SUMMARY
Tuberculosis (TB) is an ancient disease with an enormous global impact. Despite declining global incidence, the diagnosis, phenotyping, and epidemiological investigation of TB require significant clinical microbiology laboratory resources. Current methods for the detection and characterization of Mycobacterium tuberculosis consist of a series of laboratory tests varying in speed and performance, each of which yields incremental information about the disease. Since the sequencing of the first M. tuberculosis genome in 1998, genomic tools have aided in the diagnosis, treatment, and control of TB. Here we summarize genomics-based methods that are positioned to be introduced in the modern clinical TB laboratory, and we highlight how recent advances in genomics will improve the detection of antibiotic resistance-conferring mutations and the understanding of M. tuberculosis transmission dynamics and epidemiology. We imagine the future TB clinic as one that relies heavily on genomic interrogation of the M. tuberculosis isolate, allowing for more rapid diagnosis of TB and real-time monitoring of outbreak emergence.
INTRODUCTION
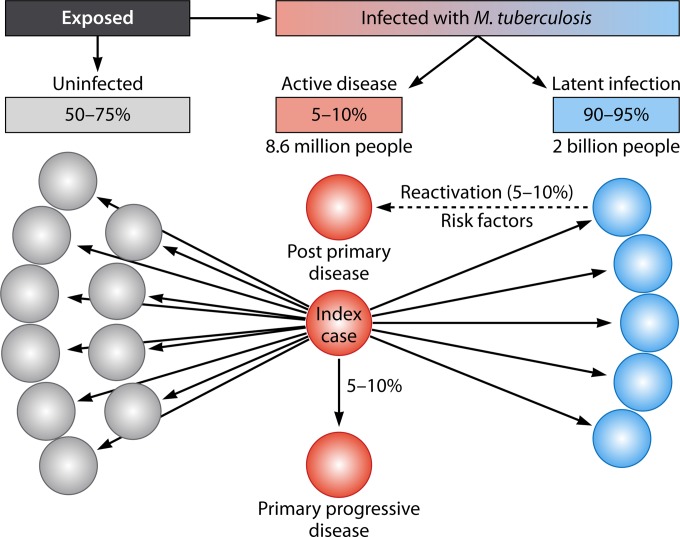
Tuberculosis (TB) is a major global health problem and the second leading cause of death from an infectious disease, after HIV (1). It is an ancient disease that has been with humankind for thousands of years, and our relationship with its causative agent, Mycobacterium tuberculosis, is so intimate that pathogen and host have coevolved (2–4). M. tuberculosis spreads through the inhalation of infectious aerosols, and newly infected individuals progress to one of two states: a symptomatic and potentially infectious state, known as active TB, or an asymptomatic, noninfectious state called latent TB infection (LTBI) (Fig. 1). Approximately one-third of the global human population has LTBI (1), and although LTBI does not manifest with any clinical symptoms, it comes with the risk of developing into active TB disease. Active TB typically presents as pulmonary disease but can also present as extrapulmonary or disseminated disease (5, 6).
FIG 1.
Infectious spread of M. tuberculosis and resulting disease. An infectious individual with pulmonary TB can spread disease via aerosols—bacilli are expelled during coughing, talking, singing, and other activities, establishing infection in approximately 25 to 50% of close contacts. Most newly infected contacts develop LTBI (90 to 95%), though a small percentage (5 to 10%) immediately develop primary progressive active disease. Among LTBI cases, 5 to 10% of individuals will have reactivated disease over their lifetime, in what is termed postprimary disease. Risk factors associated with reactivation include immunodeficiency caused by HIV coinfection, use of immunosuppressive drugs, and diabetes mellitus; socioeconomic status; and smoking. Active disease, either primary or postprimary, may present as pulmonary disease or extrapulmonary disease, depending on age, the presence or absence of underlying disease, the genotype of the M. tuberculosis strain, and immune status. Colored circles represent contacts of an “index case.” Red represents active disease, blue represents latent infection, gray represents no disease, black solid lines represent person-to-person aerosol transmission, and the black dashed line represents reactivation of latent infection.
As with other airborne diseases, the spread of M. tuberculosis is facilitated by high population densities and crowded indoor environments that optimize the aerosol transmission of the pathogen. Host factors, such as immune suppression, smoking, poor nutrition, diabetes, and respiratory comorbidities, also play a role by increasing the risk of transitioning from latent to active TB (6). This is particularly an issue in the developing world, where overcrowding, malnutrition, and HIV infection all contribute to a high burden of disease. Worldwide, the majority of cases occur in Southeast Asia and the Western Pacific Regions (56%), Africa (25%), India (24%), and China (11%) (1). Globally, the mortality rate of TB has decreased in the last decade, and the incidence of active disease is steadily but slowly declining (approximately 1 to 3% per year) (7). However, the burden of disease remains substantial, especially in low-income countries—in 2013, an estimated 9 million people developed TB, and 1.5 million died from the disease (1)—while the emergence of multidrug-resistant (MDR) TB threatens to reverse the gains we have achieved to date.
The rapid and accurate clinical laboratory diagnosis and characterization of M. tuberculosis are pivotal to battling TB (7). This is complicated by the fact that M. tuberculosis belongs to a group of genetically related species collectively termed the Mycobacterium tuberculosis complex (MTBC). The predominantly human pathogens within the MTBC, in addition to M. tuberculosis, include M. africanum, M. canettii, M. bovis, and the related M. bovis bacillus Calmette-Guérin strain used in the TB vaccine. Recent genomic investigations suggest that M. canettii is substantially more recombinogenic than other mycobacteria and may not be grouped correctly within the MTBC (8, 9). The remaining members of the complex largely infect animals (3) but have been documented as having caused human infections—these include M. microti, M. caprae, M. orygis, and M. pinnipedii (10–13).
Many classical microbiological techniques for species identification and phenotyping are of limited utility for the MTBC due to the genetic homogeneity of its members (14), with most of the species in the MTBC sharing 99.9% nucleotide identity and identical 16S rRNA sequences (15). In contrast, genomics has been integral in revealing the population structure of the MTBC, as recently reviewed by Coscolla and Gagneux (16). Seven lineages of human-adapted MTBC organisms have been described and strongly correlate with geography. M. tuberculosis lineages 2 (East Asian, including Beijing strains) and 4 (Euro-American) are widely distributed globally, lineages 1 (Indo-Oceanic) and 3 (East African-Indian) are restricted to eastern Africa and parts of Asia, and lineage 7 (Ethiopian) is restricted to a small region in the Horn of Africa. Lineages 5 and 6 comprise M. africanum isolates designated West Africa 1 and 2, respectively. Molecular and genomic investigations have also added approximately 50 new members to the list of over 150 nontuberculous mycobacteria (NTMs) since 2006, including several that have been documented as causing human disease (17), and have pointed to seals as a potential vector for the introduction of TB to the New World in the Holocene era (4).
The genomic revolution offers new opportunities for diagnosis and characterization of the MTBC. With the advent of rapid and cost-effective platforms for whole-genome sequencing (WGS) (reviewed in reference 18), many of the multiple time-consuming steps in the clinical mycobacteriology laboratory, including detection, species identification, and antimicrobial resistance phenotyping, can be replaced with a single WGS analysis. This review focuses on describing the genomic tools that have aided in the identification, diagnosis, treatment, and control of TB thus far and the potential future impact of WGS technologies on detection of antibiotic resistance-conferring mutations and TB epidemiology in the context of the clinical microbiology laboratory. While WGS-based advances, to date, have largely been in laboratories in high-income, low-burden settings, we also discuss the role that these technologies may play now and in the future in low-income regions. The general principles behind WGS in the clinical microbiology laboratory and the differences between available platforms are not covered here; instead, we refer the reader to a recent comprehensive review of these topics by Didelot et al. (19).
ROLE OF GENOMICS IN THE DIAGNOSIS OF M. TUBERCULOSIS
The modern TB laboratory's role is the detection and species identification of mycobacteria, as well as drug susceptibility testing to optimize each patient's treatment plan. For pulmonary TB, the most common form of active TB, diagnosis is typically made with a sputum sample from the patient. In the clinical laboratory, the sputum sample is decontaminated to enhance the recovery of mycobacteria, including M. tuberculosis. This treated sputum sample is concentrated, fixed onto a slide, stained with acid-fast stains, and visualized microscopically for the presence of mycobacteria in what is known as the acid-fast bacillus (AFB) smear test. Unfortunately, this test has a high limit of detection, requiring at least 105 CFU/ml in the sputum sample to be able to consistently detect mycobacteria, and the test does not distinguish between different species of mycobacteria. In resource-limited settings, the AFB smear test may be the only test available, often without the added steps of decontamination and concentration. However, for modern TB laboratories with biosafety level 3 facilities, the treated sputum sample will also be cultured using selective media for mycobacteria. Although culture-based methods are the gold standard for the diagnosis of mycobacterial disease, mycobacteria grow slowly, meaning that culture can require 6 to 8 weeks. After growth of the mycobacteria, phenotypic methods have traditionally been used to categorize and differentiate some Mycobacterium species, but lateral flow immunoassays specifically for the MTBC (20), as well as molecular methods, such as nonamplified DNA probes, line probe assays, or DNA sequencing, are more commonly used to definitively identify M. tuberculosis and nontuberculosis mycobacteria to the species level (21–23). Additionally, nucleic acid amplification tests for M. tuberculosis can be applied directly to the sputum sample for the immediate diagnosis of TB independent of the culturing process.
Advances in M. tuberculosis Detection
In an effort to improve the rapid identification and characterization of M. tuberculosis, several molecular tools have been developed based on our knowledge of the genome of M. tuberculosis, which was first completely sequenced in 1998 (14). For rapid TB diagnosis, the WHO has endorsed the use of the Xpert MTB/RIF test (Cepheid), an automated real-time PCR-based test that simultaneously detects the presence of M. tuberculosis and common mutations for rifampin resistance, considered a marker of multidrug resistance. The GeneXpert platform can provide a result within hours directly from a sputum sample and has a higher sensitivity than the AFB smear test, being able to detect as few as 102 CFU/ml (1, 24, 25), though the sensitivity is lower than those of the gold standard of culture and of the microscopic observation drug susceptibility (MODS) assay (26). The platform has been used successfully in low- and middle-income countries; however, its utility is debated for remote and/or highly underresourced settings, where a lack of reliable electricity, limited technical support, and other environmental factors exclude all but the simplest diagnostic methods.
Further methods are being developed that will allow for the rapid, sensitive, and specific detection of M. tuberculosis directly from the patient sample, be it sputum, cerebrospinal fluid, or another specimen type. Selective enrichment is one such method. Studies have shown the effectiveness of ligand-coated magnetic beads to capture and concentrate M. tuberculosis in sputum samples prior to microscopic examination (27–29). This is useful for TB diagnosis in low-income countries, where decontamination and centrifugation to concentrate M. tuberculosis may not be feasible due to a lack of resources. Elitas et al. showed a label-free approach based on the dielectric properties of bacterial cells (30). Enrichment uses dielectrophoresis on three-dimensional (3D) carbon electrode arrays to discriminate bacteria based on small changes in their intrinsic properties, such as membrane integrity (30). Further developments in nanotechnology and microfluidics offer the possibility of rapid and accurate detection of M. tuberculosis from sputum samples (reviewed by Wang et al. [31]). The development of next-generation sequencing technology also offers the potential to identify the presence of M. tuberculosis directly from a sputum sample. Recently, Pallen and colleagues used shotgun metagenomics to detect MTBC sequences directly from eight sputum samples, without an initial culture step (32). Unprocessed sputum was subjected to differential lysis to enrich microbial DNA, and the resulting nucleic acids were sequenced on an Illumina MiSeq instrument (32). By comparing the sequence reads to the H37Rv reference genome, Pallen and colleagues were able to assign seven of the eight isolates to an MTBC species and lineage (32). With the possibility of longer read lengths through emerging nanopore sequencing technology and single-cell genomics, a very low concentration of genomic DNA may be sufficient not only to diagnose the presence of M. tuberculosis directly from patient samples (32–34) but also to provide insight into genomic determinants of antibiotic susceptibility (20).
Genomics for Predicting Antibiotic Susceptibility of M. tuberculosis
Following positive identification of M. tuberculosis, a specimen will undergo antibiotic susceptibility testing. The results are critical for establishing an appropriate treatment regimen for a given patient, particularly with the recent emergence of MDR and extensively drug-resistant (XDR) TB. Globally, 480,000 cases of MDR TB are estimated to have occurred in 2013, and rapid testing for MDR is a WHO priority action for TB control (1). A culture-based phenotypic assay can be used to determine the susceptibility to and/or MICs for first- and second-line drugs, including isoniazid, rifampin, pyrazinamide, and ethambutol, but may take up to 2 weeks to complete. Line probe assays, targeting antibiotic resistance mutations revealed by genomics, can accelerate this process. These are DNA strip-based tests that use nucleic acid amplification and reverse hybridization methods for the rapid detection of mutations associated with drug resistance and can be used directly on sputum samples or cultured isolates (35). Currently, two products targeting mutations relevant to resistance to first-line drugs are commercially available: the INNO-LiPA Rif.TB test (Innogenetics NV, Belgium) and the “Hain test,” i.e., the GenoType MTBDRplus assay (Hain Lifescience GmbH, Germany) (35–37). A notable drawback to these assays is that they are limited to detecting a specific set of mutations in the rpoB, katG, and inhA genes (e.g., the INNO-LiPA Rif.TB test detects only nine mutations in rpoB [36]) and cannot identify novel mutations conferring resistance (37), although certain assays do contain probes for wild-type alleles—observing a resistance phenotype and a wild-type allele would suggest a novel mutation but would not provide further details. Recently, Hain Lifescience introduced the GenoType MTBDRsl assay, which extends resistance typing to gyrA mutations conferring fluoroquinolone resistance, rrs mutations conferring aminoglycoside resistance, and embB mutations conferring ethambutol resistance. This new assay enables the identification of XDR TB strains but, again, is incapable of characterizing novel mutations.
WGS is emerging as a promising technology for resistance typing of M. tuberculosis. Early sequencing studies first targeted specific M. tuberculosis genes known to confer resistance to first- and second-line drugs. A study by Campbell et al. sequenced nine loci associated with antibiotic resistance in 314 clinical isolates, 52% of which were MDR and 3% of which were XDR, and compared the phenotypic data with the sequencing data (38). They found that the sequencing data were able to capture the observed resistance phenotype frequently in the case of MDR isolates, with a sensitivity and specificity of 90.8% and 94.7%, respectively. However, accurate sequence-based typing of XDR isolates was more challenging, with only 40.0% sensitivity and 99.3% specificity (38). Recent studies used WGS on hundreds of clinical isolates to identify known and novel antibiotic resistance-conferring mutations (39–41). The largest of these, a study of 1,000 Russian isolates, 48% of which were MDR, identified 238 single nucleotide polymorphisms (SNPs) associated with drug resistance (42). At the other end of the spectrum, a 2014 study by Eldholm et al. examined the emergence of resistance within a single XDR TB patient sampled serially over a 42-month period (43). They observed a total of 35 mutations in the nine clinical isolates sequenced, 15 of which became fixed in the population (43). Twelve of the mutations were determined to play a role in drug resistance, and the laboratory phenotyping results were concordant with these mutations (43). For five of the seven antibiotics for which resistance arose, two independent resistance mutations were observed (43).
WGS has also been used to examine the potential for and mechanism of resistance to novel therapeutic agents. In 2005, Andries et al. sequenced the genomes of M. tuberculosis and M. smegmatis isolates before and after treatment with the investigational compound R207910 (now called bedaquiline) (44). They detected resistance-conferring mutations in the atpE gene, suggesting that the novel compound targeted the Fo subunit of the ATP synthase protein (44). However, WGS is not the only step in such an analysis; candidate resistance-conferring mutations should be confirmed with Sanger sequencing or a PCR-based single nucleotide variant (SNV) assay and then must be followed up with in vitro functional assays to confirm the link between the observed mutation and phenotype.
WGS studies of drug-resistant isolates have expanded our knowledge of drug resistance mutations but have also shown the difficulty in data interpretation. In the case of ethambutol resistance, a study showed that a low-level resistance that would be undetected by current phenotypic antibiotic susceptibility testing may accumulate within the genome of M. tuberculosis (41). In contrast, another study reported that phenotypic testing indicated isoniazid resistance, but WGS failed to identify the mutation responsible (45). Mutations conferring drug resistance are known to come at the cost of a loss in an organism's fitness; however, recent genomic studies revealed that compensatory mutations can occur which overcome the loss of fitness and create a “perfect storm” of resistant isolates without reduced fitness (42, 46–48). Furthermore, it has been shown that M. tuberculosis strains of different lineages can differ in their virulence, acquiring mutations conferring antibiotic resistance, a delayed host immune response, enhanced transmissibility, and more severe disease at different rates (49–51). The success of some of the more virulent Beijing and Euro-American strains, for example, has been attributed to the genomic diversity of these specific lineages (16). Understanding the emergence of compensatory mutations, their interactions with other mutations (epistasis), and the impact of these on treatment decisions is an emerging area into which WGS can provide important insights, as recently reviewed by Trauner et al. (52).
A recent case study by Köser et al. (53) showed retrospectively how WGS could be used in a clinical setting to examine drug resistance in a patient simultaneously infected with two strains of XDR M. tuberculosis. The WGS-based prediction of drug resistance was concordant with the phenotypic testing results done at a reference laboratory, but the WGS data were generated in a matter of days, versus the weeks required at the reference laboratory. Furthermore, the WGS analysis also identified resistance to five additional antibiotics not tested by the reference laboratory (53). These data suggest that WGS will allow faster identification of drug resistance, allowing physicians to more rapidly determine the most effective treatment plan for the patient and potentially limit the further spread of drug-resistant strains. However, before WGS can replace standard phenotypic testing, more data on the concordance between SNPs and resistance phenotypes, including the role of compensatory mutations, are needed. Furthermore, although Köser et al. were able to accurately predict the phenotype in a case of mixed infection, this is likely to not always be the case. In a mixed infection with susceptible and resistant bacteria, the observed in vivo phenotype may be difficult to predict from WGS data, especially if the sample being sequenced contains a large proportion of genomic DNA from the susceptible bacteria relative to the resistant bacteria. A sample collected early in the infection or one that has been cultured in the laboratory may have a ratio of susceptible to resistant bacteria that does not reflect the host infection, where the resistant mutants may eventually dominate if the treatment regimen is tailored only for the susceptible isolate. Still, WGS does offer a promising alternative to current growth-based susceptibility testing methods, whose performance is variable due to differences in the physiochemical environment in which the testing is performed and the protocols used for inoculum preparation and testing, and whose in vitro results may not reflect the in vivo clinical reality (54).
Currently, many mutations conferring antibiotic resistance are known for M. tuberculosis, and more continue to be identified (42, 55, 56); however, in order for WGS to be implemented as a routine clinical tool for resistance typing, a centralized and freely accessible database must be created to catalog resistance- and heightened virulence-conferring mutations, similar to the Stanford HIV Drug Resistance Database (57). A legacy database, TBDReaMDB (58), exists, as does the more recent MUBII-TB-DB (59). Both compile reported genetic polymorphisms conferring drug resistance in M. tuberculosis, with TBDReaMDB including mutations reported until 2010 and MUBII-TB-DB including data through 2014. The use of such databases in a clinical setting has yet to be formalized, but the recent announcement that the Gates Foundation has funded a software project to warehouse TB genomes to facilitate and formalize resistance typing suggests that clinical use is on the horizon (Genomeweb). The success of a centralized, standard reference database will depend on continued efforts to identify emerging mechanisms of resistance, regular comparisons of genotype with phenotype, agreement of the TB community to utilize this database globally, and dedicated maintenance of the resource.
GENOMICS AS A TOOL FOR TB OUTBREAK INVESTIGATION
In addition to rapid diagnosis and phenotyping, contact tracing and effective outbreak management are critical to controlling and preventing the spread of TB in settings where resources permit (60, 61). Contact tracing is a routine epidemiological practice in which persons who may have been exposed to an infectious case of TB are identified, screened for evidence of LTBI or active TB, and offered preventative or active TB therapy when appropriate (1). Outbreak management strategies are deployed when a TB outbreak has been declared, and these involve contact tracing coupled with enhanced community-wide screening activities to capture secondary cases in individuals who may not be in a patient's immediate circle of named contacts (1). Contact investigations have been enhanced by molecular techniques in recent years, and WGS is revealing new insights into how TB spreads within a contact network (62, 63).
Current Molecular Epidemiology Tools
A suspected epidemiological link between two TB cases can be confirmed by use of molecular epidemiology techniques in which the two M. tuberculosis isolates are compared at the genetic level: identical or nearly identical genetic fingerprints suggest contact between individuals or exposure to a common source. These techniques can also be used to discover new epidemiological links, revealing clusters of related cases. Molecular epidemiological techniques take advantage of known genetic variation within the M. tuberculosis genome. The majority of this genetic variation occurs as SNPs, insertions, deletions, genomic rearrangements, and transpositions, and the genome contains numerous repeat sequences, including transposable elements, variable-number tandem repeats (VNTRs), mycobacterial interspersed repetitive units (MIRUs), and direct repeat regions (64–67).
Classical DNA fingerprinting or genotyping methods exploiting variation in the M. tuberculosis genome include spoligotyping (64), IS6110 restriction fragment length polymorphism (RFLP) analysis (66), and MIRU-VNTR analysis (65) (Table 1). Of these, 24-locus MIRU-VNTR assay is considered to be the gold standard method, but it is often combined with spoligotyping and/or IS6110 RFLP analysis to further increase its discriminatory power (68). Even so, current fingerprinting methods can only identify linked cases—they cannot reveal the direction of transmission or discriminate between primary, secondary, and tertiary cases. Furthermore, cases with matching fingerprints generally trigger resource-intensive contact tracing efforts, and these techniques often cluster more distantly related isolates even where there is no direct epidemiological connection (67). WGS is an attractive alternative: by examining the whole genome, not just a small fraction of the available genetic information, it captures all the variation present, providing a high-resolution, accurate, and reproducible method for identifying fine-scale transmission events, while simultaneously providing information for use in other aspects of TB management, such as predicting antimicrobial resistance (Table 1) (53, 67).
TABLE 1.
Comparison of available molecular epidemiology tools for M. tuberculosis
| Method | Attributes | Strengths | Weaknesses |
|---|---|---|---|
| Spoligotyping | May be used directly on clinical samples or early cultures | Low DNA concentration required | Less discriminatory than other genotyping methods |
| Rapid initial PCR amplification step | Short turnaround time | False clustering | |
| Involves probe hybridization | Objective and reproducible results | Unable to resolve transmission events within outbreaks | |
| Numerical results | Fingerprint defined with a numerical code, which facilitates comparisons | Interrogates only a small fraction of the entire genome | |
| IS6110-based RFLP analysis | Cultured isolates are required to obtain sufficient DNA for RFLP analysis | High discriminatory power | High DNA concentration required |
| Restriction digestion followed by gel electrophoresis and blot transfer | Banding pattern may be difficult to interpret and standardize | ||
| Hybridization of IS6110-specific probe | Technically demanding and has a long turnaround time | ||
| Generates a band pattern fingerprint | Not suitable for strains with no or low-copy-number IS6110 | ||
| False clustering | |||
| Unable to resolve transmission events within outbreaks | |||
| Interrogates only a small fraction of the entire genome | |||
| 15- and 24-locus MIRU-VNTR analyses | Require cultured isolates | Low DNA concentration required | Interpretation of unexpected fragment sizes is challenging |
| Rapid initial PCR amplification step | Short turnaround time | False clustering | |
| Fragment lengths can be resolved using gel or capillary electrophoresis systems | Objective and reproducible results | Unable to resolve transmission events within outbreaks | |
| Medium-high discriminatory power (less than that of IS6110-RFLP assay) | Interrogate only a small fraction of the entire genome | ||
| Fingerprint defined with a numerical code, which facilitates comparisons | |||
| WGS | Currently requires culturea | Superior tool for resolving transmission dynamics | Technically demanding for sample preparation and data analysis |
| Fragmentation of genomic DNA followed by addition of sequencing adapters | Interrogates 100% (or close to 100%) of the genome | Not globally available due to higher costs | |
| Desktop-sized sequencers perform sequencing by synthesis | Yields information about virulence factors, antibiotic resistance, and epidemiology | No standardized methods for data analysis and data sharing | |
| Data analysis requires bioinformatics expertise | Current methods have long turnaround times |
Newer sequencing technologies may be able to generate whole-genome sequences without culture.
Advancing Molecular Epidemiology with WGS
The principle behind using WGS to identify transmission events is simple. Over the time scale of an outbreak, random mutations will accrue in the M. tuberculosis genome. A mutation arising in one host will be transmitted to all individuals infected by that host and will similarly be passed on to people those individuals go on to infect. By sequencing all of the M. tuberculosis genomes from outbreak cases, it is possible to identify these mutations and to determine whether they are present or absent in the M. tuberculosis isolate sequences from each individual. Combined with patient information, including the patient's infectious period and their social contacts, we can identify the most plausible source of an individual's infection. Mutations that arise over short time frames and are not yet subjected to selective pressures are known as single nucleotide variants (SNVs), while mutations that become fixed in the population at a rate of 1% or higher are known as single nucleotide polymorphisms (SNPs). Depending on how one defines the total bacterial population (whether it is the cultured M. tuberculosis strain from the patient sample or all the M. tuberculosis strains within an infected individual, within the outbreak, or within a defined M. tuberculosis genotype), a given mutation may be an SNV or an SNP. Within the context of investigating an outbreak by using whole-genome sequencing, we have chosen the more conservative definition of these mutations as SNVs, as it is not yet possible to calculate the degree of fixation of these mutations within any given genotype of M. tuberculosis, but we recognize that some of these mutations might be renamed SNPs if we had more complete genomic data or redefined the population in which these mutations were found. Regardless of the nomenclature, we might reasonably expect a zero- to five-SNV (or SNP) difference between the infectious source and an infected recipient.
Recent retrospective studies showed that WGS provides far superior resolution of transmission events than that of historical fingerprinting methods, even when both techniques are combined with the same contact tracing and social network data (Table 2) (62, 63, 69–76). A 2009 study by Niemann et al. was the first to compare the utility of WGS and DNA fingerprinting methods to epidemiologically link M. tuberculosis isolates (69). They sequenced two isolates with identical IS6110 patterns and the same alleles at 23 of 24 MIRU-VNTR loci (69), a pattern which suggests a close epidemiological relationship between the cases. However, the WGS data indicated that the isolates differed by 130 SNVs, from which the authors inferred that the two isolates could not represent a direct transmission event (69). Since this initial study, many large retrospective studies have expanded the scope and scale of their WGS investigations. In 2010, two studies used WGS to investigate transmission chains within the Harlingen outbreak in the Netherlands (70, 77). In the first, Schurch et al. sequenced the first and last isolates in a five-patient transmission chain that had been elucidated through traditional epidemiologic investigation, finding four SNVs, a tandem repeat polymorphism, and an IS6110 transposition (70). They then examined these variations in patients 2, 3, and 4, finding that the majority arose in one patient who was noncompliant with treatment (70). In the second study, they sequenced a third Harlingen outbreak isolate with the same DNA fingerprint but belonging to a different transmission chain, finding a total of eight SNVs differentiating the three isolates (77). They then screened all 104 isolates from the outbreak for the presence of these eight SNVs, which allowed them to divide the Harlingen outbreak into five distinct subclusters and to refine the transmission events within each cluster (77). The technique also indicated that a suspected relapse in one patient was actually reinfection with a slightly different outbreak strain (77). Noting the improvement in typing that WGS offered, the authors suggested that genome sequencing may become the new standard method for typing of M. tuberculosis isolates to identify transmission events in outbreaks (70, 77).
TABLE 2.
Use of whole-genome sequencing in TB outbreak investigations and to understand microevolution of M. tuberculosis
| Study focus and period | Location | No. of WGS isolates | Genome coverage (%) | No. of SNVsa | Sequencerb | Major finding(s) | Reference |
|---|---|---|---|---|---|---|---|
| Studies on transmission of TB | |||||||
| 2001–2004 | Uzbekistan | 2 | 95.9 | 1,209c | Illumina | Showed that isolates with identical IS6110 patterns differed by 130 SNVs | 69 |
| 1992–2008 | Netherlands | 3 | 95.4 | 8c | Roche/454 | Discovered that SNVs can be used to identify transmission chains in RFLP clusters | 70 |
| 2006–2009 | Canada | 36 | 99.2 | 204 | Illumina | Integration of WGS with a social network questionnaire generated a more accurate transmission network; used WGS to identify “superspreader” cases | 62 |
| 2010 | UK | 2 | 95 | 0 | Illumina | Outbreak identification with WGS; showed the utility of WGS to identify drug resistance markers | 78 |
| 1994–2011 | UK | 390 | 88.5 | 1,096 | Illumina | Rate of genetic changes was 0.5 SNV/genome/year; used WGS to identify “superspreader” cases | 63 |
| 1997–2010 | Germany | 86 | 96.4 | 85 | Roche/454 | WGS was proven to be more effective at generating clustering patterns than classical genotyping methods; used WGS to estimate a rate of genetic change of 0.4 SNV/genome/year | 71 |
| 22 mo | USA | 9 | 95.7 | 7c | Illumina | WGS allowed the identification of new epidemiological links in the transmission chain; used WGS to characterize a mutation rate of 0–2 SNVs/transmission event resulting in a secondary case | 72 |
| 2007–2012 | UK | 256 | 92.6 | 1,715 | Illumina | Patients born in low-incidence countries are more likely to have pulmonary disease and social risk factors, causing secondary cases in the UK | 79 |
| 1992–2008 | Netherlands | 199 | 95.6 | 11,879 | Roche/454 | Estimated an average mutation rate of 0.3 SNV/genome/year, with a large degree of variation, i.e., 0.4–17 SNVs/genome/year | 88 |
| 2009–2010 | China | 32 | ND | 1,790 | Illumina | Evaluated the usefulness of WGS in a high-incidence country | 73 |
| 1991–2011 | Switzerland | 69 | 98.3 | 133 | Illumina | WGS suggested a single origin for an outbreak involving 68 patients over 21 years that could be divided into 3 subclusters with epidemiological links differing by 0–11 SNVs | 76 |
| 2008–2011 | Canada | 33 | ND | 21 | Illumina | WGS revealed within-host diversity in an index case; a computational method was developed to infer transmission from phylogenetic data, accounting for within-host diversity | 80 |
| Not specified | Canada | 36 | ND | 3,523 | Illumina | WGS of M. tuberculosis strains belonging to the Manila sublineage showed better resolution than that of MIRU-VNTR assay and decreased the frequency of clustered cases | 74 |
| 1997–2013 | Canada | 61 | ND | 722 | Illumina | WGS revealed 6 subclusters of the 17-year outbreak showing unique patterns of evolution | 75 |
| Studies on microevolution of M. tuberculosis | |||||||
| 1994–2011 | UK | 390 | 88.5 | 1,096 | Illumina | Defined epidemiologically linked transmission events by using SNV cutoffs of <5 SNVs for transmission and >12 SNVs for no evidence of transmission; intrapatient variation was limited to <5 SNVs; rate of genetic changes was 0.5 SNV/genome/year | 63 |
| 2003–2010 | Spain | 36 | 98 | 28 | Illumina | Examined inter- and intrapatient variations and determined them to be equivalent | 89 |
| 2008 | Malaysia, South Africa, Thailand | 96 | ND | 1,419 | Illumina | Showed that WGS can be used to discriminate between relapse and reinfection | 86 |
| 1990–2010 | New Zealand | 10 | 98 | 747 | SOLiD | Used WGS to show that mutation rates during latent infection in humans are substantially lower than those during active disease | 93 |
ND, not defined in the report.
The Illumina, Roche/454, and SOLiD platforms all represent short-read sequencing-by-synthesis platforms and are reviewed in reference 18. Newer technologies, such as nanopore-based sequencing or the PacBio platform, have also been applied to microbial genomics but have yet to be used in a TB study.
SNVs were verified by secondary Sanger sequencing.
The following year, Gardy et al. were the first to sequence a complete outbreak, sequencing the first 32 isolates of an ongoing outbreak in British Columbia, Canada, along with 4 preoutbreak isolates from the same region with the same MIRU-VNTR pattern (62). The study revealed substantial genetic variation within the outbreak and, combined with epidemiological data derived from a social network questionnaire, provided the first nearly complete transmission network of an outbreak, which revealed the contribution of previously unrecognized “superspreaders” responsible for seeding multiple secondary infections. A second study, published in 2013, demonstrated that WGS could also link previously unsuspected cases (78). In the United Kingdom, two active TB cases with identical fingerprints were temporally and geographically linked; however, an epidemiological investigation could not identify a relationship (78). WGS showed that the isolates were completely identical, without a single SNV difference, suggesting direct transmission or transmission from a common source that has not yet been identified (78). Other work from the United Kingdom explored transmission dynamics in Oxfordshire in both United Kingdom-born and foreign-born individuals (79). Walker et al. sequenced isolates from 247 cases and found that foreign-born individuals were less likely to cause onward transmission, confirming results from earlier studies using other genotyping methods (79). They hypothesized that among the United Kingdom-born individuals, social risk factors may have led to a failure to seek health care, and physicians may not have considered a TB diagnosis; together, these factors would contribute to delayed diagnosis and increased community transmission. These social risk factors, including homelessness, emerged as drivers of several outbreaks recently characterized using WGS, including sequencing of 61 M. tuberculosis isolates from a protracted outbreak among the homeless in Toronto, Canada (75), which revealed six independent transmission chains, and of 33 isolates from an outbreak originating in a Kelowna, Canada, homeless shelter (80). In both scenarios, WGS suggested the occurrence of transmission within the shelter environment, as well as transmission from shelter clients to shelter staff.
WGS of every outbreak isolate can be prohibitively expensive, particularly for long outbreaks with hundreds of cases. Recent work suggests that limited WGS may also have utility in the clinical laboratory in such scenarios. Stucki et al. carried out WGS on three isolates from a large Swiss TB outbreak occurring over 21 years and on four unrelated control isolates (76). The whole-genome data revealed 118 SNVs present in the outbreak strains but not the control strains; 2 of these were selected as the targets of a PCR-based SNV typing assay that was then applied to 1,642 archived isolates. The assay ruled in 68 archived isolates as belonging to the outbreak; these were then analyzed using the full WGS approach (76). Using this limited WGS/SNV typing method, the authors were able to rapidly characterize the outbreak at a substantially reduced cost ($4,900) compared to that for other genotyping methods: the SNV assay was 15 times less expensive than MIRU-VNTR typing, and the targeted WGS was 20 times less expensive than performing WGS on the entire strain collection (76).
In another limited-WGS approach, a core genome multilocus sequence typing (cgMLST) scheme was recently proposed as a method to convert WGS-identified SNVs in coding regions of the genome into an allele numbering system (81). Although this method is standardized, shareable across jurisdictions, portable, and not computationally intensive, it failed to resolve outbreak clusters as effectively as approaches that use the entirety of the genome (81). However, the advantages of universally implementing such an approach at a national or international level mean that cgMLST may provide a high-level view of large trends that are not apparent with traditional genotyping, with the resulting cgMLST clusters then targeted for follow-up with more detailed SNV-based phylogenies. Approaches such as cgMLST also raise the possibility of leveraging WGS data to “back-calculate” traditional molecular epidemiology fingerprints from genomic data. SpolPred calculates a spoligotyping pattern directly from short-read sequencing data (82), and as sequencing platforms improve and are able to handle longer repetitive regions, generating in silico MIRU-VNTR patterns should also be possible, permitting comparisons between WGS isolates and historically fingerprinted isolates.
Importantly, it should be noted that the above-described genomic epidemiology studies all took available epidemiological data into account in establishing the direction of transmission. Recent work has shown that in the absence of epidemiological information, WGS does provide finer subclustering of isolates than would be possible with traditional genotyping, but it cannot identify individual transmission events with any certainty (80). The direction of transmission can be difficult to resolve given genomic data alone; to make plausible inferences requires knowledge of a patient's symptomatic and infectious periods, their relative infectiousness, and their social contacts and places of social aggregation. Even with this knowledge, delayed diagnoses and long infectious periods—common in vulnerable populations reluctant or unable to seek care—introduce within-host genetic diversity that can complicate linking sequenced isolates to each other. A second caveat is that much of the potentially informative variation in MTBC genomes lies in repetitive regions, such as the PE/PPE genes, which constitute ∼10% of the genome; however, current short-read sequencing technologies and bioinformatic algorithms struggle with these types of repetitive regions, meaning that studies to date have largely ignored variation in these regions, which may provide even deeper insight into the nature of transmission and of mutation rates.
The studies described above have largely been retrospective, examining outbreaks that have concluded or are already well under way, and given the prolonged nature of TB outbreaks, which typically span several years, it is likely that we will not see “real-time” genomic epidemiology unfold for TB the same way it has for other organisms, including hospital-acquired infections (83, 84). However, there are indications that while genomic data alone cannot identify individual transmission events, it can provide some insight into the overall dynamics of the outbreak, including whether or not the outbreak shows evidence of a superspreader (85) and which patients are most central to the outbreak network (80). These patterns can be detected within the first several isolates from an outbreak (85), suggesting that real-time WGS may be useful in understanding an outbreak and its key players, thereby informing the public health approach to managing the situation.
UNDERSTANDING THE MUTATION RATE OF M. TUBERCULOSIS
In 2013, studies from Walker et al., Roetzer et al., and Kato-Maeda et al. further demonstrated that WGS outperforms traditional fingerprinting methodologies in resolving outbreaks and identifying direct transmission events (63, 71, 72), with their work providing insights into the M. tuberculosis mutation rate in an outbreak scenario and into the expected genomic differences between related isolates (Table 2). Most notably, Walker et al. sequenced 390 isolates representing a range of epidemiological scenarios to determine whether the number of unique SNVs between isolates could be used to draw epidemiological inferences about cases (63). They determined that if two isolates differed by fewer than 5 SNVs, they likely belonged to the same transmission network, whereas if they differed by more than 12 SNVs, they could not represent a direct transmission event (63). Their study also examined the accrual of SNVs within a patient over time, finding that longitudinal isolates collected up to 8 years apart varied by fewer than five SNVs (63). Furthermore, they showed that cross-sectional diversity within a host, as revealed by isolates taken from various body sites (pulmonary versus nonpulmonary) at one time, was similarly limited to five or fewer SNVs (63). This five-SNV threshold was also shown to be useful in differentiating relapse of infection from reinfection, as cases of relapse did not differ by more than six SNVs (86). Roetzer et al. sequenced 86 isolates from a large outbreak, noting a maximum of three SNVs between epidemiologically linked patients, while Kato-Maeda et al. sequenced nine isolates from a small cluster of cases, observing zero to two SNVs between linked cases (71, 72).
These and other studies also shed light on the rate of genetic change in M. tuberculosis in a natural outbreak scenario. Walker et al. reported a mutation rate of 0.5 SNV/genome/year, in agreement with a previous study done in cynomolgus macaques (63, 87). A subsequent study by Bryant et al. also reported a similar mutation rate of approximately 0.3 SNV/genome/year (88), while Roetzer et al. calculated a rate of 0.4 SNV/genome/year (71). This consistency appears to support the use of total accumulated SNVs to rule in or rule out transmission; however, Bryant et al. noted a large degree of variation in numbers of SNVs/genome/year across their data set of 199 sequenced epidemiologically linked isolates, and they concluded that phylogenetic analysis would offer an advantage over SNV thresholds in transmission studies (88). The SNV cutoffs defined by Walker et al. were also challenged by another study, which showed that although inter- and intrapatient variations were similar, a constant rate of SNV acquisition may not be valid for all strains and situations (89). Recently, an examination of serial evolution within an XDR TB patient reported substantially higher rates, from 1.1 to as high as 7.0 SNVs/genome/year (43). This increased rate was attributed to selective sweeps driven by drug treatment, and the authors cautioned against reliance on strict SNV count thresholds in ruling transmissions in or out, noting that two serial isolates from the patient were separated by 11 SNVs (43).
INSIGHTS INTO MIXED INFECTIONS AND WITHIN-HOST GENETIC DIVERSITY ACCRUED DURING LTBI
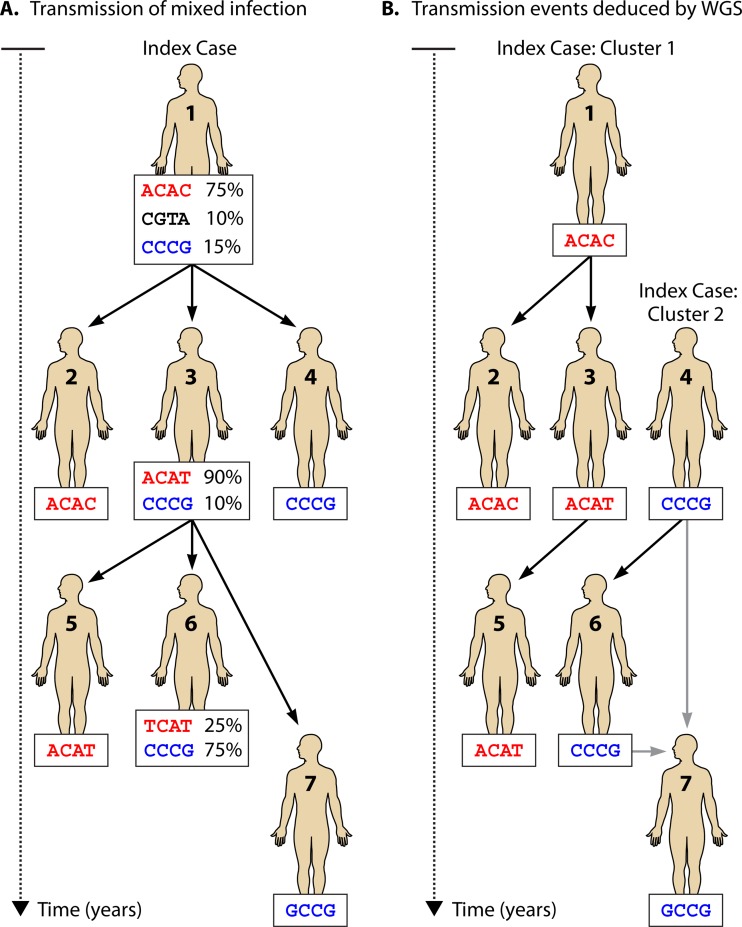
The earliest outbreak reconstructions also failed to consider the potential impact of mixed infection or within-host genetic diversity on our ability to make accurate inferences about transmission from genomic data (Fig. 2). This is a particularly important issue for TB, as LTBI and chronic, undiagnosed active disease both contribute to within-host diversity, whose magnitude depends on the time of infection and the host's immune background. Recently, efforts to account for this diversity were proposed, including a method calibrated for TB infections. Utilizing a novel Bayesian approach, Didelot and colleagues combined the phylogenic inference package BEAST with a Markov chain Monte Carlo inference of the suite of possible transmission events that could have given rise to an observed phylogenetic tree (80). This transmission network can be refined further with epidemiological data, such as time of infection, location, and smear-positive or -negative status (80). When applied to genomic data derived from a real outbreak, their TransPhylo method was able to identify the source case and the major transmission subclusters within the outbreak, and they reported a mutation rate of approximately 0.5 SNV/genome/year, in agreement with earlier reports (63, 80, 87, 88).
FIG 2.
Within-host genetic diversity can complicate transmission inferences from genomic data. (A) Hypothetical transmission scheme, starting with an index patient in whom three distinct bacterial populations are present as a result of a long period of latency, mixed infection, or undiagnosed active disease. Percentages indicate the abundance of each M. tuberculosis population. (B) Potential results of a WGS-based outbreak investigation. The index patient transmits different organisms to different secondary individuals and may potentially transmit a mixed infection, and only the most abundant genotypes are identified in each patient. Thus, in this case, WGS fails to link some secondary cases to the index patient and resolves this outbreak into two independent clusters.
The contribution of LTBI to the generation of within-host genetic diversity and its consequences for reconstruction of disease transmission are unclear. Upon initial infection of the lungs, M. tuberculosis is phagocytosed by innate immune cells, including macrophages, and initiates intracellular survival within these immune cells, modulating the immune response and leading to the formation of a granuloma (90, 91). M. tuberculosis can persist within the granuloma during LTBI, during which time its replication rate is thought to decline (92). Understanding mutation rates during LTBI is important, as LTBI treatment often consists of isoniazid treatment alone for 6 to 9 months, and this long-term monotherapy may lead to the emergence of antibiotic resistance (6).
Two studies, one in macaques and the other in humans, estimated the mutation rate of M. tuberculosis during latency, with conflicting results (87, 93). Ford et al. showed that in M. tuberculosis infections of macaques, the mutation rates of M. tuberculosis were identical during active disease, latent infection, and in vitro growth (87). They speculated that either M. tuberculosis has similar growth and mutation rates during latent infection, accounting for the similarity to the rates for active disease, or the growth rate decreases during latent infection, with a concurrent increase in the mutation rate (87). They provided evidence that the mutations that arise during latency are a result of oxidative damage, which is consistent with the exposure of the bacterium to oxidative stress during adaptation to intracellular survival within macrophages (87). In order to further characterize the mutation rate of M. tuberculosis during latency in humans, Colangeli et al. used epidemiological and phylogenetic analyses of two recently acquired M. tuberculosis isolates and two cases of reactivated latent infection (20 years after initial exposure) from a multidecade outbreak in New Zealand (93). They reported that during latent infection, the mutation rate was approximately 30-fold lower than that during active disease, in contrast to the macaque study results, and they did not report any evidence to suggest that oxidative damage increases the mutation rate of M. tuberculosis during latency (87, 93). The discrepancy in the results between these two studies may be driven by the choice of host, duration of latent infection (which was shorter in the macaque model), host-driven alterations in the immune response, and the human study's assumption that in the 2 years prior to reactivation, the replication and mutation rates gradually increased to match the rates observed in active disease (93).
UTILITY OF WGS IN COUNTRIES WITH HIGH TB BURDENS
Genomic studies of TB transmission have, to date, been performed largely in low-incidence, highly resourced settings where facilities for culture-based diagnosis are available and where the cost of WGS makes such analyses feasible. The utility of WGS in high-incidence countries has yet to be explored thoroughly, but a recent study by Luo et al. is a first step in this direction. They evaluated the effectiveness of WGS in China and suggested that WGS did allow for the resolution of some transmission chains (73). However, due to the genetic homogeneity of M. tuberculosis and the high incidence of disease in the region, transmission events were more difficult to elucidate than the case for studies in low-incidence countries (73). It is unlikely that WGS would offer much direct benefit to the control of TB outbreaks in high-incidence countries, particularly given the resource limitations of these countries; however, the information gained from WGS studies in low-incidence countries is informative with respect to transmission dynamics and may suggest screening strategies or outbreak interventions that might be useful in higher-incidence settings, where case finding may be challenging.
In the developing world, gains in TB control are most likely to come from scale-up of accurate diagnostics. While we do not foresee WGS-based diagnostics in low-income settings in the near term, the public release of increasing amounts of TB genomic data is likely to result in novel molecular diagnostics that can be used at the point of care and in the absence of culture in resource-limited laboratories. Comparative genomic analyses will likely reveal novel molecular targets specific to particular MTBC members and lineages that may be used in GeneXpert-like rapid-testing diagnostic platforms, while the discovery of new resistance-linked markers may be coupled to these detection methods for enhanced resistance surveillance efforts.
Indeed, it is in the area of drug resistance that WGS is most likely to have an impact in high-incidence countries. Sequencing of 56 TB genomes from Lisbon, Portugal, a city with high MDR rates, demonstrated the potential of genomics for identifying MTBC lineages responsible for local trends in drug-resistant TB (94). WGS-based insights into the emergence of drug resistance and the order in which mutations arise might provide a barometer for evaluating a region's resistance trends. Viewed through the molecular epidemiology lens, WGS data might also suggest the extent to which drug-resistant strains are transmitted from person to person within a region, providing data that may be used to guide local interventions.
LOOKING TO THE FUTURE: TB AS A MODEL FOR PUBLIC HEALTH GENOMICS
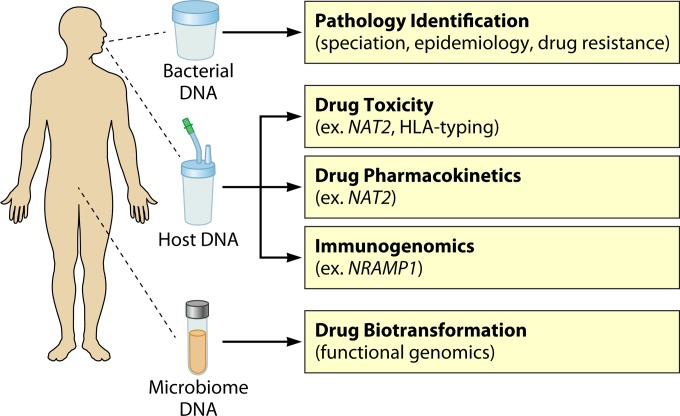
The studies reviewed here offer proof of principle that genomics can positively affect TB care from bench to bedside, from improved diagnostics to new outbreak management strategies. It is likely that TB will be the first pathogen for which a complete genomics approach can be implemented within the well-resourced public health environment, given these early successes. Indeed, the first pilot project using WGS for diagnosis, antibiotic susceptibility determination, and epidemiology was recently launched by Public Health England (https://publichealthmatters.blog.gov.uk/2014/06/16/the-genomics-of-tuberculosis). One can imagine the TB clinic and laboratory of the future as a place where genomic analyses of the pathogen, and perhaps even the host, are combined to give a complete view of an individual's infection (Fig. 3).
FIG 3.
The future TB clinic. Sputum, saliva, and fecal samples are collected from patients for genome sequencing, SNP typing, or metagenomic profiling to identify the presence of M. tuberculosis; provide strain identification and epidemiological information; identify genetic susceptibility to drug toxicity, drug pharmacokinetics, and susceptibility to reactivation of latent TB; and describe the patient-specific microbiome and its potential for oral drug biotransformation after digestion.
Advances in selective enrichment and in next-generation sample preparation methods, including single-cell genome sequencing, mean that we will be able to derive the complete genome sequence of a patient's mycobacterial infection directly from their clinical specimen within days, versus the current standard of weeks. From this sample, we will be able to identify the M. tuberculosis species infecting a patient and any genomic factors potentially associated with virulence, such as belonging to the virulent and transmissible Beijing lineage, which may affect decisions about how a patient is managed at home or in the hospital. As the quality and scope of drug resistance mutation databases increase and they become validated for use in a clinical context, we will be able to use WGS data to rapidly determine the best combination of antibiotics for each patient, thus accelerating the delivery of appropriate TB therapy. Continuous real-time genomics will enable detection of the emergence of antimicrobial resistance over the course of a patient's therapy, allowing real-time adjustments in therapy prior to the development of a more complicated drug-resistant infection requiring longer treatment. High-throughput targeted sequencing may also prove useful for large resistance typing efforts. Rather than sequencing entire genomes, next-generation sequencing platforms can be used instead for massively parallel sequencing of specific amplicons of interest—in this fashion, an instrument such as Illumina's MiSeq platform can be used to sequence thousands of amplicons spanning known resistance markers in a single run, rather than only ∼10 to 50 whole genomes. This parallelized analysis may prove useful in national or international surveys of resistance markers in which WGS of all isolates would be too costly, particularly if advances in sample preparation make culture-independent targeted sequencing easier for nonexpert laboratories.
The pathogen genome data generated in the clinical laboratory will then flow directly to TB epidemiologists, who will interpret the genomic relationships between isolates to identify emerging clusters of disease, reconstruct disease transmission patterns within a cluster or an outbreak, and monitor large-scale trends in the movement of TB genotypes within their region. Understanding outbreaks on an individual level will inform screening strategies, while developing a larger knowledge base of general trends in TB transmission will be key to creating evidence-based policies around managing outbreaks and preventing future occurrences of disease. Cross-border sharing of WGS data should be encouraged not only for identifying transmissions occurring across borders but also to reveal trends in TB dynamics in high-incidence countries, whose emigrants may progress to active disease in a low-incidence country—data from one region alone may be insufficient to draw broad conclusions, but when these data are pooled with data from many regions, we may be able to make inferences about disease occurring in other parts of the world.
We also envision a more distant future in which much more than the pathogen's DNA is analyzed. Human DNA, which can be recovered from the initial sputum or other specimen or collected using a cheek swab or saliva sample during a patient's visit to the TB clinic, can provide important insight into a patient's response to their infection and treatment plan. Antibiotic susceptibility testing data may be combined with a pharmacogenomic panel interrogating human SNPs and alleles linked to adverse drug reactions. For example, specific polymorphisms in the enzyme N-acetyltransferase 2 (NAT2) are known to increase the risk of hepatotoxicity during isoniazid treatment, as well as to affect the pharmacokinetics of the antibiotic (95–98). In addition, immunogenetic factors may also play a role in antibiotic-induced hepatotoxicity, with one study showing that the human leukocyte antigen (HLA) alleles can be predictive for isoniazid-induced hepatotoxicity (99). The treating physician would consider both of these inputs (bacterial antibiotic susceptibility and patient genetics) in designing a regimen to provide the most effective treatment while minimizing the risk of adverse reactions, such as hepatotoxicity, by using the appropriate antibiotics at the appropriate doses. As our knowledge of the human microbiome advances, a metagenomic survey of a patient's intestinal microbiota may also influence the choice of treatment if organisms known to affect the metabolism of antituberculosis drugs are detected. Although no microbe-antituberculosis drug interactions have been described to date, this new area of research is an important component of pharmacology and is rapidly advancing our understanding of how microbial xenobiotic metabolism affects treatment outcomes (100–103). These sorts of personalized treatment plans would require substantial trialing and refinement and would likely be used only in highly resourced settings, but the recent rise of personalized medicine in cancer suggests that they may one day be a possibility for the control of communicable diseases.
Analysis of specific biomarkers from the host genome may detect genetic signatures predictive of a poor response to treatment, with such patients flagged for enhanced follow-up, and host biomarkers may also be leveraged to predict which patients are at highest risk of developing primary progressive or postprimary active TB (104, 105). For example, NRAMP1 polymorphisms have been implicated in the rapid progression to primary TB and have been found in various populations spanning the globe, suggesting that these markers may have some utility in stratifying an individual's risk of developing active TB (104). This would allow for targeted preventative therapy in these individuals and, again, might be combined with pharmacogenomic data to select the optimal course of antibiotics. However, as with other polymorphism-based biomarkers, any biomarkers used in TB risk stratification must be found across a range of populations and confer a substantially increased risk to have any broad clinical decision-making utility. Azad and colleagues reviewed many innate immune polymorphisms linked to TB susceptibility, disease progression, and variable responses to tuberculin skin test (TST) and gamma interferon release assay testing (104), noting that interpretation of these biomarkers is further complicated by gene-gene interactions, HIV coinfection and other comorbidities, epigenetic modification, and variations in the infecting MTBC strain (104).
FUTURE DIRECTIONS AND CONCLUSIONS
WGS technology promises a rapid, efficient, and accurate platform for TB diagnosis, treatment planning, and epidemiology and offers the clinical laboratory the tantalizing possibility of using a single platform, a DNA sequencer, for the detailed detection and typing of a range of bacterial and viral pathogens. However, progress toward the lab of the future is not without its barriers. As with any new technology, cost and implementation are significant hurdles. Although the per-base cost of sequencing is dropping at an astonishing rate, there are numerous ancillary costs associated with establishing WGS capability in a laboratory. The required infrastructure includes a sequencing platform, equipment for high-throughput DNA library preparation, and appropriate computational power and storage for handling the large data sets arising from sequencing projects and meeting requirements related to data privacy and security. Similarly, one must also budget for library preparation and sequencing reagents, recognizing that the more “hands-off” a reagent kit is, requiring less technician time, the more expensive it will be. Finally, even the smallest sequencing group must comprise at least two trained technicians—one to handle the wet-lab aspects of preparing and sequencing the DNA and one to handle the computational, dry-lab analyses. Developing bioinformatics expertise, which must include everything from maintaining the computer hardware to understanding the various bioinformatic tools available and their associated options and parameters, is nontrivial and is frequently the rate-limiting step in laboratories' attempts to establish genomics cores.
A related personnel issue is the need for clinicians, epidemiologists, and other public health staff who have been trained in the interpretation of this new form of data and in understanding what it is and is not capable of providing. In other clinical disciplines in which genomic data are becoming increasingly common, efforts are being made to understand how users interact with online data reports in order to develop better user interfaces for services such as reporting of personal genomic data (106). These information visualization and user interface studies will inform the development of simplified reports that synthesize complex genomic information and associated risk factors into simple scores or visualizations that provide both actionable information and the necessary caveats to a clinician. Much like the GeneXpert output is simplified to clearly report diagnosis and resistance typing, we envision that WGS-based reporting will follow a similar model of a single-page report in lay language.
Sequencing platforms are also constantly changing—the current short-read platforms, which may not be able to read through long stretches of repetitive sequence, are being complemented by newer methods, such as nanopore sequencing, that can read several thousand bases of DNA in one contiguous read. As the platform landscape changes, labs must quickly adapt to assess the utility of these new methods and to integrate them into clinical practice. This will require validating new platforms for their utility in various diagnostic or characterization methods each time that the platform or the reagents change and will complicate the development of sustainable universal protocols. Similarly, updates to bioinformatic software would also merit recalibrating the system and potentially rerunning archived data sets to examine them through an updated lens.
Another critical barrier is standardization of data processing, analysis, and management in a way that is acceptable in the accredited clinical laboratory environment. The Global Microbial Identifier (GMI) project (http://www.globalmicrobialidentifier.org), an international consortium of microbial genomics researchers working within the public health sphere, was created with the vision of “develop[ing] a global system to aggregate, share, mine and use microbiological genomic data to address global public health and clinical challenges.” The GMI consortium has convened working groups to address these issues, examining the need for a minimum data and metadata format and addressing gaps in the analytical tool space, and is also working toward development of a quality assurance testing procedure that genomic clinical labs can use in their proficiency testing. Because it is organism agnostic, the work of the GMI project will have important impacts on the clinical laboratory landscape, and the policies and practices it helps to shape will be applicable across a range of pathogenic bacteria and viruses.
Despite this community-wide focus on establishing analytical standards, there will still be challenges to integrating WGS data with other clinical and epidemiological data sets and interpreting them through the appropriate lenses—these challenges must be dealt with on an organism by organism basis. In the TB community, where the consequences of incorrectly assigning a susceptibility phenotype or misinterpreting an outbreak's patterns have very real public health implications, we must work particularly hard toward reproducible and accurate analytical pipelines for interpreting WGS data, a deeper understanding of how finely genomic variation separates populations, and data-sharing approaches that will permit discovery of national and international trends in TB population genomics. As more WGS studies of TB are published, a consensus is beginning to emerge around issues such as the depth and quality of sequencing required to make confident variant calls; however, these guidelines should be formalized and shared with the community. The GMI project may be a vehicle for this, as may community-driven sites dedicated to TB genomics.
Similarly, insights derived from WGS will be translated successfully into public health interventions only if the appropriate epidemiological and clinical infrastructure is in place. The lab of the future may be capable of generating new insights into personalized treatment plans or the identification and management of emerging outbreaks, but putting this new knowledge to work will require open lines of communication between the laboratory and public-facing TB control programs, TB clinicians informed enough and confident enough around WGS data to act on their recommendations, and enough public health nurses to appropriately investigate and manage newly discovered clusters of cases.
The GMI initiative is also tackling another barrier to uptake—political involvement. Cementing new platforms into public health practice requires overcoming evidentiary thresholds of multiple stakeholders and obtaining enough buy-in from public health decision-makers to be able to roll out a new platform across an entire health care system. In the resource-constrained public health environment, a convincing case must be made to introduce a technology such as genomics, which requires a significant initial investment in infrastructure and training.
Finally, genomic testing of samples beyond the pathogen itself, i.e., samples of either the human host or his or her microbiota, introduces ethical issues. These include classical issues encountered throughout human genomics, including whether a patient's data might affect their chances of being insured or, when a family unit is being tested, issues related to paternity, but they also include issues only recently appearing on the community's radar. For example, to what degree can we rely on predictive biomarkers, such as those that can identify patients at higher risk of progressing from LTBI to active disease, and how does this influence our decision to use preventative therapy in LTBI, especially when the antibiotics used in prophylaxis carry a risk of adverse events?
Challenges aside, the genomic revolution is set to dramatically alter the clinical microbiology landscape, and we expect to see the first inroads in this area in the diagnosis, treatment, and epidemiology of tuberculosis in well-resourced settings. In order to effectively manage what is truly a global TB epidemic, interventions aimed at reducing poverty and health inequalities will be most critical. However, technological advances that create new knowledge about the biology and epidemiology of this important organism can readily be translated into sustainable interventions appropriate for lower-resource settings and will also play a role in TB control efforts. This is especially true for regions that transition from high to medium to low incidence and will require increasingly targeted approaches to reducing TB rates. Given recent successes in this area, such as with the GeneXpert system, we anticipate a future in which genomics directly contributes to a reduction in the incidence of this important global disease.
ACKNOWLEDGMENTS
We thank the British Columbia Centre for Disease Control Foundation, Genome British Columbia, and the British Columbia Lung Association for their support of our TB genomics research.
Biographies

Marta Wlodarska has 6 years of research experience in infectious disease and immunology and is currently a Postdoctoral Fellow at Novartis Institutes for Biomedical Research in Boston, MA. She obtained her B.Sc. with Honors at the University of British Columbia (UBC), Vancouver, Canada. She also completed her Ph.D. at the University of British Columbia, in the laboratory of B. Brett Finlay. Her research focused on discovering immune mechanisms required to maintain intestinal health and their relevance to enteric infection. After completing her Ph.D., she moved to the British Columbia Centre for Disease Control (BCCDC) in Vancouver to gain experience in public health and to understand the transmission dynamics of infectious disease, with a focus on the molecular epidemiology of tuberculosis.

James C. Johnston is a Clinical Assistant Professor in the Department of Medicine at UBC and a Michael Smith Foundation Scholar. He received his bachelor's degree from McGill and his M.D. from Queen's University. He returned to McGill for internal medicine training before moving to UBC for his Respiratory Medicine Fellowship. He completed postdoctoral training in tuberculosis research at the BC Centre for Disease Control and has a master's degree in public health from the Harvard School of Public Health. Dr. Johnston joined the BCCDC TB Services as the TB Evaluation Lead in 2010. His interests include TB genomics, TB screening in migrants to countries with low TB incidence, and MDR TB epidemiology and treatment.

Jennifer L. Gardy is a Senior Scientist at the British Columbia Centre for Disease Control and an Assistant Professor in UBC's School of Population and Public Health. She received a bachelor's degree in cell biology and genetics from UBC in 2000, completed a Ph.D. in computational microbial genomics at Simon Fraser University in 2006, and did 3 years of postdoctoral training in network analysis and systems biology at UBC before joining the BCCDC in 2009. Her research interests are in the use of genomics as a tool for understanding the origins and spread of communicable disease, with a particular emphasis on TB. She is particularly interested in using genomics to reconstruct outbreaks of infectious disease and translating the knowledge gained from genomics studies into real-world public health interventions.

Patrick Tang is a Medical Microbiologist at the British Columbia Centre for Disease Control and a Clinical Associate Professor in Pathology and Laboratory Medicine at the University of British Columbia. He completed his M.D./Ph.D. at UBC, followed by a medical microbiology residency at the University of Toronto. Since 2006, he has led the British Columbia TB/Mycobacteriology Laboratory and the Core Molecular/Genomics Laboratory at the BCCDC. His research interests are in the application of genomics and metagenomics to public health and infectious diseases, as well as the development of new molecular diagnostics. He uses these tools to diagnose outbreaks and unusual clinical diseases at the BCCDC. His other research projects include searching for infectious agents in chronic diseases and outbreaks, using genomics to investigate the dynamics of tuberculosis transmission, studying the microbiomes of people and animals, and even looking for better ways to test for water pollution.
REFERENCES
- 1.WHO. 2013. Global tuberculosis report 2013. WHO, Geneva, Switzerland. [Google Scholar]
- 2.Comas I, Coscolla M, Luo T, Borrell S, Holt KE, Kato-Maeda M, Parkhill J, Malla B, Berg S, Thwaites G, Yeboah-Manu D, Bothamley G, Mei J, Wei L, Bentley S, Harris SR, Niemann S, Diel R, Aseffa A, Gao Q, Young D, Gagneux S. 2013. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat Genet 45:1176–1182. doi: 10.1038/ng.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brosch R, Gordon SV, Marmiesse M, Brodin P, Buchrieser C, Eiglmeier K, Garnier T, Gutierrez C, Hewinson G, Kremer K, Parsons LM, Pym AS, Samper S, van Soolingen D, Cole ST. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc Natl Acad Sci U S A 99:3684–3689. doi: 10.1073/pnas.052548299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bos KI, Harkins KM, Herbig A, Coscolla M, Weber N, Comas I, Forrest SA, Bryant JM, Harris SR, Schuenemann VJ, Campbell TJ, Majander K, Wilbur AK, Guichon RA, Wolfe Steadman DL, Cook DC, Niemann S, Behr MA, Zumarraga M, Bastida R, Huson D, Nieselt K, Young D, Parkhill J, Buikstra JE, Gagneux S, Stone AC, Krause J. 2014. Pre-Columbian mycobacterial genomes reveal seals as a source of New World human tuberculosis. Nature 514:494–497. doi: 10.1038/nature13591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mehta JB, Dutt A, Harvill L, Mathews KM. 1991. Epidemiology of extrapulmonary tuberculosis. a comparative analysis with pre-AIDS era. Chest J 99:1134–1138. doi: 10.1378/chest.99.5.1134. [DOI] [PubMed] [Google Scholar]
- 6.Zumla A, Raviglione M, Hafner R, von Reyn CF. 2013. Tuberculosis. N Engl J Med 368:745–755. doi: 10.1056/NEJMra1200894. [DOI] [PubMed] [Google Scholar]
- 7.Raviglione M, Marais B, Floyd K, Lönnroth K, Getahun H, Migliori GB, Harries AD, Nunn P, Lienhardt C, Graham S, Chakaya J, Weyer K, Cole S, Kaufmann SH, Zumla A. 2012. Scaling up interventions to achieve global tuberculosis control: progress and new developments. Lancet 379:1902–1913. doi: 10.1016/S0140-6736(12)60727-2. [DOI] [PubMed] [Google Scholar]
- 8.Supply P, Marceau M, Mangenot S, Roche D, Rouanet C, Khanna V, Majlessi L, Criscuolo A, Tap J, Pawlik A, Fiette L, Orgeur M, Fabre M, Parmentier C, Frigui W, Simeone R, Boritsch EC, Debrie A-S, Willery E, Walker D, Quail MA, Ma L, Bouchier C, Salvignol G, Sayes F, Cascioferro A, Seemann T, Barbe V, Locht C, Gutierrez M-C, Leclerc C, Bentley SD, Stinear TP, Brisse S, Médigue C, Parkhill J, Cruveiller S, Brosch R. 2013. Genomic analysis of smooth tubercle bacilli provides insights into ancestry and pathoadaptation of Mycobacterium tuberculosis. Nat Genet 45:172–179. doi: 10.1038/ng.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boritsch EC, Supply P, Honoré N, Seeman T, Stinear TP, Brosch R. 2014. A glimpse into the past and predictions for the future: the molecular evolution of the tuberculosis agent. Mol Microbiol 93:835–852. doi: 10.1111/mmi.12720. [DOI] [PubMed] [Google Scholar]
- 10.Van Soolingen D, van der Zanden AG, de Haas PE, Noordhoek GT, Kiers A, Foudraine NA, Portaels F, Kolk AH, Kremer K, van Embden JD. 1998. Diagnosis of Mycobacterium microti infections among humans by using novel genetic markers. J Clin Microbiol 36:1840–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodríguez E, Sánchez LP, Pérez S, Herrera L, Jiménez MS, Samper S, Iglesias MJ. 2009. Human tuberculosis due to Mycobacterium bovis and M. caprae in Spain, 2004–2007. Int J Tuberc Lung Dis 13:1536–1541. [PubMed] [Google Scholar]
- 12.Van Ingen J, Rahim Z, Mulder A, Boeree MJ, Simeone R, Brosch R, van Soolingen D. 2012. Characterization of Mycobacterium orygis as M. tuberculosis complex subspecies. Emerg Infect Dis 18:653–655. doi: 10.3201/eid1804.110888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiers A, Klarenbeek A, Mendelts B, Van Soolingen D, Koëter G. 2008. Transmission of Mycobacterium pinnipedii to humans in a zoo with marine mammals. Int J Tuberc Lung Dis 12:1469–1473. [PubMed] [Google Scholar]
- 14.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream M-A, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 15.Mostowy S, Behr MA. 2005. The origin and evolution of Mycobacterium tuberculosis. Clin Chest Med 26:207–216. doi: 10.1016/j.ccm.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 16.Coscolla M, Gagneux S. 2010. Does M. tuberculosis genomic diversity explain disease diversity? Drug Discov Today Dis Mech 7:e43–e59. doi: 10.1016/j.ddmec.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tortoli E. 2014. Microbiological features and clinical relevance of new species of the genus Mycobacterium. Clin Microbiol Rev 27:727–752. doi: 10.1128/CMR.00035-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu L, Li Y, Li S, Hu N, He Y, Pong R, Lin D, Lu L, Law M. 2012. Comparison of next-generation sequencing systems. J Biomed Biotechnol 2012:251364. doi: 10.1155/2012/251364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Didelot X, Bowden R, Wilson DJ, Peto TEA, Crook DW. 2012. Transforming clinical microbiology with bacterial genome sequencing. Nat Rev Genet 13:601–612. doi: 10.1038/nrg3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin X, Zheng L, Lin L, Hu Y, Zheng F, Hu Y, Wang Q. 2013. Commercial MPT64-based tests for rapid identification of Mycobacterium tuberculosis complex: a meta-analysis. J Infect 67:369–377. doi: 10.1016/j.jinf.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 21.Roberts SA, Lowe O, Pandey S, Williamson DA, Newton S, Vaughan R. 2012. Comparison of the MGIT TBc immunochromatographic assay with the Accuprobe Gen-Probe TB assay for identification of Mycobacterium tuberculosis complex: results from a low-burden tuberculosis setting. Diagn Microbiol Infect Dis 74:415–416. doi: 10.1016/j.diagmicrobio.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 22.Scott LE, McCarthy K, Gous N, Nduna M, Van Rie A, Sanne I, Venter WF, Duse A, Stevens W. 2011. Comparison of Xpert MTB/RIF with other nucleic acid technologies for diagnosing pulmonary tuberculosis in a high HIV prevalence setting: a prospective study. PLoS Med 8:e1001061. doi: 10.1371/journal.pmed.1001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McNabb A, Adie K, Rodrigues M, Black WA, Isaac-Renton J. 2006. Direct identification of mycobacteria in primary liquid detection media by partial sequencing of the 65-kilodalton heat shock protein gene. J Clin Microbiol 44:60–66. doi: 10.1128/JCM.44.1.60-66.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helb D, Jones M, Story E, Boehme C, Wallace E, Ho K, Kop J, Owens MR, Rodgers R, Banada P, Safi H, Blakemore R, Lan NTN, Jones-López EC, Levi M, Burday M, Ayakaka I, Mugerwa RD, McMillan B, Winn-Deen E, Christel L, Dailey P, Perkins MD, Persing DH, Alland D. 2010. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J Clin Microbiol 48:229–237. doi: 10.1128/JCM.01463-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boehme CC, Nabeta P, Hillemann D, Nicol MP, Shenai S, Krapp F, Allen J, Tahirli R, Blakemore R, Rustomjee R, Milovic A, Jones M, O'Brien SM, Persing DH, Ruesch-Gerdes S, Gotuzzo E, Rodrigues C, Alland D, Perkins MD. 2010. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med 363:1005–1015. doi: 10.1056/NEJMoa0907847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walusimbi S, Bwanga F, De Costa A, Haile M, Joloba M, Hoffner S. 2013. Meta-analysis to compare the accuracy of GeneXpert, MODS and the WHO 2007 algorithm for diagnosis of smear-negative pulmonary tuberculosis. BMC Infect Dis 13:507. doi: 10.1186/1471-2334-13-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Albert H, Ademun PJ, Lukyamuzi G, Nyesiga B, Manabe Y, Joloba M, Wilson S, Perkins MD. 2011. Feasibility of magnetic bead technology for concentration of mycobacteria in sputum prior to fluorescence microscopy. BMC Infect Dis 11:125. doi: 10.1186/1471-2334-11-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghodbane R, Drancourt M. 2013. Magnetic bead protocol for culturing Mycobacterium tuberculosis from sputum specimens. J Clin Microbiol 51:1578–1579. doi: 10.1128/JCM.03428-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J, Sun Z-Q, Pei H, Zhang S-L, Zhang S-L, Wilson S, De Smet K, Cerullakandiyil N, Thayyullathil T, AL-Suwaidi Z, Song Y-Z. 2013. Increased case finding of tuberculosis from sputum and sputum deposits after magnetic bead concentration of mycobacteria. J Microbiol Methods 93:144–147. doi: 10.1016/j.mimet.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 30.Elitas M, Martinez-Duarte R, Dhar N, McKinney JD, Renaud P. 2014. Dielectrophoresis-based purification of antibiotic-treated bacterial subpopulations. Lab Chip 14:1850–1857. doi: 10.1039/C4LC00109E. [DOI] [PubMed] [Google Scholar]
- 31.Wang S, Inci F, De Libero G, Singhal A, Demirci U. 2013. Point-of-care assays for tuberculosis: role of nanotechnology/microfluidics. Biotechnol Adv 31:438–449. doi: 10.1016/j.biotechadv.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doughty EL, Sergeant MJ, Adetifa I, Antonio M, Pallen MJ. 2014. Culture-independent detection and characterisation of Mycobacterium tuberculosis and M. africanum in sputum samples using shotgun metagenomics on a benchtop sequencer. PeerJ 2:e585. doi: 10.7717/peerj.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, Di Ventra M, Garaj S, Hibbs A, Huang X, Jovanovich SB, Krstic PS, Lindsay S, Ling XS, Mastrangelo CH, Meller A, Oliver JS, Pershin YV, Ramsey JM, Riehn R, Soni GV, Tabard-Cossa V, Wanunu M, Wiggin M, Schloss JA. 2008. The potential and challenges of nanopore sequencing. Nat Biotechnol 26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Macaulay IC, Voet T. 2014. Single cell genomics: advances and future perspectives. PLoS Genet 10:e1004126. doi: 10.1371/journal.pgen.1004126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ling DI, Zwerling AA, Pai M. 2008. Rapid diagnosis of drug-resistant TB using line probe assays: from evidence to policy. Expert Rev Respir Med 2:583–588. doi: 10.1586/17476348.2.5.583. [DOI] [PubMed] [Google Scholar]
- 36.Rossau R, Traore H, De Beenhouwer H, Mijs W, Jannes G, De Rijk P, Portaels F. 1997. Evaluation of the INNO-LiPA Rif.TB assay, a reverse hybridization assay for the simultaneous detection of Mycobacterium tuberculosis complex and its resistance to rifampin. Antimicrob Agents Chemother 41:2093–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hillemann D, Rüsch-Gerdes S, Richter E. 2007. Evaluation of the GenoType MTBDRplus assay for rifampin and isoniazid susceptibility testing of Mycobacterium tuberculosis strains and clinical specimens. J Clin Microbiol 45:2635–2640. doi: 10.1128/JCM.00521-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Campbell PJ, Morlock GP, Sikes RD, Dalton TL, Metchock B, Starks AM, Hooks DP, Cowan LS, Plikaytis BB, Posey JE. 2011. Molecular detection of mutations associated with first- and second-line drug resistance compared with conventional drug susceptibility testing of Mycobacterium tuberculosis. Antimicrob Agents Chemother 55:2032–2041. doi: 10.1128/AAC.01550-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang H, Li D, Zhao L, Fleming J, Lin N, Wang T, Liu Z, Li C, Galwey N, Deng J, Zhou Y, Zhu Y, Gao Y, Wang T, Wang S, Huang Y, Wang M, Zhong Q, Zhou L, Chen T, Zhou J, Yang R, Zhu G, Hang H, Zhang J, Li F, Wan K, Wang J, Zhang X-E, Bi L. 2013. Genome sequencing of 161 Mycobacterium tuberculosis isolates from China identifies genes and intergenic regions associated with drug resistance. Nat Genet 45:1255–1260. doi: 10.1038/ng.2735. [DOI] [PubMed] [Google Scholar]
- 40.Farhat MR, Shapiro BJ, Kieser KJ, Sultana R, Jacobson KR, Victor TC, Warren RM, Streicher EM, Calver A, Sloutsky A, Kaur D, Posey JE, Plikaytis B, Oggioni MR, Gardy JL, Johnston JC, Rodrigues M, Tang PKC, Kato-Maeda M, Borowsky ML, Muddukrishna B, Kreiswirth BN, Kurepina N, Galagan J, Gagneux S, Birren B, Rubin EJ, Lander ES, Sabeti PC, Murray M. 2013. Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis. Nat Genet 45:1183–1189. doi: 10.1038/ng.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Safi H, Lingaraju S, Amin A, Kim S, Jones M, Holmes M, McNeil M, Peterson SN, Chatterjee D, Fleischmann R, Alland D. 2013. Evolution of high-level ethambutol-resistant tuberculosis through interacting mutations in decaprenylphosphoryl-β-d-arabinose biosynthetic and utilization pathway genes. Nat Genet 45:1190–1197. doi: 10.1038/ng.2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Casali N, Nikolayevskyy V, Balabanova Y, Harris SR, Ignatyeva O, Kontsevaya I, Corander J, Bryant J, Parkhill J, Nejentsev S, Horstmann RD, Brown T, Drobniewski F. 2014. Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat Genet 46:279–286. doi: 10.1038/ng.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eldholm V, Norheim G, von der Lippe B, Kinander W, Dahle UR, Caugant DA, Mannsåker T, Mengshoel AT, Dyrhol-Riise AM, Balloux F. 2014. Evolution of extensively drug-resistant Mycobacterium tuberculosis from a susceptible ancestor in a single patient. Genome Biol 15:490. doi: 10.1186/s13059-014-0490-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andries K, Verhasselt P, Guillemont J, Göhlmann HWH, Neefs J-M, Winkler H, Van Gestel J, Timmerman P, Zhu M, Lee E, Williams P, de Chaffoy D, Huitric E, Hoffner S, Cambau E, Truffot-Pernot C, Lounis N, Jarlier V. 2005. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307:223–227. doi: 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- 45.Clark TG, Mallard K, Coll F, Preston M, Assefa S, Harris D, Ogwang S, Mumbowa F, Kirenga B, O'Sullivan DM, Okwera A, Eisenach KD, Joloba M, Bentley SD, Ellner JJ, Parkhill J, Jones-López EC, McNerney R. 2013. Elucidating emergence and transmission of multidrug-resistant tuberculosis in treatment experienced patients by whole genome sequencing. PLoS One 8:e83012. doi: 10.1371/journal.pone.0083012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Vos M, Muller B, Borrell S, Black PA, van Helden PD, Warren RM, Gagneux S, Victor TC. 2013. Putative compensatory mutations in the rpoC gene of rifampin-resistant Mycobacterium tuberculosis are associated with ongoing transmission. Antimicrob Agents Chemother 57:827–832. doi: 10.1128/AAC.01541-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brandis G, Hughes D. 2013. Genetic characterization of compensatory evolution in strains carrying rpoB Ser531Leu, the rifampicin resistance mutation most frequently found in clinical isolates. J Antimicrob Chemother 68:2493–2497. doi: 10.1093/jac/dkt224. [DOI] [PubMed] [Google Scholar]
- 48.Comas I, Borrell S, Roetzer A, Rose G, Malla B, Kato-Maeda M, Galagan J, Niemann S, Gagneux S. 2012. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat Genet 44:106–110. doi: 10.1038/ng.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aguilar D, Hanekom M, Mata D, Gey van Pittius NC, van Helden PD, Warren RM, Hernandez-Pando R. 2010. Mycobacterium tuberculosis strains with the Beijing genotype demonstrate variability in virulence associated with transmission. Tuberculosis (Edinb) 90:319–325. doi: 10.1016/j.tube.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 50.Reiling N, Homolka S, Walter K, Brandenburg J, Niwinski L, Ernst M, Herzmann C, Lange C, Diel R, Ehlers S, Niemann S. 2013. Clade-specific virulence patterns of Mycobacterium tuberculosis complex strains in human primary macrophages and aerogenically infected mice. mBio 4:e00250-13. doi: 10.1128/mBio.00250-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ford CB, Shah RR, Maeda MK, Gagneux S, Murray MB, Cohen T, Johnston JC, Gardy J, Lipsitch M, Fortune SM. 2013. Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nat Genet 45:784–790. doi: 10.1038/ng.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trauner A, Borrell S, Reither K, Gagneux S. 2014. Evolution of drug resistance in tuberculosis: recent progress and implications for diagnosis and therapy. Drugs 74:1063–1072. doi: 10.1007/s40265-014-0248-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Köser CU, Bryant JM, Becq J, Török ME, Ellington MJ, Marti-Renom MA, Carmichael AJ, Parkhill J, Smith GP, Peacock SJ. 2013. Whole-genome sequencing for rapid susceptibility testing of M. tuberculosis. N Engl J Med 369:290–292. doi: 10.1056/NEJMc1215305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim SJ. 2005. Drug-susceptibility testing in tuberculosis: methods and reliability of results. Eur Respir J 25:564–569. doi: 10.1183/09031936.05.00111304. [DOI] [PubMed] [Google Scholar]
- 55.Maruri F, Sterling TR, Kaiga AW, Blackman A, van der Heijden YF, Mayer C, Cambau E, Aubry A. 2012. A systematic review of gyrase mutations associated with fluoroquinolone-resistant Mycobacterium tuberculosis and a proposed gyrase numbering system. J Antimicrob Chemother 67:819–831. doi: 10.1093/jac/dkr566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Laurenzo D, Mousa SA. 2011. Mechanisms of drug resistance in Mycobacterium tuberculosis and current status of rapid molecular diagnostic testing. Acta Trop 119:5–10. doi: 10.1016/j.actatropica.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 57.Shafer RW. 2006. Rationale and uses of a public HIV drug-resistance database. J Infect Dis 194(Suppl 1):S51–S58. doi: 10.1086/505356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sandgren A, Strong M, Muthukrishnan P, Weiner BK, Church GM, Murray MB. 2009. Tuberculosis drug resistance mutation database. PLoS Med 6:e2. doi: 10.1371/journal.pmed.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Flandrois J-P, Lina G, Dumitrescu O. 2014. MUBII-TB-DB: a database of mutations associated with antibiotic resistance in Mycobacterium tuberculosis. BMC Bioinformatics 15:107. doi: 10.1186/1471-2105-15-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kasaie P, Andrews JR, Kelton WD, Dowdy DW. 2014. Timing of tuberculosis transmission and the impact of household contact tracing. An agent-based simulation model. Am J Respir Crit Care Med 189:845–852. doi: 10.1164/rccm.201310-1846OC. [DOI] [PubMed] [Google Scholar]
- 61.Shrivastava SR, Shrivastava PS, Ramasamy J. 2014. Assessing the utility of contact tracing in reducing the magnitude of tuberculosis. Infect Ecol Epidemiol 4:10.3402/iee.v4.25265. doi: 10.3402/iee.v4.25265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gardy JL, Johnston JC, Sui SJH, Cook VJ, Shah L, Brodkin E, Rempel S, Moore R, Zhao Y, Holt R, Varhol R, Birol I, Lem M, Sharma MK, Elwood K, Jones SJM, Brinkman FSL, Brunham RC, Tang P. 2011. Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N Engl J Med 364:730–739. doi: 10.1056/NEJMoa1003176. [DOI] [PubMed] [Google Scholar]
- 63.Walker TM, Ip CLC, Harrell RH, Evans JT, Kapatai G, Dedicoat MJ, Eyre DW, Wilson DJ, Hawkey PM, Crook DW, Parkhill J, Harris D, Walker AS, Bowden R, Monk P, Smith EG, Peto TEA. 2013. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis 13:137–146. doi: 10.1016/S1473-3099(12)70277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Streicher EM, Victor TC, van der Spuy G, Sola C, Rastogi N, van Helden PD, Warren RM. 2007. Spoligotype signatures in the Mycobacterium tuberculosis complex. J Clin Microbiol 45:237–240. doi: 10.1128/JCM.01429-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rüsch-Gerdes S, Willery E, Savine E, de Haas P, van Deutekom H, Roring S, Bifani P, Kurepina N, Kreiswirth B, Sola C, Rastogi N, Vatin V, Gutierrez MC, Fauville M, Niemann S, Skuce R, Kremer K, Locht C, van Soolingen D. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive-unit–variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol 44:4498–4510. doi: 10.1128/JCM.01392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Torrea G, Levee G, Grimont P, Martin C, Chanteau S, Gicquel B. 1995. Chromosomal DNA fingerprinting analysis using the insertion sequence IS6110 and the repetitive element DR as strain-specific markers for epidemiological study of tuberculosis in French Polynesia. J Clin Microbiol 33:1899–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Köser CU, Ellington MJ, Cartwright EJP, Gillespie SH, Brown NM, Farrington M, Holden MTG, Dougan G, Bentley SD, Parkhill J, Peacock SJ. 2012. Routine use of microbial whole genome sequencing in diagnostic and public health microbiology. PLoS Pathog 8:e1002824. doi: 10.1371/journal.ppat.1002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sola C, Filliol I, Legrand E, Lesjean S, Locht C, Supply P, Rastogi N. 2003. Genotyping of the Mycobacterium tuberculosis complex using MIRUs: association with VNTR and spoligotyping for molecular epidemiology and evolutionary genetics. Infect Genet Evol 3:125–133. doi: 10.1016/S1567-1348(03)00011-X. [DOI] [PubMed] [Google Scholar]
- 69.Niemann S, Köser CU, Gagneux S, Plinke C, Homolka S, Bignell H, Carter RJ, Cheetham RK, Cox A, Gormley NA, Kokko-Gonzales P, Murray LJ, Rigatti R, Smith VP, Arends FPM, Cox HS, Smith G, Archer JAC. 2009. Genomic diversity among drug sensitive and multidrug resistant isolates of Mycobacterium tuberculosis with identical DNA fingerprints. PLoS One 4:e7407. doi: 10.1371/journal.pone.0007407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schurch AC, Kremer K, Daviena O, Kiers A, Boeree MJ, Siezen RJ, van Soolingen D. 2010. High-resolution typing by integration of genome sequencing data in a large tuberculosis cluster. J Clin Microbiol 48:3403–3406. doi: 10.1128/JCM.00370-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roetzer A, Diel R, Kohl TA, Rückert C, Nübel U, Blom J, Wirth T, Jaenicke S, Schuback S, Rüsch-Gerdes S, Supply P, Kalinowski J, Niemann S. 2013. Whole genome sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis outbreak: a longitudinal molecular epidemiological study. PLoS Med 10:e1001387. doi: 10.1371/journal.pmed.1001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kato-Maeda M, Ho C, Passarelli B, Banaei N, Grinsdale J, Flores L, Anderson J, Murray M, Rose G, Kawamura LM, Pourmand N, Tariq MA, Gagneux S, Hopewell PC. 2013. Use of whole genome sequencing to determine the microevolution of Mycobacterium tuberculosis during an outbreak. PLoS One 8:e58235. doi: 10.1371/journal.pone.0058235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Luo T, Yang C, Peng Y, Lu L, Sun G, Wu J, Jin X, Hong J, Li F, Mei J, DeRiemer K, Gao Q. 2014. Whole-genome sequencing to detect recent transmission of Mycobacterium tuberculosis in settings with a high burden of tuberculosis. Tuberculosis (Edinb) 94:434–440. doi: 10.1016/j.tube.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jamieson FB, Teatero S, Guthrie JL, Neemuchwala A, Fittipaldi N, Mehaffy C. 2014. Whole-genome sequencing of the Mycobacterium tuberculosis Manila sublineage results in less clustering and better resolution than mycobacterial interspersed repetitive-unit–variable-number tandem-repeat (MIRU-VNTR) typing and spoligotyping. J Clin Microbiol 52:3795–3798. doi: 10.1128/JCM.01726-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mehaffy C, Guthrie JL, Alexander DC, Stuart R, Rea E, Jamieson FB. 2014. Marked microevolution of a unique Mycobacterium tuberculosis strain in 17 years of ongoing transmission in a high risk population. PLoS One 9:e112928. doi: 10.1371/journal.pone.0112928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stucki D, Ballif M, Bodmer T, Coscolla M, Maurer A-M, Droz S, Butz C, Borrell S, Längle C, Feldmann J, Furrer H, Mordasini C, Helbling P, Rieder HL, Egger M, Gagneux S, Fenner L. 30 October 2014. Tracking a tuberculosis outbreak over 21 years: strain-specific single-nucleotide polymorphism typing combined with targeted whole-genome sequencing. J Infect Dis doi: 10.1093/infdis/jiu601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schürch AC, Kremer K, Kiers A, Daviena O, Boeree MJ, Siezen RJ, Smith NH, van Soolingen D. 2010. The tempo and mode of molecular evolution of Mycobacterium tuberculosis at patient-to-patient scale. Infect Genet Evol 10:108–114. doi: 10.1016/j.meegid.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 78.Török ME, Reuter S, Bryant J, Köser CU, Stinchcombe SV, Nazareth B, Ellington MJ, Bentley SD, Smith GP, Parkhill J, Peacock SJ. 2013. Rapid whole-genome sequencing for investigation of a suspected tuberculosis outbreak. J Clin Microbiol 51:611–614. doi: 10.1128/JCM.02279-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Walker TM, Lalor MK, Broda A, Ortega LS, Morgan M, Parker L, Churchill S, Bennett K, Golubchik T, Giess AP, Del Ojo Elias C, Jeffery KJ, Bowler ICJW, Laurenson IF, Barrett A, Drobniewski F, McCarthy ND, Anderson LF, Abubakar I, Thomas HL, Monk P, Smith EG, Walker AS, Crook DW, Peto TEA, Conlon CP. 2014. Assessment of Mycobacterium tuberculosis transmission in Oxfordshire, UK, 2007–12, with whole pathogen genome sequences: an observational study. Lancet Respir Med 2:285–292. doi: 10.1016/S2213-2600(14)70027-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Didelot X, Gardy J, Colijn C. 2014. Bayesian inference of infectious disease transmission from whole-genome sequence data. Mol Biol Evol 31:1869–1879. doi: 10.1093/molbev/msu121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kohl TA, Diel R, Harmsen D, Rothgänger J, Meywald Walter K, Merker M, Weniger T, Niemann S. 2014. Whole genome based Mycobacterium tuberculosis surveillance: a standardized, portable and expandable approach. J Clin Microbiol 52:2479–2486. doi: 10.1128/JCM.00567-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Coll F, Mallard K, Preston MD, Bentley S, Parkhill J, McNerney R, Martin N, Clark TG. 2012. SpolPred: rapid and accurate prediction of Mycobacterium tuberculosis spoligotypes from short genomic sequences. Bioinformatics 28:2991–2993. doi: 10.1093/bioinformatics/bts544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Halachev MR, Chan JZ-M, Constantinidou CI, Cumley N, Bradley C, Smith-Banks M, Oppenheim B, Pallen MJ. 2014. Genomic epidemiology of a protracted hospital outbreak caused by multidrug-resistant Acinetobacter baumannii in Birmingham, England. Genome Med 6:70. doi: 10.1186/s13073-014-0070-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Snitkin ES, Zelazny AM, Thomas PJ, Stock F, NISC Comparative Sequencing Program Group, Henderson DK, Palmore TN, Segre JA. 2012. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med 4:148–116. doi: 10.1126/scitranslmed.3004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Colijn C, Gardy J. 2014. Phylogenetic tree shapes resolve disease transmission patterns. Evol Med Public Health 2014:96–108. doi: 10.1093/emph/eou018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bryant JM, Harris SR, Parkhill J, Dawson R, Diacon AH, van Helden P, Pym A, Mahayiddin AA, Chuchottaworn C, Sanne IM, Louw C, Boeree MJ, Hoelscher M, McHugh TD, Bateson ALC, Hunt RD, Mwaigwisya S, Wright L, Gillespie SH, Bentley SD. 2013. Whole-genome sequencing to establish relapse or re-infection with Mycobacterium tuberculosis: a retrospective observational study. Lancet Respir Med 1:786–792. doi: 10.1016/S2213-2600(13)70231-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ford CB, Lin PL, Chase MR, Shah RR, Iartchouk O, Galagan J, Mohaideen N, Ioerger TR, Sacchettini JC, Lipsitch M, Flynn JL, Fortune SM. 2011. Use of whole genome sequencing to estimate the mutation rate of Mycobacterium tuberculosis during latent infection. Nat Genet 43:482–486. doi: 10.1038/ng.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bryant JM, Schürch AC, van Deutekom H, Harris SR, de Beer JL, de Jager V, Kremer K, van Hijum SA, Siezen RJ, Borgdorff M, Bentley SD, Parkhill J, van Soolingen D. 2013. Inferring patient to patient transmission of Mycobacterium tuberculosis from whole genome sequencing data. BMC Infect Dis 13:110. doi: 10.1186/1471-2334-13-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pérez-Lago L, Comas I, Navarro Y, González-Candelas F, Herranz M, Bouza E, García-de-Viedma D. 2014. Whole genome sequencing analysis of intrapatient microevolution in Mycobacterium tuberculosis: potential impact on the inference of tuberculosis transmission. J Infect Dis 209:98–108. doi: 10.1093/infdis/jit439. [DOI] [PubMed] [Google Scholar]
- 90.Rohde K, Yates RM, Purdy GE, Russell DG. 2007. Mycobacterium tuberculosis and the environment within the phagosome. Immunol Rev 219:37–54. doi: 10.1111/j.1600-065X.2007.00547.x. [DOI] [PubMed] [Google Scholar]
- 91.O'Garra A, Redford PS, McNab FW, Bloom CI, Wilkinson RJ, Berry MPR. 2013. The immune response in tuberculosis. Annu Rev Immunol 31:475–527. doi: 10.1146/annurev-immunol-032712-095939. [DOI] [PubMed] [Google Scholar]
- 92.Galagan JE. 2014. Genomic insights into tuberculosis. Nat Rev Genet 15:307–320. doi: 10.1038/nrg3664. [DOI] [PubMed] [Google Scholar]
- 93.Colangeli R, Arcus VL, Cursons RT, Ruthe A, Karalus N, Coley K, Manning SD, Kim S, Marchiano E, Alland D. 2014. Whole genome sequencing of Mycobacterium tuberculosis reveals slow growth and low mutation rates during latent infections in humans. PLoS One 9:e91024. doi: 10.1371/journal.pone.0091024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Perdigão J, Silva H, Machado D, Macedo R, Maltez F, Silva C, Jordao L, Couto I, Mallard K, Coll F, Hill-Cawthorne GA, McNerney R, Pain A, Clark TG, Viveiros M, Portugal I. 2014. Unraveling Mycobacterium tuberculosis genomic diversity and evolution in Lisbon, Portugal, a highly drug resistant setting. BMC Genomics 15:991. doi: 10.1186/1471-2164-15-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huang Y-S, Chern H-D, Su W-J, Wu J-C, Lai S-L, Yang S-Y, Chang F-Y, Lee S-D. 2002. Polymorphism of the N-acetyltransferase 2 gene as a susceptibility risk factor for antituberculosis drug-induced hepatitis. Hepatology 35:883–889. doi: 10.1053/jhep.2002.32102. [DOI] [PubMed] [Google Scholar]
- 96.Matsumoto T, Ohno M, Azuma J. 2014. Future of pharmacogenetics-based therapy for tuberculosis. Pharmacogenomics 15:601–607. doi: 10.2217/pgs.14.38. [DOI] [PubMed] [Google Scholar]
- 97.Parkin DP, Vandenplas S, Botha FJ, Vandenplas ML, Seifart HI, van Helden PD, van der Walt BJ, Donald PR, van Jaarsveld PP. 1997. Trimodality of isoniazid elimination: phenotype and genotype in patients with tuberculosis. Am J Respir Crit Care Med 155:1717–1722. doi: 10.1164/ajrccm.155.5.9154882. [DOI] [PubMed] [Google Scholar]
- 98.Ohno M, Yamaguchi I, Yamamoto I, Fukuda T, Yokota S, Maekura R, Ito M, Yamamoto Y, Ogura T, Maeda K, Komuta K, Igarashi T, Azuma J. 2000. Slow N-acetyltransferase 2 genotype affects the incidence of isoniazid and rifampicin-induced hepatotoxicity. Int J Tuberc Lung Dis 4:256–261. [PubMed] [Google Scholar]
- 99.Sharma SK, Balamurugan A, Saha PK, Pandey RM, Mehra NK. 2002. Evaluation of clinical and immunogenetic risk factors for the development of hepatotoxicity during antituberculosis treatment. Am J Respir Crit Care Med 166:916–919. doi: 10.1164/rccm.2108091. [DOI] [PubMed] [Google Scholar]
- 100.Haiser HJ, Gootenberg DB, Chatman K, Sirasani G, Balskus EP, Turnbaugh PJ. 2013. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science 341:295–298. doi: 10.1126/science.1235872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wallace BD, Wang H, Lane KT, Scott JE, Orans J, Koo JS, Venkatesh M, Jobin C, Yeh L-A, Mani S, Redinbo MR. 2010. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 330:831–835. doi: 10.1126/science.1191175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK. 2009. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci U S A 106:14728–14733. doi: 10.1073/pnas.0904489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung Y-M, Wu Y, Schauer P, Smith JD, Allayee H, Tang WHW, DiDonato JA, Lusis AJ, Hazen SL. 2011. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Azad AK, Sadee W, Schlesinger LS. 2012. Innate immune gene polymorphisms in tuberculosis. Infect Immun 80:3343–3359. doi: 10.1128/IAI.00443-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Abel L, El-Baghdadi J, Bousfiha AA, Casanova J-L, Schurr E. 2014. Human genetics of tuberculosis: a long and winding road. Philos Trans R Soc Lond B Biol Sci 369:20130428. doi: 10.1098/rstb.2013.0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shaer O, Nov O. 2014. HCI for personal genomics. Interactions 21:32–37. doi: 10.1145/2656622. [DOI] [Google Scholar]