SUMMARY
Many studies report the high prevalence of multiply drug-resistant (MDR) strains. Because MDR infections are often significantly harder and more expensive to treat, they represent a growing public health threat. However, for different pathogens, different underlying mechanisms are traditionally used to explain these observations, and it is unclear whether each bacterial taxon has its own mechanism(s) for multidrug resistance or whether there are common mechanisms between distantly related pathogens. In this review, we provide a systematic overview of the causes of the excess of MDR infections and define testable predictions made by each hypothetical mechanism, including experimental, epidemiological, population genomic, and other tests of these hypotheses. Better understanding the cause(s) of the excess of MDR is the first step to rational design of more effective interventions to prevent the origin and/or proliferation of MDR.
INTRODUCTION
Bacterial pathogens that are resistant to multiple drugs represent a growing public health threat, because multiply drug-resistant (MDR) infections are challenging and expensive to treat (1–3), and few antimicrobial compounds, and still fewer antimicrobial agents using novel mechanisms of action, are in clinical development (4). Using both published and unpublished data (5–8), we show that there is often a positive correlation within a bacterial population between resistance to one drug and resistance to one or more other drugs. The high frequency of MDR isolates among resistant strains represents a scientific puzzle: why do resistance determinants aggregate in certain strains of bacteria?
Different underlying mechanisms are traditionally used to explain these observations in different pathogens and are rarely critically assessed or tested. It is unclear whether each bacterial taxon has its own mechanism(s) or to what extent the pathways to the accumulation of multiple resistances are shared among pathogens. Understanding the causes of MDR is necessary for constructing appropriate models to aid in the design and evaluation of potential interventions.
Here we describe the scope of the phenomenon of MDR in bacteria and enumerate possible mechanisms for the appearance and proliferation of MDR bacteria. Importantly, we also propose experimental, epidemiological, population genomic, and other study designs that can help to clarify the roles of these potential mechanisms driving excess MDR.
THE EXCESS OF MDR IN BACTERIA
For many bacterial pathogens, the frequency of MDR pathogens exceeds the product of the frequencies of individual resistance traits. While this phenomenon of excess MDR has rarely been the focus of epidemiological studies, those studies which report the relevant data very often find positive correlations between resistance phenotypes: isolates resistant to one drug are more likely to be resistant to others. Below we briefly review such findings for a range of pathogens, including novel analyses performed for this report.
Streptococcus pneumoniae
McCormick et al. (9) and Link-Gelles et al. (8) studied invasive pneumococcal disease isolates from population-based surveillance in different eras and reported that dual resistance to penicillin and erythromycin is more common than the product of the proportions of resistance to penicillin and erythromycin in the United States. We have expanded their analysis, using the same data set from three epidemiologically distinct periods. We find significant positive correlations between resistance to drugs, including penicillin, erythromycin, tetracycline, clindamycin, trimethoprim-sulfamethoxazole, ceftazidime, and levofloxacin (Table 1).
TABLE 1.
Correlation coefficients of resistance to different drugs used to treat Streptococcus pneumoniae infections in the United Statesa
| Period (n) | Drug | Correlation coefficentb |
||||||
|---|---|---|---|---|---|---|---|---|
| ERY | TET | CLI | SXT | TAX | CAZ | LVX | ||
| Pre-PCV7 (7,571) | PEN | 0.610*** | 0.370*** | 0.266*** | 0.700*** | 0.454*** | 0.037** | |
| ERY | 0.427*** | 0.399*** | 0.574*** | 0.311*** | 0.015 | |||
| TET | 0.574*** | 0.338*** | 0.173*** | 0.009 | ||||
| CLI | 0.229*** | 0.078*** | 0.037** | |||||
| SXT | 0.397*** | 0.029* | ||||||
| TAX | 0.021 | |||||||
| Intermediate PCV7 coverage (16,735) | PEN | 0.613*** | 0.359*** | 0.288*** | 0.626*** | 0.437*** | 0.299*** | 0.026*** |
| ERY | 0.427*** | 0.440*** | 0.559*** | 0.369*** | 0.291*** | 0.045*** | ||
| β-TET | 0.668*** | 0.321*** | 0.230*** | 0.291*** | 0.052*** | |||
| CLI | 0.234*** | 0.198*** | 0.301*** | 0.040*** | ||||
| SXT | 0.389*** | 0.277*** | 0.035*** | |||||
| TAX | 0.727*** | 0.026*** | ||||||
| CAZ | −0.011 | |||||||
| High PCV7 coverage (6,785) | PEN | 0.579*** | 0.460*** | 0.422*** | 0.576*** | 0.440*** | 0.326*** | 0.030* |
| ERY | 0.565*** | 0.577*** | 0.588*** | 0.443*** | 0.338*** | 0.049*** | ||
| TET | 0.819*** | 0.399*** | 0.516*** | 0.415*** | 0.026* | |||
| CLI | 0.343*** | 0.516*** | 0.393*** | 0.035** | ||||
| SXT | 0.454*** | 0.336*** | 0.045*** | |||||
| TAX | 0.727*** | 0.014 | ||||||
| CAZ | 0.019 | |||||||
PCV7, 7-valent pneumococcal conjugate vaccine; PEN, penicillin; ERY, erythromycin; TET, tetracycline; CLI, clindamycin; SXT, trimethoprim-sulfamethoxazole; TAX, cefotaxime; CAZ, ceftazidime; LVX, levofloxacin.
***, P < 0.001; **, P < 0.01; *, P < 0.1.
Enterobacteriaceae
In the United Kingdom, sulfonamide-resistant strains of Escherichia coli were more likely to have resistance to antibiotics, including ampicillin, chloramphenicol, kanamycin, streptomycin, tetracycline, and trimethoprim, than sulfonamide-susceptible strains in both in 1991 and 1999 (6). The proportion of sulfonamide-resistant strains that were resistant to at least two other antibiotics of different chemical classes was also higher than that of sulfonamide-susceptible strains (sulfonamide resistant versus sulfonamide susceptible, 88.1% versus 17.8% in 1991 and 83.0% versus 28.4% in 1999) (6). In the United States, there was a significant association between resistance to fluoroquinolones and plasmid-mediated gentamicin resistance (10). Using data from a tertiary care hospital in the United States, we found that most correlation coefficients of resistance to different drugs used to treat Klebsiella pneumoniae, E. coli, and Pseudomonas aeruginosa infections are significantly positive (Tables 2 to 4).
TABLE 2.
Correlation coefficients of resistance to different drugs used to treat Klebsiella pneumoniae infection in a general hospital in the United Statesa
| Drug | Correlation coefficientb |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | SAM | FEP | FOX | CAZ | CRO | CIP | GEN | IPM | LVX | NIT | SXT | |
| AMK | 0.031 | 0.304*** | 0.266*** | 0.218*** | 0.451*** | 0.442*** | 0.425*** | 0.174*** | 0.247*** | 0.432*** | 0.098** | 0.279*** |
| AMP | 0.098** | 0.042 | 0.064* | 0.066* | 0.067* | 0.073* | 0.042 | 0.027 | 0.072* | 0.319*** | 0.077* | |
| SAM | 0.406*** | 0.417*** | 0.623*** | 0.645*** | 0.558*** | 0.342*** | 0.279*** | 0.542*** | 0.183*** | 0.466*** | ||
| FEP | 0.356*** | 0.603*** | 0.605*** | 0.458*** | 0.270*** | 0.447*** | 0.452*** | 0.081** | 0.366*** | |||
| FOX | 0.410*** | 0.400*** | 0.436*** | 0.227*** | 0.379*** | 0.446*** | 0.180*** | 0.213*** | ||||
| CAZ | 0.909*** | 0.683*** | 0.414*** | 0.367*** | 0.668*** | 0.160*** | 0.447*** | |||||
| CRO | 0.696*** | 0.464*** | 0.382*** | 0.672*** | 0.165*** | 0.489*** | ||||||
| CIP | 0.410*** | 0.308*** | 0.982*** | 0.174*** | 0.472*** | |||||||
| GEN | 0.207*** | 0.375*** | 0.103*** | 0.374*** | ||||||||
| IPM | 0.314*** | 0.086** | 0.232*** | |||||||||
| LVX | 0.169*** | 0.483*** | ||||||||||
| NIT | 0.127*** | |||||||||||
Data source, WHONET (n = 1,095). AMK, amikacin; AMP, ampicillin; SAM, ampicillin-sulbactam; FEP, cefepime; FOX, cefoxitin; CAZ, ceftazidime; CRO, ceftriaxone; CIP, ciprofloxacin; GEN, gentamicin; IPM, imipenem; LVX, levofloxacin; NIT, nitrofurantoin; SXT, trimethoprim-sulfamethoxazole.
***, P < 0.001; **, P < 0.01; *, P < 0.1.
TABLE 4.
Correlation coefficients of resistance to different drugs used to treat Pseudomonas aeruginosa infection in a general hospital in the United Statesa
| Drug | Correlation coefficientb |
|||||||
|---|---|---|---|---|---|---|---|---|
| CAZ | IPM | ATM | CIP | LVX | GEN | TOB | AMK | |
| PIP | 0.574*** | 0.305*** | 0.343*** | 0.208*** | 0.183*** | 0.158*** | 0.185*** | 0.067 |
| CAZ | 0.286*** | 0.331*** | 0.212*** | 0.163*** | 0.221*** | 0.162*** | 0.234*** | |
| IPM | 0.010 | 0.292*** | 0.243*** | 0.333*** | 0.415*** | 0.161*** | ||
| ATM | −0.022 | 0.001 | 0.000 | 0.040 | 0.004 | |||
| CIP | 0.759*** | 0.348*** | 0.353*** | 0.126** | ||||
| LVX | 0.244*** | 0.259*** | 0.102* | |||||
| GEN | 0.673*** | 0.575*** | ||||||
| TOB | 0.292*** | |||||||
Data source, WHONET (n = 614). PIP, piperacillin; ATM, aztreonam; TOB, tobramycin.
***, P < 0.001; **, P < 0.01; *, P < 0.1.
TABLE 3.
Correlation coefficients of resistance to different drugs used to treat Escherichia coli infection in a general hospital in the United Statesa
| Drug | Correlation coefficientb |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | SAM | FEP | FOX | CAZ | CRO | CIP | GEN | IPM | LVX | NIT | SXT | |
| AMK | −0.009 | 0.012 | 0.084*** | 0.032 | 0.023 | 0.012 | 0.050** | 0.009 | 0.101*** | 0.050** | −0.017 | 0.002 |
| AMP | 0.540*** | 0.096*** | 0.130*** | 0.132*** | 0.177*** | 0.130*** | 0.144*** | −0.025 | 0.132*** | −0.128*** | 0.207*** | |
| SAM | 0.138*** | 0.218*** | 0.188*** | 0.137*** | 0.117*** | 0.221*** | 0.010 | 0.119*** | −0.083*** | 0.100*** | ||
| FEP | 0.197*** | 0.622*** | 0.531*** | 0.263*** | 0.148*** | 0.172*** | 0.263*** | 0.074*** | 0.072*** | |||
| FOX | 0.334*** | 0.352*** | 0.281*** | 0.071*** | 0.103*** | 0.289*** | 0.086*** | −0.022 | ||||
| CAZ | 0.728*** | 0.273*** | 0.142*** | 0.161*** | 0.273*** | 0.084*** | 0.058** | |||||
| CRO | 0.374*** | 0.121*** | 0.113*** | 0.376*** | 0.080*** | 0.124*** | ||||||
| CIP | 0.302*** | 0.054** | 0.994*** | 0.037 | 0.176*** | |||||||
| GEN | 0.003 | 0.299*** | 0.028 | 0.202*** | ||||||||
| IPM | 0.054** | −0.007 | 0.032 | |||||||||
| LVX | 0.039* | 0.178*** | ||||||||||
| NIT | 0.027 | |||||||||||
Data source, WHONET (n = 2,731).
***, P < 0.001; **, P < 0.01; *, P < 0.1.
Neisseria gonorrhoeae
Fluoroquinolone resistance is associated with resistance to penicillin and tetracycline in N. gonorrhoeae in the United States during 2002 to 2007 (7). Ota et al. (11) also reported that in Ontario, Canada, in 2006, fluoroquinolone-resistant strains were more likely to be resistant to penicillin, tetracycline, and erythromycin than fluoroquinolone-sensitive strains (fluoroquinolone-resistant versus fluoroquinolone-sensitive, 98.4% versus 89.4% for penicillin, 98.0% versus 81.1% for tetracycline, and 66.2% versus 14.8% for erythromycin). We found that resistance to penicillin, tetracycline, and fluoroquinolones are positively correlated in the United States, using data from CDC's Gonococcal Isolate Surveillance Project (GISP) 2011 Annual Report (12) (correlation coefficients of 0.371, 0.416, and 0.452 for penicillin and tetracycline, penicillin and ciprofloxacin, and tetracycline and ciprofloxacin, respectively [P values could not be calculated due to the lack of sample size information]).
Mycobacterium tuberculosis
We calculated the correlation coefficients of resistance to different drugs used to treat tuberculosis (TB) by using the data from 114 countries in Anti-Tuberculosis Drug Resistance in the World: Fourth Global Report (13, 14). We found strong positive correlations between resistance to isoniazid, rifampin, ethambutol, and streptomycin (Table 5).
TABLE 5.
Correlation coefficients of resistance to different TB drugs among pretreatment casesa
| Drug | Correlation coefficientb |
||
|---|---|---|---|
| RMP | EMB | STM | |
| INH | 0.683 | 0.554 | 0.559 |
| RMP | 0.621 | 0.481 | |
| EMB | 0.417 | ||
INH, isoniazid; RMP, rifampin; EMB, ethambutol; STM, streptomycin.
All of the cases have P values of < 0.001 (n = 18,619).
Staphylococcus aureus
We found positive correlations between resistance to antibiotics used to treat S. aureus infection, including penicillin, erythromycin, clindamycin, tetracycline, levofloxacin, gentamicin, and trimethoprim, using data from a tertiary care hospital in the United States (Table 6). The significant positive correlations between resistance to drugs used to treat S. aureus infection were also identified in the province of British Columbia, Canada, in 2012 (15) (correlation coefficients of 0.229, 0.550, 0.054, and 0.052 for methicillin versus clindamycin, erythromycin, trimethoprim-sulfamethoxazole, and tetracycline, respectively [n = 5,214; P < 0.001 for all cases]).
TABLE 6.
Correlation coefficients of resistance to different drugs used to treat Staphylococcus aureus infection in a general hospital in the United Statesa
| Drug | Correlation coefficientb |
|||||||
|---|---|---|---|---|---|---|---|---|
| OXA | ERY | CLI | TCY | GEN | LVX | SXT | RIF | |
| PEN | 0.380*** | 0.350*** | 0.153*** | 0.050* | 0.046* | 0.248*** | 0.056** | 0.027 |
| OXA | 0.614*** | 0.415*** | 0.017 | 0.094*** | 0.733*** | 0.047* | 0.123*** | |
| ERY | 0.420*** | 0.052* | 0.065** | 0.603*** | 0.054** | 0.094*** | ||
| CLI | 0.124*** | 0.094*** | 0.520*** | 0.101*** | 0.085*** | |||
| TCY | 0.256*** | 0.036 | 0.185*** | 0.006 | ||||
| GEN | 0.111*** | 0.304*** | 0.109*** | |||||
| LVX | 0.109*** | 0.140*** | ||||||
| SXT | 0.064** | |||||||
Data source, WHONET (n = 2,377). OXA, oxacillin; TCY, tetracycline; RIF, rifampin.
***, P < 0.001; **, P < 0.01; *, P < 0.1.
In summary, these data show positive correlation between resistance to different drugs and the higher-than-expected proportion of MDR for Gram-positive and Gram-negative bacteria and mycobacteria. The universality of excess MDR, involving resistance conferred by a range of genetic and biochemical mechanisms, raises the question of a shared mechanism(s) driving these commonly observed patterns.
It should be noted that the association between resistance to multiple drugs and a higher-than-expected proportion of MDR shown here is based purely on phenotype. Certainly, the genetic causes of resistance to antibiotics or classes of antibiotics can vary even within a species (for example, different classes of extended-spectrum beta-lactamases in species of the Enterobacteriaceae [16] or the efflux pump [mef] and ribosomal methylase [erm] mechanisms of macrolide resistance in streptococcal species [17]). However, the aggregation of resistance phenotypes within certain subgroups of a species is a clinical problem and a scientific phenomenon worthy of understanding, even if the genetic causes may vary.
MECHANISMS LEADING TO EXCESS MDR
The excess of MDR could be caused by unexpectedly high rates of origin, high rates of spread of MDR strains or determinants, or both. A major complicating factor is the possibility of horizontal gene transfer, which can disseminate resistance to multiple antibiotics in a single step. However, it is conceptually useful to separate the explanations for MDR bacteria into two phases: origin and spread. It should be noted that the explanations we list here for each phenomenon are not mutually exclusive.
Explanations for the Origin of MDR Strains
Single biochemical mechanism conferring resistance to multiple drugs.
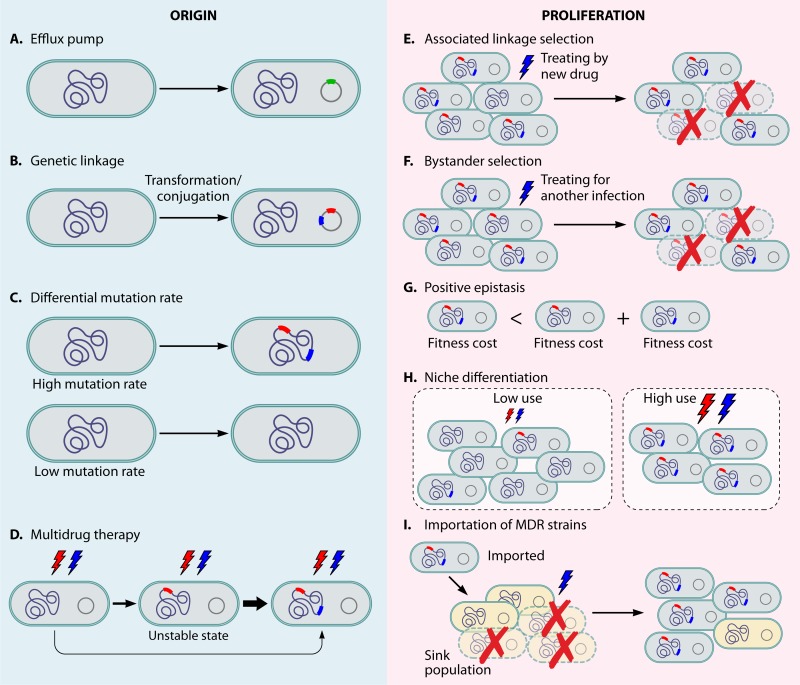
The simplest explanation for observing an excess of MDR is that a single biochemical mechanism confers resistance to more than one drug (Fig. 1A). An example is that of bacterial efflux pumps (18, 19), which extrude antibiotics out of cells such that the intracellular antibiotic concentration decreases and resistance to the antibiotics occurs. Some efflux systems are antibiotic specific, but others confer resistance to multiple drug classes (19). Typically, efflux pumps provide low-level drug resistance (20, 21). Another example is cell wall thickening in S. aureus that resulted in resistance to vancomycin and daptomycin, antibiotics with relatively large molecular sizes (22).
FIG 1.
The mechanisms for the origin and proliferation of MDR strains. Red and blue lightning bolts indicate treatments with drug A and drug B. Green rectangles represent efflux pump genes. Red and blue rectangles represent drug A and drug B resistance determinants, respectively. (A) Efflux pump. Bacteria obtaining efflux pumps (green square) that extrude more than one antibiotic out of cells confers MDR. (B) Genetic linkage. If two resistance determinants are located in the same horizontally transferred element, when a strain acquires one resistance phenotype, it acquires both. (C) Differential mutation rate. Highly mutable lineages have higher frequencies of acquiring multiple drug resistance determinants than those that are not highly mutable. (D) Multidrug therapy with accelerated treatment failure in resistant infections. If treatment fails and singly resistant strains emerge, they are likely to obtain second drug resistance and be replaced. The thickness of the arrows reflects the relative transition probabilities between states. (E) Associated linkage selection. Resistance to a new drug (blue) occurs on a background of resistance to an older drug (red) following a change in treatment practices from drug A to drug B. The resistance to the older drug continues spreading because of the linkage to the resistance of the new drug and the selective pressure from the usage of the new drug. (F) Bystander selection. Resistance in one bacterium is advantageous because it allows strains of that species to survive when the host is treated for another infection with a drug that also kills that species. For example, if drug A is used to treat this species and drug B is used to treat another infection in the same patients, MDR strains survive and strains resistant only to drug A are killed. (G) Positive epistasis. If the cost of MDR is smaller than the total cost of each resistance determinant on its own, MDR strains may outcompete strains with a limited number of resistance elements and spread more quickly. (H) Niche differentiation. Multiple unrelated drug classes may be used more frequently in certain population subgroups, resulting in an excess of MDR when the high- and low-use subgroups/settings are considered together. (I) Importation of MDR strains. The MDR strains from a high-drug-use “source” population are introduced into a lower-drug-use “sink” population and are able to spread as a result of competing successfully with pan-susceptible strains due to their resistance to the drug (blue) used in the sink population.
(i) Testable predictions.
Standard bacterial genetics (knockout and complementation or overexpression), combined with measurement of corresponding strains' MICs (the lowest concentration of an antibiotic that inhibits growth of a microorganism) for various drugs, can confirm or reject the hypothesis that a single biochemical mechanism confers multiple resistance. In the case where this is the only mechanism conferring resistance, the phenotypes should be found together without exception. For example, the deletion of the efflux pump gene ifrA in Mycobacterium smegmatis decreased the MIC (that is, increased the susceptibility) to multiple drugs, and the overexpression of the same gene increased resistance to multiple drugs (23).
Another testable hypothesis is that the diversity and intensity of antimicrobial use in settings such as hospitals select for genetic changes that expand the substrate specificity of resistance mechanisms. In vitro evolution of plasmid-borne TEM beta-lactamases with alternating exposure to a penicillin and a cephalosporin was shown to select for dual specificity for these two related compounds; moreover, the sequence changes that evolved mimicked those observed in human isolates, while exposure of plasmid-bearing bacteria to only one of the two antimicrobial compounds produced sequence changes in the beta-lactamase that were not previously observed in human isolates (24).
(ii) Practical implications.
Development of efficient efflux pump inhibitors may lead to the renewed effectiveness of several drugs in treating infections for which this mechanism is important (25, 26). For example, verapamil, an efflux pump inhibitor, has been shown to increase antituberculosis drug efficacy in mice (27). It should, however, be noted that efflux pump inhibitors should have limited effects on the host to have clinical utility. Those clinically available, such as verapamil and reserpine, have host pharmacological effects that limit their utility as antimicrobial therapies (25, 28).
Genetic linkage.
MDR strains may arise because determinants of resistance to multiple drug classes are genetically linked, because they are either physically close on bacterial chromosomes (thus coinherited vertically and potentially cotransformed when shared horizontally) or on the same horizontally transmitted element, such as a plasmid or conjugative transposon (Fig. 1B). In such cases, when a strain acquires one resistance phenotype, it acquires many.
Horizontally transmitted elements, such as transposons, integrons, and plasmids, are often modular and can incorporate new elements over time (29, 30). Transposons contain genes encoding transposases that facilitate incorporation to and from other genomic regions (18). Integrons accumulate gene cassettes with specific recombination sites through site-specific recombinases encoded by the integrons themselves. Transposons and integrons may appear on the chromosome or on a plasmid. These elements may be transmitted within and between species through transformation, transduction, and/or conjugation (reviewed in reference 18).
The genetic mechanisms (cassette-based recombination, conjugation, and transposition) underlying these mobile elements are increasingly well understood (29, 30); however, the selective pressures that keep these resistance elements together (despite possible fitness costs for some of the genes involved) are less clear.
(i) Examples.
Conjugative transposon Tn1545 in S. pneumoniae contains genes for resistance to multiple antibiotics, including kanamycin, macrolide-lincosamide-streptogramin B-type antibiotics, and tetracycline, and is capable of transferring to a new bacterial host cell via conjugation or transposition to another genomic region (31). Integrons in Enterobacteriaceae carry genes encoding resistance to unrelated antibiotics such as beta-lactams, aminoglycosides, sulfonamides, and chloramphenicol (32). There are also examples of plasmids without integrons in Enterobacteriaceae carrying antibiotic resistance genes for multiple drug classes, such as aminoglycosides, beta-lactams, tetracycline, chloramphenicol, and sulfamethoxazole (33).
(ii) Testable predictions.
In contrast with multiple resistance mechanisms encoded by a single gene or operon, these determinants are not necessarily always found in combination with one another but are encoded by genetic determinants that are adjacent on the chromosome or plasmid. This prediction can be tested by knockout, complementation, and/or sequencing of chromosomes or plasmids. Deleting each gene and comparing the MICs before and after deletion can test whether genes on the same chromosome or plasmid confer resistance to different drugs. If genes conferring resistance to each drug are known, sequencing of chromosomes or plasmids can confirm the genetic linkage of these genes on the same chromosome or plasmid.
(iii) Practical implications.
The initial appearance of multiple-drug resistance on a single genetic element likely occurs when bacteria resistant to the component drugs come into proximity or contact, such that horizontal transfer and recombination can occur. In principle, an antibiotic management strategy might be designed to reduce the probability of such contacts. However, theoretical studies of one such approach, known as antimicrobial cycling, have shown that cycling reduces such contacts only under restricted conditions (34); in practice, cycling strategies are difficult to implement and to study (35). Recently, an alternative strategy, “adjustable cycling,” in which treatment is changed when not effective in patients, has been proposed and shown in a theoretical model to suppress the emergence of MDR in most settings (36).
The clinical impact of these linked determinants depends on their persistence as a linked group. Improved understanding of the selective pressures that preserve and allow the proliferation of these multidrug resistance elements may improve our ability to reduce their frequency.
Highly mutable or recombinogenic bacterial lineages.
Bacterial lineages vary in their rates of mutation. Such variation is often due to variation in genes involved in DNA proofreading, such as the mismatch repair system (37). Additionally, certain bacterial lineages vary in their ability to accept and integrate transforming DNA (38). Because mutation and recombination are the sources of drug resistance genes and alleles, more highly mutable or highly “recombinogenic” lineages should have higher frequencies of multiple-drug resistance determinants, so that in the population as a whole, these lineages would likely contribute disproportionately to the frequency of multiple-drug resistance (Fig. 1C).
(i) Examples.
Such a process has been proposed for the accumulation of MDR in highly mutable lineages, such as the Beijing lineage, in tuberculosis (39). Combining mathematical modeling (40) with experimental measurements of resistance mutation rates, Ford et al. (39) argued that higher mutation rates in some strains could lead to the presence of multiply drug-resistant variants within a patient at the start of treatment. Such preexisting resistant variants could increase the risk of treatment failure.
In S. pneumoniae, resistance to multiple drugs is more common in lineages showing evidence of higher rates of recombination (41), a finding that has been supported by population genomic studies (38, 42, 43).
(ii) Complexities.
Antibiotic exposure at sublethal concentrations can induce elevated mutation rates in bacteria (44, 45). Therefore, antibiotic exposure, which is generally seen as a selective force, may also play a role in the generation of mutations. As a consequence, mutation rates during treatment (especially when drug concentrations are low) may be elevated compared to those measured in antibiotic-free settings. This mutation rate increase is not specific to mutations conferring resistance to the drug administered but rather is a general elevation due to DNA damage caused by reactive oxygen species created by the bacteria under antibiotic stress (45).
Mutation and recombination rates are also traits under selection in their own right. Selection in changing environments (46–48) and specifically for resistance to multiple antibiotics (49) may provide second-order selection for elevated mutation rates. Thus, high mutation rates may be both a cause and a consequence of multiple-drug resistance.
(iii) Testable predictions.
A straightforward test of the hypothesis that higher rates of mutation and recombination lead to faster accumulation of multiple-drug resistance is to assess whether present-day MDR strains show elevated rates of mutation or recombination compared to those of non-MDR strains. It may be possible to identify susceptible ancestral strains to currently observed MDR strains by phylogenetic analysis, and the mutational or recombinational assays could then be performed on these ancestral strains if historical samples are available.
If the variation in recombination rate contributes to the high level of MDR, strains with higher recombination rate are expected to acquire MDR with a higher probability. Recombination events can be inferred in a bacterial lineage from the existence of tracts of high densities of single-nucleotide polymorphisms (SNPs), indicating import of genetic material from a diverged source (43). Those sites at which recombination has not occurred can then be used to estimate a phylogeny and molecular clock by conventional means, and inferred recombination events can be mapped onto the tree topology for estimating the ratio of recombination to mutation rate (for an example, see reference 43). These analyses may be performed using the software packages ClonalFrame and Gubbins (43, 50, 51). It should be noted that this approach assumes that the genomes under analysis are closely related, that there has not been time for extensive clustering of SNPs to have been produced in genes under selection, and that anomalous concentrations of SNPs are therefore best explained by recombination with a divergent donor strain that is not in the data set.
If variation in mutation rate leads to the excess of MDR, these MDR strains are expected to show higher substitution rates at synonymous sites or other neutral sites such as pseudogenes, because neutral theory predicts that the substitution rate of neutral sites equals the mutation rate (52). Population genetic tools, such as the PAML package (53), can be used to estimate lineage-specific substitution rates when sequences from both an outgroup and strains of interest are available. However, the amounts of homologous recombination thought to occur in many bacterial species presents significant challenges to phylogenetic and population genetic analyses (54) and may produce considerable biases in estimates of mutation rate (55).
While elevated mutation or recombination rates in MDR lineages (39) are evidence in favor of this mechanism, the absence of higher rates in MDR strains does not disprove it. For example, recombination or mutation rates may have varied over the evolutionary history of the lineage, such that multiple resistance accumulated while the lineage was highly mutable/susceptible to recombination, but this trait may have changed since the resistance determinants were acquired; this is a particular risk for highly changeable lineages (56).
(iv) Practical implications.
The practice of combination treatment with multiple drug classes, described a century ago by Ehrlich (57), remains a standard approach for treatment of infections, such as tuberculosis, in which resistance occurs primarily by mutation and mutants resistant to any single drug are expected to be present in many infected hosts (58). If it were possible to assess the genetic background, and hence the likely mutation rate, of a pathogen infecting an individual, it is possible that the number or dose of drugs would be increased in treating highly mutable lineages to counteract the risk of preexisting MDR strains in a predominantly drug-susceptible infection. While fanciful at the present, as rapid pathogen genome sequencing becomes more common in diagnostic microbiology, the practicality of this approach may also increase (59, 60).
Multidrug therapy with accelerated treatment failure in resistant infections.
As noted in the preceding section, multidrug therapy has long been recommended for treating infections in which resistance is acquired by mutation to ensure that mutants that are resistant to one drug are killed by other drugs in the “cocktail” (57). This strategy is motivated by the idea that the probability of obtaining mutants resistant to multiple drugs with different mechanisms is much smaller than that of obtaining singly resistant mutants, and using drug combinations is more likely to kill all the bacteria, preventing the emergence of drug resistance. In particular, if the product of the expected frequency of bacteria resistant to all drugs being used (P) and the bacterial population size (N) is much less than one (NP ≪ 1), most hosts will harbor no bacteria which are resistant to every drug being taken. This calculation is typically made on the assumptions that the infecting inoculum is susceptible to all drugs in the regimen and that any resistant mutants will arise during the course of replication of the bacterial population within the host, at a predictable frequency (40). If, however, the infecting inoculum is already composed of bacteria resistant to one or more of the drugs used, the frequency of bacteria resistant to all the other drugs will be considerably higher than assumed and possibly high enough to make NP ≥ 1, indicating that a mutant resistant to all drugs used may be present and able to replicate. In simple terms, the emergence of multiple resistance is much more likely from a singly resistant precursor than from a pan-susceptible precursor; the first resistance mutation may form a “slippery slope” facilitating the emergence and selection of further resistance mutations. In this mechanism, under multidrug therapy, the singly resistant state is unstable, and those strains will be replaced by strains that have acquired mutations for additional drug resistance, leading to an excess of MDR (61, 62) (Fig. 1D).
(i) Example.
As drug resistance in M. tuberculosis is generally caused by spontaneous mutations (63) and the population of M. tuberculosis within a patient is large enough to generate drug-resistant mutants (64), multidrug chemotherapy is typically used for treating tuberculosis (58) in order to reduce the probability of emerging drug resistance.
One problem that has been observed in treatment of tuberculosis is the existence of mixed infections with a subpopulation of resistant organisms, which may be undetected when treatment is initiated but is rapidly selected by first-line treatment tailored to the assumed full susceptibility of the infecting strain (61, 62).
Even starting from a susceptible inoculum, singly resistant strains (or in rare cases, even doubly resistant strains [40]) could be present due to patient noncompliance with the treatment regimen, such as interrupted antibiotic treatment or insufficient dosage (65). Directly observed therapy for tuberculosis improves patient adherence and decreases the frequency of acquired drug resistance (66).
(ii) Testable predictions.
Evidence for the importance of this mechanism would include a high frequency of MDR (but not singly resistant strains) developing during multidrug therapy of infections for which the drug susceptibility of the initial infection was unknown or mismeasured, perhaps due to a mixed infection or a reinfection (61, 62). If genetic mechanisms of drug resistance are known, sequences of bacteria isolated at different time points from the same patients could be used to test this hypothesis. It is expected under this mechanism that singly resistant strains were present at the beginning of treatment and MDR emerged later on, and thus a gene(s) or mutation(s) conferring single-drug resistance is expected to be found in sequences of isolates from the beginning of treatment. Of course, if the singly resistant infection were known at the time of treatment initiation, it would be inappropriate to treat it as if it were pan-susceptible. Thus, to know about this problem (in a timely fashion) is to try to prevent it, and only retrospective investigations are likely to find evidence for this problem (61, 62).
(iii) Practical implications.
Standard WHO-recommended retreatment regimens for tuberculosis have historically called for addition of a single drug to a failing four-drug regimen. Such an approach resulted in many treatment failures due to the acquisition of resistance to the single effective drug that was added (67, 68).
In settings where this mechanism plays a role, in practice mainly tuberculosis, there is a special value of rapid diagnostics that can assess drug susceptibility at baseline, such as GeneXpert MTB/Rif (69) or rapid pathogen genome sequencing, because such diagnostics could confirm that the majority population of the infecting organism has susceptibility to enough drugs to prevent the emergence of further resistance (perhaps requiring addition or substitution of drugs in the standard regimen, when resistance to one or more drugs is found at baseline). This diagnostic information would not only improve patient outcomes but also reduce the rate at which new multiresistant strains are generated. In the absence of such baseline diagnostics, the use of extra drugs in a multidrug regimen might be warranted when the presence of resistance to one or more drugs in the patient is suspected due to population history or patient risk factors. Because singly drug-resistant strains are more likely to arise due to patient noncompliance with the treatment regimen, directly observed therapy may help to reduce the possibility of the excess of MDR due to treatment failure.
Explanations for the Proliferation of MDR Strains
Once MDR bacteria have emerged in one or more hosts, their proliferation depends on their ability to survive and be transmitted to other hosts, and changes in their relative frequency reflect natural selection—their differential survival and transmission compared to other lineages. In this section, we consider five mechanisms that can provide a selective advantage to MDR strains, leading to increases in their frequency.
Associated linkage selection.
The proliferation of a gene (and the strain harboring it) need not be the consequence of direct selection upon that gene and the trait it encodes but may result from selection of others that are inherited along with it. This is the phenomenon of linkage, and it is especially hard to disentangle in partially clonal organisms like most bacteria. If resistance to a new drug occurs on a genetic background of resistance to older drugs following a change in treatment practices, then resistance to the older drugs can continue spreading because of the linkage to the resistance to the new drug, selected by use of the new drug (Fig. 1E). Resistance to new drugs is likely to arise on the background of resistance to the older drugs because the frequency of older drug resistance is likely to be high due to longstanding selection pressure imposed by usage of the older drugs (70). This can be exacerbated if particular genetic backgrounds are more able to tolerate the fitness costs of resistance determinants, either because the fitness costs are lower in these backgrounds or because these backgrounds have higher fitness to begin with and hence can better tolerate a given fitness cost (see also “Positive epistasis between drug resistance determinants or between resistance determinants and genetic background” below).
(i) Examples.
Regarding E. coli, although sulfonamide prescriptions in the United Kingdom decreased greatly, the proportion of sulfonamide-resistant E. coli did not decline (6). One possible explanation is the close linkage between sulfonamide resistance genes and other antibiotic resistance determinants and their continued selection through the usage of other antibiotics. sulII genes encoding sulfonamide resistance in E. coli are located on plasmids carrying several resistance determinants (6). A follow-up study 5 years later reported the persistence of sulfonamide resistance and the continued association between sulfonamide resistance and resistance to other drugs (71).
In S. pneumoniae infections, antibiotics other than penicillin, such as azithromycin (72), erythromycin, trimethoprim-sulfamethoxazole (73, 74), and cephalosporins (75), may select more efficiently for penicillin-resistant strains than penicillin itself. While the MICs associated with resistance to azithromycin are farther above clinically achievable concentrations, penicillin dosing can be increased to combat less-susceptible strains and still have limited effects on toxicity experienced by the patient. As clinically achievable levels of macrolides (such as azithromycin) kill susceptible but not resistant bacteria while clinically achievable levels of penicillin may have some inhibitory effect on resistant strains, macrolides are more selective killers of susceptible bacteria than penicillin. If most resistance is multidrug resistance, then penicillin resistance will be coselected with macrolide resistance. This is exacerbated in drugs with a long half-life (e.g., azithromycin), which allows them to sustain high concentrations for a longer period.
(ii) Testable predictions.
Associated linkage selection should be tested as a mechanism for persistence of resistance to previously used drugs after the use of such drugs is reduced in a population. If the persisting lineages are resistant to both the previously used drug and the drug(s) that is used in its place, then this could be evidence of associated linkage selection. Within individuals, the selective role of one drug in promoting resistance to another, unrelated drug may be assessed by comparing the prevalence of resistance to the second drug in individuals treated with the first, as in the S. pneumoniae examples described above.
In ecological studies of the relationship between antimicrobial use and resistance, associations between use of one drug and population-level resistance to another are expected when associated linkage selection is at play. However, given the positive correlations across populations between high use of some antimicrobial classes and high use of other classes, such data may also reflect direct effects of each drug class on resistance to itself. For this reason, individual-level data, such as those described above for S. pneumoniae, are more informative.
Using sequence data from good-quality longitudinal samples documenting the emergence of resistance, phylogenetic analysis could also be used to test this hypothesis. If resistance to the new drug arose in a background of resistance to older drugs, we would see in phylogenetic trees that the strains with the new drug resistance would coalesce with the older drug-resistant strains more recently than the coalescence between the older drug-resistant and older drug-susceptible strains. Such inference may be more complicated when resistance is encoded by mobile genetic elements.
(iii) Practical implications.
In principle, recommendations for drug choice for treating a particular infection could be changed when resistance to a currently used drug is still low, so the new drug resistance is less likely to happen in the genetic background of resistance to older drugs. The practicality of this idea is limited by the paucity of alternative classes of drugs available, and it should be noted that such a policy (i.e., recommending a new first-line treatment when the frequency of resistance to the current treatment exceeds 5%) (76) has not prevented the success of MDR strains of N. gonorrhoeae in the United States. (The effectiveness may be reduced by importation of MDR strains; see “Importation of MDR strains and geographic source-sink dynamics” below.) The potential of this approach, i.e., changing drug choice recommendations when resistance to a currently used drug is low, likely depends on factors such as the relative fitness advantages of different resistance patterns. Modeling may help determine any circumstances in which this strategy is advantageous.
Perhaps a more effective strategy, when drugs vary in their tendency to select MDR strains within a host during treatment, is to reduce selection for resistance by choosing a drug regimen that is less likely to select for MDR strains while maintaining treatment efficacy. For example, it has been suggested that choosing amoxicillin-clavulanate over azithromycin can accomplish these twin goals in treatment of acute otitis media (77). Here, as described above, the explanation seems to be that MICs of strains resistant to amoxicillin-clavulanate are typically close to clinically achievable concentrations, so there may be some effect of this combination on even the “resistant” strains, whereas azithromycin-resistant strains have MICs far above in vivo concentrations and hence are little affected by treatment.
There has been much discussion of the need for new diagnostics which would permit rapid assessment of the resistance phenotype of an infection, permitting tailoring of the treatment to the individual resistance phenotype, rather than the “conservative” approach of empirical therapy with a drug that is statistically likely to be effective in the absence of knowledge of the susceptibility profile of a particular patient's infection (78). Predictions of theoretical models may help to assess the effects of such diagnostics and treatment protocols on the selection of singly and multiply resistant strains. One model of the impact of the GeneXpert MTB/RIF diagnostic for tuberculosis infection and rifampin resistance indicated that the system's use would reduce the absolute prevalence of MDR TB, but because the projected effect on reducing the prevalence of drug-susceptible forms of TB was more dramatic, the relative proportion of MDR in the TB population would increase (2). This finding emphasizes the point that reducing disease burden overall, specifically MDR disease burden, is the major goal of public health interventions. While the proportion of disease burden that is MDR may be easier to measure, reducing this proportion should rarely if ever be a goal in itself, given that one can reduce MDR and non-MDR disease and have either an increase or decrease in the proportion of MDR.
Bystander selection.
When a drug is used to treat infection with a particular species, other species carried by that same host (“bystanders”) may be affected by the treatment, and multidrug resistant variants of these bystander species may have an advantage under a varying regimen of treatments experienced by different hosts (Fig. 1F).
(i) Examples.
Selection for resistance to antimicrobials in commensal organisms, such as the normal flora of the digestive and upper respiratory tracts, must occur by this mechanism, since commensals by definition are not causing infections yet are subject to selection by systemic antibiotics (79, 80). During broad-spectrum antimicrobial therapy, the proportion of resistant strains of commensal organisms in the gut may increase (81). The selective agent need not be even an antimicrobial used for treatment but may be another selective agent that favors a trait linked to drug resistance in commensals. For example, exposure of commensal gut flora to mercury during installation and removal of dental fillings selected for drug-resistant strains that were also mercury resistant (82). Genes conferring resistance to environmental hazards such as heavy metals are often transferred between lineages together with antibiotic resistance genes on plasmids (83).
Penicillins and tetracyclines were recommended for treating gonorrhea in the United States prior to 1993 and then were replaced by fluoroquinolones and later by cephalosporins. During the period of the rise in the proportion of fluoroquinolone-resistant N. gonorrhoeae strains, the MDR strains resistant to fluoroquinolones, penicillin, and tetracycline increased faster than other fluoroquinolone-resistant types even though penicillins and tetracyclines were no longer recommended for treatment. It has been suggested that during the same period, although penicillins and tetracyclines were not recommended for treating gonorrhea, patients with asymptomatic gonorrhea might have been treated with penicillins and/or tetracyclines for infections other than gonorrhea, and therefore MDR N. gonorrhoeae strains were selected due to their resistance to penicillin and tetracycline (7), a suggestion that has also been proposed in the United Kingdom (84).
While bystander selection is most often considered for commensal or asymptomatically infecting organisms, it may also occur when an infection is undiagnosed or misdiagnosed, and treatment is directed at the (nonexistent) infection the patient is thought to have. A major driver of fluoroquinolone resistance in M. tuberculosis may be the use of fluoroquinolone monotherapy among individuals with unrecognized pulmonary tuberculosis being treated for presumptive community-acquired pneumonia (85). This can lead to multiple resistance if the M. tuberculosis strains were already resistant to other drugs or later become so during treatment.
(ii) Testable predictions.
In assessing the relationship between antimicrobial use and resistance in a particular pathogen, it may be more relevant to consider total prescriptions than to consider prescriptions for treatment of that pathogen, a practice that has been common in studies of commensal colonizing bacteria (62, 86, 87) but has not to our knowledge been examined, for example, in the setting of sexually transmitted diseases. If bacteria of species A resistant to drugs that are no longer used for treatment of species A infections are found to persist especially in settings where these drugs are used for treatment of other infections, and resistance mutations are deleterious, bystander selection would be a likely hypothesis to explain their persistence. More generally, a prediction of this mechanism is that when comparing bacterial populations across space and/or time, the prevalence of particular resistance phenotypes should be positively correlated across different named bacterial species, a phenomenon that has been called coresistance (88).
(iii) Practical implications.
The existence of bystander selection is cited as a reason to prefer narrow-spectrum antimicrobials that can target the infecting pathogens without affecting bystanders. For example, current U.S. guidelines for treatment of uncomplicated urinary tract infections recommend narrow-spectrum agents for this reason, which is broadly termed avoidance of “collateral damage” (89).
Much of the clinical judgment around antimicrobial stewardship and avoiding unnecessary use comes from a concern about the bystander effects on the individual's commensal flora (90). While they are correct for broad-spectrum antibiotics, such concerns are not evidence based in the case of narrow-spectrum anti-infective agents, including many antituberculosis drugs and antiviral drugs. There are many reasons not to overuse such drugs, including side effects and cost. However, inadvertent oseltamivir treatment of an individual not infected with influenza virus, for example, is unlikely to have any effect on resistance: if the infection is not there and the bystander flora are not affected by oseltamivir, there is no species on which bystander selection can act.
Positive epistasis between drug resistance determinants or between resistance determinants and genetic background.
Fitness is defined as the ability to survive and leave offspring in the population. The presence of resistance has been shown in some (but not all) cases to lead to a “fitness cost,” reducing the growth and survival of resistant strains in the absence of antibiotics (91). A fundamental factor determining the success of combinations of these resistance genes is how they interact in epistasis. Epistasis occurs when the combined fitness effect of multiple alleles from different loci is different from the sum of the individual allele effects. Epistasis is widespread in eukaryotes (92, 93), bacteria (94), and viruses (95, 96). If the cost of MDR in the absence of antimicrobial use is smaller than the total cost of each resistance determinant on its own (positive epistasis), MDR strains may outcompete strains with a limited number of resistance elements and spread more quickly (Fig. 1G). [In a continuous-time model, positive epistasis is defined by comparing the fitness cost of MDR and the “sum” of the fitness cost of each resistance determinant; in a discrete-time model, assuming that the costs of resistance to two drugs are c1 and c2, no epistasis means that the cost of MDR is equal to 1 − (1 − c1)(1 − c2).]
Alternatively, if drugs that are used have interactions with each other (97) and the combined effect of multiple drugs is higher than the total of the individual effects (“synergistic effects”), the selective pressure of MDR is greater than would be expected if the effects of drugs were additive. This may create epistatic fitness interactions between resistance determinants in the setting of multiple-drug treatment if resistance to one drug is more fitness enhancing in a strain which is already resistant to other drugs. This process, if it occurs, may lead to a disproportionate increase in the frequency of MDR strains relative to that of singly resistant strains.
(i) Examples.
Positive epistasis is pervasive among alleles conferring resistance to different antibiotics (quinolone, rifampin, and streptomycin) in E. coli and may explain the high level of MDR in E. coli (98). Additionally, in P. aeruginosa the cost of acquiring streptomycin resistance mutations is greater in a rifampin-sensitive background than in a rifampin-resistant background (99), indicating positive epistasis and suggesting that strains resistant to both streptomycin and rifampin are selectively more favored than strains with only streptomycin resistance even when rifampin is not used for treatment.
Moreover, epistasis may exist between the genetic background and drug resistance determinants. As an example, the fitness costs of resistance mutations in the genetic background of Beijing lineage M. tuberculosis are smaller than those in other genetic backgrounds, or compensatory mutations are in easier reach for them, possibly explaining the association between Beijing TB genotype and MDR (100). In a mouse model of gonococcal infection, the fitness cost of fluoroquinolone resistance mutations depends on genetic background (101).
(ii) Testable predictions.
Phylogenetic methods (95, 102) have been developed to test for the presence of epistasis in general, based on the idea that sites with epistatic interactions will tend to show correlated substitutions within phylogenies. These approaches can in principle be applied to the context of multiple-drug resistance, although this may be hampered by the presence of horizontal transfer, which makes it hard to know whether cooccurrence of mutations is genuinely independent or merely through introduction from the same source by recombination. Likewise, experimental measurements of the fitnesses of different genotypes (103) could also be used for detecting epistasis between resistance determinants. In such settings, it is important to distinguish between epistatic interactions in the drug-exposed and drug-free settings, which may not always be the same.
(iii) Practical implications.
In principle, if positive epistasis between resistance to different drugs could be detected experimentally before the emergence of MDR, we might avoid using combinations of drugs for which resistance determinants may have positive epistasis and increase the spread of MDR. However, the downside of such a choice would be to reduce the usage of highly synergistic combinations of drugs, which may be valuable therapeutically (97).
Niche differentiation: aggregation of multiple drug selection pressures within specific populations.
The use of multiple unrelated drug classes is higher in certain population subgroups, such as young children and the elderly (104) and sexually active persons with a high incidence of sexually transmitted infection (105). Use is also higher in certain settings, such as hospitals and long-term-care facilities, than in the general population. Antibiotic use also varies geographically, both within (5) and between (86) countries. The prevalence of resistance to each of these drugs may be higher in the high-use settings/subgroups, resulting in an excess of MDR when the high- and low-use subgroups/settings are considered together (Fig. 1H). In population genetic terms, this is an example of the Wahlund effect, in which associations between allele frequencies are created when two partially or fully distinct populations are considered together (106, 107).
(i) Examples.
High levels of use of multiple antimicrobial classes in hospitals are thought to account for the high prevalence of multiresistant organisms in these settings, with particularly high levels of use and resistance in intensive care units (108).
Antibiotic use, including the use of classes such as macrolides, penicillins, and cephalosporins, is higher in children under 5 than in older children or adults in the United States (87, 104). Young age is a risk factor for resistance to multiple drugs in common colonizing bacteria such as S. pneumoniae (109). A notable “exception that proves the rule” in the case of S. pneumoniae is resistance to fluoroquinolones, which appears sporadically and is only very weakly associated with resistance to other drug classes (Table 1). Fluoroquinolones are not commonly used in children due to side effects and hence do not contribute to the common selective force for multiple resistance in children, who appear to be a “core group” for S. pneumoniae transmission (110, 111). Thus, this mechanism would predict exactly the pattern observed: high levels of correlated MDR to all drug classes except fluoroquinolones.
(ii) Testable predictions.
A signal of this mechanism is population admixture, such that the multiresistant strains form a subpopulation genetically distinct from the susceptible ones, even after excluding sites encoding resistance mechanisms. Such admixture may be detected by using F-statistics (112, 113), principal-component analysis (114), or clustering methods (115) on genetic data after resistance-determining sites have been removed.
(iii) Practical implications.
Identification of specific populations at risk for multiple resistance may aid in the selection of empirical antimicrobial regimens that maximize the probability of treatment success. In situations where the highly treated group is also a “core group” that is a source of transmission to other groups, the avoidance of a particular antimicrobial class in the core group may preserve treatment options for the noncore group, as they are unlikely to be infected with a strain resistant to that class. Such an argument has been suggested as an additional reason to avoid fluoroquinolone use in children, in order to preserve the effectiveness of this class for treating adults (3).
Importation of MDR strains and geographic source-sink dynamics.
High levels of antimicrobial use in certain populations may increase the prevalence of resistance to many different drug classes. If some of the above-described mechanisms are operative, this may lead to an excess of MDR strains within the population, or perhaps MDR strains will simply be at a high frequency given the frequency of each resistance determinant in this population. Either way, the MDR strains in this “source” population may then be introduced into other, “sink” populations, where they spread, competing successfully with pan-susceptible strains because they have resistance to the drug(s) used in these recipient populations (Fig. 1I). The result is that most strains in the sink populations are either pan-susceptible or multiply resistant. As in other cases of population admixture, the sink population reflects a mix of native and imported strains, and the imported strains create an association between resistance to one drug and resistance to others. This process is different from the one described above because it can happen in a single, truly well-mixed population, as long as there is some importation of strains from another, largely independent population.
(i) Example.
With N. gonorrhoeae, East Asian strains are thought to have entered western North America and spread eastward due to travelers bringing MDR strains (11, 116). The countries that are highly connected to the Pacific Rim show similar epidemiological patterns (116).
(ii) Testable predictions.
If MDR strains are imported, they have a genetic background different from that of native strains. In this case, similar to niche differentiation in the mechanism described above, genetic differentiation between multiply resistant strains and native strains is expected to be high and could be examined by the methods listed for that mechanism after excluding resistance determinant sites. If genetic data from parental and admixed populations are available, the admixture proportion from each parental population can be estimated by maximum-likelihood methods (117) and Bayesian approaches (118, 119). Epidemiologically, populations that have similar exposure to the source populations are expected to show similar patterns of drug resistance. It is also expected that the frequency of travel between source populations and sink populations is related to the appearance of MDR strains in sink populations.
From an individual risk factor perspective, studies can assess the extent to which MDR strains are associated with migration from (120, 121) or travel to high-resistance areas. This mechanism predicts that when MDR is rare, such associations should be strong, but as the MDR strains spread endemically within the “sink” population, the association may decline (7).
(iii) Practical implications.
To the extent that multiple-drug resistance in many populations is a consequence of importation from the highest-use populations, international coordination of antimicrobial control policies becomes increasingly important. Moreover, heightened surveillance for MDR strains among travelers may be appropriate, as a means of detecting and delaying further spread of such strains. Once the MDR strains become widespread in a new population, however, such measures may be of little value.
CONCLUSIONS
Excess MDR is observed in many bacterial species. We have considered nine possible explanatory genetic and epidemiological mechanisms. We first considered mechanisms for the appearance of MDR strains, including individual biochemical mechanisms responsible for the MDR phenotype (e.g., efflux pumps), genetic linkage, differential mutation or recombination rate, and multidrug therapy with accelerated treatment failure with multidrug resistance. Mechanisms for the proliferation of MDR strains include associated linkage selection, bystander selection, positive epistasis, niche differentiation, and importation of MDR from a high-use population, followed by spread in a “recipient” population.
We have documented the phenomenon of MDR frequencies exceeding those expected from the product of individual resistance frequencies by counting individual patient isolates with each phenotype. It is worth emphasizing that from an evolutionary perspective, many bacterial isolates with a given phenotype, such as MDR, may result from a small number of “origin” events (122). In this case, the origin of MDR may not occur at a particularly high rate, and the high prevalence of MDR must be attributed mainly to successful spread of MDR strains. On the other hand, in S. pneumoniae, even within what appeared to be a clonal lineage, there were multiple events of gain and loss of resistance determinants both by point mutations and by horizontal gene transfer (43). In the extreme case, there may be very little clonal spread of a particular resistance phenotype but rather repeated appearance in multiple lineages, as appears to occur for fluoroquinolone resistance in S. pneumoniae (123, 124).
These mechanisms are not mutually exclusive; several mechanisms may contribute to the origin, and several to the dissemination, of MDR bacteria. In our classification we have tried to separate mechanisms that reflect different kinds of events at the patient level: those mechanisms listed for “origin” generally refer to ways in which MDR organisms arise within a patient, while those listed for “dissemination” generally refer to the process of spread between patients. Seen at a higher level of generality, one could argue that observing an excess of MDR reflects either a higher rate of formation of such strains or a higher fitness of such strains than expected from the observed rates of formation and proliferation of the individual drug resistance alleles. From a broad, population genetic perspective, several of these processes, which we have distinguished, could be combined under the heading of epistasis (92). For example, we have described treatment failure leading to multiple-drug resistance as a distinct process for the origin of MDR strains. It could alternatively be described as enhanced fitness of resistance to a second drug in strains already resistant to a first drug, leading to the survival of dual-resistant strains under combination therapy. Similarly, one could recast bystander selection as a form of epistasis. MDR strains have a fitness advantage higher than the sum of fitness advantages of singly resistant strains in the environment where individuals are treated with a range of antibiotics. Nonetheless, because they involve different kinds of events at the patient level (such as multidrug therapy or treatment targeting another infection) and since the testable predictions and practical implications of these mechanisms vary, we have chosen to consider them separately. The unifying perspective of population genetics offers a complementary approach that notes the common features of these mechanisms.
If the genetic mechanism of resistance in a “bug-drug” combination is point mutations, then a differential mutation rate and multidrug therapy with treatment failure are most likely to be factors that lead to the excess of drug resistance. As mentioned above, drug resistance in M. tuberculosis is from point mutations, and both a differential mutation rate and multidrug therapy with treatment failure played roles in MDR in TB (39). On the other hand, if the genetic mechanism of resistance is horizontal transmission of plasmids or mobile elements, then a differential recombination rate, genetic linkage, and associated linkage selection may play more important roles, because resistance determinants that are located in the same mobile elements or plasmids are likely to be transmitted together. As described above, conjugative transposons in S. pneumoniae contain MDR determinants, and both genetic linkage and recombination rate play important roles in the excess of MDR S. pneumoniae (31, 38).
The increasing ease and economy of genomic methods are making these tools increasingly important for studying MDR mechanisms. Hypotheses involving differential mutation/recombination rate (see “Highly mutable or recombinogenic bacterial lineages” above) can be tested by estimating historic mutation or recombination rates for lineages with and without MDR from sequencing data. If the genetic mechanisms of drug resistance are known, sequencing of bacteria can be used to test for multiple mechanisms: the presence of a single element conferring MDR (see “Single biochemical mechanism conferring resistance to multiple drugs”), genetic linkage between drug resistance determinants (see “Genetic linkage”), the presence of a single resistance determinant in strains at the beginning of treatment in longitudinal samples from a patient (see “Multidrug therapy with accelerated treatment failure in resistant infections”), the correlation in the presence of drug resistance determinants in sequences of different bacterial species across space or time (see “Bystander selection”), and signs of population admixture (see “Niche differentiation: aggregation of multiple drug selection pressures within specific populations” and “Importation of MDR strains and geographic source-sink dynamics”). Furthermore, phylogenetic methods (125) can be used to test whether resistance to the new drug arose in a background of resistance to older drugs; if this is the case, strains resistant to the new drug should be more closely related to those resistant to the old, compared with suitable susceptible controls (see “Associated linkage selection”). Positive epistasis could be detected by examining whether substitution rates between different resistance mutations are correlated (see “Positive epistasis between drug resistance determinants or between resistance determinants and genetic background”). While the potential of these methods is clear, they are crucially dependent on adequate sampling and quality metadata (such as time and place of isolation, reason for obtaining an isolate [carriage surveillance, outbreak-driven surveillance, or clinical culture], patient characteristics, and disease manifestation). Such epidemiological context is invaluable in aiding the interpretation of conclusions from phylogenies or other genomic analyses (126).
Through understanding these genetic and epidemiological mechanisms of MDR and using the tools we have suggested here to test each hypothetical mechanism, we will be able to identify the cause(s) of the excess of MDR and design more effective interventions to prevent the origin and/or proliferation of MDR. Although some of our practical suggestions are limited by the availability of alternative drugs, developing and applying quick and sensitive diagnostic tools and using narrow-spectrum antibiotics (for example, fidaxomicin is a narrow-spectrum antibiotic for Clostridium difficile [127]) may be two general ways to reduce the frequency of MDR strains. Although development of pathogen-specific antibiotics is unlikely, it is increasingly of interest and will potentially provide more opportunity to reduce the spread of MDR.
ACKNOWLEDGMENTS
We are grateful to Lauren M. Childs for reading the manuscript and offering suggestions.
This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number U54GM088558 (H.-H.C., T.C., Y.H.G., W.P.H. and M.L.), the National Institute of General Medical Sciences of the National Institutes of Health under award number R01GM103525 (T.F.O.), National Institutes of Health grant R01-AI106786-01 (W.P.H.), and National Institutes of Health grant K08-AI104767 (Y.H.G.).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript, and the content is solely the responsibility of the authors and does not necessarily represent the official views of the funders.
REFERENCES
- 1.Bolan GA, Sparling PF, Wasserheit JN. 2012. The emerging threat of untreatable gonococcal infection. N Engl J Med 366:485–487. doi: 10.1056/NEJMp1112456. [DOI] [PubMed] [Google Scholar]
- 2.Menzies NA, Cohen T, Lin HH, Murray M, Salomon JA. 2012. Population health impact and cost-effectiveness of tuberculosis diagnosis with Xpert MTB/RIF: a dynamic simulation and economic evaluation. PLoS Med 9:e1001347. doi: 10.1371/journal.pmed.1001347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pichichero ME, Casey JR. 2007. Emergence of a multiresistant serotype 19A pneumococcal strain not included in the 7-valent conjugate vaccine as an otopathogen in children. JAMA 298:1772–1778. doi: 10.1001/jama.298.15.1772. [DOI] [PubMed] [Google Scholar]
- 4.Coates AR, Halls G, Hu Y. 2011. Novel classes of antibiotics or more of the same? Br J Pharmacol 163:184–194. doi: 10.1111/j.1476-5381.2011.01250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States, 2013. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 6.Enne VI, Livermore DM, Stephens P, Hall LM. 2001. Persistence of sulphonamide resistance in Escherichia coli in the UK despite national prescribing restriction. Lancet 357:1325–1328. doi: 10.1016/S0140-6736(00)04519-0. [DOI] [PubMed] [Google Scholar]
- 7.Goldstein E, Kirkcaldy RD, Reshef D, Berman S, Weinstock H, Sabeti P, Del Rio C, Hall G, Hook EW, Lipsitch M. 2012. Factors related to increasing prevalence of resistance to ciprofloxacin and other antimicrobial drugs in Neisseria gonorrhoeae, United States. Emerg Infect Dis 18:1290–1297. doi: 10.3201/eid1808.111202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Link-Gelles R, Thomas A, Lynfield R, Petit S, Schaffner W, Harrison L, Farley MM, Aragon D, Nicols M, Kirley PD, Zansky S, Jorgensen J, Juni BA, Jackson D, Moore MR, Lipsitch M. 2013. Geographic and temporal trends in antimicrobial nonsusceptibility in Streptococcus pneumoniae in the post-vaccine era in the United States. J Infect Dis 208:1266–1273. doi: 10.1093/infdis/jit315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCormick AW, Whitney CG, Farley MM, Lynfield R, Harrison LH, Bennett NM, Schaffner W, Reingold A, Hadler J, Cieslak P, Samore MH, Lipsitch M. 2003. Geographic diversity and temporal trends of antimicrobial resistance in Streptococcus pneumoniae in the United States. Nat Med 9:424–430. doi: 10.1038/nm839. [DOI] [PubMed] [Google Scholar]
- 10.Hwang TJ, Hooper DC. 2014. Association between fluoroquinolone resistance and resistance to other antimicrobial agents among Escherichia coli urinary isolates in the outpatient setting: a national cross-sectional study. J Antimicrob Chemother 69:1720–1722. doi: 10.1093/jac/dku029. [DOI] [PubMed] [Google Scholar]
- 11.Ota KV, Jamieson F, Fisman DN, Jones KE, Tamari IE, Ng LK, Towns L, Rawte P, Di Prima A, Wong T, Richardson SE. 2009. Prevalence of and risk factors for quinolone-resistant Neisseria gonorrhoeae infection in Ontario. Can Med Assoc J 180:287–290. doi: 10.1503/cmaj.080222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Centers for Disease Control and Prevention. 2012. Gonococcal Isolate Surveillance Project (GISP) 2011 annual report. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 13.Izu A, Cohen T, Degruttola V. 2013. Bayesian estimation of mixture models with prespecified elements to compare drug resistance in treatment-naive and experienced tuberculosis cases. PLoS Computat Biol 9:e1002973. doi: 10.1371/journal.pcbi.1002973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.World Health Organization. 2008. Anti-tuberculosis drug resistance in the world: fourth global report. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 15.BC Centre for Disease Control. 2013. Antimicrobial resistance trends in the province of British Columbia, 2012. BC Centre for Disease Control, Vancouver, Canada. [Google Scholar]
- 16.Bradford PA. 2001. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev 14:933–951. doi: 10.1128/CMR.14.4.933-951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberts MC, Sutcliffe J, Courvalin P, Jensen LB, Rood J, Seppala H. 1999. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob Agents Chemother 43:2823–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alekshun MN, Levy SB. 2007. Molecular mechanisms of antibacterial multidrug resistance. Cell 128:1037–1050. doi: 10.1016/j.cell.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 19.Poole K. 2005. Efflux-mediated antimicrobial resistance. J Antimicrob Chemother 56:20–51. doi: 10.1093/jac/dki171. [DOI] [PubMed] [Google Scholar]
- 20.Chang TM, Lu PL, Li HH, Chang CY, Chen TC, Chang LL. 2007. Characterization of fluoroquinolone resistance mechanisms and their correlation with the degree of resistance to clinically used fluoroquinolones among Escherichia coli isolates. J Chemother 19:488–494. doi: 10.1179/joc.2007.19.5.488. [DOI] [PubMed] [Google Scholar]
- 21.Pumbwe L, Chang A, Smith RL, Wexler HM. 2006. Clinical significance of overexpression of multiple RND-family efflux pumps in Bacteroides fragilis isolates. J Antimicrob Chemother 58:543–548. doi: 10.1093/jac/dkl278. [DOI] [PubMed] [Google Scholar]
- 22.Cui L, Tominaga E, Neoh HM, Hiramatsu K. 2006. Correlation between reduced daptomycin susceptibility and vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob Agents Chemother 50:1079–1082. doi: 10.1128/AAC.50.3.1079-1082.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li XZ, Zhang L, Nikaido H. 2004. Efflux pump-mediated intrinsic drug resistance in Mycobacterium smegmatis. Antimicrob Agents Chemother 48:2415–2423. doi: 10.1128/AAC.48.7.2415-2423.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blazquez J, Morosini MI, Negri MC, Baquero F. 2000. Selection of naturally occurring extended-spectrum TEM beta-lactamase variants by fluctuating beta-lactam pressure. Antimicrob Agents Chemother 44:2182–2184. doi: 10.1128/AAC.44.8.2182-2184.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neyfakh AA, Bidnenko VE, Chen LB. 1991. Efflux-mediated multidrug resistance in Bacillus subtilis: similarities and dissimilarities with the mammalian system. Proc Natl Acad Sci U S A 88:4781–4785. doi: 10.1073/pnas.88.11.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pages JM, Amaral L. 2009. Mechanisms of drug efflux and strategies to combat them: challenging the efflux pump of Gram-negative bacteria. Biochim Biophys Acta 1794:826–833. doi: 10.1016/j.bbapap.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 27.Gupta S, Tyagi S, Almeida DV, Maiga MC, Ammerman NC, Bishai WR. 2013. Acceleration of tuberculosis treatment by adjunctive therapy with verapamil as an efflux inhibitor. Am J Respir Crit Care Med 188:600–607. doi: 10.1164/rccm.201304-0650OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ughachukwu P, Unekwe P. 2012. Efflux pump-mediated resistance in chemotherapy. Ann Med Health Sci Res 2:191–198. doi: 10.4103/2141-9248.105671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall RM. 2012. Integrons and gene cassettes: hotspots of diversity in bacterial genomes. Ann N Y Acad Sci 1267:71–78. doi: 10.1111/j.1749-6632.2012.06588.x. [DOI] [PubMed] [Google Scholar]
- 30.Roberts AP, Mullany P. 2009. A modular master on the move: the Tn916 family of mobile genetic elements. Trends Microbiol 17:251–258. doi: 10.1016/j.tim.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 31.Courvalin P, Carlier C. 1986. Transposable multiple antibiotic resistance in Streptococcus pneumoniae. Mol Gen Genet 205:291–297. doi: 10.1007/BF00430441. [DOI] [PubMed] [Google Scholar]
- 32.Nordmann P, Dortet L, Poirel L. 2012. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med 18:263–272. doi: 10.1016/j.molmed.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 33.Huang XZ, Frye JG, Chahine MA, Glenn LM, Ake JA, Su W, Nikolich MP, Lesho EP. 2012. Characteristics of plasmids in multi-drug-resistant Enterobacteriaceae isolated during prospective surveillance of a newly opened hospital in Iraq. PLoS One 7:e40360. doi: 10.1371/journal.pone.0040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bergstrom CT, Lo M, Lipsitch M. 2004. Ecological theory suggests that antimicrobial cycling will not reduce antimicrobial resistance in hospitals. Proc Natl Acad Sci U S A 101:13285–13290. doi: 10.1073/pnas.0402298101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fridkin SK. 2003. Routine cycling of antimicrobial agents as an infection-control measure. Clin Infect Dis 36:1438–1444. doi: 10.1086/375082. [DOI] [PubMed] [Google Scholar]
- 36.Abel Zur Wiesch P, Kouyos R, Abel S, Viechtbauer W, Bonhoeffer S. 2014. Cycling empirical antibiotic therapy in hospitals: meta-analysis and models. PLoS Pathog 10:e1004225. doi: 10.1371/journal.ppat.1004225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.LeClerc JE, Li B, Payne WL, Cebula TA. 1996. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 274:1208–1211. doi: 10.1126/science.274.5290.1208. [DOI] [PubMed] [Google Scholar]
- 38.Croucher NJ, Finkelstein JA, Pelton SI, Mitchell PK, Lee GM, Parkhill J, Bentley SD, Hanage WP, Lipsitch M. 2013. Population genomics of post-vaccine changes in pneumococcal epidemiology. Nat Genet 45:656–663. doi: 10.1038/ng.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ford CB, Shah RR, Maeda MK, Gagneux S, Murray MB, Cohen T, Johnston JC, Gardy J, Lipsitch M, Fortune SM. 2013. Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nat Genet 45:784–790. doi: 10.1038/ng.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Colijn C, Cohen T, Ganesh A, Murray M. 2011. Spontaneous emergence of multiple drug resistance in tuberculosis before and during therapy. PLoS One 6:e18327. doi: 10.1371/journal.pone.0018327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanage WP, Fraser C, Tang J, Connor TR, Corander J. 2009. Hyper-recombination, diversity, and antibiotic resistance in pneumococcus. Science 324:1454–1457. doi: 10.1126/science.1171908. [DOI] [PubMed] [Google Scholar]
- 42.Chewapreecha C, Harris SR, Croucher NJ, Turner C, Marttinen P, Cheng L, Pessia A, Aanensen DM, Mather AE, Page AJ, Salter SJ, Harris D, Nosten F, Goldblatt D, Corander J, Parkhill J, Turner P, Bentley SD. 2014. Dense genomic sampling identifies highways of pneumococcal recombination. Nat Genet 46:305–309. doi: 10.1038/ng.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, McGee L, von Gottberg A, Song JH, Ko KS, Pichon B, Baker S, Parry CM, Lambertsen LM, Shahinas D, Pillai DR, Mitchell TJ, Dougan G, Tomasz A, Klugman KP, Parkhill J, Hanage WP, Bentley SD. 2011. Rapid pneumococcal evolution in response to clinical interventions. Science 331:430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baquero F, Negri MC, Morosini MI, Blazquez J. 1998. Antibiotic-selective environments. Clin Infect Dis 27(Suppl 1):S5–S11. doi: 10.1086/514916. [DOI] [PubMed] [Google Scholar]
- 45.Kohanski MA, DePristo MA, Collins JJ. 2010. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Molecular cell 37:311–320. doi: 10.1016/j.molcel.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leigh EG., Jr 1973. The evolution of mutation rates. Genetics 73(Suppl):1–18. [PubMed] [Google Scholar]
- 47.Palmer ME, Lipsitch M. 2006. The influence of hitchhiking and deleterious mutation upon asexual mutation rates. Genetics 173:461–472. doi: 10.1534/genetics.105.049445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tenaillon O, Taddei F, Radmian M, Matic I. 2001. Second-order selection in bacterial evolution: selection acting on mutation and recombination rates in the course of adaptation. Res Microbiol 152:11–16. doi: 10.1016/S0923-2508(00)01163-3. [DOI] [PubMed] [Google Scholar]
- 49.Mao EF, Lane L, Lee J, Miller JH. 1997. Proliferation of mutators in A cell population. J Bacteriol 179:417–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Parkhill J, Harris SR. 20 November 2014. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Didelot X, Falush D. 2007. Inference of bacterial microevolution using multilocus sequence data. Genetics 175:1251–1266. doi: 10.1534/genetics.106.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kimura M. 1968. Evolutionary rate at the molecular level. Nature 217:624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- 53.Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 54.Feil EJ, Holmes EC, Bessen DE, Chan MS, Day NP, Enright MC, Goldstein R, Hood DW, Kalia A, Moore CE, Zhou J, Spratt BG. 2001. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc Natl Acad Sci U S A 98:182–187. doi: 10.1073/pnas.98.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Castillo-Ramirez S, Harris SR, Holden MT, He M, Parkhill J, Bentley SD, Feil EJ. 2011. The impact of recombination on dN/dS within recently emerged bacterial clones. PLoS Pathog 7:e1002129. doi: 10.1371/journal.ppat.1002129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Giraud A, Radman M, Matic I, Taddei F. 2001. The rise and fall of mutator bacteria. Curr Opin Microbiol 4:582–585. doi: 10.1016/S1369-5274(00)00254-X. [DOI] [PubMed] [Google Scholar]
- 57.Ehrlich P. 1913. Chemotherapeutics: scientific principles, methods and results. Lancet ii:445–451. [Google Scholar]
- 58.Mitchison DA. 1992. The Garrod Lecture. Understanding the chemotherapy of tuberculosis—current problems. J Antimicrob Chemother 29:477–493. [DOI] [PubMed] [Google Scholar]
- 59.Koser CU, Ellington MJ, Cartwright EJ, Gillespie SH, Brown NM, Farrington M, Holden MT, Dougan G, Bentley SD, Parkhill J, Peacock SJ. 2012. Routine use of microbial whole genome sequencing in diagnostic and public health microbiology. PLoS Pathog 8:e1002824. doi: 10.1371/journal.ppat.1002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koser CU, Fraser LJ, Ioannou A, Becq J, Ellington MJ, Holden MT, Reuter S, Torok ME, Bentley SD, Parkhill J, Gormley NA, Smith GP, Peacock SJ. 2014. Rapid single-colony whole-genome sequencing of bacterial pathogens. J Antimicrob Chemother 69:1275–1281. doi: 10.1093/jac/dkt494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hingley-Wilson SM, Casey R, Connell D, Bremang S, Evans JT, Hawkey PM, Smith GE, Jepson A, Philip S, Kon OM, Lalvani A. 2013. Undetected multidrug-resistant tuberculosis amplified by first-line therapy in mixed infection. Emerg Infect Dis 19:1138–1141. doi: 10.3201/eid1907.130313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Rie A, Victor TC, Richardson M, Johnson R, van der Spuy GD, Murray EJ, Beyers N, Gey van Pittius NC, van Helden PD, Warren RM. 2005. Reinfection and mixed infection cause changing Mycobacterium tuberculosis drug-resistance patterns. Am J Respir Crit Care Med 172:636–642. doi: 10.1164/rccm.200503-449OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Almeida Da Silva PE, Palomino JC. 2011. Molecular basis and mechanisms of drug resistance in Mycobacterium tuberculosis: classical and new drugs. J Antimicrob Chemother 66:1417–1430. doi: 10.1093/jac/dkr173. [DOI] [PubMed] [Google Scholar]
- 64.Shimao T. 1987. Drug resistance in tuberculosis control. Tubercle 68:5–18. doi: 10.1016/S0041-3879(87)80014-4. [DOI] [PubMed] [Google Scholar]
- 65.Lipsitch M, Levin BR. 1998. Population dynamics of tuberculosis treatment: mathematical models of the roles of non-compliance and bacterial heterogeneity in the evolution of drug resistance. Int J Tuberc Lung Dis 2:187–199. [PubMed] [Google Scholar]
- 66.Weis SE, Slocum PC, Blais FX, King B, Nunn M, Matney GB, Gomez E, Foresman BH. 1994. The effect of directly observed therapy on the rates of drug resistance and relapse in tuberculosis. N Engl J Med 330:1179–1184. doi: 10.1056/NEJM199404283301702. [DOI] [PubMed] [Google Scholar]
- 67.Furin J, Gegia M, Mitnick C, Rich M, Shin S, Becerra M, Drobac P, Farmer P, Hurtado R, Joseph JK, Keshavjee S, Kalandadze I. 2012. Eliminating the category II retreatment regimen from national tuberculosis programme guidelines: the Georgian experience. Bull World Health Organ 90:63–66. doi: 10.2471/BLT.11.092320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lew W, Pai M, Oxlade O, Martin D, Menzies D. 2008. Initial drug resistance and tuberculosis treatment outcomes: systematic review and meta-analysis. Ann Intern Med 149:123–134. doi: 10.7326/0003-4819-149-2-200807150-00008. [DOI] [PubMed] [Google Scholar]
- 69.Boehme CC, Nabeta P, Hillemann D, Nicol MP, Shenai S, Krapp F, Allen J, Tahirli R, Blakemore R, Rustomjee R, Milovic A, Jones M, O'Brien SM, Persing DH, Ruesch-Gerdes S, Gotuzzo E, Rodrigues C, Alland D, Perkins MD. 2010. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med 363:1005–1015. doi: 10.1056/NEJMoa0907847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Leavis HL, Bonten MJ, Willems RJ. 2006. Identification of high-risk enterococcal clonal complexes: global dispersion and antibiotic resistance. Curr Opin Microbiol 9:454–460. doi: 10.1016/j.mib.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 71.Bean DC, Livermore DM, Papa I, Hall LM. 2005. Resistance among Escherichia coli to sulphonamides and other antimicrobials now little used in man. J Antimicrob Chemother 56:962–964. doi: 10.1093/jac/dki332. [DOI] [PubMed] [Google Scholar]
- 72.Barkai G, Greenberg D, Givon-Lavi N, Dreifuss E, Vardy D, Dagan R. 2005. Community prescribing and resistant Streptococcus pneumoniae. Emerg Infect Dis 11:829–837. doi: 10.3201/eid1106.050198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arason VA, Kristinsson KG, Sigurdsson JA, Stefansdottir G, Molstad S, Gudmundsson S. 1996. Do antimicrobials increase the carriage rate of penicillin resistant pneumococci in children? Cross sectional prevalence study. BMJ 313:387–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Goldstein FW. 1999. Penicillin-resistant Streptococcus pneumoniae: selection by both beta-lactam and non-beta-lactam antibiotics. J Antimicrob Chemother 44:141–144. doi: 10.1093/jac/44.2.141. [DOI] [PubMed] [Google Scholar]
- 75.Samore MH, Lipsitch M, Alder SC, Haddadin B, Stoddard G, Williamson J, Sebastian K, Carroll K, Ergonul O, Carmeli Y, Sande MA. 2006. Mechanisms by which antibiotics promote dissemination of resistant pneumococci in human populations. Am J Epidemiol 163:160–170. doi: 10.1093/aje/kwj021. [DOI] [PubMed] [Google Scholar]
- 76.Kirkcaldy RD, Kidd S, Weinstock HS, Papp JR, Bolan GA. 2013. Trends in antimicrobial resistance in Neisseria gonorrhoeae in the USA: the Gonococcal Isolate Surveillance Project (GISP), January 2006-June 2012. Sex Transm Infect 89(Suppl 4):iv5–iv10. doi: 10.1136/sextrans-2013-051162. [DOI] [PubMed] [Google Scholar]
- 77.Dagan R, Johnson CE, McLinn S, Abughali N, Feris J, Leibovitz E, Burch DJ, Jacobs MR. 2000. Bacteriologic and clinical efficacy of amoxicillin/clavulanate vs. azithromycin in acute otitis media. Pediatr Infect Dis J 19:95–104. doi: 10.1097/00006454-200002000-00002. [DOI] [PubMed] [Google Scholar]
- 78.Institute of Medicine Forum on Microbial Threats. 2010. Appendix A, contributed manuscripts. Antibiotic resistance: implications for global health and novel intervention strategies: workshop summary. National Academies Press, Washington, DC. [PubMed] [Google Scholar]
- 79.Fantin B, Duval X, Massias L, Alavoine L, Chau F, Retout S, Andremont A, Mentre F. 2009. Ciprofloxacin dosage and emergence of resistance in human commensal bacteria. J Infect Dis 200:390–398. doi: 10.1086/600122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lacey RW, Lord VL, Howson GL, Luxton DE, Trotter IS. 1983. Double-blind study to compare the selection of antibiotic resistance by amoxycillin or cephradine in the commensal flora. Lancet ii:529–532. [DOI] [PubMed] [Google Scholar]
- 81.Carlet J. 2012. The gut is the epicentre of antibiotic resistance. Antimicrob Resist Infect Control 1:39. doi: 10.1186/2047-2994-1-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Summers AO, Wireman J, Vimy MJ, Lorscheider FL, Marshall B, Levy SB, Bennett S, Billard L. 1993. Mercury released from dental “silver” fillings provokes an increase in mercury- and antibiotic-resistant bacteria in oral and intestinal floras of primates. Antimicrob Agents Chemother 37:825–834. doi: 10.1128/AAC.37.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Silveira E, Freitas AR, Antunes P, Barros M, Campos J, Coque TM, Peixe L, Novais C. 2014. Co-transfer of resistance to high concentrations of copper and first-line antibiotics among Enterococcus from different origins (humans, animals, the environment and foods) and clonal lineages. J Antimicrob Chemother 69:899–906. doi: 10.1093/jac/dkt479. [DOI] [PubMed] [Google Scholar]
- 84.Ison CA, Deal C, Unemo M. 2013. Current and future treatment options for gonorrhoea. Sex Transm Infect 89(Suppl 4):iv52–iv56. doi: 10.1136/sextrans-2012-050913. [DOI] [PubMed] [Google Scholar]
- 85.Chen TC, Lu PL, Lin CY, Lin WR, Chen YH. 2011. Fluoroquinolones are associated with delayed treatment and resistance in tuberculosis: a systematic review and meta-analysis. Int J Infect Dis 15:e211–216. doi: 10.1016/j.ijid.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 86.Goossens H, Ferech M, Vander Stichele R, Elseviers M, ESAC Project Group . 2005. Outpatient antibiotic use in Europe and association with resistance: a cross-national database study. Lancet 365:579–587. doi: 10.1016/S0140-6736(05)70799-6. [DOI] [PubMed] [Google Scholar]
- 87.Hicks LA, Chien YW, Taylor TH Jr, Haber M, Klugman KP, Active Bacterial Core Surveillance T . 2011. Outpatient antibiotic prescribing and nonsusceptible Streptococcus pneumoniae in the United States, 1996–2003. Clin Infect Dis 53:631–639. doi: 10.1093/cid/cir443. [DOI] [PubMed] [Google Scholar]
- 88.Perez-Trallero E, Garcia-de-la-Fuente C, Garcia-Rey C, Baquero F, Aguilar L, Dal-Re R, Garcia-de-Lomas J. 2005. Geographical and ecological analysis of resistance, coresistance, and coupled resistance to antimicrobials in respiratory pathogenic bacteria in Spain. Antimicrob Agents Chemother 49:1965–1972. doi: 10.1128/AAC.49.5.1965-1972.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gupta K, Hooton TM, Naber KG, Wullt B, Colgan R, Miller LG, Moran GJ, Nicolle LE, Raz R, Schaeffer AJ, Soper DE, Infectious Diseases Society of America, European Society for Microbiology and Infectious Diseases . 2011. International clinical practice guidelines for the treatment of acute uncomplicated cystitis and pyelonephritis in women: a 2010 update by the Infectious Diseases Society of America and the European Society for Microbiology and Infectious Diseases. Clin Infect Dis 52:e103–e120. doi: 10.1093/cid/ciq257. [DOI] [PubMed] [Google Scholar]
- 90.Stewardson AJ, Huttner B, Harbarth S. 2011. At least it won't hurt: the personal risks of antibiotic exposure. Curr Opin Pharmacol 11:446–452. doi: 10.1016/j.coph.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 91.Andersson DI, Hughes D. 2010. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol 8:260–271. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- 92.Corbett-Detig RB, Zhou J, Clark AG, Hartl DL, Ayroles JF. 2013. Genetic incompatibilities are widespread within species. Nature 504:135–137. doi: 10.1038/nature12678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, Chen Y, Cheng X, Chua G, Friesen H, Goldberg DS, Haynes J, Humphries C, He G, Hussein S, Ke L, Krogan N, Li Z, Levinson JN, Lu H, Menard P, Munyana C, Parsons AB, Ryan O, Tonikian R, Roberts T, Sdicu AM, Shapiro J, Sheikh B, Suter B, Wong SL, Zhang LV, Zhu H, Burd CG, Munro S, Sander C, Rine J, Greenblatt J, Peter M, Bretscher A, Bell G, Roth FP, Brown GW, Andrews B, Bussey H, Boone C. 2004. Global mapping of the yeast genetic interaction network. Science 303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 94.Remold SK, Lenski RE. 2004. Pervasive joint influence of epistasis and plasticity on mutational effects in Escherichia coli. Nat Genet 36:423–426. doi: 10.1038/ng1324. [DOI] [PubMed] [Google Scholar]
- 95.Kryazhimskiy S, Dushoff J, Bazykin GA, Plotkin JB. 2011. Prevalence of epistasis in the evolution of influenza A surface proteins. PLoS genetics 7:e1001301. doi: 10.1371/journal.pgen.1001301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sanjuan R, Moya A, Elena SF. 2004. The contribution of epistasis to the architecture of fitness in an RNA virus. Proc Natl Acad Sci U S A 101:15376–15379. doi: 10.1073/pnas.0404125101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yeh P, Tschumi AI, Kishony R. 2006. Functional classification of drugs by properties of their pairwise interactions. Nat Genet 38:489–494. doi: 10.1038/ng1755. [DOI] [PubMed] [Google Scholar]
- 98.Trindade S, Sousa A, Xavier KB, Dionisio F, Ferreira MG, Gordo I. 2009. Positive epistasis drives the acquisition of multidrug resistance. PLoS Genet 5:e1000578. doi: 10.1371/journal.pgen.1000578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ward H, Perron GG, Maclean RC. 2009. The cost of multiple drug resistance in Pseudomonas aeruginosa. J Evol Biol 22:997–1003. doi: 10.1111/j.1420-9101.2009.01712.x. [DOI] [PubMed] [Google Scholar]
- 100.Hanekom M, Gey van Pittius NC, McEvoy C, Victor TC, Van Helden PD, Warren RM. 2011. Mycobacterium tuberculosis Beijing genotype: a template for success. Tuberculosis 91:510–523. doi: 10.1016/j.tube.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 101.Kunz AN, Begum AA, Wu H, D'Ambrozio JA, Robinson JM, Shafer WM, Bash MC, Jerse AE. 2012. Impact of fluoroquinolone resistance mutations on gonococcal fitness and in vivo selection for compensatory mutations. J Infect Dis 205:1821–1829. doi: 10.1093/infdis/jis277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nasrallah CA, Huelsenbeck JP. 2013. A phylogenetic model for the detection of epistatic interactions. Mol Biol Evol 30:2197–2208. doi: 10.1093/molbev/mst108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lenski RE. 1991. Quantifying fitness and gene stability in microorganisms. Biotechnology 15:173–192. [DOI] [PubMed] [Google Scholar]
- 104.Grijalva CG, Nuorti JP, Griffin MR. 2009. Antibiotic prescription rates for acute respiratory tract infections in US ambulatory settings. JAMA 302:758–766. doi: 10.1001/jama.2009.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Steen R, Chersich M, Gerbase A, Neilsen G, Wendland A, Ndowa F, Akl EA, Lo YR, de Vlas SJ. 2012. Periodic presumptive treatment of curable sexually transmitted infections among sex workers: a systematic review. AIDS 26:437–445. doi: 10.1097/QAD.0b013e32834ed991. [DOI] [PubMed] [Google Scholar]
- 106.Hartl DL, Clark AG. 1997. Principles of population genetics, 3rd ed Sinauer Associates, Sunderland, MA. [Google Scholar]
- 107.Wahlund S. 1928. Zusammensetzung von Populationen und Korrelationserscheinungen vom Standpunkt der Vererbungslehre aus betrachtet. Hereditas 11:65–106. [Google Scholar]
- 108.Brusselaers N, Vogelaers D, Blot S. 2011. The rising problem of antimicrobial resistance in the intensive care unit. Ann Intens Care 1:47. doi: 10.1186/2110-5820-1-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Huang SS, Hinrichsen VL, Stevenson AE, Rifas-Shiman SL, Kleinman K, Pelton SI, Lipsitch M, Hanage WP, Lee GM, Finkelstein JA. 2009. Continued impact of pneumococcal conjugate vaccine on carriage in young children. Pediatrics 124:e1–e11. doi: 10.1542/peds.2008-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Centers for Disease Control Prevention. 2005. Direct and indirect effects of routine vaccination of children with 7-valent pneumococcal conjugate vaccine on incidence of invasive pneumococcal disease–United States, 1998–2003. MMWR Morb Mortal Wkly Rep 54:893–897. [PubMed] [Google Scholar]
- 111.Walter ND, Taylor TH Jr, Dowell SF, Mathis S, Moore MR, Active Bacterial Core Surveillance System Team . 2009. Holiday spikes in pneumococcal disease among older adults. N Engl J Med 361:2584–2585. doi: 10.1056/NEJMc0904844. [DOI] [PubMed] [Google Scholar]
- 112.Weir BS, Cockerham CC. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370. doi: 10.2307/2408641. [DOI] [PubMed] [Google Scholar]
- 113.Wright S. 1951. The genetical structure of populations. Ann Eugenics 15:323–354. [DOI] [PubMed] [Google Scholar]
- 114.Patterson N, Price AL, Reich D. 2006. Population structure and eigenanalysis. PLoS Genet 2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tapsall J. 2009. Multidrug-resistant Neisseria gonorrhoeae. Can Med Assoc J 180:268–269. doi: 10.1503/cmaj.081721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang J. 2003. Maximum-likelihood estimation of admixture proportions from genetic data. Genetics 164:747–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Corander J, Marttinen P. 2006. Bayesian identification of admixture events using multilocus molecular markers. Mol Ecol 15:2833–2843. doi: 10.1111/j.1365-294X.2006.02994.x. [DOI] [PubMed] [Google Scholar]
- 119.Marttinen P, Baldwin A, Hanage WP, Dowson C, Mahenthiralingam E, Corander J. 2008. Bayesian modeling of recombination events in bacterial populations. BMC Bioinformatics 9:421. doi: 10.1186/1471-2105-9-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Holzknecht BJ, Hardardottir H, Haraldsson G, Westh H, Valsdottir F, Boye K, Karlsson S, Kristinsson KG, Gudlaugsson O. 2010. Changing epidemiology of methicillin-resistant Staphylococcus aureus in Iceland from 2000 to 2008: a challenge to current guidelines. J Clin Microbiol 48:4221–4227. doi: 10.1128/JCM.01382-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Moonan PK, Teeter LD, Salcedo K, Ghosh S, Ahuja SD, Flood J, Graviss EA. 2013. Transmission of multidrug-resistant tuberculosis in the USA: a cross-sectional study. Lancet Infect Dis 13:777–784. doi: 10.1016/S1473-3099(13)70128-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Grad YH, Kirkcaldy RD, Trees D, Dordel J, Harris SR, Goldstein E, Weinstock H, Parkhill J, Hanage WP, Bentley S, Lipsitch M. 2014. Genomic epidemiology of Neisseria gonorrhoeae with reduced susceptibility to cefixime in the USA: a retrospective observational study. Lancet Infect Dis 14:220–226. doi: 10.1016/S1473-3099(13)70693-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fuller JD, Low DE. 2005. A review of Streptococcus pneumoniae infection treatment failures associated with fluoroquinolone resistance. Clin Infect Dis 41:118–121. doi: 10.1086/430829. [DOI] [PubMed] [Google Scholar]
- 124.Pletz MW, McGee L, Jorgensen J, Beall B, Facklam RR, Whitney CG, Klugman KP. 2004. Levofloxacin-resistant invasive Streptococcus pneumoniae in the United States: evidence for clonal spread and the impact of conjugate pneumococcal vaccine. Antimicrob Agents Chemother 48:3491–3497. doi: 10.1128/AAC.48.9.3491-3497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Grad YH, Lipsitch M. 2014. Epidemiologic data and pathogen genome sequences: a powerful synergy for public health. Genome Biol 15:538. doi: 10.1186/s13059-014-0538-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sears P, Ichikawa Y, Ruiz N, Gorbach S. 2013. Advances in the treatment of Clostridium difficile with fidaxomicin: a narrow spectrum antibiotic. Ann N Y Acad Sci 1291:33–41. doi: 10.1111/nyas.12135. [DOI] [PubMed] [Google Scholar]