SUMMARY
In nature, selenium is actively cycled between oxic and anoxic habitats, and this cycle plays an important role in carbon and nitrogen mineralization through bacterial anaerobic respiration. Selenium-respiring bacteria (SeRB) are found in geographically diverse, pristine or contaminated environments and play a pivotal role in the selenium cycle. Unlike its structural analogues oxygen and sulfur, the chalcogen selenium and its microbial cycling have received much less attention by the scientific community. This review focuses on microorganisms that use selenate and selenite as terminal electron acceptors, in parallel to the well-studied sulfate-reducing bacteria. It overviews the significant advancements made in recent years on the role of SeRB in the biological selenium cycle and their ecological role, phylogenetic characterization, and metabolism, as well as selenium biomineralization mechanisms and environmental biotechnological applications.
INTRODUCTION
The Geochemical Selenium Cycle
Selenium was discovered by the father of Swedish chemistry, Jöns Jacob Berzelius, in 1817. It is an important essential trace element in living systems and has an uneven distribution in the Earth's crust. Selenium is placed next to sulfur in the list of important elements, i.e., H, O, C, N, P, S, and Se are the dominant components of all living systems (1). The selenium cycle is complex (Fig. 1) because this element has a wide range of oxidation states, from −II to +VI, and is available in different chemical (inorganic and organic) and physical (solid, liquid, and gas) forms. Moreover, selenium occurs as 6 stable isotopes, 74Se, 76Se, 77Se, 78Se, 80Se, and 82Se, among which 80Se and 78Se are the most common forms found on Earth (2). The distribution of different species of selenium may vary in the environment depending on the prevailing redox conditions. In general, selenium oxyanions (SeO42− and SeO32−) are highly soluble, stable, and potentially mobile in oxic natural environments. On the other hand, the solubilities of elemental selenium and metal selenides are limited, and they are less mobile under environmental conditions.
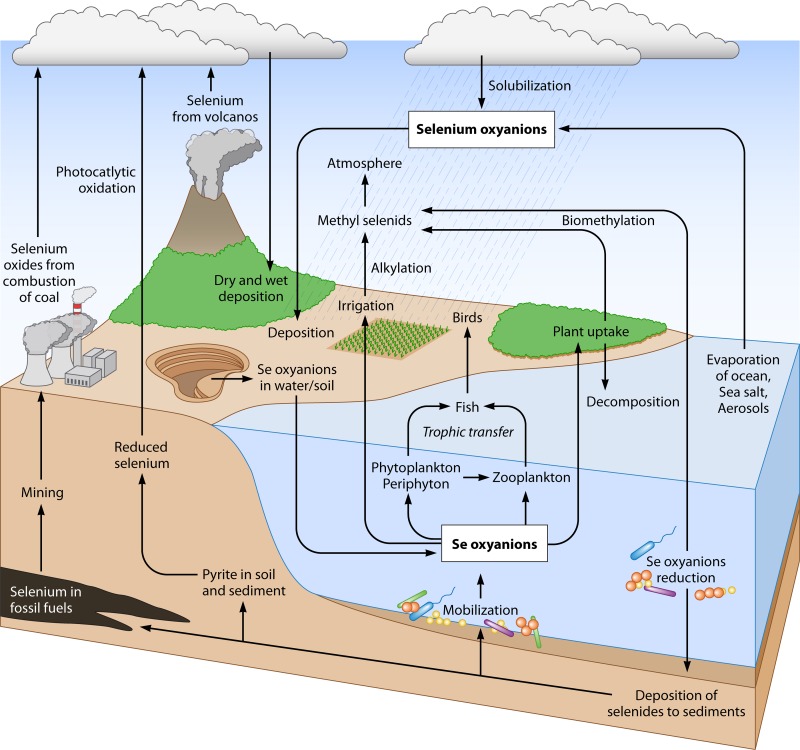
FIG 1.
Global selenium cycle in nature. The largest reservoirs of selenium on Earth are sulfide ores, pyrite, and high-sulfur coals. Geologic and anthropogenic sources release selenium as SeO42− into the environment. Selenium, an essential element, is assimilated from selenate or selenite by microbes and plants at the base of the food web and subsequently by animals. Selenium is then assimilated into organoselenides (i.e., selenoproteins) in living organisms. Decomposition of dead organisms releases selenium back into the environment. Mining operations, combustion of fossil fuels, agriculture, volcanic eruptions, and nuclear fuel cycle operations release selenium into the atmosphere, soil, and water in soluble forms (i.e., SeO42− and SeO32−). Microorganisms play a key role in the cycling of selenium compounds in nature.
The most oxidized forms, such as selenate (SeO42−) and selenite (SeO32−), are predominantly encountered in oxygenated environments, i.e., surface waters. Both selenate and selenite are highly soluble and exhibit high bioavailability and toxicity. In anoxic or anaerobic environments, formation of elemental selenium (Se0) is predicted due to the prevailing reducing conditions (3, 4). Although elemental selenium can exist in different allotropic forms (crystalline, metallic, and amorphous), it has commonly been considered an unavailable form in natural environments due to its low solubility (3). However, elemental Se in colloidal form can still be transported in the environment and become bioavailable to aquatic organisms (4).
Under highly reducing conditions, elemental selenium can be further reduced to selenide [Se(−II)], which in turn binds with metals and organics to form metal selenides and organoselenides, respectively. In natural settings, selenide is often observed as metal selenides in mineral phases of rocks and sediments. Organoselenium compounds contain selenide mostly as selenocysteine in proteins. The methylated forms of selenium also contain selenide and are volatile in nature. Additionally, selenide can also form volatile and highly toxic H2Se, a structural analogue of H2S. Thus, elemental selenium is rarely found in nature, and selenium occurs only as selenide in minerals (2), mainly in association with natural sulfides, such as pyrite (FeS2), chalcopyrite (CuFeS2), and sphalerite (ZnS) (5, 6).
Selenium in the Environment
Selenium is found in all natural environments, including rocks, soils, water bodies, and the atmosphere. It is released from Se-rich sources, such as phosphatic rocks, organic-rich black shales, and coals, through complex biogeochemical cycling processes (Fig. 1). Thus, selenium is unevenly distributed over the surface of the Earth.
Natural selenium contamination can originate from weathering of seleniferous soils and rocks. In most soils, selenium concentrations are very low, in the range of 0.01 to 2 mg kg−1. However, in seleniferous soils, selenium concentrations of up to 1,200 mg kg−1 have been reported (7). In natural waters, the dissolved selenium concentrations are reported to be in the range of <0.1 to 100 μg liter−1, although groundwaters with elevated selenium concentrations have been reported, with concentrations of 275 μg liter−1 in seleniferous aquifers in China and up to 1,000 μg liter−1 in Montana (7). High selenium concentrations of up to 2,000 μg liter−1 have been found in some saline lake waters (7).
Anthropogenic selenium pollution is generated by mining, refinement of metals, flue gases, and other industrial activities (Fig. 1). High rates of embryonic deformities and deaths in waterfowl and other wildlife due to Se contamination were recorded in the Kesterson National Wildlife Reservoir in California. Excessive phosphate mining activities in the Blackfoot River Watershed in Idaho have substantially increased the selenium levels in the river (8). The reductive microbial selenium transformations of the selenium cycle, which converts water-soluble and toxic selenium oxyanions (SeO42− and SeO32−) into sparingly soluble elemental selenium or metal selenides, are the basis of promising approaches to bioremediation (5, 9, 10).
Role of Selenium in Cellular Metabolism
Selenium was long regarded as a toxin, with the biological toxicity of selenium recognized for the first time in 1856, when it was found to be responsible for “alkali disease,” now termed “selenosis.” The symptoms of selenosis include abnormal hair loss, broken nails, nail sloughing, thick nails, and skin lesions (11). The beneficial effect of this element was not recognized until 1957, when selenium was found to prevent liver necrosis in rats (12). Today, selenium is well recognized as an essential trace element with importance in several physiological functions, such as biosynthesis of selenocysteine (the 21st amino acid), coenzyme Q, glutathione peroxidase, and thioredoxin reductase (13, 14). The margin of safety is very narrow: there is only 1 order of magnitude between the essential and toxic levels of selenium (5, 13, 14). Thus, a small change in selenium levels can cause damage to living organisms and may contribute to ecological damage.
In bacteria and archaea, selenium is readily metabolized and involved in a range of metabolic functions that include assimilation, methylation, detoxification, and anaerobic respiration (9, 15, 16). The essential trace requirement of selenium was identified for stable operation of anaerobic digesters treating food waste and operating at high ammonia concentrations (17). Acetate oxidation to hydrogen and subsequent conversion of hydrogen and CO2 to methane through the syntrophic association between acetate-oxidizing bacteria and hydrogenotrophic methanogens form a dominant methanogenic pathway in anaerobic digesters, particularly in the presence of high concentrations of ammonia and volatile fatty acids (17–19). Selenium in the form of selenocysteine has been identified in the active site of formate dehydrogenases and is important for formate oxidation (20, 21). Accumulation of formate, an intermediate of propionate oxidation, triggers a feedback inhibition resulting in propionate accumulation (22), which may lead to process failure in anaerobic digesters. Supplementation with trace amounts of selenium (200 μg liter−1) allowed successful operation of anaerobic digesters at higher organic loading rates (17). Critical selenium levels required for stable reactor operation at moderate loading rates amounted to 0.16 mg Se kg−1 fresh matter feed (17).
BIOCHEMICAL SELENIUM CYCLE
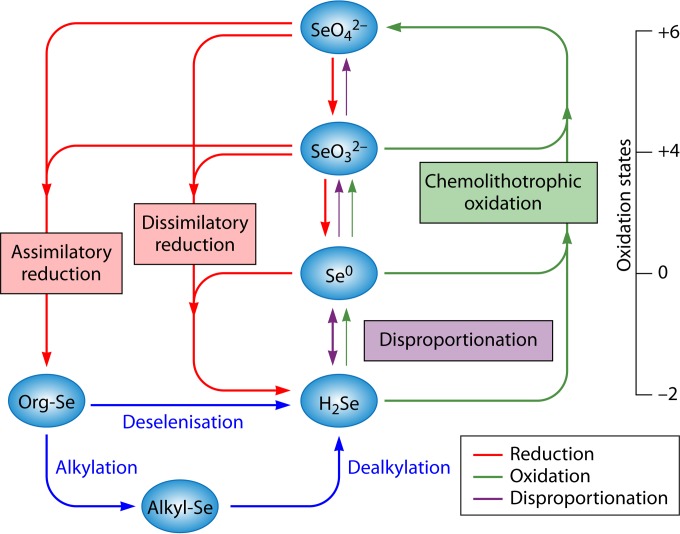
The existence of the selenium cycle in nature was proposed as early as 1964, by Shrift (23). The biochemical cycle of selenium is receiving increased attention because selenium is an essential trace element, selenium pollution can cause significant ecological damage, and selenium-respiring bacteria (SeRB) are widespread and metabolically active, thus affecting the C, N, and P cycles in nature. In nature, the transformation (oxidation and reduction) of selenium is mediated by both chemical and biological mechanisms. It is becoming increasingly evident that microorganisms play a pivotal role in the selenium cycle in the environment by performing both oxidation and reduction reactions (Fig. 2). Selenium metabolism is found in all domains of life, including Bacteria (24), Archaea (25), and Eukarya (26), as well as in viruses (27). Selenium is incorporated into amino acids, such as cysteine, and then into selenium-containing proteins (25).
FIG 2.
Selenium transformations in nature. Geologic and anthropogenic sources release selenium as SeO42− into the environment. SeRB are ubiquitous in natural settings and have a key role to play in the biochemical selenium cycle. SeRB have the ability to use selenate and/or selenite as a terminal electron acceptor in anaerobic respiration. The reduction of selenium oxyanions (SeO42− and SeO32−) is coupled to the degradation of organic matter in anaerobic sediments. Both SeO42− and SeO32− can be reduced to Se0 by bacteria under anaerobic conditions. Some bacteria reduce SeO42− and can incorporate it into organic compounds, i.e., selenoproteins. Reduction of SeO42− to Se0 appears to be a major sink for Se oxyanions in anoxic sediments. Reduction of Se(VI) or Se(IV) by SeRB (i.e., Sulfurospirillum barnesii and Bacillus arseniciselenatis) results in the production of insoluble elemental Se0. The opposite processes, such as oxidation of elemental selenium, can occur in soils and sediments, albeit at lower rates. Further reduction of elemental Se0 by other microorganisms (i.e., Bacillus selenitireducens) can lead to the formation of selenide. The selenide is then oxidized aerobically by selenium-oxidizing bacteria (SeOB), such as Thiobacillus, Thiothrix, etc. Alkylation reactions, which produce volatile (CH3)2Se and (CH3)2Se2, and dealkylation are also important processes mediated by microorganisms in soil and water (5, 9).
Selenium-Reducing Bacteria
Dissimilatory reduction of selenium oxyanions (SeO42− and SeO32−) is significantly important in the environment and involves conservation of metabolic energy for microorganisms (28). Members of both the Archaea and Bacteria domains can use selenium oxyanions (SeO42− and SeO32−) as terminal electron acceptors and reduce soluble selenate and selenite to insoluble elemental selenium via dissimilatory reduction under anaerobic conditions (29). Under aerobic or microaerophilic conditions, selenium oxyanions can also be reduced to elemental selenium by various bacterial strains, through either detoxification (30) or redox homeostasis in phototrophic bacteria (31), but these are not discussed in detail in this review. Elemental selenium can be reduced further microbiologically to soluble selenide, which in combination with metal ions forms insoluble metal selenides. Selenide can also be emitted as the volatile and highly reactive H2Se gas, but this is spontaneously and rapidly oxidized to elemental selenium in the presence of oxygen (9). Organoselenium and methylated selenium species also contain selenide.
Selenium-Oxidizing Bacteria
Oxidation of selenide and elemental selenium back to selenite or selenate by selenium-oxidizing bacteria (SeOB) completes the other half of the selenium cycle (Fig. 2). Experimental evidence on the role of microorganisms in the oxidation of either selenide or elemental selenium was fragmented when the selenium cycle was initially proposed (23). Oxidation of elemental selenium by autotrophic soil bacteria was reported as early as 1923. Addition of elemental selenium to a fresh soil was associated with oxidation of the elemental selenium and an increase in soil acidity. When this soil was added to a culture medium with elemental selenium as the only source of energy, the medium became cloudy due to the growth of a rod-shaped bacterium. This observation was reported as a news item in Science by Lipman and Waksman (32), but the details of this work and the identity of the selenium-oxidizing bacterium were not published. Subsequent studies have discovered the role of microorganisms in the oxidation of elemental selenium in experiments performed using soil slurries and bacterial cultures.
Sarathchandra and Watkinson were the first to report on bacterial oxidation of elemental selenium to SeO42− or SeO32− by Bacillus megaterium (33). SeO32− was found to be the predominant product, while SeO42− was produced only in trace amounts by this bacterium. Later, evidence for the oxidation of elemental selenium to SeO42− or SeO32− was demonstrated in soil slurries by Dowdle and Oremland (34). Production of either SeO42− or SeO32− was inhibited when the soil slurries were either autoclaved or amended with metabolic inhibitors, such as formalin, antibiotics, azide, and 2,4-dinitrophenol. On the other hand, addition of acetate, glucose, or sulfide enhanced the oxidation of selenium, which indicated the involvement of chemoheterotrophs or chemoautotrophic thiobacilli. Cultures of Thiobacillus ASN-1 and Leptothrix MnB1 and a heterotrophic soil enrichment oxidized selenium, with SeO42− as the major end product (34).
Almost at the same time, Losi and Frankenberger reported microbial oxidation and solubilization of precipitated elemental selenium in soil (35). They demonstrated that oxidation of elemental selenium in soils is largely mediated by biotic mechanisms and that use of an inorganic carbon source (NaHCO3) favored selenium oxidation over glucose oxidation, again implying chemoautotrophic oxidation. However, the oxidation of elemental selenium occurred at low rates and yielded both SeO42− and SeO32− as the end products. Oxidation rates are a function of the dissolved oxygen concentration and might vary widely in environmental settings, i.e., creeks, rivers, lakes, or ponds. In general, the oxidation rates are 3 to 4 orders of magnitude lower than those in the reductive part of the selenium cycle (34). This can result in the build-up of proportionately larger levels of either elemental selenium or selenides in sediments.
SELENIUM-REDUCING BACTERIA
Selenate-Respiring Bacteria
The reduction potential of the SeO42−/SeO32− couple is +0.48 V (Table 1) (1). Based on comparison of calculated free energies, using H2 as the electron donor, the reduction of SeO42− to SeO32− falls after the Mn(IV)/Mn(II) reaction and between the NO3−/N2 and NO3−/NH4+ redox reactions (15, 36). The reduction of SeO32− to elemental selenium falls between the reduction of ferric hydroxide [Fe(OH)3] and arsenate (AsO43−) but occurs at a higher redox potential than that required for sulfate reduction. The free energies for reduction of SeO42− and SeO32− coupled to H2 oxidation (15, 36) are given as follows:
| (1) |
| (2) |
TABLE 1.
Important inorganic forms of selenium, with oxidation states and redox potentials, along with other terminal electron acceptors of anaerobic respiration
| Redox couple | Oxidation state | Half-reaction | E0′ (V)a |
|---|---|---|---|
| O2/H2O | 0/−2 | O2 + 4e− + 4H+ → 2H2O | +0.81 |
| NO3−/N2 | +5/0 | 2NO3− + 10e− + 12H+ → N2 + 6H2O | +0.75 |
| MnO2/Mn2+ | +4/+2 | MnO2 + 2e− + 4H+ → Mn2+ + 2H2O | +0.53 |
| SeO42−/SeO32− | +6/+4 | SeO42− + 2e− + 2H+ → SeO32− + H2O | +0.48 |
| NO3−/NH4+ | +5/−3 | NO3− + 8e− + 10H+ → NH4+ + 3H2O | +0.36 |
| SeO32−/Se0 | +4/0 | SeO32− + 4e− + 6H+ → S0 + 3H2O | +0.21 |
| Fe3+/Fe2+ | +3/+2 | Fe(OH)3 + 1e− + 3H+ → Fe2+ + 3H2O | +0.10 |
| SO42−/SO32− | +6/+4 | SO42− + 2e− + 2H+ → SO32− + H2O | −0.516 |
| Se0/HSe− | 0/−2 | Se0 + 2e− + H+ → HSe− | −0.73 |
The above equations show that selenium oxyanion reduction can be a significant mechanism for certain microorganisms to conserve energy in natural environments. The reduction of SeO42− should occur at a slightly lower redox potential than that required for nitrate reduction but at a higher redox potential than that needed for sulfate reduction. Thermodynamic calculations show that SeO42− reduction is energetically favorable for microorganisms. This means that the reduction of SeO42− to SeO32− yields −575 kJ mol−1 of acetate and −343 kJ mol−1 of lactate (15, 37, 38). The stoichiometric equations for the reduction of SeO42− to SeO32−, using acetate and lactate as electron donors, are as follows:
| (3) |
| (4) |
Selenate is reducible and can be used as a terminal electron acceptor by microorganisms to support growth under anoxic or anaerobic conditions. Consequently, microorganisms may play an important role in the biogeochemical selenium cycle. Evidence for dissimilatory reduction of SeO42− was first reported by Oremland et al. (39) for experiments performed using sediment slurries. Dissimilatory reduction of SeO42− was linked to the production of stoichiometric amounts of elemental selenium. SeO42− reduction was inhibited by the presence of other electron acceptors, such as O2, NO3−, CrO42−, or MnO2, but not by SO42− or FeOOH (39). Addition of electron donors (i.e., H2 or acetate) to sediment slurries accelerated the reduction of SeO42− to elemental selenium. An uncharacterized bacterium isolated from the sediments exhibited growth by coupling acetate oxidation to SeO42− reduction and produced elemental selenium and CO2 as respiratory end products (39). Almost at the same time, Macy et al. (37) discovered a new mode of bacterial respiration by using an anaerobic coculture and 14C-labeled acetate. They isolated an anaerobic coculture from agricultural drainage waters in the San Joaquin Valley (CA) that was capable of reducing SeO42− and SeO32− to elemental selenium. The coculture comprised a strictly anaerobic Gram-positive rod which reduced SeO32− to elemental selenium and a Pseudomonas sp. which can respire SeO42− to SeO32−. The cells were then cultured anaerobically in a minimal medium supplemented with acetate plus SeO42−. The coculture oxidized [14C]acetate to 14CO2, with concomitant reduction of SeO42− to SeO32− and, finally, to elemental selenium.
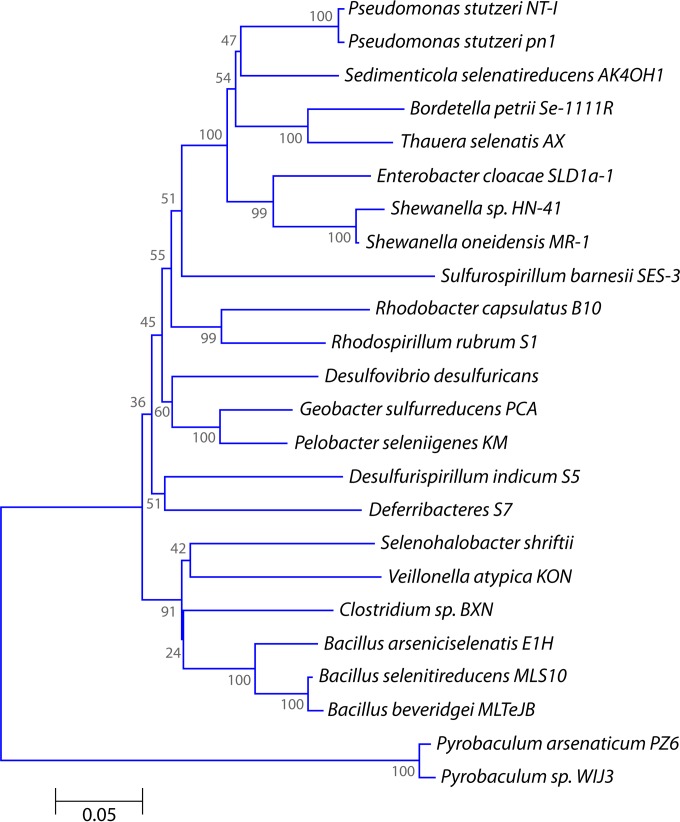
Subsequent studies showed that phylogenetically diverse groups of microorganisms are able to couple dissimilatory SeO42− reduction with anaerobic growth (37, 40–44). Interestingly, SeO42−-reducing bacteria not only are phylogenetically diverse (Fig. 3) but are able to couple growth to reduction of a wide range of electron acceptors (Table 2) (15, 16, 38, 41, 45, 46). Bacteria belonging to the Gammaproteobacteria are able to grow by coupling SeO42− reduction to the oxidation of aliphatic (i.e., acetate, ethanol, and lactate) and aromatic (i.e., benzoate, 3-hydroxybenzoate, and 4-hydroxybenzoate) compounds. These bacteria were isolated from the Arthur Kill and the Kesterson Reservoir (28). Narasingarao and Häggblom enriched and isolated dissimilatory SeO42−-respiring bacteria from geographically and characteristically different sediment samples from Chennai (India) and New Jersey by using SeO42− as the sole terminal electron acceptor (47, 48). Four bacterial strains were isolated that could grow via dissimilatory SeO42− respiration and belonged to metabolically and taxonomically diverse taxa of the Gammaproteobacteria, Deltaproteobacteria, Deferribacteres, and Chrysiogenetes. Sedimenticola selenatireducens was isolated using 4-hydroxybenzyoate and SeO42− as the sole electron donor and acceptor, respectively (48). Although Desulfurispirillum indicum was isolated using pyruvate and SeO42− as the sole electron donor and acceptor, respectively, the strain was able to couple the oxidation of other short-chain organic acids, such as lactate and acetate, to SeO42− reduction (49). The D. indicum strain demonstrated metabolic flexibility to respire SeO42− to SeO32− and then to elemental selenium under anaerobic conditions. It was also capable of dissimilatory reduction of SeO32−, AsO43−, and NO3− during anaerobic growth.
FIG 3.
Phylogenetic tree of selenium oxyanion-respiring microorganisms, based on 16S rRNA gene sequences. The tree was constructed by the neighbor-joining method (143), using MEGA6 software (144). The bootstrap values are given as percentages at the nodes.
TABLE 2.
Selenate- and selenite-respiring bacteria capable of producing selenium nanoparticles by respiration of either selenate or selenite through dissimilatory metal reduction
| Organism | Environment | Taxonomic affiliation | Electron donor(s) | Terminal electron acceptor(s)a | End product of Se respiration, Se nanosphere size (nm) | Reference(s) |
|---|---|---|---|---|---|---|
| Bacillus arseniciselenatis strain E1H | Mono Lake sediment, eastern California | Gram positive; Firmicutes | Lactate | SeO42−, AsO43− | Se0 | 43 |
| Bacillus beveridgei strain MLTeJB | Mono Lake sediment, eastern California | Gram positive; Firmicutes | SeO42−, SeO32−, TeO32−, TeO42−, AsO43−, NO3− | Se0 | 46 | |
| Bacillus selenitireducens strain MLS10 | Mono Lake sediment, eastern California | Gram positive; Firmicutes | Lactate, glucose | SeO32− | Se0, 200–400 | 43 |
| Bordetella petrii | Enriched from sediment of the River Saale, Jena, Germany | Gram negative; Betaproteobacteria | Acetate | SeO42−, NO3− | Se0 | 45 |
| Clostridium sp. BXM | Enriched from paddy soil, China | Gram positive; Firmicutes | Lactate | SeO42−, SeO32−, SO42−, SO32−, S0 | Se0, 500 | 74 |
| Desulfovibrio desulfuricans | Sediments of San Francisco Bay | Gram negative; Deltaproteobacteria | Lactate | SeO42−, SeO32− | Se0, Se(−II) | 60 |
| Desulfurispirillum indicum sp. S5 | Estuarine sediment, Chennai, India | Gram negative; Chrysiogenetes | Pyruvate, lactate, acetate | SeO42−, SeO32−, AsO43−, NO3− | Se0 | 49 |
| Deferribacteres S7 | Estuarine sediment, Chennai, India | Gram negative; Deferribacteres | Acetate | SeO42− | SeO32− | 47 |
| Enterobacter cloacae SLD1a-1 | Freshwater samples from San Luis Drain | Gram negative; Gammaproteobacteria | Glucose | SeO42−, SeO32−, NO3− | Se0 | 85 |
| Pelobacter seleniigenes KM7 | Enrichment culture from Kearny Marsh sediment (New Jersey) | Gram negative; Deltaproteobacteria | Acetate, pyruvate, lactate, citrate | SeO42−, Fe(III), NO3−, AQDS, S0 | Se0 | 47 |
| Pseudomonas stutzeri pn1 | Laboratory contaminant | Gram negative; Gammaproteobacteria | Acetate | SeO42− | SeO32− | 47 |
| Pseudomonas stutzeri NT-I | Drainage water from a selenium refinery plant, Japan | Gram negative; Gammaproteobacteria | Lactate | SeO42− | Se0, 200 | 87 |
| Pyrobaculum sp. WIJ3 | Isolated from hot springs of Pisciarelli Solfatara, Italy | Archaea | H2 | SeO42−, AsO43−, S0 | Se0 | 29 |
| Pyrobaculum arsenaticum PZ6 | Isolated from hot springs of Pisciarelli Solfatara, Italy | Archaea | H2 | SeO42−, AsO43−, S0 | Se0 | 29 |
| Geobacter sulfurreducens PCA | Enrichment from sediments of ditch, Norman, OK | Gram negative; Deltaproteobacteria | Acetate, H2 | SeO32− Fe(III) | Se0 | 54, 81 |
| Rhodobacter capsulatus B10 | Freshwater | Gram negative; Alphaproteobacteria | Photosynthesis | SeO32− | Se0 | 31 |
| Rhodospirillum rubrum | Freshwater | Gram negative; Alphaproteobacteria | photosynthesis | SeO32− | Se0 | 31 |
| Sedimenticola selenatireducens AK4OH1 | Enrichment from sediments of Kesterson Reservoir, CA | Gram negative; Gammaproteobacteria | 4-Hydroxybenzoate, 3-hydroxybenzoate, acetate, pyruvate, lactate | SeO42−, NO3−, NO2− | SeO32−, Se0 | 28, 47 |
| Selenihalanaerobacter shriftii | Dead sea sediments, offshore of Masada, Israel | Gram positive; low G+C | Glucose, glycerol | SeO42−, SeO32−, NO3− | Se0, 200–400 | 44, 78 |
| Shewanella sp. HN-41 | Isolated from the coastal wetlands of Haenam Jeollanam-do, Republic of Korea | Gram negative; Gammaproteobacteria | Lactate | SeO32− | Se0, 150–200 | 145 |
| Shewanella oneidensis MR-1 | Isolated from Oneida Lake, NY | Gram negative; Gammaproteobacteria | Lactate, pyruvate, formate | SeO32−, NO3−, Fe3+ | Se0 | 55, 79 |
| Sulfurospirillum barnesii SES-3 | SeO42−-contaminated freshwater marsh, Nevada | Gram negative; Deltaproteobacteria | Lactate, H2 | SeO42−, SeO32−, NO3−, AsO43−, Fe(III) | Se0, 200–400 | 15, 78 |
| Thauera selenatis AX | Isolated from selenate-contaminated drainage water of San Joaquin Valley | Betaproteobacteria | H2 | SeO42−, SeO32−, NO3− | Se0, 120; Se(−II) | 80 |
| Veillonella atypica | Isolated from subgingival dental plaque | Firmicutes | H2 | SeO32− | Se0, Se(−II) | 53 |
AQDS, anthraquinone-2,6-disulfonate.
Selenite-Respiring Bacteria
Reduction of SeO32− to Se0 has been observed in a wide variety of microorganisms. SeO32− reduction by microorganisms can be categorized broadly into detoxification and anaerobic respiration. Detoxification of SeO32− to Se0 in microorganisms is achieved by various mechanisms, such as Painter-type reactions (31, 50–52), the thioredoxin reductase system, and sulfide- and siderophore-mediated reduction. The details of different mechanisms proposed for microbial detoxification by reduction of SeO32− to Se0 nanospheres are discussed below. Although reduction of SeO42− to elemental selenium was shown to be an environmentally significant process, only a few SeO32−-respiring bacteria have been isolated (9). Certain SeO42−-reducing bacteria have been shown to perform dissimilatory SeO32− reduction as well (Table 2). The reduction of SeO32− to Se0 is energetically favorable and yields −529.5 kJ mol−1 of acetate and −164 kJ mol−1 of lactate (42). The stoichiometric equations for the reduction of SeO32− to Se0 coupled to the incomplete oxidation of the electron donor lactate to acetate (equation 5) or pyruvate (equation 6) are as follows:
| (5) |
| (6) |
High-rate SeO32− reduction was achieved when Veillonella atypica cells were supplemented with H2 as the electron donor (53). Interestingly, little reduction was observed using lactate as the electron donor, and no reduction at all was achieved using acetate or formate as the electron donor (53). The colorless SeO32− was reduced sequentially to red insoluble selenium and, finally, to colorless aqueous selenide. Pearce et al. (54) compared SeO32− reduction capabilities of Geobacter sulfurreducens, Shewanella oneidensis, and V. atypica. All three bacterial cultures transformed SeO32− to Se0 when grown in a medium containing Se oxyanions and an electron donor. Among the three strains, it was found that V. atypica was the most efficient SeO32− reducer. It was proposed that SeO32− reduction involves the formation of a crystalline intermediate in G. sulfurreducens and S. oneidensis MR-1 but not in V. atypica cells. The Se oxyanion reduction rate and the nature of the bioreduced Se form are influenced by the type of microorganism and the bioreduction mechanism. In some microorganisms, respiratory reductases of anaerobic respiration, such as nitrite reductase and sulfite reductase, are active in SeO32− reduction. Until now, detailed investigation of SeO32− reduction via respiratory electron transport pathways was limited to a single study using S. oneidensis MR-1 (55). More details on the roles of respiratory reductases and the electron transport pathway in S. oneidensis MR-1 for SeO32− reduction are described below.
Se0-Respiring Bacteria
In environmental settings, selenium is often observed as metal selenide minerals in rocks and sediments (1), but not as elemental selenium. However, the mechanisms of selenide incorporation during diagenesis of sedimentary rocks are not clearly understood (56). Formation of selenide from Se0 reduction is thermodynamically unfavorable through abiotic mechanisms such as disproportionation (56), as shown by the following:
| (7) |
Therefore, it is likely that biotic mechanisms, such as assimilatory and dissimilatory selenium reduction, contribute to the occurrence of selenide in environmental settings.
In assimilatory metabolism, selenium is incorporated into amino acids and then into selenoproteins, mainly in the form of selenide. Release of selenide by the decomposition of plants and animals may be a source of environmental selenide. The methylated forms of selenium (dimethyl selenide and dimethyl diselenide) also contain selenide. Biomethylation is a widely studied metabolic process which deals with the formation of both volatile and nonvolatile compounds of metal and metalloids (31, 51). Selenium methylation by the members of the Gammaproteobacteria was observed in freshwater sediments (52). Microbial methylation of selenium forms volatile compounds, such as dimethyl selenide, dimethyl diselenide, dimethyl selenyl sulfide, hydrogen selenide, and methaneselenol (53, 57). Microbial selenium methylation is significant in environmental settings, although it is a slow process (54). For additional details, the reader is referred to excellent reviews on microbial methylation of metalloids (58, 59). Demethylation is carried out by methylotrophic methanogens in anoxic sediments and may proceed via pathways that have been established for growth on the structural analogue dimethyl sulfide (60, 61). In addition, insoluble elemental selenium is reduced to soluble hydrogen selenide in the cell cytoplasm by thiol-mediated reduction (51).
There are limited studies on the microbial reduction of selenium oxyanions beyond elemental selenium. So far, there are only a few bacterial species that are known to be able to extend the selenium reduction pathway beyond insoluble elemental selenium. At least some of the SeO42−- or SeO32−-respiring bacteria may have the capability to reduce elemental selenium as well (Table 2). Formation of selenide was observed when cells of Salmonella enterica serovar Heidelberg (62) or cell extracts of Micrococcus lactilyticus (63), Clostridium pasteurianum, and Desulfovibrio desulfuricans were exposed to SeO32− (64). Zehr and Oremland (65) showed that the sulfate-reducing bacterium D. desulfuricans and anoxic estuarine sediments reduce trace amounts of 75SeO32− to 75Se(−II) in the presence of sulfate.
Herbel et al. (56) supplied elemental selenium as a terminal electron acceptor to SeRB and estuarine sediments. Bacillus selenitireducens, a SeO32−-respiring bacterium, produced significant amounts of selenide from Se0 or SeO32−. Microbial reduction of Se0 to Se(−II) occurred in estuarine sediments but not in formalin-killed control sediments. The aqueous selenide was then precipitated as FeSe in the medium (56). The free energies for dissimilatory reduction of Se0 to HSe− with the incomplete oxidation of the electron donor lactate to acetate are given as follows:
| (8) |
| (9) |
Surprisingly, the reduction of Se0 to Se(−II) was not observed in the case of the SeO42−-respiring bacteria, i.e., Sulfurospirillum barnesii, Bacillus arseniciselenatis, and Selenihalanaerobacter shriftii. Transformation of SeO32− to HSe− by the SeO32−-reducing bacterium V. atypica was exploited for the production of metal chalcogenides (i.e., CdSe and ZnSe) (53, 66). Formation of HSe− was observed only after the complete reduction of SeO32− by V. atypica or B. selenitireducens. The observed biphasic SeO32− reduction, with SeO32− reduction to Se0 followed by Se0 reduction to HSe− (56), suggests that the reduction of elemental selenium is energetically favorable to microorganisms only if other, more favorable electron acceptors, including SeO32−, are not available for anaerobic respiration (53, 56). However, it remains puzzling why Se0 reduction to selenide was not observed in SeO42−-respiring bacteria.
Fractionation of Selenium Isotopes by Selenium-Respiring Bacteria
Studies using Se isotopes demonstrated that isotope fractionation occurs as a result of abiotic and microbial reduction processes. For the abiotic processes, Se isotope fractionation occurs during chemical reduction of selenate to selenite but not during selenite adsorption onto ferric hydroxide and Se0 oxidation (67). Abiotic reduction of Se(VI) to Se(IV) induced a fractionation of −5.5‰.
Se isotope fractionation was found to be significant during bacterial respiration of Se oxyanions, and the extent of fractionation depends on the bacterial species and the metabolic state of microorganisms. Herbel et al. (68) discovered significant isotope fractionation during dissimilatory reduction of selenate or selenite to elemental selenium by SeRB, i.e., B. selenitireducens, B. arseniciselenatis, and Sulfurospirillum barnesii. The reduction reaction products were enriched with the lighter isotopes relative to reactants: 80Se/76Se fractionation levels of −8‰, −6‰, and −8.4‰ were observed during reduction of Se(IV) to Se0 by B. selenitireducens, B. arseniciselenatis, and S. barnesii, respectively. B. arseniciselenatis and S. barnesii induced isotopic fractionation levels of −5‰ and −4‰, respectively, during the reduction of Se(VI) to Se(IV). Se stable isotope fractionation due to reduction of Se oxyanions by natural microbial communities was determined in microcosm experiments using sediment slurries without the addition of an electron donor. Isotope fractionation levels of −3.9‰ to −4.7‰ were observed during the reduction of Se(VI) to Se(IV), and levels of −8.3‰ to −8.6‰ were observed during the subsequent reduction of Se(IV) to Se0 (69).
Ecology of Selenium-Reducing Bacteria
Selenium-respiring bacteria have been detected in pristine and contaminated water and soil samples collected from geographically diverse environments. Dissimilatory selenate reduction to elemental selenium was measured in situ in sediments from irrigated agricultural drainage regions of western Nevada at ambient Se oxyanion levels (63). Selenate reduction was observed without any lag period. The selenate removal rates ranged from 14 to 155 μmol m−2 day−1 for the sediments. Subsequently, radioisotopic methods were used to measure bacterial dissimilatory selenate reduction in situ (70, 71). Sediments of the agricultural drainage system of western Nevada were incubated with [78Se]selenate in the laboratory. Most of the selenate reduction was confined to the surface regions of the sediments in close proximity to denitrification regions and spatially separated from the sulfate reduction regions in the deeper sediment profile.
There is still a limited understanding of the abundance and true ecological role of SeRB in situ in natural environments. Recently, Williams et al. (10), by employing molecular microbial ecology tools, provided evidence for the reduction of SeO42− to elemental selenium coupled to acetate oxidation in a field-scale experiment carried out in a uranium-contaminated aquifer near Rifle, CO. Bioreduced elemental selenium, with a diameter of 50 to 60 nm, accumulated within the biofilms formed on the tubing used to circulate acetate-amended groundwater through the soil. Phylogenic analysis of the biofilms showed a dominance of Dechloromonas sp. and Thauera sp. strains in the community (10).
Dissimilatory selenate reduction is constitutive and widespread in natural environments, where selenium oxyanions are not normally present in significant amounts. Rapid appearance of SeRB in enrichments suggests that they are ubiquitous and metabolically active in nature. Dissimilatory selenate reduction potentials were determined for sediments with salinities ranging from that of fresh water (salinity = 1 g liter−1) to hypersaline (salinity = 320 g liter−1) and with pH values ranging from 7.1 to 9.8 (63). Significant bacterial transformation of 75SeO42− to 75Se0 occurred in sediments with salinity values ranging from 1 to 250 g liter−1.
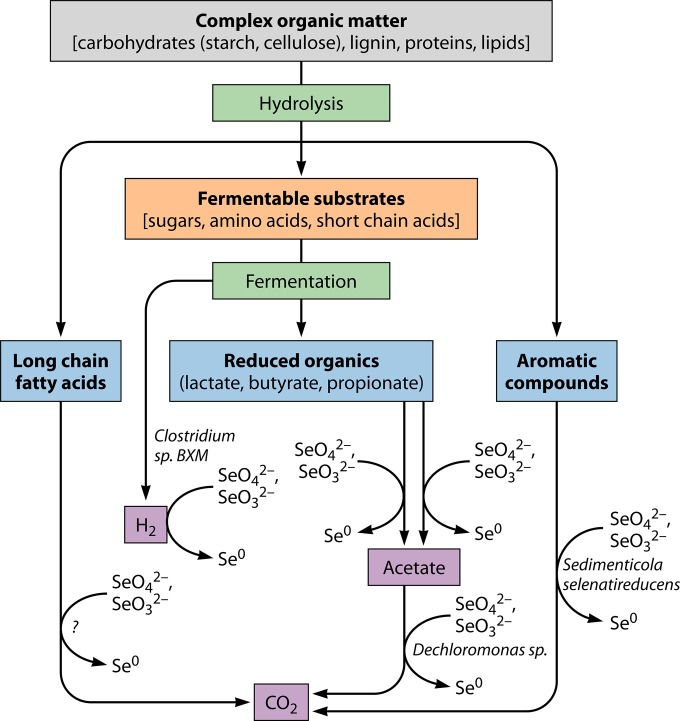
Bacteria that can couple the oxidation of inorganic (i.e., hydrogen) and organic (i.e., aliphatic and aromatic compounds) compounds to the reduction of selenium oxyanions are obtained from different environments, which indicates their ecological role in the carbon cycle (Fig. 4). SeRB metabolize fermentation products and other organics, such as aromatic compounds, by oxidizing them to carbon dioxide, with SeO42− and/or SeO32− as the electron acceptor. Dechloromonas sp. and S. selenatireducens can both couple the oxidation of aliphatic and aromatic compounds to dissimilatory reduction of selenium oxyanions (28). Effective oxidation of organic matter in anaerobic natural environments requires the contribution of microbial communities, such as fermenting and metal-reducing microorganisms (72, 73). Dissimilatory reduction of selenium oxyanions by a fermentative bacterium, i.e., Clostridium sp. BXM, has also been demonstrated (74). However, there is no information available on the ability of SeRB to oxidize long-chain fatty acids.
FIG 4.
Carbon and electron flow in the oxidation of complex organic matter coupled to reduction of selenium oxyanions in anoxic water and sediment environments. The complex organic matter is hydrolyzed by hydrolytic bacteria. The fermentable substrates are oxidized by fermentative bacteria into hydrogen and a range of fermentation products (i.e., acetate, lactate, propionate, and butyrate). These fermentation products and others are consumed by coupling to reduction of SeO42− and SeO32− by dissimilatory reducing bacteria. Bacteria capable of coupling oxidation of aromatic compounds (i.e., benzoate, 4-hydroxybenzoate, and 3-hydroxybenzoate) to SeO42− reduction have been isolated from estuarine sediments by enrichment (28, 48). The question mark shows an unidentified process and unidentified microorganisms.
In the absence of other thermodynamically favorable electron acceptors, the selenium cycle can contribute to organic matter oxidation. Like other dissimilatory reducing bacteria (i.e., iron- and sulfate-reducing bacteria), SeRB can use various other electron acceptors for cell maintenance and growth (Table 2). For example, electron acceptors such as nitrate, nitrite, arsenate, and chromate are reduced by some known SeRB. SeRB can even ferment substrates in the absence of inorganic electron acceptors (74). In addition, many SeRB can use organics (i.e., fumarate) as terminal electron acceptors for growth.
MECHANISMS OF SELENIUM OXYANION REDUCTION
Reduction of Se oxyanions is widespread in natural environments. It is mediated by diverse groups of microorganisms under both aerobic and anaerobic conditions. Several sulfate-reducing bacteria (65, 75) and Wolinella succinogenes (76) were reported to reduce significant amounts of SeO42− under anaerobic conditions without coupling bioreduction to growth. Also, some phototrophic bacteria can reduce SeO42− in the stationary phase under anaerobic growth conditions (62, 64). However, anaerobic respiration appears to be the most environmentally significant process for the selenium cycle (9, 15). Hence, the present review focuses on the biochemistry of anaerobic respiration of selenium oxyanions, which is relatively well investigated.
Selenate Reduction
A number of microorganisms have evolved the biochemical machinery to use SeO42− as a terminal electron acceptor in anaerobic respiration. Under anaerobic conditions, SeO42− is reduced sequentially to selenite and then to insoluble elemental selenium. Respiration of SeO42− is often associated with the formation of brick red-colored elemental selenium (Fig. 5). The overall process of microbial SeO42− reduction leading to the formation of Se0 (77) is given in equations 10 and 11:
| (10) |
| (11) |
FIG 5.
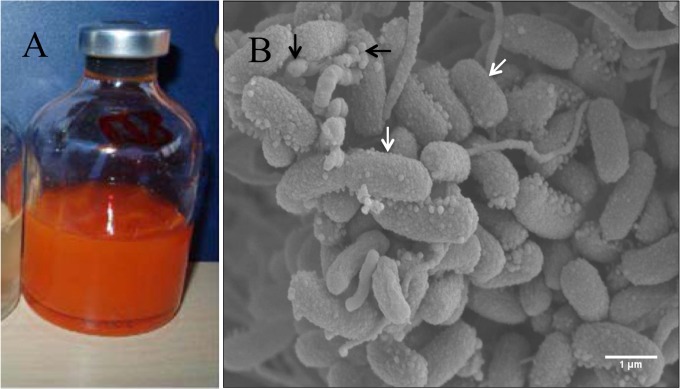
Reduction of SeO42− to elemental selenium by immobilized cells of Sulfurospirillum barnesii cells (120) and anaerobic granules (121). (A) Orange-red elemental selenium produced by S. barnesii cells immobilized on polyacrylamide beads during 7 days of incubation at 30°C. (The image was reprinted from reference 120 with permission.) (B) Environmental scanning electron microscope image of anaerobic granules collected from a mesophilic (30°C) UASB reactor performing SeO42− bioreduction, using lactate as the electron donor. White arrows show bacterial cells. Black arrows show elemental selenium nanospheres on the surfaces of microorganisms. Bar = 1 μm. (Modified from reference 121 with permission of the publisher [copyright 2008 American Society of Agronomy, Crop Science Society of America, and Soil Science Society of America].)
Imaging of SeO42−-respiring cells by transmission electron microscopy localized selenium nanospheres both inside the cell and in the extracellular medium (78, 79). Since selenium is an essential trace element used in the synthesis of selenoproteins, selenium must enter cells for its assimilation. However, the reduction inside the cytoplasm leads to the formation of selenium precipitates, which may pose a burden on cells for export outside the cell. In general, dissimilatory reduction of selenium oxyanions through anaerobic respiration can be divided into a two-step process involving formation of elemental selenium and fabrication of selenium nanospheres. The enzymes and electron transport pathways involved in SeO42− reduction have been investigated relatively well compared to the assembly and secretion of selenium nanospheres. The enzymes involved in SeO42− respiration were investigated primarily in two Gram-negative bacteria (Thauera selenatis and Enterobacter cloacae SLD1a-1) and one Gram-positive bacterium (Bacillus selenatarsenatis SF-1).
T. selenatis.
T. selenatis is a Gram-negative betaproteobacterium that uses SeO42− as an electron acceptor and was isolated from the seleniferous waters of the Joaquin Valley in California (80). Accumulation of red elemental selenium nanospheres has been observed in the cytoplasm and in extracellular spent medium (81). Biochemical analysis demonstrated that the selenate reductase (Ser) is located in the periplasmic space. The selenate-to-selenite reduction reaction is catalyzed by a trimeric molybdoenzyme, SerABC selenate reductase, and occurs in the periplasmic compartment of T. selenatis (Fig. 6). The enzyme consists of a catalytic unit (SerA; 96 kDa), an iron-sulfur protein (SerB; 40 kDa), a heme b protein (SerC; 23 kDa), and a molybdenum cofactor (82). SerA and SerB were found to contain a cysteine-rich motif. Electron paramagnetic resonance (EPR) analysis showed the presence of two types of iron-sulfur (i.e., [3Fe-4S] and [4Fe-4S]) clusters (81). Coordination of one [3Fe-4S] and three [4Fe-4S] clusters was predicted by the modeling of data obtained by EPR analysis. Electron transfer to periplasmic cytochrome c4 is proposed to be via quinol cytochrome c oxidoreductase (QCR) (83). Inhibitors of selenate reduction were used to decipher the electron transport pathway. The growth of T. selenatis was only partially inhibited in the presence of myxothiazol, a QCR inhibitor, indicating the existence of an alternate pathway for the electron transfer to SerABC. Complete inhibition of SeO42− reduction was achieved in the presence of both myxothiazol and HQNO (2-n-heptyl-4-hydroxyquinoline N-oxide), suggesting the involvement of both a QCR and a quinol dehydrogenase (QDH) in SeO42− reduction (83). SerD is a cytoplasmic protein and might act as a chaperone assembly protein involved in the insertion of the molybdenum cofactor into SerA (84).
FIG 6.
Model showing the proposed electron transport pathway involved in SeO42− reduction in Thauera selenatis (77). The membrane-bound quinol cytochrome c oxidoreductase (QCR) supplies electrons for SeO42− reduction. The electrons are shunted to periplasmic cytochrome c4 (cytc4) and then to SerABC, located in the periplasmic compartment (77, 81, 149). The reduction of SeO42− to SeO32− occurs in the periplasmic compartment. Subsequently, it was proposed that SeO32− crosses the inner cytoplasmic membrane and enters the cytoplasm through an unknown mechanism. However, SeO32− is thought to enter the cell through the sulfate permease system in E. coli (89). Reduction of SeO32− to elemental selenium occurs in the cytoplasm via thiol-mediated detoxification. Elemental selenium binds to the SefA protein to form a selenium nanosphere, which is subsequently exported to the extracellular environment. The process by which SefA-Se is exported from the cytoplasm to the extracellular environment remains unknown. The identification of a gene (sefB) encoding a putative S-adenosylmethionine (SAM)-dependent methyltransferase might also provide a mechanism for SeO32− detoxification via volatilization to methylated selenides (R-Se-R). OM, outer membrane; CM, cell membrane. The schematic drawing was created by using information from references 77 and 89. Dotted lines and question marks show unidentified processes.
This model explains the reduction of SeO42− to SeO32− in the periplasmic space but does not provide evidence on further reduction of SeO32− to elemental selenium. SeO32− formed in the periplasmic place is presumed to be transported to the cytoplasm via a sulfate transporter and is reduced to elemental selenium in the cytoplasm (77). Reduction of SeO32− to elemental selenium was hypothesized to occur by thiol-mediated reduction in the cytoplasm. Recently, a new 95-kDa protein, SefA (selenium factor A), was isolated from the elemental Se secreted from T. selenatis cells into the extracellular medium (77). Evidence shows that the SefA protein is involved in stabilization of selenium nanospheres by preventing their aggregation and may possibly aid in the secretion process. For additional details on biomineralization of selenium and the assembly of Se0 nanospheres in T. selenatis, the reader is referred to the recent review by Butler et al. (81).
E. cloacae SLD1a-1.
E. cloacae SLD1a-1 is a SeO42−-respiring bacterium isolated from the selenium-rich waters of the San Luis Drain in California (85). In SLD1a-1, intracellular accumulation of selenium nanospheres was not observed during SeO42− respiration. Biochemical studies suggested that SeO42− reductase of E. cloacae SLD1a-1 is a membrane-bound trimeric complex with a catalytic subunit of 100 kDa (85, 86). The SeO42− reductase activity of E. cloacae SLD1a-1 was inhibited by tungstate but activated by molybdate, suggesting that the enzyme is a molybdoenzyme. Molybdenum has been detected in the purified enzyme of E. cloacae SLD1a-1. The SeO42− reductase is positioned in the inner cell membrane such that the active site faces the periplasmic compartment (Fig. 7A). The reduction of SeO42− to elemental selenium occurs in the periplasmic compartment, and the selenium nanospheres are expelled into the extracellular environment (85).
FIG 7.
Models showing the proposed electron transport pathways involved in SeO42− reduction in Enterobacter cloacae SLD1a-1 (A) and Bacillus selenatarsenatis SF-1 (B). In E. cloacae SLD1a-1, the active site of the membrane-bound SeO42− reductase faces the periplasmic compartment (85, 86). The reduction of SeO42− was proposed to occur in the periplasmic space, and the resultant elemental selenium is expelled away from the periplasmic space to the extracellular environment. In B. selenatarsenatis SF-1, a membrane-bound SrdBAC catalyzes SeO42− reduction to elemental selenium. The electrons are shunted from the quinol pool to SeO42− via SrdC, SrdB (an iron-sulfur protein), and SrdA. Selenate is reduced by the molybdenum-containing subunit SrdA (88). The produced selenium nanoparticles are released into the extracellular medium. OM, outer membrane; CM, cell membrane. Dotted lines and question marks show unidentified processes.
B. selenatarsenatis SF-1.
B. selenatarsenatis SF-1 is a Gram-positive bacterium that was isolated from the effluent sediments of a glass manufacturing plant by using lactate and SeO42− as the electron donor and acceptor, respectively (40, 87). The strain shows a stoichiometric relationship between cell growth, lactate consumption, and SeO42− reduction. The SeO42− reductase of B. selenatarsenatis SF-1 is a membrane-bound, trimeric molybdoenzyme (Fig. 7B). The electrons from the quinol pool (QH2) are channeled to the catalytic subunit (SrdA) via SrdC and an iron-sulfur protein, SrdB (Fig. 7B). SeO42− receives the electrons from SrdA via the molybdenum cofactor (88). The active site faces outside the cell, and the end products of SeO42− respiration, elemental selenium nanospheres, are released into the extracellular medium. In both E. cloacae SLD1a-1 and B. selenatarsenatis SF-1, reduction of SeO42− occurs primarily either in the periplasmic space or outside the cell, and the biogenic selenium nanospheres are released into the extracellular medium.
Selenite Reduction
Reduction of SeO32− to elemental selenium was widely recognized to be mediated by thiols in the cytoplasm as part of a microbial detoxification strategy (51, 89). SeO32− reacts with GSH and forms selenodiglutathione (GS-Se-SG), which is further reduced to selenopersulfide of glutathione (GS-Se−) by NADPH-glutathione reductase. The selenopersulfide of glutathione is an unstable intermediate and undergoes a hydrolysis reaction to form Se0 and reduced GSH.
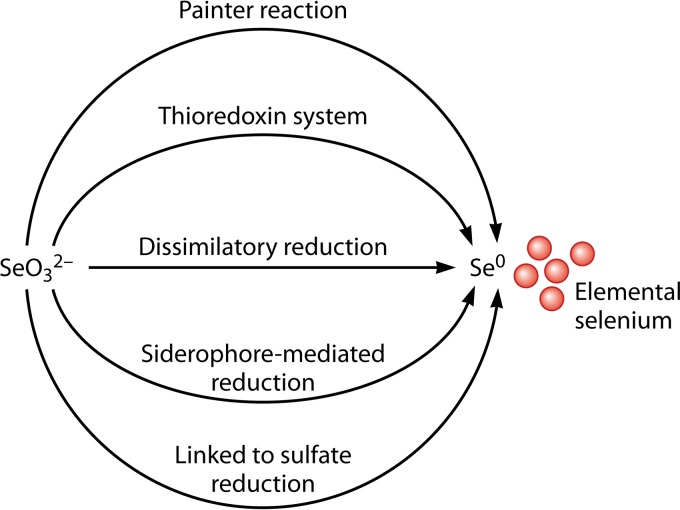
Different mechanisms have been proposed for the reduction of SeO32− to Se0 in microorganisms, including (i) Painter-type reactions, (ii) the thioredoxin reductase system, (iii) siderophore-mediated reduction, (iv) sulfide-mediated reduction, and (v) dissimilatory reduction (90) (Fig. 8). Painter observed a high reactivity between SeO32− and thiols and demonstrated the formation of selenotrisulfide (RS-Se-SR) (50). Formation of RS-Se-SR was confirmed in vivo in Escherichia coli cells exposed to SeO32− by using 77Se nuclear magnetic resonance (77Se-NMR) (82), giving the following equation:
| (12) |
FIG 8.
Selenite reduction mechanisms in microorganisms. The microbial reduction of selenite to elemental selenium may occur via (i) Painter-type reactions with thiol (SH) groups of glutathione and proteins (50, 51, 90–92), (ii) thioredoxin and the thioredoxin reductase system (93, 94), (iii) abiotic reduction coupled by H2S to sulfate reduction (95, 96), (iv) siderophore-mediated reduction (95, 96), and (v) dissimilatory reduction (9, 15, 16).
Subsequently, Ganther proposed an analogous Painter-type reaction between the reduced GSH and SeO32− and showed the formation of selenotrisulfide of glutathione (GS-Se-SG) (50, 91). The selenotrisulfide was later renamed selenodiglutathione, which is converted to the selenopersulfide anion (GS-Se−) by glutathione reductase. Kessi and Hanselmaan (51) modified the reactions of Painter and Ganther because of production of superoxide anions during abiotic reduction of selenite by glutathione in E. coli cells grown in the presence of SeO32− (50, 92). In aerobic bacteria and some anaerobic bacteria, superoxide anions are removed by superoxide dismutase and catalase enzymes to protect cells from oxidative stress. GS-Se− is an unstable compound and undergoes a hydrolysis reaction to form GSH and Se0, as follows:
| (13) |
| (14) |
| (15) |
The levels of reduced thioredoxin [Trx(SH)2] and thioredoxin reductase (93) were increased in E. coli cells grown in medium containing SeO32−. The reduced thioredoxin reacts with selenodiglutathione and forms oxidized thioredoxin (Trx-S2), reduced glutathione, and selenopersulfide anion. Elemental selenium is released from the reactive selenopersulfide anion as shown in equation 15. Thus, reduced thioredoxin and thioredoxin reductase were hypothesized to be involved in the reduction of selenite and selenodiglutathione, respectively (94), as follows:
| (16) |
| (17) |
In addition, SeO32− can react with the reactive biogenic sulfide abiotically and yields both elemental selenium and sulfur (95, 96):
| (18) |
An iron siderophore, pyridine-2,6-bis(thiocarboxylic acid) (PDTC; [C7H3O2S2]2−), produced by Pseudomonas stutzeri KC, was proposed for the detoxification of selenite through reduction and formation of insoluble Se0 precipitates (97). A hydrolysis reaction was proposed for the release of dipicolinic acid ([C7H3O4]2−) and H2S from PDTC (equation 19). The hydrolyzed product of PDTC, H2S, acts as the reducing agent and forms Se0 (90):
| (19) |
Apart from glutathione-mediated reduction, respiratory reductases (i.e., nitrite reductase, sulfite reductase, and hydrogenase 1) can support SeO32− reduction in some microorganisms (e.g., T. selenatis AX, Rhizobium sullae strain HCNT1, and Clostridium pasteurianum) (98–100).
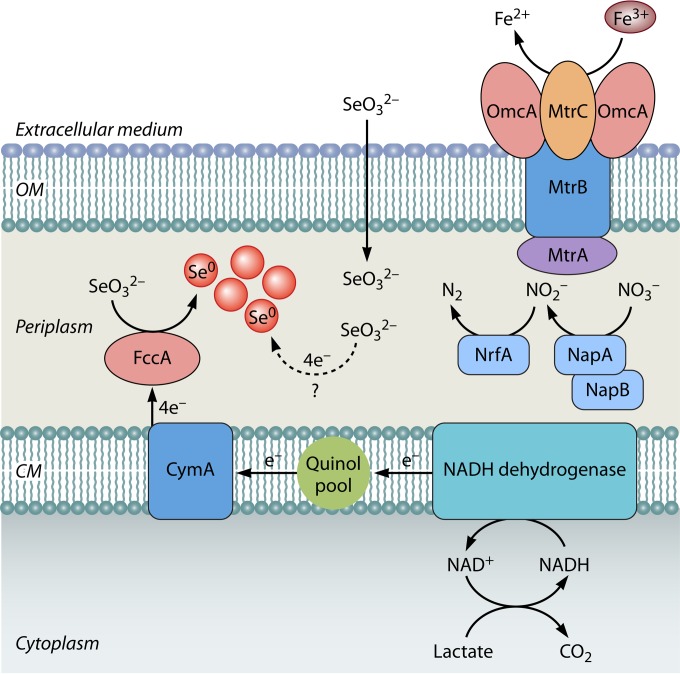
Selenite reduction-deficient mutants of S. oneidensis MR-1 were incapable of anaerobic growth with Fe(III), NO3−, NO2−, SO32−, Mn(IV), or fumarate as the sole terminal electron acceptor, which suggested a link between selenite reduction and anaerobic respiration (101). Recently, Li et al. (55) experimentally demonstrated the existence of an anaerobic respiratory pathway for SeO32− reduction in S. oneidensis MR-1 (Fig. 9). Mutant strains defective in periplasmic terminal reductases, i.e., nitrate reductase (NapA), nitrite reductase (NrfA), and fumarate reductase (FccA), and periplasmic mediators of anaerobic respiration, i.e., MtrA and DmsE, were tested for SeO32− reduction ability (55). The ΔnapA and ΔnrfA mutant strains reduced selenite at the same rate as wild-type S. oneidensis MR-1. Selenite reduction was decreased 60% in the ΔfccA mutant compared to the wild-type strain. Severe inhibition of SeO32− reduction was observed in the ΔcymA mutant; CymA channels electrons from the quinol pool to several respiratory reductases in S. oneidensis MR-1 during anaerobic growth. But the deletion of another electron-shunting molecule, SirCD, did not decrease SeO32− reduction. These results suggested a possible involvement of FccA and CymA in catalyzing SeO32− reduction and shunting electrons from the quinol pool to FccA, respectively. It is still unclear if SeO32− reduction in S. oneidensis MR-1 is linked to growth or is just a fortuitous process that occurs without supporting growth. It is possible that S. oneidensis MR-1 grows by anaerobic respiration of other electron acceptors, i.e., Fe(III), nitrate, or nitrite, and executes SeO32− reduction for detoxification, wherein FccA removes the selenite that has entered the periplasm to avoid its entry into the cytoplasm and toxicity.
FIG 9.
Schematic of proposed electron transport pathway of Shewanella oneidensis MR-1 for anaerobic reduction of SeO32−. The oxidation of lactate releases electrons in the form of NADH, which are channeled to CymA through NADH dehydrogenase and the quinol pool. Electron flow from CymA to SeO32− enables formation of elemental selenium in the periplasmic compartment. OM, outer membrane; CM, cell membrane. Dotted lines and question marks show as yet unidentified processes. (Based on data from reference 55.)
LOCATION OF Se0 SYNTHESIS AND EXPULSION
The formation of an amorphous red elemental nanoprecipitate has been found as the stable end product of Se oxyanion reduction by microorganisms. Following microbial reduction, the elemental selenium grows in the form of selenium nanospheres. The size, shape, and surface properties are important in governing the fate of selenium nanospheres in the environment. Microbiologically produced Se0 selenospheres exhibit colloidal properties, which makes it difficult to achieve complete selenium removal, the ultimate objective of bioremediation or wastewater treatment. The colloidal Se0 can still be mobile and be discharged with the treated waters and has potential for mobility in aqueous environments. Removal of colloidal Se0 from bioreactor effluents is necessary to meet discharge limits and to reduce environmental pollution (102). The colloidal properties of Se0 nanospheres are majorly determined by the organic layer associated with them. The nature and origin of components of the organic coating of Se0 nanoparticles might be related to the location of their formation.
There are still unresolved questions on the microbial genesis of intracellular and/or extracellular Se0 nanospheres, particularly regarding the secretion of intracellularly synthesized Se0 nanospheres. Divergent hypotheses have been formulated to explain the emergence of selenium nanospheres both inside the cell and in the extracellular medium. Selenium-containing protrusions on the cell surface and selenium particles in the extracellular medium were observed in E. cloacae SLD1a-1 exposed to SeO42− or SeO32− (85, 86). It was proposed that the reduction reaction occurs through a membrane-associated reductase. Se nanospheres formed by the anaerobic respiration of SeO42− or SeO32− were released into the medium surrounding E. cloacae SLD1a-1 cells via a rapid expulsion process.
Large amounts of selenium-containing particles were observed in the extracellular medium of the photosynthetic bacterium Rhodospirillum rubrum after SeO32− reduction (31). It is proposed that this bacterium is efficient at transporting these selenium particles out of the cells via a vesicular secretion system. In contrast, release of elemental selenium particles into the medium surrounding D. desulfuricans was proposed to be due to cell lysis (76). However, there is not enough experimental evidence in support of either vesicular secretion or cell lysis for the release of intracellularly formed Se0 nanospheres, and it is desirable to further elucidate the mechanisms behind the location of reduction sites and the transportation mechanisms.
It is emerging that during dissimilatory reduction, extracellular accumulation of Se0 nanospheres is much more than the intracellular accumulation. The difference was much more pronounced when SeO32− reduction was performed using nitrate-respiring cells (78) or E. coli amended with exogenous redox mediators (103). Such a larger accumulation of Se0 nanospheres outside the cell cannot be explained by intracellular synthesis followed by secretion. The large size of Se0 particles also suggests that either cell lysis (76) or disassembly of particles to the size of a macromolecular complex (31) prior to secretion is the only possible way out for Se0 particles formed inside the cytoplasm. Both processes are, however, detrimental to bacteria and might not be part of a microbial respiration process.
Biochemical studies of SeRB show that SeO42− reductases are mainly located in the periplasmic space, which essentially supports the idea that the majority of the SeO42− reduction occurs either in the periplasm or outside the cell. Dissimilatory reduction of SeO32− is also mediated by a periplasmic reductase system (55), which indicates that the majority of the SeO42− and SeO32− reduction to Se0 occurs either in the periplasm or outside the cell envelope. Some of the SeO32− bypasses dissimilatory reduction in the periplasmic space and enters the cytoplasm, where it is reduced to intracellularly accumulated Se0 particles by the thiol-mediated detoxification mechanism. This can be an important issue in SeRB which respire SeO42− but not SeO32−, wherein the bulk of Se0 particle formation occurs in the cytoplasm. In this case, the export of Se0 particles out of the cells is needed. For example, T. selenatis, which does not use SeO32− as a respiratory substrate, appears to have developed a biochemical machinery for export of intracellularly synthesized Se0 nanospheres (77).
It seems possible that the internal and external Se0 nanospheres are formed by different and independent mechanisms. The extracellular accumulation of Se0 nanospheres appears to be linked directly to an electron transport pathway and anaerobic respiration (78). Intracellular accumulation might be the result of a detoxification of SeO32− that has escaped from the respiratory electron transport pathway in true SeRB and entered the cytoplasm via sulfate, nitrite, or an independent, as yet unidentified transporter.
BIOTECHNOLOGICAL APPLICATIONS OF SELENIUM-REDUCING BACTERIA
Microbial Selenium Reduction for Wastewater Treatment
Selenium is widely used in many industrial processes and products (i.e., electronics, glass manufacturing, pigments, stainless steel, metallurgical additives, photoelectric cells, and pesticides), which leads to the generation of SeO42−- and/or SeO32−-containing wastewaters (5, 104, 105). The combustion of selenium-bearing coals in power plants results in the release of SeO42− and/or SeO32− into the flue gas or fly ash (5, 6). In natural environments, oxidation of selenium-containing pyrite results in the occurrence of elevated amounts of SeO42− along with sulfate in acid mine drainage and groundwater (96). Selenium contamination of wetlands of the San Joaquin Valley, CA, is due to the release of agricultural drainage waters containing SeO42− and/or SeO32− (106). The solid wastes (i.e., mine tailings) and liquid effluents from coal and phosphate mining activities are highly enriched with selenium compared to the Earth's crust and surface waters.
Chemical coprecipitation with iron salts is often used to remove SeO42−/SeO32− from industrial wastewaters. However, this method generates sludge with selenium in a nonrecyclable form, which requires additional handling (107). Alternative, environmentally sustainable technologies are needed to remove selenium from these industrial wastewaters, preferably in a recoverable form. Dissimilatory reduction by microorganisms can be used for the removal of water-soluble forms of selenium (i.e., SeO42− and SeO32−) by converting them into insoluble elemental selenium. Recovery of selenium is seriously considered in industrial wastewater treatment to partially offset the costs incurred by treatment. However, several studies have demonstrated that the bacterially recovered selenium is not in pure form and often contains different selenium species, heavy metals, and organics (5). Selenium recovery is still challenging, because bacterially produced Se0 exhibits colloidal properties and requires development of novel methods for separation of colloidal Se0 from the treated wastewater (102). Nevertheless, selenium-respiring bacteria can be applied beneficially not only for removing but also for recovering selenium for reuse purposes.
Bioreactors for Selenium Oxyanion Removal
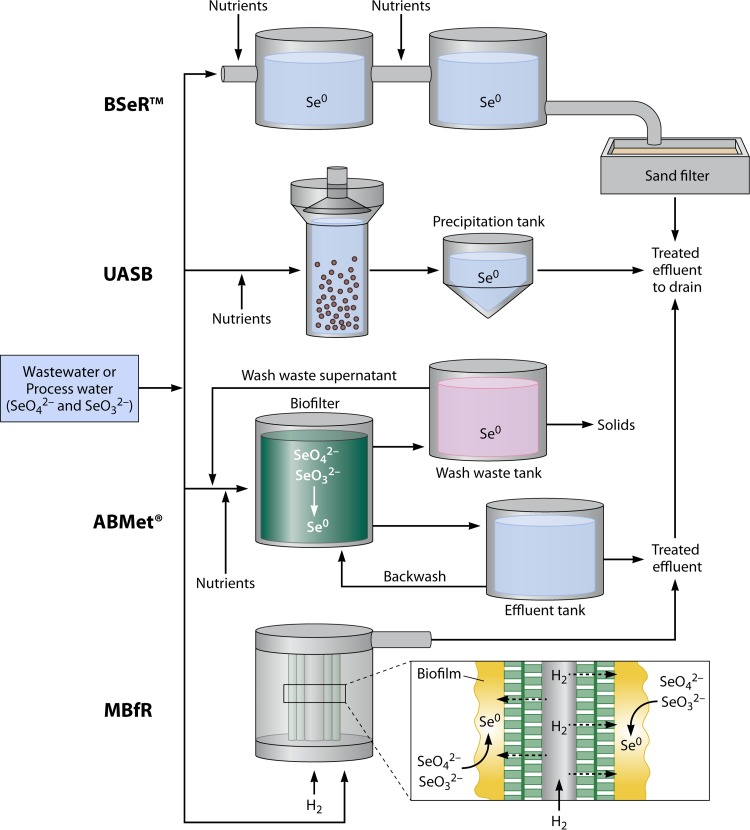
Microbial reduction of Se oxyanions to insoluble elemental selenium has been applied successfully for removal of selenium from wastewater or process water by using different types of bioreactors and process configurations (Fig. 10). Microbial communities in the form of microbial biofilms and biogranules show promise for the development of novel biotechnological applications and immobilization of metals (108, 109). Microorganisms and microbial communities are widely studied for bioremediation through bioreduction of oxidized contaminants and oxyanions of metals and metalloids (9, 110–116). Organic waste substrates (i.e., molasses) or defined organic substrates (i.e., acetate and lactate) have been added to bioreactors as carbon sources and electron donors to facilitate microbial growth and reduction of selenium oxyanions. Selenium removal processes have been developed using biofilm reactors (BSeR), membrane biofilm reactors (MBfRs), upflow anaerobic sludge bed (UASB) reactors, and biofilters (ABMet).
FIG 10.
Overview of the various biotechnological processes (BSer process, UASB reactor, ABMet, and MBfR) used to remove SeO42−, SeO32−, and heavy metals from wastewater or process water. In the BSer process, SeO42− and SeO32− are reduced to elemental selenium by biofilms of propriety microorganisms, using molasses as the carbon source and electron donor (117). In the UASB reactor, carbon (i.e., adipic acid, acetic acid, ethanol, or formic acid) and phosphorus sources are added to facilitate microbial growth and reduction of Se oxyanions to elemental selenium. ABMet consists of a biofilter tank, backwash effluent tank, wash waste tank, and nutrient dosage tank. SeO42− and SeO32− are reduced to insoluble Se0 in a biofilter, and a backwash is performed to remove elemental selenium from the biofilter. Molasses-based nutrients were added to self-sustain biofilm growth and microbial reduction of Se oxyanions (124, 125). In MBfRs, selenium oxyanions are bioreduced to elemental selenium by the autotrophic biofilm, using H2 as the electron donor (104).
BSeR process for selenium removal.
The BSeR technology uses biofilms developed on granular activated carbon for removal of selenium, and the BSeR system basically consists of a series of anaerobic fixed-film biofilm reactors (Fig. 10). A pilot-scale study (1,892 liters) was tested at Garfield Wetlands-Kessler Springs, UT, with water containing 1,870 μg SeO42− liter−1 and 49 μg SeO32− liter−1 (117). Agriculture-grade molasses was added to serve as an electron donor for SeO42− and SeO32− reduction (117). The effluent of the biofilm reactors was passed through a slow sand filter. The effluent selenium levels were below 2 μg liter−1, which is much lower than the prescribed discharge limit of 50 μg liter−1. Reduction of SeO42− and SeO32− was visually evident in the process pipes as a reddish pink color due to the formation of amorphous selenium. Back flushing was used to recover almost 97% of the elemental selenium from the BSeR setup for reuse purposes in the agricultural market (117). It is likely that heavy metals are also removed by the BSeR reactor configuration, along with selenium, and will thus also accumulate in the reactor. It is unclear in what concentration ranges heavy metals are present in the recovered selenium from the BSeR system and how to deal with the heavy metals if the recovered Se is to be used in agriculture.
Selenium removal by UASB reactors.
Removal of SeO42− in UASB reactors was investigated under methanogenic, denitrifying, or sulfate-reducing conditions (6). Anaerobic granular sludge from a UASB reactor removed SeO42− from synthetic wastewater under methanogenic (118, 119), sulfate-reducing (119, 120), and denitrifying (120, 121) conditions. SeO42− was most likely removed by sulfate-reducing bacteria under sulfate-reducing conditions, while enrichment of selenium-respiring bacteria enabled reduction of SeO42− under methanogenic conditions (118). The speciation of selenium associated with the anaerobic biofilms was determined using nondestructive X-ray absorption near-edge structure (XANES) spectroscopy and solid-phase microextraction gas chromatography-mass spectrometry during biological treatment (122, 123). The use of UASB reactors under methanogenic conditions is particularly promising because the process allows recovery of water, selenium, and energy.
Flue gas desulfurization (FGD) systems, typically wet lime stone scrubbers, are increasingly installed along with coal-fired power stations in order to mitigate air pollution and to meet SO2 emission standards. The FGD scrubber liquids contain selenium in the range of 0.5 to 2 mg liter−1, along with significantly larger amounts of sulfate (119). A UASB reactor was used for biological removal of selenium oxyanions and other heavy metals from the FGD outlet water. Carbon (i.e., adipic acid, acetic acid, ethanol, or formic acid) and phosphorus (2 mg liter−1 PO43−-P) sources were added to facilitate microbial growth and reduction of selenium oxyanions. The influent water had a selenium concentration of 953 μg liter−1, which was reduced to below 26 μg Se liter−1 in the treated effluent of the UASB reactor.
ABMet process for selenium removal.
The ABMet technology uses a biofilter to remove selenium from the FGD outlet water. ABMet consists of a biofilter tank, a backwash effluent tank, a wash waste tank, and a nutrient dosage tank. SeO42− and SeO32− are reduced to elemental selenium in the biofilter tank, and a backwash step is performed to remove elemental selenium from the biofilter tank (124, 125). A molasses-based nutrient solution is added to sustain biofilm growth and microbial reduction of selenium oxyanions in the biofilter tank (125).
Selenium removal in membrane biofilm reactors.
Membrane biofilm reactors were evaluated for removing SeO42− from drinking water and FGD brine by using H2 as an electron donor (104, 105, 126). The H2-based membrane biofilm reactor (MBfR) contains bundles of hollow-fiber membranes that deliver H2 directly to the biofilm which develops on the outside walls of the membrane fibers. This system supports the growth of autotrophic bacteria in biofilms by coupling the oxidation of an inorganic electron donor (H2) to the bioreduction of various soluble electron acceptors (i.e., NO3−, NO2−, SeO42−, and ClO4−) (126–128). The MBfR system appears to be a more efficient and simpler means of delivering H2 directly to the biofilm. The dissimilatory selenium-reducing bacteria of the biofilm convert influent SeO42− to elemental selenium. The MBfR system is promising, but it has not been implemented at an industrial scale, probably because of the cost of the electron donor and because of practical implementation issues.
Microbial methylation for conversion of soluble selenium oxyanions into volatile selenium compounds has been proposed for bioremediation of selenium-contaminated agricultural drainage water and seleniferous soils (59, 129–133). Microbial volatilization opens perspectives for recovering selenium via the gas phase, which is thus free from cells and other metal contaminants. For a long time, microbial volatilization was considered a slow process (129, 130, 133–135) that was unsuitable for industrial applications. Interestingly, high-rate volatilization of SeO42−, SeO32−, and Se0 was reported for a Pseudomonas stutzeri NT-1 culture under aerobic conditions (131), thus opening new avenues for high-rate applications. However, the toxicity of volatile Se compounds requires careful assessment of the full-scale application of this approach.
Microbial Reduction for Nanomanufacturing
Selenium has excellent semiconducting and photoconducting properties and is routinely used in rectifiers, solar cells, photographic exposure meters, and xerography. Applications of selenium can be broadened further if it is available in nanostructures (136). Therefore, research efforts are directed toward the synthesis, characterization, and modification of Se nanomaterials, i.e., Se0, metal selenide, and other Se nanostructures. Different shapes of elemental selenium have been produced, i.e., nanowires, nanoribbons, nanotubes, and nanospheres, by use of chemical methods (137). Also, microorganisms synthesize a variety of nanosized biominerals with unique physical and chemical properties and in varied morphologies (138–140). Therefore, several selenium-respiring microorganisms have been investigated for applications in the green synthesis of nanomaterials (Fig. 11). The use of microbial metabolism in nanomanufacturing is a relatively unexplored area with huge potential for the development of cost-effective and environmentally sustainable processes for large-scale production of nanomaterials (139).
FIG 11.
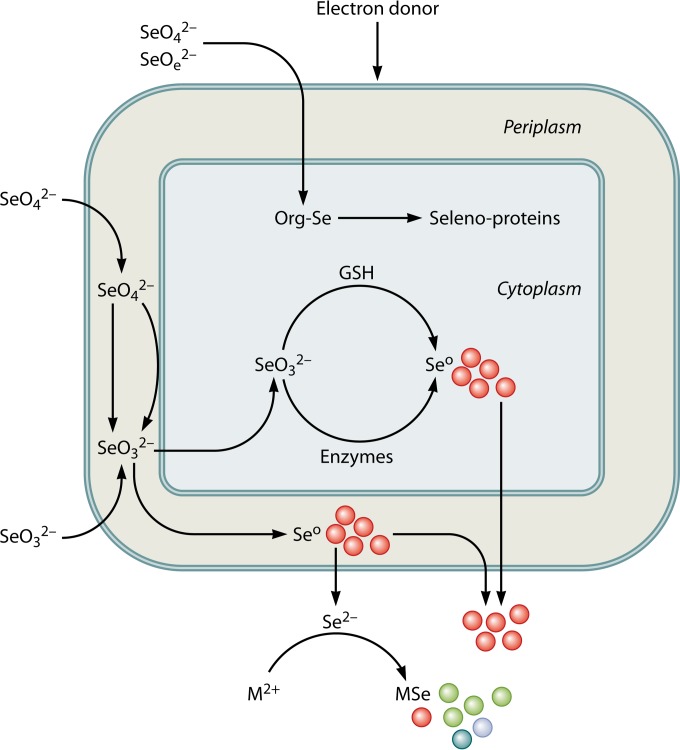
Mechanisms of microbial fabrication of functional selenium nanomaterials. The synthesis is catalyzed by microbial transformation of selenium oxyanions, SeO42− and SeO32−, using various electron donors. Elemental Se is produced as spherical particles in the nanometer size range, localized either inside or outside the cell. Recent research showed that nanoscale semiconductor materials (also called quantum dots) can be synthesized by using microbial metabolism (53, 54, 66). Selenium-respiring microorganisms are used to reduce selenium oxyanions to produce selenide, which combines with metal cations (i.e., Cd and Zn) and forms metal selenide (i.e., CdSe and ZnSe) quantum dots. Org-Se, organoselenium; M2+, Cd and Zn; MSe, metal selenide.
Almost a decade ago, a study by Oremland et al. (78) noted that the spectral properties of microbially produced nanospheres of elemental selenium were substantially different from those of their chemically synthesized counterparts. It appears that the assembly of nanospheres achieved by microorganisms cannot be achieved with today's chemical synthetic routes (78). Evidence is emerging on the possible role of proteins in the fabrication and stabilization of selenium nanoparticles (77, 141, 142). The biological components associated with SeO32− reduction influence the crystallization and growth of metal selenide particles by decreasing the crystal growth rate and increasing particle stability (66). Control of the nanoparticle size is critical, however, in manufacturing elemental selenium or chalcogenide nanoparticles by use of bacteria (53, 54, 77).
The challenges in microbial production of either selenium nanospheres or metal selenide quantum dots include control of the particle size and the polydispersity index of the particles. Since the microbial synthesis of nanoparticles follows the Ostwald ripening process, the size of the nanospheres produced through microbial respiration increases with time. In general, the sizes of microbially synthesized selenium nanoparticles and quantum dots exceed 100 nm and 20 nm, respectively. Therefore, there is a need to understand the mechanisms governing the formation and growth of nanoparticles during microbial respiration of metal(loid) oxyanions. Addition of stabilizing agents, such as glutathione or 2-mercaptoethanol, along with metal chlorides, to microbially produced selenide was proposed to control particle size and to attain quantum confinement (53, 66).
CONCLUSIONS AND OUTLOOK
Selenium-respiring bacteria are ubiquitous in environmental settings, where they use selenium oxyanions and certain other compounds as terminal electron acceptors in the degradation of organic compounds. The accumulated evidence shows that the existence of the selenium cycle may contribute to the carbon and nitrogen cycle in nature. The dissimilatory reduction of selenium oxyanions and selenium reduction mechanisms operating in dissimilatory selenium-respiring bacteria were reviewed. The processes of selenium oxyanion reduction and microbial selenium biomineralization are complex, and divergent mechanisms operate in phylogenetically diverse microorganisms. Biochemical evidence shows that SeO42− reduction predominantly occurs either in the periplasmic space of Gram-negative bacteria or on the outside of the cell membrane of Gram-positive bacteria. Although SeO32− reduction is predominantly observed to occur intracellularly, in the cytoplasm, a recent study showed evidence for its reduction in the periplasmic space of Shewanella oneidensis MR-1. Extracellular reduction is advantageous for bacteria, as intracellular reduction is associated with the additional burden of packaging and exporting of biominerals outside the cell. Certainly, additional studies are needed to improve our understanding of the location of reduction sites, electron-shunting pathways, and export mechanisms (in cases of intracellular reduction) in microbial selenium biomineralization. The physiology and biochemistry of SeO42− reduction have been studied primarily with a few model strains, such as Bacillus selenatarsenatis SF-1, Enterobacter cloacae DL1-1a, and Thauera selenatis. Similar model organisms used to study the biochemical aspects of SeO32− reduction are Shewanella oneidensis MR-1 and Veillonella atypica. Future studies using advanced molecular microbial ecology tools and high-throughput screening techniques may be applied to provide further insights into functionally active true selenium-respiring bacteria, selenium biomineralization mechanisms in natural habitats, and isolation of efficient candidates for biotechnology applications.
ACKNOWLEDGMENTS
We thank our past and present coworkers of UNESCO-IHE, Wageningen University, and BARC, as well as our collaborators.
We also thank our national and international granting agencies, in particular the BioMatch project (project 103922), funded by an EU Marie Curie International Incoming Fellowship (MC-IIF).
Biographies

Y. V. Nancharaiah is a Scientific Officer at the Biofouling and Biofilm Processes Group of the Water and Steam Chemistry Division of Bhabha Atomic Research Centre (BARC), India. Dr. Nancharaiah was trained in biochemistry at Andhra University (India) and then obtained his Ph.D. in biochemistry from the University of Madras (India) and the Bhabha Atomic Research Centre, where he first began his research on microbial biofilms and biotransformation of redox-active metal(loid)s by microbial communities. He worked as a visiting researcher at the Technical University of Munich, Germany (August 2001 to June 2002), Brookhaven National Laboratory, USA (October 2009 to January 2010), and Arizona State University, USA (October to December 2010). His main research interests include microbial transformation of organic and inorganic contaminants by mixed microbial communities for development of sustainable environmental biotechnologies. His awards include an ASM Visiting Research Professor Award (2009), Indo-U.S. Research Fellow Award (2010), DAE Group Achievement Award (2011), and Marie Curie International Incoming Fellow-Experienced Researcher Award (2012).

P. N. L. Lens is Professor of Environmental Biotechnology at the Pollution Prevention and Resource Recovery Chair Group of the Department of Environmental Engineering and Water Technology of UNESCO-IHE. Previously, he was on the faculty of the Sub-Department of Environmental Technology at Wageningen University (1999–2006). He has also held visiting faculty appointments at the Universities of Louvain La Neuve (UCL) and Leuven (KUL). Professor Lens was trained in environmental sanitation and then obtained his Ph.D. in environmental engineering at University Gent (Belgium). He is founding Editor-in-Chief of the review journal Re/Views in Environmental Science and Biotechnology and founding Editor of the IWA Publishing book series Integrated Environmental Technology. His research focuses on biofilms, sulfur biotechnology, selenium bioreduction, natural treatment systems, anaerobic wastewater, and waste gas treatment for resource recovery and reuse. He has (co)authored over 350 scientific publications and edited nine book volumes. His awards include the IWA Publishing Award (2002), a Marie Curie Excellence Grant (2004), and a nomination as an IWA fellow (2010).
REFERENCES
- 1.Madigan MT, Martinko JM, Stahl D, Clark DP. 2010. Brock biology of microorganisms, 13th ed. Pearson Benjamin-Cummings, San Francisco, CA. [Google Scholar]
- 2.Boyd R. 2011. Selenium stories. Nat Chem 3:570. doi: 10.1038/nchem.1076. [DOI] [PubMed] [Google Scholar]
- 3.Minaev VS, Timoshenkov SP, Kalugin VV. 2005. Structural and phase transformations in condensed selenium. J Optoelectron Adv Mater 7:1717–1741. [Google Scholar]
- 4.Buchs B, Evangelou MW, Winkel LH, Lenz M. 2013. Colloidal properties of nanoparticular biogenic selenium govern environmental fate and bioremediation effectiveness. Environ Sci Technol 47:2401–2407. doi: 10.1021/es304940s. [DOI] [PubMed] [Google Scholar]
- 5.Lenz M, Lens PNL. 2009. The essential toxin: the changing perception of selenium in environmental sciences. Sci Total Environ 407:3620–3633. doi: 10.1016/j.scitotenv.2008.07.056. [DOI] [PubMed] [Google Scholar]
- 6.Yudovich YE, Ketris MP. 2005. Selenium in coal: a review. Int J Coal Geol 67:112–126. doi: 10.1016/j.coal.2005.09.003. [DOI] [Google Scholar]
- 7.Fordyce FM. 2013. Selenium deficiency and toxicity in the environment. Essent Med Geol 16:375–416. doi: 10.1007/978-94-007-4375-5_16. [DOI] [Google Scholar]
- 8.Myers T. 2013. Remediation scenarios for selenium contamination, Blackfoot watershed, southeast Idaho, USA. Hydrogeol J 104:309–339. doi: 10.1007/s10040-013-0953-8. [DOI] [Google Scholar]
- 9.Stolz JF, Basu P, Santini JM, Oremland RS. 2006. Arsenic and selenium in microbial metabolism. Annu Rev Microbiol 60:107–130. doi: 10.1146/annurev.micro.60.080805.142053. [DOI] [PubMed] [Google Scholar]
- 10.Williams KH, Wilkins MJ, N′Guessan AL, Arey B, Dodova E, Dohnalkova A, Holmes D, Lovley DR, Long PE. 2013. Field evidence of selenium bioreduction in a uranium-contaminated aquifer. Environ Microbiol Rep 5:444–452. doi: 10.1111/1758-2229.12032. [DOI] [PubMed] [Google Scholar]
- 11.Hatfield DL, Tsuji PA, Carlson BA, Gladyshev VN. 2014. Selenium and selenocysteine: roles in cancer, health, and development. Trends Biochem Sci 39:112–120. doi: 10.1016/j.tibs.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwarz K, Foltz CM. 1957. Selenium as an integral part of factor 3 against dietary liver degeneration. J Am Chem Soc 79:3292–3293. [PubMed] [Google Scholar]
- 13.Navarro-Alarcon M, Cabrera-Vique C. 2008. Selenium in food and the human body: a review. Sci Total Environ 400:115–141. doi: 10.1016/j.scitotenv.2008.06.024. [DOI] [PubMed] [Google Scholar]
- 14.Rahmanto AS, Davies MJ. 2012. Selenium-containing amino acids as direct and indirect antioxidants. IUBMB Life 64:863–871. doi: 10.1002/iub.1084. [DOI] [PubMed] [Google Scholar]
- 15.Stolz JF, Oremland RS. 1999. Bacterial respiration of arsenic and selenate. FEMS Microbiol Rev 23:615–627. doi: 10.1111/j.1574-6976.1999.tb00416.x. [DOI] [PubMed] [Google Scholar]
- 16.Stolz JF, Basi P, Oremland RS. 2002. Microbial transformation of elements: the case of arsenic and selenium. Int Microbiol 5:201–207. doi: 10.1007/s10123-002-0091-y. [DOI] [PubMed] [Google Scholar]
- 17.Banks CJ, Zhang Y, Jiang Y, Heaven S. 2012. Trace element requirements for stable food waste digestion at elevated ammonia concentrations. Bioresour Technol 104:127–135. doi: 10.1016/j.biortech.2011.10.068. [DOI] [PubMed] [Google Scholar]
- 18.Karakashev D, Batstone DJ, Trably E, Angelidaki I. 2006. Acetate oxidation is the dominant methanogenic pathway from acetate in the absence of Methanosaetaceae. Appl Environ Microbiol 72:5138–5141. doi: 10.1128/AEM.00489-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayumi D, Mochimaru H, Yoshioka H, Sakata S, Maeda H, Miyagawa Y, Ikarashi M, Takeuchi M, Kamagata Y. 2011. Evidence for syntrophic acetate oxidation coupled to hydrogenotrophic methanogenesis in the high-temperature petroleum reservoir of Yabase oil field (Japan). Environ Microbiol 13:1995–2006. doi: 10.1111/j.1462-2920.2010.02338.x. [DOI] [PubMed] [Google Scholar]
- 20.Soboh B, Pinske C, Kuhns M, Waclawek M, Ihling C, Trchounian K, Trchounian A, Sinz A, Sawers G. 2011. The respiratory molybdo-selenoprotein formate dehydrogenases of Escherichia coli have hydrogen:benzyl viologen oxidoreductase activity. BMC Microbiol 11:173. doi: 10.1186/1471-2180-11-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaw FL, Mulholland F, Gall GL, Porcelli I, Hart DJ, Pearson BM, van Vliet AHM. 2012. Selenium-dependent biogenesis of formate dehydrogenase in Campylobacter jejuni is controlled by the fdhTU accessory genes. J Bacteriol 194:3814–3823. doi: 10.1128/JB.06586-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong X, Plugge CM, Stams AJM. 1994. Anaerobic degradation of propionate by a mesophilic acetogenic bacterium in coculture and triculture with different methanogens. Appl Environ Microbiol 60:2834–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shrift A. 1964. A selenium cycle in nature. Nature 201:1304–1305. doi: 10.1038/2011304a0. [DOI] [PubMed] [Google Scholar]
- 24.Forchhammer K, Leinfelder W, Boesmiller K, Veprek B, Bock A. 1991. Selenocysteine synthase from Escherichia coli. Nucleotide sequence of the gene (selA) and purification of the protein. J Biol Chem 266:6318–6323. [PubMed] [Google Scholar]
- 25.Turanov AA, Xu X-M, Carlson BA, Yoo M-H, Gladyshev VN, Hatfield DL. 2011. Biosynthesis of selenocysteine, the 21st amino acid in the genetic code, and a novel pathway for cysteine biosynthesis. Adv Nutr 2:122–128. doi: 10.3945/an.110.000265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu XM, Turanov AA, Carlson BA, Yoo MH, Everley RA, Nandakumar R, Sorokina I, Gygi SP, Hatfield DL. 2007. Biosynthesis of selenocysteine on its tRNA in eukaryotes. PLoS Biol 5:e4. doi: 10.1371/journal.pbio.0050004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shisler JL, Senkevich TG, Berry MJ, Moss B. 1998. Ultraviolet-induced cell death blocked by a selenoprotein from a human dermatotropic poxvirus. Science 279:102–105. doi: 10.1126/science.279.5347.102. [DOI] [PubMed] [Google Scholar]
- 28.Knight VK, Nijenhuis I, Kerkhof LJ, Mäggblom MM. 2002. Degradation of aromatic compounds coupled to selenate reduction. Geomicrobiol J 19:77–86. doi: 10.1080/014904502317246183. [DOI] [Google Scholar]
- 29.Huber R, Sacher M, Vollmann A, Huber H, Rose D. 2000. Respiration of arsenate and selenate by hyperthermophilic archaea. Syst Appl Microbiol 23:305–314. doi: 10.1016/S0723-2020(00)80058-2. [DOI] [PubMed] [Google Scholar]
- 30.Tejo Prakash N, Sharma N, Prakash R, Raina KK, Fellowes J, Pearce CI, Lloyd JR, Pattrick RA. 2009. Aerobic microbial manufacturing of nanoscale selenium: exploiting nature's bio-nanomineralization potential. Biotechnol Lett 31:1857–1862. doi: 10.1007/s10529-009-0096-0. [DOI] [PubMed] [Google Scholar]
- 31.Kessi J, Ramuz M, Wehrli E, Spycher M, Bachofen R. 1999. Reduction of selenite and detoxification of elemental selenium by the phototrophic bacterium Rhodospirillum rubrum. Appl Environ Microbiol 65:4734–4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lipman JG, Waksman SA. 1923. The oxidation of selenium by a new group of autotrophic microorganisms. Science 57:60. doi: 10.1126/science.57.1463.60. [DOI] [PubMed] [Google Scholar]
- 33.Sarathchandra SU, Watkinson JH. 1981. Oxidation of elemental selenium to selenite by Bacillus megaterium. Science 211:600–601. doi: 10.1126/science.6779378. [DOI] [PubMed] [Google Scholar]
- 34.Dowdle PR, Oremland RS. 1998. Microbial oxidation of elemental selenium in soil slurries and bacterial cultures. Environ Sci Technol 32:3749–3755. doi: 10.1021/es970940s. [DOI] [Google Scholar]
- 35.Losi ME, Frankenberger WT. 1998. Microbial oxidation and solubilization of precipitated elemental selenium in soil. J Environ Qual 27:836–843. [Google Scholar]
- 36.Newman DK, Ahmann D, Morel FMM. 1998. A brief review of microbial arsenate reduction. Geomicrobiol J 15:255–268. doi: 10.1080/01490459809378082. [DOI] [Google Scholar]
- 37.Macy JM, Michel TA, Kirsch DG. 1989. Selenate reduction by a Pseudomonas species: a new mode of anaerobic respiration. FEMS Microbiol Lett 61:195–198. doi: 10.1111/j.1574-6968.1989.tb03577.x. [DOI] [PubMed] [Google Scholar]
- 38.Laverman AM, Blum JS, Schaefer JK, Phillips EJP, Lovley DR, Oremland RS. 1995. Growth of strain SES-3 with arsenate and other diverse electron acceptors. Appl Environ Microbiol 61:3556–3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oremland RS, Hollibaugh JT, Maest AS, Presser TS, Miller LG, Culbertson CW. 1989. Selenate reduction to elemental selenium by anaerobic bacteria in sediments and culture: biogeochemical significance of a novel sulfate-independent respiration. Appl Environ Microbiol 55:2333–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujita M, Ike M, Nishimoto S, Takahashi K, Kashiwa M. 1997. Isolation and characterization of a novel selenate-reducing bacterium, Bacillus sp SF-1. J Ferment Bioeng 83:517–522. doi: 10.1016/S0922-338X(97)81130-0. [DOI] [Google Scholar]
- 41.Knight VK, Blakemore RP. 1998. Reduction of diverse electron acceptors by Aeromonas hydrophila. Arch Microbiol 169:239–248. doi: 10.1007/s002030050567. [DOI] [PubMed] [Google Scholar]
- 42.Oremland RS, Switzer Blum J, Culbertson CW, Visscher PT, Miller LG, Dowdle P, Strohmaier FE. 1994. Isolation, growth, and metabolism of an obligately anaerobic, selenate-respiring bacterium, strain SES-3. Appl Environ Microbiol 60:3011–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Switzer Blum J, Bindi AB, Buzzelli J, Stolz JF, Oremland RS. 1998. Bacillus arsenoselenatis sp. nov., and Bacillus selenitireducens sp. nov.; two haloalkaliphiles from Mono Lake, California, which respire oxyanions of selenium and arsenic. Arch Microbiol 171:19–30. doi: 10.1007/s002030050673. [DOI] [PubMed] [Google Scholar]
- 44.Switzer Blum J, Stolz JF, Ohren A, Oremland RS. 2001. Selenihalanaerobacter shriftii gen. nov. sp. nov., a halophilic anaerobe from Dead Sea sediments that respires selenate. Arch Microbiol 175:208–219. doi: 10.1007/s002030100257. [DOI] [PubMed] [Google Scholar]
- 45.von Wintzingerode F, Schattke A, Siddiqui RA, Rösick U, Göbel R, Gross UB. 2001. Bordetella petrii sp. nov., isolated from an anaerobic bioreactor, and emended description of the genus Bordetella. Int J Syst Evol Microbiol 51:1257–1265. doi: 10.1099/00207713-51-4-1257. [DOI] [PubMed] [Google Scholar]
- 46.Baesman SM, Stolz JF, Kulp TR, Oremland RS. 2009. Enrichment and isolation of Bacillus beveridgei sp. nov., a facultative anaerobic haloalkaliphile from Mono Lake, California, that respires oxyanions of tellurium, selenium, and arsenic. Extremophiles 13:695–705. doi: 10.1007/s00792-009-0257-z. [DOI] [PubMed] [Google Scholar]
- 47.Narasingarao P, Häggblom MM. 2007. Identification of anaerobic selenate-respiring bacteria from aquatic sediments. Appl Environ Microbiol 73:3519–3527. doi: 10.1128/AEM.02737-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Narasingarao P, Häggblom MM. 2006. Sedimenticola selenatireducens, gen. nov., sp. nov., an anaerobic selenate-respiring bacterium isolated from estuarine sediment. Syst Appl Microbiol 29:382–388. doi: 10.1016/j.syapm.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 49.Rauschenbach I, Narasingarao P, Häggblom MM. 2011. Desulfurispirillum indicum sp. nov., a selenate- and selenite-respiring bacterium isolated from an estuarine canal. Int J Syst Evol Microbiol 61:654–658. doi: 10.1099/ijs.0.022392-0. [DOI] [PubMed] [Google Scholar]
- 50.Painter EP. 1941. The chemistry and toxicity of selenium compounds with special reference to the selenium problem. Chem Rev 28:179–213. doi: 10.1021/cr60090a001. [DOI] [Google Scholar]
- 51.Kessi J, Hanselmann KW. 2004. Similarities between the abiotic reduction of selenite with glutathione and the dissimilatory reaction mediated by Rhodospirillum rubrum and Escherichia coli. J Biol Chem 279:50662–50669. doi: 10.1074/jbc.M405887200. [DOI] [PubMed] [Google Scholar]
- 52.Kessi J. 2006. Enzymic systems proposed to be involved in the dissimilatory reduction of selenite in the purple non-sulfur bacteria Rhodospirillum rubrum and Rhodobacter capsulatus. Microbiology 152:731–743. doi: 10.1099/mic.0.28240-0. [DOI] [PubMed] [Google Scholar]
- 53.Pearce CI, Coker VS, Charnock JM, Pattrick RAD, Mosselmans JFW, Law N, Beveridge TJ, Lloyd JR. 2008. Microbial manufacture of chalcogenide-based nanoparticles via the reduction of selenite using Veillonella atypica: an in situ EXAFS study. Nanotechnology 19:155603. doi: 10.1088/0957-4484/19/15/155603. [DOI] [PubMed] [Google Scholar]
- 54.Pearce CI, Pattrick RAD, Law N, Charnock JM, Coker VS, Fellowes JW, Oremland RS, Lloyd JR. 2009. Investigating different mechanisms for biogenic selenite transformations: Geobacter sulfurreducens, Shewanella oneidensis and Veillonella atypica. Environ Technol 30:1313–1326. doi: 10.1080/09593330902984751. [DOI] [PubMed] [Google Scholar]
- 55.Li DB, Cheng YY, Wu C, Li WW, Li N, Yang ZC, Tong ZH, Yu HQ. 2014. Selenite reduction by Shewanella oneidensis MR-1 is mediated by fumarate reductase in periplasm. Sci Rep 4:3755. doi: 10.1038/srep03735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Herbel MJ, Blum JS, Oremland RS, Borglin SE. 2003. Reduction of elemental selenium to selenide: experiments with anoxic sediments and bacteria that respire Se-oxyanions. Geomicrobiol J 20:587–602. doi: 10.1080/713851163. [DOI] [Google Scholar]
- 57.Doran JW, Alexander M. 1976. Microbial formation of volatile selenium compounds in soil. Soil Sci Soc Am J 40:687–690. doi: 10.2136/sssaj1976.03615995004000050025x. [DOI] [Google Scholar]
- 58.Bentley R, Chasteen TG. 2002. Microbial methylation of metalloids: arsenic, antimony, and bismuth. Microbiol Mol Biol Rev 66:250–271. doi: 10.1128/MMBR.66.2.250-271.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chasteen TG, Bentley R. 2003. Biomethylation of selenium and tellurium: microorganisms and plants. Chem Rev 103:1–25. doi: 10.1021/cr010210+. [DOI] [PubMed] [Google Scholar]
- 60.Oremland RS, Zehr JP. 1986. Formation of methane and carbon dioxide from dimethylselenide in anoxic sediments and by a methanogenic bacterium. Appl Environ Microbiol 52:1031–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kiene R, Oremland RS, Catena A, Miller L, Capone DG. 1986. Metabolism of reduced methylated sulfur compounds in anaerobic sediments and by a pure culture of an estuarine methanogen. Appl Environ Microbiol 52:1037–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McCready RGL, Campbell JN, Payne JI. 1966. Selenite reduction by Salmonella heidelberg. Can J Microbiol 12:703–714. doi: 10.1139/m66-097. [DOI] [PubMed] [Google Scholar]
- 63.Steinberg NA, Oremland RS. 1990. Dissimilatory selenate reduction potentials in a diversity of sediment types. Appl Environ Microbiol 56:3550–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Woolfolk CA, Whiteley HR. 1962. Reduction of inorganic compounds with molecular hydrogen by Micrococcus lactilyticus. J Bacteriol 84:647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zehr JP, Oremland RS. 1987. Reduction of selenate to selenide by sulfate-reducing bacteria: experiments with cell suspensions and estuarine sediments. Appl Environ Microbiol 53:1365–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fellowes JW, Pattrick RA, Lloyd JR, Charnock JM, Coker VS, Mosselmans JF, Weng TC, Pearce CI. 2013. Ex situ formation of metal selenide quantum dots using bacterially derived selenide precursors. Nanotechnology 24:145603. doi: 10.1088/0957-4484/24/14/145603. [DOI] [PubMed] [Google Scholar]
- 67.Johnson TM, Herbel MJ, Bullen TD, Zawislanski PT. 1999. Selenium isotope ratios as indicators of selenium sources and oxyanions reduction. Geochim Cosmochim Acta 63:2775–2783. doi: 10.1016/S0016-7037(99)00279-3. [DOI] [Google Scholar]
- 68.Herbel MJ, Johnson TM, Oremland RS, Bullen TD. 2000. Fractionation of selenium isotopes during bacterial respiratory reduction of selenium oxyanions. Geochim Cosmochim Acta 64:3701–3709. doi: 10.1016/S0016-7037(00)00456-7. [DOI] [Google Scholar]
- 69.Ellis AS, Johnson TM, Herbel MJ, Bullen TD. 2003. Stable isotope fractionation of selenium by natural microbial consortia. Chem Geol 195:119–129. doi: 10.1016/S0009-2541(02)00391-1. [DOI] [Google Scholar]
- 70.Oremland RS, Steinberg NA, Maest AS, Miller LG, Hollibaugh JT. 1990. Measurement of in situ rates of selenate removal by dissimilatory bacterial reduction in sediments. Environ Sci Technol 24:1157–1164. doi: 10.1021/es00078a001. [DOI] [Google Scholar]
- 71.Oremland RS, Steinberg NA, Presser TS, Miller LG. 1991. In situ bacterial selenate reduction in the agricultural drainage systems of western Nevada. Appl Environ Microbiol 57:615–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lovley DR. 2002. Dissimilatory metal reduction: from early life to bioremediation. ASM News 68:231–237. [Google Scholar]
- 73.Lovley DR. 2008. The microbe electric: conversion of organic matter to electricity. Curr Opin Biotechnol 19:564–571. doi: 10.1016/j.copbio.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 74.Bao P, Huang H, Hu Z-Y, Häggblom MM, Zhu Y-G. 2013. Impact of temperature, CO2 fixation and nitrate reduction on selenium reduction, by a paddy soil Clostridium strain. J Appl Microbiol 114:703–712. doi: 10.1111/jam.12084. [DOI] [PubMed] [Google Scholar]
- 75.Maiers DT, Wichlacz PL, Thompson DL, Bruhn DF. 1988. Selenate reduction by bacteria from a selenium-rich environment. Appl Environ Microbiol 54:2591–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tomei FA, Barton LL, Lemanski CL, Zocco TG, Fink NH, Sillerud LO. 1995. Transformation of selenate and selenite to elemental selenium by Desulfovibrio desulfuricans. J Ind Microbiol 14:329–336. doi: 10.1007/BF01569947. [DOI] [Google Scholar]
- 77.Debieux CM, Dridge EJ, Mueller CM, Splatt P, Paszkiewicz K, Knight I, Florance H, Love J, Titball RW, Lewis RJ, Richardson DJ, Butler CS. 2011. A bacterial process for selenium nanosphere assembly. Proc Natl Acad Sci U S A 108:13480–13485. doi: 10.1073/pnas.1105959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Oremland RS, Herbel MJ, Blum JS, Langley S, Beveridge TJ, Ajayan PM, Sutto T, Ellis AV, Curran S. 2004. Structural and spectral features of selenium nanospheres produced by Se-respiring bacteria. Appl Environ Microbiol 70:52–60. doi: 10.1128/AEM.70.1.52-60.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Klonowska A, Heulin T, Vermeglio A. 2005. Selenite and tellurite reduction by Shewanella oneidensis. Appl Environ Microbiol 71:5607–5609. doi: 10.1128/AEM.71.9.5607-5609.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Macy JM, Rech S, Auling G, Dorsch M, Stackebrandt E, Sly LI. 1993. Thauera selenatis gen. nov., sp. nov., a member of the beta subclass of proteobacteria with a novel type of anaerobic respiration. Int J Syst Bacteriol 43:135–142. doi: 10.1099/00207713-43-1-135. [DOI] [PubMed] [Google Scholar]
- 81.Butler CS, Debieux CM, Dridge EJ, Splatt P, Wright M. 2012. Biomineralization of selenium by the selenate-respiring bacterium Thauera selenatis. Biochem Soc Trans 40:1239–1243. doi: 10.1042/BST20120087. [DOI] [PubMed] [Google Scholar]
- 82.Rabenstein DL, Tan K-S. 1988. 77Se NMR studies of bis(alkylthio)selenides of biological thiols. Magn Reson Chem 26:1079–1085. doi: 10.1002/mrc.1260261209. [DOI] [Google Scholar]
- 83.Lowe EC, Bydder S, Hartshorne RS, Tape HLU, Dridge EJ, Debieux CM, Paszkiewicz K, Singleton I, Lewis RJ, Santini JM, Richardson DJ, Butler CS. 2010. Quinol-cytochrome c oxidoreductase and cytochrome c4 mediate electron transfer during selenate respiration in Thauera selenatis. J Biol Chem 285:18433–18442. doi: 10.1074/jbc.M110.115873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Krafft T, Browen A, Theis F, Macy JM. 2000. Cloning and sequencing of the genes encoding the periplasmic-cytochrome B-containing selenate reductase of Thauera selenatis. DNA Seq 10:365–377. [DOI] [PubMed] [Google Scholar]
- 85.Losi ME, Frankenberger WT. 1997. Reduction of selenium oxyanions by Enterobacter cloacae SLD1a-1: isolation and growth of the bacterium and its expulsion of selenium nanoparticles. Appl Environ Microbiol 63:3079–3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ridley H, Watts CA, Richardson DJ, Butler CS. 2006. Resolution of distinct membrane-bound enzymes from Enterobacter cloacae SLD1a-1 that are responsible for selective reduction of nitrate and selenate oxyanions. Appl Environ Microbiol 72:5173–5180. doi: 10.1128/AEM.00568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yamamura S, Yamashita M, Fujimoto N, Kuroda M, Kashiwa M, Sei K, Fujita M, Ike M. 2007. Bacillus selenatarsenatis sp. nov., a selenate- and arsenate-reducing bacterium isolated from the effluent drain of a glass-manufacturing plant. Int J Syst Evol Microbiol 57:1060–1064. doi: 10.1099/ijs.0.64667-0. [DOI] [PubMed] [Google Scholar]
- 88.Kuroda M, Yamashita M, Miwa E, Imao K, Fujimoto N, Ono H, Nagano K, Sei K, Ike M. 2011. Molecular cloning and characterization of the srdBCA operon, encoding the respiratory selenate reductase complex, from the selenate-reducing bacterium Bacillus selenatarsenatis SF-1. J Bacteriol 193:2141–2148. doi: 10.1128/JB.01197-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Turner RJ, Weiner JH, Taylor DE. 1998. Selenium metabolism in Escherichia coli. Biometals 11:223–237. doi: 10.1023/A:1009290213301. [DOI] [PubMed] [Google Scholar]
- 90.Zannoni D, Borsetti F, Harrison JJ, Turner RJ. 2008. The bacterial response to the chalcogen metalloids Se and Te. Adv Microb Physiol 53:1–72. doi: 10.1016/S0065-2911(07)53001-8. [DOI] [PubMed] [Google Scholar]
- 91.Ganther HE. 1971. Reduction of the selenotrisulfide derivative of glutathione to a persulfide analog by glutathione reductase. Biochemistry 10:4089–4098. doi: 10.1021/bi00798a013. [DOI] [PubMed] [Google Scholar]
- 92.Bébien MG, Kirsch J, Méjean V, Verméglio A. 2002. Involvement of a putative molybdenum enzyme in the reduction of selenate by Escherichia coli. Microbiology 148:3865–3872. [DOI] [PubMed] [Google Scholar]
- 93.Yamada A, Miyashita M, Inoue K, Matsunaga T. 1997. Extracellular reduction of selenite by a novel marine photosynthetic bacterium. Appl Environ Microbiol 48:367–372. [DOI] [PubMed] [Google Scholar]
- 94.Björnstedt M, Kumar S, Holmgren A. 1992. Selenodiglutathione is a highly efficient oxidant of reduced thioredoxin and a substrate for mammalian thioredoxin reductase. J Biol Chem 267:8030–8034. [PubMed] [Google Scholar]
- 95.Hockin SL, Gadd GM. 2003. Linked redox precipitation of sulfur and selenium under anaerobic conditions by sulfate-reducing bacterial biofilms. Appl Environ Microbiol 69:7063–7072. doi: 10.1128/AEM.69.12.7063-7072.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pettine M, Gennan F, Campanella L, Casentini B, Marani D. 2012. The reduction of selenium(IV) by hydrogen sulfide in aqueous solutions. Geochim Cosmochim Acta 83:37–47. doi: 10.1016/j.gca.2011.12.024. [DOI] [Google Scholar]
- 97.Zawadzka AM, Crawford RL, Paszczynski AJ. 2006. Pyridine-2,6-bis(thiocarboxylic acid) produced by Pseudomonas stutzeri KC reduces and precipitates selenium and tellurium oxyanions. Appl Environ Microbiol 72:3119–3129. doi: 10.1128/AEM.72.5.3119-3129.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Basaglia M, Toffanin A, Baldan E, Bottegal M, Shapleigh JP, Casella S. 2007. Selenite-reducing capacity of the copper-containing nitrite reductase of Rhizobium sullae. FEMS Microbiol Lett 269:124–130. doi: 10.1111/j.1574-6968.2006.00617.x. [DOI] [PubMed] [Google Scholar]
- 99.Harrison G, Curie C, Laishley EJ. 1984. Purification and characterization of an inducible dissimilatory type sulphite reductase from Clostridium pasteurianum. Arch Microbiol 138:72–78. doi: 10.1007/BF00425411. [DOI] [PubMed] [Google Scholar]
- 100.DeMoll-Decker H, Macy JM. 1993. The periplasmic nitrite reductase of Thauera selenatis may catalyze the reduction of selenite to elemental selenium. Arch Microbiol 160:241–247. [Google Scholar]
- 101.Taratus EM, Eubanks SG, DiChristina TJ. 2000. Design and application of a rapid screening technique for isolation of selenite reduction-deficient mutants of Shewanella putrefaciens. Microbiol Res 155:79–85. doi: 10.1016/S0944-5013(00)80041-5. [DOI] [PubMed] [Google Scholar]
- 102.Staicu LC, van Hullebusch ED, Lens PN, Pilon-Smits EA, Oturan MA. 20 September 2014. Electrocoagulation of colloidal biogenic selenium. Environ Sci Pollut Res Int doi: 10.1007/s11356-014-3592-2. [DOI] [PubMed] [Google Scholar]
- 103.Wang X, Liu G, Zhou J, Wang J, Jin R, Lv H. 2011. Quinone-mediated reduction of selenite and tellurite by Escherichia coli. Bioresour Technol 102:3268–3271. doi: 10.1016/j.biortech.2010.11.078. [DOI] [PubMed] [Google Scholar]
- 104.Chung J, Nerenberg R, Rittmann BR. 2006. Bioreduction of selenate using a hydrogen-based membrane biofilm reactor. Environ Sci Technol 40:1664–1671. doi: 10.1021/es051251g. [DOI] [PubMed] [Google Scholar]
- 105.Chung J, Ryu H, Abbaszadegan M, Rittmann BE. 2006. Community structure and function in a H2-based membrane biofilm reactor capable of bioreduction of selenate and chromate. Appl Microbiol Biotechnol 72:1330–1339. doi: 10.1007/s00253-006-0439-x. [DOI] [PubMed] [Google Scholar]
- 106.Presser TS. 1994. The Kesterson effect. Environ Manage 18:437–454. doi: 10.1007/BF02393872. [DOI] [Google Scholar]
- 107.Soda S, Kashiwa M, Kagami T, Kuroda M, Yamashita M, Ike M. 2011. Laboratory-scale bioreactors for soluble selenium removal from selenium refinery wastewater using anaerobic sludge. Desalination 279:433–438. doi: 10.1016/j.desal.2011.06.031. [DOI] [Google Scholar]
- 108.Rittmann BE. 2006. Microbial ecology to manage processes in environmental biotechnology. Trends Biotechnol 24:261–266. doi: 10.1016/j.tibtech.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 109.van Hullebusch ED, Zandvoort MH, Lens PNL. 2003. Metal immobilisation by biofilms: mechanisms and analytical tools. Rev Environ Sci Biotechnol 2:9–33. doi: 10.1023/B:RESB.0000022995.48330.55. [DOI] [Google Scholar]
- 110.Nancharaiah YV, Venugopalan VP. 2011. Denitrification of synthetic concentrated nitrate wastes by aerobic granular sludge under anoxic conditions. Chemosphere 85:683–688. doi: 10.1016/j.chemosphere.2011.06.077. [DOI] [PubMed] [Google Scholar]
- 111.Nancharaiah YV, Joshi JM, Mohan TVK, Venugopalan VP, Narasimhan SV. 2006. Aerobic granular biomass: a novel biomaterial for efficient uranium removal. Curr Sci 91:503–509. [Google Scholar]
- 112.Nancharaiah YV, Venugopalan VP, Francis AJ. 2012. Removal and biotransformation of U(VI) and Cr(VI) by aerobically grown mixed microbial granules. Desalination Water Treat 1–3:90–95. doi: 10.1080/19443994.2012.664269. [DOI] [Google Scholar]
- 113.Gao W, Francis AJ. 2008. Reduction of uranium(VI) to uranium(IV) by clostridia. Appl Environ Microbiol 74:4580–4584. doi: 10.1128/AEM.00239-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Francis AJ. 2008. Biotransformation of uranium and other actinides in radioactive wastes. J Alloys Compd 271–273:78–84. doi: 10.1016/S0925-8388(98)00028-0. [DOI] [Google Scholar]
- 115.Nancharaiah YV, Dodge C, Venugopalan VP, Narasimhan SV, Francis AJ. 2010. Immobilization of Cr(VI) and its reduction to Cr(III) phosphate by granular biofilms comprising a mixture of microbes. Appl Environ Microbiol 76:2433–2438. doi: 10.1128/AEM.02792-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lloyd JR, Renshaw JC. 2005. Bioremediation of radioactive waste: radionuclide-microbe interactions in laboratory and field-scale studies. Curr Opin Biotechnol 16:254–260. doi: 10.1016/j.copbio.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 117.USEPA. 2001. Final report—selenium treatment/removal alternatives demonstration project. http://www.epa.gov/nrmrl/std/mwt/a3/mwtp191.pdf.
- 118.Astratinei V, van Hullebusch ED, Lens P. 2006. Bioconversion of selenate in methanogenic anaerobic granular sludge. J Environ Qual 35:1873–1883. doi: 10.2134/jeq2005.0443. [DOI] [PubMed] [Google Scholar]
- 119.Lenz M, van Hullebusch ED, Hommes G, Corvini PFX, Lens PNL. 2008. Selenate removal in methanogenic and sulfate-reducing upflow anaerobic sludge bed reactors. Water Res 42:2184–2194. doi: 10.1016/j.watres.2007.11.031. [DOI] [PubMed] [Google Scholar]
- 120.Lenz M, Enright AM, O'Flaherty V, van Aelst AC, Lens PN. 2009. Bioaugmentation of UASB reactors with immobilized Sulfurospirillum barnesii for simultaneous selenate and nitrate removal. Appl Microbiol Biotechnol 83:377–388. doi: 10.1007/s00253-009-1915-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lenz M, Smit M, Binder P, van Aelst AC, Lens PNL. 2008. Biological alkylation and colloid formation of selenium in methanogenic UASB reactors. J Environ Qual 37:1691–1700. doi: 10.2134/jeq2007.0630. [DOI] [PubMed] [Google Scholar]
- 122.Lenz M, van Huellebusch ED, Farges F, Nikitenko S, Borca CN, Grolimund D, Lens PN. 2008. Selenium speciation assessed by X-ray absorption spectroscopy of sequentially extracted anaerobic biofilms. Environ Sci Technol 42:7587–7593. doi: 10.1021/es800811q. [DOI] [PubMed] [Google Scholar]
- 123.Lenz M, van Huellebusch ED, Farges F, Nikitenko S, Corvini PF, Lens PN. 2011. Combined speciation analysis by X-ray absorption near-edge structure spectroscopy, ion chromatography, and solid-phase microextraction gas chromatography-mass spectrometry to evaluate biotreatment of concentrated selenium wastewaters. Environ Sci Technol 45:1067–1073. doi: 10.1021/es1022619. [DOI] [PubMed] [Google Scholar]
- 124.Sonstegard J, Harwood J, Pickett T, Kennedy W. ABMeT: setting the standard for selenium removal. http://www.epa.gov/region1/npdes/merrimackstation/pdfs/ar/AR-144.pdf. [Google Scholar]
- 125.Sonstegard J, Pickett T, Harwood J, Johnson D. 2008. Full scale operation of GE ABMet® biological technology for the removal of selenium from FGD wastewaters. IWC-08-31. Int Water Conf, San Antonio, TX: http://www.gewater.com/images/abmet/IWC_08-31-GE_Full_Scale_Operation_of_Biological_Technology_for_Selenium_Removal_in_FGD.pdf. [Google Scholar]
- 126.Van Ginkel SW, Yang Z, Kim BO, Sholin M, Rittmann BE. 2011. The removal of selenate to low ppb levels from flue gas desulfurization brine using the H2-based membrane biofilm reactor (MBfR). Bioresour Technol 102:6360–6364. doi: 10.1016/j.biortech.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 127.Lai CY, Yang X, Tang Y, Rittmann BE, Zhao HP. 2014. Nitrate shaped the selenate-reducing microbial community in a hydrogen-based biofilm reactor. Environ Sci Technol 48:3395–3402. doi: 10.1021/es4053939. [DOI] [PubMed] [Google Scholar]
- 128.Chung J, Rittmann BE, Wright WF, Bowman RH. 2007. Simultaneous bio-reduction of nitrate, perchlorate, selenate, chromate, arsenate, and dibrochloropropane using a hydrogen-membrane biofilm reactor. Biodegradation 18:199–209. doi: 10.1007/s10532-006-9055-9. [DOI] [PubMed] [Google Scholar]
- 129.Van Fleet-Stadler V, Chasteen TG, Pickering IJ, George GN, Prince RC. 2000. Fate of selenate and selenite metabolized by Rhodobacter sphaeroides. Appl Environ Microbiol 66:4849–4853. doi: 10.1128/AEM.66.11.4849-4853.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Favre-Bonté S, Ranjard L, Colinon C, Prigent-Combaret C, Nazaret S, Cournoyer B. 2005. Freshwater selenium-methylating bacterial thiopurine methyltransferases: diversity and molecular phylogeny. Environ Microbiol 77:153–164. doi: 10.1111/j.1462-2920.2004.00670.x. [DOI] [PubMed] [Google Scholar]
- 131.Kagami T, Narita T, Kuroda M, Notaguchi E, Yamashita M, Sei K, Soda S, Ike M. 2013. Effective selenium volatilization under aerobic conditions and recovery from the aqueous phase by Pseudomonas stutzeri NT-1. Water Res 47:1361–1368. doi: 10.1016/j.watres.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 132.Vriens B, Lenz M, Charlet L, Berg M, Winkel LHE. 2014. Natural wetland emissions of methylated trace elements. Nat Commun 5:3035. doi: 10.1038/ncomms4035. [DOI] [PubMed] [Google Scholar]
- 133.Dungan RS, Frankenberger WT. 1999. Microbial transformations of selenium and the bioremediation of seleniferous environments. Bioremediat J 3:171–188. doi: 10.1080/10889869991219299. [DOI] [Google Scholar]
- 134.Frankenberger WT, Arshad M. 2001. Bioremediation of selenium-contaminated sediments and water. BioFactors 14:241–254. doi: 10.1002/biof.5520140130. [DOI] [PubMed] [Google Scholar]
- 135.Ranjard L, Nazaret Cournoyer B. 2003. Freshwater bacteria can methylate selenium through the thiopurine methyltransferase pathway. Appl Environ Microbiol 69:3784–3790. doi: 10.1128/AEM.69.7.3784-3790.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Qin D, Zhou J, Luo C, Liu Y, Han L, Cao Y. 2006. Surfactant-assisted synthesis of size-controlled trigonal Se/Te allow nanowires. Nanotechnology 17:674. doi: 10.1088/0957-4484/17/3/010. [DOI] [Google Scholar]
- 137.Kumar A, Sevonkaev I, Goia D. 2014. Synthesis of selenium particles with various morphologies. J Colloid Interface Sci 416:119–123. doi: 10.1016/j.jcis.2013.10.046. [DOI] [PubMed] [Google Scholar]
- 138.Gadd GM. 2010. Metals, minerals and microbes: geomicrobiology and bioremediation. Microbiology 156:609–643. doi: 10.1099/mic.0.037143-0. [DOI] [PubMed] [Google Scholar]
- 139.Lloyd JR, Byrne JM, Coker VS. 2011. Biotechnological synthesis of functional nanomaterials. Curr Opin Biotechnol 22:509–515. doi: 10.1016/j.copbio.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 140.Lloyd JR, Pearce CI, Coker VS, Pattrick RA, van der Laan G, Cutting R, Vaughan DJ, Paterson-Beedle M, Mikheenko IP, Yong P, Macaskie LE. 2008. Biomineralization: linking the fossil record to the production of high value functional materials. Geobiology 6:285–297. doi: 10.1111/j.1472-4669.2008.00162.x. [DOI] [PubMed] [Google Scholar]
- 141.Lenz M, Kolvenbach B, Gygax B, Moes S, Corvini PF. 2011. Shedding light on selenium biomineralization: proteins associated with bionanomaterials. Appl Environ Microbiol 77:4676–4680. doi: 10.1128/AEM.01713-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Dobias J, Suvorova EI, Bernier-Latmani R. 2011. Role of proteins in controlling selenium nanoparticle size. Nanotechnology 22:195605. doi: 10.1088/0957-4484/22/19/195605. [DOI] [PubMed] [Google Scholar]
- 143.Tamura K, Nei M, Kumar S. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A 101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tam K, Hu CT, Lee JH, Lai M, Chang CH, Rheem Y, Chen W, Hur HG, Myung NV. 2010. Growth mechanism of amorphous selenium nanoparticles synthesized by Shewanella sp. HN-41. Biosci Biotechnol Biochem 74:696–700. doi: 10.1271/bbb.90454. [DOI] [PubMed] [Google Scholar]
- 146.Muyzer G, Stams AFM. 2008. The ecology and biotechnology of sulphate-reducing bacteria. Nat Rev Microbiol 6:441–454. doi: 10.1038/nrmicro1892. [DOI] [PubMed] [Google Scholar]
- 147.Stumm W, Morgan JJ. 2006. Aquatic chemistry: chemical equilibria and rates in natural waters. John Wiley & Sons, New York, NY. [Google Scholar]
- 148.Doran JW. 1982. Microorganisms and the biological cycling of selenium. Adv Microb Ecol 6:1–32. doi: 10.1007/978-1-4615-8318-9_1. [DOI] [Google Scholar]
- 149.Dridge EJ, Watts CA, Jepson BJ, Line K, Santini JM, Richardson DJ, Butler CS. 2007. Investigation of the redox centres of periplasmic selenate reductase from Thauera selenatis by EPR spectroscopy. Biochem J 408:19–28. doi: 10.1042/BJ20070669. [DOI] [PMC free article] [PubMed] [Google Scholar]