SUMMARY
Curcuminoids, components of the rhizome of turmeric, show several beneficial biological activities, including anticarcinogenic, antioxidant, anti-inflammatory, and antitumor activities. Despite their numerous pharmaceutically important properties, the low natural abundance of curcuminoids represents a major drawback for their use as therapeutic agents. Therefore, they represent attractive targets for heterologous production and metabolic engineering. The understanding of biosynthesis of curcuminoids in turmeric made remarkable advances in the last decade, and as a result, several efforts to produce them in heterologous organisms have been reported. The artificial biosynthetic pathway (e.g., in Escherichia coli) can start with the supplementation of the amino acid tyrosine or phenylalanine or of carboxylic acids and lead to the production of several natural curcuminoids. Unnatural carboxylic acids can also be supplemented as precursors and lead to the production of unnatural compounds with possibly novel therapeutic properties. In this paper, we review the natural conversion of curcuminoids in turmeric and their production by E. coli using an artificial biosynthetic pathway. We also explore the potential of other enzymes discovered recently or already used in other similar biosynthetic pathways, such as flavonoids and stilbenoids, to increase curcuminoid yield and activity.
INTRODUCTION
Curcuminoids are polyphenolic compounds, more specifically diarylheptanoids (C6-C7-C6), isolated from the rhizome of turmeric (Curcuma longa Linn.), which gives it its yellow color (1). These compounds have been used in traditional medicine and as food additives, especially as a curry spice, preservative, coloring (E100), and flavoring agent in Asian countries such as China and India (2, 3). The rhizome of turmeric contains a mixture of curcuminoids in which curcumin, also known as diferuloyl-methane, is the main active chemical constituent (4). Other curcuminoids present in such mixtures include demethoxycurcumin and bisdemethoxycurcumin. Curcuminoids are abundantly present in C. longa but also occur in other Curcuma species. However, since different Curcuma species are difficult to distinguish due to their similar morphologies and their different names given in Latin and by Japanese, Chinese, and Indian cultures, pharmacological studies are very scarce (5). Nevertheless, curcumin, and in some cases bisdemethoxycurcumin and demethoxycurcumin, has also been isolated from Curcuma mangga (6, 7), Curcuma xanthorrhiza (8, 9), Curcuma zedoaria (10, 11), Curcuma phaeocaulis (5), Curcuma aromatica (5), and other species such as Etlingera elatior (12). Other type of complex curcuminoids, cassumunin and cassumunarin, were isolated from Zingiber cassumunar (13, 14). Curcuminoids, depending on the cultivar, constitute 2 to 4% of the dry weight of the dried rhizome (3, 15, 16). The commercial-grade curcumin is isolated from the powdered dry rhizome of C. longa and consists of a mixture of curcumin (∼77%), demethoxycurcumin (∼18%), and bisdemethoxycurcumin (∼5%) (17, 18). Generally, the term curcumin is used to represent all the three curcuminoids found in the turmeric extract.
“Biological” Relevance of Curcuminoids
Curcumin was discovered and isolated almost 2 centuries ago by Vogel and Pelletier (19); however, its biological properties were not identified until the middle of the 20th century, when Schraufstätter and Bernt (20) reported its antibacterial activity. Since then, the biosynthesis and properties of curcumin and other curcuminoids have attracted much interest from the scientific community. Besides antibacterial potential (21–23), these compounds present several properties that are beneficial to human health (Table 1). Therapeutic properties of curcuminoids have been reviewed in several papers (2, 17, 18, 24–29).
TABLE 1.
Some therapeutic applications of curcumin/curcuminoids
| Type of biological activity | Therapeutic effectsa | References |
|---|---|---|
| Anticancer | Suppress cancer cell proliferation—inhibit NF-κB and consequently downregulate NF-κB-regulated gene products such as COX-2 protein and other proteins that are associated with carcinogenesis, tumor initiation, promotion, and metastasis in a wide variety of cancers | 30–40 |
| Induce apoptosis (programmed cell death type I) by activation of caspase-8, BID cleavage, cytochrome c release, and downregulation of Bcl-2 expression | ||
| Induce autophagy (programmed cell death type II) regulated by simultaneous inhibition of the Akt/mTOR/p70S6K pathway and stimulation of the ERK1/2 pathway | ||
| Cholesterol lowering | Decrease LDL cholesterol, total cholesterol, and triglycerides and raise HDL (good) cholesterol | 41, 42 |
| Upregulate CYP7A1, a rate-limiting enzyme in the biosynthesis of bile acid from cholesterol in liver that is involved in the decrease of cholesterol; this induction of CYP7A1 increases the conversion of cholesterol into bile acids and its excretion | ||
| Antidiabetic | Suppress expression of hepatic gluconeogenesis genes (PEPCK and G6Pase) similarly to insulin | 43–46 |
| Activate AMPK and downregulate ACC; also implicated in glucose transport and shown to suppress the key gluconeogenic genes PEPCK and G6Pase | ||
| Anti-inflammatory | Inhibit NF-кB, COX-2, 5-LOX, iNOS, and other molecules that mediate inflammatory effect | 40, 47–52 |
| Antioxidant | Protect biomembranes against lipid peroxidative damage by scavenging the reactive free radicals involved in the peroxidation | 31, 49, 50, 53–55 |
| Anti-Alzheimer's disease | Reduce amyloid plaques and accumulated β-amyloid aggregates | 56–59 |
| Suppress pro-oxidant, proinflammatory, and JNK-mediated toxic amyloid aggregate effects | ||
| Anti-HIV | Inhibit HIV replication | 60–62 |
| Inhibit HIV-1 and HIV-2 proteases | ||
| Wound healing | Protect tissue from oxidative damage due to their antioxidant properties | 63, 64 |
| Increase formation of granulation tissue and biosynthesis of extracellular matrix proteins, especially lower in diabetic wounds | ||
| Anti-Parkinson's disease | Reduce aggregation of α-synuclein | 65, 66 |
| Protect dopaminergic neurons from apoptosis by inhibition of JNK |
NF-κB, nuclear transcription factor κB; COX-2, cyclooxygenase-2; BID, BH3-interacting domain death agonist; LDL, low-density lipoprotein; HDL, high-density lipoprotein; CYP7A1, cholesterol 7a-hydroxylase; 5-LOX, 5-lipoxygenase; iNOS, inducible nitric oxide synthase; JNK, Jun N-terminal kinase.
Even though curcumin, demethoxycurcumin, and bisdemethoxycurcumin have been extensively studied, whether the three analogues exhibit equal therapeutic activity has not been completely elucidated. Although curcumin was found to be most potent/active in some systems (55), in other cases it was concluded that the activities of the three analogues are very similar (67), while in other systems bisdemethoxycurcumin was found to exhibit the highest cytotoxic activity (10, 31, 32, 68). Furthermore, the mixture of all three curcuminoids has been suggested to have a more effective bioprotectant activity than any one alone due to synergistic effects (69).
Bioavailability of Curcuminoids
Nowadays, curcumin is considered a safe, novel, and promising drug for the prevention and treatment of cancer, chronic inflammation, and other diseases (70, 71). Several clinical trials have been conducted to confirm its effectiveness (36, 70, 72–74) and have revealed that curcumin is safe for humans at a high oral dosage of 10 to 12 g per day (70, 74).
In August 2009, Sabinsa Corporation received a “generally regarded as safe” (GRAS) status for the branded ingredient Curcumin C3 Complex (69) after a review of safety and toxicology data by Soni & Associates, Inc. In February 2013, due to curcumin's health benefits and the results obtained in clinical trials, the U.S. Food and Drug Administration issued a “no-objection letter” to Sabinsa for the self-affirmed GRAS status of its Curcumin C3 Complex for use in food and beverages (75).
Although the therapeutic applications of curcuminoids appear very promising, to date clinical trials are still preliminary, and the true value of curcumin as a therapeutic agent for human diseases remains elusive (70). It is imperative that well-designed clinical trials, with improved formulations of curcuminoids or novel routes of administration (70), be conducted in the near future. Animal studies and phase I and II clinical trials conducted so far showed that curcumin and other curcuminoids exhibit a poor bioavailability, which is the main reason why curcumin has not yet been approved as a therapeutic agent (76). The poor bioavailability of curcuminoids appears to be due to their poor absorption and distribution, rapid metabolism, and rapid systemic elimination (excretion) (76). These compounds are insoluble and unstable in water, and, in general, after administration their presence in serum is very low or undetectable (76). Several approaches have been evaluated to improve the bioavailability of curcumin and the other curcuminoids. The use of adjuvants that can block metabolism of the curcuminoids is one of the most studied ways to improve their bioavailability (77). For example, the coadministration of piperine proved to highly increase curcumin serum concentration, absorption, and bioavailability in rats and humans (76, 78). Other promising approaches to improve curcumin bioavailability include the use of liposomes (79–81), micelles (82–84), nanoparticles (25, 85–91), phospholipid complexes (92, 93), and curcuminoid derivatives and structural analogues (58, 94, 95).
An efficient application of curcumin requires specific lesion-oriented delivery methods to guarantee the needed concentrations in tissues. Noninvasive and invasive drug delivery methods can be used (96). Noninvasive methods are usually painless and easy to use, and the most common routes of administration include peroral, topical, and transmucosal (nasal, vaginal, rectal, and urethral) routes (i.e., oral lozenges, vaginal suppository tablets, and pomades). Invasive methods penetrate the cutaneous barrier to reach either the circulatory system or the diseased tissue and include, for example, intravenous, intramuscular, and subcutaneous methods.
To solve the curcuminoid bioavailability problem, new methods for its production and delivery at the intended body site need to be developed. According to Hsu and Cheng (70) it is very likely that to obtain active curcumin concentrations in tissues, it is necessary to apply the drugs locally or topically using a targeted drug delivery system. Controlled in situ drug delivery and production, using combinatorial biosynthesis and a host microorganism, is a very promising approach. In this paper we review the production of curcuminoids in C. longa and by Escherichia coli using an artificial biosynthetic pathway. We also suggest new approaches using metabolic engineering and synthetic biology tools to increase the curcuminoid yield and activity.
CURCUMINOID PRODUCTION IN CURCUMA LONGA
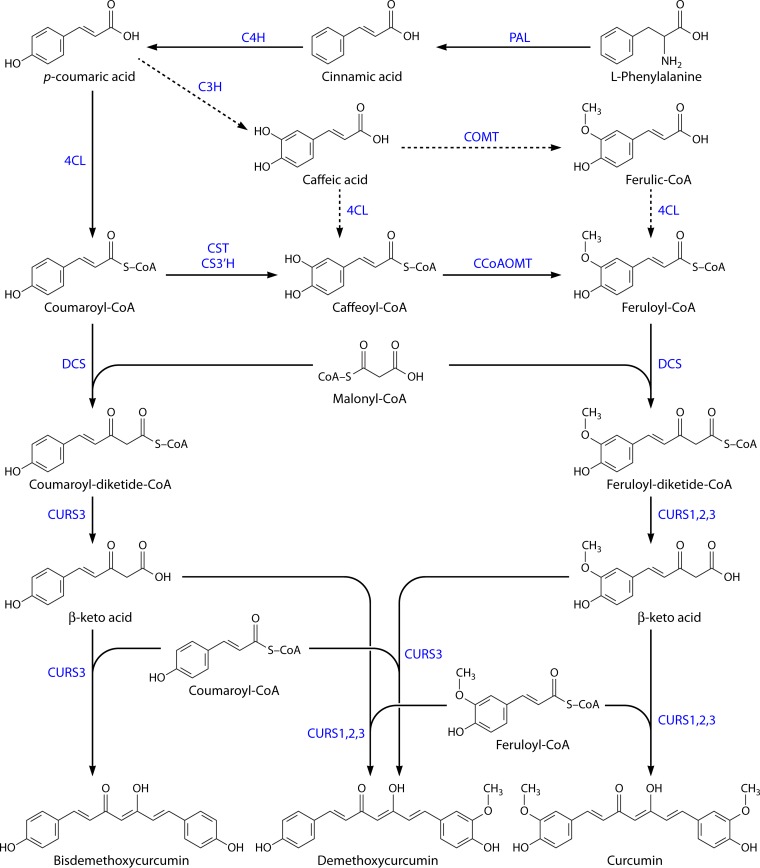
Curcuminoids are produced by type III polyketide synthases (PKS) in plants and consist of two phenylpropanoid units chemically derived from the amino acid phenylalanine and connected by a central carbon unit derived from malonyl-coenzyme A (malonyl-CoA) (97). Until the work by Ramirez-Ahumada et al. (98), very little was known about curcuminoid biosynthesis in turmeric. Those authors identified the enzymes in the curcuminoid biosynthetic pathway and confirmed the involvement of the phenylpropanoid pathway in the production of these compounds in plants. The first enzyme identified was phenylalanine ammonia lyase (PAL), which is involved in amino acid metabolism, but also in the biosynthesis of plant natural products, as the branch point between primary and secondary metabolism (99). Other enzymes identified include p-coumaroyl shikimate transferase (CST), p-coumaroyl quinate transferase (CQT), caffeic acid O-methyltransferase (COMT), and caffeoyl-CoA O-methyltransferase (CCOMT or CCoAOMT). The results for polyketide synthase (PKS) in in vitro assays showed detectable curcuminoid synthase activity in the extracts from turmeric. However, the authors were not able to characterize the enzyme(s) responsible for this reaction using anion-exchange column chromatography (98). Furthermore, Katsuyama et al. (97) proposed a pathway for curcuminoid biosynthesis in the herb C. longa, which included two type III PKS (Fig. 1). One of the PKS, named diketide-CoA synthase (DCS), catalyzes the formation of feruloyl-diketide-CoA from feruloyl-CoA and malonyl-CoA. The other PKS, named curcumin synthase 1 (CURS1), catalyzes two reactions. First, it catalyzes the hydrolysis of feruloyl-diketide-CoA in a β-keto acid. Second, using the β-keto acid and other molecule of feruloyl-CoA, it catalyzes the formation of curcumin. β-Keto acid is not detected in vitro because it is not released from the enzyme (100, 101). Both DCS and CURS1 accept p-coumaroyl-CoA, but at low efficiency, and are capable of synthesizing bisdemethoxycurcumin. The asymmetric curcuminoid demethoxycurcumin can also be produced from p-coumaroyl-diketide-CoA and feruloyl-CoA or from feruloyl-diketide-CoA and p-coumaroyl-CoA. DCS and CURS1 share 62% amino acid sequence identity and a conserved Cys-His-Asn catalytic triad (97).
FIG 1.
Curcuminoid biosynthetic pathway in Curcuma longa. Cinnamic acid is synthesized from phenylalanine by phenylalanine ammonia lyase (PAL) and converted to coumaric acid by cinnamate-4-hydroxylase (C4H). Then, 4-coumarate-CoA ligase (4CL) converts coumaric acid to coumaroyl-CoA, and p-coumaroyl shikimate transferase (CST), p-coumaroyl 5-O-shikimate 3′-hydroxylase (CS3′H), and caffeoyl-CoA O-methyltransferase (CCoAOMT) convert it to feruloyl-CoA. Coumaroyl-CoA and feruloyl-CoA are then converted by diketide-CoA synthase (DCS) to diketide-CoAs by condensation with malonyl-CoA. In the end, curcumin synthases (CURSs) catalyze the formation of curcuminoids by condensing the diketide-CoAs with coumaroyl-CoA and feruloyl-CoA. Depending on the combination, different curcuminoids are produced, namely, bisdemethoxycurcumin, demethoxycurcumin, and curcumin. The route indicated by dashed arrows corresponds to a less central phenylpropanoid pathway and may not occur in vivo in C. longa.
The same authors identified and characterized two other type III PKS from turmeric, named CURS2 and CURS3 (102). In vitro analysis revealed that CURS2 preferred feruloyl-CoA as the substrate, while CURS3 used both feruloyl-CoA and p-coumaroyl-CoA (Table 2). CURS2 and CURS3 share 81% identity, and CURS2 and CURS3 share 78% and 81% identity with CURS1 and 62% and 66% with DCS, respectively. The existence of these three curcumin synthases with distinctive substrate specificities might explain the distribution of the three curcuminoids (curcumin, demethoxycurcumin, and bisdemethoxycurcumin) in C. longa. Before it was known that C. longa had different CURSs with different specificities and that they could utilize both CoA esters of coumaric acid and ferulic acid, Ramirez-Ahumada et al. (98) proposed that bisdemethoxycurcumin (synthesized only from coumaroyl-CoA) would be transformed in demethoxycurcumin and then in curcumin by hydroxylation followed by O-methylation. This hypothesis was never confirmed and was almost discarded after the finding of curcumin synthase, which was able to use feruloyl-CoA to produce curcumin.
TABLE 2.
Kinetic parameters of DCS, CURS1, CURS2, and CURS3 from C. longa
| EC no. | Enzymea | GenBank accession no. | Substrate | Km (μM) | kcat (s−1) | kcat/Km (s−1 M−1) | Reference |
|---|---|---|---|---|---|---|---|
| 2.3.1.218 | DCS | AB495006 | Feruloyl-CoA | – | 0.020 | – | 97b |
| Malonyl-CoA | 8.4 | 0.011 | 1,329 | ||||
| 2.3.1.217/2.3.1.219 | CURS1 | AB495007 | Feruloyl-CoA | 18 | 0.018 | 1,001 | 97 |
| Coumaroyl-CoA | 189 | 0.014 | 75 | ||||
| CURS2 | AB506762 | Feruloyl-CoA | 4.3 | 0.007 | 1,622 | 102 | |
| Coumaroyl-CoA | 89 | 0.016 | 176 | ||||
| CURS3 | AB506763 | Feruloyl-CoA | 2.2 | 0.003 | 2,017 | 102 | |
| Coumaroyl-CoA | 3.4 | 0.006 | 1,742 |
DCS, diketide-CoA synthase; CURS, curcumin synthase/demethoxycurcumin synthase.
Kinetic properties of DCS for feruloyl-CoA were calculated according to its allosteric properties (sigmoidal curve). The S50, kcat, and Hill slope values were 46 μM, 0.02 s−1, and 1.8, respectively.
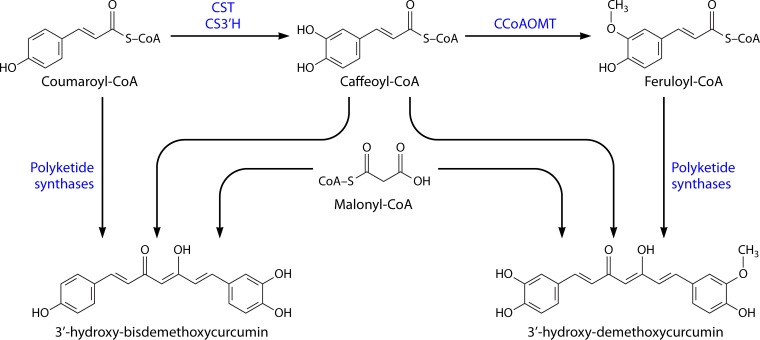
In addition to the well-known curcuminoids, other diarylheptanoids can also be found in turmeric. The biosynthetic pathway of these diarylheptanoids has not yet been elucidated and several routes have been hypothesized. Xie et al. (104), after using metabolic profiling analysis, proposed that the biosynthetic pathway belongs to a different metabolite module (Fig. 2) and that they are not intermediates in curcuminoid biosynthesis that belong to another biosynthetic module (Fig. 1).
FIG 2.
Biosynthetic production pathway of some diarylheptanoids in Curcuma longa. CST, coumaroyl shikimate transferase; CS3′H, p-coumaroyl 5-O-shikimate 3′-hydroxylase; CCoAOMT, caffeoyl-CoA O-methyltransferase.
According to Xie et al. (104), caffeoyl-CoA in combination with coumaroyl-CoA or feruloyl-CoA is used to produce 3′-hydroxy-bisdemethoxycurcumin and 3′-hydroxy-demethoxycurcumin, respectively. The polyketide synthases responsible for these reactions have specificities different from the ones from the other module (Fig. 1), which cannot use caffeoyl-CoA as a substrate and produce the major curcuminoids. Furthermore, Xie et al. (104) reported that some PKS, instead of using caffeoyl-CoA, use 5-hydroxy-feruloyl-CoA or feruloyl-CoA and coumaroyl-CoA to produce 5′-hydroxy-curcumin and 5′-hydroxy-demethoxycurcumin, respectively.
Dihydrocurcuminoids, which are dihydro derivatives of curcuminoids (103), have also been identified in C. longa, for example, dihydrodemethoxycurcumin, dihydrobisdemethoxycurcumin, and dihydrocurcumin. Kita et al. (105) considered two hypotheses for the production of these dihydrocurcuminoids. In one hypothesis, an enzyme accepts one dihydrophenylpropanoid and one phenylpropanoid instead of the two phenylpropanoids used in the production of the major curcuminoids. The other hypothesis considers the reduction of a double bond of the major curcuminoids by a reductase.
HETEROLOGOUS PRODUCTION OF CURCUMINOIDS
Curcuminoids, like many plant secondary metabolites, accumulate at low quantities over very long growth periods in plants and are difficult and expensive to isolate and hard to synthesize chemically. These reasons, in combination with the wide benefits of curcuminoids and their application potential, have led to an increased interest in the last 10 years, and attempts to implement the heterologous biosynthesis of curcuminoids have been reported (106). One of the approaches to produce curcuminoids is combinatorial biosynthesis, which consists of combining enzyme-encoding genes from different species and designing a new set of gene clusters to produce bioactive compounds in a heterologous host (107, 108). Heterologous production of curcuminoids in microorganisms is highly advantageous, since they can grow on inexpensive substrates and, compared to plants, are easier to manipulate and have very rapid production cycles (109), allowing curcuminoids to be produced faster and maybe in larger amounts. Large-scale microbial fermentation and downstream purification can also be more easily attained, since microbes usually do not have competing pathways to transgenic metabolism (110). Heterologous production in plants has the advantage of requiring the introduction of only one or two genes, since the other genes from the phenylpropanoid pathway are already present in the plant kingdom (111). Also, the engineering of plants can lead to an increase in antioxidant activities and in disease resistance in the transgenic plants and can extend the postharvest shelf life of some fruits, such as in cases where resveratrol is engineered in plants (111, 112). However, introducing genetically modified crops has a high cost, and there is public resistance to the acceptance of these food products. Regulations for genetically modified microorganisms are simpler than those for crops (112). Thus, taking into consideration all the pros and cons, the commercial application of heterologous production of curcuminoids by microorganisms is more attractive.
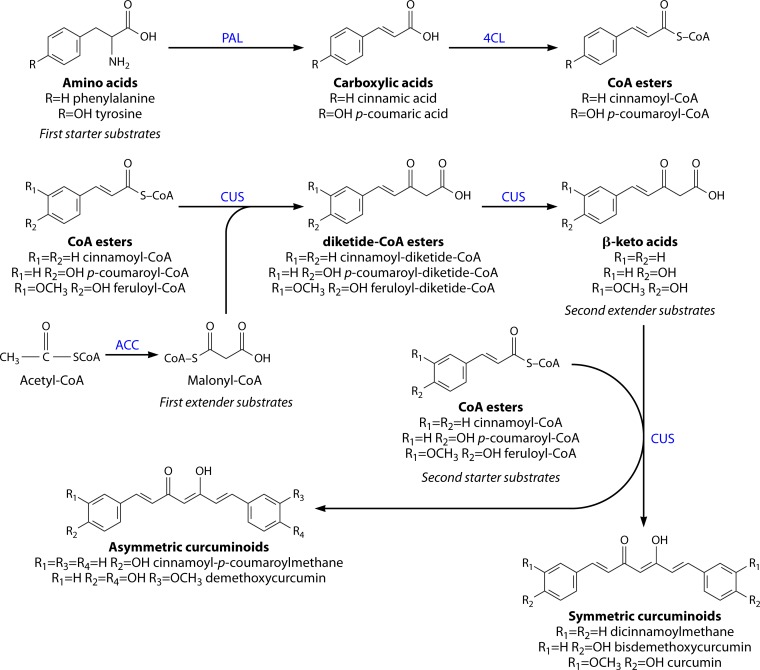
Although DCS and CURS are the enzymes responsible for the last steps of the curcuminoid pathway in C. longa, these enzymes were discovered only in 2009 (97). At that time, Katsuyama et al. (106) had already implemented in E. coli an artificial biosynthetic pathway to produce curcuminoids. Those authors identified a type III PKS from rice, Oryza sativa. This enzyme was named curcuminoid synthase (CUS) (GenBank accession number AK109558) and was the first identified type III PKS capable of catalyzing the “one-pot” synthesis of bisdemethoxycurcumin from two molecules of coumaroyl-CoA (starter substrate) and one molecule of malonyl-CoA (extender substrate). CUS itself catalyzes both steps catalyzed by DCS and CURS in C. longa, disobeying the traditional model of head-to-tail polyketide assembly by only catalyzing condensation reactions without a cyclization (113). Morita et al. (114) proposed that CUS has a unique downward-expanding active-site architecture that allows accommodation of two coumaroyl molecules (coumaroyl-CoA and coumaroyl β-keto acid) and one malonyl-CoA molecule and a putative nucleophilic water molecule that forms hydrogen bond networks at the active-site center. These unique structural features are the basis for the “one-pot” synthesis of bisdemethoxycurcumin. Thus, the use of CUS in the artificial biosynthetic production of curcuminoids is simpler than the DCS/CURS system. The Cys-His-Asn catalytic triad is conserved in CUS (113, 114), and the enzyme shares 51% amino acid identity with DCS and 45% identity with CURS1. Although CUS also accepts cinnamoyl-CoA and feruloyl-CoA as a substrate to produce dicinnamoylmethane and curcumin, it prefers coumaroyl-CoA. To date, curcuminoids have not been reported in O. sativa and CUS activity in vivo remains to be identified. Hypothetically, the rice CUS may produce curcuminoids, albeit in undetectable amounts (100). In addition to curcuminoid synthesis, CUS also proved to be able to produce gingerol derivatives (100, 115).
The report of the artificial curcuminoid biosynthetic pathway in E. coli, designed by Katsuyama et al. (106), is the first study demonstrating the production of curcuminoids in a heterologous organism. This pathway (Fig. 3) starts with the reaction of phenylalanine ammonia lyase (PAL) from the yeast Rhodotorula rubra, which converts l-phenylalanine to cinnamic acid. This enzyme was previously shown to have tyrosine ammonia lyase (TAL) activity (116, 117). Using this enzymatic feature, tyrosine can be used as a precursor and further converted to coumaric acid. The carboxylic acids are converted to CoA esters by 4-coumarate-CoA ligase (4CL) from Lithospermum erythrorhizon (Le4CL1) and then to curcuminoids by CUS from O. sativa. Acetyl-CoA carboxylase (ACC) from Corynebacterium glutamicum was also overexpressed to increase the intracellular pool of malonyl-CoA in E. coli. The enzymes and microorganisms in this pathway were chosen after the successful production of flavonoid and stilbene compounds, whose pathway included some of the same enzymes (118, 119). The recombinant E. coli was cultivated in M9 medium with the supplementation of tyrosine or/and phenylalanine (3 mM), the precursors of the phenylpropanoid pathway. Bisdemethoxycurcumin, dicinnamoylmethane, and cinnamoyl-p-coumaroylmethane were produced. In another experiment, phenylpropanoid acids (p-coumaric acid, cinnamic acid, and ferulic acid [1 mM]) were directly supplied to test a system with only 4CL, CUS, and ACC genes. The goal was to increase the CoA ester concentration in the cell by removing the PAL step where the amino acids are converted to carboxylic acids. E. coli produced 91 ± 23 mg/liter of bisdemethoxycurcumin, 84 ± 15 mg/liter of dicinnamoylmethane, and 113 ± 22 mg/liter of curcumin. Furthermore, rice bran pitch was used for curcumin production. Rice bran pitch is an industrial waste obtained from rice bran in the production of rice oil (120). It is known that rice bran pitch is rich in ferulic acid. Starting with 11 ± 1.4 mg of ferulic acid extracted from 500 mg of rice bran pitch, the E. coli strain engineered by Katsuyama et al. (106), harboring 4CL, CUS, and ACC genes, produced 57 ± 21 mg/liter of curcumin.
FIG 3.
Biosynthesis of curcuminoids by recombinant Escherichia coli, using tyrosine and/or phenylalanine as starter substrates that are converted to carboxylic acids by PAL (phenylalanine ammonia lyase). The carboxylic acids, which can also be added directly to the medium, are converted into the corresponding CoA esters by 4CL (4-coumarate-CoA ligase) (top), which is followed by several reactions catalyzed by curcuminoids synthase (CUS) (bottom). Malonyl-CoA is overproduced by ACC (acetyl-CoA carboxylase). (Adapted from references 106 and 100.)
Using the same pathway (4CL, CUS, and ACC), 15 asymmetric curcuminoids (9 of these compounds are not found in nature) were produced by adding two different unnatural carboxylic acids simultaneously (analogues of p-coumaric acid) (121). This precursor-directed biosynthesis system is possible due to the unusually broad, promiscuous substrate specificities of the enzymes involved in the pathway, which allows production of unnatural novel polyketides. This way of producing unnatural curcuminoids may provide novel drug candidates.
Curcuminoid production in E. coli was also used to develop a reporter assay for the screening of PAL enzyme efficiency (122). PALs were coexpressed with 4CL1 from Arabidopsis thaliana (At4CL1) and CUS from O. sativa in E. coli. The yellow color of the product allowed the development of a microplate-based assay to measure dicinnamoylmethane. Using this system, the authors screened three PALs (PAL1, PAL3, and PAL4) from Trifolium pratense. PAL1 showed the best results by producing 0.36 g/liter of dicinnamoylmethane. This combination of enzymes allowed production of around 3.4 times more dicinnamoylmethane from phenylalanine than that obtained by Katsuyama et al. (106) using PAL from R. rubra and 4CL from L. erythrorhizon. Furthermore, the ACC enzyme was not used, which means that, at least in this case, the naturally present malonyl-CoA was enough for a successful production. In addition, by feeding 2-fluorol-phenylalanine, Wang and colleagues produced three curcuminoids: dicinnamoylmethane, 6,6′-difluoro-dicinnamoylmethane, and 6-fluoro-dicinnamoylmethane, a new unnatural compound (122).
OPTIMIZATION OF THE CURCUMINOID HETEROLOGOUS PATHWAY AND ALTERNATIVE HOSTS
The success of a biosynthetic pathway depends significantly on the use of appropriate synthetic enzymes. Curcuminoid production can be improved by exploring alternative enzymes from other microorganisms compatible with the heterologous host that allow higher curcuminoid yield or more specific enzymes that allow production of the desired curcuminoid with fewer undesired by-products.
To our knowledge, heterologous synthesis of curcuminoids has so far been accomplished only in E. coli. Saccharomyces cerevisiae, which is also easy to grow and manipulate and is well characterized, has been used only to produce other polyketides, such as resveratrol, naringenin, and pinocembrim, among others (Table 3) (115, 123–132). However, as a eukaryote, S. cerevisiae presents some unique advantages over E. coli for the design and construction of a biosynthetic pathway for the production of curcuminoids. First, it has a food-grade status (GRAS organism) which allows its use in human nutrition and pharmaceuticals. Furthermore, S. cerevisiae does not lack the posttranslational machinery like E. coli, having intracellular compartments similar to those of plant cells (126). Also, membrane proteins such as cytochrome P450 (C4H) would be more adequately expressed in a eukaryotic organism. These questions have been properly addressed and reviewed recently (133), and emerging strategies have been employed to overcome the problems that occur when expressing eukaryotic proteins in a bacterial host. Nevertheless, in some cases the use of a eukaryotic organism like S. cerevisiae would be simpler.
TABLE 3.
Production titers and yields of polyketides obtained by heterologous production in microorganisms or extracted from plants
| Organism | Polyketide | Substrate | Titer (mg/liter) | Yield (g polyketide/g substrate) | Reference(s) |
|---|---|---|---|---|---|
| C. longa | Curcuminoids | Curcuminoids in dry turmeric | 0.5–0.7 | 15, 134–136 | |
| E. coli | Curcuminoids | ||||
| Curcumin | Ferulic acid | 113 | 0.58 | 106 | |
| Bisdemethoxy curcumin | p-Coumaric acid | 91 | 0.55 | 106 | |
| Dicinnamoyl methane | Phenylalanine | 360 | 0.73 | 122 | |
| Cinnamoyl-p-coumaroyl methane | Tyrosine + phenylalanine | 19 | 0.02 | 106 | |
| Flavonoids | |||||
| Naringenin | p-Coumaric acid | 474 | 1.11 | 137 | |
| Tyrosine | 57 | 0.10 | 116 | ||
| Pinocembrin | Cinnamic acid | 480 | 1.08 | 138 | |
| Phenylalanine | 58 | 0.12 | 116 | ||
| Stillbenoids | |||||
| Resveratrol | p-Coumaric acid | 171 | 1.04 | 119 | |
| S. cerevisiae | Flavonoids | ||||
| Naringenin | Phenylalaninea | 109 | 132 | ||
| Pinocembrin | Cinnamic acid | 16.3 | 0.11 | 127 | |
| Stillbenoids | |||||
| Resveratrol | p-Coumaric acid | 391 | 0.16 | 139 |
Phenylalanine-overproducing strain.
Tyrosine/Phenylalanine-Overproducing Strains
E. coli and S. cerevisiae lack the CoA-ester starter substrates needed for curcuminoid production, and the naturally produced amino acids (phenylalanine and tyrosine) that are converted to the phenylpropanoic acids are not produced in sufficient amounts, which represents a limiting step (126, 140, 141). Although the amino acids and phenylpropanoic acids can be supplemented to the culture medium, the development of strains capable of converting glucose or other simple carbon sources to curcuminoids represents an important strategy for the engineering process. This approach eliminates the need to add expensive precursors to the medium, thus decreasing the production cost. Large-scale production also would be more suitable. This medium simplification would also be very advantageous in cases where curcuminoids are to be used as drugs that need to be produced and delivered in situ.
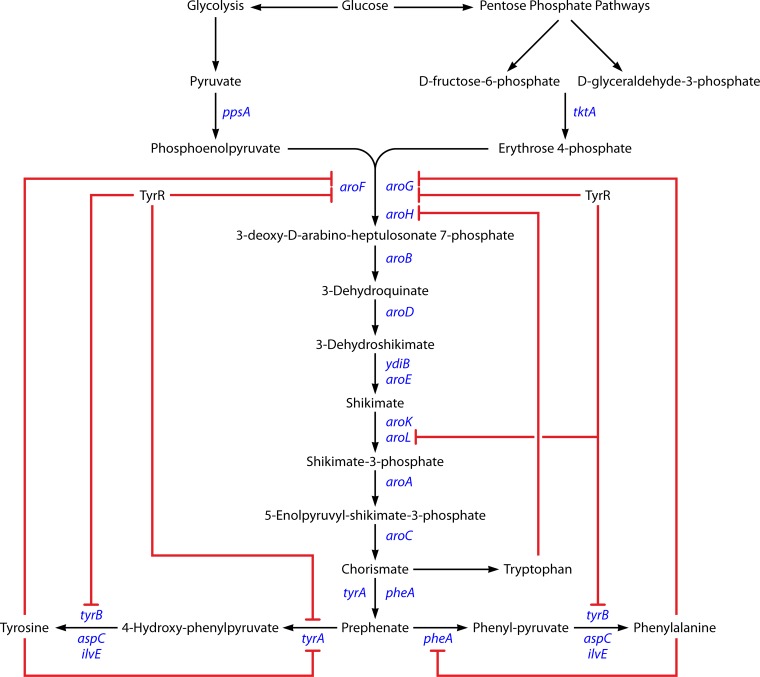
The aromatic amino acid biosynthesis in E. coli is a very complex, highly regulated branched pathway (Fig. 4). This pathway is regulated at the transcription and allosteric levels. One of the most regulated steps of the pathway is the one catalyzed by 3-deoxy-d-arabino-heptulosonate 7-phosphate (DAHP) synthase (142). The three isoenzymes of this DAHP synthase (encoded by aroH, aroF, and aroG) are feedback inhibited by the end products. Another regulatory point is present at the chorismate branch point with the enzymes chorismate mutase and prephenate dehydratase (pheA and tyrA). These bifunctional enzymes are feedback regulated by their end products, phenylalanine and tyrosine. In addition to the allosteric inhibition, it is necessary to take into account the transcriptional control mediated by the protein TyrR (tyrosine repressor). In case of amino acid overproduction, TyrR can repress aroF, aroG, tyrA, and tyrB (143). As a result, the elimination of TyrR-mediated control was shown to be an important target for the successful overexpression of amino acids (144). Also, the sequences of the repressed genes were modified so that their products were no longer sensitive to feedback inhibition. The overexpression of feedback inhibition-resistant derivatives of the aroG (aroGfbr) and tyrA (tyrAfbr) genes proved to help in amino acid overexpression (145–149).
FIG 4.
Biosynthesis of aromatic amino acids in Escherichia coli. Red lines show the regulation points.
Engineering the central carbon metabolism to increase the availability of the two main precursors, phosphoenolpyruvate (PEP) and erythrose 4-phosphate (E4P), has been another approach to overexpress these amino acids. Several strategies have been tested, for example, the overexpression of ppsA (encoding PEP synthase) and tktA (encoding transkelotase) (147–149).
Some studies related to production of phenylpropanoic acids (140, 141, 150–152) and flavonoids (153) have used tyrosine- or phenylalanine-overproducing strains, and all obtained better results with these strains than with the wild type. All of them used similar approaches, in which they overexpressed aroGfbr and tyrAfbr, and in some cases ppsA and tktA, and deleted tyrR (ΔtyrR). To direct the pathway only to tyrosine production, they also deleted the pheA gene (ΔpheA). Huang et al. (151) engineered a well-developed phenylalanine overproducer strain to obtain a tyrosine-overproducing strain. The phenylalanine-overproducing strain E. coli ATCC 31884 overexpresses aroGfbr, aroFfbr, and pheAfbr and suppresses tyrR. This strain was transformed into a tyrosine-overproducing strain by disruption of the pheAfbr and tyrA loci and insertion of tyrAfbr.
Also in S. cerevisiae, aromatic amino acid biosynthesis is subject to strong feedback inhibition by tyrosine and phenylalanine. To overcome this problem, Koopman et al. (132) introduced a tyrosine feedback inhibition-resistant derivative of aro4 (aro4fbr) in combination with the deletion of aro3, the other allele of DAHP synthase. Those authors also eliminated the competing activity of phenylpyruvate decarboxylase (which yields phenyl acetaldehyde instead of phenylalanine) by deleting the aro10, pdc5, and pdc6 genes that encode it. This engineered S. cerevisiae strain allowed significantly higher production of naringenin than the wild-type strain.
From Tyrosine/Phenylalanine to Coumaric Acid, Caffeic Acid, Ferulic Acid, and Their Corresponding CoA Esters
In the last years, several studies have used the enzymes involved in the phenylpropanoid pathway to produce compounds other than curcuminoids, such as phenylpropanoic acids (coumaric acid, caffeic acid, and ferulic acid) (140, 141, 151, 152, 154–157), stilbenoids (155, 158), and flavonoids (116, 117, 150, 155, 159–162). In the design and construction of those pathways, some enzymes from other organisms were used that could also be considered for the optimization of the heterologous production of curcuminoids.
Due to its valuable pharmacological properties of caffeic acid, among them antioxidant (163), anti-inflammatory (164), and anticancer (165) properties, the caffeic acid biosynthetic pathway has been intensively studied in the last decade. Berner et al. (154) studied the caffeic acid biosynthetic pathway in the actinomycete Saccharothrix espanaensis and heterologously expressed it in Streptomyces fradiae XKS, a polyketide synthase-defective mutant strain that is considered to be a very convenient host for expression experiments (166). Streptomyces, applied in the production of a wide range of secondary metabolites, including polyketides, can be another candidate for the heterologous production of plant-specific polyketides (167). Berner et al. (154) concluded that l-tyrosine (and not l-phenylalanine) is converted to p-coumaric acid by sam8, encoding TAL (SeTAL). This is not in accordance with the general phenylpropanoid pathway in plants, but it allows a simpler pathway that can be integrated in the heterologous production of curcuminoids by E. coli. Other authors also reported their findings on other bacterial TALs. Studies of Rhodobacter capsulatus TAL (RcTAL) and Rhodobacter sphaeroides TAL (RsTAL) catalytic efficiencies showed a clear preference of these enzymes for tyrosine instead of phenylalanine (Table 4). These TAL enzymes that allow p-coumaric acid to be produced without requiring the C4H step, where this enzyme converts cinnamic acid to coumaric acid, are highly advantageous since one can avoid the use of the C4H enzyme, which is not successfully expressed in prokaryotic organisms, thus creating a blockage in the biosynthetic pathways (161). C4H is a membrane-bound cytochrome P450-dependent hydroxylase, and its expression in bacteria is very challenging due to protein instability and insolubility, as well as the lack of cytochrome P450 reductase activity that is necessary for P450 function (redox partners) (108, 168). After producing coumaric acid, caffeic acid is directly converted by sam5, encoding a 4-coumarate 3-hydroxylase (C3H or Coum3H). C3H can also convert coumaroyl-CoA to caffeoyl-CoA (169). Moreover, Berner et al. (154) identified sam7, encoding a caffeoyl-CoA ligase (4CL) that catalyzes the ligation of caffeic acid to CoA, leading to caffeoyl-CoA. This route, producing caffeic acid and then caffeoyl-CoA, can be very useful to ultimately produce curcumin since caffeoyl-CoA can be converted in feruloyl-CoA by CCoAOMT (Fig. 1) and curcumin is produced from two molecules of feruloyl-CoA and one of malonyl-CoA.
TABLE 4.
Kinetic parameters of phenylalanine ammonia lyases/tyrosine ammonia lyases from different organisms toward l-phenylalanine and l-tyrosine
| EC no. | Enzyme | Organism | Substrate | Km (μM) | kcat (s−1) | kcat/Km (s−1 M−1) | Reference |
|---|---|---|---|---|---|---|---|
| 4.3.1.24 | Phenylalanine ammonia lyase | Rhodotorula rubra | l-Phe | 446 | 157a | ||
| l-Tyr | 220 | ||||||
| 4.3.1.23 | Tyrosine ammonia lyase | Saccharothrix espanaensis | l-Phe | 2860 | 0.0038 | 1.3 | 154 |
| l-Tyr | 15.5 | 0.015 | 968 | ||||
| Rhodobacter sphaeroides | l-Phe | 560 | 0.01 | 18 | 170 | ||
| l-Tyr | 60 | 0.02 | 333 | ||||
| Rhodobacter capsulatus | l-Phe | 560 | 0.04 | 57 | 170 | ||
| l-Tyr | 160 | 0.06 | 375 | ||||
| 4.3.1.25 | Phenylalanine/tyrosine ammonia lyase | Rhodotorula glutinis | l-Phe | 183 | 1.6 | 8.7 × 103 | 171b |
| l-Tyr | 615 | 0.53 | 8.6 × 102 |
kcat and kcat/Km were not determined. However, the enzyme showed a Vmax of 0.169 μmol min−1 mg−1 for l-Phe and 0.033 μmol min−1 mg−1 for l-Tyr.
Kinetic properties at pH 8.5. At pH 9.5 the RgTAL enzyme showed kcat, Km and kcat/Km values of 1.5 s−1, 126 μM, and 1.2 × 104 s−1 M−1, respectively, for l-Phe and 0.93 s−1, 68 μM, and 1.4 × 104 s−1 M−1, respectively, for l-Tyr.
The pathway described by Berner et al. (154) was afterwards reconstituted in E. coli by Choi et al. (155) and Kang et al. (141) to produce phenylpropanoic acids such as p-coumaric acid, caffeic acid, and ferulic acid. In their pathways, the authors used SeTAL and sam5 (C3H) from S. espanaensis and COMT from Arabidopsis thaliana. Although Choi et al. (155) reported positive results, ferulic acid production in the end was only 7.1 mg/liter. The pathway created by Kang et al. (141) contained a codon-optimized tal gene that improved the production of p-coumaric acid and ferulic acid but not that of caffeic acid. The authors also tested a tyrosine-overproducing strain that produced 974 mg/liter p-coumaric acid, 150 mg/liter caffeic acid, and 196 mg/liter ferulic acid in shake flasks after 36 h of cultivation.
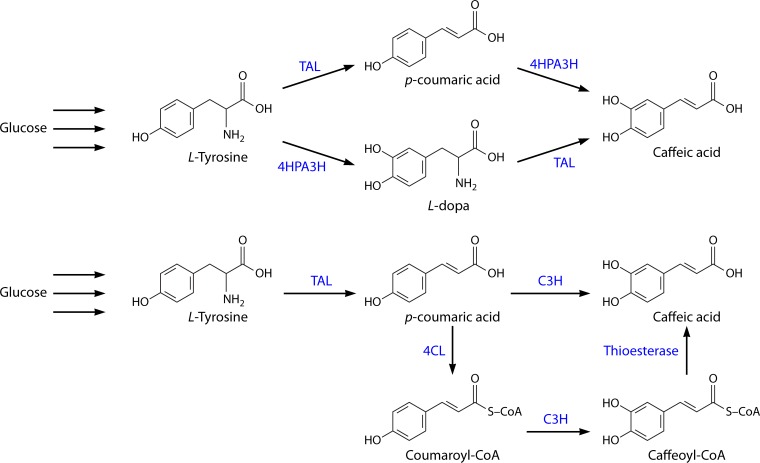
Lin and Yan (140) and Huang et al. (151) tested a different approach for caffeic acid production. Since one of the most challenging steps in the recreation of the plant phenylpropanoid pathway in E. coli is the one carried out by the cytochrome P450-dependent hydroxylases (172), the search for alternative enzymes compatible with E. coli is crucial. With that in mind, Lin and Yan (140) characterized an E. coli native hydroxylase complex, hydroxyphenylacetate 3-hydroxylase (4HPA3H), and concluded that this enzyme, besides converting l-tyrosine to l-dopa, was also able to efficiently convert p-coumaric acid to caffeic acid (Fig. 5, top). Furthermore, the authors found that RsTAL and RcTAL were able to accept tyrosine and l-dopa as substrates, with RcTAL being slightly more active toward both substrates. Hence, they were able to design an artificial dual and promiscuous pathway that used both p-coumaric acid and l-dopa as intermediates, resulting in the production of 50.2 mg/liter caffeic acid in shake flasks after 48 h of cultivation from simple carbon sources. This production was also obtained by alleviating feedback inhibition and redirecting carbon flux into tyrosine biosynthesis. Huang et al. (151) explored the catalytic potential of 4HPA3H and were able to produce 3.82 g/liter caffeic acid from 3.5 g/liter p-coumaric acid in a wild-type E. coli strain. When using a tyrosine-overproducing strain, 767 mg/liter caffeic acid was produced in 72 h using shake flasks. In this work, the authors used Rhodotorula glutinis TAL (RgTAL), which was reported to be the most active TAL among all the identified ones (150, 173–175) (Table 4).
FIG 5.
Pathway for biosynthesis of caffeic acid using the approaches described by Lin and Yan (140) and Huang et al. (151) (top) and Zhang and Stephanopoulos (152) (bottom). TAL, tyrosine ammonia lyase; C3H, 4-coumarate 3-hydroxylase; 4HPA3H, hydroxyphenylacetate 3-hydroxylase; 4CL, 4-coumarate-CoA ligase.
Zhang and Stephanopoulos (152) also used RgTAL for caffeic acid biosynthesis, in an experiment using two parallel routes and a tyrosine overproducer strain (Fig. 5, bottom). The authors cloned 4CL from Petroselinum crispum (Pc4CL1), that converts coumaric acid to coumaroyl-CoA, and C3H from S. espanaensis, which converts coumaroyl-CoA to caffeoyl-CoA or coumaric acid to caffeic acid. Caffeoyl-CoA is converted to caffeic acid by E. coli endogenous thioesterases (176). They obtained good results with the TAL and C3H route, i.e., 106 mg/liter caffeic acid in a 2-liter bioreactor. However, the introduction of the alternative biosynthesis route through coumaroyl-CoA and caffeoyl-CoA did not lead to increased caffeic acid production from glucose. On the contrary, it introduced an extra metabolic burden and led to lower production. The problem could be due to inactivity or low activity of E. coli endogenous thioesterases against caffeoyl-CoA. The authors suggested that low copies of TAL gene were sufficient to obtain high-titer production and that fine-tuning the C3H gene copy number should be pursued since the precursors were accumulating instead of being converted to caffeic acid.
Furuya et al. (156) found that Rhodopseudomonas palustris cytochrome P450 CYP199A2 possesses hydroxylation activity toward p-coumaric acid (and cinnamic acid) and subjected it to site-directed mutagenesis to create mutants with novel and improved catalytic properties. E. coli coexpressed CYP199A2 with the redox partners' putidaredoxin reductase (pdr) from Pseudomonas putida and palustrisredoxin (pux) from R. palustris. In this study, the authors constructed a CYP199A2 F185L mutant that showed 5.5 times higher hydroxylation activity toward p-coumaric acid (3.3 g/liter) than the wild type, reaching 2.8 g/liter caffeic acid after 24 h in a glycerol medium. Furuya and Kino (177) produced caffeic acid in E. coli from p-coumaric acid using 4HPA3H from Pseudomonas aeruginosa strain PAO1. In a whole-cell reaction, 10 mM p-coumaric acid was converted to caffeic acid in 2 h. Since an initial p-coumaric acid concentration higher than 10 to 20 mM inhibited the reaction, during the 24-h fermentation, dimethyl sulfoxide (DMSO) containing 20 mM p-coumaric acid was repeatedly added to the reaction mixture (at 2, 8, and 14 h). The reaction with no energy sources produced 4 times more caffeic acid than the one with glucose. However, the highest yield obtained (10.2 g/liter) was in a glycerol medium, thus showing once again that glycerol strongly enhanced the productivity. To our knowledge, this is the highest production level attained so far.
Santos et al. (150) and Choi et al. (155) optimized the heterologous pathway for the production of flavonoids and stilbenoids (naringenin and resveratrol) from glucose. Santos et al. (150) used codon-optimized RgTAL, after discarding RsTAL. To convert p-coumaric acid to coumaroyl-CoA, the authors compared P. crispus 4CL (Pc4CL1) and Streptomyces coelicolor 4CL2 (Sc4CL2) efficiencies and concluded that combining these enzymes with TAL enzyme completely abolished accumulation of coumaric acid, which was highly produced when TAL enzyme was expressed alone. Since codon-optimized Pc4CL1 performed slightly better than Sc4CL2 (i.e., there was a higher p-coumaric acid yield in combination with TAL), this enzyme was chosen to continue the optimization of the flavonoid production pathway. In the past, Sc4CL2 has proven to be more efficient when the conversion of cinnamic acid is also needed (116, 117, 162), allowing the avoidance of the C4H step. This bacterial enzyme efficiently converts cinnamic acid (Table 5), which is usually a very poor substrate for plant 4CLs (178). Choi et al. (155) used SeTAL and Sc4CL2 in the first part of the flavonoid and stilbenoid pathway and concluded that Sc4CL2 is specific for coumaric acid and that it could not use caffeic acid and ferulic acid as a substrate (Table 5). This conclusion is important, since when using promiscuous enzymes in the curcuminoid pathway (Fig. 3), the final product is a mixture of curcuminoids. This enzyme could help to direct the pathway to a specific curcuminoid if desired. Kim et al. (153) produced flavonoids through comparison of three 4CL enzymes from three different organisms: P. crispum (Pc4CL2), O. sativa (Os4CL3), and S. coelicolor (Sc4CL2). The authors concluded that Os4CL3 was by far the one that allowed higher titers of flavonoids, followed by Pc4CL2. Furthermore, in their study the authors deleted the isocitrate dehydrogenase gene, icdA, to increase the amount of CoA in E. coli and consequently accelerated the production of p-coumaroyl-CoA. This deletion allowed for a higher yield of flavonoids.
TABLE 5.
Kinetic parameters of 4-coumarate-CoA ligases (EC 6.2.1.12) from different organismsa
| Organism | Enzyme | Substrate | Km (μM) | Vmaxb | Vmax/Kmb | kcat (s−1) | kcat/Km (s−1 M−1) | Reference(s) |
|---|---|---|---|---|---|---|---|---|
| Petroselinum crispum | Pc4CL1 | Cinnamic acid | 2,850 | 0.008 nmol s−1 | 0.003 μl s−1 | — | — | 179 |
| p-Coumaric acid | 14 | 0.020 nmol s−1 | 1.429 μl s−1 | |||||
| Caffeic acid | 20 | 0.017 nmol s−1 | 0.850 μl s−1 | |||||
| Ferulic acid | 33 | 0.018 nmol s−1 | 0.545 μl s−1 | |||||
| Arabidopsis thaliana | At4CL1 | Cinnamic acid | 6,320 | 11.6 × 10−4 nmol s−1 ml−1 | 0.184 μs−1 | — | — | 180 |
| p-Coumaric acid | 38 | 11.3 × 10−4 nmol s−1 ml−1 | 29.737 μs−1 | |||||
| Caffeic acid | 11 | 3.1 × 10−4 nmol s−1 ml−1 | 28.182 μs−1 | |||||
| Ferulic acid | 199 | 6.0 × 10−4 nmol s−1 ml−1 | 3.015 μs−1 | |||||
| At4CL2 | Cinnamic acid | 6,630 | 0.6 × 10−4 nmol s−1 ml−1 | 0.009 μs−1 | — | — | ||
| p-Coumaric acid | 252 | 2.8 × 10−4 nmol s−1 ml−1 | 1.111 μs−1 | |||||
| Caffeic acid | 20 | 2.1 × 10−4 nmol s−1 ml−1 | 10.500 μs−1 | |||||
| Ferulic acid | NC | — | — | |||||
| At4CL3 | Cinnamic acid | 2,070 | 3.1 × 10−4 nmol s−1 ml−1 | 0.150 μs−1 | — | — | ||
| p-Coumaric acid | 23 | 1.9 ×10−4 nmol s−1 ml−1 | 8.261 μs−1 | |||||
| Caffeic acid | 374 | 2.5 × 10−4 nmol s−1 ml−1 | 0.668 μs−1 | |||||
| Ferulic acid | 166 | 1.6 × 10−4 nmol s−1 ml−1 | 0.964 μs−1 | |||||
| At4CL4 | p-Coumaric acid | 432 | 129.6 nmol s−1 | 300 μl s−1 | — | — | 181 | |
| Caffeic acid | 186 | 204.6 nmol s−1 | 1,100 μl s−1 | |||||
| Ferulic acid | 26 | 171.6 nmol s−1 | 6,600 μl s−1 | |||||
| At4CL1 | p-Coumaric acid | 66.7 | — | — | 0.248 | 5,365 | 130 | |
| At4CL2 | Cinnamic acid | 6,642 | 203 nmol s−1 mg−1 | 31 μl s−1 mg−1 | 12.5 | 1,897 | 182, 183 | |
| p-Coumaric acid | 233 | 475 nmol s−1 mg−1 | 2,039 μl s−1 mg−1 | 29.5 | 126,609 | |||
| Caffeic acid | 22 | 236 nmol s−1 mg−1 | 10,727 μl s−1 mg−1 | 14.6 | 663,636 | |||
| Ferulic acid | ND | ND | ND | ND | ND | |||
| At4CL2 mutant (M293P + K320L) | Cinnamic acid | 286 | 190 nmol s−1 mg−1 | 664 μl s−1 mg−1 | 11.8 | 41,259 | ||
| p-Coumaric acid | 22 | 321 nmol s−1 mg−1 | 14,591 μl s−1 mg−1 | 20.0 | 909,091 | |||
| Caffeic acid | 41 | 267 nmol s−1 mg−1 | 6,512 μl s−1 mg−1 | 16.5 | 402,439 | |||
| Ferulic acid | 30 | 247 nmol s−1 mg−1 | 82,333 μl s−1 mg−1 | 15.3 | 510,000 | |||
| Oryza sativa | Os4CL1 | Cinnamic acid | 9.4 | 0.100 nmol s−1 mg−1 | 10.638 μl s−1 mg−1 | 0.006 | 663 | 184 |
| p-Coumaric acid | 11.9 | 0.130 nmol s−1 mg−1 | 10.924 μl s−1 mg−1 | 0.008 | 681 | |||
| Caffeic acid | 29.3 | 0.140 nmol s−1 mg−1 | 4.778 μl s−1 mg−1 | 0.009 | 296 | |||
| Ferulic acid | 8.3 | 0.110 nmol s−1 mg−1 | 13.253 μl s−1 mg−1 | 0.007 | 824 | |||
| Os4CL2 | Cinnamic acid | 21.7 | 0.299 nmol s−1 mg−1 | 13.779 μl s−1 mg−1 | 0.019 | 867 | ||
| p-Coumaric acid | 16.8 | 0.629 nmol s−1 mg−1 | 37.440 μl s−1 mg−1 | 0.040 | 2,351 | |||
| Caffeic acid | 27.6 | 0.708 nmol s−1 mg−1 | 25.652 μl s−1 mg−1 | 0.045 | 1,612 | |||
| Ferulic acid | 2.2 | 0.613 nmol s−1 mg−1 | 278.636 μl s−1 mg−1 | 0.039 | 17,295 | |||
| Os4CL3 | Cinnamic acid | 28.2 | 3.010 nmol s−1 mg−1 | 106.737 μl s−1 mg−1 | 0.182 | 6,458 | ||
| p-Coumaric acid | 4.9 | 4.700 nmol s−1 mg−1 | 959.184 μl s−1 mg−1 | 0.285 | 58,521 | |||
| Caffeic acid | 10.9 | 4.210 nmol s−1 mg−1 | 386.239 μl s−1 mg−1 | 0.255 | 23,330 | |||
| Ferulic acid | 3.5 | 4.930 nmol s−1 mg−1 | 1,408.571 μl s−1 mg−1 | 0.299 | 84,943 | |||
| Os4CL4 | Cinnamic acid | 15.7 | 0.350 nmol s−1 mg−1 | 22.293 μl s−1 mg−1 | 0.021 | 1,361 | ||
| p-Coumaric acid | 3.9 | 0.770 nmol s−1 mg−1 | 197.436 μl s−1 mg−1 | 0.047 | 12,176 | |||
| Caffeic acid | 5.8 | 0.590 nmol s−1 mg−1 | 101.724 μl s−1 mg−1 | 0.036 | 6,206 | |||
| Ferulic acid | 4.6 | 0.520 nmol s−1 mg−1 | 113.043 μl s−1 mg−1 | 0.032 | 6,956 | |||
| Os4CL5 | Cinnamic acid | 54.4 | 0.300 nmol s−1 mg−1 | 5.515 μl s−1 mg−1 | 0.017 | 312 | ||
| p-Coumaric acid | 10.3 | 0.830 nmol s−1 mg−1 | 80.583 μl s−1 mg−1 | 0.048 | 4,660 | |||
| Caffeic acid | 26.1 | 0.240 nmol s−1 mg−1 | 9.195 μl s−1 mg−1 | 0.014 | 535 | |||
| Ferulic acid | 6.9 | 0.590 nmol s−1 mg−1 | 85.507 μl s−1 mg−1 | 0.034 | 4,871 | |||
| Streptomyces coelicolor A3(2) | Sc4CL2 | Cinnamic acid | 190 | — | — | 0.475 | 2,499 | 178 |
| p-Coumaric acid | 131 | 0.202 | 1,545 | |||||
| Caffeic acid | ND | ND | ND | |||||
| Ferulic acid | NC | NC | NC |
—, information was not available; ND, not determined, due to very low activity; NC, no conversion, no enzymatic activity.
It was not possible to convert all the data to the same units for comparison purposes. The units are indicated for each case.
Lin et al. (175), aiming to produce coumarins using a biosynthetic pathway for the first time, used RgTAL since it possesses high activity toward tyrosine compared with the RcTAL (Table 4) used in their previous work (140). They tested three different 4CLs (Pc4CL2, At4CL1, and At4CL2) (Table 6). When broad substrate specificity was needed, Pc4CL2 was used to synthesize the CoA esters of coumaric acid and ferulic acid, since it was shown in the past that it can accept several phenylpropanoid acids as the substrate (160, 185). When higher catalytic efficiency toward coumaric acid was required, the enzyme At4CL1 was used (Table 6). E. coli 4HPA3H was also used to hydroxylate coumaric acid to caffeic acid, as was shown in other studies (140, 151) (Fig. 5, top). The 4-coumarate-CoA ligase used in this case was At4CL2 due to its higher affinity and catalytic activity toward caffeic acid than Pc4CL2 and At4CL1. CCoAOMT1 from A. thaliana was used to convert caffeoyl-CoA to feruloyl-CoA. Also with the goal of obtaining feruloyl-CoA, but from ferulic acid, Watts et al. (158) used the recently reported 4-coumarate-CoA ligase At4CL4. This enzyme was shown to prefer ferulic acid as a substrate (181) (Table 5). However, the expected production of stilbene was not detectable by high-pressure liquid chromatography (HPLC). This does not imply that At4CL4 was not working, since the problem could be in the stilbene synthase step. Feruloyl-CoA conversion could be slow, causing an accumulation of feruloyl-CoA, which can be converted again to ferulic acid due to a balance between CoA ligase and endogenous E. coli thioesterases (158, 176). Since the production of feruloyl-CoA would be highly advantageous in curcumin production, further characterization of At4CL4 should be considered. At4CL2 and its mutant (Table 5) also seem to be very interesting options to be explored in the future.
TABLE 6.
Comparison of kinetic parameters of 4-coumarate-CoA ligases (EC 6.2.1.12) from different organismsa
| Organism | Enzyme | Substrate | Km (μM) | Sp act (nmol s−1 mg−1) | Sp act/Km (μl s−1 mg−1) |
|---|---|---|---|---|---|
| Petroselinum crispum | Pc4CL2 | p-Coumaric acid | 11.8 | 1.983 | 168.051 |
| Caffeic acid | 8.4 | 1.533 | 182.540 | ||
| Ferulic acid | 11.2 | 1.367 | 122.054 | ||
| Arabidopsis thaliana | At4CL1 | p-Coumaric acid | 28.7 | 11.550 | 403.439 |
| Caffeic acid | 15.7 | 5.267 | 335.478 | ||
| Ferulic acid | 40.8 | 6.800 | 166.667 | ||
| At4CL2 | p-Coumaric acid | 45.1 | 10.433 | 231.330 | |
| Caffeic acid | 7.9 | 8.483 | 1,073.797 | ||
| Ferulic acid | ND |
Data are from reference 175. ND, not determined due to very low activity.
As mentioned above, although S. cerevisiae has never been engineered to produce curcuminoids, it has been used to produce other polyketides. Therefore, these already engineered and validated enzymatic steps may be used in the future for successful production of curcuminoids by the yeast. Furthermore, CUS has already been successfully expressed in S. cerevisiae to produce gingerol derivatives (115).
Again, an attractive advantage of the yeast compared to E. coli is its ability to functionally express plant cytochrome P450 monooxygenases. C4H from A. thaliana and Glycine max was successfully cloned and expressed in S. cerevisiae in several studies (123, 127, 128, 131). Although C4H was functionally expressed, it was still a rate-limiting step. Since the endogenous CPR activity was not enough to support the C4H expression, CPR1 (NADPH-cytochrome P450 reductase) from S. cerevisiae or CPR from a Populus hybrid (P. trichocarpa × P. deltoids) was overexpressed.
Other enzymes from the phenylpropanoid pathway, such as PAL from Rhodosporidium toruloides and a Populus hybrid, RsTAL, RcTAL, RgTAL, 4CL2 from Nicotiana tabacum, 4CL216 from a Populus hybrid, 4CL from G. max, Le4CL1, At4CL1, and Pc4CL2, were also successfully expressed in yeast (115, 123–132, 174). The At4CL1 gene seems to be the best option to produce resveratrol in S. cerevisiae (139), since its yield was more than 65 times higher than when using 4CL216 from Populus hybrid (125) and 4CL2 from Nicotiana tabacum (124) in similar experiments using Vitis vinifera STS (stilbene synthase). Also, the resveratrol yield in S. cerevisiae was 3.4 times higher than the maximum obtained with E. coli using At4CL1, but using Arachis hypogaea STS (158). Shin et al. (186) used At4CL1 and A. hypogaea STS in S. cerevisiae and obtained very small amounts of resveratrol compared to those obtained by Sydor et al. (139). This proves that the combination between 4CL and STS, as well as the host, is very important to obtain excellent results and that the enzymes from some organisms work better together than those from others. Although flavonoid and stilbenoid production has been proven in some cases to be more efficient in bacteria (124, 127, 138), there are studies suggesting that S. cerevisiae is an adequate host for the expression of aromatic plant compounds (139).
Type III Polyketide Synthases and the Biosynthesis of Curcuminoids
In the last decade, several advances have been made in curcuminoid biosynthesis as a result of the identification of DCS and multiple CURSs from C. longa and of CUS from O. sativa. However, DCS and CURS enzymes have been used to synthesize curcuminoids only in vitro, never in vivo. CUS was chosen to produce curcuminoids in vivo (106, 121, 122) since it was the first polyketide synthase involved in curcuminoid biosynthesis to be reported. Also, CUS has the advantage of being able to produce curcuminoids directly from the CoA esters, unlike CURSs, which need DCS to convert the CoA ester to the corresponding diketide-CoA ester. However, this advantage does not mean that an approach using DCS and CURS will not lead to higher yields, and this option should continue to be the object of further research.
Due to the immense pharmaceutical value of curcuminoids, other genes encoding type III polyketide synthase (PKS) curcumin/curcuminoid synthases are being sought. Brand et al. (187) characterized a type III PKS from Wachendorfia thyrsiflora, WtPKS1 (GenBank accession number AY727928.1). W. thyrsiflora is known to synthesize phenylphenalenones which are thought to be synthesized from curcuminoids. The authors also characterized another PKS in W. thyrsiflora, WtPKS2 (unpublished work) (GenBank accession number AAW50922), that synthesizes a methylketone product, which is presumably derived from the corresponding diketide-CoA via hydrolysis and decarboxylation. Based on their high homology with C. longa DCS and CURS1 (WtPKS1 shares 63% identity to DCS and WtPKS2 shares 62% identity to CURS1) (97), it is assumed that WtPKS1 is a diketide synthase, WtPKS2 is a curcuminoid synthase, and curcuminoids are synthesized by an enzyme system similar to the one with DCS and CURS1, not only in W. thyrsiflora but also in other plants.
Also, with the goal of finding uncharacterized type III PKS in C. longa, Resmi and Soniya (103) used reverse transcription-PCR (RT-PCR) genomic screening. This helped them to identify cDNAs that encode type III PKS proteins. One of the type III PKS, ClPKS10 (GenBank accession number JN017185), proved to possess all the possible requirements for catalyzing curcuminoid biosynthesis; namely, it shares 93% identity with CURS1 and 81% identity with CURS2, and the putative active-site environment (amino acids needed for coumaroyl-CoA binding site and cyclization) showed the same characteristics found in the reported CURSs. However, ClPKS10 showed high expression levels in leaf tissues, low levels in shoots, and very low levels in rhizomes and roots. This is in contrast with the reported curcuminoid expression pattern, which can indicate possible curcuminoid biosynthesis in aerial parts also. Dihydro derivatives of curcuminoids (dihydrocurcuminoids) also showed higher expression levels in leaves than in the rhizome (105). Ramirez-Ahumada et al. (98) also concluded that the highest curcuminoid synthase activity from turmeric extracts was found in leaves, followed by shoots and rhizome, which proves that curcuminoids and their analogues can be synthesized not only in the rhizomes but also in the roots and the leaves. Future work to characterize ClPKS10, WtPKS1, and WtPKS2 through in vitro and in vivo assays is required to confirm the curcuminoid synthase activity.
Malonyl-CoA Availability
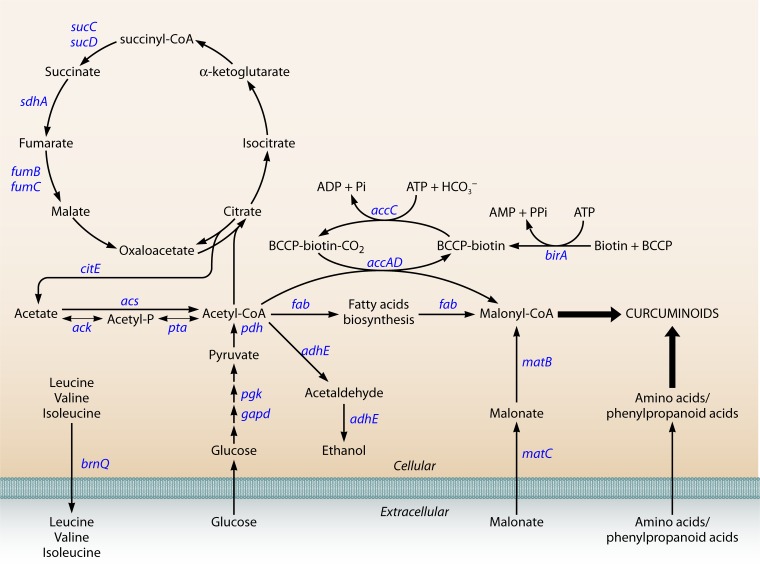
Although it has been demonstrated that some curcuminoid synthases are able to catalyze two or more reactions from CoA esters and to efficiently produce curcuminoids heterologously, the presence of the precursor malonyl-CoA is still essential for these reactions to succeed. It is believed that the biosynthetic production of flavonoids, stilbenoids, and polyketides is controlled by the limited intracellular pool of malonyl-CoA (116, 137, 188). In these pathways, three molecules of malonyl-CoA are needed to obtain the final product, whereas for curcuminoid production, only one molecule is needed. Malonyl-CoA is naturally synthesized in microorganisms, but it is used for the production of fatty acids and phospholipids, leaving only a very limited amount available for the production of secondary metabolites (189). Therefore, it is essential to engineer the microbial host to achieve metabolic balance between the need for malonyl-CoA for growth and secondary metabolite production (137). In the last decade, several metabolic engineering approaches were developed to increase the intracellular pool of malonyl-CoA. One of them is the overexpression of acetyl-CoA carboxylase (ACC), which converts acetyl-CoA into malonyl-CoA (106, 116, 188, 190–192) (Fig. 6). Leonard et al. (188) and Xu et al. (190) overexpressed ACC encoded by accABCD from Photorhabdus luminescens and E. coli, respectively, and obtained an increase in the flavanone and fatty acid production. The ACC reaction is divided into two partial reactions (Fig. 6). In the first reaction, biotin is attached to the biotin carboxyl carrier protein (BCCP) (encoded by accB) and is carboxylated via ATP consumption by the biotin carboxylase AccC. The carboxyl group is then transferred to acetyl-CoA by carboxyltransferase (AccAD). Since ACC requires biotinylation by the action of biotin ligase (BLP) to be completely functional, BLP (encoded by birA) overexpression was also studied using P. luminescens (188, 193) and E. coli (188) BLP. Coexpression of ACC and BPL, both from P. luminescens, was found to be the best approach to improve the flavonoid yield (188). The use of the acetyl-CoA pathway to increase the supply of malonyl-CoA has also been explored (188, 192). When glucose is used as a carbon source, acetyl-CoA is converted to acetate under aerobic conditions, which is toxic for E. coli and inhibits its growth (194). By overexpressing E. coli acetate assimilation pathways, it is possible to increase the production of the desired compound and, at the same time, reduce acetate accumulation. The first pathway includes two enzymes, acetate kinase (ACK) and phosphotransacetylase (PTA) (195), and the second pathway includes acetyl-CoA synthase (ACS). According to Leonard et al. (188), ACS overexpression resulted in a more efficient acetate assimilation than ACK-PTA overexpression. The better results obtained with ACS are likely due to the reversibility of the ACK-PTA pathway (196). ACS coexpression with ACC increased flavonoid production up to 1,221%.
FIG 6.
Metabolic pathways connecting malonyl-CoA with curcuminoid biosynthesis in Escherichia coli.
Another strategy to increase the malonyl-CoA supply is the overexpression of the Rhizobium trifolii matB and matC genes, encoding malonate synthase and malonate carrier protein, respectively (138, 150, 197). These enzymes allow the transport of supplemented malonate into the cell and its subsequent conversion to malonyl-CoA. Santos et al. (150) obtained an increase of flavonoid synthesis up to 59% with this approach, while Leonard et al. (138) obtained up to 250%.
Other strategies include the deletion of the genes encoding succinate dehydrogenase (sdhA), acetaldehyde dehydrogenase (adhE), amino acid transporter (brnQ), citrate lyase (citE) (193), fumarase (FUM) (encoded by fumB and fumC), and succinyl-CoA synthetase (SUCOAS) (encoded by sucC and sucD) and overexpression of phosphoglycerate kinase (PGK), glyceraldehyde-3-phosphate dehydrogenase (GAPD), and pyruvate dehydrogenase (PDH) (encoded by aceEF and lpdA) (137). Leonard et al. (138) added the fatty acid inhibitor cerulenin to the fermentation to downregulate fatty acid biosynthesis (FAB) initiation. Cerulenin represses both fabB and fabF and led to an increase of flavonoid production by over 900%. Santos et al. (150) also obtained a significant gain with cerulenin supplementation. However, the cost of cerulenin may be excessive if industrial-scale fermentations are foreseen (150). Therefore, a strategy that focuses on rerouting native metabolic flows and uses stoichiometric modeling (193) to improve malonyl-CoA availability is more appropriate for increasing curcuminoid production.
Other Limitations
Although several studies in the last years made significant progress in proving the practicality of curcuminoid production in E. coli, the established protocols still present some disadvantages that could be prohibitive during process scale-up. One of them is the dependence on precursor feeding, which may be solved by engineering strains capable of converting cheaper substrates such as glucose. A second limitation is that fermentation often needs two separate cultivation steps to obtain high titers of curcuminoids. Usually strains are first grown in a rich medium like LB (Luria broth) in order to produce large amounts of biomass and reach a suitable protein production level. Then, after reaching the exponential phase, the cells are harvested and transferred to minimal medium, such as M9 medium, where the substrates are added and the curcuminoids are produced. This strategy was pursued in curcuminoid production in vivo (106, 121, 122) and also for other phenolic compounds (116, 117, 137, 138, 141, 151, 153, 155, 192, 193). The use of plasmids imposes a metabolic burden on the host strain that usually reduces the growth rate of the cell due to the expression of plasmid-borne resistance and replication of the plasmids. Therefore, the two-step fermentation strategy is used to compensate for the metabolic burden associated with protein overexpression and poor growth observed in minimal medium. Although this strategy is feasible at the laboratory scale, the separation of biomass is much more difficult and expensive in large-scale fermentations (150). Therefore, the development of vigorous strains that allow an efficient production in a single medium formulation is essential (150). The use of MOPS (morpholinepropanesulfonic acid) minimal medium (198) has demonstrated the successful production of flavonoids in a one-step fermentation (150, 197) with no apparent growth deficiencies. The delay of IPTG (isopropyl-β-d-thiogalactopyranoside) induction can also alleviate the metabolic burden. Induction should be performed at the exponential phase, since after that point, although bacteria are still growing, the production titers decrease (150, 197). During induction, the temperature should be maintained or decreased to 26 to 30°C for optimal enzyme synthesis and substrate conversion. The compounds possibly used as substrates (caffeic acid, coumaric acid, and ferulic acid) present some toxicity to the cells at high concentrations (152, 158), and cases where a lower concentration was added at the beginning of the experiment and after it was almost consumed more was added showed better results (151) and should be considered, as well as fed-batch fermentations.
The success of the design and construction of heterologous pathways and their expression depends on many factors. These factors include cell growth characteristics but also the expression levels and biological activity of the proteins of interest. The host, the codon biases among different organisms, and the strengths of promoters linked to the potential toxicity of the protein to the host are topics that should always be considered in recombinant protein expression. A stable, robust, and well-characterized host cell capable of growing on minimal and inexpensive carbon sources is crucial for laboratory and large-scale production (199). However, robust hosts are usually able to inactive or rid themselves of the foreign DNA to minimize the metabolic burden. Several studies proved that genome reductions (removal of transposons, insertion sequence elements, cryptic phages, integrase genes, damaged genes, and genes with unknown function) improve metabolic efficiency and even electroporation efficiency and accurate propagation of foreign DNA that was unstable in other strains (200–203). These strains should be considered for curcuminoid production. The vector(s) used to carry the curcuminoid pathway should also be stable and consistent in copy number to ensure that all the cells in the culture have the plasmid and produce curcuminoids. Plasmids should also have copy numbers as low as possible to minimize the burden on the host cell and the ability to carry large sequences of DNA. A better option is to integrate the genes of interest in the chromosome due to the increased stability. This insertion can dramatically impact the expression level of the heterologous genes; however, the promoter and ribosome binding strength can be adapted, as well as the stability of the mRNA and the resulting protein (199, 204). When expressing multiple heterologous genes, several approaches can be used. For example, different inducible promoters can be used for each gene. This approach has the disadvantage of adding multiple inducers to the medium, increasing the cost of production. It is also possible to use the same inducible promoter for each gene but to vary the promoter strength or to use nonnative T7 RNA polymerase to control the expression (106, 122). Grouping multiple and related genes into operons is also a good approach for regulating several genes simultaneously using the same promoter (149, 205).
The choice of the right promoter with adequate strength is very important, especially when the construction of the metabolic pathway involves the introduction of more than one heterologous gene, such as in the curcuminoid production case. Imbalances can lead to over- or underproduction of enzymes and accumulation of intermediate metabolites which may result in suboptimal titers. Codon bias is also a subject to take into consideration, since rare codons in E. coli are often abundant in heterologous genes, and expression of those genes can lead to translational errors, frameshifting events, stalling, or premature translational termination, especially when transcripts containing rare codons accumulate in large quantities (206). This codon bias can be solved using synthetic DNA with codon optimization or by cotransformation of the host with a plasmid harboring rare tRNA, thus increasing the copy number of the limiting tRNA (159). So far, in heterologous production of curcuminoids by E. coli using plant genes (106, 121, 122), no approaches to solve codon bias have been used, which proves that successful production can be achieved using plant cDNA. Since this is the simplest and least expensive approach, it is always worth testing it first. Production of flavonoids and stilbenes was also achieved recently without codon optimization (116, 117, 153, 158, 160, 161). In addition to the importance of the Shine-Dalgarno sequence in translation efficiency, the first codons downstream of the initiation codon also act as a translation enhancer, and depending on this sequence, gene expression can vary considerably (207–209). Therefore, to achieve production-scalable titers, codon optimization of recombinant plant proteins and improvement of catalytic activity, and also the identification of the best combination of plasmid copy number with the right promoters to achieve optimal instead of maximal expression levels, need to be carefully studied.
The heterologous production of curcuminoids is very recent; however, the titers and yields obtained so far are in the same range of those obtained for flavonoids and stilbenoids that have been thoroughly studied (Table 3). Additionally, yields similar to those obtained from plants can be obtained by heterologous production in E. coli, thus suggesting heterologous production as a promising alternative to obtain these compounds. Moreover, the isolation of curcuminoids from plants is challenging, and curcumin is usually sold as a mixture that contains a significant percentage of other curcuminoids (bisdemethoxycurcumin and demethoxycurcumin) (17, 18). Indeed, these curcuminoids present in smaller amounts in plants are difficult and expensive to isolate, and therefore heterologous production in E. coli could be a viable alternative for their production through the use of a combination of specific enzymes that could drive the production to a specific curcuminoid.
FUTURE PERSPECTIVES
In the last decade, there have been remarkable advances in understanding the biosynthetic pathway of curcuminoid production in C. longa and its heterologous production in E. coli. However, continuous efforts toward exploiting new heterologous hosts and finding the most adequate synthetic enzymes and plasmids are needed. Several enzymes should be studied to find the best candidates for each step of the curcuminoid production pathway to adequately transform the metabolic intermediates into the desired products. Accumulation of intermediates should be avoided, since this can be toxic to the heterologous host and lead to a decrease of the desired final product production.
Microbial production of curcuminoids is still at an early stage and is limited by the identification of some curcuminoid synthases, while PKS diversity is barely touched. Deeper study is extremely important and would allow the identification of novel enzymes that catalyze unnatural reactions and consequently produce more unnatural, potentially valuable curcuminoids. The huge accumulation of genome information from a variety of organisms due to the development of molecular biology techniques and bioinformatic prediction of catalytic properties of gene products will allow combination of the best enzymes to generate novel biosynthetic pathways for curcuminoid production in several organisms.
The curcuminoid titers and yields obtained so far using E. coli are very promising, and we believe that, using synthetic biology approaches and metabolic engineering tools, these can be further improved to make heterologous production competitive with the current process of extraction from plants. Although the current levels of production of curcuminoids are probably below those required for viable industrial production, process optimization toward scale-up should be considered. To date, curcuminoids have been produced only by using two separate cultivation media, which represents a limitation when considering bioreactor fermentations. Therefore, it is crucial to optimize the fermentation conditions, including media and operating parameters. As mentioned above, heterologous production of curcuminoids is advantageous compared to extraction from plants, as the former can be easily controlled in a bioreactor and is not subject to unpredictable factors such as weather that may affect plant cultivation. Also, in contrast to the extraction process, heterologous production in microorganisms is not seasonal. In summary, we believe that after adequate optimization efforts, curcuminoids can be produced by microorganisms in bioreactors, providing the same or larger amounts as reported for plant extraction, in a shorter period of time and a smaller space, while using a process that is less expensive and harmless to the environment.
ACKNOWLEDGMENTS
We acknowledge financial support from the Strategic Project PEst-OE/EQB/LA0023/2013, project reference RECI/BBB-EBI/0179/2012 (project number FCOMP-01-0124-FEDER-027462), project SYNBIOBACTHER (PTDC/EBB-BIO/102863/2008), and a doctoral grant (SFRH/BD/51187/2010) to J. L. Rodrigues, funded by Fundação para a Ciência e a Tecnologia. We thank the MIT-Portugal Program for support given to J. L. Rodrigues.
Biographies

J. L. Rodrigues is a Ph.D. student at the Center of Biological Engineering at the University of Minho (MIT Portugal Program). She obtained her degree in biological engineering at the University of Minho in 2010. The main focus of her thesis was the study of E. coli heat shock promoters with the goal of using them in synthetic biology approaches. From 2008 to 2010 she was a research assistant in a project that studied bioremediation of soils contaminated with heavy metals. Under the scope of her Ph.D. work, during one year she was a visiting student at the Chemical Engineering Department at MIT, where she worked on the design and construction of a biosynthetic pathway for the production of curcuminoids in E. coli. Her research interests include the use of recombinant microorganisms to express new biosynthetic pathways and also the use of heat shock for their expression.

K. L. J. Prather is a Theodore T. Miller Career Development Associate Professor of Chemical Engineering at MIT. She obtained a Bachelor of Science degree in chemical engineering from MIT in 1994 and a Ph.D. degree from the University of California, Berkeley, in 1999. At Berkeley, she worked under the supervision of Professor Jay D. Keasling on the development of expression vectors for metabolic engineering. Professor Prather joined the faculty of MIT after 4 years in BioProcess Research and Development at Merck Research Labs (Rahway, NJ), first as a Senior Research Biochemical Engineer and then as a Research Fellow. While at Merck, she worked on projects in the areas of biocatalysis for small-molecule transformations, high-yield production of plasmids as DNA vaccines, and mammalian cell line development for production of therapeutic proteins. Her current research interests are centered on the design and assembly of recombinant microorganisms for the production of small molecules, with a particular focus on novel biosynthetic pathway design.

L. D. Kluskens is a Principal Investigator at the University of Minho (Portugal). He completed his Master in Food Science and Technology in 1997 at Wageningen University, The Netherlands. He did his Ph.D. work at the Laboratory of Microbiology at Wageningen University on hydrolytic enzymes from hyperthermophilic bacteria, and in 2002 he joined the Biomade Technology Foundation in Groningen, The Netherlands, as a researcher, working on the enzymatic stabilization of therapeutic peptides. Since May 2008 he has worked as a researcher (first as an assistant researcher and since 2014 as a principal researcher) at the Centre of Biological Engineering at the University of Minho, Braga, funded by the Fundação para a Ciência e a Tecnología (FCT), where he focuses on the biotechnological aspects of the application of phage particles for medical purposes and pathogen elimination.

L. R. Rodrigues is an Assistant Professor at the University of Minho (Portugal). She received her degree in biological engineering in 1997, her M.Sc. in bioprocess engineering in 2001, and her Ph.D. in chemical and biological engineering in 2005 from the University of Minho. In 2006 to 2007, she was a postdoctoral research fellow at the three different universities (University of Minho, IPATIMUP-University of Porto, and University of Lund). Parts of her graduate, Ph.D., and postdoctoral research were conducted in international labs and included the production and use of bioactive molecules for biomedical applications, including antibiofilm strategies and breast cancer diagnosis and prevention. In 2007, she joined the faculty of the Department of Biological Engineering at the University of Minho as an invited assistant professor (until 2011). Since 2010, she has been a member of the Bioprocess Systems Biology Group, where she established her research group within the field of synthetic biology. Her current research interests are synthetic biology, cancer diagnosis and prevention, bioprocess development, and functional food.
REFERENCES
- 1.Jayaprakasha GK, Jagan Mohan Rao L, Sakariah KK. 2005. Chemistry and biological activities of C. longa. Trends Food Sci Technol 16:533–548. doi: 10.1016/j.tifs.2005.08.006. [DOI] [Google Scholar]
- 2.Sharma RA, Gescher AJ, Steward WP. 2005. Curcumin: the story so far. Eur J Cancer 41:1955–1968. doi: 10.1016/j.ejca.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 3.Tayyem RF, Heath DD, Al-Delaimy WK, Rock CL. 2006. Curcumin content of turmeric and curry powders. Nutr Cancer 55:126–131. doi: 10.1207/s15327914nc5502_2. [DOI] [PubMed] [Google Scholar]
- 4.Srinivasan KR. 1953. A chromatographic study of the curcuminoids in Curcuma longa, L. J Pharm Pharmacol 5:448–457. doi: 10.1111/j.2042-7158.1953.tb14007.x. [DOI] [PubMed] [Google Scholar]
- 5.Tohda C, Nakayama N, Hatanaka F, Komatsu K. 2006. Comparison of anti-inflammatory activities of six Curcuma rhizomes: a possible curcuminoid-independent pathway mediated by Curcuma phaeocaulis extract. Evid Based Complement Alternat Med 3:255–260. doi: 10.1093/ecam/nel008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abas F, Lajis NH, Shaari K, Israf DA, Stanslas J, Yusuf UK, Raof SM. 2005. A labdane diterpene glucoside from the rhizomes of Curcuma mangga. J Nat Prod 68:1090–1093. doi: 10.1021/np0500171. [DOI] [PubMed] [Google Scholar]
- 7.Malek SNA, Lee GS, Hong SL, Yaacob H, Wahab NA, Faizal Weber J-F, Shah SAA. 2011. Phytochemical and cytotoxic investigations of Curcuma mangga rhizomes. Molecules 16:4539–4548. doi: 10.3390/molecules16064539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruslay S, Abas F, Shaari K, Zainal Z, Sirat H, Israf DA, Lajis NH. 2007. Characterization of the components present in the active fractions of health gingers (Curcuma xanthorrhiza and Zingiber zerumbet) by HPLC-DAD-ESIMS. Food Chem 104:1183–1191. doi: 10.1016/j.foodchem.2007.01.067. [DOI] [Google Scholar]
- 9.Uehara S-I, Yasuda I, Akiyama K, Morita H, Takeya K, Itokawa H. 1987. Diarylheptanoids from the rhizomes of Curcuma xanthorrhiza and Alpinia officinarum. Chem Pharm Bull 35:3298–3304. doi: 10.1248/cpb.35.3298. [DOI] [Google Scholar]
- 10.Syu W-J, Shen C-C, Don M-J, Ou J-C, Lee G-H, Sun C-M. 1998. Cytotoxicity of curcuminoids and some novel compounds from Curcuma zedoaria. J Nat Prod 61:1531–1534. doi: 10.1021/np980269k. [DOI] [PubMed] [Google Scholar]
- 11.Lobo R, Prabhu KS, Shirwaikar A, Shirwaikar A. 2009. Curcuma zedoaria Rosc. (white turmeric): a review of its chemical, pharmacological and ethnomedicinal properties. J Pharm Pharmacol 61:13–21. doi: 10.1211/jpp/61.01.0003. [DOI] [PubMed] [Google Scholar]
- 12.Mohamad H, Lajis NH, Abas F, Ali AM, Sukari MA, Kikuzaki H, Nakatani N. 2005. Antioxidative constituents of Etlingera elatior. J Nat Prod 68:285–288. doi: 10.1021/np040098l. [DOI] [PubMed] [Google Scholar]
- 13.Masuda T, Jitoe A. 1994. Antioxidative and antiinflammatory compounds from tropical gingers: isolation, structure determination, and activities of cassumunins A, B, and C, new complex curcuminoids from Zingiber cassumunar. J Agric Food Chem 42:1850–1856. doi: 10.1021/jf00045a004. [DOI] [Google Scholar]
- 14.Masuda T, Jitoe A, Mabry TJ. 1995. Isolation and structure determination of cassumunarins A, B, and C: new anti-inflammatory antioxidants from a tropical ginger, Zingiber cassumunar. J Am Oil Chem Soc 72:1053–1057. doi: 10.1007/BF02660721. [DOI] [Google Scholar]
- 15.Bagchi A. 2012. Extraction of curcumin. J Environ Sci Toxicol Food Technol 1:1–16. [Google Scholar]
- 16.Ravindran P, Babu KN, Sivaraman K. 2007. Turmeric: the genus Curcuma. CRC Press, Boca Raton, FL. [Google Scholar]
- 17.Aggarwal BB, Kumar A, Bharti AC. 2003. Anticancer potential of curcumin: preclinical and clinical studies. Anticancer Res 23:363–398. [PubMed] [Google Scholar]
- 18.Goel A, Kunnumakkara AB, Aggarwal BB. 2008. Curcumin as “curecumin”: from kitchen to clinic. Biochem Pharmacol 75:787–809. doi: 10.1016/j.bcp.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 19.Vogel A, Pelletier J. 1815. Examen chimique de la racine de Curcuma. J Pharmacol 1:289–300. [Google Scholar]
- 20.Schraufstätter E, Bernt H. 1949. Antibacterial action of curcumin and related compounds. Nature 164:456–457. doi: 10.1038/164456a0. [DOI] [PubMed] [Google Scholar]
- 21.Negi P, Jayaprakasha G, Jagan Mohan Rao L, Sakariah K. 1999. Antibacterial activity of turmeric oil: a byproduct from curcumin manufacture. J Agric Food Chem 47:4297–4300. doi: 10.1021/jf990308d. [DOI] [PubMed] [Google Scholar]
- 22.Kim M-K, Choi G-J, Lee H-S. 2003. Fungicidal property of Curcuma longa L. rhizome-derived curcumin against phytopathogenic fungi in a greenhouse. J Agric Food Chem 51:1578–1581. doi: 10.1021/jf0210369. [DOI] [PubMed] [Google Scholar]
- 23.Reddy RC, Vatsala PG, Keshamouni VG, Padmanaban G, Rangarajan PN. 2005. Curcumin for malaria therapy. Biochem Biophys Res Commun 326:472–474. doi: 10.1016/j.bbrc.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 24.Strimpakos AS, Sharma RA. 2008. Curcumin: preventive and therapeutic properties in laboratory studies and clinical trials. Antioxid Redox Signal 10:511–546. doi: 10.1089/ars.2007.1769. [DOI] [PubMed] [Google Scholar]
- 25.Shimatsu A, Kakeya H, Imaizumi A, Morimoto T, Kanai M, Maeda S. 2012. Clinical application of “curcumin,” a multi-functional substance. Anti-Aging Med 9:43–51. [Google Scholar]
- 26.Liu D, Chen Z. 2013. The effect of curcumin on breast cancer cells. J Breast Cancer 16:133–137. doi: 10.4048/jbc.2013.16.2.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bisht K, Wagner K-H, Bulmer AC. 2010. Curcumin, resveratrol and flavonoids as anti-inflammatory, cyto-and DNA-protective dietary compounds. Toxicology 278:88–100. doi: 10.1016/j.tox.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 28.Aggarwal BB, Sung B. 2009. Pharmacological basis for the role of curcumin in chronic diseases: an age-old spice with modern targets. Trends Pharmacol Sci 30:85–94. doi: 10.1016/j.tips.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 29.Prasad S, Gupta SC, Tyagi AK, Aggarwal BB. 2014. Curcumin, a component of golden spice: from bedside to bench and back. Biotechnol Adv 32:1053–64. doi: 10.1016/j.biotechadv.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 30.Kuttan R, Bhanumathy P, Nirmala K, George MC. 1985. Potential anticancer activity of turmeric (Curcuma longa). Cancer Lett 29:197–202. doi: 10.1016/0304-3835(85)90159-4. [DOI] [PubMed] [Google Scholar]
- 31.Ruby A, Kuttan G, Dinesh Babu K, Rajasekharan K, Kuttan R. 1995. Anti-tumour and antioxidant activity of natural curcuminoids. Cancer Lett 94:79–83. doi: 10.1016/0304-3835(95)03827-J. [DOI] [PubMed] [Google Scholar]
- 32.Anto RJ, Mukhopadhyay A, Denning K, Aggarwal BB. 2002. Curcumin (diferuloylmethane) induces apoptosis through activation of caspase-8, BID cleavage and cytochrome c release: its suppression by ectopic expression of Bcl-2 and Bcl-xl. Carcinogenesis 23:143–150. doi: 10.1093/carcin/23.1.143. [DOI] [PubMed] [Google Scholar]
- 33.Bharti AC, Donato N, Singh S, Aggarwal BB. 2003. Curcumin (diferuloylmethane) down-regulates the constitutive activation of nuclear factor-κB and IκBα kinase in human multiple myeloma cells, leading to suppression of proliferation and induction of apoptosis. Blood 101:1053–1062. doi: 10.1182/blood-2002-05-1320. [DOI] [PubMed] [Google Scholar]
- 34.Aoki H, Takada Y, Kondo S, Sawaya R, Aggarwal BB, Kondo Y. 2007. Evidence that curcumin suppresses the growth of malignant gliomas in vitro and in vivo through induction of autophagy: role of Akt and extracellular signal-regulated kinase signaling pathways. Mol Pharmacol 72:29–39. doi: 10.1124/mol.106.033167. [DOI] [PubMed] [Google Scholar]
- 35.Anuchapreeda S, Tima S, Duangrat C, Limtrakul P. 2008. Effect of pure curcumin, demethoxycurcumin, and bisdemethoxycurcumin on WT1 gene expression in leukemic cell lines. Cancer Chemother Pharmacol 62:585–594. doi: 10.1007/s00280-007-0642-1. [DOI] [PubMed] [Google Scholar]
- 36.Dhillon N, Aggarwal BB, Newman RA, Wolff RA, Kunnumakkara AB, Abbruzzese JL, Ng CS, Badmaev V, Kurzrock R. 2008. Phase II trial of curcumin in patients with advanced pancreatic cancer. Clin Cancer Res 14:4491–4499. doi: 10.1158/1078-0432.CCR-08-0024. [DOI] [PubMed] [Google Scholar]
- 37.Yallapu MM, Ebeling MC, Khan S, Sundram V, Chauhan N, Gupta BK, Puumala SE, Jaggi M, Chauhan SC. 2013. Novel curcumin loaded magnetic nanoparticles for pancreatic cancer treatment. Mol Cancer Ther 12:1471–1480. doi: 10.1158/1535-7163.MCT-12-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yallapu MM, Maher DM, Sundram V, Bell MC, Jaggi M, Chauhan SC. 2010. Curcumin induces chemo/radio-sensitization in ovarian cancer cells and curcumin nanoparticles inhibit ovarian cancer cell growth. J Ovarian Res 3:11. doi: 10.1186/1757-2215-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skommer J, Wlodkowic D, Pelkonen J. 2006. Cellular foundation of curcumin-induced apoptosis in follicular lymphoma cell lines. Exp Hematol 34:463–474. doi: 10.1016/j.exphem.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 40.Singh S, Aggarwal BB. 1995. Activation of transcription factor NF-κB is suppressed by curcumin (diferuloylmethane). J Biol Chem 270:24995–25000. doi: 10.1074/jbc.270.42.24995. [DOI] [PubMed] [Google Scholar]
- 41.Rao DS, Sekhara NC, Satyanarayana M, Srinivasan M. 1970. Effect of curcumin on serum and liver cholesterol levels in the rat. J Nutr 100:1307–1315. [DOI] [PubMed] [Google Scholar]
- 42.Kim M, Kim Y. 2010. Hypocholesterolemic effects of curcumin via up-regulation of cholesterol 7a-hydroxylase in rats fed a high fat diet. Nutr Res Pract 4:191–195. doi: 10.4162/nrp.2010.4.3.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arun N, Nalini N. 2002. Efficacy of turmeric on blood sugar and polyol pathway in diabetic albino rats. Plant Foods Hum Nutr 57:41–52. doi: 10.1023/A:1013106527829. [DOI] [PubMed] [Google Scholar]
- 44.Peeyush KT, Gireesh G, Jobin M, Paulose C. 2009. Neuroprotective role of curcumin in the cerebellum of streptozotocin-induced diabetic rats. Life Sci 85:704–710. doi: 10.1016/j.lfs.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 45.Kumar TP, Antony S, Gireesh G, George N, Paulose C. 2010. Curcumin modulates dopaminergic receptor, CREB and phospholipase c gene expression in the cerebral cortex and cerebellum of streptozotocin induced diabetic rats. J Biomed Sci 17:43. doi: 10.1186/1423-0127-17-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim T, Davis J, Zhang AJ, He X, Mathews ST. 2009. Curcumin activates AMPK and suppresses gluconeogenic gene expression in hepatoma cells. Biochem Biophys Res Commun 388:377–382. doi: 10.1016/j.bbrc.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 47.Srimal R, Dhawan B. 1973. Pharmacology of diferuloyl methane (curcumin), a non-steroidal anti-inflammatory agent. J Pharm Pharmacol 25:447–452. doi: 10.1111/j.2042-7158.1973.tb09131.x. [DOI] [PubMed] [Google Scholar]
- 48.Mukhopadhyay A, Basu N, Ghatak N, Gujral P. 1982. Anti-inflammatory and irritant activities of curcumin analogues in rats. Agents Actions 12:508–515. doi: 10.1007/BF01965935. [DOI] [PubMed] [Google Scholar]
- 49.Motterlini R, Foresti R, Bassi R, Green CJ. 2000. Curcumin, an antioxidant and anti-inflammatory agent, induces heme oxygenase-1 and protects endothelial cells against oxidative stress. Free Radical Biol Med 28:1303–1312. doi: 10.1016/S0891-5849(00)00294-X. [DOI] [PubMed] [Google Scholar]
- 50.Sandur SK, Ichikawa H, Pandey MK, Kunnumakkara AB, Sung B, Sethi G, Aggarwal BB. 2007. Role of pro-oxidants and antioxidants in the anti-inflammatory and apoptotic effects of curcumin (diferuloylmethane). Free Radical Biol Med 43:568–580. doi: 10.1016/j.freeradbiomed.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Menon VP, Sudheer AR. 2007. Antioxidant and anti-inflammatory properties of curcumin, p 105–125. In Aggarwal BB, Surh Y-J, Shishodia S (ed), The molecular targets and therapeutic uses of curcumin in health and disease. Springer, New York, NY. [DOI] [PubMed] [Google Scholar]
- 52.Huang M-T, Lysz T, Ferraro T, Abidi TF, Laskin JD, Conney AH. 1991. Inhibitory effects of curcumin on in vitro lipoxygenase and cyclooxygenase activities in mouse epidermis. Cancer Res 51:813–819. [PubMed] [Google Scholar]
- 53.Sharma O. 1976. Antioxidant activity of curcumin and related compounds. Biochem Pharmacol 25:1811–1812. doi: 10.1016/0006-2952(76)90421-4. [DOI] [PubMed] [Google Scholar]
- 54.Toda S, Miyase T, Arichi H, Tanizawa H, Takino Y. 1985. Natural antioxidants. III. Antioxidative components isolated from rhizome of Curcuma longa L. Chem Pharm Bull 33:1725–1728. [DOI] [PubMed] [Google Scholar]
- 55.Ahsan H, Parveen N, Khan NU, Hadi SM. 1999. Pro-oxidant, anti-oxidant and cleavage activities on DNA of curcumin and its derivatives demethoxycurcumin and bisdemethoxycurcumin. Chem Biol Interact 121:161–175. doi: 10.1016/S0009-2797(99)00096-4. [DOI] [PubMed] [Google Scholar]
- 56.Lim GP, Chu T, Yang F, Beech W, Frautschy SA, Cole GM. 2001. The curry spice curcumin reduces oxidative damage and amyloid pathology in an Alzheimer transgenic mouse. J Neurosci 21:8370–8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ringman JM, Frautschy SA, Cole GM, Masterman DL, Cummings JL. 2005. A potential role of the curry spice curcumin in Alzheimer's disease. Curr Alzheimer Res 2:131–136. doi: 10.2174/1567205053585882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ryu EK, Choe YS, Lee K-H, Choi Y, Kim B-T. 2006. Curcumin and dehydrozingerone derivatives: synthesis, radiolabeling, and evaluation for β-amyloid plaque imaging. J Med Chem 49:6111–6119. doi: 10.1021/jm0607193. [DOI] [PubMed] [Google Scholar]
- 59.Garcia-Alloza M, Borrelli L, Rozkalne A, Hyman B, Bacskai B. 2007. Curcumin labels amyloid pathology in vivo, disrupts existing plaques, and partially restores distorted neurites in an Alzheimer mouse model. J Neurochem 102:1095–1104. doi: 10.1111/j.1471-4159.2007.04613.x. [DOI] [PubMed] [Google Scholar]
- 60.Jordan W, Drew C. 1996. Curcumin—a natural herb with anti-HIV activity. J Natl Med Assoc 88:333. [PMC free article] [PubMed] [Google Scholar]
- 61.Sui Z, Salto R, Li J, Craik C, Ortiz de Montellano PR. 1993. Inhibition of the HIV-1 and HIV-2 proteases by curcumin and curcumin boron complexes. Biorg Med Chem 1:415–422. doi: 10.1016/S0968-0896(00)82152-5. [DOI] [PubMed] [Google Scholar]
- 62.Gandapu U, Chaitanya R, Kishore G, Reddy RC, Kondapi AK. 2011. Curcumin-loaded apotransferrin nanoparticles provide efficient cellular uptake and effectively inhibit HIV-1 replication in vitro. PLoS One 6:e23388. doi: 10.1371/journal.pone.0023388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sidhu GS, Mani H, Gaddipati JP, Singh AK, Seth P, Banaudha KK, Patnaik GK, Maheshwari RK. 1999. Curcumin enhances wound healing in streptozotocin induced diabetic rats and genetically diabetic mice. Wound Repair Regen 7:362–374. doi: 10.1046/j.1524-475X.1999.00362.x. [DOI] [PubMed] [Google Scholar]
- 64.Gopinath D, Ahmed MR, Gomathi K, Chitra K, Sehgal P, Jayakumar R. 2004. Dermal wound healing processes with curcumin incorporated collagen films. Biomaterials 25:1911–1917. doi: 10.1016/S0142-9612(03)00625-2. [DOI] [PubMed] [Google Scholar]
- 65.Pandey N, Strider J, Nolan WC, Yan SX, Galvin JE. 2008. Curcumin inhibits aggregation of α-synuclein. Acta Neuropathol 115:479–489. doi: 10.1007/s00401-007-0332-4. [DOI] [PubMed] [Google Scholar]
- 66.Pan J, Li H, Ma J-F, Tan Y-Y, Xiao Q, Ding J-Q, Chen S-D. 2012. Curcumin inhibition of JNKs prevents dopaminergic neuronal loss in a mouse model of Parkinson's disease through suppressing mitochondria dysfunction. Transl Neurodegen 1:1–9. doi: 10.1186/2047-9158-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sreejayan Rao MNA. 1997. Nitric oxide scavenging by curcuminoids. J Pharm Pharmacol 49:105–107. doi: 10.1111/j.2042-7158.1997.tb06761.x. [DOI] [PubMed] [Google Scholar]
- 68.Thapliyal R, Maru G. 2001. Inhibition of cytochrome P450 isozymes by curcumins in vitro and in vivo. Food Chem Toxicol 39:541–547. doi: 10.1016/S0278-6915(00)00165-4. [DOI] [PubMed] [Google Scholar]
- 69.Majeed M, Badmaev V, Rajendran R. November 2002. Bioprotectant composition, method of use and extraction process of curcuminoids. EP patent 0,839,037.
- 70.Hsu C-H, Cheng A-L. 2007. Clinical studies with curcumin, p 471–480. In Aggarwal BB, Surh Y-J, Shishodia S (ed), The molecular targets and therapeutic uses of curcumin in health and disease. Springer, New York, NY. [Google Scholar]
- 71.Li R, Qiao X, Li Q, He R, Ye M, Xiang C, Lin X, Guo D. 2011. Metabolic and pharmacokinetic studies of curcumin, demethoxycurcumin and bisdemethoxycurcumin in mice tumor after intragastric administration of nanoparticle formulations by liquid chromatography coupled with tandem mass spectrometry. J Chromatogr B 879:2751–2758. doi: 10.1016/j.jchromb.2011.07.042. [DOI] [PubMed] [Google Scholar]
- 72.Cheng A-L, Hsu C-H, Lin J-K, Hsu M-M, Ho Y-F, Shen T-S, Ko J-Y, Lin J-T, Lin B, Wu M-S, Yu H, Jee S-H, Chen G, Chen T, Chen C, Lai M, Pu Y, Pan M, Wang Y, Tsai C, Hsieh C. 2001. Phase I clinical trial of curcumin, a chemopreventive agent, in patients with high-risk or pre-malignant lesions. Anticancer Res 21:2895–2900. [PubMed] [Google Scholar]
- 73.Sharma RA, Euden SA, Platton SL, Cooke DN, Shafayat A, Hewitt HR, Marczylo TH, Morgan B, Hemingway D, Plummer SM. 2004. Phase I clinical trial of oral curcumin biomarkers of systemic activity and compliance. Clin Cancer Res 10:6847–6854. doi: 10.1158/1078-0432.CCR-04-0744. [DOI] [PubMed] [Google Scholar]
- 74.Vareed SK, Kakarala M, Ruffin MT, Crowell JA, Normolle DP, Djuric Z, Brenner DE. 2008. Pharmacokinetics of curcumin conjugate metabolites in healthy human subjects. Cancer Epidemiol Biomarkers Prevention 17:1411–1417. doi: 10.1158/1055-9965.EPI-07-2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thomas J, Tarka S, Soni M. 2013. Expert panel statement: determination of the generally recognized as safe (GRAS) status of curcumin (Curcumin C3 Complex) as a food ingredient. GRAS notice (GRN) no. 460 Accessed 17 June 2014 http://www.fda.gov/Food/IngredientsPackagingLabeling/GRAS/.
- 76.Anand P, Kunnumakkara AB, Newman RA, Aggarwal BB. 2007. Bioavailability of curcumin: problems and promises. Mol Pharm 4:807–818. doi: 10.1021/mp700113r. [DOI] [PubMed] [Google Scholar]
- 77.Cruz-Correa M, Shoskes DA, Sanchez P, Zhao R, Hylind LM, Wexner SD, Giardiello FM. 2006. Combination treatment with curcumin and quercetin of adenomas in familial adenomatous polyposis. Clin Gastroenterol Hepatol 4:1035–1038. doi: 10.1016/j.cgh.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 78.Shoba G, Joy D, Joseph T, Majeed M, Rajendran R, Srinivas P. 1998. Influence of piperine on the pharmacokinetics of curcumin in animals and human volunteers. Planta Med 64:353–356. doi: 10.1055/s-2006-957450. [DOI] [PubMed] [Google Scholar]
- 79.Li L, Ahmed B, Mehta K, Kurzrock R. 2007. Liposomal curcumin with and without oxaliplatin: effects on cell growth, apoptosis, and angiogenesis in colorectal cancer. Mol Cancer Ther 6:1276–1282. doi: 10.1158/1535-7163.MCT-06-0556. [DOI] [PubMed] [Google Scholar]
- 80.Li L, Braiteh FS, Kurzrock R. 2005. Liposome-encapsulated curcumin. In vitro and in vivo effects on proliferation, apoptosis, signaling, and angiogenesis. Cancer 104:1322–1331. doi: 10.1002/cncr.21300. [DOI] [PubMed] [Google Scholar]
- 81.Kunwar A, Barik A, Pandey R, Priyadarsini KI. 2006. Transport of liposomal and albumin loaded curcumin to living cells: an absorption and fluorescence spectroscopic study. Biochim Biophys Acta 1760:1513–1520. doi: 10.1016/j.bbagen.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 82.Suresh D, Srinivasan K. 2007. Studies on the in vitro absorption of spice principles—curcumin, capsaicin and piperine in rat intestines. Food Chem Toxicol 45:1437–1442. doi: 10.1016/j.fct.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 83.Ma Z, Shayeganpour A, Brocks DR, Lavasanifar A, Samuel J. 2007. High-performance liquid chromatography analysis of curcumin in rat plasma: application to pharmacokinetics of polymeric micellar formulation of curcumin. Biomed Chromatogr 21:546–552. doi: 10.1002/bmc.795. [DOI] [PubMed] [Google Scholar]
- 84.Letchford K, Liggins R, Burt H. 2008. Solubilization of hydrophobic drugs by methoxy poly(ethylene glycol)-block-polycaprolactone diblock copolymer micelles: theoretical and experimental data and correlations. J Pharm Sci 97:1179–1190. doi: 10.1002/jps.21037. [DOI] [PubMed] [Google Scholar]
- 85.Bisht S, Feldmann G, Soni S, Ravi R, Karikar C, Maitra A, Maitra A. 2007. Polymeric nanoparticle-encapsulated curcumin (“nanocurcumin”): a novel strategy for human cancer therapy. J Nanobiotechnol 5:1–18. doi: 10.1186/1477-3155-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tiyaboonchai W, Tungpradit W, Plianbangchang P. 2007. Formulation and characterization of curcuminoids loaded solid lipid nanoparticles. Int J Pharm 337:299–306. doi: 10.1016/j.ijpharm.2006.12.043. [DOI] [PubMed] [Google Scholar]
- 87.Shaikh J, Ankola D, Beniwal V, Singh D, Kumar M. 2009. Nanoparticle encapsulation improves oral bioavailability of curcumin by at least 9-fold when compared to curcumin administered with piperine as absorption enhancer. Eur J Pharm Sci 37:223–230. doi: 10.1016/j.ejps.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 88.Anand P, Nair HB, Bokyung S, Kunnumakkara AB, Yadav VR, Tekmal RR, Aggarwal BB. 2010. Design of curcumin-loaded PLGA nanoparticles formulation with enhanced cellular uptake, and increased bioactivity in vitro and superior bioavailability in vivo. Biochem Pharmacol 79:330–338. doi: 10.1016/j.bcp.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 89.Mulik RS, Mönkkönen J, Juvonen RO, Mahadik KR, Paradkar AR. 2010. Transferrin mediated solid lipid nanoparticles containing curcumin: Enhanced in vitro anticancer activity by induction of apoptosis. Int J Pharm 398:190–203. doi: 10.1016/j.ijpharm.2010.07.021. [DOI] [PubMed] [Google Scholar]
- 90.Kim TH, Jiang HH, Youn YS, Park CW, Tak KK, Lee S, Kim H, Jon S, Chen X, Lee KC. 2011. Preparation and characterization of water-soluble albumin-bound curcumin nanoparticles with improved antitumor activity. Int J Pharm 403:285–291. doi: 10.1016/j.ijpharm.2010.10.041. [DOI] [PubMed] [Google Scholar]
- 91.Basniwal RK, Buttar HS, Jain V, Jain N. 2011. Curcumin nanoparticles: preparation, characterization, and antimicrobial study. J Agric Food Chem 59:2056–2061. doi: 10.1021/jf104402t. [DOI] [PubMed] [Google Scholar]
- 92.Liu A, Lou H, Zhao L, Fan P. 2006. Validated LC/MS/MS assay for curcumin and tetrahydrocurcumin in rat plasma and application to pharmacokinetic study of phospholipid complex of curcumin. J Pharm Biomed Anal 40:720–727. doi: 10.1016/j.jpba.2005.09.032. [DOI] [PubMed] [Google Scholar]
- 93.Maiti K, Mukherjee K, Gantait A, Saha BP, Mukherjee PK. 2007. Curcumin–phospholipid complex: preparation, therapeutic evaluation and pharmacokinetic study in rats. Int J Pharm 330:155–163. doi: 10.1016/j.ijpharm.2006.09.025. [DOI] [PubMed] [Google Scholar]
- 94.Ohori H, Yamakoshi H, Tomizawa M, Shibuya M, Kakudo Y, Takahashi A, Takahashi S, Kato S, Suzuki T, Ishioka C. 2006. Synthesis and biological analysis of new curcumin analogues bearing an enhanced potential for the medicinal treatment of cancer. Mol Cancer Ther 5:2563–2571. doi: 10.1158/1535-7163.MCT-06-0174. [DOI] [PubMed] [Google Scholar]
- 95.Mosley CA, Liotta DC, Snyder JP. 2007. Highly active anticancer curcumin analogues, p 77–103. In Aggarwal BB, Surh Y-J, Shishodia S (ed), The molecular targets and therapeutic uses of curcumin in health and disease Springer, New York, NY. [DOI] [PubMed] [Google Scholar]
- 96.Helson L. 2013. Curcumin (diferuloylmethane) delivery methods: a review. BioFactors 39:21–26. doi: 10.1002/biof.1080. [DOI] [PubMed] [Google Scholar]
- 97.Katsuyama Y, Kita T, Funa N, Horinouchi S. 2009. Curcuminoid biosynthesis by two type III polyketide synthases in the herb Curcuma longa. J Biol Chem 284:11160–11170. doi: 10.1074/jbc.M900070200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ramirez-Ahumada M, Timmermann BN, Gang DR. 2006. Biosynthesis of curcuminoids and gingerols in turmeric (Curcuma longa) and ginger (Zingiber officinale): identification of curcuminoid synthase and hydroxycinnamoyl-CoA thioesterases. Phytochemistry 67:2017–2029. doi: 10.1016/j.phytochem.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 99.Dixon RA, Paiva NL. 1995. Stress-induced phenylpropanoid metabolism. Plant Cell 7:1085–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Katsuyama Y, Matsuzawa M, Funa N, Horinouchi S. 2007. In vitro synthesis of curcuminoids by type III polyketide synthase from Oryza sativa. J Biol Chem 282:37702–37709. doi: 10.1074/jbc.M707569200. [DOI] [PubMed] [Google Scholar]
- 101.Katsuyama Y, Miyazono K-I, Tanokura M, Ohnishi Y, Horinouchi S. 2011. Structural and biochemical elucidation of mechanism for decarboxylative condensation of β-keto acid by curcumin synthase. J Biol Chem 286:6659–6668. doi: 10.1074/jbc.M110.196279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Katsuyama Y, Kita T, Horinouchi S. 2009. Identification and characterization of multiple curcumin synthases from the herb Curcuma longa. FEBS Lett 583:2799–2803. doi: 10.1016/j.febslet.2009.07.029. [DOI] [PubMed] [Google Scholar]
- 103.Resmi M, Soniya E. 2012. Molecular cloning and differential expressions of two cDNA encoding type III polyketide synthase in different tissues of Curcuma longa L. Gene 491:278–283. doi: 10.1016/j.gene.2011.09.025. [DOI] [PubMed] [Google Scholar]
- 104.Xie Z, Ma X, Gang DR. 2009. Modules of co-regulated metabolites in turmeric (Curcuma longa) rhizome suggest the existence of biosynthetic modules in plant specialized metabolism. J Exp Bot 60:87–97. doi: 10.1093/jxb/ern263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kita T, Imai S, Sawada H, Seto H. 2009. Isolation of dihydrocurcuminoids from cell clumps and their distribution in various parts of turmeric (Curcuma longa). Biosci Biotech Biochem 73:1113–1117. doi: 10.1271/bbb.80871. [DOI] [PubMed] [Google Scholar]
- 106.Katsuyama Y, Matsuzawa M, Funa N, Horinouchi S. 2008. Production of curcuminoids by Escherichia coli carrying an artificial biosynthesis pathway. Microbiology 154:2620–2628. doi: 10.1099/mic.0.2008/018721-0. [DOI] [PubMed] [Google Scholar]
- 107.Horinouchi S. 2009. Combinatorial biosynthesis of plant medicinal polyketides by microorganisms. Curr Opin Chem Biol 13:197–204. doi: 10.1016/j.cbpa.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 108.Horinouchi S. 2008. Combinatorial biosynthesis of non-bacterial and unnatural flavonoids, stilbenoids and curcuminoids by microorganisms. J Antibiot 61:709–728. doi: 10.1038/ja.2008.85. [DOI] [PubMed] [Google Scholar]
- 109.Lussier F-X, Colatriano D, Wiltshire Z, Page JE, Martin VJ. 2012. Engineering microbes for plant polyketide biosynthesis. Comput Struct Biotechnol J 3:1–11. doi: 10.5936/csbj.201210020.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chemler JA, Koffas MA. 2008. Metabolic engineering for plant natural product biosynthesis in microbes. Curr Opin Biotechnol 19:597–605. doi: 10.1016/j.copbio.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 111.Jeandet P, Delaunois B, Aziz A, Donnez D, Vasserot Y, Cordelier S, Courot E. 2012. Metabolic engineering of yeast and plants for the production of the biologically active hydroxystilbene, resveratrol. J Biomed Biotechnol 2012:579089. doi: 10.1155/2012/579089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Halls C, Yu O. 2008. Potential for metabolic engineering of resveratrol biosynthesis. Trends Biotechnol 26:77–81. doi: 10.1016/j.tibtech.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 113.Miyazono Ki. Um J, Imai FL, Katsuyama Y, Ohnishi Y, Horinouchi S, Tanokura M. 2011. Crystal structure of curcuminoid synthase CUS from Oryza sativa. Proteins 79:669–673. doi: 10.1002/prot.22888. [DOI] [PubMed] [Google Scholar]
- 114.Morita H, Wanibuchi K, Nii H, Kato R, Sugio S, Abe I. 2010. Structural basis for the one-pot formation of the diarylheptanoid scaffold by curcuminoid synthase from Oryza sativa. Proc Natl Acad Sci U S A 107:19778–19783. doi: 10.1073/pnas.1011499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Katsuyama Y, Ohnishi Y, Horinouchi S. 2010. Production of dehydrogingerdione derivatives in Escherichia coli by exploiting a curcuminoid synthase from Oryza sativa and a β-oxidation pathway from Saccharomyces cerevisiae. Chembiochem 11:2034–2041. doi: 10.1002/cbic.201000379. [DOI] [PubMed] [Google Scholar]
- 116.Miyahisa I, Kaneko M, Funa N, Kawasaki H, Kojima H, Ohnishi Y, Horinouchi S. 2005. Efficient production of (2S)-flavanones by Escherichia coli containing an artificial biosynthetic gene cluster. Appl Microbiol Biotechnol 68:498–504. doi: 10.1007/s00253-005-1916-3. [DOI] [PubMed] [Google Scholar]
- 117.Hwang EI, Kaneko M, Ohnishi Y, Horinouchi S. 2003. Production of plant-specific flavanones by Escherichia coli containing an artificial gene cluster. Appl Environ Microbiol 69:2699–2706. doi: 10.1128/AEM.69.5.2699-2706.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Katsuyama Y, Funa N, Horinouchi S. 2007. Precursor-directed biosynthesis of stilbene methyl ethers in Escherichia coli. Biotechnol J 2:1286–1293. doi: 10.1002/biot.200700098. [DOI] [PubMed] [Google Scholar]
- 119.Katsuyama Y, Funa N, Miyahisa I, Horinouchi S. 2007. Synthesis of unnatural flavonoids and stilbenes by exploiting the plant biosynthetic pathway in Escherichia coli. Chem Biol 14:613–621. doi: 10.1016/j.chembiol.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 120.Kikuzaki H, Hisamoto M, Hirose K, Akiyama K, Taniguchi H. 2002. Antioxidant properties of ferulic acid and its related compounds. J Agric Food Chem 50:2161–2168. doi: 10.1021/jf011348w. [DOI] [PubMed] [Google Scholar]
- 121.Katsuyama Y, Hirose Y, Funa N, Ohnishi Y, Horinouchi S. 2010. Precursor-directed biosynthesis of curcumin analogs in Escherichia coli. Biosci Biotech Biochem 74:641–645. doi: 10.1271/bbb.90866. [DOI] [PubMed] [Google Scholar]
- 122.Wang S, Zhang S, Zhou T, Zeng J, Zhan J. 2013. Design and application of an in vivo reporter assay for phenylalanine ammonia-lyase. Appl Microbiol Biotechnol 97:7877–7885. doi: 10.1007/s00253-013-5122-4. [DOI] [PubMed] [Google Scholar]
- 123.Trantas E, Panopoulos N, Ververidis F. 2009. Metabolic engineering of the complete pathway leading to heterologous biosynthesis of various flavonoids and stilbenoids in Saccharomyces cerevisiae. Metab Eng 11:355–366. doi: 10.1016/j.ymben.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 124.Beekwilder J, Wolswinkel R, Jonker H, Hall R, de Vos CR, Bovy A. 2006. Production of resveratrol in recombinant microorganisms. Appl Environ Microbiol 72:5670–5672. doi: 10.1128/AEM.00609-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Becker JV, Armstrong GO, Merwe MJ, Lambrechts MG, Vivier MA, Pretorius IS. 2003. Metabolic engineering of Saccharomyces cerevisiae for the synthesis of the wine-related antioxidant resveratrol. FEMS Yeast Res 4:79–85. doi: 10.1016/S1567-1356(03)00157-0. [DOI] [PubMed] [Google Scholar]
- 126.Jiang H, Wood KV, Morgan JA. 2005. Metabolic engineering of the phenylpropanoid pathway in Saccharomyces cerevisiae. Appl Environ Microbiol 71:2962–2969. doi: 10.1128/AEM.71.6.2962-2969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yan Y, Kohli A, Koffas MA. 2005. Biosynthesis of natural flavanones in Saccharomyces cerevisiae. Appl Environ Microbiol 71:5610–5613. doi: 10.1128/AEM.71.9.5610-5613.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Leonard E, Yan Y, Lim KH, Koffas MA. 2005. Investigation of two distinct flavone synthases for plant-specific flavone biosynthesis in Saccharomyces cerevisiae. Appl Environ Microbiol 71:8241–8248. doi: 10.1128/AEM.71.12.8241-8248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhang Y, Li S-Z, Li J, Pan X, Cahoon RE, Jaworski JG, Wang X, Jez JM, Chen F, Yu O. 2006. Using unnatural protein fusions to engineer resveratrol biosynthesis in yeast and mammalian cells. J Am Chem Soc 128:13030–13031. doi: 10.1021/ja0622094. [DOI] [PubMed] [Google Scholar]
- 130.Wang Y, Halls C, Zhang J, Matsuno M, Zhang Y, Yu O. 2011. Stepwise increase of resveratrol biosynthesis in yeast Saccharomyces cerevisiae by metabolic engineering. Metab Eng 13:455–463. doi: 10.1016/j.ymben.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 131.Shin S-Y, Jung S-M, Kim M-D, Han NS, Seo J-H. 2012. Production of resveratrol from tyrosine in metabolically engineered Saccharomyces cerevisiae. Enzyme Microb Technol 51:211–216. doi: 10.1016/j.enzmictec.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 132.Koopman F, Beekwilder J, Crimi B, van Houwelingen A, Hall RD, Bosch D, van Maris AJ, Pronk JT, Daran J-M. 2012. De novo production of the flavonoid naringenin in engineered Saccharomyces cerevisiae. Microb Cell Fact 11:155. doi: 10.1186/1475-2859-11-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sahdev S, Khattar SK, Saini KS. 2008. Production of active eukaryotic proteins through bacterial expression systems: a review of the existing biotechnology strategies. Mol Cell Biochem 307:249–264. [DOI] [PubMed] [Google Scholar]
- 134.Popuri AK, Pagala B. 2013. Extraction of curcumin from turmeric roots. Int J Innovative Res Stud 2:289–299. [Google Scholar]
- 135.Paulucci VP, Couto RO, Teixeira CC, Freitas LAP. 2013. Optimization of the extraction of curcumin from Curcuma longa rhizomes. Brazil J Pharmacog 23:94–100. doi: 10.1590/S0102-695X2012005000117. [DOI] [Google Scholar]
- 136.Gaikar VG, Dandekar DV. May 2001. Process for extraction of curcuminoids from curcuma species. US patent 6,224,877.
- 137.Xu P, Ranganathan S, Fowler ZL, Maranas CD, Koffas MAG. 2011. Genome-scale metabolic network modeling results in minimal interventions that cooperatively force carbon flux towards malonyl-CoA. Metab Eng 13:578–587. doi: 10.1016/j.ymben.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 138.Leonard E, Yan Y, Fowler ZL, Li Z, Lim C-G, Lim K-H, Koffas MA. 2008. Strain improvement of recombinant Escherichia coli for efficient production of plant flavonoids. Mol Pharm 5:257–265. doi: 10.1021/mp7001472. [DOI] [PubMed] [Google Scholar]
- 139.Sydor T, Schaffer S, Boles E. 2010. Considerable increase in resveratrol production by recombinant industrial yeast strains with use of rich medium. Appl Environ Microbiol 76:3361–3363. doi: 10.1128/AEM.02796-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lin Y, Yan Y. 2012. Biosynthesis of caffeic acid in Escherichia coli using its endogenous hydroxylase complex. Microb Cell Fact 11:42. doi: 10.1186/1475-2859-11-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kang S-Y, Choi O, Lee JK, Hwang BY, Uhm T-B, Hong Y-S. 2012. Artificial biosynthesis of phenylpropanoic acids in a tyrosine overproducing Escherichia coli strain. Microb Cell Fact 11:153. doi: 10.1186/1475-2859-11-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sprenger GA. 2007. From scratch to value: engineering Escherichia coli wild type cells to the production of l-phenylalanine and other fine chemicals derived from chorismate. Appl Microbiol Biotechnol 75:739–749. doi: 10.1007/s00253-007-0931-y. [DOI] [PubMed] [Google Scholar]
- 143.Pittard J, Camakaris H, Yang J. 2005. The TyrR regulon. Mol Microbiol 55:16–26. doi: 10.1111/j.1365-2958.2004.04385.x. [DOI] [PubMed] [Google Scholar]
- 144.Bongaerts J, Krämer M, Müller U, Raeven L, Wubbolts M. 2001. Metabolic engineering for microbial production of aromatic amino acids and derived compounds. Metab Eng 3:289–300. doi: 10.1006/mben.2001.0196. [DOI] [PubMed] [Google Scholar]
- 145.Lütke-Eversloh T, Stephanopoulos G. 2005. Feedback inhibition of chorismate mutase/prephenate dehydrogenase (TyrA) of Escherichia coli: generation and characterization of tyrosine-insensitive mutants. Appl Environ Microbiol 71:7224–7228. doi: 10.1128/AEM.71.11.7224-7228.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Berry A. 1996. Improving production of aromatic compounds in Escherichia coli by metabolic engineering. Trends Biotechnol 14:250–256. doi: 10.1016/0167-7799(96)10033-0. [DOI] [PubMed] [Google Scholar]
- 147.Draths K, Pompliano D, Conley D, Frost J, Berry A, Disbrow G, Staversky R, Lievense J. 1992. Biocatalytic synthesis of aromatics from d-glucose: the role of transketolase. J Am Chem Soc 114:3956–3962. doi: 10.1021/ja00036a050. [DOI] [Google Scholar]
- 148.Lütke-Eversloh T, Stephanopoulos G. 2007. l-Tyrosine production by deregulated strains of Escherichia coli. Appl Microbiol Biotechnol 75:103–110. doi: 10.1007/s00253-006-0792-9. [DOI] [PubMed] [Google Scholar]
- 149.Juminaga D, Baidoo EE, Redding-Johanson AM, Batth TS, Burd H, Mukhopadhyay A, Petzold CJ, Keasling JD. 2012. Modular engineering of L-tyrosine production in Escherichia coli. Appl Environ Microbiol 78:89–98. doi: 10.1128/AEM.06017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Santos CNS, Koffas M, Stephanopoulos G. 2011. Optimization of a heterologous pathway for the production of flavonoids from glucose. Metab Eng 13:392–400. doi: 10.1016/j.ymben.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 151.Huang Q, Lin Y, Yan Y. 2013. Caffeic acid production enhancement by engineering a phenylalanine over-producing Escherichia coli strain. Biotechnol Bioeng 110:3188–3196. doi: 10.1002/bit.24988. [DOI] [PubMed] [Google Scholar]
- 152.Zhang H, Stephanopoulos G. 2013. Engineering E. coli for caffeic acid biosynthesis from renewable sugars. Appl Microbiol Biotechnol 97:3333–3341. doi: 10.1007/s00253-012-4544-8. [DOI] [PubMed] [Google Scholar]
- 153.Kim M-J, Kim B-G, Ahn J-H. 2013. Biosynthesis of bioactive O-methylated flavonoids in Escherichia coli. Appl Microbiol Biotechnol 97:7195–7204. doi: 10.1007/s00253-013-5020-9. [DOI] [PubMed] [Google Scholar]
- 154.Berner M, Krug D, Bihlmaier C, Vente A, Müller R, Bechthold A. 2006. Genes and enzymes involved in caffeic acid biosynthesis in the actinomycete Saccharothrix espanaensis. J Bacteriol 188:2666–2673. doi: 10.1128/JB.188.7.2666-2673.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Choi O, Wu C-Z, Kang SY, Ahn JS, Uhm T-B, Hong Y-S. 2011. Biosynthesis of plant-specific phenylpropanoids by construction of an artificial biosynthetic pathway in Escherichia coli. J Ind Microbiol Biotechnol 38:1657–1665. doi: 10.1007/s10295-011-0954-3. [DOI] [PubMed] [Google Scholar]
- 156.Furuya T, Arai Y, Kino K. 2012. Biotechnological production of caffeic acid by bacterial cytochrome P450 CYP199A2. Appl Environ Microbiol 78:6087–6094. doi: 10.1128/AEM.01103-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Vannelli T, Xue Z, Breinig S, Qi WW, Sariaslani FS. 2007. Functional expression in Escherichia coli of the tyrosine-inducible tyrosine ammonia-lyase enzyme from yeast Trichosporon cutaneum for production of p-hydroxycinnamic acid. Enzyme Microb Technol 41:413–422. doi: 10.1016/j.enzmictec.2007.03.013. [DOI] [Google Scholar]
- 158.Watts KT, Lee PC, Schmidt-Dannert C. 2006. Biosynthesis of plant-specific stilbene polyketides in metabolically engineered Escherichia coli. BMC Biotechnol 6:22. doi: 10.1186/1472-6750-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Leonard E, Yan Y, Koffas MAG. 2006. Functional expression of a P450 flavonoid hydroxylase for the biosynthesis of plant-specific hydroxylated flavonols in Escherichia coli. Metab Eng 8:172–181. doi: 10.1016/j.ymben.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 160.Leonard E, Chemler J, Lim KH, Koffas MA. 2006. Expression of a soluble flavone synthase allows the biosynthesis of phytoestrogen derivatives in Escherichia coli. Appl Microbiol Biotechnol 70:85–91. doi: 10.1007/s00253-005-0059-x. [DOI] [PubMed] [Google Scholar]
- 161.Watts KT, Lee PC, Schmidt-Dannert C. 2004. Exploring recombinant flavonoid biosynthesis in metabolically engineered Escherichia coli. Chembiochem 5:500–507. doi: 10.1002/cbic.200300783. [DOI] [PubMed] [Google Scholar]
- 162.Miyahisa I, Funa N, Ohnishi Y, Martens S, Moriguchi T, Horinouchi S. 2006. Combinatorial biosynthesis of flavones and flavonols in Escherichia coli. Appl Microbiol Biotechnol 71:53–58. doi: 10.1007/s00253-005-0116-5. [DOI] [PubMed] [Google Scholar]
- 163.Mori H, Iwahashi H. 2009. Antioxidant activity of caffeic acid through a novel mechanism under uva irradiation. J Clin Biochem Nutr 45:49–55. doi: 10.3164/jcbn.08-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Chao P, Hsu C, Yin M. 2009. Anti-inflammatory and anti-coagulatory activities of caffeic acid and ellagic acid in cardiac tissue of diabetic mice. Nutr Metab 6:1–8. doi: 10.1186/1743-7075-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Prasad NR, Karthikeyan A, Karthikeyan S, Reddy BV. 2011. Inhibitory effect of caffeic acid on cancer cell proliferation by oxidative mechanism in human HT-1080 fibrosarcoma cell line. Mol Cell Biochem 349:11–19. doi: 10.1007/s11010-010-0655-7. [DOI] [PubMed] [Google Scholar]
- 166.Mulert U, Luzhetskyy A, Hofmann C, Mayer A, Bechthold A. 2004. Expression of the landomycin biosynthetic gene cluster in a PKS mutant of Streptomyces fradiae is dependent on the coexpression of a putative transcriptional activator gene. FEMS Microbiol Lett 230:91–97. doi: 10.1016/S0378-1097(03)00861-9. [DOI] [PubMed] [Google Scholar]
- 167.Park SR, Yoon JA, Paik JH, Park JW, Jung WS, Ban Y-H, Kim EJ, Yoo YJ, Han AR, Yoon YJ. 2009. Engineering of plant-specific phenylpropanoids biosynthesis in Streptomyces venezuelae. J Biotechnol 141:181–188. doi: 10.1016/j.jbiotec.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 168.Hotze M, Schröder G, Schröder J. 1995. Cinnamate 4-hydroxylase from Catharanthus roseus and a strategy for the functional expression of plant cytochrome P450 proteins as translational fusions with P450 reductase in Escherichia coli. FEBS Lett 374:345–350. doi: 10.1016/0014-5793(95)01141-Z. [DOI] [PubMed] [Google Scholar]
- 169.Kneusel RE, Matern U, Nicolay K. 1989. Formation of trans-caffeoyl-CoA from trans-4-coumaroyl-CoA by Zn2+-dependent enzymes in cultured plant cells and its activation by an elicitor-induced pH shift. Arch Biochem Biophys 269:455–462. doi: 10.1016/0003-9861(89)90129-X. [DOI] [PubMed] [Google Scholar]
- 170.Xue Z, McCluskey M, Cantera K, Sariaslani FS, Huang L. 2007. Identification, characterization and functional expression of a tyrosine ammonia-lyase and its mutants from the photosynthetic bacterium Rhodobacter sphaeroides. J Ind Microbiol Biotechnol 34:599–604. doi: 10.1007/s10295-007-0229-1. [DOI] [PubMed] [Google Scholar]
- 171.Xue Z, McCluskey M, Cantera K, Ben-Bassat A, Sariaslani FS, Huang L. 2007. Improved production of p-hydroxycinnamic acid from tyrosine using a novel thermostable phenylalanine/tyrosine ammonia lyase enzyme. Enzyme Microb Technol 42:58–64. doi: 10.1016/j.enzmictec.2007.07.025. [DOI] [Google Scholar]
- 172.Kim YH, Kwon T, Yang HJ, Kim W, Youn H, Lee JY, Youn B. 2011. Gene engineering, purification, crystallization and preliminary X-ray diffraction of cytochrome P450 p-coumarate-3-hydroxylase (C3H), the Arabidopsis membrane protein. Protein Expr Purif 79:149–155. doi: 10.1016/j.pep.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 173.Gatenby AA, Sariaslani S, Tang X-S, Qi WW, Vannelli T. April 2002. Bioproduction of para-hydroxycinnamic acid. US patent 6,368,837.
- 174.Vannelli T, Wei Qi W, Sweigard J, Gatenby AA, Sariaslani FS. 2007. Production of p-hydroxycinnamic acid from glucose in Saccharomyces cerevisiae and Escherichia coli by expression of heterologous genes from plants and fungi. Metab Eng 9:142–151. doi: 10.1016/j.ymben.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 175.Lin Y, Sun X, Yuan Q, Yan Y. 2013. Combinatorial biosynthesis of plant-specific coumarins in bacteria. Metab Eng 18:69–77. doi: 10.1016/j.ymben.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 176.Beuerle T, Pichersky E. 2002. Enzymatic synthesis and purification of aromatic coenzyme A esters. Anal Biochem 302:305–312. doi: 10.1006/abio.2001.5574. [DOI] [PubMed] [Google Scholar]
- 177.Furuya T, Kino K. 2014. Catalytic activity of the two-component flavin-dependent monooxygenase from Pseudomonas aeruginosa toward cinnamic acid derivatives. Appl Microbiol Biotechnol 98:1145–1154. doi: 10.1007/s00253-013-4958-y. [DOI] [PubMed] [Google Scholar]
- 178.Kaneko M, Ohnishi Y, Horinouchi S. 2003. Cinnamate:coenzyme A ligase from the filamentous bacterium Streptomyces coelicolor A3 (2). J Bacteriol 185:20–27. doi: 10.1128/JB.185.1.20-27.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Knobloch K-H, Hahlbrock K. 1977. 4-Coumarate:CoA ligase from cell suspension cultures of Petroselinum hortense Hoffm: partial purification, substrate specificity, and further properties. Arch Biochem Biophys 184:237–248. doi: 10.1016/0003-9861(77)90347-2. [DOI] [PubMed] [Google Scholar]
- 180.Ehlting J, Büttner D, Wang Q, Douglas CJ, Somssich IE, Kombrink E. 1999. Three 4-coumarate: coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J 19:9–20. doi: 10.1046/j.1365-313X.1999.00491.x. [DOI] [PubMed] [Google Scholar]
- 181.Hamberger B, Hahlbrock K. 2004. The 4-coumarate:CoA ligase gene family in Arabidopsis thaliana comprises one rare, sinapate-activating and three commonly occurring isoenzymes. Proc Natl Acad Sci U S A 101:2209–2214. doi: 10.1073/pnas.0307307101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Schneider K, Hövel K, Witzel K, Hamberger B, Schomburg D, Kombrink E, Stuible H-P. 2003. The substrate specificity-determining amino acid code of 4-coumarate:CoA ligase. Proc Natl Acad Sci U S A 100:8601–8606. doi: 10.1073/pnas.1430550100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Schneider K. 2005. Mechanismus der Substratselektion durch Adenylat bildende Enzyme: Analyse der 4-Cumarat: CoA-Ligase (4CL) und ausgewählter 4CL-ähnlicher Enzyme aus Arabidopsis thaliana. Ph.D. thesis Cuvillier Verlag, Göttingen, Germany. [Google Scholar]
- 184.Gui J, Shen J, Li L. 2011. Functional characterization of evolutionarily divergent 4-coumarate:coenzyme A ligases in rice. Plant Physiol 157:574–586. doi: 10.1104/pp.111.178301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lozoya E, Hoffmann H, Douglas C, Schulz W, Scheel D, Hahlbrock K. 1988. Primary structures and catalytic properties of isoenzymes encoded by the two 4-coumarate:CoA ligase genes in parsley. Eur J Biochem 176:661–667. doi: 10.1111/j.1432-1033.1988.tb14328.x. [DOI] [PubMed] [Google Scholar]
- 186.Shin S-Y, Han NS, Park Y-C, Kim M-D, Seo J-H. 2011. Production of resveratrol from p-coumaric acid in recombinant Saccharomyces cerevisiae expressing 4-coumarate: coenzyme A ligase and stilbene synthase genes. Enzyme Microb Technol 48:48–53. doi: 10.1016/j.enzmictec.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 187.Brand S, Hölscher D, Schierhorn A, Svatoš A, Schröder J, Schneider B. 2006. A type III polyketide synthase from Wachendorfia thyrsiflora and its role in diarylheptanoid and phenylphenalenone biosynthesis. Planta 224:413–428. doi: 10.1007/s00425-006-0228-x. [DOI] [PubMed] [Google Scholar]
- 188.Leonard E, Lim K-H, Saw P-N, Koffas MA. 2007. Engineering central metabolic pathways for high-level flavonoid production in Escherichia coli. Appl Environ Microbiol 73:3877–3886. doi: 10.1128/AEM.00200-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Takamura Y, Nomura G. 1988. Changes in the intracellular concentration of acetyl-CoA and malonyl-CoA in relation to the carbon and energy metabolism of Escherichia coli K12. J Gen Microbiol 134:2249–2253. [DOI] [PubMed] [Google Scholar]
- 190.Xu P, Gu Q, Wang W, Wong L, Bower AG, Collins CH, Koffas MA. 2013. Modular optimization of multi-gene pathways for fatty acids production in E. coli. Nat Commun 4:1–8. doi: 10.1038/ncomms2425. [DOI] [PubMed] [Google Scholar]
- 191.Xu P, Vansiri A, Bhan N, Koffas MA. 2012. ePathBrick: a synthetic biology platform for engineering metabolic pathways in E. coli. ACS Synthet Biol 1:256–266. doi: 10.1021/sb300016b. [DOI] [PubMed] [Google Scholar]
- 192.Malla S, Koffas MA, Kazlauskas RJ, Kim B-G. 2012. Production of 7-O-methyl aromadendrin, a medicinally valuable flavonoid, in Escherichia coli. Appl Environ Microbiol 78:684–694. doi: 10.1128/AEM.06274-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Fowler ZL, Gikandi WW, Koffas MA. 2009. Increased malonyl coenzyme A biosynthesis by tuning the Escherichia coli metabolic network and its application to flavanone production. Appl Environ Microbiol 75:5831–5839. doi: 10.1128/AEM.00270-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Luli GW, Strohl WR. 1990. Comparison of growth, acetate production, and acetate inhibition of Escherichia coli strains in batch and fed-batch fermentations. Appl Environ Microbiol 56:1004–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.El-Mansi E, Holms W. 1989. Control of carbon flux to acetate excretion during growth of Escherichia coli in batch and continuous cultures. J Gen Microbiol 135:2875–2883. [DOI] [PubMed] [Google Scholar]
- 196.Prüss BM, Wolfe AJ. 1994. Regulation of acetyl phosphate synthesis and degradation, and the control of flagellar expression in Escherichia coli. Mol Microbiol 12:973–984. doi: 10.1111/j.1365-2958.1994.tb01085.x. [DOI] [PubMed] [Google Scholar]
- 197.Wu J, Du G, Zhou J, Chen J. 2013. Metabolic engineering of Escherichia coli for (2S)-pinocembrin production from glucose by a modular metabolic strategy. Metab Eng 16:48–55. doi: 10.1016/j.ymben.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 198.Neidhardt FC, Bloch PL, Smith DF. 1974. Culture medium for enterobacteria. J Bacteriol 119:736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Keasling JD. 2008. Synthetic biology for synthetic chemistry. ACS Chem Biol 3:64–76. doi: 10.1021/cb7002434. [DOI] [PubMed] [Google Scholar]
- 200.Pósfai G, Plunkett G, Fehér T, Frisch D, Keil GM, Umenhoffer K, Kolisnychenko V, Stahl B, Sharma SS, De Arruda M. 2006. Emergent properties of reduced-genome Escherichia coli. Science 312:1044–1046. doi: 10.1126/science.1126439. [DOI] [PubMed] [Google Scholar]
- 201.Kolisnychenko V, Plunkett G, Herring CD, Fehér T, Pósfai J, Blattner FR, Pósfai G. 2002. Engineering a reduced Escherichia coli genome. Genome Res 12:640–647. doi: 10.1101/gr.217202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Sharma SS, Blattner FR, Harcum SW. 2007. Recombinant protein production in an Escherichia coli reduced genome strain. Metab Eng 9:133–141. doi: 10.1016/j.ymben.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Sharma SS, Campbell JW, Frisch D, Blattner FR, Harcum SW. 2007. Expression of two recombinant chloramphenicol acetyltransferase variants in highly reduced genome Escherichia coli strains. Biotechnol Bioeng 98:1056–1070. doi: 10.1002/bit.21491. [DOI] [PubMed] [Google Scholar]
- 204.Keasling JD. 2012. Synthetic biology and the development of tools for metabolic engineering. Metab Eng 14:189–195. doi: 10.1016/j.ymben.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 205.Furuya T, Kino K. 2009. Discovery of 2-naphthoic acid monooxygenases by genome mining and their use as biocatalysts. Chemsuschem 2:645–649. doi: 10.1002/cssc.200900054. [DOI] [PubMed] [Google Scholar]
- 206.McNulty DE, Claffee BA, Huddleston MJ, Kane JF. 2003. Mistranslational errors associated with the rare arginine codon CGG in Escherichia coli. Protein Expr Purif 27:365–374. doi: 10.1016/S1046-5928(02)00610-1. [DOI] [PubMed] [Google Scholar]
- 207.Valdivia E, Isaksson LA. 2004. A codon window in mRNA downstream of the initiation codon where NGG codons give strongly reduced gene expression in Escherichia coli. Nucleic Acids Res 32:5198–5205. doi: 10.1093/nar/gkh857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Stenström CM, Jin H, Major LL, Tate WP, Isaksson LA. 2001. Codon bias at the 3′-side of the initiation codon is correlated with translation initiation efficiency in Escherichia coli. Gene 263:273–284. doi: 10.1016/S0378-1119(00)00550-3. [DOI] [PubMed] [Google Scholar]
- 209.Stenström CM, Isaksson LA. 2002. Influences on translation initiation and early elongation by the messenger RNA region flanking the initiation codon at the 3′ side. Gene 288:1–8. doi: 10.1016/S0378-1119(02)00501-2. [DOI] [PubMed] [Google Scholar]