutaneous leishmaniasis. Trop Med Int Health 13:1159–1171. doi: 10.1111/j.1365-3156.2008.02121.x. [DOI] [PubMed] [Google Scholar]
- 117.Rotureau B, Ravel C, Couppie P, Pratlong F, Nacher M, Dedet JP, Carme B. 2006. Use of PCR-restriction fragment length polymorphism analysis to identify the main new world Leishmania species and analyze their taxonomic properties and polymorphism by application of the assay to clinical samples. J Clin Microbiol 44:459–467. doi: 10.1128/JCM.44.2.459-467.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cupolillo E, Grimaldi JG, Momen H, Beverley SM. 1995. Intergenic region typing (IRT): a rapid molecular approach to the characterization and evolution of Leishmania. Mol Biochem Parasitol 73:145–155. doi: 10.1016/0166-6851(95)00108-D. [DOI] [PubMed] [Google Scholar]
- 119.Davila AM, Momen H. 2000. Internal-transcribed-spacer (ITS) sequences used to explore phylogenetic relationships within Leishmania. Ann Trop Med Parasitol 94:651–654. doi: 10.1080/00034980050152085. [DOI] [PubMed] [Google Scholar]
- 120.Rahbarian N, Mesgarian A, Mahmoudi RM, Hajaran H, Shahbazi F, Mesgarian Z, Taghipour N. 2009. Identification of Leishmania species isolated from human cutaneous leishmaniasis using PCR method. J Res Health Sci 9:48–51. [PubMed] [Google Scholar]
- 121.Uliana SR, Nelson K, Beverley SM, Camargo EP, Floeter-Winter LM. 1994. Discrimination amongst Leishmania by polymerase chain reaction and hybridization with small subunit ribosomal DNA derived oligonucleotides. J Eukaryot Microbiol 41:324–330. doi: 10.1111/j.1550-7408.1994.tb06085.x. [DOI] [PubMed] [Google Scholar]
- 122.Schulz A, Mellenthin K, Schönian G, Fleischer B, Drosten C. 2003. Detection, differentiation, and quantitation of pathogenic Leishmania organisms by a fluorescence resonance energy transfer-based real-time PCR assay. J Clin Microbiol 41:1529–1535. doi: 10.1128/JCM.41.4.1529-1535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Meredith SEO, Zijlstra EE, Schoone GJ, Kroon CCM, van Eys GJJM, Schaeffer KU, el-Hassan AM, Lawyer PG. 1993. Development and application of the polymerase chain reaction for the detection and identification of Leishmania parasites in clinical material. Arch Inst Pasteur Tunis 70:419–431. [PubMed] [Google Scholar]
- 124.van Eys GJ, Schoone GJ, Kroon NC, Ebeling SB. 1992. Sequence analysis of small subunit ribosomal RNA genes and its use for detection and identification of Leishmania parasites. Mol Biochem Parasitol 51:133–142. doi: 10.1016/0166-6851(92)90208-2. [DOI] [PubMed] [Google Scholar]
- 125.de Lima AC, Zampieri RA, Tomokane TY, Laurenti MD, Silveira FT, Corbett CE, Floeter-Winter LM, Gomes CM. 2011. Leishmania sp. identification by PCR associated with sequencing of target SSU rDNA in paraffin-embedded skin samples stored for more than 30 years. Parasitol Res 108:1525–1531. doi: 10.1007/s00436-010-2208-0. [DOI] [PubMed] [Google Scholar]
- 126.Savani ES, Nunes VL, Galati EA, Castilho TM, Araujo FS, Ilha IM, Camargo MC, D'Auria SR, Floeter-Winter LM. 2005. Occurrence of co-infection by Leishmania (Leishmania) chagasi and Trypanosoma (Trypanozoon) evansi in a dog in the state of Mato Grosso do Sul, Brazil. Mem Inst Oswaldo Cruz 100:739–741. doi: 10.1590/S0074-02762005000700011. [DOI] [PubMed] [Google Scholar]
- 127.Deborggraeve S, Laurent T, Espinosa D, Van der Auwera G, Mbuchi M, Wasunna M, El-Safi S, Al-Basheer AA, Arévalo J, Miranda-Verástegui C, Leclipteux T, Mertens P, Dujardin JC, Herdewijn P, Büscher P. 2008. A simplified and standardized polymerase chain reaction format for the diagnosis of leishmaniasis. J Infect Dis 198:1565–1572. doi: 10.1086/592509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cruz I, Canavate C, Rubio JM, Morales MA, Chicharro C, Laguna F, Jimenez-Mejias M, Sirera G, Videla S, Alvar J. 2002. A nested polymerase chain reaction (Ln-PCR) for diagnosing and monitoring Leishmania infantum infection in patients co-infected with human immunodeficiency virus. Trans R Soc Trop Med Hyg 96(Suppl 1):S185–S189. doi: 10.1016/S0035-9203(02)90074-X. [DOI] [PubMed] [Google Scholar]
- 129.Simpson AG, Stevens JR, Lukeš J. 2006. The evolution and diversity of kinetoplastid flagellates. Trends Parasitol 22:168–174. doi: 10.1016/j.pt.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 130.Aphasizhev R, Aphasizheva I. 2011. Mitochondrial RNA processing in trypanosomes. Res Microbiol 162:655–663. doi: 10.1016/j.resmic.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Singh N, Curran MD, Middleton D, Rastogi AK. 1999. Characterization of kinetoplast DNA minicircles of an Indian isolate of Leishmania donovani. Acta Trop 73:313–319. doi: 10.1016/S0001-706X(99)00036-4. [DOI] [PubMed] [Google Scholar]
- 132.Degrave W, Fernandes O, Campbell D, Bozza M, Lopes U. 1994. Use of molecular probes and PCR for detection and typing of Leishmania—a mini-review. Mem Inst Oswaldo Cruz 89:463–469. doi: 10.1590/S0074-02761994000300032. [DOI] [PubMed] [Google Scholar]
- 133.Gomes Rodrigues EH, da Silva Soares FC, Werkhauser RP, de Brito ME, Fernandes O, Coutinho Abath FG, Brandao A. 2013. The compositional landscape of minicircle sequences isolated from active lesions and scars of American cutaneous leishmaniasis. Parasit Vectors 6:228. doi: 10.1186/1756-3305-6-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Noyes HA, Reyburn H, Bailey JW, Smith D. 1998. A nested-PCR-based schizodeme method for identifying Leishmania kinetoplast minicircle classes directly from clinical samples and its application to the study of the epidemiology of Leishmania tropica in Pakistan. J Clin Microbiol 36:2877–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Aransay AM, Scoulica E, Tselentis Y. 2000. Detection and identification of Leishmania DNA within naturally infected sand flies by seminested PCR on minicircle kinetoplastic DNA. Appl Environ Microbiol 66:1933–1938. doi: 10.1128/AEM.66.5.1933-1938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Anders G, Eisenberger CL, Jonas F, Greenblatt CL. 2002. Distinguishing Leishmania tropica and Leishmania major in the Middle East using the polymerase chain reaction with kinetoplast DNA-specific primers. Trans R Soc Trop Med Hyg 96(Suppl 1):S87–S92. doi: 10.1016/S0035-9203(02)90057-X. [DOI] [PubMed] [Google Scholar]
- 137.Mahboudi F, Abolhassani M, Tehrani SR, Azimi M, Asmar M. 2002. Differentiation of Old and New World Leishmania species at complex and species levels by PCR. Scand J Infect Dis 34:756–758. doi: 10.1080/0036554021000026930. [DOI] [PubMed] [Google Scholar]
- 138.Mahboudi F, Abolhassan M, Yaran M, Mobtaker H, Azizi M. 2001. Identification and differentiation of Iranian Leishmania species by PCR amplification of kDNA. Scand J Infect Dis 33:596–598. doi: 10.1080/00365540110026746. [DOI] [PubMed] [Google Scholar]
- 139.Brenière SF, Telleria J, Bosseno MF, Buitrago R, Bastrenta B, Cuny G, Bañuls AL, Brewster S, Barker DC. 1999. Polymerase chain reaction-based identification of New World Leishmania species complexes by specific kDNA probes. Acta Trop 73:283–293. doi: 10.1016/S0001-706X(99)00025-X. [DOI] [PubMed] [Google Scholar]
- 140.Fernandes O, Bozza M, Pascale JM, de Miranda AB, Lopes UG, Degrave WM. 1996. An oligonucleotide probe derived from kDNA minirepeats is specific for Leishmania (Viannia). Mem Inst Oswaldo Cruz 91:279–284. doi: 10.1590/S0074-02761996000300005. [DOI] [PubMed] [Google Scholar]
- 141.Wirth DF, Pratt DM. 1982. Rapid identification of Leishmania species by specific hybridization of kinetoplast DNA in cutaneous lesions. Proc Natl Acad Sci U S A 79:6999–7003. doi: 10.1073/pnas.79.22.6999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Das Gupta S, Ghosh DK, Majumder HK. 1991. A cloned kinetoplast DNA mini-circle fragment from a Leishmania spp. specific for post-kala-azar dermal leishmaniasis strains. Parasitology 102:187–191. doi: 10.1017/S0031182000062478. [DOI] [PubMed] [Google Scholar]
- 143.Rodgers MR, Popper SJ, Wirth DF. 1990. Amplification of kinetoplast DNA as a tool in the detection and diagnosis of Leishmania. Exp Parasitol 71:267–275. doi: 10.1016/0014-4894(90)90031-7. [DOI] [PubMed] [Google Scholar]
- 144.Pirmez C, da Silva Trajano V, Paes-Oliveira Neto M, da-Cruz AM, Goncalves-da-Costa SC, Catanho M, Degrave W, Fernandes O. 1999. Use of PCR in diagnosis of human American tegumentary leishmaniasis in Rio de Janeiro, Brazil. J Clin Microbiol 37:1819–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.de Pita-Pereira D, Alves CR, Souza MB, Brazil RP, Bertho AL, de Figueiredo BA, Britto CC. 2005. Identification of naturally infected Lutzomyia intermedia and Lutzomyia migonei with Leishmania (Viannia) braziliensis in Rio de Janeiro (Brazil) revealed by a PCR multiplex non-isotopic hybridisation assay. Trans R Soc Trop Med Hyg 99:905–913. doi: 10.1016/j.trstmh.2005.06.019. [DOI] [PubMed] [Google Scholar]
- 146.Salotra P, Sreenivas G, Pogue GP, Lee N, Nakhasi HL, Ramesh V, Negi NS. 2001. Development of a species-specific PCR assay for detection of Leishmania donovani in clinical samples from patients with kala-azar and post-kala-azar dermal leishmaniasis. J Clin Microbiol 39:849–854. doi: 10.1128/JCM.39.3.849-854.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rocha MN, Margonari C, Presot IM, Soares RP. 2010. Evaluation of 4 polymerase chain reaction protocols for cultured Leishmania spp. typing. Diagn Microbiol Infect Dis 68:401–409. doi: 10.1016/j.diagmicrobio.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 148.De Bruijn MH, Barker DC. 1992. Diagnosis of New World leishmaniasis: specific detection of species of the Leishmania braziliensis complex by amplification of kinetoplast DNA. Acta Trop 52:45–58. doi: 10.1016/0001-706X(92)90006-J. [DOI] [PubMed] [Google Scholar]
- 149.Vergel C, Walker J, Saravia NG. 2005. Amplification of human DNA by primers targeted to Leishmania kinetoplast DNA and post-genome considerations in the detection of parasites by a polymerase chain reaction. Am J Trop Med Hyg 72:423–429. [PubMed] [Google Scholar]
- 150.Lopez M, Inga R, Cangalaya M, Echevarria J, Llanos-Cuentas A, Orrego C, Arévalo J. 1993. Diagnosis of Leishmania using the polymerase chain reaction: a simplified procedure for field work. Am J Trop Med Hyg 49:348–356. [DOI] [PubMed] [Google Scholar]
- 151.Miranda A, Carrasco R, Paz H, Pascale JM, Samudio F, Saldana A, Santamaria G, Mendoza Y, Calzada JE. 2009. Molecular epidemiology of American tegumentary leishmaniasis in Panama. Am J Trop Med Hyg 81:565–571. doi: 10.4269/ajtmh.2009.08-0265. [DOI] [PubMed] [Google Scholar]
- 152.Bhattarai NR, Dujardin JC, Rijal S, De Doncker S, Boelaert M, Van der Auwera G. 2010. Development and evaluation of different PCR-based typing methods for discrimination of Leishmania donovani isolates from Nepal. Parasitology 137:947–957. doi: 10.1017/S0031182009991752. [DOI] [PubMed] [Google Scholar]
- 153.Laurent T, Rijal S, Yardley V, Croft S, De Doncker S, Decuypere S, Khanal B, Singh R, Schönian G, Kuhls K, Chappuis F, Dujardin JC. 2007. Epidemiological dynamics of antimonial resistance in Leishmania donovani: genotyping reveals a polyclonal population structure among naturally-resistant clinical isolates from Nepal. Infect Genet Evol 7:206–212. doi: 10.1016/j.meegid.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 154.Rijal S, Ostyn B, Uranw S, Rai K, Bhattarai NR, Dorlo TP, Beijnen JH, Vanaerschot M, Decuypere S, Dhakal SS, Das ML, Karki P, Singh R, Boelaert M, Dujardin JC. 2013. Increasing failure of miltefosine in the treatment of kala-azar in Nepal and the potential role of parasite drug resistance, reinfection, or noncompliance. Clin Infect Dis 56:1530–1538. doi: 10.1093/cid/cit102. [DOI] [PubMed] [Google Scholar]
- 155.Volpini AC, Passos VM, Oliveira GC, Romanha AJ. 2004. PCR-RFLP to identify Leishmania (Viannia) braziliensis and L. (Leishmania) amazonensis causing American cutaneous leishmaniasis. Acta Trop 90:31–37. doi: 10.1016/j.actatropica.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 156.Nicolas L, Milon G, Prina E. 2002. Rapid differentiation of Old World Leishmania species by LightCycler polymerase chain reaction and melting curve analysis. J Microbiol Methods 51:295–299. doi: 10.1016/S0167-7012(02)00099-4. [DOI] [PubMed] [Google Scholar]
- 157.Pita-Pereira D, Lins R, Oliveira MP, Lima RB, Pereira BA, Moreira OC, Brazil RP, Britto C. 2012. SYBR Green-based real-time PCR targeting kinetoplast DNA can be used to discriminate between the main etiologic agents of Brazilian cutaneous and visceral leishmaniases. Parasit Vectors 5:15. doi: 10.1186/1756-3305-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Adams ER, Schoone GJ, Ageed AF, Safi SE, Schallig HD. 2010. Development of a reverse transcriptase loop-mediated isothermal amplification (LAMP) assay for the sensitive detection of Leishmania parasites in clinical samples. Am J Trop Med Hyg 82:591–596. doi: 10.4269/ajtmh.2010.09-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Karani M, Sotiriadou I, Plutzer J, Karanis P. 2014. Bench-scale experiments for the development of a unified loop-mediated isothermal amplification (LAMP) assay for the in vitro diagnosis of Leishmania species' promastigotes. Epidemiol Infect 142:1671–1677. doi: 10.1017/S0950268813002677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Takagi H, Itoh M, Islam MZ, Razzaque A, Ekram AR, Hashighuchi Y, Noiri E, Kimura E. 2009. Sensitive, specific, and rapid detection of Leishmania donovani DNA by loop-mediated isothermal amplification. Am J Trop Med Hyg 81:578–582. doi: 10.4269/ajtmh.2009.09-0145. [DOI] [PubMed] [Google Scholar]
- 162.Verma S, Avishek K, Sharma V, Negi NS, Ramesh V, Salotra P. 2013. Application of loop-mediated isothermal amplification assay for the sensitive and rapid diagnosis of visceral leishmaniasis and post-kala-azar dermal leishmaniasis. Diagn Microbiol Infect Dis 75:390–395. doi: 10.1016/j.diagmicrobio.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 163.Luyo-Acero GE, Uezato H, Oshiro M, Takei K, Kariya K, Katakura K, Gomez-Landires E, Hashiguchi Y, Nonaka S. 2004. Sequence variation of the cytochrome b gene of various human infecting members of the genus Leishmania and their phylogeny. Parasitology 128:483–491. doi: 10.1017/S0031182004004792. [DOI] [PubMed] [Google Scholar]
- 164.Foulet F, Botterel F, Buffet P, Morizot G, Rivollet D, Deniau M, Pratlong F, Costa JM, Bretagne S. 2007. Detection and identification of Leishmania species from clinical specimens by using a real-time PCR assay and sequencing of the cytochrome b gene. J Clin Microbiol 45:2110–2115. doi: 10.1128/JCM.02555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kato H, Uezato H, Gomez EA, Terayama Y, Calvopina M, Iwata H, Hashiguchi Y. 2007. Establishment of a mass screening method of sand fly vectors for Leishmania infection by molecular biological methods. Am J Trop Med Hyg 77:324–329. [PubMed] [Google Scholar]
- 166.Kato H, Caceres AG, Mimori T, Ishimaru Y, Sayed AS, Fujita M, Iwata H, Uezato H, Velez LN, Gomez EA, Hashiguchi Y. 2010. Use of FTA cards for direct sampling of patients' lesions in the ecological study of cutaneous leishmaniasis. J Clin Microbiol 48:3661–3665. doi: 10.1128/JCM.00498-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kato H, Caceres AG, Gomez EA, Mimori T, Uezato H, Marco JD, Barroso PA, Iwata H, Hashiguchi Y. 2008. Molecular mass screening to incriminate sand fly vectors of Andean-type cutaneous leishmaniasis in Ecuador and Peru. Am J Trop Med Hyg 79:719–721. [PubMed] [Google Scholar]
- 168.Rivier D, Bovay P, Shah R, Didisheim S, Mauel J. 1999. Vaccination against Leishmania major in a CBA mouse model of infection: role of adjuvants and mechanism of protection. Parasite Immunol 21:461–473. doi: 10.1046/j.1365-3024.1999.00244.x. [DOI] [PubMed] [Google Scholar]
- 169.Victoir K, De Doncker S, Cabrera L, Alvarez E, Arévalo J, Llanos-Cuentas A, Le Ray D, Dujardin JC. 2003. Direct identification of Leishmania species in biopsies from patients with American tegumentary leishmaniasis. Trans R Soc Trop Med Hyg 97:80–87. doi: 10.1016/S0035-9203(03)90031-9. [DOI] [PubMed] [Google Scholar]
- 170.Victoir K, Bañuls AL, Arévalo J, Llanos-Cuentas A, Hamers R, Noel S, De Doncker S, Le Ray D, Tibayrenc M, Dujardin JC. 1998. The gp63 gene locus, a target for genetic characterization of Leishmania belonging to subgenus Viannia. Parasitology 117:1–13. [PubMed] [Google Scholar]
- 171.Guerbouj S, Victoir K, Guizani I, Seridi N, Nuwayri-Salti N, Belkaid M, Ismail RB, Le Ray D, Dujardin JC. 2001. gp63 gene polymorphism and population structure of Leishmania donovani complex: influence of the host selection pressure? Parasitology 122:25–35. doi: 10.1017/S0031182000007125. [DOI] [PubMed] [Google Scholar]
- 172.Mauricio IL, Gaunt MW, Stothard JR, Miles MA. 2001. Genetic typing and phylogeny of the Leishmania donovani complex by restriction analysis of PCR amplified gp63 intergenic regions. Parasitology 122:393–403. doi: 10.1017/S0031182001007466. [DOI] [PubMed] [Google Scholar]
- 173.Mauricio IL, Howard MK, Stothard JR, Miles MA. 1999. Genomic diversity in the Leishmania donovani complex. Parasitology 119:237–246. doi: 10.1017/S0031182099004710. [DOI] [PubMed] [Google Scholar]
- 174.Seridi N, Belkaid M, Quispe-Tintaya W, Zidane C, Dujardin JC. 2008. Application of PCR-RFLP for the exploration of the molecular diversity of Leishmania infantum in Algeria. Trans R Soc Trop Med Hyg 102:556–563. doi: 10.1016/j.trstmh.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 175.Yao C, Luo J, Storlie P, Donelson JE, Wilson ME. 2004. Multiple products of the Leishmania chagasi major surface protease (MSP or GP63) gene family. Mol Biochem Parasitol 135:171–183. doi: 10.1016/j.molbiopara.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 176.Victoir K, Dujardin JC. 2002. How to succeed in parasitic life without sex? Asking Leishmania. Trends Parasitol 18:81–85. doi: 10.1016/S1471-4922(01)02199-7. [DOI] [PubMed] [Google Scholar]
- 177.Gupta S, Anderson RM. 1999. Population structure of pathogens: the role of immune selection. Parasitol Today 15:497–501. doi: 10.1016/S0169-4758(99)01559-8. [DOI] [PubMed] [Google Scholar]
- 178.Quispe-Tintaya KW, Ying X, Dedet JP, Rijal S, De Bolle X, Dujardin JC. 2004. Antigen genes for molecular epidemiology of leishmaniasis: polymorphism of cysteine proteinase B and surface metalloprotease glycoprotein 63 in the Leishmania donovani complex. J Infect Dis 189:1035–1043. doi: 10.1086/382049. [DOI] [PubMed] [Google Scholar]
- 179.Laurent T, Van der Auwera G, Hide M, Mertens P, Quispe-Tintaya W, Deborggraeve S, De Doncker S, Leclipteux T, Bañuls AL, Büscher P, Dujardin JC. 2009. Identification of Old World Leishmania spp. by specific polymerase chain reaction amplification of cysteine proteinase B genes and rapid dipstick detection. Diagn Microbiol Infect Dis 63:173–181. doi: 10.1016/j.diagmicrobio.2008.10.015. [DOI] [PubMed] [Google Scholar]
- 180.Hide M, Bañuls AL. 2008. Polymorphisms of cpb multicopy genes in the Leishmania (Leishmania) donovani complex. Trans R Soc Trop Med Hyg 102:105–106. doi: 10.1016/j.trstmh.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 181.Chaouch M, Fathallah-Mili A, Driss M, Lahmadi R, Ayari C, Guizani I, Ben Said M, BenAbderrazak S. 2013. Identification of Tunisian Leishmania spp. by PCR amplification of cysteine proteinase B (cpb) genes and phylogenetic analysis. Acta Trop 125:357–365. doi: 10.1016/j.actatropica.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 182.Kuru T, Jirata D, Genetu A, Barr S, Mengistu Y, Aseffa A, Gedamu L. 2007. Leishmania aethiopica: identification and characterization of cathepsin L-like cysteine protease genes. Exp Parasitol 115:283–290. doi: 10.1016/j.exppara.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 183.Pollock KG, McNeil KS, Mottram JC, Lyons RE, Brewer JM, Scott P, Coombs GH, Alexander J. 2003. The Leishmania mexicana cysteine protease, CPB28, induces potent Th2 responses. J Immunol 170:1746–1753. doi: 10.4049/jimmunol.170.4.1746. [DOI] [PubMed] [Google Scholar]
- 184.Garcia AL, Kindt A, Quispe-Tintaya KW, Bermudez H, Llanos A, Arévalo J, Bañuls AL, De Doncker S, Le Ray D, Dujardin JC. 2005. American tegumentary leishmaniasis: antigen-gene polymorphism, taxonomy and clinical pleomorphism. Infect Genet Evol 5:109–116. doi: 10.1016/j.meegid.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 185.Montalvo AM, Monzote L, Fraga J, Montano I, Muskus C, Marin M, De Doncker S, Velez ID, Dujardin JC. 2008. PCR-RFLP and RAPD for typing neotropical Leishmania. Biomedica 28:597–606. doi: 10.7705/biomedica.v28i4.66. [DOI] [PubMed] [Google Scholar]
- 186.Hide M, Bañuls AL. 2006. Species-specific PCR assay for L. infantum/L. donovani discrimination. Acta Trop 100:241–245. doi: 10.1016/j.actatropica.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 187.Lopez L, Robayo M, Vargas M, Velez ID. 2012. Thermotherapy. An alternative for the treatment of American cutaneous leishmaniasis. Trials 13:58. doi: 10.1186/1745-6215-13-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Chaouch M, Mhadhbi M, Adams ER, Schoone GJ, Limam S, Gharbi Z, Darghouth MA, Guizani I, BenAbderrazak S. 2013. Development and evaluation of a loop-mediated isothermal amplification assay for rapid detection of Leishmania infantum in canine leishmaniasis based on cysteine protease B genes. Vet Parasitol 198:78–84. doi: 10.1016/j.vetpar.2013.07.038. [DOI] [PubMed] [Google Scholar]
- 189.Hartl FU, Hayer-Hartl M. 2002. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 190.Folgueira C, Cañavate C, Chicharro C, Requena JM. 2007. Genomic organization and expression of the HSP70 locus in New and Old World Leishmania species. Parasitology 134:369–377. doi: 10.1017/S0031182006001570. [DOI] [PubMed] [Google Scholar]
- 191.Fraga J, Montalvo AM, De Doncker S, Dujardin JC, Van der Auwera G. 2010. Phylogeny of Leishmania species based on the heat-shock protein 70 gene. Infect Genet Evol 10:238–245. doi: 10.1016/j.meegid.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 192.Giorgobiani E, Lawyer PG, Babuadze G, Dolidze N, Jochim RC, Tskhvaradze L, Kikaleishvili K, Kamhawi S. 2012. Incrimination of Phlebotomus kandelakii and Phlebotomus balcanicus as vectors of Leishmania infantum in Tbilisi, Georgia. PLoS Negl Trop Dis 6:e1609. doi: 10.1371/journal.pntd.0001609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Zhang CY, Lu XJ, Du XQ, Jian J, Shu L, Ma Y. 2013. Phylogenetic and evolutionary analysis of Chinese Leishmania isolates based on multilocus sequence typing. PLoS One 8:e63124. doi: 10.1371/journal.pone.0063124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Garcia L, Kindt A, Bermudez H, Llanos-Cuentas A, De Doncker S, Arévalo J, Quispe Tintaya KW, Dujardin JC. 2004. Culture-independent species typing of neotropical Leishmania for clinical validation of a PCR-based assay targeting heat shock protein 70 genes. J Clin Microbiol 42:2294–2297. doi: 10.1128/JCM.42.5.2294-2297.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Montalvo AM, Fraga J, Monzote L, Montano I, De Doncker S, Dujardin JC, Van der Auwera G. 2010. Heat-shock protein 70 PCR-RFLP: a universal simple tool for Leishmania species discrimination in the New and Old World. Parasitology 137:1159–1168. doi: 10.1017/S0031182010000089. [DOI] [PubMed] [Google Scholar]
- 196.Montalvo AM, Fraga J, Maes I, Dujardin JC, Van der Auwera G. 2012. Three new sensitive and specific heat-shock protein 70 PCRs for global Leishmania species identification. Eur J Clin Microbiol Infect Dis 31:1453–1461. doi: 10.1007/s10096-011-1463-z. [DOI] [PubMed] [Google Scholar]
- 197.Alves da Silva L, dos Santos de Sousa C, da Graça GC, Porrozzi R, Cupolillo E. 2010. Sequence analysis and PCR-RFLP profiling of the hsp70 gene as a valuable tool for identifying Leishmania species associated with human leishmaniasis in Brazil. Infect Genet Evol 10:77–83. doi: 10.1016/j.meegid.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 198.Garcia AL, Tellez T, Parrado R, Rojas E, Bermudez H, Dujardin JC. 2007. Epidemiological monitoring of American tegumentary leishmaniasis: molecular characterization of a peridomestic transmission cycle in the Amazonian lowlands of Bolivia. Trans R Soc Trop Med Hyg 101:1208–1213. doi: 10.1016/j.trstmh.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 199.Garcia AL, Parrado R, De Doncker S, Bermudez H, Dujardin JC. 2007. American tegumentary leishmaniasis: direct species identification of Leishmania in non-invasive clinical samples. Trans R Soc Trop Med Hyg 101:368–371. doi: 10.1016/j.trstmh.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 200.Cardoso da Graça G, Volpini AC, Romero GA, Oliveira Neto MP, Hueb M, Porrozzi R, Boite MC, Cupolillo E. 2012. Development and validation of PCR-based assays for diagnosis of American cutaneous leishmaniasis and identification of the parasite species. Mem Inst Oswaldo Cruz 107:664–674. doi: 10.1590/S0074-02762012000500014. [DOI] [PubMed] [Google Scholar]
- 201.Fraga J, Veland N, Montalvo AM, Praet N, Boggild AK, Valencia BM, Arévalo J, Llanos-Cuentas A, Dujardin JC, Van der Auwera G. 2012. Accurate and rapid species typing from cutaneous and mucocutaneous leishmaniasis lesions of the New World. Diagn Microbiol Infect Dis 74:142–150. doi: 10.1016/j.diagmicrobio.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 202.Montalvo AM, Fraga J, El Safi S, Gramiccia M, Jaffe CL, Dujardin JC, Van der Auwera G. 2014. Direct Leishmania species typing in Old World clinical samples: evaluation of 3 sensitive methods based on the heat-shock protein 70 gene. Diagn Microbiol Infect Dis 80:35–39. doi: 10.1016/j.diagmicrobio.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 203.Arora SK, Gupta S, Bhardwaj S, Sachdeva N, Sharma NL. 2008. An epitope-specific PCR test for diagnosis of Leishmania donovani infections. Trans R Soc Trop Med Hyg 102:41–45. doi: 10.1016/j.trstmh.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 204.Requena JM, Chicharro C, Garcia L, Parrado R, Puerta CJ, Canavate C. 2012. Sequence analysis of the 3′-untranslated region of HSP70 (type I) genes in the genus Leishmania: its usefulness as a molecular marker for species identification. Parasit Vectors 5:87. doi: 10.1186/1756-3305-5-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Fernandes O, Murthy VK, Kurath U, Degrave WM, Campbell DA. 1994. Mini-exon gene variation in human pathogenic Leishmania species. Mol Biochem Parasitol 66:261–271. doi: 10.1016/0166-6851(94)90153-8. [DOI] [PubMed] [Google Scholar]
- 206.Marfurt J, Nasereddin A, Niederwieser I, Jaffe CL, Beck HP, Felger I. 2003. Identification and differentiation of Leishmania species in clinical samples by PCR amplification of the miniexon sequence and subsequent restriction fragment length polymorphism analysis. J Clin Microbiol 41:3147–3153. doi: 10.1128/JCM.41.7.3147-3153.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Hu RV, Kent AD, Adams ER, van der Veer C, Sabajo LO, Mans DR, de Vries HJ, Schallig HD, Lai A, Fat RF. 2012. First case of cutaneous leishmaniasis caused by Leishmania (Viannia) braziliensis in Suriname. Am J Trop Med Hyg 86:825–827. doi: 10.4269/ajtmh.2012.11-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Mosimann V, Neumayr A, Hatz C, Blum JA. 2013. Cutaneous leishmaniasis in Switzerland: first experience with species-specific treatment. Infection 41:1177–1182. doi: 10.1007/s15010-013-0500-5. [DOI] [PubMed] [Google Scholar]
- 209.Marfurt J, Niederwieser I, Makia ND, Beck HP, Felger I. 2003. Diagnostic genotyping of Old and New World Leishmania species by PCR-RFLP. Diagn Microbiol Infect Dis 46:115–124. doi: 10.1016/S0732-8893(03)00040-3. [DOI] [PubMed] [Google Scholar]
- 210.Zelazny AM, Fedorko DP, Li L, Neva FA, Fischer SH. 2005. Evaluation of 7SL RNA gene sequences for the identification of Leishmania spp. Am J Trop Med Hyg 72:415–420. [PubMed] [Google Scholar]
- 211.Stevenson LG, Fedorko DP, Zelazny AM. 2010. An enhanced method for the identification of Leishmania spp. using real-time polymerase chain reaction and sequence analysis of the 7SL RNA gene region. Diagn Microbiol Infect Dis 66:432–435. doi: 10.1016/j.diagmicrobio.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Bousslimi N, Ben-Ayed S, Ben-Abda I, Aoun K, Bouratbine A. 2012. Natural infection of North African gundi (Ctenodactylus gundi) by Leishmania tropica in the focus of cutaneous leishmaniasis, Southeast Tunisia. Am J Trop Med Hyg 86:962–965. doi: 10.4269/ajtmh.2012.11-0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Ocampo CB, Ferro MC, Cadena H, Gongora R, Perez M, Valderrama-Ardila CH, Quinnell RJ, Alexander N. 2012. Environmental factors associated with American cutaneous leishmaniasis in a new Andean focus in Colombia. Trop Med Int Health 17:1309–1317. doi: 10.1111/j.1365-3156.2012.03065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Ferro C, Marin D, Gongora R, Carrasquilla MC, Trujillo JE, Rueda NK, Marin J, Valderrama-Ardila C, Alexander N, Perez M, Munstermann LE, Ocampo CB. 2011. Phlebotomine vector ecology in the domestic transmission of American cutaneous leishmaniasis in Chaparral, Colombia. Am J Trop Med Hyg 85:847–856. doi: 10.4269/ajtmh.2011.10-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Castilho TM, Camargo LM, Mahon-Pratt D, Shaw JJ, Floeter-Winter LM. 2008. A real-time polymerase chain reaction assay for the identification and quantification of American Leishmania species on the basis of glucose-6-phosphate dehydrogenase. Am J Trop Med Hyg 78:122–132. [PubMed] [Google Scholar]
- 216.Castilho TM, Shaw JJ, Floeter-Winter LM. 2003. New PCR assay using glucose-6-phosphate dehydrogenase for identification of Leishmania species. J Clin Microbiol 41:540–546. doi: 10.1128/JCM.41.2.540-546.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Zhang WW, Miranda-Verastegui C, Arévalo J, Ndao M, Ward B, Llanos-Cuentas A, Matlashewski G. 2006. Development of a genetic assay to distinguish between Leishmania Viannia species on the basis of isoenzyme differences. Clin Infect Dis 42:801–809. doi: 10.1086/500326. [DOI] [PubMed] [Google Scholar]
- 218.Greenblatt CL, Schnur LF, Bar-Gal GK, Ermolaev H, Peleg N, Barrett MP. 2002. Polymorphism among alleles of the 6-phosphogluconate dehydrogenase gene from Leishmania major and Leishmania tropica. Mol Biochem Parasitol 125:185–188. doi: 10.1016/S0166-6851(02)00213-X. [DOI] [PubMed] [Google Scholar]
- 219.Wortmann G, Hochberg L, Houng HH, Sweeney C, Zapor M, Aronson N, Weina P, Ockenhouse CF. 2005. Rapid identification of Leishmania complexes by a real-time PCR assay. Am J Trop Med Hyg 73:999–1004. [PubMed] [Google Scholar]
- 220.Piarroux R, Fontes M, Perasso R, Gambarelli F, Joblet C, Dumon H, Quilici M. 1995. Phylogenetic relationships between Old World Leishmania strains revealed by analysis of a repetitive DNA sequence. Mol Biochem Parasitol 73:249–252. doi: 10.1016/0166-6851(95)00097-K. [DOI] [PubMed] [Google Scholar]
- 221.Minodier P, Piarroux R, Gambarelli F, Joblet C, Dumon H. 1997. Rapid identification of causative species in patients with Old World leishmaniasis. J Clin Microbiol 35:2551–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Haouas N, Garrab S, Gorcii M, Khorchani H, Chargui N, Ravel C, Mezhoud H, Babba H. 2010. Development of a polymerase chain reaction-restriction fragment length polymorphism assay for Leishmania major/Leishmania killicki/Leishmania infantum discrimination from clinical samples, application in a Tunisian focus. Diagn Microbiol Infect Dis 68:152–158. doi: 10.1016/j.diagmicrobio.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 223.Mimori T, Sasaki J, Nakata M, Gomez EA, Uezato H, Nonaka S, Hashiguchi Y, Furuya M, Saya H. 1998. Rapid identification of Leishmania species from formalin-fixed biopsy samples by polymorphism-specific polymerase chain reaction. Gene 210:179–186. doi: 10.1016/S0378-1119(97)00663-X. [DOI] [PubMed] [Google Scholar]
- 224.Hajjaran H, Mohebali M, Teimouri A, Oshaghi MA, Mirjalali H, Kazemi-Rad E, Shiee MR, Naddaf SR. 2014. Identification and phylogenetic relationship of Iranian strains of various Leishmania species isolated from cutaneous and visceral cases of leishmaniasis based on N-acetylglucosamine-1-phosphate transferase gene. Infect Genet Evol 26:203–212. doi: 10.1016/j.meegid.2014.05.026. [DOI] [PubMed] [Google Scholar]
- 225.Haralambous C, Antoniou M, Pratlong F, Dedet JP, Soteriadou K. 2008. Development of a molecular assay specific for the Leishmania donovani complex that discriminates L. donovani/Leishmania infantum zymodemes: a useful tool for typing MON-1. Diagn Microbiol Infect Dis 60:33–42. doi: 10.1016/j.diagmicrobio.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 226.Zauli-Nascimento RC, Miguel DC, Yokoyama-Yasunaka JK, Pereira LI, Pelli de Oliveira MA, Ribeiro-Dias F, Dorta ML, Uliana SR. 2010. In vitro sensitivity of Leishmania (Viannia) braziliensis and Leishmania (Leishmania) amazonensis Brazilian isolates to meglumine antimoniate and amphotericin B. Trop Med Int Health 15:68–76. doi: 10.1111/j.1365-3156.2009.02414.x. [DOI] [PubMed] [Google Scholar]
- 227.Croan DG, Morrison DA, Ellis JT. 1997. Evolution of the genus Leishmania revealed by comparison of DNA and RNA polymerase gene sequences. Mol Biochem Parasitol 89:149–159. doi: 10.1016/S0166-6851(97)00111-4. [DOI] [PubMed] [Google Scholar]
- 228.Hadighi R, Mohebali M, Boucher P, Hajjaran H, Khamesipour A, Ouellette M. 2006. Unresponsiveness to Glucantime treatment in Iranian cutaneous leishmaniasis due to drug-resistant Leishmania tropica parasites. PLoS Med 3:e162. doi: 10.1371/journal.pmed.0030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Luis L, Ramirez A, Aguilar CM, Eresh S, Barker DC, Mendoza-Leon A. 1998. The genomic fingerprinting of the coding region of the beta-tubulin gene in Leishmania identification. Acta Trop 69:193–204. doi: 10.1016/S0001-706X(97)00128-9. [DOI] [PubMed] [Google Scholar]
- 230.Fraga J, Montalvo AM, Van der Auwera G, Maes I, Dujardin JC, Requena JM. 2013. Evolution and species discrimination according to the Leishmania heat-shock protein 20 gene. Infect Genet Evol 18:229–237. doi: 10.1016/j.meegid.2013.05.020. [DOI] [PubMed] [Google Scholar]
- 231.Miranda A, Samudio F, Saldana A, Castillo J, Brandao A, Calzada JE. 2014. The calmodulin intergenic spacer as molecular target for characterization of Leishmania species. Parasit Vectors 7:35. doi: 10.1186/1756-3305-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Restrepo CM, De La Guardia C, Sousa OE, Calzada JE, Fernandez PL, Lleonart R. 2013. AFLP polymorphisms allow high resolution genetic analysis of American tegumentary leishmaniasis agents circulating in Panama and other members of the Leishmania genus. PLoS One 8:e73177. doi: 10.1371/journal.pone.0073177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Mirzaei A, Schweynoch C, Rouhani S, Parvizi P, Schönian G. 2014. Diversity of Leishmania species and of strains of Leishmania major isolated from desert rodents in different foci of cutaneous leishmaniasis in Iran. Trans R Soc Trop Med Hyg 108:502–512. doi: 10.1093/trstmh/tru085. [DOI] [PubMed] [Google Scholar]
- 234.Adaui V, Maes I, Huyse T, Van den Broeck F, Talledo M, Kuhls K, De Doncker S, Maes L, Llanos-Cuentas A, Schönian G, Arévalo J, Dujardin JC. 2011. Multilocus genotyping reveals a polyphyletic pattern among naturally antimony-resistant Leishmania braziliensis isolates from Peru. Infect Genet Evol 11:1873–1880. doi: 10.1016/j.meegid.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 235.Alam MZ, Kuhls K, Schweynoch C, Sundar S, Rijal S, Shamsuzzaman AKM, Raju BVS, Salotra P, Dujardin JC, Schönian G. 2009. Multilocus microsatellite typing (MLMT) reveals genetic homogeneity of Leishmania donovani strains in the Indian subcontinent. Infect Genet Evol 9:24–31. doi: 10.1016/j.meegid.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 236.Kuhls K, Alam MZ, Cupolillo E, Ferreira GE, Mauricio IL, Oddone R, Feliciangeli MD, Wirth T, Miles MA, Schönian G. 2011. Comparative microsatellite typing of new world Leishmania infantum reveals low heterogeneity among populations and its recent Old World origin. PLoS Negl Trop Dis 5:e1155. doi: 10.1371/journal.pntd.0001155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Oddone R, Schweynoch C, Schönian G, de Sousa CS, Cupolillo E, Espinosa D, Arévalo J, Noyes H, Mauricio I, Kuhls K. 2009. Development of a multilocus microsatellite typing approach for discriminating strains of Leishmania (Viannia) species. J Clin Microbiol 47:2818–2825. doi: 10.1128/JCM.00645-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Alam MZ, Kovalenko DA, Kuhls K, Nasyrova RM, Ponomareva VI, Fatullaeva AA, Razakov SA, Schnur LF, Schönian G. 2009. Identification of the agent causing visceral leishmaniasis in Uzbeki and Tajiki foci by analysing parasite DNA extracted from patients' Giemsa-stained tissue preparations. Parasitology 136:981–986. doi: 10.1017/S0031182009006465. [DOI] [PubMed] [Google Scholar]
- 239.Bhattacharyya R, Singh R, Hazra TK, Majumder HK. 1993. Application of polymerase chain reaction with specific and arbitrary primers to identification and differentiation of Leishmania parasites. FEMS Microbiol Lett 114:99–104. doi: 10.1111/j.1574-6968.1993.tb06557.x. [DOI] [PubMed] [Google Scholar]
- 240.Eisenberger CL, Jaffe CL. 1999. Leishmania: identification of Old World species using a permissively primed intergenic polymorphic-polymerase chain reaction. Exp Parasitol 91:70–77.(Erratum, 92: 159–160.) [DOI] [PubMed] [Google Scholar]
- 241.Vuylsteke M, Peleman JD, van Eijk MJT. 2007. AFLP technology for DNA fingerprinting. Nat Protoc 2:1387–1398. doi: 10.1038/nprot.2007.175. [DOI] [PubMed] [Google Scholar]
- 242.Mueller UG, Wolfenbarger LL. 1999. AFLP genotyping and fingerprinting. Trends Ecol Evol 14:389–394. doi: 10.1016/S0169-5347(99)01659-6. [DOI] [PubMed] [Google Scholar]
- 243.Kumar A, Boggula VR, Misra P, Sundar S, Shasany AK, Dube A. 2010. Amplified fragment length polymorphism (AFLP) analysis is useful for distinguishing Leishmania species of visceral and cutaneous forms. Acta Trop 113:202–206. doi: 10.1016/j.actatropica.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 244.Wall EC, Watson J, Armstrong M, Chiodini PL, Lockwood DN. 2012. Epidemiology of imported cutaneous leishmaniasis at the Hospital for Tropical Diseases, London, United Kingdom: use of polymerase chain reaction to identify the species. Am J Trop Med Hyg 86:115–118. doi: 10.4269/ajtmh.2012.10-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Olliaro P, Vaillant M, Arana B, Grogl M, Modabber F, Magill A, Lapujade O, Buffet P, Alvar J. 2013. Methodology of clinical trials aimed at assessing interventions for cutaneous leishmaniasis. PLoS Negl Trop Dis 7:e2130. doi: 10.1371/journal.pntd.0002130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Reveiz L, Maia-Elkhoury AN, Nicholls RS, Romero GA, Yadon ZE. 2013. Interventions for American cutaneous and mucocutaneous leishmaniasis: a systematic review update. PLoS One 8:e61843. doi: 10.1371/journal.pone.0061843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Lavergne RA, Iriart X, Martin-Blondel G, Chauvin P, Menard S, Fillaux J, Cassaing S, Roques-Malecaze C, Arnaud S, Valentin A, Magnaval JF, Marchou B, Berry A. 2014. Contribution of molecular diagnosis to the management of cutaneous leishmaniasis in travellers. Clin Microbiol Infect 20:O528–O530. doi: 10.1111/1469-0691.12487. [DOI] [PubMed] [Google Scholar]
- 248.Buffet PA, Rosenthal E, Gangneux JP, Lightburne E, Couppie P, Morizot G, Lachaud L, Marty P, Dedet JP. 2011. Therapy of leishmaniasis in France: consensus on proposed guidelines. Presse Med 40:173–184. doi: 10.1016/j.lpm.2010.09.023. [DOI] [PubMed] [Google Scholar]
- 249.Vanaerschot M, Decuypere S, Downing T, Imamura H, Stark O, De Doncker S, Roy S, Ostyn B, Maes L, Khanal B, Boelaert M, Schönian G, Berriman M, Chappuis F, Dujardin JC, Sundar S, Rijal S. 2012. Genetic markers for SSG resistance in Leishmania donovani and SSG treatment failure in visceral leishmaniasis patients of the Indian subcontinent. J Infect Dis 206:752–755. doi: 10.1093/infdis/jis424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Coelho AC, Boisvert S, Mukherjee A, Leprohon P, Corbeil J, Ouellette M. 2012. Multiple mutations in heterogeneous miltefosine-resistant Leishmania major population as determined by whole genome sequencing. PLoS Negl Trop Dis 6:e1512. doi: 10.1371/journal.pntd.0001512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Downing T, Stark O, Vanaerschot M, Imamura H, Sanders M, Decuypere S, De Doncker S, Maes I, Rijal S, Sundar S, Dujardin JC, Berriman M, Schönian G. 2012. Genome-wide SNP and microsatellite variation illuminate population-level epidemiology in the Leishmania donovani species complex. Infect Genet Evol 12:149–159. doi: 10.1016/j.meegid.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Strazzulla A, Cocuzza S, Pinzone MR, Postorino MC, Cosentino S, Serra A, Cacopardo B, Nunnari G. 2013. Mucosal leishmaniasis: an underestimated presentation of a neglected disease. Biomed Res Int 2013:805108. doi: 10.1155/2013/805108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]