Abstract
BACKGROUND
Metabolomics, the systematic analysis of low molecular weight biochemical compounds in a biological specimen, has been increasingly applied to biomarker discovery.
CONTENT
Because no single analytical method can accommodate the chemical diversity of the entire metabolome, various methods such as nuclear magnetic resonance spectroscopy (NMR) and mass spectrometry (MS) have been employed, with the latter coupled to an array of separation techniques including gas and liquid chromatography. Whereas NMR can provide structural information and absolute quantification for select metabolites without the use of exogenous standards, MS tends to have much higher analytical sensitivity, enabling broader surveys of the metabolome. Both NMR and MS can be used to characterize metabolite data either in a targeted manner or in a nontargeted, pattern-recognition manner. In addition to technical considerations, careful sample selection and study design are important to minimize potential confounding influences on the metabolome, including diet, medications, and comorbitidies. To this end, metabolite profiling has been applied to human biomarker discovery in small-scale interventions, in which individuals are extremely well phenotyped and able to serve as their own biological controls, as well as in larger epidemiological cohorts. Understanding how metabolites relate to each other and to established risk markers for diseases such as diabetes and renal failure will be important in evaluating the potential value of these metabolites as clinically useful biomarkers.
SUMMARY
Applied to both experimental and epidemiological study designs, metabolite profiling has begun to highlight the breadth metabolic disturbances that accompany human disease. Experimental work in model systems and integration with other functional genomic approaches will be required to establish a causal link between select biomarkers and disease pathogenesis.
Although the number of potential cardiovascular biomarkers continues to grow, many provide only limited improvement over established metrics because they participate in pathways that are already known to be associated with cardiovascular disease (e.g., inflammation, thrombosis/hemostasis, and cholesterol transport). Therefore, much interest in biomarker research has been directed toward the application of unbiased methods to disease phenotyping. Metabolomics, or metabolite profiling, refers to the systematic analysis of metabolites—i.e., low molecular weight biochemicals including sugars, amino acids, organic acids, nucleotides, and lipids—in a biological specimen (1–3). Downstream of transcriptional, translational, and posttranslational processes (Fig. 1), metabolites serve as the most proximal reporters of alterations in the body in response to a disease process. Furthermore, with an estimated 3000–5000 detectable serum metabolites, the human metabolome is potentially more tractable for interrogation than are other, more informationally complex “omic” data sets (4).
Fig. 1. The conceptual relationship of the genome, transcriptome, proteome, and metabolome.
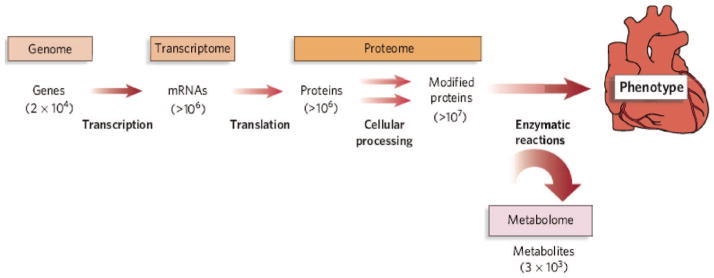
Informational complexity increases from genome to transcriptome to proteome. The estimated number of entities of each type of molecule in a typical cell is indicated in parentheses. Reproduced with permission from (36).
Metabolomics has been increasingly applied to biomarker discovery, and the results have demonstrated both feasibility and flexibility across physiological, interventional, and epidemiological human studies. Ongoing advances in analytical chemistry and computing power will no doubt lead to continued improvement of the breadth and throughput of such discovery efforts. In addition to technical considerations, however, the successful application of metabolite profiling to biomarker research requires equal attention to issues of sample selection and study design. For example, initial work demonstrating the broad metabolomic sequelae of diabetes and renal failure, as well as the intercorrelation of metabolites, has highlighted the potential for confounding in metabolomic investigations of cardiovascular disease. Conversely, by highlighting select metabolic perturbations across different clinical contexts, such studies may also reveal shared pathways to disease. This review provides an overview of metabolomics applied to biomarker discovery, with an emphasis on the insights gained and lessons learned from recent human studies.
Metabolomics Technologies
Endogenous metabolites span a variety of compound classes, with significant differences in size and polarity, across a wide range of concentrations. As a consequence, no single analytical method is able to accommodate the chemical diversity of the entire metabolome. Although various methodologies have been employed, 2 core technologies have prevailed as the workhorses of metabolite profiling: nuclear magnetic resonance spectroscopy (NMR)4 and mass spectrometry (MS), with the latter coupled to an array of separation techniques including gas chromatography (GC) and liquid chromatography (LC). Although capable of providing complementary and overlapping coverage of the metabolome, each of these methods has important differences in relative strengths and weaknesses (Table1) (5).
Table 1.
Metabolomics technologies.
| Chromatography | None | GC | • Ideal for volatile nonpolar analytes | |
| LC | • Ideal for ionizable analytes in solution | |||
| Other | • Direct infusion (no chromatography) | |||
| • Capillary electrophoresis | ||||
| • Thin-layer chromatography | ||||
| ↓ | ↓ | |||
| MS | ||||
| Analytical method | NMR | TOF | Triple quadrupole | Ion trap |
| Metabolite identification | Chemical shift | m/z (Flight time) | m/z (Filters for m/z) | m/z (Trapping frequency) |
| Advantages | • Robust | • Mass accuracy | • Sensitivity | • Sensitivity |
| • No sample destruction | • Wide mass range | • Can perform tandem MS | • Can perform tandem MS | |
| • No chromatography or analyte ionization | • Dynamic range | |||
| • Unambiguous identification of abundant analytes | ||||
| Disadvantages | • Limited sensitivity | • Limited dynamic range | • Poor mass accuracy | • Limited dynamic range |
| • Isotopic standards required for absolute quantitation | • Isotopic standards required for absolute quantitation | • Isotopic standards required for absolute quantitation | ||
NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY
NMR uses the magnetic properties of select atomic nuclei (e.g., 1H, 13C, or 31P) to determine the structure and abundance of metabolites in a biological specimen. In a strong magnetic field, a given NMR-active nucleus absorbs electromagnetic radiation at a characteristic frequency. Because this NMR signal is influenced in an identifiable way by the nature of neighboring atoms, chemical shifts in its resonance frequency can be used to assign local molecular structure. By measuring of all frequencies, metabolite identity can be determined, although this is not feasible for all compounds in a complex mixture. Stronger magnetic fields permit higher information content and increased analytical sensitivity, but at increased cost. NMR can be applied to in vivo tissues or to biological fluids obtained from humans or cells in culture. In addition to its usefulness for structure elucidation, this technology is advantageous because it is nondestructive and requires relatively little sample preparation, does not require chromatographic separation or ionization of analytes, and is able to provide information on flux through metabolic pathways. Unlike MS, NMR can be used to perform absolute quantification without the use of isotope-labeled standards. However, because of the combination of relatively low analytical sensitivity and high data complexity, unambiguous identification and quantification by use of NMR is limited to abundant metabolites—typically <100 analytes in human biofluids.
MASS SPECTROMETRY
MS tends to have much higher analytical sensitivity than NMR, permitting the measurement of hundreds to thousands of metabolite peaks. Although experimental samples can be infused directly into the mass spectrometer, most platforms employ upfront GC or LC to separate analytes over a fixed period of time. In GC, the sample is vaporized and undergoes chromatography in the gas phase; this method is particularly suited for volatile, nonpolar metabolites. LC is best suited for nonvolatile metabolites in solution, and various column chemistries are available to facilitate separation of different analyte classes (e.g., polar vs non-polar). After chromatography, metabolites enter the mass spectrometer and undergo ionization. As the name implies, MS resolves metabolites on the basis of mass, or more precisely, the mass-to-charge ratio (m/z) of their respective ions. Several distinct methods exist to separate ions according to their m/z.
TOF-MS
TOF mass spectrometers apply a fixed electric field to accelerate ions and then measure the time they take to reach the detector: lighter ions will arrive at the detector first. Important advantages of TOF instruments include excellent mass accuracy and wide m/z range, although the ability to scan relatively higher m/z molecules is more important for proteomic applications than for metabolite profiling.
Quadrupole MS
Quadrupole mass spectrometers apply oscillating electrical fields between 4 parallel rods to selectively stabilize the flight path of ions of select m/z, thus serving as a mass-selective filter. The potentials across the rods can be adjusted to sweep across a range of m/z values, or to settle only on select ion masses of interest. Although the mass accuracy and m/z range of quadrupole-MS instruments are generally inferior to those of TOF instruments, the linear dynamic range for relative quantification is generally better with quadrupole instruments. Furthermore, in triple-quadrupole MS, the first quadrupole can act as a mass filter for select “precursor” ions, the second quadrupole can be used as a collision cell to allow collision-induced dissociation of these precursor ions, and the third quadrupole can either allow all fragments to pass to the detector or serve as a mass filter for a single product fragment. Operated in the latter mode, triple-quadrupole instruments permit tandem MS spectrometry, with subsequent improvements in both the analytical sensitivity and specificity for preselected metabolites of interest.
Ion trap MS
Ion trap mass spectrometers, which can have different structural configurations, apply an electrical field to accumulate and hold ions of a specific m/z. Trapped metabolites can then be ejected from the ion trap, or fragmented in the ion trap for tandem MS analysis; m/z is determined on the basis of the radio frequency potential required to retain ions in the trap. By accumulating metabolites of interest, ion traps maximize analytical sensitivity, but provide less robust quantification across a narrower dynamic range.
Instruments that combine mass analyzers are available, e.g., quadrupole-TOF and ion-trap TOF instruments, in which the upstream module can be used to select and fragment ions of interest, and the downstream TOF component provides high-resolution mass spectra. Furthermore, alternative methodologies for m/z discrimination are available, including Sector mass spectrometers and Fourier transform ion cyclotron resonance mass spectrometers. To date, these instruments have been applied less frequently for human biomarker discovery.
TARGETED VS PATTERN-RECOGNITION (NONTARGETED) ANALYSES
Both NMR and MS can be used to characterize metabolite data either in a targeted manner, or in a nontargeted, pattern-recognition manner (6–7). As previously noted, because of limited analytical sensitivity and data complexity NMR can assign definitive metabolite identities to only a subset of peaks arising from a biological specimen. Similarly, although MS can generate hundreds, and in some cases thousands, of metabolite peaks from human biofluids, chromatographic elution time and m/z are often insufficient to confidently assign peak identities. For targeted approaches, the user focuses on a predefined set of metabolites of known identity—typically several dozen to hundreds—for detection and quantification. Tandem MS has proven to be an invaluable tool for such approaches: e.g., using a triple-quadrupole mass spectrometer, the investigator can selectively monitor for specific precursor/product ion combinations corresponding to known metabolites, as confirmed empirically by collision-induced–dissociation analyses of commercial standards. For pattern-recognition analyses, the user measures as many peaks as possible, although the underlying identities of the species giving rise to the peaks are not generally known. In these studies the investigators generally adopt a strategy of identifying qualitative differences in peak profiles between experimental groups.
Although the targeted approach generates a narrower view of the metabolome that is biased toward a predefined set of analytes, researchers have more confidence in the end results because they know what is giving rise to the signals. Mass spectrometers are more analytically sensitive when operated in a targeted fashion, acquiring data only for specific m/z. The nontargeted approach generates rich signatures, but provides little immediate insight into underlying biological processes and can lead to overfitting of data. Also, spurious results can occur if the identity of the peaks is unknown. For example, in one study pattern-recognition techniques were applied to 1H NMR spectra of human serum to aid in the noninvasive diagnosis of chronic coronary artery disease (8). In this study, however, the metabolites that generated the spectroscopy peaks were not unambiguously identified, and the findings ultimately proved to be confounded by the effects of statin therapy (9–10).
Metabolomics Research on Human Samples
Although samples derived across a spectrum of experimental models are amenable to metabolite profiling, human plasma or serum confers several important advantages for metabolomic cardiovascular biomarker discovery. As previously noted, no single analytical method can provide comprehensive coverage of the metabolome. Therefore, several aliquots of a given experimental sample may have to be distributed across different profiling methods to achieve broad analyte coverage. Even more sample is required for other laboratory tests, e.g., clinical measurements of lipids, troponin, and insulin, which may be important confounders or internal controls for a given experiment. Whereas sufficient plasma is readily obtained from human study participants, phlebotomy on laboratory mice typically yields a sample limited to a few hundred microliters and is associated with relatively greater hemodynamic and physiologic stress. Collection of larger volumes of blood from a mouse often requires that the animal be killed, introducing a dramatic perturbation that may confound investigation of various cardiovascular phenotypes, e.g., blood pressure and ischemia, and precluding longitudinal follow-up. By contrast, phlebotomy is expected to have little to no effect on the cardiovascular profile of humans, aside from the occasional vasovagal reaction.
In addition to practical considerations, there are important biological advantages to the use of human samples for metabolite profiling. Exogenous inputs make important contributions to the circulating metabolome and are difficult to duplicate across species. First, food intake impacts the metabolome, both directly with the absorption of dietary metabolites and indirectly as hormones like insulin act to modulate metabolic pathways (11). In some cases, discriminating metabolites between individuals with and without disease may reflect an imbalance in long-term dietary habits but would be obscured in animal models with homogeneous and/or artificial (from a human perspective) diets. Second, there is increasing awareness of how gut bacteria impact the metabolome by metabolizing and modifying both dietary inputs and constituents of the enterohepatic circulation (e.g., bile acids) (12). One study applied nontargeted LC-MS–based metabolite profiling to plasma obtained from genetically identical mice that did or did not (germ-free) have gut bacteria (13). A total of 197 metabolites were found to be unique to 1 of the 2 mouse populations, and many more had significant differences in abundance between the 2 groups. Because humans, mice, and other animals harbor fundamentally different gut flora, the study of human samples is the most direct method for circumventing this potential biological confounder.
Human Metabolomics Biomarker Studies
Circulating biomarkers that have been successfully incorporated into cardiology practice include diagnostic markers of acute changes, e.g., troponin I and troponin T for myocardial infarction and B-type natriuretic peptide for decompensated congestive heart failure, as well as prognostic markers of adverse cardiovascular outcomes, e.g., LDL cholesterol. The discovery of both kinds of biomarkers requires the ability to differentiate subtle clinical phenotypes, both acutely and over time, and to control for a wide range of confounding variables, including diet, medications, and comorbidities. Human metabolomic studies have begun to address these challenges, using both experimental and epidemiological study designs.
EXPERIMENTAL STUDIES
Because metabolomic studies are at risk for numerous confounders and because of the inherent unpredictability of the onset of pathological states, controlled human studies offer important advantages for metabolite biomarker discovery. Clinical cardiology is uniquely suited for such investigation, including experiments in which serial sampling of study participants before and after a controlled perturbation allows each individual to serve as his or her own biological control. In addition to attenuating noise attributable to interindividual variability, such studies allow more precise assessment of the kinetics, and even tissue specificity, of metabolite changes.
This type of strategy has been exploited in several cardiometablic diseases. In a human model of “planned myocardial infarction” (PMI), investigators profiled plasma drawn serially from 36 individuals with hypertrophic obstructive cardiomyopathy undergoing left-heart catheter-guided alcohol septal ablation (14). Alterations in several metabolites were detected as early as 10 min after PMI, and profiling of plasma obtained directly from the coronary sinus confirmed myocardial origin for select metabolites. In validation experiments, a PMI-derived plasma signature of aconitic acid, hypoxanthine, trimethylamine-N-oxide, and threonine differentiated with high diagnostic accuracy individuals presenting to the emergency room with spontaneous myocardial infarctions from controls undergoing diagnostic coronary angiography. More recently, to better understand the metabolomic response to nonpathologic cardiovascular exertion, we profiled plasma obtained from individuals undergoing exercise. Metabolite profiles of individuals running on a treadmill and riding a stationary bike demonstrated highly concordant changes, including plasma indicators of glyceogenolysis (glucose-6-phosphate), tricarboxylic acid cycle span 2 expansion (succinate, malate, and fumarate), and lipolysis (glycerol) (15). Among the 8 individuals who underwent bicycle ergometry, multisite blood sampling—superior vena cava, pulmonary artery, and radial artery—permitted instantaneous assessment of metabolite gradients in distinct vascular beds, confirming glycerol release from exercising tissue. Interestingly, plasma glycerol concentrations had a positive correlation with acute exercise fitness, a relation that held in a subsequent profiling experiment of 25 individuals who completed the 26.2-mile Boston marathon. Furthermore, plasma glycerol concentrations had a negative correlation with resting heart rate, an independent predictor of exercise capacity, in 302 individuals in the Framingham Heart Study (FHS).
Thus, human metabolomic studies can shed light on controlled, highly phenotyped perturbations, as well as generate hypotheses for more heterogeneous populations. This approach is certainly not restricted to research in cardiology. Metabolomic investigations of plasma obtained immediately before and after hemodialysis have identified novel markers of uremia (16–17). Investigators profiling plasma before and after oral glucose challenge have sought to identify novel markers of insulin resistance (18–19). Most recently, investigators have applied targeted LC-MS–based metabolite profiling to plasma before and after gastric bypass surgery (20–21). In a study of equivalent weight loss achieved either by gastric bypass surgery or dietary changes, only gastric bypass surgery was associated with decreases in branched-chain amino acids (BCAAs) and BCAA oxidation products (21). Notably, these authors previously identified a BCAA-related signature that differentiates obese vs lean humans and showed that BCAA may contribute to the development of obesity-associated insulin resistance (22). These findings raise the possibility that decreases in circulating BCAAs may contribute to the relatively greater improvements in glucose homeostasis associated with surgical vs diet-induced weight loss.
EPIDEMIOLOGICAL STUDIES
In addition to discrete perturbational studies, metabolite profiling in large, well-phenotyped human cohorts is an alternative strategy to address concerns about confounding. Epidemiological approaches are particularly suited for the study of chronic human diseases that are not amenable to acute physiological modeling, for which biomarkers are needed more for screening and determining prognosis than for diagnosis. Large, adequately powered studies are necessary because the predictive effects of new biomarkers may be smaller than those observed with classic risk factors and because multiple biomarkers are often studied concurrently. Ongoing improvements in metabolomics technologies now enable sufficient throughput to make such studies feasible. For example, present techniques coupling LC with a triple quadrupole mass spectrometer can acquire data on hundreds of analytes in a run time of 30 min per sample. At present, similar studies are not feasible with proteomic applications.
In an initial proof-of-principle study in 2 large population-based cohorts, we found that BCAA and aromatic amino acid concentrations had a significant association with future type 2 diabetes up to 12 years after initial plasma profiling. These findings were essentially unchanged after adjustment for established clinical risk factors (23). The initial discovery experiment used a nested, case-control design in the FHS with 189 individuals who ultimately developed type 2 diabetes and 189 control individuals who were matched for age, body mass index, and fasting glucose and did not develop type 2 diabetes. A validation experiment in 326 individuals from the Malmo Diet and Cancer Study employed the same matching scheme. Because the study design enriched the control population for high-risk features such as obesity and increased fasting glucose, we also performed metabolite profiling on 400 randomly selected controls from FHS. Although the association between metabolites and incident diabetes was significant across each experiment, the strength of association was attenuated in the case vs random cohort analysis. Whereas amino acid increases led to large improvements in model fit and discrimination (c statistics) between cases and matched controls, changes in these parameters were less significant between cases and random controls. These findings suggest that amino acid profiling might have greater value in high-risk individuals and underscore how study design modulates study outcome and interpretation; although metabolomics technologies are new, old lessons in epidemiology still apply.
Metabolite profiling in well-phenotyped clinical cohorts provides an opportunity to integrate metabolite data with a wide range of demographic and clinical data. Such efforts may illuminate novel risk associations, and they also demonstrate the diversity of potential confounders that mandate judicious interpretation of metabolomic data. For example, given the emerging associations between BCAA concentrations and obesity, insulin resistance, and incident diabetes, BCAA are likely to be associated with other cardiometabolic phenotypes as well. Indeed, in a metabolite-profiling study of more than 600 individuals undergoing cardiac catheterization, BCAA concentrations were independently associated with death or myocardial infarction during follow-up (24). The authors appropriately adjusted their statistical analyses for an initial imbalance in diabetes, but their findings may still represent a subtle manifestation of how insulin resistance modulates profiles of circulating amino acids. Because of the kidneys’ fundamental role in small-molecule handling, renal function is another important potential confounder of metabolite biomarker discovery. Dunn et al. applied a GC-MS–based platform to measure 272 metabolite peaks in plasma obtained from 52 individuals with heart failure and 57 age-matched controls, and identified pseudouridine as the most significant discriminator of disease status (25). However, pseudouridine was positively correlated with serum creatinine, and cases had significantly higher serum creatinine concentrations than controls, highlighting the potential confounding influence of renal function. Indeed, in a study of 41 individuals with chronic kidney disease, 52 of the 117 measured metabolites were either positively or negatively associated with estimated glomerular filtration rate (26). Future efforts to fully annotate the metabolome as a function of estimated glomerular filtration rate will be particularly important in cardiology research given the marked cardiovascular risk associated with even moderate renal impairment (27). Similar studies to cross-reference metabolite concentrations against other clinical variables such as age, sex, body mass index, and blood pressure will ultimately serve as an important resource for a range of metabolomic applications, including biomarker discovery.
Metabolite Profiling and Multimarker Approaches
The majority of current biomarkers for cardiovascular screening fall along pathways already known to be associated with cardiovascular disease, such as inflammation and cholesterol biosynthesis. Consequently, available biomarkers often provide information that is correlated with what is already known or being measured. Although correlated biomarkers can underscore the importance of a biological pathway, they may not provide a substantial increase in diagnostic or predictive value. This point has been highlighted by several studies. Pepe and Thompson performed simulations using 2 hypothetical cancer biomarkers (28). Assuming an area under the curve of 0.80 with 1 biomarker alone, they showed that the addition of a second biomarker raised the area under the curve to 0.88 if the 2 biomarkers were weakly correlated, but to only 0.83 if the 2 biomarkers were moderately correlated. This result translates into a diagnostic sensitivity of 80% with 2 weakly correlated biomarkers, compared with a diagnostic sensitivity of 70% with 2 moderately correlated biomarkers (assuming a false-positive rate of 20%). The implication is that an additional 10 individuals for every 100 people destined to develop disease would be identified with the use of less correlated biomarkers, a clinically meaningful difference.
Wang and colleagues recently explored factors that influence whether new biomarkers can add to diagnosis and risk prediction above and beyond established markers (29). They performed a simulation based on adding 1–100 hypothetical biomarkers to a traditional risk model for cardiovascular disease. Traditional risk models (including cholesterol concentrations, age, and hypertension) have an approximate area under the curve of 0.75 across populations. For the simulation performed by Wang et al., each of the hypothetical biomarkers had a similar magnitude of association with cardiovascular events similar to established cardiovascular biomarker such as C-reactive protein (CRP) or B-type natriuretic peptide. These investigators demonstrated that a key determinant of improvement in the c statistic is the degree of correlation between biomarkers. With a set of biomarkers that has a mean marker–marker correlation of r =0.4 (moderately correlated), >50 biomarkers are needed before the c statistic is increased by 0.05. By contrast, when the average marker–marker correlation is r =0.05 (weakly correlated), <10 biomarkers are needed to raise the c statistic by 0.05.
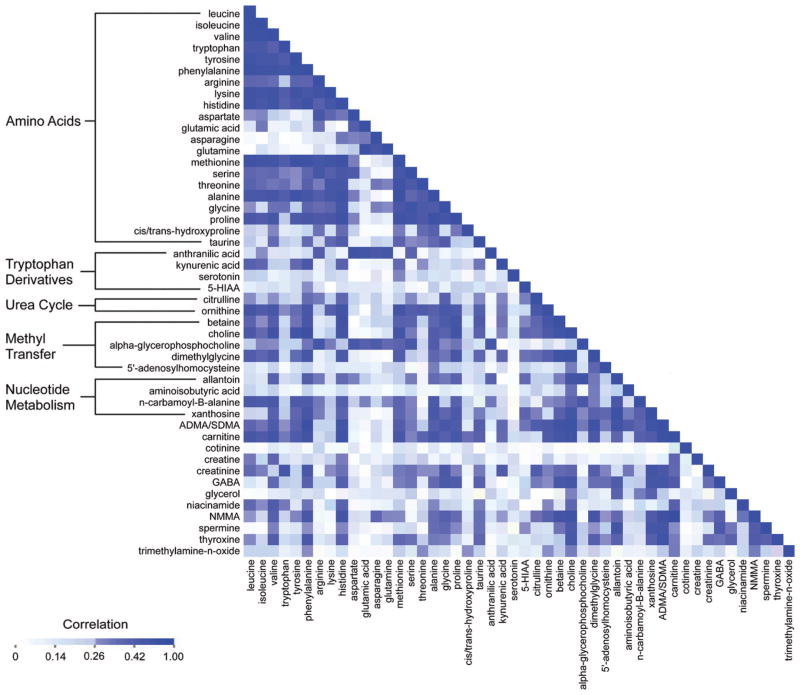
The intercorrelation for 48 plasma metabolite concentrations in 189 fasting individuals in the FHS is shown in Fig. 2. Mean correlations within groups of molecules were highest for urea cycle metabolites (r = 0.49), metabolites involved in nucleotide metabolism (r =0.38), amino acids (r =0.34), and methyl-transfer metabolites (r = 0.34). As more metabolites are measured in large cohorts, similar correlation clusters, both expected and unexpected, are likely to emerge. Understanding how metabolites relate with each other and with established risk markers will be important in assessing their value as potential biomarkers and may also lead to previously unappreciated connections between metabolic pathways.
Fig. 2. Correlation matrix for plasma metabolite concentrations in the FHS.
Age- and sex-adjusted Pearson correlation coefficients for metabolite concentrations in 189 individuals in the FHS. 5-HIAA, 5-hydroxyindoleacetic acid; ADMA/SDMA, asymmetric and symmetric dimethylarginine; GABA, gamma-aminobutyric acid; NMMA, N-monomethyl-arginine.
From Association to Causation
Several limitations to metabolomic biomarker discovery warrant mention. Although metabolite profiling of clinical plasma samples can generate metabolic “snapshots,” it does not provide information on pathway flux—for example, metabolite profiling cannot be used to determine whether a metabolite is increased in plasma because it is being produced in excess or because of a downstream block. Furthermore, the development of clinically useful biomarkers will require a more comprehensive understanding of how exogenous factors impact the metabolome, e.g., how diet and medications impact metabolite markers of interest, and what intra- and interindividual variability is normal for these metabolites. Finally, although adequately powered clinical studies enable investigators to incorporate strategies to minimize confounding, e.g., multivariable adjustment or population stratification by known risk factors, such studies cannot be used to establish a causal link between disease biomarkers and disease pathogenesis.
To this end, metabolomic findings derived in human studies can be further interrogated in model systems. Using nontargeted LC-MS–based metabolite profiling, Wang et al. profiled plasma from 75 individuals who experienced myocardial infarction, stroke, or death in the ensuing 3 years and 75 age-and sex-matched controls who did not (30). Of 18 analytes that were significantly different between cases and controls, 3 demonstrated significant correlations among one another, suggesting a potential common biochemical pathway. Using a variety of analytical methods, these investigators identified the metabolites as betaine, choline, and trimethylamine-N-oxide, all metabolites of dietary phosphatidylcholine. Dietary supplementation of choline was sufficient to promote atherosclerosis in mice, and suppression of intestinal bacteria responsible for the conversion of phosphatidylcholine to choline inhibited this atherogenesis. In addition to reinforcing the interaction between diet, gut bacteria, and the metabolome, this study demonstrates how metabolomic biomarker discovery can elucidate novel pathways to disease.
Integrating genomic and metabolomic data in humans is an alternative strategy to establish a causal link between metabolite biomarkers and disease. Analyses demonstrating that single-nucleotide polymorphisms that modulate plasma LDL cholesterol concentrations are independently associated with incident cardiovascular disease are consistent with the known causal role of LDL cholesterol in atherogenesis (31). By contrast, genetic loci associated with plasma CRP concentrations (including in the CRP locus) had no association with coronary heart disease, arguing against a causal association between CRP and cardiovascular disease (32). Recent work has begun to explore the genetic determinants of plasma metabolite levels in large human cohorts (33–35). As ongoing studies further delineate the genetic determinants of plasma metabolite profiles, efforts to triangulate gene–metabolite–disease associations will provide insight into if and how metabolite markers contribute to disease pathogenesis.
Summary
Emerging technologies now permit higher resolution phenotyping of biological specimens. However, whereas robust technologies capable of genomic and transcriptomic profiling are now well established, no single analytical method provides comprehensive coverage of the human metabolome. Thus, investigators have employed various technologies, including NMR and MS (coupled to upfront LC or GC), and used alternative analytical strategies, i.e., targeted vs pattern recognition. Metabolite profiling has demonstrated feasibility and flexibility for biomarker discovery in both small, extremely well-phenotyped human interventions and larger, epidemiological human cohorts. Although both approaches seek to minimize confounding by uncontrolled clinical variables, further efforts to understand the diversity of inputs to the metabolome will be an important resource for future metabolomic biomarker studies. In parallel, investigation in model systems and integration with other functional genomic approaches in humans will provide insight into the pathophysiologic interactions between metabolite markers and disease.
Footnotes
Nonstandard abbreviations: NMR, nuclear magnetic resonance spectroscopy; MS, mass spectrometry; GC, gas chromatography; LC, liquid chromatography; PMI, planned myocardial infarction; FHS, Framingham Heart Study; BCAA, branched-chain amino acids; CRP, C-reactive protein.
Author Contributions: All authors confirmed they have contributed to the intellectual content of this paper and have met the following 3 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; and (c) final approval of the published article.
Authors’ Disclosures or Potential Conflicts of Interest: No authors declared any potential conflicts of interest.
References
- 1.Lindon JC, Holmes E, Bollard ME, Stanley EG, Nicholson JK. Metabonomics technologies and their applications in physiological monitoring, drug safety assessment and disease diagnosis. Biomarkers. 2004;9:1–31. doi: 10.1080/13547500410001668379. [DOI] [PubMed] [Google Scholar]
- 2.Dunn WB, Bailey NJ, Johnson HE. Measuring the metabolome: current analytical technologies. Analyst. 2005;130:606–25. doi: 10.1039/b418288j. [DOI] [PubMed] [Google Scholar]
- 3.Mayr M. Metabolomics: ready for the prime time? Circ Cardiovasc Genet. 2008;1:58–65. doi: 10.1161/CIRCGENETICS.108.808329. [DOI] [PubMed] [Google Scholar]
- 4.Psychogios N, Hau DD, Peng J, Guo AC, Mandal R, Bouatra S, et al. The human serum metabolome. PLoS One. 2011;6:e16957. doi: 10.1371/journal.pone.0016957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griffiths WJ. Metabolomics, metabonomics and metabolite profiling. Cambridge: RSC Publishing; 2008. [Google Scholar]
- 6.Lewis GD, Asnani A, Gerszten RE. Application of metabolomics to cardiovascular biomarker and pathway discovery. J Am Coll Cardiol. 2008;52:117–23. doi: 10.1016/j.jacc.2008.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bain JR, Stevens RD, Wenner BR, Ilkayeva O, Muoio DM, Newgard CB. Metabolomics applied to diabetes research: moving from information to knowledge. Diabetes. 2009;58:2429–43. doi: 10.2337/db09-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brindle JT, Antti H, Holmes E, Tranter G, Nicholson JK, Bethell HW, et al. Rapid and noninvasive diagnosis of the presence and severity of coronary heart disease using 1H-NMR-based metabonomics. Nat Med. 2002;8:1439–44. doi: 10.1038/nm1202-802. [DOI] [PubMed] [Google Scholar]
- 9.Nielsen E-MD, Hansen L, Carstensen B, Echwald SM, Drivsholm T, Glumer C, et al. The E23K variant of Kir6. 2 associates with impaired post-OGTT serum insulin response and increased risk of type 2 diabetes. Diabetes. 2003;52:573–7. doi: 10.2337/diabetes.52.2.573. [DOI] [PubMed] [Google Scholar]
- 10.Kirschenlohr HL, Griffin JL, Clarke SC, Rhydwen R, Grace AA, Schofield PM, et al. Proton NMR analysis of plasma is a weak predictor of coronary artery disease. Nat Med. 2006;12:705–10. doi: 10.1038/nm1432. [DOI] [PubMed] [Google Scholar]
- 11.O’Sullivan A, Gibney MJ, Brennan L. Dietary intake patterns are reflected in metabolomic profiles: potential role in dietary assessment studies. Am J Clin Nutr. 2011;93:314–21. doi: 10.3945/ajcn.110.000950. [DOI] [PubMed] [Google Scholar]
- 12.Nicholson JK, Holmes E, Wilson ID. Gut microorganisms, mammalian metabolism and personalized health care. Nat Rev Microbiol. 2005;3:431–8. doi: 10.1038/nrmicro1152. [DOI] [PubMed] [Google Scholar]
- 13.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106:3698–703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis GD, Wei R, Liu E, Yang E, Shi X, Martinovic M, et al. Metabolite profiling of blood from individuals undergoing planned myocardial infarction reveals early markers of myocardial injury. J Clin Invest. 2008;118:3503–12. doi: 10.1172/JCI35111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis GD, Farrell L, Wood MJ, Martinovic M, Arany Z, Rowe GC, et al. Metabolic signatures of exercise in human plasma. Sci Transl Med. 2010;2:33ra7. doi: 10.1126/scitranslmed.3001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rhee EP, Souza A, Farrell L, Pollak MR, Lewis GD, Steele DJ, et al. Metabolite profiling identifies markers of uremia. J Am Soc Nephrol. 2010;21:1041–51. doi: 10.1681/ASN.2009111132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato E, Kohno M, Yamamoto M, Fujisawa T, Fujiwara K, Tanaka N. Metabolomic analysis of human plasma from haemodialysis patients. Eur J Clin Invest. 2011;41:241–55. doi: 10.1111/j.1365-2362.2010.02398.x. [DOI] [PubMed] [Google Scholar]
- 18.Shaham O, Wei R, Wang TJ, Ricciardi C, Lewis GD, Vasan RS, et al. Metabolic profiling of the human response to a glucose challenge reveals distinct axes of insulin sensitivity. Mol Syst Biol. 2008;4:214. doi: 10.1038/msb.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao X, Peter A, Fritsche J, Elcnerova M, Fritsche A, Haring HU, et al. Changes of the plasma metabolome during an oral glucose tolerance test: is there more than glucose to look at? Am J Physiol Endocrinol Metab. 2009;296:E384–93. doi: 10.1152/ajpendo.90748.2008. [DOI] [PubMed] [Google Scholar]
- 20.Mutch DM, Fuhrmann JC, Rein D, Wiemer JC, Bouillot JL, Poitou C, et al. Metabolite profiling identifies candidate markers reflecting the clinical adaptations associated with Roux-en-Y gastric bypass surgery. PLoS One. 2009;4:e7905. doi: 10.1371/journal.pone.0007905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laferrere B, Reilly D, Arias S, Swerdlow N, Gorroochurn P, Bawa B, et al. Differential metabolic impact of gastric bypass surgery versus dietary intervention in obese diabetic subjects despite identical weight loss. Sci Transl Med. 2011;3:80re2. doi: 10.1126/scitranslmed.3002043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9:311–26. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–53. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shah SH, Bain JR, Muehlbauer MJ, Stevens RD, Crosslin DR, Haynes C, et al. Association of a peripheral blood metabolic profile with coronary artery disease and risk of subsequent cardiovascular events. Circ Cardiovasc Genet. 2010;3:207–14. doi: 10.1161/CIRCGENETICS.109.852814. [DOI] [PubMed] [Google Scholar]
- 25.Dunn WB, Deepak SM, Buch MH, McDowell G, Spasic G, Ellis DI, et al. Serum metabolomics reveals many novel metabolic markers of heart failure, including pseudouridine and 2-oxoglutarate. Metabolomics. 2007;3:413–26. [Google Scholar]
- 26.Toyohara T, Akiyama Y, Suzuki T, Takeuchi Y, Mishima E, Tanemoto M, et al. Metabolomic profiling of uremic solutes in CKD patients. Hypertens Res. 2010;33:944–52. doi: 10.1038/hr.2010.113. [DOI] [PubMed] [Google Scholar]
- 27.Go AS, Chertow GM, Fan D, McCulloch CE, Hsu CY. Chronic kidney disease and the risks of death, cardiovascular events, and hospitalization. N Engl J Med. 2004;351:1296–305. doi: 10.1056/NEJMoa041031. [DOI] [PubMed] [Google Scholar]
- 28.Pepe MS, Thompson ML. Combining diagnostic test results to increase accuracy. Biostatistics. 2000;1:123–40. doi: 10.1093/biostatistics/1.2.123. [DOI] [PubMed] [Google Scholar]
- 29.Wang TJ. Assessing the role of circulating, genetic, and imaging biomarkers in cardiovascular risk prediction. Circulation. 2011;123:551–65. doi: 10.1161/CIRCULATIONAHA.109.912568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kathiresan S, Melander O, Anevski D, Guiducci C, Burtt NP, Roos C, et al. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med. 2008;358:1240–9. doi: 10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
- 32.Elliott P, Chambers JC, Zhang W, Clarke R, Hopewell JC, Peden JF, et al. Genetic Loci associated with C-reactive protein levels and risk of coronary heart disease. JAMA. 2009;302:37–48. doi: 10.1001/jama.2009.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gieger C, Geistlinger L, Altmaier E, Hrabe de Angelis M, Kronenberg F, Meitinger T, et al. Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet. 2008;4:e1000282. doi: 10.1371/journal.pgen.1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Illig T, Gieger C, Zhai G, Romisch-Margl W, Wang-Sattler R, Prehn C, et al. A genome-wide perspective of genetic variation in human metabolism. Nat Genet. 2010;42:137–41. doi: 10.1038/ng.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suhre K, Shin SY, Petersen AK, Mohney RP, Meredith D, Wagele B, et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477:54–60. doi: 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gerszten RE, Wang TJ. The search for new cardiovascular biomarkers. Nature. 2008;451:949–52. doi: 10.1038/nature06802. [DOI] [PubMed] [Google Scholar]