Abstract
Numerous reports describe phenomena of transgene silencing in plants, yet the underlying genetic and molecular mechanisms are poorly understood. We observed that regeneration of Arabidopsis thaliana plants transgenic for the rolB gene of Agrobacterium rhizogenes results in a selection for transgene silencing. Transgene silencing could be monitored in this system by reversion of the visible RolB phenotype. We report a phenotypic, molecular, and genetic characterization of a meiotically reversible transgene silencing phenomenon observed in a rolB transgenic line. In this line, the rolB gene is expressed strongly and uniformly in seedlings, but in the course of further development, the rolB gene is silenced erratically at a frequency that depends on the dosage of rolB. The silenced state is mitotically stable, while complete resetting of rolB gene expression occurs in seedlings of the following generation. The silencing of rolB correlates with a dramatic reduction of steady-state rolB transcripts, while rolB nuclear run-off transcripts are only moderately reduced. Therefore, rolB gene silencing seems to act predominantly at the posttranscriptional level. The process of rolB gene silencing was found to be affected by two extragenic modifier loci that influence both the frequency and the timing of rolB gene silencing during plant development. These genetic data demonstrate a direct involvement of defined plant genes in this form of gene silencing.
Full text
PDF

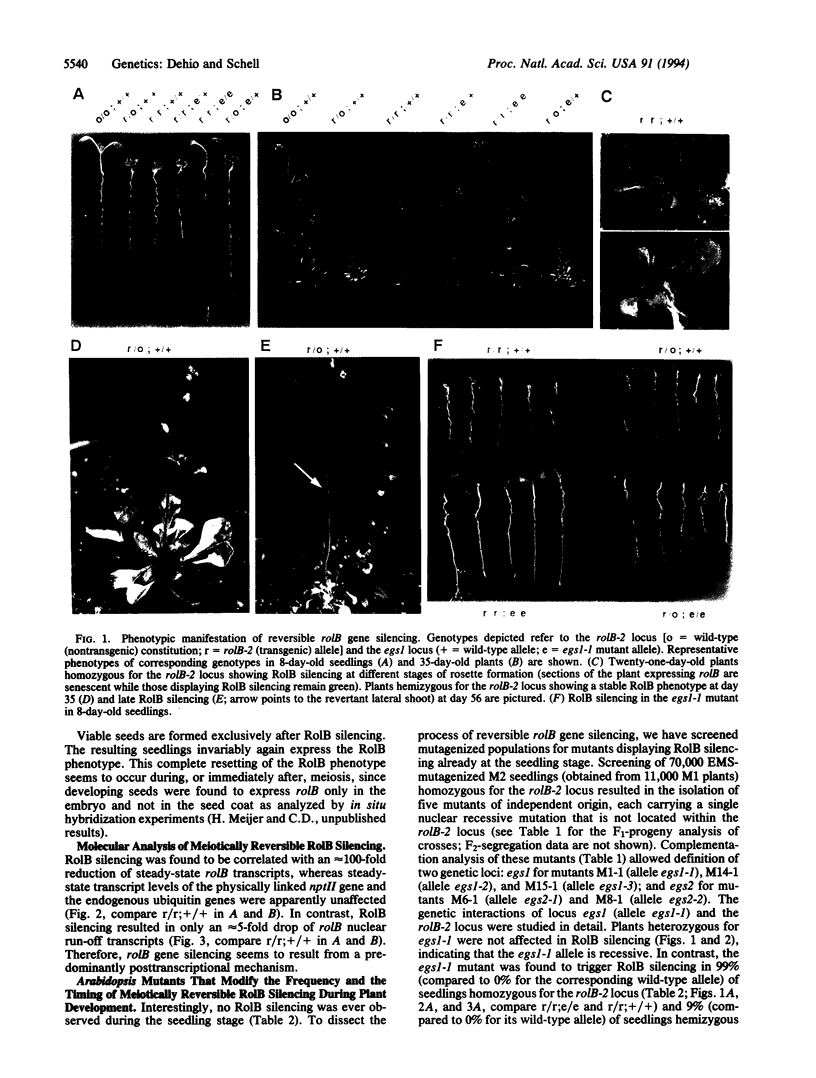


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Dehio C., Schell J. Stable expression of a single-copy rolA gene in transgenic Arabidopsis thaliana plants allows an exhaustive mutagenic analysis of the transgene-associated phenotype. Mol Gen Genet. 1993 Nov;241(3-4):359–366. doi: 10.1007/BF00284689. [DOI] [PubMed] [Google Scholar]
- Hart C. M., Fischer B., Neuhaus J. M., Meins F., Jr Regulated inactivation of homologous gene expression in transgenic Nicotiana sylvestris plants containing a defense-related tobacco chitinase gene. Mol Gen Genet. 1992 Nov;235(2-3):179–188. doi: 10.1007/BF00279359. [DOI] [PubMed] [Google Scholar]
- Linn F., Heidmann I., Saedler H., Meyer P. Epigenetic changes in the expression of the maize A1 gene in Petunia hybrida: role of numbers of integrated gene copies and state of methylation. Mol Gen Genet. 1990 Jul;222(2-3):329–336. doi: 10.1007/BF00633837. [DOI] [PubMed] [Google Scholar]
- Matzke M. A., Primig M., Trnovsky J., Matzke A. J. Reversible methylation and inactivation of marker genes in sequentially transformed tobacco plants. EMBO J. 1989 Mar;8(3):643–649. doi: 10.1002/j.1460-2075.1989.tb03421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meins F., Jr Habituation: heritable variation in the requirement of cultured plant cells for hormones. Annu Rev Genet. 1989;23:395–408. doi: 10.1146/annurev.ge.23.120189.002143. [DOI] [PubMed] [Google Scholar]
- Meyer P., Heidmann I., Niedenhof I. Differences in DNA-methylation are associated with a paramutation phenomenon in transgenic petunia. Plant J. 1993 Jul;4(1):89–100. doi: 10.1046/j.1365-313x.1993.04010089.x. [DOI] [PubMed] [Google Scholar]
- Meyerowitz E. M. Arabidopsis, a useful weed. Cell. 1989 Jan 27;56(2):263–269. doi: 10.1016/0092-8674(89)90900-8. [DOI] [PubMed] [Google Scholar]
- Napoli C., Lemieux C., Jorgensen R. Introduction of a Chimeric Chalcone Synthase Gene into Petunia Results in Reversible Co-Suppression of Homologous Genes in trans. Plant Cell. 1990 Apr;2(4):279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otten L., De Greve H., Hernalsteens J. P., Van Montagu M., Schieder O., Straub J., Schell J. Mendelian transmission of genes introduced into plants by the Ti plasmids of Agrobacterium tumefaciens. Mol Gen Genet. 1981;183(2):209–213. doi: 10.1007/BF00270619. [DOI] [PubMed] [Google Scholar]
- Pröls F., Meyer P. The methylation patterns of chromosomal integration regions influence gene activity of transferred DNA in Petunia hybrida. Plant J. 1992 Jul;2(4):465–475. doi: 10.1046/j.1365-313x.1992.t01-20-00999.x. [DOI] [PubMed] [Google Scholar]
- Schmülling T., Schell J., Spena A. Single genes from Agrobacterium rhizogenes influence plant development. EMBO J. 1988 Sep;7(9):2621–2629. doi: 10.1002/j.1460-2075.1988.tb03114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. J., Watson C. F., Bird C. R., Ray J., Schuch W., Grierson D. Expression of a truncated tomato polygalacturonase gene inhibits expression of the endogenous gene in transgenic plants. Mol Gen Genet. 1990 Dec;224(3):477–481. doi: 10.1007/BF00262443. [DOI] [PubMed] [Google Scholar]
- Spena A., Schmülling T., Koncz C., Schell J. S. Independent and synergistic activity of rol A, B and C loci in stimulating abnormal growth in plants. EMBO J. 1987 Dec 20;6(13):3891–3899. doi: 10.1002/j.1460-2075.1987.tb02729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trezzini G. F., Horrichs A., Somssich I. E. Isolation of putative defense-related genes from Arabidopsis thaliana and expression in fungal elicitor-treated cells. Plant Mol Biol. 1993 Jan;21(2):385–389. doi: 10.1007/BF00019954. [DOI] [PubMed] [Google Scholar]
- de Carvalho F., Gheysen G., Kushnir S., Van Montagu M., Inzé D., Castresana C. Suppression of beta-1,3-glucanase transgene expression in homozygous plants. EMBO J. 1992 Jul;11(7):2595–2602. doi: 10.1002/j.1460-2075.1992.tb05324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]






