Significance
Maintenance and reprogramming of pluripotency are among the most important issues in stem cell biology and regenerative medicine. Pluripotency is governed by several key transcription factors regulating transcription of other factors. Among these, regulation of OCT4 transcription by NANOG (from Irish myth-ology Tír na nÓg) is a critical interaction. We present here the crystal structure of human NANOG homeodomain in complex with the OCT4 promoter DNA and, through a series of ration-ally designed mutations, we identify key functional residues in the protein–DNA interaction, protein stability, and maintenance of mouse ESC self-renewal. Furthermore, we describe a mutation, NANOG L122A, which enhances DNA binding affinity, protein stability, mouse ESC self-renewal, and reprogramming into ground state from primed state pluripotency.
Keywords: NANOG, crystal structure, pluripotent stem cells, DNA-binding, reprogramming
Abstract
NANOG (from Irish mythology Tír na nÓg) transcription factor plays a central role in maintaining pluripotency, cooperating with OCT4 (also known as POU5F1 or OCT3/4), SOX2, and other pluripotency factors. Although the physiological roles of the NANOG protein have been extensively explored, biochemical and biophysical properties in relation to its structural analysis are poorly understood. Here we determined the crystal structure of the human NANOG homeodomain (hNANOG HD) bound to an OCT4 promoter DNA, which revealed amino acid residues involved in DNA recognition that are likely to be functionally important. We generated a series of hNANOG HD alanine substitution mutants based on the protein–DNA interaction and evolutionary conservation and determined their biological activities. Some mutant proteins were less stable, resulting in loss or decreased affinity for DNA binding. Overexpression of the orthologous mouse NANOG (mNANOG) mutants failed to maintain self-renewal of mouse embryonic stem cells without leukemia inhibitory factor. These results suggest that these residues are critical for NANOG transcriptional activity. Interestingly, one mutant, hNANOG L122A, conversely enhanced protein stability and DNA-binding affinity. The mNANOG L122A, when overexpressed in mouse embryonic stem cells, maintained their expression of self-renewal markers even when retinoic acid was added to forcibly drive differentiation. When overexpressed in epiblast stem cells or human induced pluripotent stem cells, the L122A mutants enhanced reprogramming into ground-state pluripotency. These findings demonstrate that structural and biophysical information on key transcriptional factors provides insights into the manipulation of stem cell behaviors and a framework for rational protein engineering.
NANOG (from Irish mythology Tír na nÓg, Land of Eternal Youth) is a key transcription factor regulating pluripotency in mammalian early embryos and pluripotent stem cells. Cooperating with other master regulators of pluripotency, NANOG plays a central role in pluripotency (1–3) and forms autoregulatory loops to maintain ES cell (ESC) identity (4–7). NANOG was initially identified from its ability to confer mouse (m)ESC self-renewal without dependence on leukemia inhibitory factor (LIF) when overexpressed in mESCs (8, 9). Disruption of the NANOG gene in mESCs compromises their pluripotency (8); however, mESCs can maintain their self-renewal without NANOG (10). NANOG expression marks fully reprogrammed induced pluripotent stem cells (iPSCs) during reprogramming mammalian somatic cells into a pluripotent state (11); however, NANOG is dispensable for generating iPSCs from somatic cells both exogenously (12) and endogenously (13, 14). In contrast, NANOG robustly promotes reprogramming of epiblast stem cells (EpiSCs) (15, 16) or human iPSCs (HiPSCs), which have primed state pluripotency with distinct gene expression patterns and cell signaling dependence, into ground-state pluripotency (17–20).
NANOG is composed of 305 amino acids, including a central homeodomain (HD). HDs are ∼60 amino acid DNA-binding domains that are found in the highly conserved HOX genes (also known as homeotic genes), and in other transcription factors that are less evolutionarily conserved (21). The HD structure is composed of an unstructured N-terminal arm, a bundle of three α-helices (H1–H3), and loops joining the helices (22). The N-terminal arm sits in the DNA minor groove and helix H3 forms an extensive DNA contact interface in the major groove. The interactions of helix H3 are critical in determining the specificity to the core consensus sequence (23, 24). Although NANOG is highly diversified among vertebrates (e.g., full-length hNANOG has ∼60% sequence identity with full-length mNANOG), the NANOG HD shares high similarity among species (e.g., hNANOG HD has ∼84% sequence identity with mNANOG HD). In contrast, NANOG HD is distinct from other HD protein families and the amino acid similarity to the closely related NK-2 gene family is less than 50% (25). Although a previous study describing the structure of mNANOG HD provided hints into residues that may be involved in NANOG–DNA interactions (26), the exact mapping of NANOG–DNA interactions leading to unique functions of NANOG in self-renewal and reprogramming of PSCs remained unknown.
Here we present the crystal structure of hNANOG HD in complex with a 12-bp DNA element derived from the OCT4 promoter. Using a combination of biophysical and cell-based assays, we investigated the key structural features of NANOG interactions with a physiological promoter DNA. These experiments uncovered a functionally enhanced mutant, NANOG L122A, which showed higher affinity for OCT4 promoter DNA, greater stability of the purified complex, enhanced mESC self-renewal against forced differentiation by retinoic acid (RA), and enhanced reprogramming efficiency of EpiSCs or HiPSCs into ground-state pluripotency.
Results
Structure of the hNANOG HD–DNA Complex.
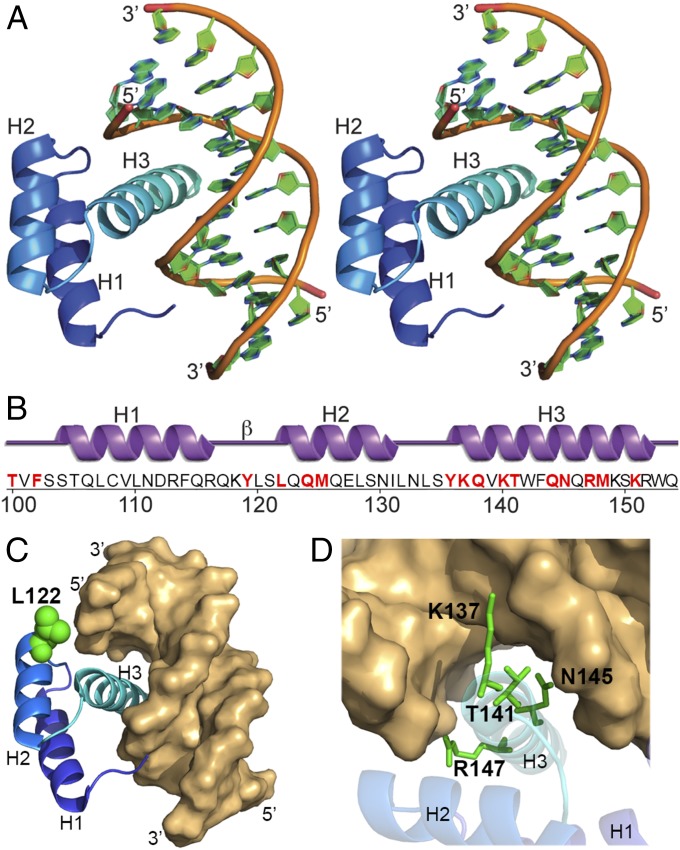
The crystal structure of the hNANOG HD–DNA complex was determined by molecular replacement to a resolution of 3.3 Å using the crystal structure of mNANOG HD (PDB ID code 2VI6) (26) as the search model. Data collection, model, and refinement statistics are summarized in Table S1. There are two hNANOG HD–DNA complexes in the crystallographic asymmetric unit. The final model (Fig. 1 A and B) includes residues 100–154 of the full-length protein in one hNANOG HD molecule and residues 100–153 in the second hNANOG HD molecule, as well as the 12 bp DNA. Overall, the structure matches the canonical HD-type fold consisting of three α-helices, with H1 and H2 being antiparallel. Helices H1 and H2 position the longest helix H3 in a manner that promotes its insertion into the major groove of the DNA (26). Helices H1 and H2 interact with the DNA minor/major groove, which is half of the duplex DNA, whereas H3 has some interactions with the DNA backbone and helix H1.
Fig. 1.
Crystal structure of hNANOG–OCT4 promoter DNA complex. (A) Stereo ribbon diagram of the hNANOG HD in complex with OCT4 promoter DNA. Helices H1, H2, and H3 are labeled. (B) Diagram showing the secondary structure elements of the hNANOG HD superimposed on its primary sequence. The α-helices (H1–H3) and β-turn (β) are indicated. The residues used for making alanine mutants are highlighted in red and boldface type. (C) The L122 residue in hNANOG HD that enhance interaction with OCT4 promoter DNA is shown in green. (D) Residues in hNANOG HD (K137, T141, N145, and R147) that are critical for interaction with OCT4 promoter DNA are shown in green.
Structural Comparisons and Protein–DNA Interactions.
The hNANOG and mNANOG HDs are almost identical; their structures can be superimposed with an RMSD of ∼0.7 Å over the entire HD, with a structure-based sequence identity of 87%. Comparison of the crystal structure of our hNANOG HD in complex with DNA with the crystal structure of the apo-mNANOG HD shows that DNA binding does not result in any significant conformational changes in NANOG HD. Both hNANOG HD and mNANOG HD are structurally similar to other HDs with and without bound DNA (26). For example, the RMSDs between hNANOG HD and octamer-binding transcription factor 1 or ventral nervous system defective HDs are 0.88 Å and 0.94 Å, respectively.
Interactions of hNANOG HD residues with OCT4 promoter DNA were analyzed using the NUCPLOT (27) (Fig. S1A) and PDBePISA (Proteins, Interfaces, Structures and Assemblies) servers (Fig. S1B). Because of the limited resolution of the crystal structure, only residues that directly interact with DNA were identified and not any water-mediated interactions. The interactions are essentially identical for both protein–DNA complexes in the asymmetric unit, including hydrogen bonds and nonbonded contacts with the backbone sugars and phosphate groups of the DNA. Aromatic residue interactions include F102, Y119, and Y136 that contact phosphates in the DNA backbone. Basic residues include K137, K140, R147, and K151, which form the majority of the contacts as might be expected for a protein–DNA complex. The other interacting residues include T100, L122, Q124, M125, Q138, T141, Q144, N145, and M148. The L122 residue is ∼4.5 Å from the DNA but has a favorable interfacial buried surface area (BSA) percentage based on PISA analysis (Fig. 1C and Fig. S1B). Featured residues for hNANOG HD interaction with the OCT4 promoter DNA are depicted in Fig. 1D and Fig. S1 C and D.
Mutational Analysis of hNANOG HD and Binding Affinity for OCT4 Promoter DNA.
To investigate the key features of hNANOG HD interactions with the OCT4 promoter DNA, we designed a series of single-point mutations to alanine based on the interfacial residues with DNA derived from the NUCPLOT and PDBePISA analysis (i.e., T100A, F102A, Y119A, L122A, Q124A, M125A, Y136A, K137A, Q138A, K140A, T141A, Q144A, N145A, R147A, M148A, and K151A). Although most of the hNANOG HD mutants could be expressed and purified, T100A, Y119A, or Q138A could not be expressed or purified.
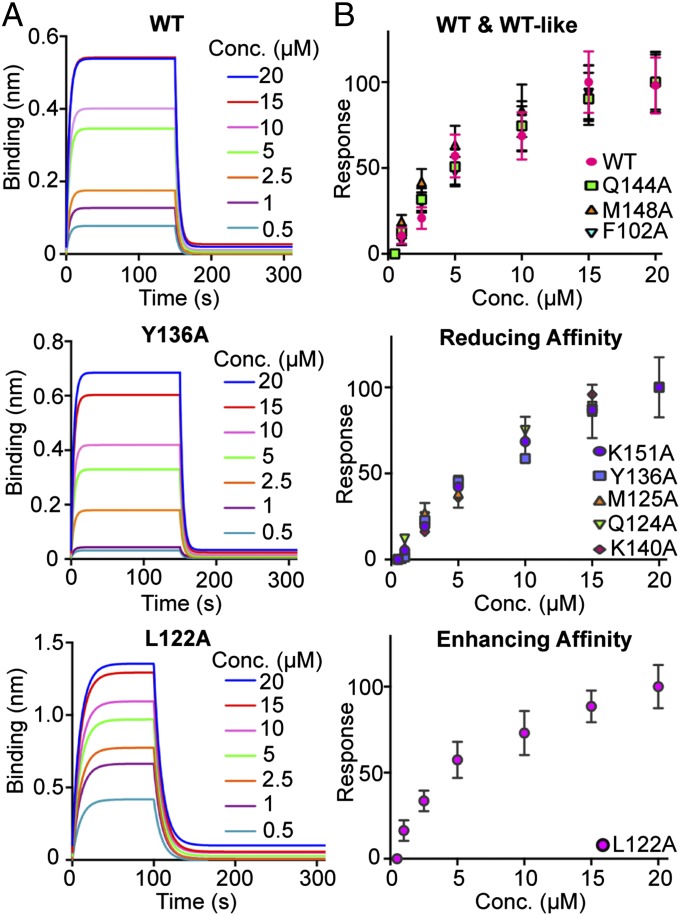
We examined the effect of the hNANOG HD mutations on binding affinity for OCT4 Promoter DNA by bio-layer interferometry (BLI) (Fig. 2, Fig. S2, and Table S2). The binding of WT hNANOG HD to the biotinylated OCT4 promoter DNA is specific with a KD ∼5.9 µM. This observed affinity is low compared with DNA binding affinities reported for other HDs determined by EMSA (28), but it is reasonable given that the hNANOG HD–DNA interaction was challenged by high salt concentration in the buffer to reduce nonspecific binding to a minimum (SI Materials and Methods). The L122A mutation in helix H2 unexpectedly increased hNANOG HD binding affinity (KD ∼1.4 µM) nearly fourfold. As expected, most mutations weakened association: Q124A, M125A, Y136A, K140A, and K151A decreased DNA binding and K137A, T141A, N145A, or R147A abrogated binding to the OCT4 promoter DNA. The mutations F102A, Q144A, and M148A had a minor effect on binding to OCT4 promoter DNA (i.e., WT-like behavior).
Fig. 2.
BLI analysis of hNANOG HD mutants binding to the OCT4 promoter DNA. (A) Dose–response curves of hNANOG WT, Y136A, and L122A as representatives of WT-like, or mutants that weaken or enhance DNA binding affinity, respectively. These curves show the binding response (expressed as nanometer shift) to the biotinylated 12-bp fragment of the DNA of a range of hNANOG HD concentrations (20 µM to 0.5 µM). (B) Steady-state analysis to determine the equilibrium dissociation constants (KD) of the protein–DNA interaction divided in three categories: WT-like, and mutants that weaken or enhance DNA binding affinity. The error bars represent three different binding cycles expressed as percent of the maximal response after sensor regeneration with 1 M MgCl2. The equilibrium dissociation constants (KD) and kinetic constant rates for all of the mutants are summarized in Fig. S2 and Table S2.
Protein Stability of NANOG HD Mutants in Purified Complex or in mESCs.
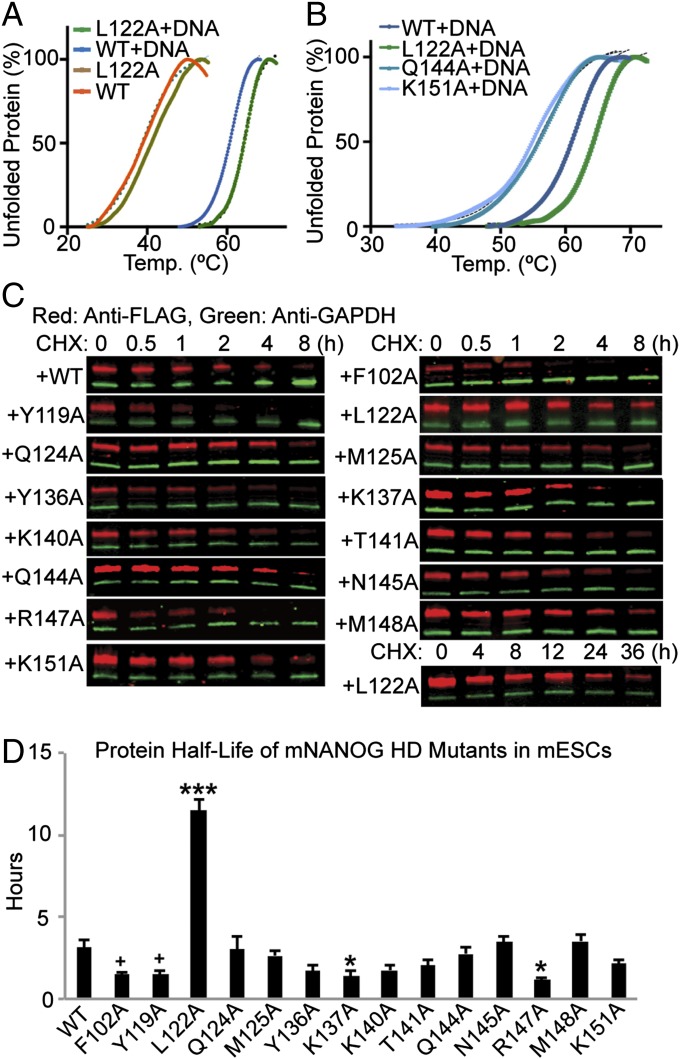
We investigated the thermal stability of recombinant hNANOG HD mutants by differential scanning fluorimetry (DSF) (Fig. 3 A and B and Table S2). Interestingly, the L122A mutant in complex with OCT4 promoter DNA was more stable by 3 °C compared with hNANOG WT, whereas Q144A and K151A caused a consistent reduction in melting temperature (Tm) by around 6 °C. DNA has a significant effect on protein stability. It is noteworthy that the addition of the DNA stabilizes the hNANOG WT by a 20 °C thermal shift. Consistent with the BLI experiments, no DSF melting curve could be obtained upon addition of the DNA for K137A, T141A, N145A, or R147A, which lose DNA binding specificity, suggesting that these mutations destabilize the hNANOG HD structure hampering DNA binding.
Fig. 3.
Protein stability of NANOG HD mutants in purified protein complex or in mESCs. (A) DSF analysis of the hNANOG HD WT and mutants alone or in complex with OCT4 promoter DNA. The addition of the DNA stabilizes hNANOG HD WT by a thermal shift of around 20 °C. (B) Melting curves of WT and hNANOG HD mutants in presence of the DNA, showing a 3 °C increase in stability for the L122A variant, and around 6 °C reduction in stability for the Q144A and K151A variants. Melting curves are presented as mean values of the percentage of unfolded protein in function of the temperature (°C); values in the table represent mean ± SEM for n = 3; n.d., not determined. Melting temperature (Tm) of all of the mutants are summarized in Table S2. (C) Protein expression of exogenous mNANOG WT or mutants fused with N-terminal 3×FLAG tag in mESCs treated with cycloheximide (CHX) detected by Western blotting. Green and red bands in each sample show the amount of GAPDH and exogenous mNANOG mutants, respectively. The half-life of GAPDH is more than 72 h. (D) Protein half-life of each mNANOG mutants in mESCs. n = 4, values are mean + SEM. P values of Dunnett’s post hoc test against WT were shown as follows: +P < 0.1, *P < 0.05, ***P < 0.001.
We also examined the lifetime of NANOG mutants in mammalian cells using translationally inhibited mESCs with cycloheximide. In these experiments, we used full-length mNANOG as a proxy for hNANOG for practical considerations of our experimental set-up. Although both full-length proteins are 305 residues, because of small insertions within the respective proteins, the residue numbering of the mNANOG differs slightly from the hNANOG at certain positions. However, we numbered the residues with respect to hNANOG HD (offset by 1 in the mNANOG HD). We calculated amounts of exogenous full-length mNANOG protein tagged with N-terminal 3×FLAG sequence from Western blotting analysis (Fig. 3 C and D and Table S2). The exogenous WT mNANOG protein half-life in mESCs was 3.1 ± 0.5 h, which is consistent with previous studies of human and mouse NANOG (either exogenous or endogenous) in ESCs (29–31). The half-life of some mNANOG mutants (e.g., F102A, Y119A, K137A, and R147A) was significantly shorter than WT. In contrast, the half-life of L122A, which showed stronger DNA binding, was 11.5 ± 0.7 h. These results suggested that the protein lifetime of mNANOG mutants in mESCs were similar to the thermal stability of purified hNANOG mutants, which was regulated by their DNA-binding strength.
Constitutive Expression of mNANOG Mutants in mESCs.
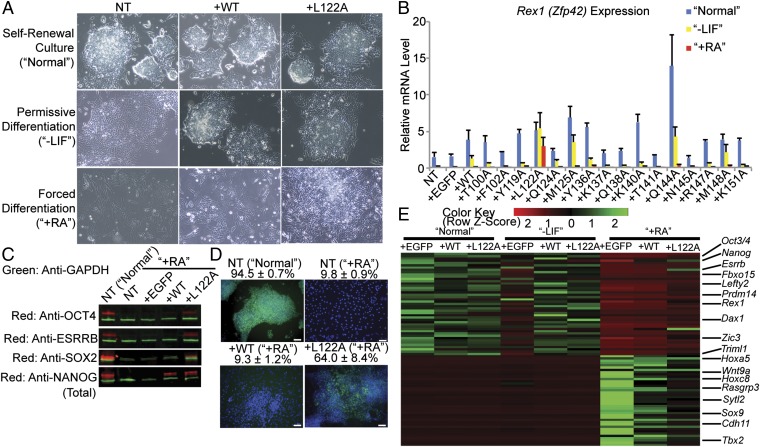
We examined the role of each NANOG mutant in the self-renewal of PSCs. Previous studies showed that overexpressing WT mNANOG in mESCs repressed spontaneous differentiation when LIF was withdrawn from the culture medium (8, 9). We used similar assays to examine the functionality of each mNANOG mutant. First, we examined the expression levels and cell proliferation in mESC transfected with each mNANOG mutant in LIF-containing medium (i.e., “Normal” culture conditions) (Figs. S3 and S4A), confirming that all of the transfectants similarly expressed these mutants and grew stably. When these mESC transfectants were cultured in medium without LIF (i.e., “−LIF” culture conditions), which is a permissive differentiation condition, some transfectants (i.e., T100A, F102A, Y119A, K137A, Q138A, K140A, T141A, N145A, R147A, and K151A), as well as EGFP-transfectant or nontransfected (NT) mESCs, showed differentiated colony morphologies (Fig. 4A and Fig. S4B) and lower gene expression of self-renewal markers [i.e., Rex1 (Zfp42), endogenous Nanog, Oct4 (Pou5f1), Sox2, Dax1 (Nr0b1), or Esrrb (Nr3b2 or Errβ)] than WT transfectants (Fig. 4B and Fig. S5A). Conversely, the gene expression of early differentiation markers (i.e., Fgf5 and T) was relatively higher in these transfectants. These results suggested that these mutants lost the WT mNANOG functions and that these residues were important in mNANOG transcriptional activity.
Fig. 4.
The effects of overexpressing mNANOG mutants on mESC self-renewal. (A) Typical colony morphologies of NT mESCs or mESC transfectants overexpressing WT or L122A mNANOG mutant in Normal, −LIF, or +RA culture conditions. The images were taken 5 d after seeding. (B) Rex1 expression detected by RT-qPCR in mESC transfectants overexpressing each mNANOG mutant in these culture conditions. The amount of an undifferentiated mESC sample was set as 1.0. n = 4, values are mean + SEM. (C) Protein expression of OCT4, SOX2, NANOG (total), ESRRB, and GAPDH in NT mESCs in Normal or +RA conditions or in mESC transfectants overexpressing EGFP, WT, or L122A in +RA conditions detected by Western blotting. Green and red bands in each sample show the amounts of GAPDH and OCT4, ESRRB, SOX2, or total NANOG, respectively. (D) OCT4 protein expression detected by immunocytochemistry in NT mESCs in Normal or +RA conditions or in mESC transfectants overexpressing WT or L122A in +RA conditions. Secondary antibodies were labeled with AlexaFluor488 (green). Nuclei were stained with DAPI (blue). (Scale bars, 100 µm.) The percentage of OCT4+ cells was analyzed from five randomly taken images for each sample, which contain at least 100 cells in each image. n = 5, values are mean + SEM. (E) Heatmap image of RNA-seq data illustrating gene expression profiles for the panel of genes that were differentially expressed between Normal and +RA culture conditions in EGFP-overexpressing mESCs.
When these mESC transfectants were cultured with the addition of high concentration of RA (5 µM) without LIF (i.e., “+RA” culture condition), which is a forced differentiation condition for mESCs, all of the transfectants except for L122A transfectant showed flattened cell morphologies (Fig. 4A and Fig. S4B) and suppressed gene expression of self-renewal markers (Fig. 4B and Fig. S5A). L122A transfectants in +RA showed distinct dense cell morphologies and markedly higher gene expression of self-renewal markers compared with other transfectants. Thus, we focused on this condition for further experiments. The protein expression of OCT4, ESRRB, and SOX2 detected by Western blotting assays was apparent only in L122A transfectants in +RA (Fig. 4C). The ratio of OCT4 protein-positive cells in L122A transfectants in +RA was 64 ± 8% detected by immunocytochemistry; however, NT mESCs or WT transfectants in this condition were less than 10% (Fig. 4D). We examined global gene-expression patterns by RNA-seq experiments (Fig. 4E). In +RA, the expression patterns of the RA-responsive genes were slightly different in WT transfectants and more different in L122A transfectants compared with EGFP transfectants. These results suggested that L122A transfectants showed partial resistant activity against RA-induced differentiation, which was not found in WT mNANOG transfectants.
NANOG L122A Enhanced Reprogramming to Ground-State Pluripotency.
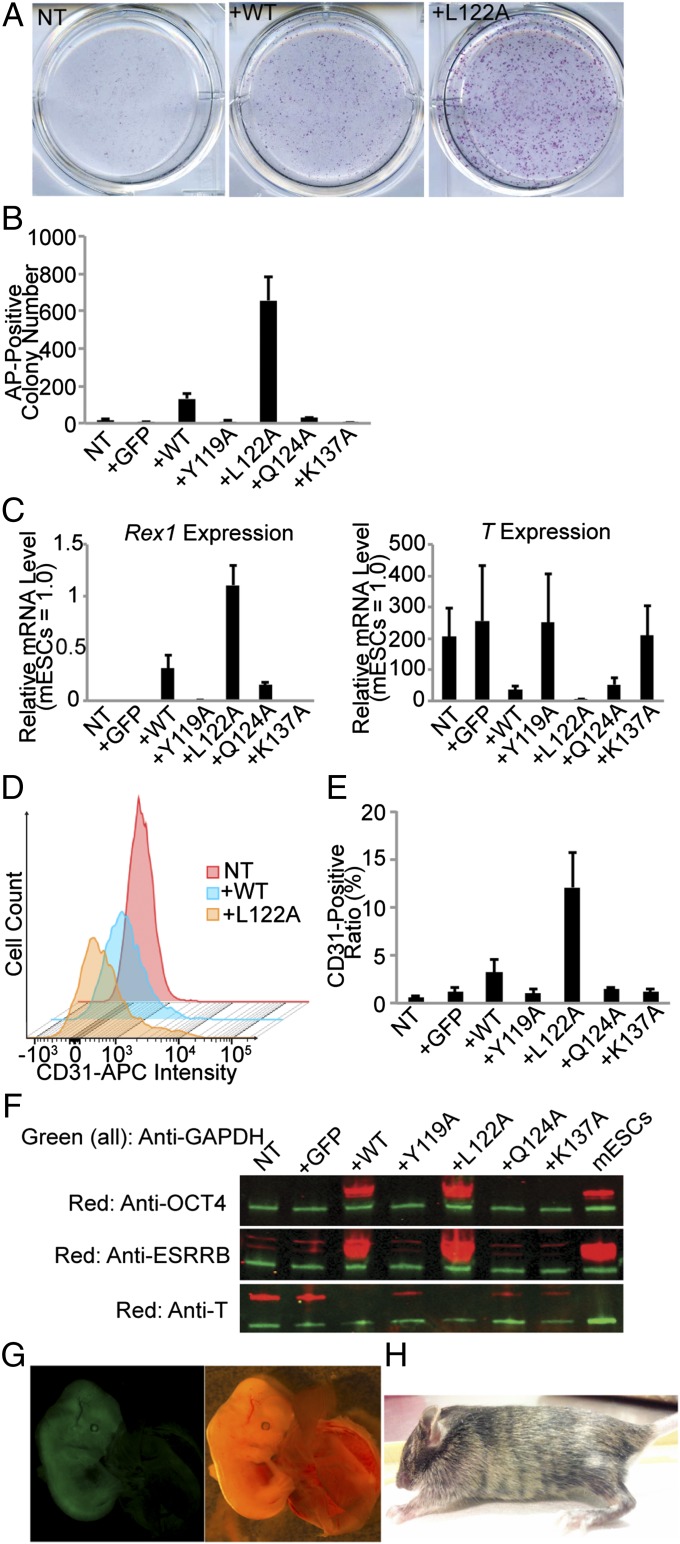
We then examined the effect of L122A in cellular reprogramming. The assay system in which EpiSCs reprograms into ground-state pluripotency in the medium supplemented with LIF and the inhibitors of MEK and GSK3β (“2i+LIF”-containing medium) is a robust but inefficient experimental model to demonstrate mNANOG functions (17, 32). We used this assay system to test whether L122A could be more potent than WT mNANOG. We generated mEpiSC transfectants, which constitutively express GFP or mNANOG WT or mutants (Y119A, L122A, Q124A, or K137A). For reprogramming, these transfectants were cultured in 2i+LIF-containing medium for 5 d. An alkaline phosphatase (AP) activity, which is a marker for ground-state pluripotency (15, 33), was evaluated in these cells (Fig. 5 A and B). L122A transfectant produced much more AP+ colonies (657 ± 124) than WT (132 ± 30) or Q124A (30 ± 4) transfectants. Gene expression of ground-state pluripotency markers [i.e., Oct4, Rex1, Nanog (endogenous), Dax1, Esrrb, Klf4, and Stella (Dppa3)] and primed markers (i.e., T and Fgf5) in these conditions was analyzed by quantitative RT-PCR (RT-qPCR) (Fig. 5C and Fig. S5B). L122A transfectants up-regulated the gene expression of all of the ground-state pluripotency markers analyzed more than WT transfectants. Conversely, T expression was more effectively suppressed in L122A transfectants than in WT transfectants. A cell surface marker, CD31 (PECAM-1), which distinguishes ground-state PSCs from EpiSCs (18, 34), was analyzed in these cells by flow cytometry (Fig. 5 D and E). L122A transfectants had much more CD31+ cells (12.1 ± 3.6%) than WT transfectants (3.2 ± 1.3%). The protein expression of OCT4, ESRRB, and T was analyzed by Western blotting (Fig. 5F). L122A transfectants expressed more OCT4 and ESRRB proteins and less T proteins than those in WT transfectants. These results suggested that L122A mNANOG mutant was more potent in reprogramming mEpiSCs into ground state pluripotency than WT mNANOG.
Fig. 5.
The effect of L122A on EpiSC reprogramming into ground-state pluripotency. (A) AP-staining (shown in red) of NT, WT-overexpressing, or L122A-overexpressing EpiSCs cultured in LIF2i medium for 5 d. The images were taken from a whole well of a six-well plate. (B) AP+ colony numbers counted in a well of a six-well plate. n = 4, values are mean + SEM. (C) Gene expression of Rex1, Klf4, or T in these conditions detected by RT-qPCR. The amount of an undifferentiated mESC sample was set as 1.0. n = 4, values are mean + SEM. (D) Flow cytometry of these EpiSCs using anti-CD31 (PECAM) antibody. The secondary antibody used was conjugated with Allophycocyanin (APC). (E) CD31+ cell ratio calculated from flow cytometry. n = 4, values are mean + SEM. (F) Protein expression of OCT4, ESRRB, T, or GAPDH in the EpiSC transfectants detected by Western blotting analysis. Green and red bands in each sample show the amounts of GAPDH and OCT4, ESRRB, or T, respectively. (G) Contribution of GFP-marked reprogrammed EpiSCs transfected with hNANOG L122A to embryonic day 13.5 chimeric mouse embryo. (H) Chimeric mice obtained by injection of reprogrammed EpiSCs (agouti) transfected with hNANOG L122A into C57BL/6 blastocysts (black) show coat-color contribution.
We examined the developmental potential of reprogrammed L122A transfectants. EGFP-marked EpiSCs (derived from 129Sv mouse line with agouti fur) were reprogrammed with PiggyBac-transposon–based plasmid vectors carrying hNANOG L122A 2A-peptide-fused with mCherry in 2i+LIF culture medium. Then, the integrated plasmid sequences were excised by transfecting “excision-only” transposase and sorting mCherry− population. After being injected into C57/B6 mouse blastocysts, the mCherry− reprogrammed cells efficiently contributed to chimeric embryos at embryonic day 13.5 evaluated by EGFP fluorescence (Fig. 5G) (6 out of 10 embryos analyzed) and chimeric live-born mice at 4-wk-old evaluated by mixed fur coat color with agouti and black (Fig. 5H) (6 out of 8 live pups). These results confirmed that the EpiSCs reprogramed with L122A had developmental potential in chimeric mice.
Finally, we examined whether hNANOG L122A could enhance the characteristics of putative ground-state or naïve pluripotency in HiPSCs. We generated HiPSC transfectants, which constitutively express mCherry or hNANOG WT or mutants (Y119A, L122A, or Q124A) fused with mCherry. Colony-forming activity from single cells dissociated with Trypsin/EDTA in 2i+LIF medium was examined on these transfectants. L122A transfectants enhanced AP+ colony-forming activity compared with WT transfectants (Fig. S6 A and B). Gene expression of ground-state and primed pluripotency markers in these transfectants cultured in 2i+LIF culture medium for 5 d was analyzed by RT-qPCR (Fig. S6C). L122A transfectants significantly up-regulated the gene expression of KLF2, endogenous NANOG, OCT4, PRDM14, and TFCP2L1 more than WT transfectants. These results suggested that L122A enhanced the characteristics of putative ground state or naïve pluripotency in HiPSCs.
Discussion
In this study, we describe the crystal structure of the hNANOG HD in complex with a 12-bp fragment of the OCT4 promoter DNA. Through alanine scanning, we evaluate the effect of key amino acids in the NANOG HD–DNA interface on DNA-binding affinity, protein stability in the purified complex, protein lifetime in mESCs, and mESC self-renewal. From these results (summarized in Table S2), we classify the role of each of these residues as outlined in SI Discussion.
The L122A mutant showed higher binding affinity to OCT4 promoter DNA, higher thermal stability in the complex, greater protein lifetime in mESCs, enhanced resistance against RA-induced differentiation in mESCs, and enhanced reprogramming ability to ground-state pluripotency from EpiSCs and HiPSCs. These results suggest that enhanced DNA-binding activity of L122A lessens the off rate of NANOG from DNA to favor the transcriptionally active state in mammalian stem cells. Conversely, WT NANOG’s relatively weak binding activity and modest protein stability may contribute to prompt and proper differentiation of epiblast or germ cells in developing embryos (10, 35) and to heterogeneous and fluctuating expression in PSC culture, which has been mainly explained by feedback loops in transcription networks (36, 37).
In conclusion, we demonstrate that an engineered key transcriptional factor based on structural and biophysical information improved their performance in stem cell self-renewal and reprogramming. Several key transcriptional factors have been identified for regulating stem cell behavior and for reprogramming somatic cells into specific cell lineages. Using our approach for these factors may be beneficial to stem cell biology and regenerative medicine.
Materials and Methods
The hNANOG HD construct (residues 94–162) was overexpressed in Escherichia coli (BL21Star, DE3; Invitrogen) and purified as a complex with a 12-bp fragment of the NANOG binding site in the OCT4 promoter (5′-GGCCCATTCAAG-3′/3′-CCGGGTAAGTTC-5′). The cloning, expression, purification, and crystallization of hNANOG HD–DNA complex were carried out using standard protocols in the R.J.F. laboratory and at the Joint Center for Structural Genomics (JCSG; www.jcsg.org). mESCs, RF8 line (a gift from R. V. Farese Jr., Harvard University, Boston) were cultured in FCS-based culture medium supplemented with LIF without feeder cells. EPiSCs, EpiSC-5 line (a gift from Paul Teser, Case Western Reserve University, Cleveland) were cultured in N2B27 medium (StemCells) supplemented with basic FGF (10 ng/mL; Millipore) and Activin A (10 ng/mL; R&D Systems). All of the protocols of mouse experiments were approved by the Institutional Animal Care and Use Committee at University of California, San Francisco. Details of the materials, methods, and associated references are in SI Materials and Methods. See Table S3 for the DNA oligos and primers used for each mutation.
Supplementary Material
Acknowledgments
We thank all members of the Joint Center for Structural Genomics (JCSG) for their general contributions to the structural work; K. Essex, Y. Miyake, and D. Singer for administrative support; S. Sami and C. Kime for technical assistance; and the Gladstone Flow Cytometry, Stem Cell, Genomics, Transgenic, and Bioinformatics Cores for technical support. This work was funded by the National Institute of Health (NIH), National Institutes of General Medical Sciences (NIGMS), Protein Structure Initiative Grants U54 GM094586 (to the JCSG) and U01 GM094614 (to R.J.F.). Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource (SSRL), a Directorate of the Stanford Linear Accelerator Center National Accelerator Laboratory and an Office of Science User Facility operated for the US Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the US Department of Energy’s Office of Biological and Environmental Research, and by the NIH, National Center for Research Resources, Biomedical Technology Program (Grant P41RR001209), and NIGMS; Postdoctoral Fellowship for Research Abroad of Japan Society for the Promotion of Science (JSPS) and University of California, San Francisco’s Program for Breakthrough Biomedical Research (to Y.H.); and the National Heart, Lung, and Blood Institute/NIH (Grant UO1HL098179), the L. K. Whittier Foundation, and the Roddenberry Foundation (to S.Y.). The Gladstone Institutes received support from a National Center for Research Resources Grant RR18928.
Footnotes
Conflict of interest statement: S.Y. is a scientific advisor of iPS Academia Japan without salary.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4RBO). Expression profiling by high throughput sequencing has been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE66127).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1502855112/-/DCSupplemental.
References
- 1.Loh YH, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38(4):431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 2.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122(6):947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang J, et al. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10(3):353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- 4.Rodda DJ, et al. Transcriptional regulation of nanog by OCT4 and SOX2. J Biol Chem. 2005;280(26):24731–24737. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]
- 5.Pan G, Li J, Zhou Y, Zheng H, Pei D. A negative feedback loop of transcription factors that controls stem cell pluripotency and self-renewal. FASEB J. 2006;20(10):1730–1732. doi: 10.1096/fj.05-5543fje. [DOI] [PubMed] [Google Scholar]
- 6.Zhou Q, Chipperfield H, Melton DA, Wong WH. A gene regulatory network in mouse embryonic stem cells. Proc Natl Acad Sci USA. 2007;104(42):16438–16443. doi: 10.1073/pnas.0701014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132(6):1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitsui K, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113(5):631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 9.Chambers I, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113(5):643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 10.Chambers I, et al. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450(7173):1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 11.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448(7151):313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 13.Schwarz BA, Bar-Nur O, Silva JC, Hochedlinger K. Nanog is dispensable for the generation of induced pluripotent stem cells. Curr Biol. 2014;24(3):347–350. doi: 10.1016/j.cub.2013.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carter AC, Davis-Dusenbery BN, Koszka K, Ichida JK, Eggan K. Nanog-independent reprogramming to iPSCs with canonical factors. Stem Cell Rev. 2014;2(2):119–126. doi: 10.1016/j.stemcr.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brons IG, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448(7150):191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 16.Tesar PJ, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448(7150):196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 17.Silva J, et al. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138(4):722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rugg-Gunn PJ, et al. Cell-surface proteomics identifies lineage-specific markers of embryo-derived stem cells. Dev Cell. 2012;22(4):887–901. doi: 10.1016/j.devcel.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gillich A, et al. Epiblast stem cell-based system reveals reprogramming synergy of germline factors. Cell Stem Cell. 2012;10(4):425–439. doi: 10.1016/j.stem.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takashima Y, et al. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell. 2014;158(6):1254–1269. doi: 10.1016/j.cell.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gehring WJ, Affolter M, Bürglin T. Homeodomain proteins. Annu Rev Biochem. 1994;63:487–526. doi: 10.1146/annurev.bi.63.070194.002415. [DOI] [PubMed] [Google Scholar]
- 22.Gehring WJ, et al. Homeodomain-DNA recognition. Cell. 1994;78(2):211–223. doi: 10.1016/0092-8674(94)90292-5. [DOI] [PubMed] [Google Scholar]
- 23.Berger MF, et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133(7):1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noyes MB, et al. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell. 2008;133(7):1277–1289. doi: 10.1016/j.cell.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harvey RP. NK-2 homeobox genes and heart development. Dev Biol. 1996;178(2):203–216. doi: 10.1006/dbio.1996.0212. [DOI] [PubMed] [Google Scholar]
- 26.Jauch R, Ng CK, Saikatendu KS, Stevens RC, Kolatkar PR. Crystal structure and DNA binding of the homeodomain of the stem cell transcription factor Nanog. J Mol Biol. 2008;376(3):758–770. doi: 10.1016/j.jmb.2007.11.091. [DOI] [PubMed] [Google Scholar]
- 27.Luscombe NM, Laskowski RA, Thornton JM. NUCPLOT: A program to generate schematic diagrams of protein-nucleic acid interactions. Nucleic Acids Res. 1997;25(24):4940–4945. doi: 10.1093/nar/25.24.4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weiler S, et al. Site-directed mutations in the vnd/NK-2 homeodomain. Basis of variations in structure and sequence-specific DNA binding. J Biol Chem. 1998;273(18):10994–11000. doi: 10.1074/jbc.273.18.10994. [DOI] [PubMed] [Google Scholar]
- 29.Moretto-Zita M, et al. Phosphorylation stabilizes Nanog by promoting its interaction with Pin1. Proc Natl Acad Sci USA. 2010;107(30):13312–13317. doi: 10.1073/pnas.1005847107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramakrishna S, et al. PEST motif sequence regulating human NANOG for proteasomal degradation. Stem Cells Dev. 2011;20(9):1511–1519. doi: 10.1089/scd.2010.0410. [DOI] [PubMed] [Google Scholar]
- 31.Abranches E, Bekman E, Henrique D. Generation and characterization of a novel mouse embryonic stem cell line with a dynamic reporter of Nanog expression. PLoS ONE. 2013;8(3):e59928. doi: 10.1371/journal.pone.0059928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silva J, et al. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008;6(10):e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Oosten AL, Costa Y, Smith A, Silva JC. JAK/STAT3 signalling is sufficient and dominant over antagonistic cues for the establishment of naive pluripotency. Nat Commun. 2012;3:817. doi: 10.1038/ncomms1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robson P, Stein P, Zhou B, Schultz RM, Baldwin HS. Inner cell mass-specific expression of a cell adhesion molecule (PECAM-1/CD31) in the mouse blastocyst. Dev Biol. 2001;234(2):317–329. doi: 10.1006/dbio.2001.0274. [DOI] [PubMed] [Google Scholar]
- 35.Yamaguchi S, Kimura H, Tada M, Nakatsuji N, Tada T. Nanog expression in mouse germ cell development. Gene Expr Patterns. 2005;5(5):639–646. doi: 10.1016/j.modgep.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 36.Kalmar T, et al. Regulated fluctuations in nanog expression mediate cell fate decisions in embryonic stem cells. PLoS Biol. 2009;7(7):e1000149. doi: 10.1371/journal.pbio.1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacArthur BD, et al. Nanog-dependent feedback loops regulate murine embryonic stem cell heterogeneity. Nat Cell Biol. 2012;14(11):1139–1147. doi: 10.1038/ncb2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.